Abstract
Accurate titration of adeno-associated viral (AAV) vector genome copies is critical for ensuring correct and reproducible dosing in both preclinical and clinical settings. Quantitative PCR (qPCR) is the current method of choice for titrating AAV genomes because of the simplicity, accuracy, and robustness of the assay. However, issues with qPCR-based determination of self-complementary AAV vector genome titers, due to primer–probe exclusion through genome self-annealing or through packaging of prematurely terminated defective interfering (DI) genomes, have been reported. Alternative qPCR, gel-based, or Southern blotting titering methods have been designed to overcome these issues but may represent a backward step from standard qPCR methods in terms of simplicity, robustness, and precision. Droplet digital PCR (ddPCR) is a new PCR technique that directly quantifies DNA copies with an unparalleled degree of precision and without the need for a standard curve or for a high degree of amplification efficiency; all properties that lend themselves to the accurate quantification of both single-stranded and self-complementary AAV genomes. Here we compare a ddPCR-based AAV genome titer assay with a standard and an optimized qPCR assay for the titration of both single-stranded and self-complementary AAV genomes. We demonstrate absolute quantification of single-stranded AAV vector genomes by ddPCR with up to 4-fold increases in titer over a standard qPCR titration but with equivalent readout to an optimized qPCR assay. In the case of self-complementary vectors, ddPCR titers were on average 5-, 1.9-, and 2.3-fold higher than those determined by standard qPCR, optimized qPCR, and agarose gel assays, respectively. Droplet digital PCR-based genome titering was superior to qPCR in terms of both intra- and interassay precision and is more resistant to PCR inhibitors, a desirable feature for in-process monitoring of early-stage vector production and for vector genome biodistribution analysis in inhibitory tissues.
Introduction
Clinical successes in adeno-associated viral (AAV) vector-mediated gene therapy (Kaplitt et al., 2007; Bainbridge et al., 2008; Eberling et al., 2008; Hauswirth et al., 2008; Maguire et al., 2008, 2009; Christine et al., 2009; Simonelli et al., 2010) and the approval of the first AAV-based gene therapy product (Glybera) in Europe, highlight the need for a greater sophistication of AAV vector manufacturing and characterization assays in order to meet industry standards and the demands of commercial production (Dolgin, 2012).
Perhaps the most critical lot release assay for AAV vector preparations is the AAV genome titer assay, because genome copy numbers are universally used for dosing purposes in both preclinical and clinical studies. In the past, the predominant method for determining genome titers was the DNA dot-blot (Samulski et al., 1989). With the advent of real-time quantitative PCR (qPCR), AAV genome titer assays were soon developed that took advantage of the enhanced accuracy, relative simplicity, and robustness of the technique (Clark et al., 1999), and at present qPCR is routinely used for AAV genome titer determination. During production of a self-complementary AAV (scAAV) vector for a clinical trial, a large discrepancy (10-fold) was noted between the qPCR-determined titers and those determined by dot-blot and UV spectrophotometry (Allay et al., 2011; Fagone et al., 2011). The explanation for this discrepancy was that the qPCR assay was designed to target a region close to the hairpin inverted terminal repeat (ITR) of the self-complementary genome. Therefore, the first-order kinetics of the scAAV genome self-annealing outcompeted annealing of the primer and probe and interfered with the overall efficiency of the PCR amplification reaction and the accurate determination of genome titer. A subsequent paper by Fagone and colleagues (2011) reported that the degree of error due to this effect was related to the proximity of the PCR target sequence to the hairpin ITR of the scAAV genome and that physical separation of the hairpin ITR from the target sequence by restriction enzyme digestion resulted in elevated titers that approximated dot-blot titers. This finding led to a proposal for a new assay based on restriction enzyme digestion of the hairpin ITR before qPCR and also to an alternative agarose gel-based assay that is independent of qPCR. A further implication of the study was that standard qPCR could be used to quantify scAAV genomes provided that amplicons distal to the hairpin ITR were employed. More recently, Wang and colleagues (2012) have proposed that packaging of terminal genome fragments (“DI AAV genomes”) into the vector capsid, which was previously demonstrated using single-molecule sequencing (Kapranov et al., 2012), can lead to the overreporting of genome titers. A Southern blot assay was introduced such that these terminal genome fragments would be excluded from contributing to genome titers. Quantitative PCR-based titration was not ruled out, however, as long as targeting of terminal (i.e., hairpin ITR-distal) regions was avoided. Thus, it is evident that quantification of scAAV genomes is problematic and that the researcher must either resort to alternative assays, which may be cumbersome and prone to operator error, or exercise extreme care in the design and validation of qPCR assays.
Although qPCR is robust under ideal conditions, significant impairment of DNA amplification efficiency can result from poor design of primer pairs, the presence of inhibitors in the PCR reaction or as noted previously for scAAV vectors, secondary structure in the template. Another potential problem with the technique is the dependence on a DNA standard curve that itself can be incorrectly calibrated. Either separately or combined, these issues can result in the misreporting of AAV genome titers. Digital PCR (dPCR) was first introduced in the 1990s (Sidransky et al., 1992; Vogelstein and Kinzler, 1999) but only with modern engineering advances has the technique become practical for routine use. Modern versions of dPCR involve compartmentalization of individual DNA molecules such that thousands of nanoliter-scale PCRs can be run in parallel. Standard “TaqMan” primer–probe sets are used so that positive and negative reactions can be scored via fluorescence and an absolute count of DNA copies can be obtained through Poisson statistical manipulation of the data. Many of the problems noted previously for qPCR are of less concern with dPCR because even low-efficiency PCRs are scored as positive and because no standard curve is required (Sanders et al., 2011; Hoshino and Inagaki, 2012; Pinheiro et al., 2012). The technique is known for an un-paralleled degree of precision and sensitivity in a variety of DNA applications (Hindson et al., 2011; Sedlak and Jerome, 2013) and these features alone would seem to make it a highly suitable candidate for the titration of clinical-grade AAV preparations. A digital PCR technology called droplet digital PCR (ddPCR) has been introduced (Hindson et al., 2011; Pinheiro et al., 2012) that allows for high-throughput sample processing as well as a high level of sample compartmentalization. In this technology, a 20-μl real-time PCR reaction is divided into ∼15,000 oil-encased droplets and up to 96 such reactions can be accommodated on a 96-well plate. After cycling the droplets are scored as positive or negative, using fluorescence-based flow cytometry.
We hypothesized that the ability to directly quantify DNA copies without the need for a standard curve or for high-efficiency amplification would allow us to apply a universal technique for the titration of both single-stranded (ss) and scAAV genomes without fear of underreporting. Here we demonstrate that an rAAV genome titer assay based on ddPCR is practical, reports equivalent or higher titers than both qPCR and DNA gel-based assays, and translates the precision for which dPCR is known to the titering of AAV vectors. Furthermore, we demonstrate enhanced resistance of the ddPCR assay to PCR inhibitors in comparison with qPCR, a result that will be useful in the assessment of crude preparation titers, as well as in biodistribution studies.
Materials and Methods
Vectors
All vector preparations were prepared by triple transfection of HEK293 cells followed by purification by either the iodixanol gradient method as described by Lock and colleagues (2010a) or over CsCl gradients as described previously (Lock et al., 2012).
Primers and probes
The following primers and probes were used in these studies:
-
Cytomegalovirus (CMV) promoter assay
Forward: 5′-CATCTACGTATTAGTCATCGCTATTACCA-3′
Reverse: 5′-GAAATCCCCGTGAGTCAAACC-3′
Probe: 5′-6FAM-TCAATGGGCGTGGATAG-MGBNFQ-3′
-
Human thyroxine-binding globulin (TBG) promoter assay
Forward: 5′-AAACTGCCAATTCCACTGCTG-3′
Reverse: 5′-CCATAGGCAAAAGCACCAAGA-3′
Probe: 5′-6FAM-TTGGCCCAATAGTGAGAACTTTTTCCTGC-TAMRA-3′
-
Enhanced green fluorescent protein (eGFP) assay
Forward: 5′-AGCAAAGACCCCAACGAGAA-3′
Reverse: 5′-GGCGGCGGTCACGAA-3′
Probe: 5′-6FAM-CGCGATCACATGGTCCTGCTGG-TAMRA-3′
-
Bovine growth hormone (BGH) polyadenylation signal assay
Forward: 5′-GCCAGCCATCTGTTGT-3′
Reverse: 5′-GGAGTGGCACCTTCCA-3′
Probe: 5′-6FAM-TCCCCCGTGCCTTCCTTGACC-TAMRA-3′
-
Normal rabbit β-globin (nRBG) polyadenylation signal assay
Forward: 5′-GCCAAAAATTATGGGGACAT-3′
Reverse: 5′-ATTCCAACACACTATTGCAATG-3′
Probe: 5′-6FAM-ATGAAGCCCCTTGAGCATCTGACTTCT-TAMRA-3′
-
Simian virus 40 (SV40) polyadenylation signal assay
Forward: 5′-AGCAATAGCATCACAAATTTCACAA-3′
Reverse: 5′-CCAGACATGATAAGATACATTGATGAGTT-3′
Probe: 5′-6FAM-AGCATTTTTTTCACTGCATTCTAGTTGTGGTTTGTC-TAMRA-3′
qPCR genome titer assays
For the standard qPCR genome titer assay, purified vectors were diluted 10-fold and treated with DNase I (Roche Applied Science, Indianapolis, IN) at 400 U/ml for 30 min at 37°C. Treated samples were then diluted a further 10,000-fold in dilution buffer (1×PCR buffer [Applied Biosystems, Foster City, CA] plus 2-ng/μl sheared salmon sperm DNA [Invitrogen, Carlsbad, CA]). Five microliters was added to a 50-μl reaction along with TaqMan universal PCR master mix (Applied Biosystems) and TaqMan primers and probes directed against various regions of the transgene cassette (Applied Biosystems; 0.3 μM and 0.2 μM final concentration, respectively). The plasmid standard curves were prepared by restriction enzyme linearization and purification with a QIAquick PCR purification kit (Qiagen, Valencia, CA). The linearized DNA was quantified by UV spectrophotometry and 10-fold serially diluted from 108 to 10 copies per 5 μl. Diluted standard curves were assembled into 50-μl PCR volumes as for the vector samples. The samples were PCR amplified with an SDS-7500 real-time PCR instrument (Applied Biosystems), using a two-step cycling protocol (40 cycles: 95°C for 15 sec, 60°C for 1 min) preceded by a 10-min incubation at 95°C.
The optimized qPCR genome titer assay was similar to the standard assay, except that after the DNase I digestion, samples were treated with proteinase K (Qiagen; 0.2 mg/ml, 55°C, 30 min) followed by heat inactivation at 95°C for 15 min. In addition, all dilutions were made in the presence of 0.1% Pluronic F-68 (Invitrogen). Genome copy number per milliliter (GC/ml) was calculated for both standard and optimized assays according to the volume of diluted sample loaded in each qPCR (5 μl) and the total dilution factor. Because positive- and negative-sense genomes are packaged into separate single-stranded vector capsids, two vector particles are required to give the equivalent signal of one molecule of the double-stranded plasmid standard; therefore a multiplication factor of two was included. For self-complementary vectors, where each capsid contains both positive and negative target sequences, the 2-fold multiplication was not applied.
Droplet digital PCR genome titer assay
Vector aliquots were treated with DNase I as described previously for the qPCR assays and then diluted to within the linear range of the ddPCR assay (1 to 105 copies), using the same dilution buffer as in the qPCR assays but with the addition of 0.05% Pluronic F-68 (Invitrogen). The reaction mixtures were assembled with the recommended ddPCR supermix (Bio-Rad, Hercules, CA), the same TaqMan primers and probes used in the qPCR genome titer assays (final concentrations of 900 and 250 nM, respectively), and template (5 μl) in a final volume of 20 μl. Each reaction was then loaded into the sample well of an eight-well disposable cartridge (Bio-Rad) along with 70 μl of droplet generation oil (Bio-Rad), and droplets were formed in a droplet generator (Bio-Rad). Droplets were then transferred to a 96-well PCR plate, heat-sealed with foil, and amplified to the end point with a conventional thermal cycler (95°C for 10 min, followed by 42 cycles of 95°C for 30 sec, 60°C for 1 min, and 72°C for 15 sec followed by a final 98°C heat treatment for 10 min). The PCR plate was subsequently scanned on a QX100 droplet reader (Bio-Rad) and the data were analyzed with QuantaSoft software (Bio-Rad). The copies per microliter readout from the QX100 reader was converted to genome copies per milliliter according to the formula X=[(aY)(1000/b)]D, where X is GC/ml, a is volume of the ddPCR (20 μl), Y is ddPCR readout copies per microliter, b is volume of diluted vector in the ddPCR (5 μl), and D is total dilution applied to the test material. Single-stranded vector capsids containing either a positive or negative strand will each give a ddPCR signal and count as a positive event. Hence, multiplication by a factor of two is not necessary for either single-stranded or self-complementary vectors with this assay.
Agarose gel genome titer assay
The native agarose gel genome titer assay for scAAV vectors was performed such that it conformed as closely as possible to the original method described by Fagone and colleagues (2011). Briefly, 1–3×1010 GC of each vector as determined by the standard qPCR assay was incubated in 0.5% sodium dodecyl sulfate (SDS) at 95°C for 10 min and then cooled slowly to room temperature (0.6°C/min). Twenty percent of the denatured vector samples (2–6×109 GC) and high DNA mass ladder (Invitrogen) were mixed with loading buffer and the reference plasmid (ApoI-digested pLITMUS 29; New England BioLabs, Ipswich, MA) as originally described and then loaded onto a 1.0% agarose gel for electrophoresis in Tris–acetate–EDTA (TAE) buffer at 5 V/cm. The gel was stained for 2 hr in an equal gel volume of 3×GelRed solution (Biotium, Hayward, CA) and imaged without destaining, using a G:BOX Chemi XL imager and GeneSnap software (Syngene, Frederick, MD). Band volumes were determined with the GeneTools software package (Syngene) and normalized to the reference band. The amount of scAAV vector genome DNA was determined by regression analysis, and used for the calculation of vector genome titer, based on genome size.
Unpurified vector interference assay
An unpurified bulk (crude harvest) vector preparation (lot WL716S) from concentrated production culture medium (Lock et al., 2010a) was 10-fold serially diluted and each dilution was spiked with a purified vector preparation (lot WL717S) containing a PCR target sequence not present in the bulk vector preparation. A total of 3.48×103 spike copies were present in the final PCR for both the optimized qPCR and ddPCR genome titer assays. The qPCR and ddPCR PCR reactions were assembled with identical eGFP primers and probes and subjected to cycling and analysis as described previously. For comparison purposes, the detection of the spiked vector by each assay was expressed as a percentage of the initial amount spiked.
Results and Discussion
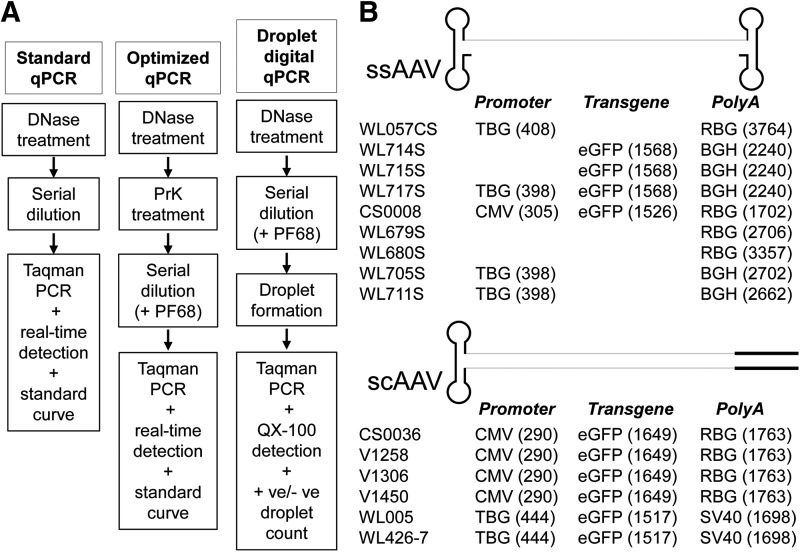
qPCR assays for determining AAV genome titers have become the standard of the field because of their simplicity, accuracy, and robustness. However, findings of the underestimation of AAV self-complementary genome titers by qPCR (Fagone et al., 2011; Wang et al., 2012) led us to reexamine our AAV titering method and to explore the potential of digital PCR in AAV genome copy number determination. Our standard qPCR assay involves DNase I treatment of the sample followed by serial dilution, heat denaturation of the capsid to release the vector genome, and qPCR titration against a linearized plasmid standard curve (Fig. 1A). It has been noted that considerable loss of vector can occur during the titering process through binding to plastic surfaces at low vector dilutions and that this can be overcome by inclusion of a surfactant in the vector preparation itself or in dilution buffers (Bennicelli et al., 2008). Optimization assays were therefore performed that examined the effect of including various concentrations of Pluronic F-68 (PF-68) in the dilution buffers and also of pretreatment of the capsid with proteinase K followed by heat inactivation (data not shown). Our initial results indicated that inclusion of PF-68 at 0.1% or pretreatment with proteinase K followed by heat inactivation could increase average titers over our standard qPCR assay by 2.9- and 2.4-fold, respectively, with some modest improvement when the two treatments were combined (3.5-fold). Consequently, the final optimized qPCR assay developed for our study employed both treatments (Fig. 1A). For digital PCR, dilutions were performed in the presence of PF-68 (0.05%) and samples were diluted serially to well within the dynamic range of the ddPCR instrument according to Bio-Rad specifications (1 to 1×105 DNA copies). PCR was performed with the same primer–probe sets used for the qPCR assays. Proteinase K treatment was not performed on ddPCR samples because of the concern that the positive- and negative-sense strands of single-stranded vector genomes packaged into individual capsids would reanneal once released from the capsids and therefore would not partition into individual droplets. Because the digital PCR output is an absolute measurement, no plasmid calibration curve was necessary (Fig. 1A).
FIG. 1.
(A) Flow diagram depicting the three different PCR-based AAV genome titer assays. (B) Graphical representation of the single-stranded (ssAAV) and self-complementary sc(AAV) vectors used in the study. The line drawings represent the two genome configurations with inverted terminal repeats (ITRs) depicted as lariat structures with the hairpin ITR of the scAAV genome oriented to the left. The lot number of each vector is given below the drawings, and the various TaqMan assays used appear below, with the target sequence position indicated in parentheses. +ve, positive; −ve, negative; BGH, bovine growth hormone; CMV, cytomegalovirus; eGFP, enhanced green fluorescent protein; PF68, Pluronic F-68; PrK, proteinase K; QX-100, QX100 droplet reader; RBG, rabbit β-globin; scAAV, self-complementary AAV; ssAAV, single-stranded AAV; SV40, simian virus 40; TBG, human thyroxine-binding globulin.
Ultimately our goal was to compare all three genome titer assays (standard qPCR, optimized qPCR, and ddPCR) across a range of AAV vectors. Initially, however, we set out to calibrate two plasmid standards using ddPCR (Table 1). For the nRBG standard, the ddPCR-determined copies were initially 50% lower than those predicted by spectrophotometric determination using two different primer–probe sets (nRBG and eGFP). In an attempt to improve the agreement between ddPCR- and spectrophotometry-determined copy numbers, the PCR cycling parameters were altered from the manufacturer's recommendations by increasing the cycle number and adding an extension step at 72°C. This change resulted in a modest increase in the ddPCR titer although it was still 30% lower than expected. For the BGH assay standard, copy numbers detected by ddPCR using the modified conditions closely matched the copy number calculated from spectrophotometric readings at 260 nm. The discrepancy between the nRBG and BGH ddPCR results indicates an error in the nRBG spectrophotometric copy number determination and highlights the problem of using plasmid standard curves for qPCR titer determination, because they themselves can be a source of error.
Table 1.
Plasmid Standard Copy Number Validation by Droplet Digital PCR
| Plasmid DNA standard | Calculated copiesa | Assay | ddPCR-detected copies |
|---|---|---|---|
| RBG (pAAV.CBA.PI.eGFP.RBG) | 1,000 | nRBG | 488 |
| 1,000 | eGFP | 502 | |
| 10,000 | nRBG | 6,920b | |
| 10,000 | nRBG | 6,313b | |
| 1,000 | nRBG | 677b | |
| BGH (pAAV.CMV.eGFP.BGH) | 1,000 | CMV2 | 1,096b |
| 1,000 | eGFP | 1,014b | |
| 1,000 | eGFP | 1,063b | |
| 10,000 | eGFP | 11,373b |
Determined by spectrophotometry.
Modified PCR cycling conditions.
BGH, bovine growth hormone poly(A); CMV, cytomegalovirus promoter; ddPCR, droplet digital PCR; eGFP, enhanced green fluorescent protein; RGB, rabbit β-globin poly(A).
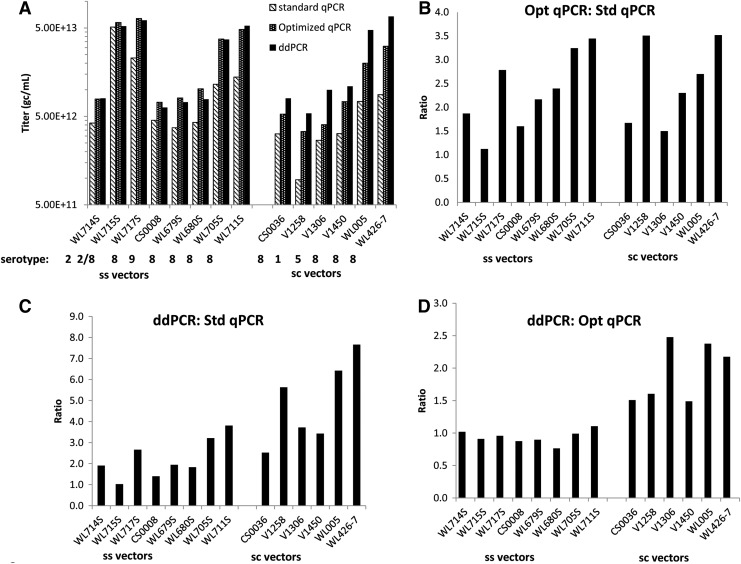
A series of AAV vectors containing both single-stranded and self-complementary genomes (Fig. 1B) were titered using the three PCR assays and the titers were compared (Fig. 2). For single-stranded AAV vectors of various serotypes, the optimized qPCR and ddPCR genome titer assays showed 1.5- to 3.8-fold increases over the standard qPCR assay regardless of serotype, although for one high-titer lot (lot WL715S) no increase was observed (Fig. 2A and C). Ratios of ddPCR to optimized qPCR titers were consistently close to 1 for single-stranded vectors (Fig. 2D). For self-complementary vectors, the optimized qPCR and ddPCR assays gave up to 4- and 8-fold higher titers than the standard qPCR assay, respectively (Fig. 2B and C). Most notably, the ddPCR assay gave 1.5- to 2.5-fold higher titers than the optimized qPCR assay (Fig. 2D) for the six scAAV vectors tested. The possibility that the high ddPCR titer obtained for scAAV vectors was due to residual plasmid DNA was assessed. Self-complementary AAV vectors were titered by the ddPCR method, using the eGFP assay in the presence or absence of DNase I, and in all cases the titers were similar (Supplementary Table S1; supplementary data are available online at http://online.liebertpub.com/hgtb). In addition, 5×1010 copies of a qPCR plasmid standard were included in a DNase I digestion reaction either treated or left untreated with one-tenth of the DNase I (4 units) usually used in the assay. This plasmid copy number was reduced to background levels in the DNase I-treated reactions, whereas the untreated control returned the expected number of copies (data not shown). Extrapolating upward, this result demonstrates that at least 5×1011 plasmid copies can be cleared in the DNase I digestion reaction containing 40 units of enzyme, which is the equivalent copy level that would be present in the reaction when assaying an scAAV vector with a titer of 5×1013 GC/ml. The data show therefore that large amounts of excess plasmid DNA were not present in the vector preparations tested. In addition, because plasmid DNA amounts that would result in the highest titers we have obtained are easily cleared in the DNase I digestion reaction, unpackaged plasmid DNA is evidently not contributing to ddPCR-derived GC titers. In the case of ssAAV vectors, the higher titers observed for the optimized and ddPCR assays most likely trace to the inclusion of PF-68 surfactant in the dilution buffers. However, for scAAV vectors the higher ddPCR assay titers obtained in comparison with the optimized qPCR assay appear to be specific for this genome type and are thought to reflect the relative tolerance of ddPCR for inefficient PCR amplification, as discussed in more detail below.
FIG. 2.
PCR-based AAV genome titer assay comparison. (A) Titer in genome copies per milliliter (GC/ml) for several vector lots of various serotypes, as indicated below the x axis. Averages of titers obtained with assays targeting the appropriate poly(A) signal sequence are shown. (B–D) Ratios of titers obtained for the various lots: (B) optimized versus standard qPCR; (C) ddPCR versus standard qPCR; (D) ddPCR versus optimized qPCR. Primer–probe sets targeting the vector poly(A) signal sequence were used in all PCR-based assays for this analysis. ddPCR, droplet digital PCR.
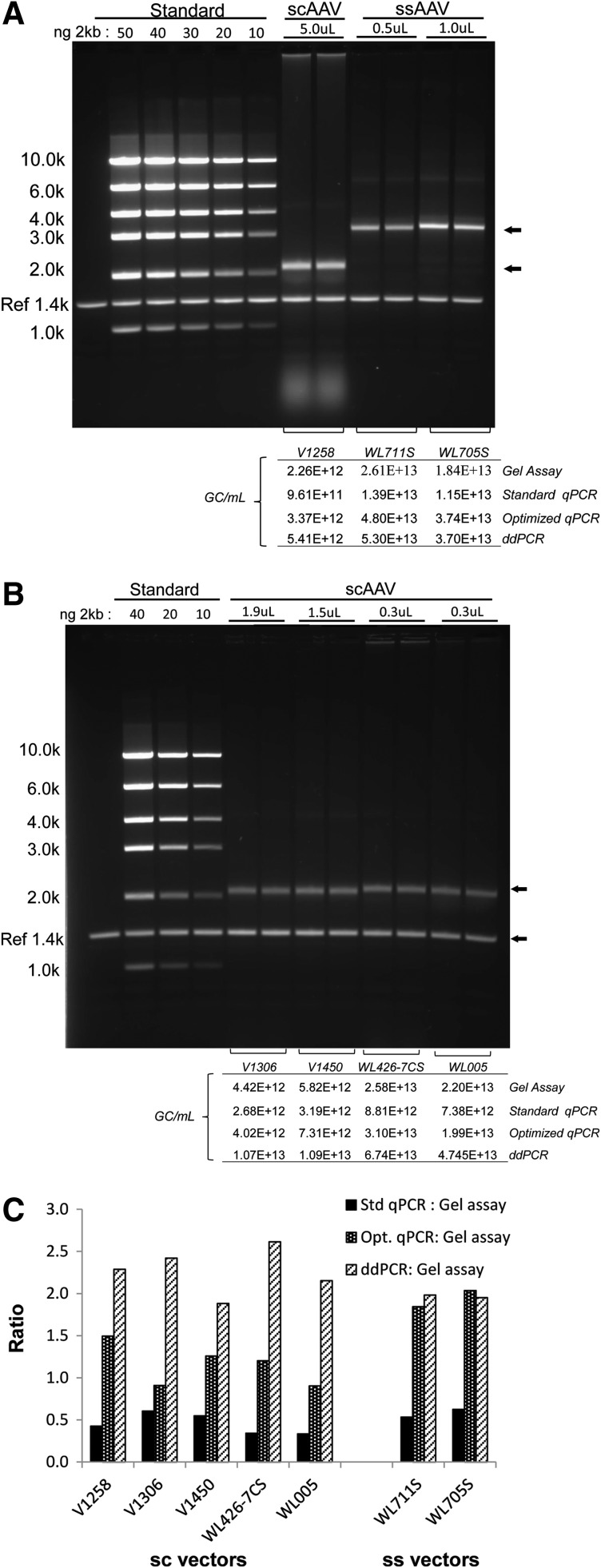
A native agarose gel assay was developed as an alternative to qPCR for the determination of scAAV vector genome titers (Fagone et al., 2011). Given the discrepancy we found between the ddPCR and optimized qPCR-derived titers for scAAV vectors, we decided to corroborate the results, using the native agarose gel assay (Fig. 3). The gel assay titers obtained for scAAV vector lots were reproducibly equivalent to optimized qPCR assay titers and in general 2- to 3-fold higher than those obtained with the standard qPCR assay. The ddPCR assay, however, produced titers that were 2- to 3-fold higher than the gel assay titers. Overall, therefore, in the case of scAAV vectors there was good agreement between the gel assay and the optimized qPCR assay, but the ddPCR assay gave higher titers than both. Two ssAAV vectors were also run on a native agarose gel (Fig. 3B) and, contrary to the initial findings of Fagone and colleagues (2011), the genomes appeared to migrate uniformly. This unexpected result is most likely attributable to the fact that the thermal cycling machine used in the gel assay to denature the vector capsid and reanneal the genomes before loading the gel required longer ramping times than those published, therefore resulting in increased annealing of positive and negative strands. The calculated gel assay titers for these vector lots, however, were still ∼2-fold lower than the optimized qPCR and ddPCR titers and more comparable to the standard qPCR assay titers (Fig. 3C). This discrepancy may be due to some degree of interference with single-strand genome annealing caused by undissociated capsid protein, which may result in retardation of the DNA migration on the gel and thus less DNA in the genomic DNA band. Although not attempted here, such retardation might be resolved by pretreating the DNA with proteinase K before electrophoresis. Although our results appear to highlight a drawback of the agarose gel genome titer assay, that is, the inability to titer both scAAV and ssAAV vectors on the same gel system using the same standards, it is possible that with further optimization of annealing and sample pretreatment, the estimation of ssAAV genome titers with native gel systems may improve.
FIG. 3.
Native agarose gel genome titer assay. (A) The assay was performed according to the published method of Fagone and colleaguges (2011) and as described in Materials and Methods. A dilution series of DNA mass markers was run alongside duplicate lanes of one scAAV and two ssAAV vector samples and used to quantify the full-length double-stranded genome band (arrows). An internal reference marker (Ref 1.4k) was run in each lane and used to normalize loading between lanes. The gels were quantified and gel titers were calculated as described in Materials and Methods. The amount (nanograms) of the 2-kb standard fragment and the volume (microliters) of each vector loaded are indicated above the respective lanes. The table below the lanes shows the calculated titer for the vectors analyzed by the gel assay and compares them with titers calculated by each of the PCR-based assays using primer–probe sets targeting the poly(A) signal sequence. (B) Four additional scAAV vectors (duplicate lanes) and standards as described previously were run on native agarose gels. The calculated gel assay titers in comparison with PCR-based assay titers [poly(A) signal sequence] are shown in the table below the lanes. (C) Ratios between titers obtained by PCR-based assays and the native gel assay for scAAV and ssAAV vector lots are shown.
Fagone and colleagues (2011) have also reported that primer–probe sets with targets located less than 1000 bp from the hairpin ITR of scAAV genomes tend to underreport the genome titer in comparison with other types of genome titer assay and showed that primer–probe sets situated closer to the hairpin exhibited a greater amount of inhibition. It has been hypothesized that the inhibition results from the exclusion of primers and probes from the template due to the rapid reannealing of the hairpin genome. Such exclusion is presumably most detrimental in the very early cycles of the PCR reaction before amplicons have become the majority template, but is enough to shift the amplification curve to the right and result in delayed attainment of the cycle threshold. ddPCR has been reported to be less dependent on the efficiency of the PCR reaction than qPCR, because there is no need to attain a cycle threshold (Hoshino and Inagaki, 2012). To assess whether ddPCR genome titers would be impacted differently than optimized qPCR titers by the scAAV genome exclusion effect, primer–probe sets at various distances from the scAAV hairpin ITR were used for both assay types (Fig. 1B and Table 2). Primer–probe sets targeted at the CMV and TBG promoters (290 and 444 bp away from the 5′ ITR, respectively) gave both ddPCR and optimized qPCR titers that were approximately 2-fold lower than those generated with primer–probe sets directed at the hairpin ITR–distal sequence elements, that is, the transgene-coding sequences and poly(A) sequences (Table 2). The decreased ddPCR and optimized qPCR titers produced with the hairpin ITR–proximal primer–probe sets are consistent with the ∼2-fold qPCR titer decreases published for primer–probe sets within 320–567 bp of the 5′ ITR hairpin (Fagone et al., 2011). Conversely, no such decrease relative to more distally targeted primer–probe sets was seen with ssAAV genomes (Table 2) using CMV or TBG primers and probes in either ddPCR or optimized qPCR assays. This latter observation supports the contention that proximity to the hairpin and not intrinsic properties of the primer–probe sets themselves was likely responsible for the titer decreases observed. No significant difference in titer was observed when comparing primer–probe sets targeting central and terminal [Fig. 1B; transgene and poly(A)] genome regions with either the ddPCR or the optimized qPCR assay, indicating that interference with primer–probe annealing was less pronounced at these genome positions. Furthermore, equivalent titers from assays targeting central and terminal genomic regions do not support the claim that packaged, terminal, defective interfering (DI) genome fragments artificially increase vector genome titers (Wang et al., 2012). It remains possible, however, that the assay targets were not entirely overlapped by the DI genomes. The fact that decreased titers are seen with hairpin-proximal primer–probe sets in both ddPCR and optimized qPCR assays demonstrates that no specific advantage is conferred by ddPCR with respect to overcoming the inhibitory effects of the neighboring scAAV 5′ ITR hairpin. In conclusion, therefore, hairpin-proximal primer–probe sets are best avoided in both types of assay. In one case (lot V1450), the ITR-proximal primer–probe set (CMV) gave similar ddPCR and optimized qPCR titers to the hairpin-distal primer–probe sets (eGFP and nRBG). This result suggests that close proximity of target sequences to the hairpin may not be the only factor in determining lower PCR genome titers and that other factors, such as the sequence context of the target or the particular design of a primer–probe set, may also contribute. Despite the variation in scAAV vector genome titers produced by hairpin proximity of the PCR target, the higher titers given by the ddPCR assay compared with the optimized qPCR assay remained consistent no matter which primer–probe set was used. Thus, although ddPCR is inhibited when the assay is targeted close to the hairpin ITR, the lack of requirement to reach a cycle threshold means that there is still an advantage over qPCR. This advantage must also hold true at hairpin-distal targets where presumably a degree of inhibition is still occurring.
Table 2.
TaqMan Assay Comparison
| Lot no. | Assay | ddPCR | OqPCR | ddPCR: OqPCR | ddPCR: Distal: Proximala | OqPCR: Distal: Proximala | |
|---|---|---|---|---|---|---|---|
| ss vectors | CS0008 | CMV2 | 7.04×1012 | ND | N/A | ||
| eGFP | 6.75×1012 | ND | N/A | 0.96 | N/A | ||
| nRBG | 6.31×1012 | 7.22×1012 | 0.87 | 0.90 | N/A | ||
| WL057CS | TBG | 3.84×1013 | 3.93×1013 | 0.98 | |||
| nRBG | 3.01×1013 | 3.06×1013 | 0.98 | 0.78 | 0.78 | ||
| WL705S | TBG | 3.39×1013 | ND | N/A | |||
| BGH | 3.68×1013 | 3.74×1013 | 0.98 | 1.09 | N/A | ||
| WL711S | TBG | 4.96×1013 | ND | N/A | |||
| BGH | 5.26×1013 | 4.80×1013 | 1.10 | 1.06 | N/A | ||
| sc vectors | CS0036 | CMV | 4.36×1012 | 2.27×1012 | 1.92 | ||
| eGFP | 8.68×1012 | 5.22×1012 | 1.66 | 1.99 | 2.30 | ||
| nRBG | 8.00×1012 | 5.30×1012 | 1.51 | 1.83 | 2.34 | ||
| V1258 | CMV | 3.44×1012 | 1.61×1012 | 2.14 | |||
| eGFP | 5.87×1012 | 3.00×1012 | 1.96 | 1.71 | 1.86 | ||
| nRBG | 5.46×1012 | 3.37×1012 | 1.62 | 1.59 | 2.09 | ||
| V1306 | CMV | 6.65×1012 | 2.93×1012 | 2.27 | |||
| eGFP | 1.14×1013 | 4.79×1012 | 2.39 | 1.72 | 1.63 | ||
| nRBG | 9.97×1012 | 4.02×1012 | 2.48 | 1.50 | 1.37 | ||
| V1450 | CMV | 1.10×1013 | 5.18×1012 | 2.12 | |||
| eGFP | 1.10×1013 | 6.48×1012 | 1.69 | 1.00 | 1.25 | ||
| nRBG | 1.09×1013 | 7.35×1012 | 1.48 | 0.99 | 1.42 | ||
| WL005 | TBG | 1.85×1013 | 1.11×1013 | 1.67 | |||
| eGFP | 4.60×1013 | 2.17×1013 | 2.11 | 2.48 | 1.97 | ||
| SV40 | 4.74×1013 | 1.99×1013 | 2.38 | 2.56 | 1.80 | ||
| WL426-7 | TBG | 4.32×1013 | 2.04×1013 | 2.12 | |||
| eGFP | 6.45×1013 | 3.93×1013 | 1.64 | 1.49 | 1.93 | ||
| SV40 | 6.74×1013 | 3.10×1013 | 2.17 | 1.56 | 1.52 |
Ratio of titers determined by distal assays [transgene/poly(A) signal] and proximal assay (promoter).
BGH, bovine growth hormone poly(A); CMW, cytomegalovirus promoter; ddPCR, droplet digital PCR; eGFP, enhanced green fluorescent protein; OqPCR, optimized qPCR; N/A, not applicable; ND, not determined; nRBG, normal rabbit β-globin poly(A); sc, self-complementary; ss, single-stranded; SV40, simian virus 40 poly(A); TBG, human thyroxine-binding globulin promoter.
A much-heralded advantage of digital PCR is the high degree of precision in estimating copy number, which is facilitated by the partitioning of the sample into many thousands of individual PCRs, thus providing a large sample size and a statistically accurate count. Although greater precision of ddPCR compared with qPCR has been demonstrated (Sanders et al., 2011), we were interested to know whether there would be an advantage in this regard for AAV genome titering. Multiple samples were run in triplicate, using both optimized qPCR and ddPCR genome titer assays with different primer–probe sets, and the intraassay precision of each method was assessed as a coefficient of variance (CV) for each triplicate set (Table 3). For the optimized qPCR assay, the median CVs were 5.35 and 4.37% for the nRBG and BGH primer–probe sets, with the high end of the range at 33.2 and 8.7%, respectively. For the ddPCR genome titer assay, the median CV for three separate primer–probe sets was 2% or lower, with a much tighter distribution over the samples tested. As part of a clinical development program, a formal interassay validation study for the optimized qPCR procedure was carried out with an ssAAV vector (lot WL057CS; Table 4). Six independent runs were performed by two operators and incorporated two separate assays at three initial sample dilutions. The interassay variance was in the range of 11.5–16.5% (mean, 13.7%), which was similar to the CV (16.5%) for a positive control vector that was run on 13 separate occasions with the standard qPCR assay. A similar validation experiment was undertaken for the ddPCR genome titer assay, again using the WL057CS vector lot (Table 4). The mean titers obtained with this assay were remarkably similar to those obtained with the optimized qPCR assay, but the interassay variance was almost 3-fold lower with a mean of 5.0% (range, 2.7–7.92%). Taken together, the results demonstrated an improved precision of the ddPCR genome titer assay compared with the qPCR assays at both intra- and interassay levels.
Table 3.
Intraassay Precision of Optimized qPCR and Droplet Digital PCR Assays
| na | Assay | Median CVb | CV range | |
|---|---|---|---|---|
| Optimized qPCR | 36 | nRBG | 5.35 | 2.11–33.23 |
| 17 | BGH | 4.37 | 1.95–8.72 | |
| ddPCR | 4 | nRBG | 2.21 | 0.60–2.84 |
| 7 | BGH | 1.65 | 0.57–2.19 | |
| 13 | eGFP | 1.81 | 1.22–5.01 |
Independent assays performed in triplicate.
Median of triplicate CV values.
BGH, bovine growth hormone poly(A); CV, coefficient of variation; ddPCR, droplet digital PCR; eGFP, enhanced green fluorescent protein; nRBG, normal rabbit β-globin poly(A).
Table 4.
Interassay Precision
| na | Assay | Sample dilution | Mean (GC/ml) | SD | CV | |
|---|---|---|---|---|---|---|
| Standard qPCR | 13 | RBG | 1× | 1.23×1013 | 2.03×1012 | 16.50 |
| Optimized qPCR | 6 | RBG | 1× | 3.06×1013 | 4.44×1012 | 14.50 |
| 100× | 2.75×1013 | 3.53×1012 | 12.80 | |||
| 1000× | 2.77×1013 | 4.57×1012 | 16.50 | |||
| 6 | TBG | 1× | 3.93×1013 | 4.53×1012 | 11.50 | |
| 100× | 3.52×1013 | 4.52×1012 | 12.80 | |||
| 1000× | 3.74×1013 | 5.31×1012 | 14.20 | |||
| ddPCR | 4 | RBG | 1× | 3.01×1013 | 1.64×1012 | 5.45 |
| 5 | 10× | 2.97×1013 | 1.01×1012 | 3.38 | ||
| 5 | 100× | 2.69×1013 | 8.40×1011 | 3.12 | ||
| 5 | 1000× | 2.67×1013 | 2.12×1012 | 7.92 | ||
| 4 | TBG | 1× | 3.84×1013 | 1.95×1012 | 5.08 | |
| 5 | 10× | 3.85×1013 | 2.81×1012 | 7.29 | ||
| 5 | 100× | 3.46×1013 | 9.30×1011 | 2.69 | ||
| 5 | 1000× | 3.59×1013 | 1.77×1012 | 4.93 |
Number of independent assays performed with ssAAV vector lot number WL057CS.
CV, coefficient of variation; ddPCR, droplet digital PCR; GC, genome copies; RBG, rabbit β-globin poly(A); TBG, human thyroxine-binding globulin promoter.
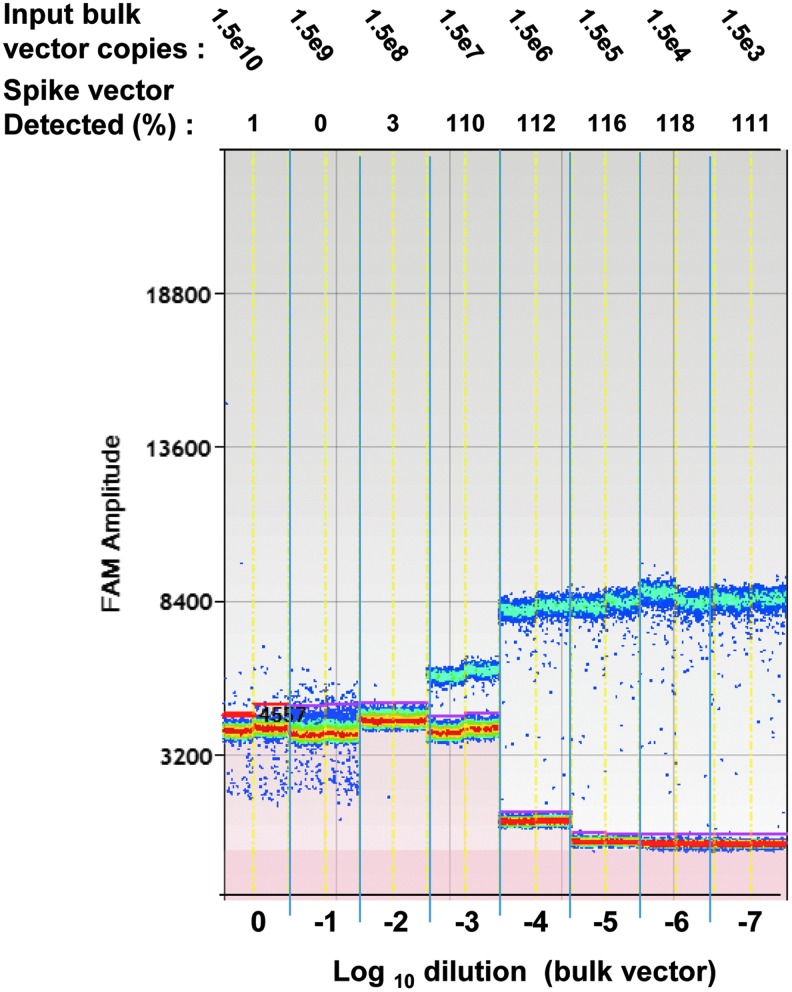
The data obtained during the course of our study indicate that qPCR titers can be significantly enhanced when a surfactant is used during vector dilution and when capsids are treated with proteinase K. However, lower amplification efficiency during qPCR cycling caused by secondary structure, rapid template self-annealing (as in the case of scAAV vector genomes), or by contaminants can affect the proper attainment of cycle threshold values. With digital PCR, sufficient amplification such that the signal exceeds the background in each individual nanoliter-scale PCR is the minimal requirement to record a positive event. Hence, PCR inhibitors such as those found in impure samples may not affect ddPCR-based assays to the same degree as qPCR assays. To test this hypothesis in the context of an AAV genome titer assay, an unpurified “bulk vector” (crude harvest) sample was serially diluted and the dilutions were spiked with a known amount of purified AAV vector from a different production lot. Detection of the spiked sample was done, using both the ddPCR and the optimized qPCR assay (Fig. 4 and Table 5). With ddPCR (Fig. 4), separation of the signal band (blue) from the background (red) is first seen in the 1000-fold dilution of the bulk material, which corresponded to 100% detection of the spiked vector. In contrast, a 10,000-fold dilution was required to achieve 80% recovery of the spiked vector in the optimized qPCR assay (Table 5) because of qPCR amplification curve suppression and the resulting delay in reaching the cycle threshold. Our preliminary data therefore indicate superior (at least 10-fold) resistance of the ddPCR genome titration to inhibition by a complex mixture of cellular macromolecules and excess contaminating AAV genomes. This result suggests an application of the assay for assessing AAV copy numbers in unpurified vector samples from early stages of production. In addition, an advantage of using ddPCR is implied for the quantitation of AAV genomes in biodistribution samples, some of which can be inhibitory to qPCR.
FIG. 4.
Bulk preparation interference with the ddPCR genome titer assay. The amplitude of the fluorescent signal after cycling is shown, with red representing low signal (baseline) and blue indicating high-level signal. Each of the ∼15,000 20-nl reactions is represented by a single dot. Dilutions of the bulk (unpurified) vector sample are shown on the x axis. Top: Number of genome copies in the bulk vector dilutions and percent recovery of the spiked genome input (3.5×103 GC).
Table 5.
Bulk Preparation Interference with Droplet Digital PCR and Optimized qPCR Assays
| log10 bulk vector dilution | ||||||||
|---|---|---|---|---|---|---|---|---|
| 0 | −1 | −2 | −3 | −4 | −5 | −6 | −7 | |
| Input bulk vector copies | 1.5×1010 | 1.5×109 | 1.5×108 | 1.5×107 | 1.5×106 | 1.5×105 | 1.5×104 | 1.5×103 |
| Spike copies | 3.5×103 | 3.5×103 | 3.5×103 | 3.5×103 | 3.5×103 | 3.5×103 | 3.5×103 | 3.5×103 |
| ddPCR % spike recovery | 1.0 | 0.0 | 3.0 | 110.0 | 112.0 | 116.0 | 118.0 | 111.0 |
| OqPCR % spike recovery | 0.0 | 0.0 | 3.6 | 4.3 | 78.0 | 116.0 | 115.0 | 111.0 |
ddPCR, droplet digital PCR; OqPCR, optimized qPCR.
qPCR has come to be viewed as the best available assay for AAV vector genome titration because it provides specificity as well as enhanced precision in comparison with earlier genome titer assays. An rAAV2 reference standard material (AAV2 RSM) was characterized by multiple independent laboratories using a common qPCR protocol (Lock et al., 2010b). In this study, significant interlaboratory variation in genome titers was observed (CV, 77.7%) and even within a laboratory, CVs as high as 60% were sometimes noted. Interestingly, when the AAV2 RSM was titered by the ddPCR assay, the titer obtained (3.6×1013 GC/ml) was similar to the reported value (3.28×1013 GC/ml), suggesting that despite the high degree of variance, the average titer obtained for the AAV2 RSM was a good estimate. In addition to the potential for variance of a qPCR assay performed in multiple laboratories, another source of error stems from the fact that several different protocols for qPCR-based titration of AAV vector genomes exist. It is clear from the data presented here that even small protocol changes, such as performing assays in the absence of surfactant or treatment with proteinase K, can lead to large discrepancies in titer. In the case of scAAV vectors, further variation in qPCR-based assays is possible because of poor selection of qPCR target regions resulting in primer–probe exclusion. Overall, therefore, there is a need for more robust, precise assays for determining AAV vector genome titers.
Alternative assays for genome titration of scAAV vectors have been proposed. Fagone and colleagues (2011) introduced two new assays, one in which the terminal hairpin was cleaved with a restriction endonuclease before qPCR (termed ED-PCR) and the second based on native agarose gel analysis, as performed in the current study (Fig. 3). Although both methods are able to overcome the inhibitory effect of the scAAV genome on PCR, they each suffer from limitations. Interassay variance was noted to be a problem for ED-PCR and such variance may be substantially compounded should different restriction enzymes be required for different vector genomes (Fagone et al., 2011). In the case of the native agarose gel assay, the amount of DNA in the full-length double-stranded vector genome band may be affected by varying degrees of gel retardation due to undissociated capsid–genome complexes or to inefficient genome reannealing. The assay is suitable only for highly purified preparations and the effects of impurities may adversely affect titer determination. Furthermore, different gel assay systems are required for ssAAV versus scAAV quantification, although data reported here suggest that this limitation could be overcome. Older methods of AAV genome quantification such as the DNA dot/slot blot and more recently, the Southern blot (Wang et al., 2012), can also overcome the problem of genome self-annealing but are particularly cumbersome and prone to operator–induced variability; they are therefore unlikely to be adopted as release assays in a commercial production environment. Direct quantification of AAV vector genomes by UV spectrophotometry (Sommer et al., 2003) is another assay, but is also limited to highly pure preparations. In contrast, we find that ddPCR represents a highly precise method to determine AAV vector genome titers that is not unduly influenced by inefficient amplification or standard curve issues. The technique delivers a universal assay for all AAV vector types without some of the limitations of qPCR or the need to resort to separate methods for scAAV vector titration. Although the technique does not eliminate the PCR inhibition seen with primer–probe sets targeted close to the terminal hairpin of scAAV vectors, the moderate increase in titers observed over our best qPCR assay, along with increased intra- and interassay precision, make the ddPCR assay highly suitable for genome copy titration of these vectors. This is especially likely to be the case in clinical or commercial vector production, where the highest degree of reproducibility and precision is required and where small titer differences could translate to fewer scaled production runs and significant financial savings. The high degree of precision of the ddPCR assay also allows for a reduced number of replicates and thus multi-sample, multi-primer–probe set analysis on a single 96-well plate. The lack of requirement for a plasmid standard curve eliminates the time, expense, and error involved in their preparation. ddPCR effectively lowers the amplification efficiency threshold of primer–probe sets and allows for meaningful, direct comparisons of results where different assay efficiencies are an issue. Substantial benefits of the assay are also suggested in terms of sensitivity, precision, and refractivity to inhibition during analysis of impure vector samples, or in biodistribution analysis of AAV vector genomes for preclinical/clinical studies.
To develop the ddPCR genome titer assay into a cGMP/GLP-compliant assay, further validation would be required with an appropriate validation vector lot specific to the final product, such that accuracy/precision, repeatability/reproducibility, as well as limits and range of detection/quantification could be established. This validation lot would be included on each run and serve as a reference point to confirm proper sample preparation and assay performance. In addition, verification and validation of the ddPCR instrumentation itself would be required, and for which installation and operational qualification (IQ/OQ) procedures will soon be available. With such validation in place, the ddPCR genome titer assay is likely to be highly suitable for use in clinical manufacturing.
Supplementary Material
Acknowledgments
The authors thank members of the Wilson laboratory and Penn Vector Core for providing vector and qPCR validation results. This work was performed with funding from the NHLBI Gene Therapy Resource Program (HHSN268200748202C), NHLBI (HL059407-14), a grant from the Bill & Melinda Gates Foundation, and a sponsored research agreement from ReGenX Biosciences.
Author Disclosure Statement
J.M.W., M.L., and M.R.A. are inventors on patents licensed to various biopharmaceutical companies, including ReGenX. J.M.W. is a consultant to ReGenX Holdings, and is a founder of, holds equity in, and receives a grant from affiliates of ReGenX Holdings.
References
- Allay J.A., Sleep S., Long S., et al. (2011). Good manufacturing practice production of self-complementary serotype 8 adeno-associated viral vector for a hemophilia B clinical trial. Hum. Gene Ther. 22, 595–604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bainbridge J.W., Smith A.J., Barker S.S., et al. (2008). Effect of gene therapy on visual function in Leber's congenital amaurosis. N. Engl. J. Med. 358, 2231–2239 [DOI] [PubMed] [Google Scholar]
- Bennicelli J., Wright J.F., Komaromy A., et al. (2008). Reversal of blindness in animal models of Leber congenital amaurosis using optimized AAV2-mediated gene transfer. Mol. Ther. 16, 458–465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christine C.W., Starr P.A., Larson P.S., et al. (2009). Safety and tolerability of putaminal AADC gene therapy for Parkinson disease. Neurology 73, 1662–1669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark K.R., Liu X., McGrath J.P., and Johnson P.R. (1999). Highly purified recombinant adeno-associated virus vectors are biologically active and free of detectable helper and wild-type viruses. Hum. Gene Ther. 10, 1031–1039 [DOI] [PubMed] [Google Scholar]
- Dolgin E. (2012). Gene therapies advance, but some see manufacturing challenges. Nat. Med. 18, 1718–1719 [DOI] [PubMed] [Google Scholar]
- Eberling J.L., Jagust W.J., Christine C.W., et al. (2008). Results from a phase I safety trial of hAADC gene therapy for Parkinson disease. Neurology 70, 1980–1983 [DOI] [PubMed] [Google Scholar]
- Fagone P., Wright J.F., Nathwani A.C., et al. (2011). Systemic errors in quantitative polymerase chain reaction titration of self-complementary adeno-associated viral vectors and improved alternative methods. Hum. Gene Ther. Methods 23, 1–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauswirth W.W., Aleman T.S., Kaushal S., et al. (2008). Treatment of leber congenital amaurosis due to RPE65 mutations by ocular subretinal injection of adeno-associated virus gene vector: Short-term results of a phase I trial. Hum. Gene Ther. 19, 979–990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hindson B.J., Ness K.D., Masquelier D.A., et al. (2011). High-throughput droplet digital PCR system for absolute quantitation of DNA copy number. Anal. Chem. 83, 8604–8610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoshino T., and Inagaki F. (2012). Molecular quantification of environmental DNA using microfluidics and digital PCR. Syst. Appl. Microbiol. 35, 390–395 [DOI] [PubMed] [Google Scholar]
- Kaplitt M.G., Feigin A., Tang C., et al. (2007). Safety and tolerability of gene therapy with an adeno-associated virus (AAV) borne GAD gene for Parkinson's disease: An open label, phase I trial. Lancet 369, 2097–2105 [DOI] [PubMed] [Google Scholar]
- Kapranov P., Chen L., Dederich D., et al. (2012). Native molecular state of adeno-associated viral vectors revealed by single-molecule sequencing. Hum. Gene Ther. 23, 46–55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lock M., Alvira M., Vandenberghe L.H., et al. (2010a). Rapid, simple, and versatile manufacturing of recombinant adeno-associated viral vectors at scale. Hum. Gene Ther. 21, 1259–1271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lock M., McGorray S., Auricchio A., et al. (2010b). Characterization of a recombinant adeno-associated virus type 2 Reference Standard Material. Hum. Gene Ther. 21, 1273–1285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lock M., Alvira M.R., and Wilson J.M. (2012). Analysis of particle content of recombinant adeno-associated virus serotype 8 vectors by ion-exchange chromatography. Hum. Gene Ther. Methods 23, 56–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maguire A.M., Simonelli F., Pierce E.A., et al. (2008). Safety and efficacy of gene transfer for Leber's congenital amaurosis. N. Engl. J. Med. 358, 2240–2248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maguire AM, High KA, Auricchio A, Wright JF, et al. (2009). Age-dependent effects of RPE65 gene therapy for Leber's congenital amaurosis: A phase 1 dose-escalation trial. Lancet 374, 1597–605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinheiro L.B., Coleman V.A., Hindson C.M., et al. (2012). Evaluation of a droplet digital polymerase chain reaction format for DNA copy number quantification. Anal. Chem. 84, 1003–1011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samulski R.J., Chang L.S., and Shenk T. (1989). Helper-free stocks of recombinant adeno-associated viruses: Normal integration does not require viral gene expression. J. Virol. 63, 3822–3828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders R., Huggett J.F., Bushell C.A., et al. (2011). Evaluation of digital PCR for absolute DNA quantification. Anal. Chem. 83, 6474–6484 [DOI] [PubMed] [Google Scholar]
- Sedlak R.H., and Jerome K.R. (2013). Viral diagnostics in the era of digital polymerase chain reaction. Diagn. Microbiol. Infect. Dis. 75, 1–4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sidransky D., Tokino T., Hamilton S.R., et al. (1992). Identification of ras oncogene mutations in the stool of patients with curable colorectal tumors. Science 256, 102–105 [DOI] [PubMed] [Google Scholar]
- Simonelli F., Maguire A.M., Testa F., et al. (2010). Gene therapy for Leber's congenital amaurosis is safe and effective through 1.5 years after vector administration. Mol. Ther. 18, 643–650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sommer J.M., Smith P.H., Parthasarathy S., et al. (2003). Quantification of adeno-associated virus particles and empty capsids by optical density measurement. Mol. Ther. 7, 122–128 [DOI] [PubMed] [Google Scholar]
- Vogelstein B., and Kinzler K.W. (1999). Digital PCR. Proc. Natl. Acad. Sci. U.S.A. 96, 9236–9241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Ling C., Song L., et al. (2012). Limitations of encapsidation of recombinant self-complementary adeno-associated viral genomes in different serotype capsids and their quantitation. Hum. Gene Ther. Methods 23, 225–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.