Abstract
Purpose: To determine whether interleukin-20 receptors (IL-20R) are expressed in trabecular meshwork cells and the effect of a T104M mutation in IL-20R2 on downstream cellular functions.
Methods: Evaluation of signal transducer and activator of transcription (STAT)3 phosphorylation and generic matrix metalloproteinase (MMP) activity in primary open angle glaucoma (POAG) dermal fibroblasts (pHDF) with the T104M IL-20R2 mutation were compared with normal human dermal fibroblasts (HDF). Expression of IL-20R1 and IL-20R2 in human trabecular meshwork (HTM) cells was determined by immunohistochemistry and western immunoblotting.
Results: A T104M mutation in IL20-R2 was identified in a large POAG family in which the GLC1C locus was originally mapped. pHDFs harboring this mutation had significantly increased phosphorylated STAT3 (pSTAT3) activity compared with normal HDFs. However, stimulation with either IL-19 or IL-20 for 15 min resulted in significantly decreased levels of pSTAT3 in pHDFs compared with controls. Generic MMP activity was significantly decreased in pHDFs compared with controls after stimulation with IL-20 for 24 h. Both IL-20R1 and IL-20R2 receptors were expressed in HTM cells by western immunoblot and immunofluorescence, and they appeared to be up-regulated in response to cytokine treatment.
Conclusions: A T104M mutation in IL-20R2 significantly impacts the function of this receptor as shown by decreased pSTAT3 levels and generic MMP activity. Reduced MMP activity may affect the ability of glaucoma patients to alter outflow resistance in response to elevated intraocular pressure.
Introduction
Glaucoma is one of the leading causes of blindness in the world with close to 80 million individuals predicted to develop this devastating disease by 2020.1 The most common form of glaucoma, primary open angle glaucoma (POAG), will affect 58.6 million individuals by 2020.2 Due to the subtlety of this disease and limited public awareness, the majority of individuals with glaucoma remain undiagnosed. Often, extensive and usually bilateral visual field loss should occur before individuals seek treatment. Unfortunately, once loss of vision has occurred, it cannot be reversed. Vision loss from glaucoma has a significant impact on health-related quality of life with the overall burden increasing as glaucomatous damage and vision loss progress.
The exact etiology of POAG remains unknown, although it is clear that it is a heterogeneous group of disorders and is frequently associated with elevated intraocular pressure (IOP). The genetic complexity of POAG is demonstrated by the mapping of more than 20 glaucoma loci,3 the identification of common risk alleles,4–9 the finding of affected individuals with mutations in multiple genes,10,11 and compound heterozygous mutations in myocilin often leading to a more severe disease.12–14 Thus, finding causative glaucoma genes is not a simple task.
Several lines of evidence suggest that inflammatory cytokines in human trabecular meshwork (HTM) cells may be involved in the onset and/or progression of glaucoma. Interleukin-6 (IL-6) is up-regulated in HTM by oxidative stress,15 elevated IOP,16 early pressure-induced optic nerve head injury,17 and mechanical stress.18 IL-6 increases outflow facility18 and is neuroprotective,19 suggesting that IL-6 may play a protective role in glaucoma. Other cytokines and growth factors, including transforming growth factor beta-1 (TGFβ1), TGFβ2, IL-6, IL-8, IL-10, IL-12, α-serum amyloid A, interferon-γ (IFNγ), and CXL9, are increased in POAG aqueous humor compared with normal controls, or from aqueous derived from patients undergoing cataract surgery.20–25 Collectively, these studies implicate altered cytokine signaling in glaucoma.
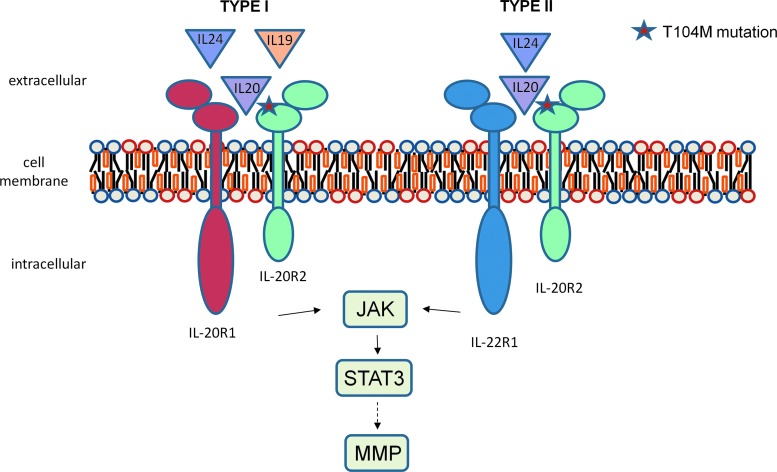
IL-6 production is induced by the IL-10 family of cytokines, that is IL-20, IL-19, and IL-24.26,27 These cytokines exert their biological activities by binding to 2 heterodimeric IL-20 receptor complexes on the cell surface: the type I receptor and the type II receptor.28–30 The interleukin-20 receptor (IL-20R)2 protein is common to both classes. Dimerization of IL-20R2 with IL-20R1 (receptor I class) mediates signaling from IL-20, IL-19, and IL-24, whereas dimerization of IL-20R2 with IL-22R1 (receptor II class) propagates signaling from IL-20 and IL-24 (Fig. 1). Despite sharing common receptor complexes, these 3 cytokines appear to have distinct roles in diverse biological processes: IL-19 directly affects immune cells; IL-20 is involved in keratinocyte biology, psoriasis, and skin biology, while IL-24 is pro-apoptotic for a variety of cancer cells.28,31–34 After binding of the cytokine to the IL-20 receptor, signals are propagated intracellularly via the Janus kinase and signal transducer and activator of transcription (JAK-STAT) signal transduction pathway.31,35 In addition, a previous study showed that matrix metalloproteinase (MMP) protein levels were increased in mouse breast cancer cells treated with IL-20, suggesting that one downstream effect of IL-20 stimulation is altered through MMP activity.36
FIG. 1.
Interleukin-20 receptor (IL-20R)2 co-receptor family. Ligands are shown above their respective receptors. The T104M mutation in IL-20R2 is indicated by a star with its impact on binding to IL-20, IL-19, and IL-24. Signaling through the class 2 receptor involves Janus kinase (JAK) and signal transducer and activator of transcription (STAT)3 activation through phosphorylation. Activation of STAT3 can lead to altered matrix metalloproteinase (MMP) levels and activity.
In this study, we identify a mutation in IL-20R2, T104M, in a large POAG family in which the GLC1C locus was originally mapped. This mutation lies in the active binding site of IL-20R2 to the cytokines, IL-19, IL-20, and IL-24. Phosphorylation of STAT3 and MMP activity in normal and glaucomatous dermal fibroblasts was also investigated. Since HTM cells are involved in the regulation of IOP under both normal and disease conditions,37 we also demonstrate that TM cells express IL-20R2 and IL-20R1.
Methods
Ethics approval and consent
Human subjects
Ethics approval was obtained from the Oregon Health & Science University (OHSU) Institutional Review Board and the Kaiser Permanente Northwest Center for Health Research Institutional Review Board. This study was conducted in accordance with the tenets of the Declaration of Helsinki with informed consent obtained from all participants.
Donor tissue
Informed consent to tissue donation was obtained from the donors or their relatives. The protocol of the study was approved by the local ethics committee and adhered to the tenets of the Declaration of Helsinki for experiments involving human tissue. For HTM cell isolation, cadaver eyes were obtained from Lions Eye Bank (Portland, OR).
Study populations
All patients and controls were of non-Hispanic European ancestry. We recruited 140 POAG subjects from Kaiser Permanente Northwest (Portland, OR) and 90 patients from the Casey Eye Institute, OHSU (Portland, OR). The age of the patients from Kaiser Permanente ranged from 34 to 93 with a mean age of 72.5±11.5 years. Of these patients, 43% had a family history of glaucoma and 60% were women. The patients from the Casey Eye Institute ranged in age from 34 to 96 with a mean age of 70.6±11.5 years; 67% were women. We enrolled 109 control subjects; 62% were women. Their age ranged from 31 to 86 with a mean of 67±13.2 years. None of the control subjects had signs of glaucoma or ocular hypertension as determined by a clinical examination by a board-certified ophthalmologist.
Clinical examination
Clinical examination and diagnosis has been previously documented.38,39 POAG was clinically defined as an optic neuropathy with the following findings: optic nerve head excavation with thinning of the neuroretinal rim, often with “Drance”-type nerve fiber layer hemorrhages, notching, pitting, significant focal loss or general loss of retinal fiber layer, and visual field defects consistent with the disc changes.39 POAG cases were enrolled in the study if they met one or more of the following criteria: (1) treatment for POAG had been instigated before our study and examination of the patient confirmed that they had POAG; (2) two or more of the following findings were present: (a) optic nerve head and/or nerve fiber layer analysis was compatible with moderately advanced glaucomatous damage (vertical cup/disc ratio >0.7)40; (b) abnormal Humphrey glaucoma hemifield test; (c) untreated IOP of 24 mmHg measured by Goldman applanation tonometry41; and (3) glaucomatous field defects on Humphrey perimetry. Exclusion criteria were diagnosis of secondary OAG, including exfoliation glaucoma, pigmentary glaucoma, trauma, uveitis, or steroid-induced glaucoma.
IOP was measured using a recently calibrated Goldmann applanation tonometer. A slit-lamp biomicroscope was used to classify optic disc appearance after pupil dilation. The sizes of the cups were evaluated using the standard method of a reference spot from a direct ophthalmoscope or using a 78-diopter lens.
Polymerase chain reaction and sequencing
Primers were designed to amplify each exon with surrounding intronic sequence in order to cover all coding sequences and 5′ and 3′ untranslated regions (UTRs). Primer3 was used to design primers through the UCSC Human Genome Browser ExonPrimer command (http://genome.ucsc.edu/cgi-bin/hgGateway). Primers were purchased from Integrated DNA Technologies, Inc. (San Diego, CA).
Genomic DNA was obtained from human blood by a standard salting-out procedure.38 Polymerase chain reaction (PCR) reagents were obtained from Roche Applied Science Division (Indianapolis, IN), and REDtaq® Polymerase was purchased from (Sigma-Aldrich, St. Louis, MO). PCR was performed on genomic DNA according to standard touchdown protocols on a Gene Amp PCR System 9700 (Applied Biosystems, Inc., Foster City, CA). Exo-SAP-IT was used to purify PCR products according to the manufacturer's instructions (USB Corporation, Cleveland, OH). Purified products separated by agarose gel electrophoresis were sequenced on an ABI 3130XL Genetic Analyzer at the Oregon Clinical and Translational Research Institute (OCTRI). Sequencing chromatograms were analyzed using Sequencer 4.9 (Gene Codes Corporation, Ann Arbor, MI).
Restriction enzyme IL-20R2 mutation detection
PCR primers were designed with the SNP of interest located mid-product (forward primer: 5′ AAGGGACGATGG ATGGTTGG 3′ reverse primer: 5′ CTAAAGGAGGGGC TGGTTGG 3′). PCR was performed according to standard touchdown protocols using 25 ng of genomic DNA added to PCR reagents and REDtaq polymerase with a 25-μL final volume and an annealing temperature of 57°C. After PCR, 0.5 μL of CviAII restriction enzyme (5,000 units/mL supplied by NEB, Ipswich, MA) was added to 20 μL of PCR product and incubated for 1 h at 25°C in a Gene Amp PCR System 9700. After digestion, the DNA fragments were then separated by agarose gel (2% containing ethidium bromide) electrophoresis at 100 volts for 1 h.
Primary cell cultures
Human TM cells were established from cadaver eyes as previously described.42–44 HTM cells from 3 individuals were evaluated (average age=30.6±14.15 years; range=22–47 years). HTM cells were cultured in medium glucose Dulbecco's modified Eagle's medium (DMEM; Invitrogen, Carlsbad, CA) containing 10% fetal bovine serum (FBS) and antibiotic-antimycotic. Primary HTM cells were used until passage 6.
Primary dermal skin fibroblasts from glaucomatous patients (pHDFs) were cultured from punch skin biopsies from 2 female patients (94099 and 93005), who were 52 and 39 years old, respectively, at the time of the biopsy. Skin biopsies were placed in flasks containing low-glucose DMEM that was supplemented with 10% FBS and 1% penicillin–streptomycin and used for approximately passage 8.45 In addition, 3 normal adult human dermal fibroblasts (HDF) were obtained from ATCC (female, 50 years old; Manassas, VA), Lifeline Cell Technology (female, 34 years old; Frederick, MD), and Promocell (male, 60 years old; Heidelberg, Germany) and cultured in the same media as pHDFs. DNA from all these adult fibroblasts was isolated and sequenced to confirm that the cells did not harbor a mutation in the IL-20R2 gene. All pHDFs and normal HDFs showed a similar spindle-shaped cell phenotype by light microscopy, and they were sub-cultured when they were 80%–100% confluent. Normal HDFs were used for approximately passage 15.
Antibodies
The following primary antibodies were used: a rabbit polyclonal anti-human IL-20R2 (NBP1-55832; Novus Biologicals, Littleton, CO), which recognizes an internal region of IL-20R2, a rabbit polyclonal anti-human IL-20R2 (NBP1-88020; Novus Biologicals), which recognizes the C-terminus of IL-20R2, a goat polyclonal anti-human IL-20R2 antibody (BAF1788; R&D Systems, Minneapolis, MN), which recognizes the extracellular region, and a mouse monoclonal anti-human IL-20R1 (MAB11761; R&D Systems).
Western immunoblotting
HTM cells were grown to confluence in 6-well plates, washed with phosphate-buffered saline (PBS), and placed in serum-free DMEM. HTM cells were treated for 24 h with IL-19 (100 ng/mL), IL-20 (100 and 200 ng/mL), and IL-24 (100 ng/mL) recombinant proteins (R&D Systems). Vehicle control was PBS. Cells were lysed in radioimmunoprecipitation (RIPA) buffer containing protease inhibitor cocktail (Sigma-Aldrich). Proteins in cell lysates were quantitated using the BCA assay kit (Pierce, Rockford, IL), and equal amounts of total protein were separated on 10% sodium dodecyl sulfate–polyacrylamide gel electrophoresis gels (BioRad Labs, Hercules, CA) under reducing conditions. After transferring to nitrocellulose, immunoblots were probed with goat anti-IL-20R2 or mouse anti-IL-20R1. Secondary antibodies were IRDye 800-conjugated anti-goat or anti-mouse (Rockland Immunochemicals, Gilbertsville, PA). Immunoblots were imaged using the Odyssey infrared imaging system (Li-Cor Biosciences, Lincoln, NE). Density of gel bands were quantitated using FIJI software (http://fiji.sc/wiki/index.php/Fiji) following background correction.
Immunofluorescence and confocal microscopy
HTM cells were cultured for 3 days on collagen-coated Bioflex membranes (FlexCell Int, Hillsborough, NC) as previously described.46,47 This provides a more compliant culture environment than plastic tissue culture flasks and is, therefore, more similar to their native environment. HTM cells were washed and treated with cytokines as described earlier. Cells were fixed with 4% paraformaldehyde (PFA) and blocked with CAS universal blocking reagent (Invitrogen). Membranes were removed from the dish, placed on a glass slide, and immunostained with rabbit anti-IL-20R2 (55832) and mouse anti-IL-20R1. For a negative control, primary antibody was substituted with PBS. Secondary antibodies were Alexa Fluor 488-conjugated anti-mouse or anti-goat and Alexa Fluor 594-conjugated anti-rabbit (Molecular Probes, Eugene, OR). Coverslips were mounted in ProLong gold containing DAPI nuclear stain (Invitrogen). Images were obtained with a Fluoview laser scanning confocal microscope (Olympus, San Diego, CA) and processed with FIJI software.
STAT3 assay
STAT3 phosphorylation, as a measure of STAT3 activation, was quantitated in normal and pHDFs in response to cytokine treatment using an ELISA assay kit (Ray Biotech, Norcross, GA). Normal HDFs and pHDFs were cultured in 12-well plates in serum-containing DMEM until they were confluent. The cells were washed with PBS and cytokines (as described earlier) in serum-free media and were added for 15 min. Cells were washed with PBS, lysed with cell lysis buffer containing 1 mM sodium vanadate and protease inhibitor cocktail (Sigma-Aldrich), and placed on ice for 30 min. Samples were centrifuged to pellet debris, and then, 100 μL of each sample was added in duplicate to the 96-well ELISA plate, which was pre-coated with a pan-STAT3 antibody. A positive control supplied with the kit was also included on the plate. Samples were incubated overnight at 4°C, extensively washed and then, one of the duplicate samples was incubated with rabbit anti-phosphorylated STAT3(Y705), while the other well was incubated with a biotinylated anti-pan-STAT3 antibody to quantitate total STAT3 in each sample. Primary antibodies were incubated for 1 h at room temperature. After another 4 wash steps, HRP-conjugated anti-rabbit IgG or HRP-streptavidin were added to the appropriate wells and incubated for 1 h at room temperature. After a final 4 wash steps, 3,3′5,5′-tetramethylbenzidine (TMB) substrate was added to each well for 30 min and then, stop solution (0.2 M sulfuric acid) was added. The absorbance of each well was immediately measured at 450 nm on a plate reader (Perkin Elmer, Waltham, MA). The levels of phosphorylated STAT3 (pSTAT3) were divided by pan-STAT3 levels, and then, data from POAG (n=2) and normal (n=3) cells were averaged±standard error of the mean (SEM). Significance was determined using ANOVA, and P<0.05 was considered significant.
MMP assay
The Sensolyte 520 generic MMP assay kit (Anaspec, Fremont, CA) was used to measure MMP activity in HDF cells. In this assay, a quenched fluorescence resonance energy transfer (FRET) peptide is used as an MMP substrate. On cleavage of the intact peptide by an MMP, fluorescence is generated and can be measured by a plate reader. Relative fluorescent units (RFUs) are directly correlated to MMP activity. The peptide used in this assay is a generic substrate and can be cut by MMPs −1, −2, −3, −7, −8, −9, −10, −12, −13, and −14. Normal HDFs and pHDFs were treated in serum-free media treated with and without cytokines as described earlier for 24 h, and conditioned media was harvested with the addition of protease inhibitor cocktail. After centrifugation (10,000 g for 15 min), MMPs were activated with 1 mM 4-aminophenylmercuric acetate (APMA) for 90 min on ice. The quenched peptide substrate was then incubated with the samples in a 96-well format for 60 min. RFUs were quantitated at excitation/emission wavelengths (490/520 nm), normalized for total protein in each sample and then, a percentage of the untreated control was calculated. Data from 2 glaucomatous individuals and 3 normal individuals were averaged (“n” stated in figure legend±SEM). ANOVA was used to determine significance.
Results
Mutation screening of the genes in the GLC1C region
The POAG gene in a large Oregon family was mapped to chromosome 3, the GLC1C locus, and later refined to a 4-cM region between D3S3637 and D3S3694.39,48 Sanger sequencing of the 49 genes in the narrowed region identified only 1 nonsynonymous mutation with a minor frequency of less than 5% in the general population that was present in all of the screened affected individuals. This variant was a T104M change in IL-20R2 (rs367923973). Screening of 230 random POAG patients and 109 controls did not find this variant. This SNP is not present in the 1000 Genomes project but has been identified by the ESP Cohort Populations with no reported frequency.
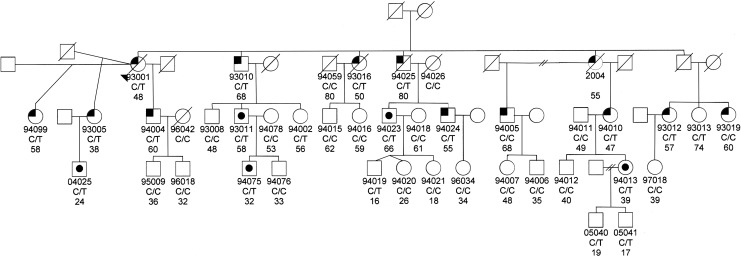
We then screened the entire GLC1C family for the T104M variant. Ten of the affected individuals had the mutation, while 2 of the affected did not (93019 and 94005; Fig. 2). These 2 individuals were diagnosed with POAG after the publication of the mapping of the GLC1C locus to chromosome 3. Neither of them have the GLC1C haplotype; therefore, they would not be expected to have the T104M mutation. Both have normal visual fields; their POAG classification is based on their ophthalmologists' diagnoses. The other one affected individual had no DNA sample, so it is unknown whether she had the T104M mutation. Altogether, 20 of the 36 family members have the T104M mutation. Of these 20, 10 are affected, 5 are glaucoma suspects based either on high IOPs or on optic nerve findings, and 5 had normal ocular findings at the time of examination (Table 1).
FIG. 2.
Pedigree of the GLC1C family. Below the symbol is the identification number of those family members for whom we have DNA and/or clinical information. Below the identification number is the IL-20R2 genotype (C/T indicates IL-20R2 mutation, C/C indicates normal IL-20R2 genotype) and age of last exam or age of diagnosis, if they have primary open angle glaucoma (POAG). No DNA was available for individual 2004; therefore, the IL-20R2 genotype is not listed. The left upper corner is black if they have been diagnosed with POAG. A black dot in the middle of the symbol indicates that they are a suspect. The genotype is not shown for those individuals for whom we have no DNA sample.
Table 1.
Summary of the Clinical Characteristics of Family Members with the IL-20R2 T104M Mutation
| Cup to disc ratio | Central corneal thickness | Maximum IOP | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Individual no. | Age at diagnosis or exam | Diagnosis | OD | OS | OD | OS | OD | OS | Visual field | Surgery |
| 93001 | 48 | POAG | 0.9 | 0.9 | 0.552 | NA | 26 | 46 | Severe loss OU | Trabeculectomy OU |
| 94099 | 58 | POAG | 0.7 | 0.4 | 0.578 | 0.568 | 21 | 28 | Superior loss OD | Trabeculectomy OU |
| 93005 | 38 | POAG | 0.7 | 0.7 | 0.544 | 0.550 | 28 | 24 | Nasal step OS | None |
| 93010 | 68 | POAG | 0.9 | 0.9 | 0.527 | 0.517 | 24 | 25 | Superior loss OD | None |
| 93012 | 57 | POAG | 0.7 | 0.6 | 0.572 | 0.592 | 22 | 23 | Nasal step OU | None |
| 93016 | 50 | POAG | 0.9 | 0.9 | NA | NA | 22 | 22 | NA | Laser trabeculoplasty OU |
| 94004 | 60 | POAG | 0.7 | 0.6 | NA | NA | 26 | 24 | NA | None |
| 94010 | 47 | POAG | 0.7 | 0.8 | 0.608 | 0.580 | 24 | 24 | Nasal step OS | None |
| 94024 | 55 | POAG | 0.5 | 0.7 | 0.599 | 0.594 | 21 | 25 | Nasal step OS | None |
| 94025 | 80 | POAG | 0.5 | 0.9 | NA | NA | 35 | 36 | NA | Laser trabeculoplasty OS |
| 02004a | 55 | POAG | 0.9 | 0.9 | NA | NA | 35 | 26 | Severe loss OU | Trabeculectomy OU |
| 04025 | 24 | Suspect | 0.3 | 0.3 | 0.590 | 0.590 | 32 | 25 | NA | None |
| 93011 | 58 | Suspect | 0.4 | 0.4 | 0.604 | 0.593 | 23 | 23 | Normal | None |
| 94075 | 32 | Suspect | 0.5 | 0.5 | 0.532 | 0.544 | 18 | 17 | NA | None |
| 94023 | 66 | Suspect | 0.5 | 0.5 | 0.532 | 0.533 | 18 | 18 | NA | None |
| 94013 | 39 | Suspect | 0.4 | 0.4 | 0.566 | 0.572 | 20 | 20 | NA | None |
| 94002 | 56 | Normal | 0.3 | 0.3 | 0.543 | 0.547 | 19 | 18 | NA | None |
| 94019 | 16 | Normal | 0.3 | 0.3 | 0.552 | 0.543 | 16 | 16 | NA | None |
| 05040 | 19 | Normal | 0.2 | 0.2 | 0.544 | 0.554 | 20 | 20 | NA | None |
| 05041 | 17 | Normal | 0.2 | 0.2 | 0.551 | 0.540 | 20 | 20 | NA | None |
| 93013 | 74 | Normal | 0.3 | 0.3 | 0.626 | 0.650 | 17 | 18 | NA | None |
No DNA is available; the genotype of this individual is inferred from her daughter and granddaughter's status.
IL-20R, interleukin-20 receptor; POAG, primary open angle glaucoma; IOP, intraocular pressure; OD, oculus dexter; OS, oculus sinister; OU, oculus uterque.
The T104M missense change affects an evolutionarily conserved amino acid [Genomic Evolutionary Rate Profiling (GERP) score 4.91] and is predicted to impact the binding site of IL-20R2 to the cytokines, IL-19, IL-20, or IL-24.49 The T104 site in IL-20R2 site binds to S111 in IL-20. Replacing the T104 with a methionine would replace the hydroxyl group that forms a hydrogen bond with S111 in IL-20 with a sulfate group, thus disrupting the bond between these 2 proteins at this site. Based on these findings, it seems highly likely that this mutation could impact the pathways involved in glaucoma.
STAT3 activity
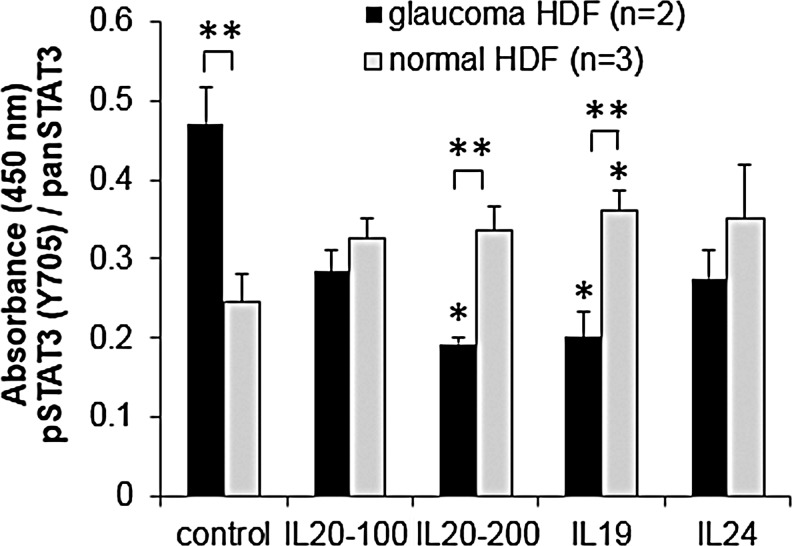
The interaction of IL-20R2 with IL-19, IL-20, or IL-24 activates the JAK-STAT cell signaling pathway.31,35 Phosphorylation of STAT3 at Y705 is a direct indicator of this activation. Therefore, we quantitated phosphorylation of STAT3 in normal and POAG HDFs (pHDFs) that have the IL-20R2 T104M mutation using an ELISA assay (Fig. 3). In unstimulated cells, basal levels of pSTAT3 were greater in pHDFs than in normal cells. In normal HDFs, there was an increase in phosphorylated STAT3(Y705) in response to all 3 cytokines. However, in pHDFs, there was a decrease in STAT3 phosphorylation on cytokine treatment.
FIG. 3.
Quantitation of STAT3 activation in POAG (n=2) and normal human dermal fibroblasts (HDF; n=3) with and without cytokine stimulation (100 and 200 signify different doses of IL-20 in ng/mL). Phosphorylated STAT3(Y705) was quantitated by ELISA assay, and absorbance data were normalized to pan-STAT3. Results show the average±standard error of the mean (SEM). ANOVA determined significance: *P<0.05 between cytokine-treated and -untreated cells; **P<0.05 comparing POAG versus normal cells with the same cytokine treatment.
MMP activity
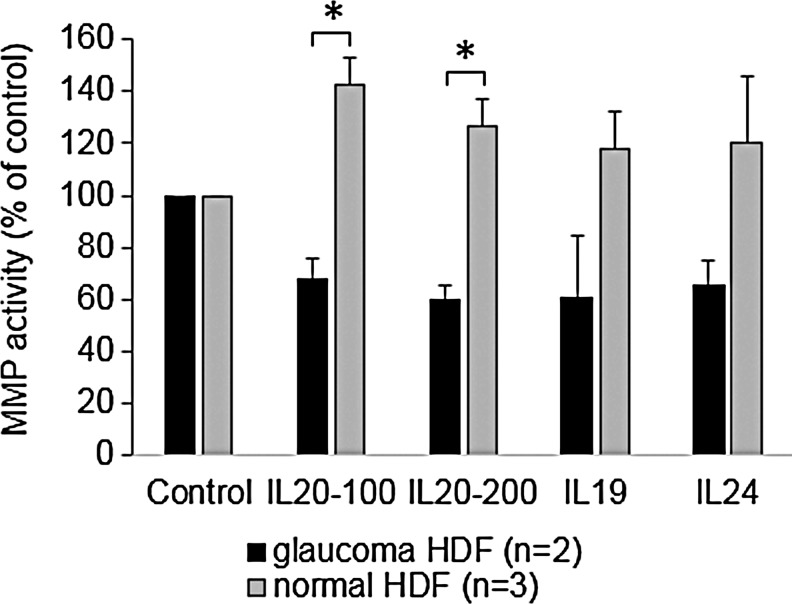
A previous study found that MMP-9 and MMP-12 were up-regulated in a mouse breast cancer cell line in response to IL-20 treatment.36 Moreover, another study showed that activation of STAT3 led to increased levels and activity of MMPs −1, −3, −7, and −9.50 Therefore, we analyzed MMP activity using a quenched FRET peptide, which only produces fluorescence after cleavage by an MMP. We chose a generic MMP substrate, which can be cleaved by multiple MMPs, including MMP-9 and MMP-12. In untreated cells, pHDFs have an increased basal MMP activity (mean RFUs=554±93.64) compared with normal HDFs (mean 247.25±55.7), although this was not significant (P-value=0.097). pHDFs and normal HDFs were then treated for 24 h with IL-19, IL-20, or IL-24 and MMP activity in conditioned media was quantitated. In normal HDFs, all cytokines increased MMP activity in the media (Fig. 4). Conversely, pHDFs decreased MMP activity compared with the untreated control.
FIG. 4.
MMP activity in the conditioned media of normal (n=3) and glaucoma (n=2) HDFs treated with cytokines for 24 h (100 and 200 signify different doses of IL-20 in ng/mL). Results are shown as a percentage of the untreated control±SEM. *P<0.03 by ANOVA.
IL-20 receptor expression in HTM cells
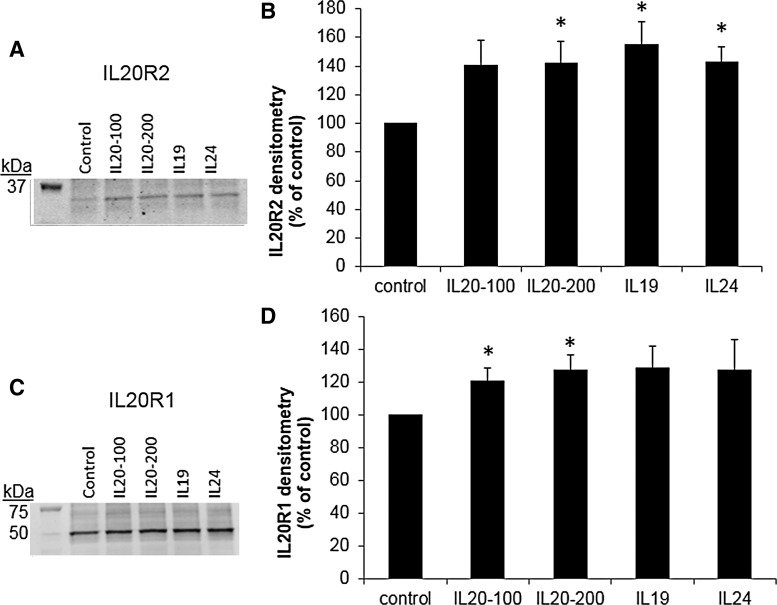
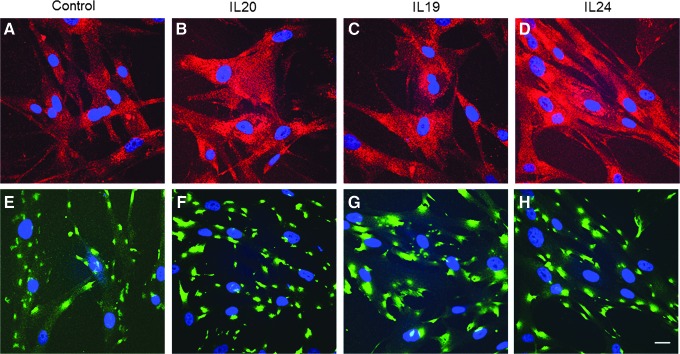
Next, we investigated whether IL-20 receptors were detected in HTM cell lysates by western immunoblot. Figure 5 shows that both IL-20R1 and IL-20R2 were detected in HTM cell lysates and were up-regulated in response to cytokine treatment for 24 h. Two other IL-20R2 antibodies showed similar results (not shown). IL-20R2 and IL-20R1 immunolocalization in HTM cells was also assessed using confocal microscopy (Fig. 6). As expected, both IL-20R1 and IL-20R2 were shown to localize to the cell membrane and cytoplasm of HTM cells. There was more immunostaining in response to cytokine treatments compared with control cells.
FIG. 5.
Western immunoblots and densitometry of human trabecular meshwork (HTM) cells treated with cytokines for 24 h (100 and 200 signify different doses of IL-20 in ng/mL). (A) A representative immunoblot of IL-20R2 protein levels in HTM cell lysates. (B) Densitometry of gel bands presented as a percentage of untreated control cells±SEM. *P<0.04 by ANOVA; n=4. (C) A representative immunoblot of IL-20R1 protein levels in HTM cell lysates. (D) Densitometry of gel bands presented as a percentage of untreated control cells±SEM. *P<0.05 by ANOVA; n=3.
FIG. 6.
Immunofluorescence and confocal microscopy of IL-20R2 and IL-20R1 in HTM cells. Representative images of IL-20R2 (A–D) and IL-20R1 (E–H) localization in HTM cells stimulated for 24 h with vehicle control (A, E), IL-20 at 100 ng/mL (B, F), IL-19 (C, G), and IL-24 (D, H). DAPI was used to stain the nuclei (blue). Scale bar=20 μm.
Discussion
In this article, we present evidence that IL-20R2 may be a glaucoma susceptibility gene and that dermal fibroblasts derived from glaucoma patients have altered STAT3 phosphorylation and MMP activity compared with normal dermal fibroblasts. IL-20R2 is a cytokine receptor that is involved in signaling by the IL-10 family cytokines, IL-19, IL-20, and IL-24.49 IL-20 and IL-24 can signal through both heterodimeric receptor types, but IL-19 signals only through the type I complex (Fig. 1). Previous studies have shown that IL-20R2 binds to the ligand, which then recruits IL-20R1 and heterodimerization occurs.30 Since T104M sits at a site critical for ligand binding,49 it is likely that cytokine binding and heterodimerization is disrupted. The T104M mutation could disrupt signaling that is initiated by all 3 cytokines.
STAT3 is phosphorylated at Y705 and S727.51 In this study, we quantitated phosphorylation at Y705 and found that dermal fibroblasts from glaucoma patients had a higher level of activation in unstimulated cells compared with cells derived from normal individuals. This suggests that the STAT3 pathway may also be utilized by other cytokines, which leads to a higher basal level of STAT3 activity in pHDFs. In addition, in response to all cytokine treatments, there was an increase in phosphorylation at Y705 in normal individuals within 15 min, whereas there was a decrease in phosphorylation in glaucoma patients. These data suggest that POAG cells are unable to launch an appropriate cell signaling response on stimulation by IL-20, IL-19, and IL-24, which supports our hypothesis that mutations in IL-20R2 may interfere with cytokine binding. These data also suggest that type I receptors are definitely being used in HDFs, as IL-19, which only uses the type I receptors, elicited a response.
A previous study correlated STAT3 activation with increased levels and activity of MMPs −1, −3, −7, and −9.50 The higher basal level of STAT3 in pHDFs is, therefore, consistent with increased basal MMP activity. In agreement with an earlier study,36 IL-20 stimulation of normal HDFs increased MMP activity. In addition, we also showed that IL-19 and IL-24 elicited a similar response. However, pHDFs showed an opposite effect compared with normal HDFs and MMP activity was decreased in conditioned media. Since MMPs are known to significantly influence extracellular matrix degradation by TM cells and lower outflow resistance,37,52,53 our data suggest that MMP activity may also be altered in TM cells. This could potentially disrupt a major mechanism by which outflow resistance is usually adjusted in response to elevated IOP.
The T104M mutation in IL-20R2 has been documented in 10 of the affected family members of the GLC1C family. The 2 affected family members that do not carry this mutation also do not have the GLC1C haplotype and, thus, would not be expected to have this variant, as it is within the GLC1C region. They may be phenocopies or there may be more than one gene for POAG segregating through this family. Since this mutation is predicted to significantly impact the protein function and has the potential to negatively affect pathways that could be involved in glaucoma, we explored the effect of this mutation on IL-20R2 activity. The STAT3 assay and MMP assay provide evidence that downstream cellular functions are significantly different in patients harboring this mutation than in normal controls.
This is the first report of the expression of IL-20R family members in HTM cells. Moreover, the western immunoblot and immunofluorescence data show that IL-20 type I receptors are up-regulated in response to cytokine treatment. These data suggest that a feedback loop may exist which up-regulates receptor subtypes on cytokine stimulation. Thus, a mutation in IL-20R2 may not only reduce STAT3 and MMP activity, but also inhibit an increase in IL-20 receptor expression.
According to the database, IL20R1 has 4 protein isoforms. Isoform 1 is the full-length protein consisting of 553 amino acids with a molecular weight of ∼64 kDa. Isoforms 2, 3, and 4 have different 5′ UTR and coding regions, leading to truncation of the N-terminus. These isoforms consist of 504, 442, and 420 amino acids, respectively. The molecular weight (∼49 kDa) of the major band detected in western immunoblots of HTM cell lysates is consistent with IL-20R1 isoform 3 or 4. Although the biological significance of this is not yet known, it could affect cell signaling, as it lacks the extracellular portion that presumably binds to the cytokine. Further studies are required to investigate the alternative splicing of IL-20R1 by TM cells.
In summary, we have demonstrated that the TM cells express IL-20 receptors. Mutation of the T104 residue in IL-20R2 leads to a reduction in STAT3 phosphorylation as well as decreased MMP activity. Thus, IL-20R2 may be a glaucoma susceptibility gene. Earlier findings suggest that a defect in the IL-10 family of cytokines, signaling through the JAK-STAT pathway, may be involved in glaucoma pathogenesis. Identification of the downstream consequences of reduced IL-20R2 activity has the potential to identify new targets for the treatment of this devastating blinding disease.
Acknowledgments
This work was supported by EY010555, EY011650 (M.K.W.), EY019643 (K.E.K.), EY003279, EY008247, EY010572 (T.S.A.), UL1TR000128, and an unrestricted grant to the Casey Eye Institute from Research to Prevent Blindness, New York, NY.
Author Disclosure Statement
All authors declare that no conflicts of interest exist.
References
- 1.Varma R., Lee P.P., Goldberg I., and Kotak S.An assessment of the health and economic burdens of glaucoma. Am. J. Ophthalmol. 152:515–522, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Quigley H.A.Glaucoma. Lancet. 377:1367–1377, 2011 [DOI] [PubMed] [Google Scholar]
- 3.Fan B.J., Wang D.Y., Lam D.S., and Pang C.P.Gene mapping for primary open angle glaucoma. Clin. Biochem. 39:249–258, 2006 [DOI] [PubMed] [Google Scholar]
- 4.Fan B.J., Wang D.Y., Pasquale L.R., Haines J.L., and Wiggs J.L.Genetic variants associated with optic nerve vertical cup-to-disc ratio are risk factors for primary open angle glaucoma in a US Caucasian population. Invest. Ophthalmol. Vis. Sci. 52:1788–1792, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ramdas W.D., van Koolwijk L.M., Ikram M.K., et al. A genome-wide association study of optic disc parameters. PLoS Genet. 6:e1000978, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ramdas W.D., van Koolwijk L.M., Lemij H.G., et al. Common genetic variants associated with open-angle glaucoma. Hum. Mol. Genet. 20:2464–2471, 2011 [DOI] [PubMed] [Google Scholar]
- 7.Macgregor S., Hewitt A.W., Hysi P.G., et al. Genome-wide association identifies ATOH7 as a major gene determining human optic disc size. Hum. Mol. Genet. 19:2716–2724, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Burdon K.P., Macgregor S., Hewitt A.W., et al. Genome-wide association study identifies susceptibility loci for open angle glaucoma at TMCO1 and CDKN2B-AS1. Nat. Genet. 43:574–578, 2011 [DOI] [PubMed] [Google Scholar]
- 9.Thorleifsson G., Walters G.B., Hewitt A.W., et al. Common variants near CAV1 and CAV2 are associated with primary open-angle glaucoma. Nat. Genet. 42:906–909, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vincent A.L., Billingsley G., Buys Y., et al. Digenic inheritance of early-onset glaucoma: CYP1B1, a potential modifier gene. Am. J. Hum. Genet. 70:448–460, 2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Melki R., Colomb E., Lefort N., Brezin A.P., and Garchon H.J.CYP1B1 mutations in French patients with early-onset primary open-angle glaucoma. J. Med. Genet. 41:647–651, 2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhou X.M., Yin Y., Fan N., et al. Single nucleotide polymorphism of MYOC affected the severity of primary open angle glaucoma. Int. J. Ophthalmol. 6:264–268, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Young T.K., Souzeau E., Liu L., et al. Compound heterozygote myocilin mutations in a pedigree with high prevalence of primary open-angle glaucoma. Mol. Vis. 18:3064–3069, 2012 [PMC free article] [PubMed] [Google Scholar]
- 14.Rose R., Balakrishnan A., Muthusamy K., Arumugam P., Shanmugam S., and Gopalswamy J.Myocilin mutations among POAG patients from two populations of Tamil Nadu, South India, a comparative analysis. Mol. Vis. 17:3243–3253, 2011 [PMC free article] [PubMed] [Google Scholar]
- 15.Mochizuki H., Murphy C.J., Brandt J.D., Kiuchi Y., and Russell P.Altered stability of mRNAs associated with glaucoma progression in human trabecular meshwork cells following oxidative stress. Invest. Ophthalmol. Vis. Sci. 53:1734–1741, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gonzalez P., Epstein D.L., and Borras T.Genes upregulated in the human trabecular meshwork in response to elevated intraocular pressure. Invest. Ophthalmol. Vis. Sci. 41:352–361, 2000 [PubMed] [Google Scholar]
- 17.Johnson E.C., Doser T., Cepurna W.O., et al. Cell proliferation and interleukin-6 type cytokine signaling are implicated by gene expression responses in early optic nerve head injury in rat glaucoma. Invest. Ophthalmol. Vis. Sci. 52:504–518, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Luna C., Li G., Liton P.B., Epstein D.L., and Gonzalez P.Alterations in gene expression induced by cyclic mechanical stress in trabecular meshwork cells. Mol. Vis. 15:534–544, 2009 [PMC free article] [PubMed] [Google Scholar]
- 19.Pucci S., Mazzarelli P., Missiroli F., Regine F., and Ricci F.Neuroprotection: VEGF, IL-6, and clusterin: the dark side of the moon. Prog. Brain Res. 173:555–573, 2008 [DOI] [PubMed] [Google Scholar]
- 20.Tripathi R.C., Li J., Chan W.F., and Tripathi B.J.Aqueous humor in glaucomatous eyes contains an increased level of TGF-beta 2. Exp. Eye Res. 59:723–727, 1994 [DOI] [PubMed] [Google Scholar]
- 21.Inatani M., Tanihara H., Katsuta H., Honjo M., Kido N., and Honda Y.Transforming growth factor-beta 2 levels in aqueous humor of glaucomatous eyes. Graefes Arch. Clin. Exp. Ophthalmol. 239:109–113, 2001 [DOI] [PubMed] [Google Scholar]
- 22.Kuchtey J., Rezaei K.A., Jaru-Ampornpan P., Sternberg P., Jr., and Kuchtey R.W.Multiplex cytokine analysis reveals elevated concentration of interleukin-8 in glaucomatous aqueous humor. Invest. Ophthalmol. Vis. Sci. 51:6441–6447, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Takai Y., Tanito M., and Ohira A.Multiplex cytokine analysis of aqueous humor in eyes with primary open-angle glaucoma, exfoliation glaucoma, and cataract. Invest. Ophthalmol. Vis. Sci. 53:241–247, 2012 [DOI] [PubMed] [Google Scholar]
- 24.Chua J., Vania M., Cheung C.M., et al. Expression profile of inflammatory cytokines in aqueous from glaucomatous eyes. Mol. Vis. 18:431–438, 2012 [PMC free article] [PubMed] [Google Scholar]
- 25.Freedman J., and Iserovich P.Pro-inflammatory cytokines in glaucomatous aqueous and encysted molteno implant blebs and their relationship to pressure. Invest. Ophthalmol. Vis. Sci. 54:4851–4855, 2013 [DOI] [PubMed] [Google Scholar]
- 26.Menon R., Ismail L., Ismail D., Merialdi M., Lombardi S.J., and Fortunato S.J.Human fetal membrane expression of IL-19 and IL-20 and its differential effect on inflammatory cytokine production. J. Matern. Fetal Neonatal Med. 19:209–214, 2006 [DOI] [PubMed] [Google Scholar]
- 27.Zhang B.F., Liu J.J., Pei D.S., et al. Potent antitumor effect elicited by RGD-mda-7, an mda-7/IL-24 mutant, via targeting the integrin receptor of tumor cells. Cancer Biother. Radiopharm. 26:647–655 [DOI] [PubMed] [Google Scholar]
- 28.Blumberg H., Conklin D., Xu W.F., et al. Interleukin 20: discovery, receptor identification, and role in epidermal function. Cell. 104:9–19, 2001 [DOI] [PubMed] [Google Scholar]
- 29.Dumoutier L., Leemans C., Lejeune D., Kotenko S.V., and Renauld J.C.Cutting edge: STAT activation by IL-19, IL-20 and mda-7 through IL-20 receptor complexes of two types. J. Immunol. 167:3545–3549, 2001 [DOI] [PubMed] [Google Scholar]
- 30.Pletnev S., Magracheva E., Kozlov S., et al. Characterization of the recombinant extracellular domains of human interleukin-20 receptors and their complexes with interleukin-19 and interleukin-20. Biochemistry. 42:12617–12624, 2003 [DOI] [PubMed] [Google Scholar]
- 31.Parrish-Novak J., Xu W., Brender T., et al. Interleukins 19, 20, and 24 signal through two distinct receptor complexes. Differences in receptor-ligand interactions mediate unique biological functions. J. Biol. Chem. 277:47517–47523, 2002 [DOI] [PubMed] [Google Scholar]
- 32.Su Z.Z., Madireddi M.T., Lin J.J., et al. The cancer growth suppressor gene mda-7 selectively induces apoptosis in human breast cancer cells and inhibits tumor growth in nude mice. Proc. Natl. Acad. Sci. U. S. A. 95:14400–14405, 1998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Saeki T., Mhashilkar A., Chada S., Branch C., Roth J.A., and Ramesh R.Tumor-suppressive effects by adenovirus-mediated mda-7 gene transfer in non-small cell lung cancer cell in vitro. Gene Ther. 7:2051–2057, 2000 [DOI] [PubMed] [Google Scholar]
- 34.Gallagher G., Dickensheets H., Eskdale J., et al. Cloning, expression and initial characterization of interleukin-19 (IL-19), a novel homologue of human interleukin-10 (IL-10). Genes Immun. 1:442–450, 2000 [DOI] [PubMed] [Google Scholar]
- 35.Chang C., Magracheva E., Kozlov S., et al. Crystal structure of interleukin-19 defines a new subfamily of helical cytokines. J. Biol. Chem. 278:3308–3313, 2003 [DOI] [PubMed] [Google Scholar]
- 36.Hsu Y.H., Hsing C.H., Li C.F., et al. Anti-IL-20 monoclonal antibody suppresses breast cancer progression and bone osteolysis in murine models. J. Immunol. 188:1981–1991, 2012 [DOI] [PubMed] [Google Scholar]
- 37.Keller K.E., Aga M., Bradley J.M., Kelley M.J., and Acott T.S.Extracellular matrix turnover and outflow resistance. Exp. Eye Res. 88:676–682, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pasutto F., Keller K.E., Weisschuh N., et al. Variants in ASB10 are associated with open-angle glaucoma. Hum. Mol. Genet. 21:1336–1349, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wirtz M.K., Samples J.R., Kramer P.L., et al. Mapping a gene for adult-onset primary open-angle glaucoma to chromosome 3q. Am. J. Hum. Genet. 60:296–304, 1997 [PMC free article] [PubMed] [Google Scholar]
- 40.Hitchings R.A., and Wheeler C.A.The optic disc in glaucoma. IV: Optic disc evaluation in the ocular hypertensive patient. Br. J. Ophthalmol. 64:232–239, 1980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pohjanpelto P.E., and Palva J.Ocular hypertension and glaucomatous optic nerve damage. Acta Ophthalmol. (Copenh.) 52:194–200, 1974 [DOI] [PubMed] [Google Scholar]
- 42.Polansky J.R., Weinreb R.N., Baxter J.D., and Alvarado J.Human trabecular cells. I. Establishment in tissue culture and growth characteristics. Invest. Ophthalmol. Vis. Sci. 18:1043–1049, 1979 [PubMed] [Google Scholar]
- 43.Ritch R., Hargett N.A., Podos S.M., Mardirossian J., and Kass M.A.Wrong drug dispensed. JAMA. 238:1628, 1977 [PubMed] [Google Scholar]
- 44.Stamer W.D., Seftor R.E., Williams S.K., Samaha H.A., and Snyder R.W.Isolation and culture of human trabecular meshwork cells by extracellular matrix digestion. Curr. Eye Res. 14:611–617, 1995 [DOI] [PubMed] [Google Scholar]
- 45.Keller K.E., Yang Y.F., Sun Y.Y., Sykes R., Acott T.S., and Wirtz M.K.Ankyrin repeat and suppressor of cytokine signaling box containing protein-10 is associated with ubiquitin-mediated degradation pathways in trabecular meshwork cells. Mol. Vis. 19:1639–1655, 2013 [PMC free article] [PubMed] [Google Scholar]
- 46.Aga M., Bradley J.M., Keller K.E., Kelley M.J., and Acott T.S.Specialized podosome- or invadopodia-like structures (PILS) for focal trabecular meshwork extracellular matrix turnover. Invest. Ophthalmol. Vis. Sci. 49:5353–5365, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Keller K.E., Sun Y.Y., Vranka J.A., Hayashi L., and Acott T.S.Inhibition of hyaluronan synthesis reduces versican and fibronectin levels in trabecular meshwork cells. PLoS One. 7:e48523, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Samples J.R., Kitsos G., Economou-Petersen E., et al. Refining the primary open-angle glaucoma GLC1C region on chromosome 3 by haplotype analysis. Clin. Genet. 65:40–44, 2004 [DOI] [PubMed] [Google Scholar]
- 49.Logsdon N.J., Deshpande A., Harris B.D., Rajashankar K.R., and Walter M.R.Structural basis for receptor sharing and activation by interleukin-20 receptor-2 (IL-20R2) binding cytokines. Proc. Natl. Acad. Sci. U. S. A. 109:12704–12709, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tsareva S.A., Moriggl R., Corvinus F.M., et al. Signal transducer and activator of transcription 3 activation promotes invasive growth of colon carcinomas through matrix metalloproteinase induction. Neoplasia. 9:279–291, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sakaguchi M., Oka M., Iwasaki T., Fukami Y., and Nishigori C.Role and regulation of STAT3 phosphorylation at Ser727 in melanocytes and melanoma cells. J. Invest. Dermatol. 132:1877–1885, 2012 [DOI] [PubMed] [Google Scholar]
- 52.Bradley J.M., Kelley M.J., Zhu X., Anderssohn A.M., Alexander J.P., and Acott T.S.Effects of mechanical stretching on trabecular matrix metalloproteinases. Invest. Ophthalmol. Vis. Sci. 42:1505–1513, 2001 [PubMed] [Google Scholar]
- 53.Bradley J.M., Vranka J., Colvis C.M., et al. Effect of matrix metalloproteinases activity on outflow in perfused human organ culture. Invest. Ophthalmol. Vis. Sci. 39:2649–2658, 1998 [PubMed] [Google Scholar]