Abstract
Background
The disaccharide trehalose is a major component of fungal spores and is released upon germination. Moreover, the sugar is well known for is protective functions, e.g. against thermal stress and dehydration. The properties and synthesis of trehalose have been well investigated in the bakers’ yeast Saccharomyces cerevisiae. In filamentous fungi, such knowledge is limited, although several gene products have been identified.
Results
Using Aspergillus niger as a model fungus, the aim of this study was to provide an overview of all genes involved in trehalose synthesis. This fungus has three potential trehalose-6-phosphate synthase encoding genes, tpsA-C, and three putative trehalose phosphate phosphatase encoding genes, tppA-C, of which two have not previously been identified. Expression of all six genes was confirmed using real-time PCR, and conserved orthologs could be identified in related Aspergilli. Using a two-hybrid approach, there is a strong indication that four of the proteins physically interact, as has previously been shown in S. cerevisiae. When creating null mutants of all the six genes, three of them, ΔtpsA, ΔtppA and ΔtppB, had lower internal trehalose contents. The only mutant with a pronounced morphological difference was ΔtppA, in which sporulation was severely reduced with abnormal conidiophores. This was also the only mutant with accumulated levels of trehalose-6-phosphate, indicating that the encoded protein is the main phosphatase under normal conditions. Besides ΔtppA, the most studied deletion mutant in this work was ΔtppB. This gene encodes a protein conserved in filamentous Ascomycota. The ΔtppB mutant displayed a low, but not depleted, internal trehalose content, and conidia were more susceptible to thermal stress.
Conclusion
A. niger contains at least 6 genes putatively involved in trehalose synthesis. Gene expressions related to germination have been quantified and deletion mutants characterized: Mutants lacking tpsA, tppA or tppB have reduced internal trehalose contents. Furthermore, tppA, under normal conditions, encodes the functional trehalose-6-phosphate-phosphatase.
Keywords: Ascomycota, Conidia, Germination, Saccharomyces cerevisiae, Stress-resistance, Targeted gene deletion
Background
Trehalose (α-D-glucopyranosyl-α-D-glucopyranoside) is a non-reducing disaccharide that is present in a wide variety of organisms. It has been isolated from plants, fungi, nematodes and insects [1-3]. In fungi, trehalose has been shown to accumulate in dispersal and survival structures such as spores (where it can constitute as much as 10% of the dry weight), sclerotia, and in yeast cells going into stationary phase [3,4] . Since the sugar is rapidly degraded when these structures germinate or resume vegetative growth, early research concluded that trehalose serves as a storage molecule [5,6]. However, later studies showed that the function of trehalose is more complex and diverse than just serving as an energy reserve; the molecule has been shown to function as a regulator of carbon metabolism [1], a signaling molecule and a protection molecule against various kinds of abiotic stress [3,7]. Several fungal species have been shown to induce trehalose production as a stress response. Examples include: Saccharomyces cerevisiae[8,9], Zygosaccharomyces bailii[10], A. nidulans[11], A. fumigatus[12], Rhizopus oryzae[13], and Botrytis cinerea[14]. Trehalose is known to protect both proteins and lipid membranes of living cells against stressors such as heat, desiccation and cold. Although the mode of bio-protection of trehalose is not fully elucidated, three main hypotheses are generally accepted, and the true mechanism is likely a combination of these. The hypotheses include: water replacement (direct interaction of trehalose with the protected structure through hydrogen bonds); mechanical entrapment (glass formation of trehalose that creates a protective coating around the structure); preferential exclusion (bulk water is ordered around trehalose and is thereby separated from the bio-molecule, which then becomes more compact and stabilized) [15,16]. The physico-chemical properties of trehalose that lie behind these hypotheses include several crystalline forms, a high glass transition temperature, and the stereochemistry of the sugar [7,15].
In fungi, trehalose is synthesized via the intermediate trehalose-6-phosphate (T6P) and involves two enzymatic steps. First, T6P is formed from one glucose-6-phosphate and one UDP-glucose catalyzed by T6P-synthase (here called TPS). In the next step, the phosphate molecule is removed by trehalose-phosphate-phosphatase (here called TPP) yielding trehalose [1,11]. The organism in which trehalose synthesis has been most thoroughly studied is S. cerevisiae. Here, four homologous gene products responsible for trehalose synthesis physically interact forming a “trehalose synthase complex”, which consists of one TPS (called Tps1), one TPP (called Tps2), and two other subunits, Tsl1 and Tps3, with proposed regulatory and stabilizing functions [6,17-19]. In filamentous fungi, the gene products involved in trehalose synthesis are not as thoroughly investigated as in S. cerevisiae, but have been studied with respect to germination [20], plant pathology [21] and human pathology [12,22].
Within Aspergilli, several individual gene products have been identified and characterized. In A. niger, two Tps1 orthologs, tpsA and tpsB, have been identified and characterized. At ambient temperature, the trehalose level of ΔtpsA mycelia was lowered compared to wild-type. In contrast to the constitutively expressed tpsA, the expression of tpsB was induced by thermal stress [23]. In the opportunistic human pathogen A. fumigatus, four Tps1 paralogs, tpsA – D have been identified [12]. When deleting these genes, the authors found that either tpsA or tpsB was sufficient to maintain normal trehalose levels, but if both genes were deleted, the resulting mutant strain was depleted of trehalose and showed slower germination rates as well as higher susceptibility to heat and oxidative stress compared to wild-type. Another notable finding was that this double mutant was hypervirulent in infected mice [12]. In A. nidulans, a Tps1 ortholog, tpsA, has been identified and deleted. In this mutant, trehalose was not accumulated, and in addition, the authors could conclude that in A. nidulans trehalose is important for resistance to continual exposure to sub-lethal stress but not to short exposure of lethal stress [11]. In contrast to S. cerevisiae, tps mutants in Aspergilli are able to utilize glucose as carbon source [11,23,24]. All identified Tps1 orthologs in Aspergilli are generally much shorter than the S. cerevisiae Tps1, around 500 amino acids compared to 1447.
Besides Tps1 orthologs, two Tps2 orthologs have been identified within the Aspergilli, one in A. nidulans[25] and one in A. fumigatus[22]: In both species they are designated orlA. The ΔorlA mutant of A. fumigatus had a pronounced phenotype with abolished asexual reproduction as well as decreased virulence. However, the phenotype could be restored to wild-type appearance by growing the mutant on media containing an osmotic stabilizer (sorbitol or glycerol). As also observed in A. nidulans, the A. fumigatus ΔorlA mutant strain contained wild-type levels of trehalose but the T6P levels were elevated [22,25].
In this study we focused on trehalose synthesis in filamentous fungi, and more specifically, in Aspergillus niger. This is a common food spoilage mould as well as an industrially important organism, utilized for production of citric acid, for instance [26]. Six genes, tpsA (ANI_1_1406074), tpsB (ANI_1_1078064), tpsC (ANI_1_1216124), tppA (ANI_1_1432094), tppB (ANI_1_48114) and tppC (ANI_1_2070064) were identified to be involved in trehalose biosynthesis. Expression of these genes was studied during conidial outgrowth. In addition, we deleted these genes and characterized the mutants in terms of trehalose and T6P content, protein interactions, and stress survival coupled to situations often occurring in foodstuff.
Methods
Software, hardware and computer-based analyses used in this study
GraphPad Prism® version 5 was used for generating figures (line drawings) and calculating mean, standard error of the mean, and significance between samples (using one or two way ANOVA and Bonferroni post-test). Adobe Illustrator CS5 and Adobe Photoshop CS6 were used for managing pictures (cropping and minor changes in contrast levels for best visualization). Bio-Rad CFX 96™ Real-Time System was used for generating gene expression data and the Bio-Rad CFX Manager™ version 1.6 software was used for analyzing the data. MacVector version 12 was used for primer design and phylogenetic analyses.
Culture maintenance, spore preparation and spore densities
The fungal strains used in this study are listed in Table 1. As wild-type, we used the A. niger N402 strain, which is also the mother strain of all generated mutants [27]. The strains were maintained on Aspergillus Minimal Media (AMM) as previously described [28]. For the MA70.15 and MA169.4 strains, AMM was supplemented with 10 mM uridine. The complemented strain (tppB+) was maintained on AMM containing Hygromycin B (0.10 mg/ml). The ΔtppA mutant was tested for sporulation both on AMM agar and on AMM agar containing 1.2 M sorbitol. Normally plates were incubated at 25°C for 14 days. All deletion mutants as well as the control strains were tested for growth in 10, 15, 20, 25, 30 and 37°C for 14 days. For trehalose measurements, conidia were harvested from plates incubated at 25°C for 5, 14, 28 and 90 days. Spore suspensions were prepared in water containing Tween 80 (0.01% v/v), were filtered through sterile Miracloth (Calbiochem), and the spore count was determined using a Bürker chamber. To estimate the number of conidia produced, a circular area of 95 mm2 was cut out from centrally inoculated AMM plates that had been incubated at 25°C for 14 days. 10 ml of water containing Tween 80 (0.01% v/v) and 10 glass beads (2 mm in diameter) were added to the agar plug, the mixture was vortexed for 10 min and spore concentrations were counted in a Bürker chamber. Three biological replicates, each calculated from the average of three technical replicates, were used for all samples.
Table 1.
Strains used in this study
| Strain | Genotype | Reference |
|---|---|---|
| N402 |
cspA1 |
[27] |
| MA70.15 |
cspA1,pyrG1, ∆kusA::amdS
+
|
[32] |
| MA169.4 |
cspA1,pyrG1, ∆kusA::DR-amdS
+
-DR |
[33] |
| J699* (∆tpsA) |
cspA1,pyrG1, ∆kusA::amdS
+
, ∆tpsA::pyrG |
This study |
| J700 (∆tpsB) |
cspA1,pyrG1, ∆kusA::amdS
+
, ∆tpsB::pyrG |
This study |
| J701 (∆tpsC) |
cspA1,pyrG1, ∆tpsC::pyrG |
This study |
| J684 (∆tppA) |
cspA1,pyrG1, ∆kusA::amdS
+
, ∆tppB::pyrG |
This study |
| J685 (∆tppB) |
cspA1,pyrG1, ∆kusA::amdS
+
, ∆tppB::pyrG |
This study |
| J702 (∆tppB2) |
cspA1,pyrG1, ∆tppB::pyrG |
This study |
| J686 (∆tppC) |
cspA1,pyrG1, ∆kusA::amdS
+
, ∆tppC::pyrG |
This study |
| J689 (pyrG+) |
cspA1, ∆kusA::amdS
+
|
[28] |
| J693 (tppB+) | cspA1,pyrG1, ∆tppB::pyrG, tppB::hph | This study |
*Strain numbers from the fungal collection at the Department of Microbiology, Swedish University of Agricultural Sciences.
Low-temperature scanning electron microscopy (SEM)
Wild-type, N402, and ΔtppA were grown for 1 week on AMM. Margins of colonies containing conidiophores were excised with a surgical blade and carefully transferred into a copper cup (diameter 10 mm, height 8 mm). Dislodging during snap freezing was prevented by gluing agar blocks in the copper cup with frozen tissue medium (KP-Cryoblock, Klinipath, Duiven, the Netherlands). The sample was snap-frozen in nitrogen slurry and immediately transferred in a vacuum transfer device to an Oxford CT1500 Cryostation attached to a JEOL 5600LV scanning electron microscope (JEOL, Tokyo, Japan). Electron micrographs were acquired from uncoated frozen samples, or after sputter-coating with gold three times during 30 s. Micrographs of uncoated samples were taken at an acceleration voltage of 2.5 kV, and consisted of 30 averaged fast scans (SCAN 2 mode). Coated samples were observed at 5 kV using F4 scans.
Extraction of nucleic acids
DNA was extracted as previously described [28]. RNA from dormant conidia and conidia in early stages of germination (0 and 3 h) was extracted according to Leeuwen and co-workers [29]. RNA from germinating spores (6 and 12 h), mycelia and sporulating mycelia (plate) were extracted according to Plumridge and co-workers [30]. As a final step in both protocols, the RNA products were purified using a Qiagen RNeasy Mini kit (RNA clean up protocol).
Two-hybrid assay
The two-hybrid assay was performed using the BACTH System Kit (Euromedex). Full-length cDNA for all six genes were amplified using primers with internal restriction sites (Table 2). After digestion of the PCR products, the inserts were ligated into linearized and dephosphorylated pKT25 and pUT18C vectors and used to transform E. coli. All ligations in this work were performed with the ReadyToGo ligation kit (GE Healthcare) and were transformed into NEB 10-β Competent E. coli cells (New England Biolabs), unless otherwise stated. Correct insertions were confirmed with vector specific primers (Table 2) followed by sequencing. Successful clones were co-transformed into electrocompetent BTH101 cells and selected on LA plates supplemented with ampicillin (100 μg/ml) and kanamycin (50 μg/ml). The protein-protein interactions were assayed according to the manufacturer’s protocol with the following modifications. One fresh colony of each interaction was transferred to 100 ml conical flasks with 5 ml LB supplemented with ampicillin 50 μg/ml, kanamycin 50 μg/ml and 0.5 mM IPTG, and incubated with shaking at 100 rpm at 20°C for 72 h. The extent of protein-protein interaction was measured with β-galactosidase assays as units/mg dry weight.
Table 2.
Primers used for cDNA synthesis, qPCR and Two-Hybrid cloning
| Primer name | Sequence 5′-3′ | Purpose |
|---|---|---|
| T12VN |
TTTTTTTTTTTTVN |
cDNA synthesis |
| tpsAF |
TGAGGGCTGTTGTGAATGAGC |
qPCR tpsA |
| tpsAR |
ACTCGGAAAGCACCAAGACACC |
|
| tpsBF |
GTGGGCAGAATCAACGGAAAG |
qPCR tpsB |
| tpsBR |
TGAACACTTGGATAGTTCGGCAAC |
|
| tpsCF |
TTGCCGATGCCTGCTTGTTG |
qPCR tpsC |
| tpsCR |
TTCGCTGGATGGAAAGTAAGACAC |
|
| tppAF |
TTGAAGACACCGTTGGGAAGAG |
qPCR tppA |
| tppAR |
GGAGCAAAAGATGAACTCAGGAGC |
|
| tppBF |
TGGACACTTACCTCTGGGATGAAG |
qPCR tppB |
| tppBR |
GCTGATGGGCATTGAGTATTTCC |
|
| tppCF |
AAAGCCAAAGCAGCCGAATC |
qPCR tppC |
| tppCR |
TGCCCGTTAGTATCCTCAGCAGAG |
|
| actF |
TCGTGACCTGACGGATTACCTC |
qPCR actin |
| actR |
TGGAAGAAGGAGCAAGAGCAGTG |
|
| pKT25F |
ACGATTTCGAGGCGGTCAAG |
Confirmation of cloned cDNA to pKT25 vector |
| pKT25R |
GATGTGCTGCAAGGCGATTAAG |
|
| pUT18CF |
TGTCTTCTACGAGAACCGTGCATAC |
Confirmation of cloned cDNA to pUT18C vector |
| pUT18CR |
CGGTGAAAACCTCTGACACATGC |
|
| tpsAFpst |
GACTCTGCAGCCGTTTCCGACAGCATGCCTT |
Cloning of tpsA cDNA |
| tpsAFbam |
TATCTGGATCCCGTTTCCGACAGCATGCCTT |
|
| tpsARkpn |
TGATCGGTACCAGCTCACTGTGCCACCTGCT |
|
| tpsBFbam |
ATCAGGATCCTCCTTTTCCAATGGCTGCCAA |
Cloning of tpsB cDNA |
| tpsBeco |
ATCAGAATTCAGCTGCAGTCATAACATAATCA |
|
| tpsCFxba |
AGTCTCTAGAATCCAGAATGACGAAGCGCAA |
Cloning of tpsC cDNA |
| tpsCReco |
ACTCGAATTCCGTCCATGTCAGGGCTCAA |
|
| tppAxbaF |
ACTGTCTAGAATCCCCCATCATGCT |
Cloning of tppA cDNA |
| tppAecoR |
AGTAGAATTCATTAACAGAACCCTCAATAC |
|
| tppBxbaF |
TACATCTAGATGTCGCCATGACCATCTACA |
Cloning of tppB cDNA |
| tppBkpnR |
TTCCGGTACCCCTTTCACTCCTTATCGTGA |
|
| tppCFsal |
TATCGTCGACCACCCCAATGACGGTCTTCAT |
Cloning of tppC cDNA |
| tppCRbam | CATAGGATCCTCAGTCATGGCTTTCTCCGT |
cDNA synthesis and Real-Time PCR
Using total RNA as template, cDNA was synthesized using a polyT primer (Table 2) and the enzyme SuperscriptIII (Invitrogen) according to the manufacturer’s protocol. Quantitative real-time PCR was performed with the BioRad CFX-96 system using the EvaGreen reagent (BioRad), gene specific primers (Table 2), and the following protocol: Initial denaturation and enzyme activation, 95°C 30 s; 40 cycles of 95°C for 2 s and 56-60°C for 8 s; plate read; and finally, melt curve analysis starting at 65°C and ending at 95°C. Relative expression for tpsA-C and tppA-C were calculated from and compared to a serially-diluted cDNA pool and normalized to the actin-encoding gene (ANI_1_106134), which has been successfully used in previous experiments [28,31] and is expressed at high levels throughout germination according to published microarray data [29]. For each growth stage, the expressions were calculated from four biological replicates, each with three technical replicates. To verify the expression, or lack thereof, in the reconstituted and null mutant of tppB, the expression in mutants was normalized against N402 as previously described [28] using the efficiency calibrated mathematical method for the relative expression ratio in real-time PCR [32].
Gene deletions and complementation
Deletion constructs for the genes, tpsA, tpsB, tppA, tppB and tppC were made using fusion PCR to replace the coding sequence with the A. oryzae pyrG gene, and used to transform the uridine auxotrophic strain MA70.15 [33] as previously described [29]. With the same technique, a mutant lacking both tpsB and tppC was created. A second deletion mutant of tppB, (ΔtppB2) was generated in a different uridine auxotrophic strain, MA169.4 [34]. Both MA70.15 and MA169.4 have deficient kusA that is the A. niger ortholog of kus70, which is required for the non-homologous end-joining pathway [35]. The tpsC deletion strain was constructed by cloning tpsC in the standard pBS-SK vector (Stratagene) using BamHI and XhoI. Next, the vector was digested with HindIII to remove 1648 bp, containing most of the coding sequence. After dephosphorylation of the vector, a HindIII digested PCR product of the A. oryzae pyrG gene was ligated into the vector, thus replacing tpsC. This deletion construct was PCR-amplified and used to transform strain MA169.4. All A. niger transformants were confirmed using PCR and sequencing. For the deletion strains where MA169.4 was used as parent strain, the kusA gene was repaired using induced recombination by repeated transfer to agar plates supplemented with fluoroacetamide 0.75 μg/ml, as described [34]. All primers for gene deletions are listed in Table 3. The ΔtppB strain was complemented as previously described [28]. Briefly, the strain was transformed with a plasmid carrying an intact copy of tppB and a cassette carrying hygromycin resistance.
Table 3.
Primers used for targeted gene deletions
| Primer name | Sequence 5′-3′ | Purpose |
|---|---|---|
| pyrGN2 |
CACATGCCTCATTTTGACCA |
Mutant confirmation |
| PyrtpsAup |
ACCGTTGGAAGGTGGGATCCTATGGATCTCAGAA |
Amplifies pyrG with 3' tpsA overhangs |
| PyrtpsAdown |
CCTTTCAGAATGAGTGTGAGCGGATAACAATTTC |
|
| tpsAup |
CCATCTGTCTAGCTCTTCATCCCC |
tpsA, upstream fragment |
| tpsApyrup |
GATCCATAGGATCCCACCTTCCAACGGTGTAGAGACTCC |
|
| tpsApyrdown |
TTATCCGCTCACACTCATTCTGAAAGGTGGGGTTTTC |
tpsA, downstream fragment |
| tpsAdown |
GCAAGATTCCCGCATCCATC |
|
| tpsAupN1 |
CAACCCCACCAGTTCTCTCAAG |
Amplification of KO-fragment |
| tpsAdownN1 |
AAAGGGAGTTCCAAGCAGCCTG |
|
| pyrtpsBup* |
ATCTGCTCTGCCTGGGATCCTATGGATCTCAGAA |
Amplifies pyrG with 3′ tpsB overhangs |
| pyrtpsBdown |
CTGCCCATCACCATGTGAGCGGATAACAATTTC |
|
| tpsBup* |
TTGAACCCTTGAAACCGAACAC |
tpsB, upstream fragment |
| tpsBpyrGup* |
GATCCATAGGATCCCAGGCAGAGCAGATACTTACCCGTC |
|
| tpsBpyrGdown |
TTATCCGCTCACATGGTGATGGGCAGACGATTG |
tpsB, downstream fragment |
| tpsBdown |
TGCTAAAGAGGGTGTGGGATTG |
|
| tpsBupN3 |
TCCCGATTGGTAGAATCCCTAAAG |
Amplification of tpsB KO-fragment |
| tpsBdownN3 |
CATGCGAAAATGACAGGAACATTC |
|
| pyrGuphind |
TAAAAGCTTCTATATTGATCCTTA |
pyrG, KO of tpsC |
| pyrGdown |
TGTGAGCGGATAACAATTTC |
|
| tpsCupN-2 |
TGCCGAATTGACGTGCGTAGAG |
Cloning of tpsC |
| tpsCdownN-2 |
TGGTGGTGAACCTTTCGTTGTTC |
|
| tpsCupN5 |
CCCTCCATACTTACTCCATACATCTCG |
Amplification of tpsC KO-fragment |
| tpsCdownN5 |
CCAGCTTGACACATCCAACATAAC |
|
| pyrtppAup |
CCTGTCCCCGCTTCAAGAAAGGGATCCTATGGATCTCAGAA |
pyrG with 3′ tppA overhangs |
| pyrtppAdown |
GAGTCATCAGTGCTGCTTTCTGCTGTGAGCGGATAACAATTTC |
|
| TppAup |
TGTTGGAAGCGTCTTTCTGCC |
tppA, upstream fragment |
| tppApyrup |
TTCTGAGATCCATAGGATCCCTTTCTTGAAGCGGGGACAGG |
|
| tppApyrdown |
GAAATTGTTATCCGCTCACAGCAGAAAGCAGCACTGATGACTC |
tppA, downstream fragment |
| tppAdown |
TGTCCGATTGGGGGTGATTG |
|
| tppAupN1 |
TGAGGAGGCGTTGTCAAAAGATAG |
Amplification of tppA KO-fragment |
| tppAdownN1 |
CGATTGGGGGTGATTGGCTTAC |
|
| pyrtppBup |
CGGTAGGTTAGGGATCCTATGGATCTCAGAA |
Amplification of A. oryzae pyrG with 3′ tppB overhangs |
| pyrtppBdown |
GTTTGTCTTGTGTGAGCGGATAACAATTTC |
|
| tppBup |
ATACCAAGCAATCGCCCAAGCCAG |
tppB, upstream fragment |
| tppBpyrGup |
TCCATAGGATCCCTAACCTACCGCCCAAAGAGAGAGC |
|
| tppBpyrGdown |
TTGTTATCCGCTCACACAAGACAAACGATGCGGAATG |
tppB, downstream fragment |
| tppBdown |
CGTATCCTGGACTTTCAGCACG |
|
| tppBupN1 |
TTTTCGACCTTGGTGGGTGCTTCC |
Amplification of tppB KO-fragment |
| tppBdownN1 |
GAGACATTGTCGGTCAGTGAGGTAG |
|
| pyrtppCup |
TGTCCTTCAGGGATCCTATGGATCTCAGAA |
pyrG with 3′ tppC overhangs |
| pyrtppCdown* |
CTGTTCAGCATTGTGAGCGGATAACAATTTC |
|
| tppCup |
ATGAGGTGATAGTCGTGGACCCAG |
tppC, upstream fragment |
| tppCpyrup |
TCCATAGGATCCCTGAAGGACAAAGACAGGCTGAAG |
|
| tppCpyrdown* |
TTGTTATCCGCTCACAATGCTGAACAGATGATCCCCAG |
tppC, downstream fragment |
| tppCdown* |
TCGAGGTAGAGGTTCCCTTTCG |
|
| tppCupN1* |
CGATAGTCTTTGCGAACAGACGG |
Amplification of tppC KO-fragment |
| tppCdownN1* |
CGAGGTAGAGGTTCCCTTTCGATG |
|
| tpsBupN1 | CCCTTTCCCGATTGGTAGAATC | Amplification of tpsB/tppC double mutant |
*Also used in the design of the tpsB/tppC double mutant.
Extraction and quantification of trehalose and trehalose-6-phosphate
Trehalose from dormant and swollen conidia, germlings and mycelia was extracted and quantified as previously described [28]. In brief, harvested fungal material was freeze-dried and homogenized using a mortar. Samples were diluted with ultra pure water, boiled, evaporated and derivatized by trimethylsilylanization before injection into the gas chromatograph–mass spectrometer (GC–MS). Relative concentrations of α-α-trehalose were calculated as the ratio to an internal standard (α-β-trehalose) and thereafter correlated to a standard curve to obtain the absolute concentrations. All trehalose measurements were performed in biological duplicates based on the average of three technical triplicates.
Extraction and quantification of T6P was performed essentially as described by [22]. Liquid cultures were inoculated with 106 spores per ml, incubated at 25°C for 3 days at 140 rpm, and all mycelia from one culture made up one sample. Three biological replicates based on the average of three technical replicates were used for all strains.
Stress tolerance and long term viability of conidia
Dormant conidia from wild-type A. niger, the additional control strain pyrG+, and the deletion mutants ΔtppB and ΔtppB2 were subjected to heat stress for 20, 60, 90 and 120 min at 55°C. Dormant conidia of wild-type, pyrG + and ΔtppB were subjected to sub-lethal salt and benzoic acid stress by being spread on AMM plates containing benzoic acid or NaCl at concentrations ranging from non-effective to total growth inhibition of the control strains. For detailed description of these stress experiments see [28]. In addition, dormant conidia from control strains and ΔtppB were subjected to oxidative stress by adding 200 mM H2O2 to freshly made conidial suspensions (approximately 250 spores/ml liquid AMM). The suspensions were incubated for 10, 20 or 40 min before being spread on AMM plates. To test long-term viability, conidial suspensions (106 conidia/ml water) were stored at 4°C for a total of 8 weeks. An aliquot of the suspension was withdrawn weekly, diluted and spread on AMM plates for enumeration.
Plates from all experiments were incubated at 25°C for 3–7 days before CFU were estimated, and all experiments were performed at least in triplicates (based on three technical replicates).
Results
Identification of genes involved in trehalose synthesis in Aspergillus niger and other fungi
Known amino acid sequences of the proteins of the trehalose synthesis complex of S. cerevisiae were used as queries to identify homologous genes in the A. niger genome by searching the databases available at NCBI using blastP (http://blast.ncbi.nlm.nih.gov). We confirmed the presence of two previously characterized tps1 orthologs: tpsA (ANI_1_1406074) and tpsB (ANI_1_1078064). Also, we could detect two genes, previously identified by sequence homology [36]; a third tps1 paralog, tpsC (ANI_1_1216124), and a tps2 ortholog, which we call tppA (ANI_1_1432094). In addition, we could identify two previously unidentified, putative tppA paralogs designated tppB (ANI_1_48114) and tppC (ANI_1_2070064). Compared to TppA, these two encoded proteins were of similar length (all three proteins have between 926 to 946 residues) and had a protein identity of 37% (250 out of 683) and 35% (241 out of 688), respectively (Figure 1). From the NCBI’s Conserved Domain Database [37] it was revealed that all three Tpp proteins contain a phosphate synthase domain approximately 200 residues from the N-terminal, and a phosphatase domain approximately 700 residues from the N-terminal (Figure 1). The Tps proteins only contain the phosphate synthase domain (data not shown). In summary, three tps1 orthologs, tpsA-C, and three tps2 orthologs, tppA-C, were identified from the A. niger genome.
Figure 1.
Protein alignment indicating the similarities between the A. niger Tpp proteins. Boxed amino acids are either identical or similar in at least two of the aligned sequences. Approximated borders of the phosphate synthase (closer to the N-terminal) and the phosphatase (closer to the C-terminal) domains are indicated in the figure.
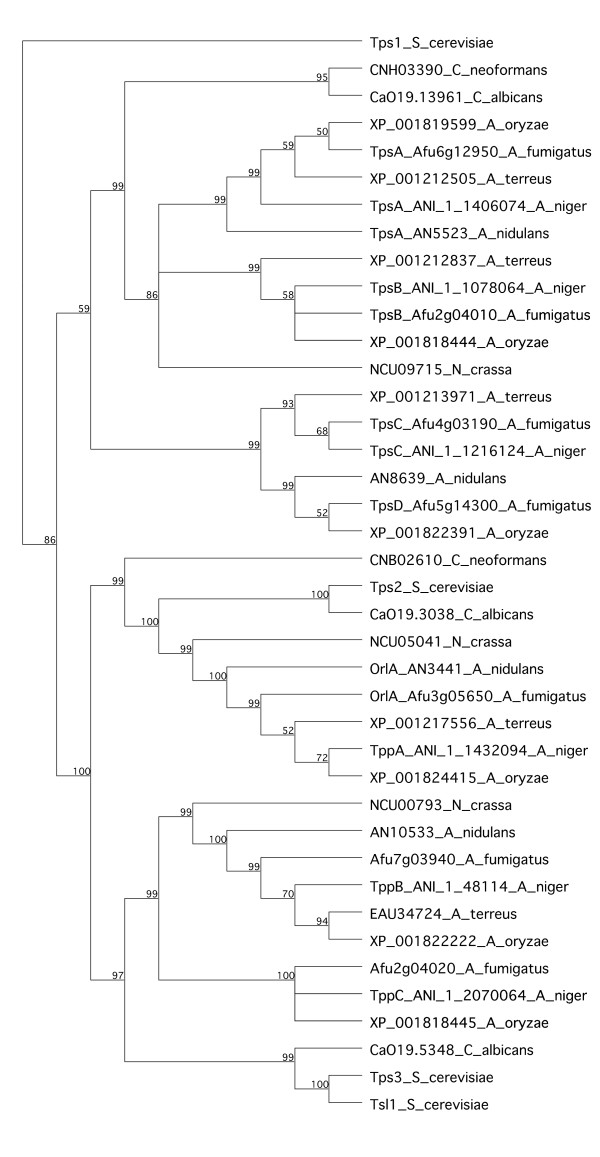
The obtained amino acid sequences of Tpp and Tps proteins were compared to those present in all known genomes of Aspergillus species, as well as other fungal species as references. For this, we used blastP at NCBI (http://blast.ncbi.nlm.nih.gov) and AspND (http://www.aspergillusgenome.org/; [38]; available August 2013). All identified fungal genomes contain at least one putative T6P synthase and trehalose-6-phosphate-phosphatases orthologous to Tps2/TppA/OrlA. In addition, TppB could be identified in all filamentous Ascomycota, whereas TppC is only present in the Aspergillus subgenera Fumigati and Circumdati. Both TppB and TppC group together with the S. cerevisiae Tps3 and Tsl1 proteins. The relationships of different gene products in some reference species are displayed as a phylogenetic tree (Figure 2). An additional observation is that, whenever present, tpsB and tppC are located adjacent on the chromosomes. The protein outside the putative trehalose synthase complex that had the highest blast score against TpsA was ANI_1_512164, encoding a glutamate carboxypeptidase, where the most similar region consisted of 30% over 50 amino acid residues. In contrast, close homologs could be identified in more distantly related species such as the bacterium Escherichia coli and the protist Dictyostelium discoideum (data not shown).
Figure 2.
Proteins in the trehalose synthesis family. Analyzed species are: A. fumigatus, A. nidulans, A. niger, A. oryzae, A. terreus, Candida albicans, Cryptococcus neoformans, Neurospora crassa and S. cerevisiae. All proteins from filamentous fungi have their accession number included. For A. niger and previously characterized gene products, given names are also included. This phylogenetic tree was built using the neighbor joining algorithm with 32 000 bootstrap replicates. Based on sequence identities, the S. cerevisiae Tps1 protein was selected by the software as outgroup. Optional settings or use of other algorithms gave identical, or very similar, results.
Two-hybrid assay to reveal putative protein-protein interactions
In order to determine whether the homologous proteins physically interact, as has been reported in S. cerevisiae[39], we performed a bacterial-based two-hybrid assay screening for interactions between all six A. niger proteins. For each protein, the full-length open reading frame was cloned into an expression vector and co-transformed into E. coli cells. All 36 possible combinations of A. niger proteins were screened, together with two clones containing different subunits of the leucine zipper GCN4 serving as a positive control and four combinations of one A. niger protein and one bacterial protein serving as negative controls. Results with no interactions were repeated at least once in an additional independent two-hybrid assay. Where interactions were detected, the assay was repeated in at least two independent assays. Results indicated that TpsB interacts with TpsA, TpsB and TppA, and that all Tps units interact with themselves (Table 4). All putative interactions involving either TppB or TppC did not score any signals above the negative controls (data not shown).
Table 4.
Protein-protein interactions assayed by Bacterial adenylate cyclase two-hybrid system
| Protein | TpsA | TpsB | TpsC | TppA |
|---|---|---|---|---|
| TpsA |
418 (210–863)* |
1746 (1582–1799) |
113 (77–135) |
71 (43–89) |
| TpsB |
1593 (1467–1832) |
1776 (1658–1988) |
441 (341–560) |
581 (322–714) |
| TpsC |
172 (101–244) |
688 (315–980) |
1214 (861–1551) |
80 (67–102) |
| TppA | 429 (167–656) | 691 (462–987) | 156 (133–198) | 83 (58–98) |
*Estimated values are in units/mg dry weight bacteria. Values in parentheses are the highest and lowest scores for each based on three to four independent assays. The positive control zip-zip (T18 and T25 fragments of the leucine zipper of GCN4) was scored to 3429 (2938–4270). Negative controls and remaining protein interactions scored at maximum 220 (zip-tpsA) but usually less than 50. Values in bold are considered true protein-protein interactions.
Gene expression during conidial outgrowth
Gene expressions were quantified during different stages of A. niger development. Preliminary results showed that due to the extractability of different structures, two RNA extraction protocols (see Methods) were required: The first included high force to break the tough cell walls of conidia and early germination structures; and, the second was more efficient for fragile structures. Notably, the second protocol was not vigorous enough to extract any RNA from spores (data not shown). Therefore, the “Plate” value in Figure 3 only represents gene expression in mycelia and conidiation structures, but not mRNA present in the produced conidia. cDNA was made with the mRNA as a template, and the relative expressions of the six putative trehalose synthesis genes, tpsA, tpsB, tpsC, tppA, tppB and tppC, were analyzed with real-time PCR.
Figure 3.
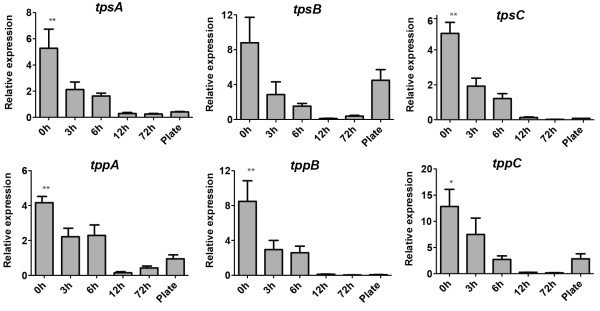
Expression of putative trehalose synthesis genes during outgrowth of A. niger conidia. The developmental stages are given on the x- axis: 0 h are dormant conidia; 3–72 h are swollen conidia, germlings or mycelia after so many hours of incubation in liquid AMM media; and Plate is the entire sporulating culture grown on AMM plates for 5 days. Error bars show standard error of the mean based on four biological replicates each calculated as the average of three technical replicates. For all genes, the expressions are normalized against the expression of actin. *Indicates that the expression at 0 h was statistically significant to the following time-points within the same group except 3 h (one-way ANOVA, P < 0.05). **Indicates that the expressions at 0 h were statistically significant to all of the following time-points within the same group (one-way ANOVA, P < 0.05).
The general expression pattern of the genes (Figure 3) was as follows: The expression was highest in still dormant conidia and had decreased by approximately 2-fold after 3 h incubation; after 6 h incubation there was a slight, but not significant, decrease; and, in 12 and 72 h mycelium the expression was very low. For tpsB, tppA and tppC, the expression was then up-regulated in sporulating colonies (5 days old), while it remained low for tpsC and tppB. One gene, tppA, deviated slightly from the described pattern: The decrease in expression after 3 h was not as profound as in the other genes, and a slight, but not significant, up-regulation could be seen in 72 h mycelium.
Targeted gene deletions of six Aspergillus niger genes
To characterize the function of the six A. niger proteins, tpsA, tpsB, tpsC, tppA, tppB and tppC were all subjected to targeted gene deletions by replacing the gene with the A. oryzae pyrG resistance cassette. A double mutant, lacking the two adjacent genes tpsB and tpsC was also constructed. All deletion mutants were confirmed with PCR using both internal and flanking primers (data not shown).
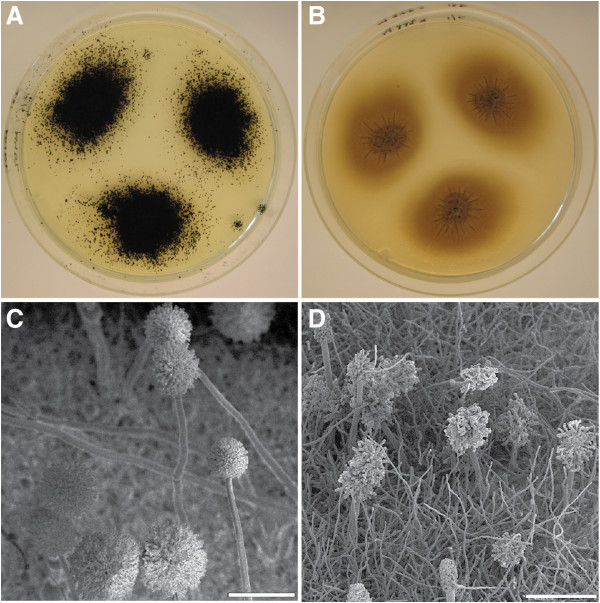
With the exception of ΔtppA, all deletion mutants showed phenotypes similar to wild-type. When culturing the wild-types and mutants at temperatures ranging from 15°C to 37°C, no strain-dependent differences in growth rates or morphologies could be observed; at 10°C no growth was observed for any strain (data not shown). The tppA mutant showed a marked reduction in the number of conidia produced compared to the other strains, giving the colonies growing on plate a whitish, and with age, light brownish appearance, compared to the black wild-type (Figure 4A,B). This phenotype was retained during aging, and under all growth conditions. When comparing the spore densities, the wild-type, N402, yielded an average of 2.44 × 106 (±0.045 × 106) spores/mm2, whereas ΔtppA yielded an average of 4.40 × 103 (±0.69 × 103) spores/mm2, i.e. a 6 × 102-fold reduction. Microscopic studies revealed that the conidiophores of ΔtppA had a clearly different appearance as is shown in Figure 4C and D. Most notably, vesicle swelling was almost completely absent and metulae were irregularly positioned (Figure 4C,D and Figure 5). However, the conidia produced showed similar size and ornamentation to wild-type (Figure 5C,F). In contrast to what has been reported in the corresponding mutant of A. fumigatus[22], it was not possible to restore wild-type morphology by growing ΔtppA on media containing an osmotic stabilizer, i.e. the described phenotype persisted in all growth conditions.
Figure 4.
Morphologies of cultures grown for 1 week on AMM. Wild-type, left (A and C), and ΔtppA right (B and D). Size bars of SEM photos are 100 μm.
Figure 5.
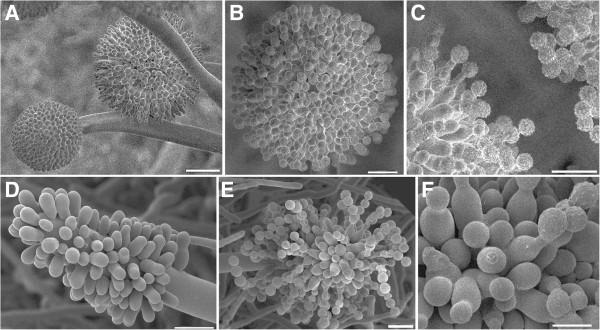
Detailed morphologies of cultures grown for 1 week on AMM. Wild-type, top (A, B and C), and ΔtppA bottom (D, E and F). Size bars: A = 20 μm, B = 10 μm, C = 10 μm, D = 10 μm, E = 10 μm, F = 5 μm.
Quantification of trehalose-6-phosphate and trehalose in wild-type and mutants
All three Tpp genes putatively encode the enzyme trehalose-6-phosphate-phosphatase. To investigate if this enzyme was absent in the Tpp deletion strains, the amount of trehalose-6-phosphate (T6P) in mycelia from wild-type, ΔtppA, ΔtppB and ΔtppC were analyzed. There were no significant differences in T6P levels between wild-type, ΔtppB or ΔtppC. In ΔtppA, however, T6P was clearly accumulated; the mycelium from this strain contained an average of 124 nmol T6P per gram dry weight compared to 18 nmol in the wild-type (Figure 6).
Figure 6.
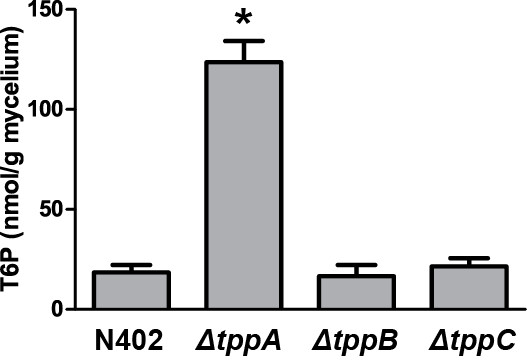
Content of T6P in mycelium dry weight of wild-type and Tpp deletion mutants. Error bars show standard error of the mean. In ΔtppA, the level of T6P was significantly higher compared to all other strains (one-way ANOVA, P < 0.05)
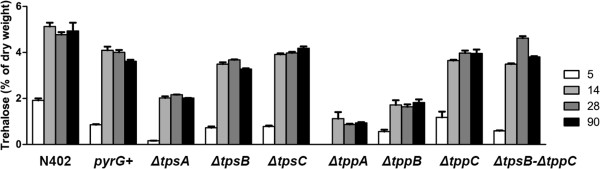
To elucidate how specific gene products influence the trehalose content of A. niger conidia in different stages of maturation, conidia were harvested from control and mutant strains after 5, 14, 28 and 90 days. In these and the following stress experiments, in addition to the wild-type N402 strain, we also included a kusA deficient strain with a repaired pyrG gene, pyrG + [28] as a control with identical genetic background as the tps and tpp deletion mutants. The dormant conidia were extracted and the trehalose levels analyzed and expressed as percentage of conidial dry weight (Figure 7). For ΔtppA it was not possible to analyze the trehalose content of 5 day conidia, as insufficient conidia were produced. For the other strains, a significant increase in trehalose was detected between the two first time points tested, 5 and 14 days. During further aging of conidia, between 14 and 90 days, no significant changes in trehalose levels were detected (with the exception of ΔtpsB-ΔtppC where the level in 28 day old conidia was 4.6% compared to 3.5 and 3.8 in 14 and 90 day old conidia, respectively).
Figure 7.
Trehalose content in mutant and wild-type conidia of different age. The numbers to the right represent how many days the colony had grown on AMM plates before conidia were harvested and analysed. Error bars show standard error of the mean.
At all time points, conidia from all mutant strains contained significantly less trehalose compared to wild-type conidia (again, with the exception of ΔtpsB-ΔtppC 28 days). When comparing the deletion mutants to the other control strain, pyrG+, significantly lower levels of trehalose were detected in strains ΔtpsA, ΔtppA and ΔtppB. After 14 days of maturation the conidial trehalose level was 50% lower in ΔtpsA compared to pyrG+, and 73 and 60% lower in ΔtppA and ΔtppB, respectively. For ΔtpsA and ΔtppA, the reduction was significant at all time points tested, and for ΔtppB, the difference was significant in 14, 28 and 90 day old conidia but not after 5 days.
Among the deletion mutants with wild-type like phenotypes, i.e. when excluding ΔtppA, ΔtppB had the lowest overall trehalose content. After 14 days of incubation, the trehalose level was 1.7% of conidial dry weight compared to 5.1 and 4.1% in wild-type N402 and pyrG+, respectively. Although the conidial trehalose content was consistently lower in ΔtppA, the extremely low number of spores produced made this strain unsuitable for studies on conidial survival. Therefore, ΔtppB was, due to its wild-type morphology, selected for additional studies to reveal whether or not a normal internal trehalose level has any impact on stress survival and growth.
Confirmation and further characterization of ΔtppB
Before subjecting the tppB deletion mutant to stress, a few confirmatory experiments were performed to ensure that the lowered trehalose content was a consequence of the deleted gene: A new deletion mutant of tppB, ΔtppB2, was generated using MA169.4 as parent strain, and on a selected transformant the ΔkusA gene was restored using acetamide. Analysis of trehalose content in 14 day old conidia from this new mutant showed that they were as low as in ΔtppB (1.54 ± 0.1% of conidia dry weight in ΔtppB2 versus 1.72 ± 0.5% in ΔtppB). Moreover, the deletions mutants were complemented by transformation of an autonomously replicating plasmid carrying the gene for hygromycin resistance as well as an intact copy of the tppB gene. Putative transformants were selected on hygromycin plates. The presence of the construct was confirmed using PCR and plasmid rescue (data not shown). In a previous study we discovered that, when using this methodology, only a fraction of conidia carry the plasmid [28]. This was also valid for tppB + conidia, where only a few percent germinated on hygromycin media (data not shown). Therefore, the mycelial trehalose content was measured after growth in liquid AMM media supplemented with hygromycin – under a continuous selection pressure to retain the plasmid, we showed that in tppB + the amount of trehalose was restored to wild-type level (Figure 8A). The expression of tppB was examined in mycelium from wild-type, ΔtppB and tppB+. In the deletion mutant, no expression was detected, whereas in the complemented strain, the levels were in the same range as in the wild-type (Figure 8B). From these experiments we concluded that the deletion of tppB causes the lowered trehalose levels in ΔtppB. However, since the plasmid carrying the wild-type version of the gene was lost in most conidia, the tppB+ strain was not included in the following experiments.
Figure 8.
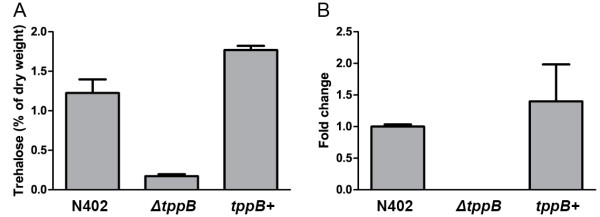
Trehalose content of mycelium (A) and relative expression of tppB (B). Error bars show standard error of the mean, based on three biological replicates, and for qPCR each biological replicate was calculated as the average of three technical replicates.
To evaluate the importance of trehalose as a stress protectant, the trehalose contents of the ΔtppB mutant and the control strains were analyzed in early stages of germination, and were subjected to lethal and sub-lethal heat and oxidative stress as well as sub-lethal salt and acid stress. The trehalose levels in ΔtppB followed the same pattern of breakdown and re-synthesis as in the control strains, but they were consistently lower in accordance with the lower initial value (Figure 9). Dormant conidia of ΔtppB were significantly less tolerant to heat stress compared to the control strains; After 60 min of heat stress, the survival of ΔtppB was 35% compared to 78% in wild-type. After an additional 60 min, the survival of ΔtppB further decreased to 2% compared to 38% in wild-type. (Figure 10). These experiments were repeated with the new independent deletion mutant, ΔtppB2, and the results were identical to those for ΔtppB (data not shown). For the other stressors tested, benzoic acid, NaCl and H2O2, as well as long-term viability where conidia were stored in water at 4°C for a total of 8 weeks, no significant differences between the mutant and the control strains could be detected (data not shown).
Figure 9.
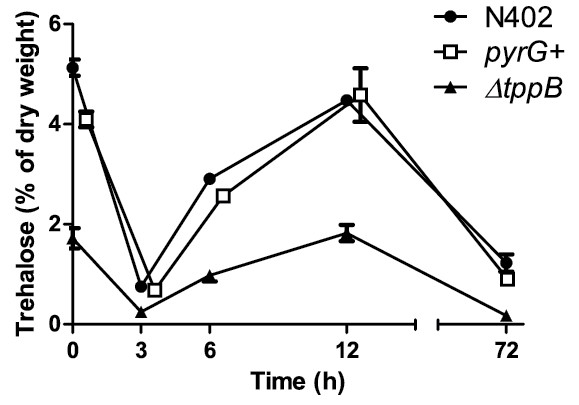
Concentration of trehalose during outgrowth of wild-type, pyrG + and ΔtppB conidia. Note the scale break between 12 and 72 h and that pyrG + observations are horizontally offset to avoid visual overlap. The error bars represent the standard error of the mean. The level of trehalose in ΔtppB was significantly different compared to wild-type for all time points except 3 h (two-way ANOVA, P < 0.0001 at 0, 6 and 12 h, and P < 0.01 at 72 h).
Figure 10.
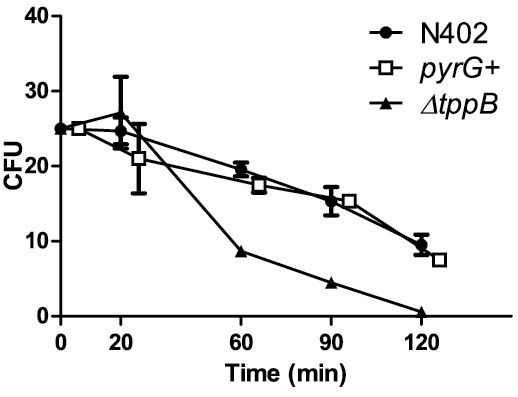
Viabilities of dormant A. niger conidia after subjection to heat stress. Conidia were held at 55°C for 20, 60, 90 and 120 min. For all strains, the numbers of counted colonies were normalized to 25 at time = 0 min to avoid differences in numbers of assayed spores. Note that pyrG + observations are horizontally offset to avoid visual overlap. There were no significant differences between the control strains (N402 and pyrG+). Except for the two first time points, the viability of ΔtppB was significantly lower than in N402 (two-way ANOVA, P < 0.01 in 60 and 90 min, and P < 0.05 in 120 min).
Discussion
In this project we have studied six genes with a putative role in trehalose synthesis in A. niger: tpsA, tpsB, tpsC, tppA, tppB and tppC. All six genes encode homologous proteins and no similar gene products within the A. niger genome could be detected. Three proteins, TpsA, TpsB and TpsC, have previously been identified as orthologs to the yeast protein Tps1. As the orthologs are conserved in related species, it is plausible that there is a functional differentiation between the paralogs, e.g. one paralog could be essential for trehalose synthesis in conidia, whereas another paralog is strictly induced by stress. This assumption is in line with the previous observation in A. niger where the expression of tpsB is stress-induced whereas tpsA is constitutively expressed [23], although our data also suggest that tpsB has a role during differentiation (see Figure 3). When deleting the trehalose-phosphate-synthase paralogs, only ΔtpsA displayed a reduced trehalose content. The lower level in this mutant is in line with a previous report using a different target strain and deletion procedure [23]. In the related fungus, A. fumigatus, a tpsA/tpsB double deletion resulted in a strain with depleted trehalose content, and in the same study, it was shown that the expressions of tpsC and –D were very low at all time points [12]. These authors evaluated their expression data using RNA extracted from hyphae, and in the present study, the A. niger tpsC was expressed at very low levels at 72 h. Thus the results from the two fungi are not contradictory, and most likely an A. niger tpsA/tpsB deletion mutant would also have a depleted trehalose content. The results from A. niger and A. fumigatus are also in accordance with findings in A. nidulans where deletion of tpsA resulted in depleted trehalose content [11], as that species does not have the tpsB paralogue. A conclusion from studying the trehalose content from these three species is that TpsA is the most important trehalose-phosphate-synthase under normal conditions, but lack of the tpsA gene can be fully compensated by TpsB in A. fumigatus and partly by at least one of TpsB or TpsC in A. niger, but not by TpsD in A. nidulans.
The deletion mutant with the most distinctive characteristics in our experiments was ΔtppA, i.e. with an abnormal morphology and reduced levels of both trehalose-6-phosphate and trehalose. The altered morphology of the strain is probably due to toxicity of T6P as indicated for the corresponding deletion mutant in A. fumigatus[22]. However, in A. niger as well as A. fumigatus and A. nidulans[12,25], mutants of tppA are not totally lacking in trehalose. Therefore, it is possible that under specific conditions, e.g. when TppA is absent, TppB, and also TppC where present, may contribute to some T6P activity. Another possibility is that the sugar can be synthesized by proteins other than Tps/Tpp, e.g. the Trehalose Phosphorylase pathway, for which putative genes have been identified and partially characterized in N. crassa[40] and A. fumigatus[22] and also exist in A. niger (ANI_1_2720024). However, it is possible to generate mutants, within the homologous Tps/Tpp group, in A. fumigatus and A. nidulans that totally lack trehalose [11,12]. Therefore, we believe that this is the only active trehalose synthesis pathway in Aspergilli. However, internal trehalose contents may not solely be dependent on the presence and expression of these six genes, as in S. cerevisiae there is a strong linkage between trehalose synthesis and the degrading trehalases [41] as well as evidences of posttranscriptional activation of the genes involved in trehalose metabolism [42,43].
Besides a putative phosphatase activity, TppB and TppC may have similar biological roles as the yeast proteins Tps3 and Tsl1, which also contain phosphatase domains – in yeasts, deletion of both genes is necessary before some reduction in internal trehalose content can be observed [17]. It is intriguing that tpsB and tppC are linked on the chromosome. We cannot explain why the conidial trehalose content in this double mutant was significantly higher after 28 days, but based on the expression patterns (see Figure 3), it is possible that the expression of the two genes are regulated by the same factors. In addition to the above-mentioned observations, some conclusions can be drawn from the gene expression data: All identified genes were expressed, indicating that the paralogs are not inactive duplicates. For tpsC and tppB, the expressions were consistently low after 6 h, indicating that the two genes may be regulated by the same mechanism. This assumption is supported by a previous observation using A. oryzae arrays where the tpsC and tppB orthologs were down-regulated in a deletion strain of atfA, a gene encoding a transcription factor [44]. To our knowledge, two previous studies describing the expression of trehalose synthesis genes in A. niger during germination, using microarray technology, or in combination with RNA sequencing, have been published [29,45]. With the exception that van Leeuwen and co-workers [29] saw a drastic drop after 2 h and then a gradual up-regulation of tpsA and tpsB, those results are in line with our findings.
The extensive measurements of internal trehalose indicate that the trehalose contents, for all strains, were low in 5 day old conidia, significantly elevated in 14 day old conidia, and then maintained at the value of 14 days (Figure 7). A plausible hypothesis is that conidia of A. niger reach full maturity, at least in terms of trehalose accumulation, sometime between 5 days and 2 weeks. Consequently it is not advisable to perform stress experiments on young conidia because their trehalose content is not necessarily typical for the final level, especially not in kusA deficient strains that seem to have slower conidial maturation in terms of trehalose content.
We found that 2 week old conidia of ΔtppB were more susceptible to heat shock than wild-type conidia, indicating that trehalose protects the spores from thermal stress. These results are in line with earlier studies in Aspergillus species [11,12,23]. However, in contrast to results from A. fumigatus and A. nidulans, we could not detect any increased sensitivity of ΔtppB to oxidative stress [11,12], salt or acid stress, or any decreased viability after long term storage. It should be noted that unlike ΔtppB in our experiments, which harbored approximately one third of wild-type trehalose content, the A. fumigatus and A. nidulans mutants were totally depleted of trehalose.
In S. cerevisiae it has been shown that, using a two-hybrid assay, the four homologous proteins physically interact. When repeating the experiments using the six identified A. niger proteins, we could observe interactions for four of six proteins. These results suggest that TppA and TpsA-C form a complex, while the phylogenetically more distant proteins, TppB and TppC, are present outside the complex. However, due to the experimental limits, it is possible that neither TppB nor TppC was correctly folded and therefore not interacting. It is notable that in S. cerevisiae, a truncated version of Tsl1 was necessary for the success of the interaction experiments [40], in contrast to our experiment in which we only used full-length proteins.
Conclusions
To conclude, in this study novel information about the six gene products involved in trehalose synthesis in A. niger has been generated. When characterizing deletion mutants, lack of the most conserved trehalose phosphate synthase tpsA, the trehalose phosphate phosphatase tppA, or the previously non-characterized tppB, resulted in lower trehalose contents. An additional insight is that the components in a putative trehalose synthesis complex differ among the Aspergilli, but some gene products are common throughout the fungal kingdom.
Competing interests
The authors declare the absence of competing interests.
Authors’ contributions
ÅS performed the majority of the laboratorial work. ÅS and PM performed all experiments with exception of the RNA extraction from dormant conidia and conidia in early stages of germination, performed by MRL, and the SEM studies, performed by JD. ÅS and PM conceived and designed the study and wrote the manuscript. All authors read and approved the final manuscript.
Contributor Information
Åsa Svanström, Email: asa.svanstrom@slu.se.
Martin Richard van Leeuwen, Email: richardvanleeuwen79@gmail.com.
Jan Dijksterhuis, Email: j.dijksterhuis@cbs.knaw.nl.
Petter Melin, Email: petter.melin@icloud.com.
Acknowledgements
Dr. Jonathan Hilmer for assistance with the T6P analysis and Dr. Su-lin Leong for proofreading the manuscript before submission, are greatly acknowledged. This work was financed by the Swedish research council Formas.
References
- Avonce N, Mendoza-Vargas A, Morett E, Iturriaga G. Insights on the evolution of trehalose biosynthesis. BMC Evol Biol. 2006;6:109. doi: 10.1186/1471-2148-6-109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iordachescu M, Imai R. Trehalose biosynthesis in response to abiotic stresses. J Integr Plant Biol. 2008;50(10):1223–1229. doi: 10.1111/j.1744-7909.2008.00736.x. [DOI] [PubMed] [Google Scholar]
- Elbein AD, Pan YT, Pastuszak I, Carroll D. New insights on trehalose: a multifunctional molecule. Glycobiology. 2003;13(4):17R–27R. doi: 10.1093/glycob/cwg047. [DOI] [PubMed] [Google Scholar]
- Thevelein JM. Regulation of trehalose mobilization in fungi. Microbiol Mol Biol Rev. 1984;48(1):42–59. doi: 10.1128/mr.48.1.42-59.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elbein AD. The metabolism of α, α-trehalose. Adv Carbohydr Chem Biochem. 1974;30:227–256. doi: 10.1016/s0065-2318(08)60266-8. [DOI] [PubMed] [Google Scholar]
- Gancedo C, Flores CL. The importance of a functional trehalose biosynthetic pathway for the life of yeasts and fungi. FEMS Yeast Res. 2004;4(4–5):351–359. doi: 10.1016/S1567-1356(03)00222-8. [DOI] [PubMed] [Google Scholar]
- Crowe JH, Hoekstra FA, Crowe LM. Anhydrobiosis. Annu Rev Physiol. 1992;54:579–599. doi: 10.1146/annurev.ph.54.030192.003051. [DOI] [PubMed] [Google Scholar]
- Wiemken A. Trehalose in yeast, stress protectant rather than reserve carbohydrate. Antonie Van Leeuwenhoek. 1990;58(3):209–217. doi: 10.1007/BF00548935. [DOI] [PubMed] [Google Scholar]
- Hottiger T, Virgilio C, Hall M, Boller T, Wiemken A. The role of trehalose synthesis for the acquisition of thermotolerance in yeast. Eur J Biochem. 1994;219(1–2):187–193. doi: 10.1111/j.1432-1033.1994.tb19929.x. [DOI] [PubMed] [Google Scholar]
- Cheng L, Moghraby J, Piper PW. Weak organic acid treatment causes a trehalose accumulation in low-pH cultures of Saccharomyces cerevisiae, not displayed by the more preservative-resistant Zygosaccharomyces bailii. FEMS Microbiol Lett. 1999;170(1):89–95. doi: 10.1111/j.1574-6968.1999.tb13359.x. [DOI] [PubMed] [Google Scholar]
- Fillinger S, Chaveroche M-K, van Dijck P, de Vries R, Ruijter G, Thevelein J, d’Enfert C. Trehalose is required for the acquisition of tolerance to a variety of stresses in the filamentous fungus Aspergillus nidulans. Microbiology. 2001;147(7):1851–1862. doi: 10.1099/00221287-147-7-1851. [DOI] [PubMed] [Google Scholar]
- Al-Bader N, Vanier G, Liu H, Gravelat FN, Urb M, Hoareau CMQ, Campoli P, Chabot J, Filler SG, Sheppard DC. Role of trehalose biosynthesis in Aspergillus fumigatus development, stress response, and virulence. Infect Immun. 2010;78(7):3007–3018. doi: 10.1128/IAI.00813-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uyar EO, Hamamci H, Turkel S. Effect of different stresses on trehalose levels in Rhizopus oryzae. J Basic Microbiol. 2010;50(4):368–372. doi: 10.1002/jobm.200900339. [DOI] [PubMed] [Google Scholar]
- Doehlemann G, Berndt P, Hahn M. Trehalose metabolism is important for heat stress tolerance and spore germination of Botrytis cinerea. Microbiol-Sgm. 2006;152:2625–2634. doi: 10.1099/mic.0.29044-0. [DOI] [PubMed] [Google Scholar]
- Jain NK, Roy I. Effect of trehalose on protein structure. Protein Sci. 2009;18(1):24–36. doi: 10.1002/pro.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lins RD, Pereira CS, Hünenberger PH. Trehalose–protein interaction in aqueous solution. Proteins Struct Funct Bioinf. 2004;55(1):177–186. doi: 10.1002/prot.10632. [DOI] [PubMed] [Google Scholar]
- Bell W, Sun WN, Hohmann S, Wera S, Reinders A, De Virgilio C, Wiemken A, Thevelein JM. Composition and functional analysis of the Saccharomyces cerevisiae trehalose synthase complex. J Biol Chem. 1998;273(50):33311–33319. doi: 10.1074/jbc.273.50.33311. [DOI] [PubMed] [Google Scholar]
- de Virgilio C, Burckert N, Bell W, Jeno P, Boller T, Wiemken A. Disruption of Tps2, the gene encoding the 100-kDa subunit of the trehalose-6-phosphate synthase phosphatase complex in Saccharomyces cerevisiae, causes accumulation of trehalose-6-phosphate and loss of trehalose-6-phopshate phosphatase activity. Eur J Biochem. 1993;212(2):315–323. doi: 10.1111/j.1432-1033.1993.tb17664.x. [DOI] [PubMed] [Google Scholar]
- Londesborough J, Vuorio O. Trehalose-6-phosphate synthase/phosphatase complex from bakers’ yeast: purification of a proteolytically activated form. J Gen Microbiol. 1991;137(2):323–330. doi: 10.1099/00221287-137-2-323. [DOI] [PubMed] [Google Scholar]
- d’Enfert C. Fungal spore germination: insights from the molecular genetics of Aspergillus nidulans and Neurospora crassa. Fungal Genet Biol. 1997;21(2):163–172. doi: 10.1006/fgbi.1997.0975. [DOI] [Google Scholar]
- Foster AJ, Jenkinson JM, Talbot NJ. Trehalose synthesis and metabolism are required at different stages of plant infection by Magnaporthe grisea. EMBO J. 2003;22(2):225–235. doi: 10.1093/emboj/cdg018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puttikamonkul S, Willger SD, Grahl N, Perfect JR, Movahed N, Bothner B, Park S, Paderu P, Perlin DS, Cramer RA Jr. Trehalose 6-phosphate phosphatase is required for cell wall integrity and fungal virulence but not trehalose biosynthesis in the human fungal pathogen Aspergillus fumigatus. Mol Microbiol. 2010;77(4):891–911. doi: 10.1111/j.1365-2958.2010.07254.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolschek MF, Kubicek CP. The filamentous fungus Aspergillus niger contains two “differentially regulated” trehalose-6-phosphate synthase-encoding genes, tpsA and tpsB. J Biol Chem. 1997;272(5):2729–2735. doi: 10.1074/jbc.272.5.2729. [DOI] [PubMed] [Google Scholar]
- Thevelein JM, Hohmann S. Trehalose synthase – guard to the gate of glycolysis in yeast. Trends Biochem Sci. 1995;20(1):3–10. doi: 10.1016/S0968-0004(00)88938-0. [DOI] [PubMed] [Google Scholar]
- Borgia PT, Miao YH, Dodge CL. The orlA gene from Aspergillus nidulans encodes a trehalose-6-phosphate phosphatase necessary for normal growth and chitin synthesis at elevated temperatures. Mol Microbiol. 1996;20(6):1287–1296. doi: 10.1111/j.1365-2958.1996.tb02647.x. [DOI] [PubMed] [Google Scholar]
- Schuster E, Dunn-Coleman N, Frisvald JC, van Dijck PW. On the safety of Aspergillus niger-a review. Appl Microbiol Biotech. 2002;59:426–435. doi: 10.1007/s00253-002-1032-6. [DOI] [PubMed] [Google Scholar]
- Bos CJ, Debets AJM, Swart K, Huybers A, Kobus G, Slakhorst SM. Genetic-analysis and the construction of master strains for assignment of genes to 6 linkage groups in Aspergillus niger. Curr Genet. 1988;14(5):437–443. doi: 10.1007/BF00521266. [DOI] [PubMed] [Google Scholar]
- Svanström Å, Melin P. Intracellular trehalase activity is required for development, germination and heat-stress resistance of Aspergillus niger conidia. Res Microbiol. 2013;164(2):91–99. doi: 10.1016/j.resmic.2012.10.018. [DOI] [PubMed] [Google Scholar]
- van Leeuwen MR, Krijgsheld P, Bleichrodt R, Menke H, Stam H, Stark J, Wosten HAB, Dijksterhuis J. Germination of conidia of Aspergillus niger is accompanied by major changes in RNA profiles. Stud Mycol. 2013;74:59–70. doi: 10.3114/sim0009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plumridge A, Melin P, Stratford M, Novodvorska M, Shunburne L, Dyer PS, Roubos JA, Menke H, Stark J, Stam H, Archer DB. The decarboxylation of the weak-acid preservative, sorbic acid, is encoded by linked genes in Aspergillus spp. Fungal Genet Biol. 2010;47(8):683–692. doi: 10.1016/j.fgb.2010.04.011. [DOI] [PubMed] [Google Scholar]
- Bohle K, Junglebloud A, Göcke Y, Dalpiaz A, Cordes C, Horn H, Hempel DC. Selection of reference genes for normalisation of specific gene quantification data of Aspergillus niger. J Biotech. 2007;132:353–358. doi: 10.1016/j.jbiotec.2007.08.005. [DOI] [PubMed] [Google Scholar]
- Pfaffl MW. A new mathematical model for relative quantification in real-time RT–PCR. Nucleic Acids Res. 2001;29(9):e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer V, Arentshorst M, El-Ghezal A, Drews A-C, Kooistra R, van den Hondel CAMJJ, Ram AFJ. Highly efficient gene targeting in the Aspergillus niger kusA mutant. J Biotechnol. 2007;128:770–775. doi: 10.1016/j.jbiotec.2006.12.021. [DOI] [PubMed] [Google Scholar]
- Carvalho N, Arentshorst M, Kwon MJ, Meyer V, Ram AFJ. Expanding the ku70 toolbox for filamentous fungi: establishment of complementation vectors and recipient strains for advanced gene analyses. Appl Microbiol Biotechnol. 2010;87(4):1463–1473. doi: 10.1007/s00253-010-2588-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudasova Z, Dudas A, Chovanec M. Non-homologous end-joining factors of Saccharomyces cerevisiae. FEMS Microbiol Rev. 2004;28:581–601. doi: 10.1016/j.femsre.2004.06.001. [DOI] [PubMed] [Google Scholar]
- Pel HJ, de Winde JH, Archer DB, Dyer PS, Hofmann G, Schaap PJ, Turner G, de Vries RP, Albang R, Albermann K, Andersen MR, Bendtsen JD, Benen JAE, van den Berg M, Breestraat S, Caddick MX, Contreras R, Cornell M, Coutinho PM, Danchin EGJ, Debets AJM, Dekker P, van Dijck A, Dijkhuizen L, Driessen AJM, d’Enfert C, Geysens S, Goosen C, Groot GSP. Genome sequencing and analysis of the versatile cell factory Aspergillus niger CBS 513.88. Nat Biotechnol. 2007;25(2):221–231. doi: 10.1038/nbt1282. [DOI] [PubMed] [Google Scholar]
- Marchler-Bauer A, Lu S, Anderson JB, Chitsaz F, Derbyshire MK, DeWeese-Scott C, Fong JH, Geer LY, Geer RC, Gonzales NR, Gwadz M, Hurwitz DI, Jackson JD, Ke Z, Lanczycki CJ, Lu F, Marchler GH, Mullokandov M, Omelchenko MV, Robertson CL, Song JS, Thanki N, Yamashita RA, Zhang D, Zhang N, Zheng C, Bryant SH. CDD a Conserved Domain Database for the functional annotation of proteins. Nucleic Acids Res. 2011;39:D225–D229. doi: 10.1093/nar/gkq1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnaud MB, Cerqueira GC, Inglis DO, Skrzypek MS, Binkley J, Chibucos MC, Crabtree J, Howarth C, Orvis J, Shah P, Wymore F, Binkley G, Miyasato SR, Simison M, Sherlock G, Wortman JR. The Aspergillus Genome Database (AspGD): recent developments in comprehensive multispecies curation, comparative genomics and community resources. Nucleic Acids Res. 2012;40(D1):D653–D659. doi: 10.1093/nar/gkr875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinders A, Bürckert N, Hohmann S, Thevelein JM, Boller T, Wiemken A, De Virgilio C. Structural analysis of the subunits of the trehalose-6-phosphate synthase/phosphatase complex in Saccharomyces cerevisiae and their function during heat shock. Mol Microbiol. 1997;24(4):687–696. doi: 10.1046/j.1365-2958.1997.3861749.x. [DOI] [PubMed] [Google Scholar]
- Shinohara ML, Correa A, Bell-Pedersen D, Dunlap JC, Loros JJ. Neurospora Clock-Controlled Gene 9 (ccg-9) encodes trehalose synthase: circadian regulation of stress responses and development. Eukaryot Cell. 2002;1(1):33–43. doi: 10.1128/EC.1.1.33-43.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jules M, Beltran G, Francois J, Parrou JL. New insights into trehalose metabolism by Saccharomyces cerevisiae:NTH2 encodes a functional cytosolic trehalase, and deletion of TPS1 reveals Ath1p-dependent trehalose mobilization. Appl Environ Microbiol. 2008;74(3):605–614. doi: 10.1128/AEM.00557-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirimburegama K, Durnez P, Keleman J, Oris E, Vergauwen R, Mergelsberg H, Thevelein JM. Nutrient-induced activation of trehalose in nutrient-starved cells of the yeast Saccharomyces cerevisiae: cAMP is not involved as second messenger. J Gen Microbiol. 1992;138:2035–2043. doi: 10.1099/00221287-138-10-2035. [DOI] [PubMed] [Google Scholar]
- Giots F, Donaton MCV, Thevelein JM. Inorganic phosphate is sensed by specific phosphate carriers and acts in concert with glucose as a nutrient signal for activation of the protein kinase A pathway in the yeast Saccharomyces cerevisiae. Mol Microbiol. 2003;47(4):1163–1181. doi: 10.1046/j.1365-2958.2003.03365.x. [DOI] [PubMed] [Google Scholar]
- Sakamoto K, Iwashita K, Yamada O, Kobayashi K, Mizuno A, Akita O, Mikami S, Shimoi H, Gomi K. Aspergillus oryzae atfA controls conidial germination and stress tolerance. Fungal Genet Biol. 2009;46(12):887–897. doi: 10.1016/j.fgb.2009.09.004. [DOI] [PubMed] [Google Scholar]
- Novodvorska M, Hayer K, Pullan ST, Wilson R, Blythe MJ, Stam H, Stratford M, Archer DB. Trancriptional landscape of Aspergillus niger at breaking of conidial dormancy revealed by RNA-sequencing. BMC Genomics. 2013;14:246. doi: 10.1186/1471-2164-14-246. [DOI] [PMC free article] [PubMed] [Google Scholar]