Abstract
Human pluripotent stem cells (hPSCs) represent a prime cell source for pharmacological research and regenerative therapies because of their extensive expansion potential and their ability to differentiate into essentially all somatic lineages in vitro. Improved methods to stably introduce multiple transgenes into hPSCs will promote, for example, their preclinical testing by facilitating lineage differentiation and purification in vitro and the subsequent in vivo monitoring of respective progenies after their transplantation into relevant animal models. To date, the establishment of stable transgenic hPSC lines is still laborious and time-consuming. Current limitations include the low transfection efficiency of hPSCs via nonviral methods, the inefficient recovery of genetically engineered clones, and the silencing of transgene expression. Here we describe a fast, electroporation-based method for the generation of multitransgenic hPSC lines by overcoming the need for any preadaptation of conventional hPSC cultures to feeder-free conditions before genetic manipulation. We further show that the selection for a single antibiotic resistance marker encoded on one plasmid allowed for the stable genomic (co-)integration of up to two additional, independent expression plasmids. The method thereby enables the straightforward, nonviral generation of valuable multitransgenic hPSC lines in a single step. Practical applicability of the method is demonstrated for antibiotic-based lineage enrichment in vitro and for sodium iodide symporter transgene-based in situ cell imaging after intramyocardial cell infusion into explanted pig hearts.
Introduction
Human pluripotent stem cells (hPSCs) including embryonic stem cells (hESCs) and induced pluripotent stem cells (hiPSCs) are considered a prime cell source for envisioned regenerative therapies because of their extensive proliferation and multilineage differentiation potential in vitro. To date, such cell-based approaches are still under development and numerous issues remain to be solved before hPSC-derived progenies can enter routine clinical applications. Stable transgene-expressing hPSC lines can be valuable tools along this path. For example, the overexpression of specific transcription or growth factors aiming at the directed differentiation into a desired cell type may overcome poor differentiation efficiencies that exist for several somatic lineages (David et al., 2009; Hartung et al., 2013). On the other hand, lineage-restricted reporter gene expression could be used to enrich for specific progenies (Boecker et al., 2004; Huber et al., 2007; Xu et al., 2008b; Kita-Matsuo et al., 2009; Ritner et al., 2011). Another application is the cell type-specific or ubiquitous expression of marker genes to monitor cell engraftment in respective animal models posttransplantation (Dai et al., 2007; Sun et al., 2009; Mauritz et al., 2011).
A multitude of methods to genetically manipulate hPSCs have been established including viral transduction (Jang et al., 2006; Wurm et al., 2011), chemical transfection (Liu et al., 2009), and electroporation (Zwaka and Thomson, 2003; Costa et al., 2005). Among integrating vector systems that could be used for the generation of stable transgenic PSC lines, classical oncoretroviral vectors are well known to become silenced in pluripotent stem cells (Yao et al., 2004). Although great transduction efficiencies of >90% and high transgene expression levels can be achieved by lentiviral vectors (Ma et al., 2003; Moore et al., 2005), murine and human PSCs were found to substantially suppress long-term expression of transgenes delivered by these vector systems (Cherry et al., 2000; Xia et al., 2007). This effect becomes manifested mainly during the first days after transduction and appears to depend on the type of viral promoter used and the site of transgene integration into the genome as well (Haas et al., 2000; Philippe et al., 2006; Xia et al., 2007; Norrman et al., 2010). Another objection is the tendency of integrating viral vectors to induce insertional mutagenesis that provokes oncogene activation or inactivation of tumor suppressor genes (Kustikova et al., 2005), thereby increasing the likelihood of malignant cell transformation. In contrast to plasmid vectors generated in bacteria, the production of viral vectors is also more time-consuming and requires higher safety levels.
Common techniques for the introduction of plasmid-based expression vectors into mammalian cells include electroporation and chemical transfection with liposomes, or polycationic complexes, being the most frequently used agents to facilitate cell entry (Pichon et al., 2010; Ziello et al., 2010). The development of improved transfection reagents such as FuGENE HD or GeneJammer, for example, allowed the generation of genetically engineered hPSC clones (Braam et al., 2008a; Liu et al., 2009), but efficiencies were relative low (Liu et al., 2009) and cell line dependent.
During electroporation a transient permeabilization of the plasma membrane occurs, thereby enforcing the uptake of exogenous nucleic acid molecules into the cell, which is in principle not limited by the size and nature of a vector (Moore et al., 2005). However, although the successful electroporation-based genetic modification of semidissociated “hPSC clumps” was achieved (Zwaka and Thomson, 2003), the method ideally requires dissociation into a single-cell suspension to substantially increase the transfection efficiency (Schinzel et al., 2011). In contrast to pluripotent mouse cells, human PSCs are highly sensitive to complete dissociation, resulting in dramatically reduced survival rates after electroporation. Prior approaches have tried to tackle this issue by adapting hPSCs to feeder-free conditions, which were established by trypsin-dependent cell dissociation for several passages before transfection. This approach, however, may lead to an inadvertent, clonal preselection of cells adapted to this treatment, thus requiring a frequent assessment of pluripotency and karyotyping (Braam et al., 2008a). The adaptation procedure is also time-consuming, thereby extending the generation of transgenic lines for several weeks (Costa et al., 2007; Braam et al., 2008b).
Here we describe a straightforward approach for the rapid, relative efficient, and simultaneous introduction of several plasmid vectors into hPSCs. Stable multitransgenic clones were established by means of single drug resistance encoded on only one of these expression vectors. The method is applicable directly to conventional feeder-based cultures and does not require any preadaptation to single-cell passaging. Multitransgenic hPSC lines were established carrying stably integrated (1) fluorescence-emitting reporters, (2) a sodium iodide symporter (NIS) facilitating noninvasive in vivo cell tracking (Acton and Kung, 2003; Templin et al., 2012), and (3) antibiotic resistance genes controlled by ubiquitous or lineage-specific promoters. We show that the transgenic lines resulting from our approach maintain their pluripotency and differentiation potential and retain a normal karyotype. In numerous clones the reporter gene expression was found to be stable after extensive passaging in the undifferentiated state, even if transgenes were encoded on cointroduced plasmids carrying no additional antibiotic resistance marker. Finally, the possibility of NIS-dependent in situ imaging after intramyocardial infusion of radionuclide-labeled cells was demonstrated and antibiotic-based purification of cardiomyocytes (CMs) was performed to demonstrate the broad practical applicability of the method.
Materials and Methods
Feeder-dependent adherent culture
Human ES cell lines hES3 (Reubinoff et al., 2000) and I3 (Amit and Itskovitz-Eldor, 2002), as well as the three human iPSC lines hCBiPS2 (cord blood endothelial cell derived; Haase et al., 2009), hFF1-iPS4 (human foreskin fibroblast derived), and hHSC_F1285_T-iPS2 (hematopoietic stem cell derived; Hartung et al., 2013), were cultured on irradiated murine embryonic fibroblasts (MEFs) in 80% KnockOut DMEM (Dulbecco's modified Eagle's medium) supplemented with 20% KnockOut Serum Replacement, 1 mM l-glutamine, 0.1 mM 2-mercaptoethanol, 1% nonessential amino acid stock (all from Life Technologies, Karlsruhe, Germany), and basic fibroblast growth factor (bFGF) at either 50 ng/ml (hES3, I3) or 4 ng/ml (hiPSCs) (supplied by the Institute for Technical Chemistry, Leibniz University Hannover, Hannover, Germany) (Chen et al., 2012). Cells were cultured in 6-well plates and passaged once per week with collagenase type IV (1 mg/ml; Invitrogen). The medium was changed daily.
Feeder-independent adherent culture
For the transfer of hESCs to feeder-independent culture conditions, cells were detached from MEFs, using collagenase type IV (1 mg/ml). After washing once with phosphate-buffered saline (PBS) without Ca2+/Mg2+ the cells were dissociated into single cells, using Accutase (PAA Laboratories, Cölbe, Germany). hESCs were seeded at 1.25×106 per 25 cm2 flask in MEF-conditioned medium (Burridge et al., 2011) and passaged twice per week. The medium was changed daily.
Plasmid vectors
The vector αMHCneoPGKhygro is a 10.9-kb construct carrying two selection cassettes: (1) the hygromycin resistance gene (hygroR) under the control of the ubiquitous phosphoglycerate kinase (PGK) promoter, allowing the establishment of stable integrants; and (2) the neomycin resistance gene (neoR) under the control of the heart-specific α-myosin heavy chain (αMHC) promoter, allowing the selection of hPSC-derived cardiomyocytes (Klug et al., 1996). Reporter gene constructs are based on CAG-eGFP (5.5 kb) with transgene expression controlled by the chicken β-actin (CBA) promoter combined with the human immediate-early cytomegalovirus (CMV-IE) enhancer (together termed the CAG promoter). CAG-nRedStar (5.6 kb) was constructed by replacing enhanced green fluorescent protein (eGFP) with red fluorescent protein (RedStar) in CAG-eGFP (Knop et al., 2002) coupled to a nuclear membrane location signal (nucmem, nRedStar) (Okita et al., 2004). The bicistronic vector CAG-rNIS-nVenus contains the rat sodium iodide symporter (rNIS) (Templin et al., 2012) coupled to the yellow nucleus-located fluorescent reporter Venus (nVenus) by an internal ribosomal entry site (IRES2).
Cell transfection and generation of transgenic clones
All transfections were performed with the Neon® system (Life Technologies). Several enzyme combinations were tested for cell harvesting. These included treatment with collagenase IV (Life Technologies) to remove the cells from their feeder layer, followed by an additional incubation step with collagenase B (Roche, Mannheim, Germany) or TrypLE (Life Technologies) for single-cell dissociation according to the manufacturers' specifications. In the final protocol the combination of collagenase IV and TrypLE was used. For single-cell preparation, 0.2% collagenase IV was added directly to the cell cultures and incubated for 5 min at 37°C. After centrifugation cells were washed once with PBS without Ca2+/Mg2+and incubated with 0.5 ml of TrypLE for 3 min at 37°C. The cells were counted in PBS without Ca2+/Mg2+ in a Thoma hemocytometer, and 1.5×106 dissociated single-cell hPSCs were pelleted at 1000 rpm for 5 min, resuspended in 110 μl of resuspension buffer R, and transferred with the corresponding circular plasmid DNA into sterile microcentrifuge tubes. The cells were electroporated with two pulses at 1000 V for 20 msec and plated onto Matrigel-coated 6-cm dishes in preincubated conditioned medium supplemented with 10 μM ROCK (Rho-associated coiled-coil kinase) inhibitor (Y-27632; supplied by the Institute for Organic Chemistry, Leibniz University Hannover) (Palecek et al., 2011). Plasmids were used in their circular form without prior linearization and were applied in equimolar amounts without exceeding 20 μg of total DNA per electroporation sample. Cell counts were performed directly after electroporation and on plated cells 48 hr after transfection by means of trypan blue exclusion in a Thoma hemocytometer; respective viable cell counts were compared with the cell number applied for transfection (i.e., 1.5×106 per sample) to calculate the percentage of cell viability/recovery. Forty-eight hours after electroporation the transient transfection efficiencies were determined for eGFP or nRedStar by flow cytometry. On parallel dishes, at 48–72 hr after transfection, hygromycin (Invitrogen) was added at 50 μg/ml (hESCs) or 200 μg/ml (hiPSCs) to select for stable transgenic clones. Overall, cells were incubated for 4 days in the presence of hygromycin, with the selection medium replaced once after 48 hr. Upcoming colonies were manually picked ∼14 days after electroporation, transferred onto irradiated feeder cells, and expanded clonally.
Characterization of multitransgenic clones and cell lines
Established clones and cell lines derived therefrom were analyzed for the expression of typical pluripotency-associated markers via immunofluorescence staining. PCR was applied to test for the genomic integrity of the plasmid αMHCneoPGKhygro (see primer sequences in Supplementary Table S1; supplementary data are available online at http://online.liebertpub.com/hgtb) in cells five passages after clone picking. Stable expression of fluorescence reporter genes in undifferentiated cell lines was monitored by flow cytometry for up to 23 passages after clone picking and at respective time points after initiation of differentiation, as indicated in Results.
Southern blot analyses
Ten micrograms of genomic DNA was cleaved with endonucleases as indicated in Results, separated on a 0.8% TAE (Tris–acetate–EDTA)–agarose gel; transferred onto Biodyne B nylon membrane (Pall Life Sciences, Dreieich, Germany); and hybridized with radiolabeled probes, using a DecaLabel kit (Fermentas, St. Leon-Rot, Germany). The hybridization probe for eGFP was an 807-bp EcoRI–BglII fragment from pCAGGS2, and for neoR a 915-bp HindIII–SacI fragment isolated from αMHCneoPGKhygro.
Karyotype analyses
After trypsinization, metaphases were prepared according to standard procedures. Fluorescence R-banding using chromomycin A3 and methyl green was performed as described in detail previously (Schlegelberger et al., 1999). At least 15 metaphases were analyzed for each clone. Karyotypes were described according to the International System for Human Cytogenetic Nomenclature (ISCN).
Induction of hPSC differentiation, cardiomyocyte selection, and quantification
To induce embryoid body (EB) formation of transgenic cell lines, undifferentiated cells were washed with a chemically defined serum-free (SF) medium and subsequently were mechanically harvested. Resulting cell clumps were transferred to ultralow attachment 6-well plates (Greiner Bio-One, Frickenhausen, Germany) and cultured in SF supplemented with 10 μM SB203580 (Graichen et al., 2008; Xu et al., 2008a). The medium was replaced every second day. Flow cytometry was performed to quantify the amount of eGFP-expressing cells and the presence of cardiomyocytes, which was tested by the expression of the intracellular cardiac-specific isoform troponin T (cTnT; see experimental details below). Selection of cardiomyocytes was initiated by the addition of Gibco G418 (200 μg/ml; Invitrogen) 2 days after the first beating foci were observed. Antibiotic selection was retained for at least 7 days (Xu et al., 2008b). To assess the moiety of cTnTpos cardiomyocytes, cell samples were fixed on the dish with 4% paraformaldehyde (Sigma-Aldrich, St. Louis, MO) for 20 min and blocked with Tris-buffered saline plus 5% donkey serum and 0.25% Triton X-100 (Sigma-Aldrich) for 20 min at room temperature. To analyze the expression of cTnT, respective primary and secondary antibodies (1:200 anti-Troponin T, cardiac isoform Ab-1, mouse monoclonal IgG [Thermo Scientific, Karlsruhe, Germany] and 1:200 donkey anti-mouse IgG labeled with DyLight 549 [Jackson ImmunoResearch Laboratories, West Grove, PA]) were diluted in PBS without Ca2+/Mg2+, supplemented with 1% bovine serum albumin (BSA; Sigma-Aldrich), and incubated for 1 hr at room temperature. For quantification, samples were photographed at appropriate magnification and the total number of 4′,6-diamidino-2-phenylindole (DAPI)-stained nuclei was determined versus the number of cTnTpos cells.
Flow cytometry
To assess eGFP or nRedStar expression cells were washed with PBS without Ca2+/Mg2+ and dissociated by treatment with trypsin for 3 min at 37°C to create a single-cell suspension. Cells were resuspended in PBS without Ca2+/Mg2+ and analyzed on a FACSCalibur system (BD Biosciences, San Jose, CA) by applying BD CellQuest software (BD Biosciences). Nontransgenic hPSCs served as control.
To analyze the expression of cTnT in differentiated cells, cultures were dissociated with collagenase B, following cell fixation and permeabilization with a Fix&Perm cell permeabilization kit (Dianova, Hamburg, Germany) and incubated with respective antibodies (1:200 anti-TnT, cardiac isoform Ab-1, mouse monoclonal IgG [Thermo Scientific] and 1:200 donkey anti-mouse IgG labeled with DyLight 549 [Jackson ImmunoResearch Laboratories]) according to the manufacturers' specifications. Stained cells were analyzed by flow cytometry, using a BD Accuri C6 (BD Biosciences), and data were analyzed with FlowJo software (Treestar, Ashland, OR).
Immunohistological staining
Undifferentiated as well as differentiated cells were fixed with 4% paraformaldehyde, washed with PBS, and treated with 5% donkey serum, 0.25% Triton X-100 diluted in Tris-buffered saline (TBS) for 20 min at room temperature. After rinsing, the cells were incubated for at least 1 hr with respective primary antibodies (1:300 anti-α-fetoprotein, mouse IgG1 [R&D Systems, Minneapolis, MN]; 1:400 anti-β-tubulin, mouse IgG2a [Millipore, Darmstadt, Germany]; 1:100 anti-GFP [cross-reacting with Venus] goat IgG [Acris, Herford, Germany]; 1:100 anti-OCT4 mouse IgG [Santa Cruz Biotechnology, Santa Cruz, CA]; 1:200 anti-SOX17, goat IgG [R&D Systems], 1:100 anti-SSEA-3 mouse IgM [Developmental Studies Hybridoma Bank, Iowa City, IA]; 1:70 anti-SSEA-4 mouse IgG [Developmental Studies Hybridoma Bank]; 1:100 anti-TRA-1-60 mouse IgM [Abcam, Cambridge, MA]; 1:100 anti-Troponin T, mouse IgG [Thermo Scientific]) diluted in PBS–1% BSA. Further incubation was performed with the respective secondary antibody (1:200 donkey anti-mouse IgG labeled with DyLight 549, 1:200 donkey anti-mouse IgG labeled with DyLight 488, 1:200 donkey anti-mouse IgM labeled with Cy3, and 1:100 donkey anti-goat IgG labeled with Cy2; all from Jackson ImmunoResearch Laboratories) for 30 min. Cells were rinsed, counterstained with DAPI (Sigma-Aldrich), and analyzed with an Axio Observer A1 fluorescence microscope (Carl Zeiss MicroImaging, Goettingen, Germany). For the detection of nVenuspos cells, pig heart tissue was embedded in Tissue-Tek (Sakura Finetek, Alphen aan den Rijn, The Netherlands) directly after the injection of 1×106 undifferentiated hCBiPS2 CAG-rNIS6 and stored at −80°C. Subsequently, 10 μm cryosections were generated with a Microm Cryostat HM 560 cryotome (Thermo Scientific).
Analysis of mRNA expression by qRT-PCR
Total RNA was prepared with an RNeasy kit (Macherey-Nagel, Düren, Germany) and reverse transcribed with SuperScript II (Life Technologies) using oligo(dT) primers according to the manufacturer's instructions. Quantitative RT-PCR (qRT-PCR) was performed in triplicate, using a Mastercycler ep realplex2 (Eppendorf, Hamburg, Germany) and ABSOLUTE QPCR SYBR green mix (ABgene, Epsom, Surrey, UK). The size of amplicons and the absence of nonspecific products were controlled by melting curves. Sequences of primers are shown in Supplementary Table S2. Relative changes in gene expression were analyzed via 2−ΔΔCt (Livak and Schmittgen, 2001) by comparing transgenic versus control cells, using Mastercycler ep realplex2 software version 2.0 (Eppendorf). Expression levels of target genes were normalized to β-actin; means±SEM of normalized gene expression levels are presented.
Ex vivo cardiac SPECT-CT imaging
NISpos-hPSCs (1×106) were incubated for 90 min with 1 MBq of 123I and vigorously washed, and 5×106 labeled cells were injected into the anterior wall of the left ventricle of an explanted pig heart, using a NOGA MyoStar intramyocardial injection catheter system (Biosense Webster/Johnson & Johnson, Diamond Bar, CA). The 123I signal was visualized through a hybrid SPECT-CT (single-photon emission computed tomography combined with computed tomography) camera with semiconductor detector technique (Discovery NM 570C; GE Healthcare, Piscataway, NJ). To mimic in vivo signal attenuation, imaging of 123I signals was performed through a dissected pig chest wall that was placed above the heart.
Statistical analysis
Results are reported as means and standard deviation of the mean. p values <0.01, indicated by double asterisks (**), were considered significant.
Results
Adaptation-free electroporation of plasmid DNA into hPSCs results in >60% transient transfection efficiency accompanied by high cell viability
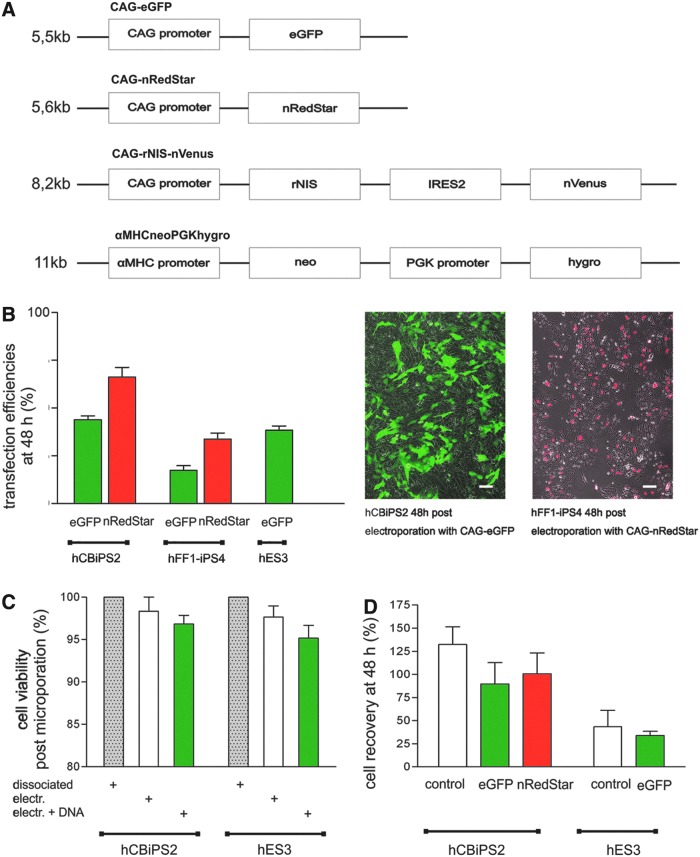
Common feeder-based hPSC cultures were used without any preadaptation and cells were routinely passaged weekly. For electroporation, cells were harvested on day 4 postpassaging to ensure log-phase growth. Applying pretested electroporation parameters, a first step of optimization was implied using various enzyme combinations to detach and dissociate hPSCs. Investigating collagenase IV, collagenase B, and TrypLE, best results regarding cell viability and transfection efficiency were achieved by combining collagenase IV followed by TrypLE treatment (data not shown; see detailed protocol in Materials and Methods). Cell survival also critically depended on the Rho-associated coiled-coil kinase (ROCK) inhibitor Y-27632 added to the culture medium postelectroporation (data not shown). To assess the transient transfection efficiency, two constitutively expressed fluorescence reporters (eGFP and nRedStar; Fig. 1A) and five independent hPSC lines (two hESC and three hiPSC lines) were tested. The application of up to 20 μg of total, circular plasmid DNA per electroporation approach resulted in balanced cell viability and transgene expression as depicted in Fig. 1B–D. At 48 hr postelectroporation we found 44±5% eGFPpos (n=7) and 63±13% nRedStarpos (n=7) cells for the iPSC line hCBiPS2, 18±3% eGFPpos (n=3) and 38±1% nRedStarpos (n=6) cells for hFF1-iPS4, and 38±4.8% eGFPpos cells for the ESC line hES3 (n=6) (Fig. 1B). Regarding the cell viability directly after electroporation, only marginal differences were observed between (1) dissociation-only controls (which included virtually no dead cells in the trypan blue assay and were set to 100% viability in Fig. 1C), (2) cells that were electroporated without DNA (98±3% hCBiPS2 and 97±6% hES3 viability), and cells that received plasmid DNA (96±8% hCBiPS2 and 95±1% hES3 viability). Next, the number of viable cells recovered at 48 hr posttreatment was compared with the number of cells initially used per electroporation approach (i.e., always 1.5×106=100%). For hCBiPS2 this recovery rate was 132±19% in electroporated controls that received no DNA, 100±22% after CAG-nRedStar transfection, and 89±22% after CAG-eGFP transfection (Fig. 1D). For hES3 the recovery rate at 48 hr was substantially lower, that is, 43±17% for non–DNA electroporated controls and 34±4% after CAG-eGFP transfection (Fig. 1D), suggesting their higher sensitivity to the protocol.
FIG. 1.
Electroporation-based transfection resulted in good cell viability and reporter gene expression. (A) Schematic presentation of vector structures used for cell line generation. CAG-eGFP and CAG-nRedStar: constitutive expression of eGFP or the codon-optimized red GFP variant RedStar fused to a nuclear membrane localization signal (nucmem, nRedStar) is controlled by the CAG promoter. CAG-rNIS-nVenus: bicistronic vector carrying the rat sodium iodide symporter (rNIS) linked via an internal ribosomal entry site (IRES2) to the yellow GFP variant Venus (nucmem, nVenus) controlled by the CAG promoter. αMHCneoPGKhygro: hygromycin resistance gene constitutively expressed under the control of the phosphoglycerate kinase (PGK) promoter and the neomycin resistance gene under the control of the cardiomyocyte-specific αMHC promoter. (B) Left: Graph depicting transfection efficiencies at 48 hr postelectroporation as determined via flow cytometry for CAG-eGFP- and CAG-nRedStar-transfected cells. Right: Fluorescence microscopy of seeded cells. On average, 44±5% eGFPpos and 63±13% nRedStarpos hCBiPS2 cells (n=7), 18±3% eGFPpos (n=3) and 38±1% nRedStarpos hFF1-iPS4 cells (n=6), as well as 38±5% eGFPpos hES3 cells (n=6) were detected. (C) Cell viability directly after cell dissociation and electroporation revealed 98±3% viable hCBiPS2 cells and 97±6% viable hES3 cells. The addition of plasmid DNA led to 96±8% viable hCBiPS2 cells and 95±1% viable hES3 cells as assessed by trypan blue staining (n=6). (D) Cell recovery at 48 hr as compared with the cell number used for transfection (i.e., 1.5×106 set to 100%). For hCBiPS2 cells, 89±22% were recovered at 48 hr after transfection with CAG-eGFP versus 100±22% in the case of CAG-nRedStar; for hES3 cells only 34±4% were recovered after transfection with the plasmid CAG-eGFP in this assay (n=5). Color images available online at www.liebertpub.com/hgtb
One-step generation of stable multitransgenic clones
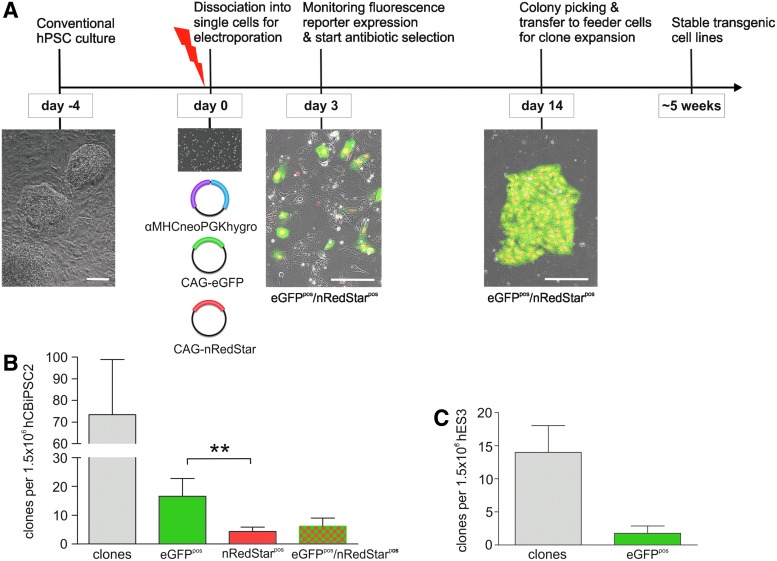
To test the possibility of generating stable multitransgenic hPSC clones in a one-step procedure (see schematic summary of the protocol in Fig. 2A), cells were coelectroporated with various vector combinations. All experiments included the vector αMHCneoPGKhygro (the only plasmid entailing constitutive hygromycin resistance, which was used for clone selection) together with one additional fluorescence reporter plasmid (either CAG-eGFP or CAG-rNIS-nVenus; see vector structure in Fig. 1A) or two additional plasmids (CAG-eGFP and CAG-nRedStar), aiming at establishing triple-transgenic cell lines. According to the electroporation approach using 1.5×106 hCBiPS2 cells, on average, 74±23 microscopically distinguishable hygromycin-resistant colonies were recovered (n=5; Fig. 2B and Table 1) at 7–10 days after initiation of hygromycin selection, resulting in an efficiency of ∼5×10−5 (Table 1). The efficiency of forming hygromycin-resistant colonies was similar for the hiPSC lines hHSC_F1285_T-iPS2 (up to ∼2×10−5) and hFF1-iPS4 (∼1×10−5), and the hESC line I3 (∼3×10−5; Table 1). However, for hES3 the average number of hygromycin-resistant colonies was 14±4 (n=5, Fig. 2C; efficiency, ∼1×10−5; Table 1) and thus ∼5-fold lower compared with hCBiPS2, putatively reflecting the lower recovery of hES3 cells at 48 hr postelectroporation (Fig. 2D), that is, before initiation of antibiotic selection.
FIG. 2.
Schema and timeline for the efficient generation of multitransgenic hPSC clones. (A) hPSCs used for transgenic modification were cultured under standard feeder-based culture conditions without the necessity for feeder-free preadaptation. For transfection cultures were dissociated into single cells and 1.5×106 cells were applied per individual electroporation approach. Cells were seeded onto Matrigel and 2–3 days posttreatment the transfection efficiency was monitored and hygromycin-based clone selection was initiated. After 7–10 days resistant colonies became visible. On day 14 colonies were picked and expanded. (B) Efficiency of antibiotic-resistant clone generation from hCBiPS2 per electroporation approach coapplying three vectors (αMHCneoPGKhygro, CAG-eGFP, and CAG-nRedStar). On average, 74±23 independent colonies were generated per 1.5×106 cells, whereof 16±6 appeared to be eGFPpos (∼30% of all hygromycin-resistant clones), 4±2 were nRedStarpos (∼6%), and 6±3 colonies (∼10%) expressed both fluorescence markers, that is, eGFPpos/nRedStarpos (n=5). (C) Efficiency of antibiotic-resistant colony generation from hES3 cells per electroporation approach coapplying two vectors (αMHCneoPGKhygro, CAG-eGFP). On average, 14±4 independent colonies were generated per 1.5×106 cells whereof 2±2 appeared eGFPpos (n=5). Color images available online at www.liebertpub.com/hgtb
Table 1.
Efficiency of (Multi-)Transgenic Colonies and Cell Line Generation
| Parental hPSC line | Cultivation method before electroporation | Designation of (co-) applied vectors | Efficiency,aabsolute number of hygromycin-resistant colonies [ ], and number of experimental repeats ( ) | Frequencyband absolute number of colonies [ ] expressing the respective fluorescence transgenes | No. of clones expanded/no. of stable fluorescence reporter lines generated, success rate ( ) |
|---|---|---|---|---|---|
| hCBiPS2 | MEF | αMHCneoPGKhygro | 3.9×10−5 [58±13] (n=7) | NA | |
| hCBiPS2 | MEF | αMHCneoPGKhygro | 3.7×10−5 [56±23] (n=9) | ||
| CAG-eGFP | 1.2×10−5 [18±12] | 59/24 (40%) | |||
| hCBiPS2 | MEF | αMHCneoPGKhygro | 5.3×10−5 [80±31] (n=6) | ||
| CAG-nRedStar | 1.3×10−5 [19±5.7] | 33/–(0%) | |||
| hCBiPS2 | MEF | αMHCneoPGKhygro | 1.1×10−5 [16±13] (n=4) | ||
| CAG-rNIS-nVenus | 0.3×10−5 [4±1.5] | 13/3 (23%) | |||
| hCBiPS2 | MEF | αMHCneoPGKhygro | 4.9×10−5 [74±23] (n=5) | GFP-only: 1.1×10−5 [16±6] | NT |
| CAG-eGFP | nRed-only: 0.3×10−5 [4±2] | NT | |||
| CAG-nRedStar | GFP and nRed: 0.4×10−5 [6±3] | 10/1 (10%) | |||
| hFF1-iPS4 | MEF | αMHCneoPGKhygro | 1.1×10−5 [17±3] (n=9) | ||
| CAG-eGFP | 0.2×10−5 [3±3] | NT | |||
| hFF1-iPS4 | Feeder-independent | αMHCneoPGKhygro | 3.1×10−5 [47] (n=1) | 0.5×10−5 [7] | NT |
| CAG-eGFP | |||||
| hFF1-iPS4 | Feeder-independent | αMHCneoPGKhygro | 3.8×10−5 [58±7] (n=2) | NR | |
| CAG-nRedStar | |||||
| hHSC_F1285_T-iPS2 | MEF | αMHCneoPGKhygro | 2.1×10−5 [33±5] (n=3) | GFP-only: 0.1×10−5 [2±1] | NT |
| CAG-eGFP | nRed-only: none | ||||
| CAG-nRedStar | GFP and nRed: none | ||||
| hHSC_F1285_T-iPS2 | Feeder-independent | αMHCneoPGKhygro | 0.7×10−5 [10±1] (n=2) | 0.2×10−5 [2.5±0.5] | NT |
| CAG-eGFP | |||||
| hHSC_F1285_T-iPS2 | Feeder-independent | αMHCneoPGKhygro | 0.2×10−5 [3] (n=1) | NR | |
| CAG-nRedStar | |||||
| hES3 | hFF/MEF | αMHCneoPGKhygro | 0.7×10−5 [11] (n=1) | NA | |
| hES3 | Feeder-independent | αMHCneoPGKhygro | 0.9×10−5 [14±4] (n=5) | ||
| CAG-eGFP | 0.1×10−5 [2±2] | NT | |||
| I3 | MEF | αMHCneoPGKhygro | 2.9×10−5 [44±19] (n=4) | GFP-only: 0.1×10−5 [2±2] | NT |
| CAG-eGFP | nRed-only: none | ||||
| CAG-nRedStar | GFP and nRed: none |
GFP, green fluorescent protein: hFF, human foreskin fibroblasts; hPSC, human pluripotent stem cell; MEF, mouse embryonic fibroblasts; n, number of independent electroporation experiments; NA, not applicable; NR, not recovered; NT, not tested.
Average efficiency of recovering hygromycin-resistant colonies (and respective average number of colonies per dish, shown in brackets) in relation to the number of cells per electroporation approach, that is, always 1.5×106.
Average efficiency of recovering hygromycin-resistant colonies (and respective average number of colonies per dish, shown in brackets) that also expressed the respective fluorescence transgene(s) (i.e. depending on the type of vector[s] used for coelectroporation) in relation to the number of cells per electroporation approach, that is, 1.5×106.
Another interesting observation was made on three-plasmid–transfected hCBiPS2 cells. Despite the equimolar transfection with nRedStar- and eGFP-encoding plasmid DNA (note that the plasmids were also equivalent in size and structure; Fig. 1) and in contrast to the higher transient transfection efficiency with CAG-nRedStar (Fig. 1B), the average number of eGFPpos colonies (among all hygromycin-resistant ones) was significantly higher than the number of nRedStarpos colonies, that is, 16±6 GFPpos versus 4±2 nRedStarpos (Fig. 2B and Table 1). We further found that, although eGFP and nRedStar were encoded on two independent plasmids, the average number of 6±3 double-fluorescent eGFPpos/nRedStarpos colonies found in this assay was even higher than the number of 4±2 colonies expressing nVenuspos only (Table 1). For three-plasmid–transfected hFF1-iPS4 and I3 cells an equivalent pattern was observed. Despite the higher transient transfection efficiency with CAG-nRedStar (see Fig. 1B for hFF1-iPS4; data not shown for I3) and equimolar transfection with nRedStar and eGFP plasmids, eGFPpos colonies were only recovered after hygromycin selection (Table 1).
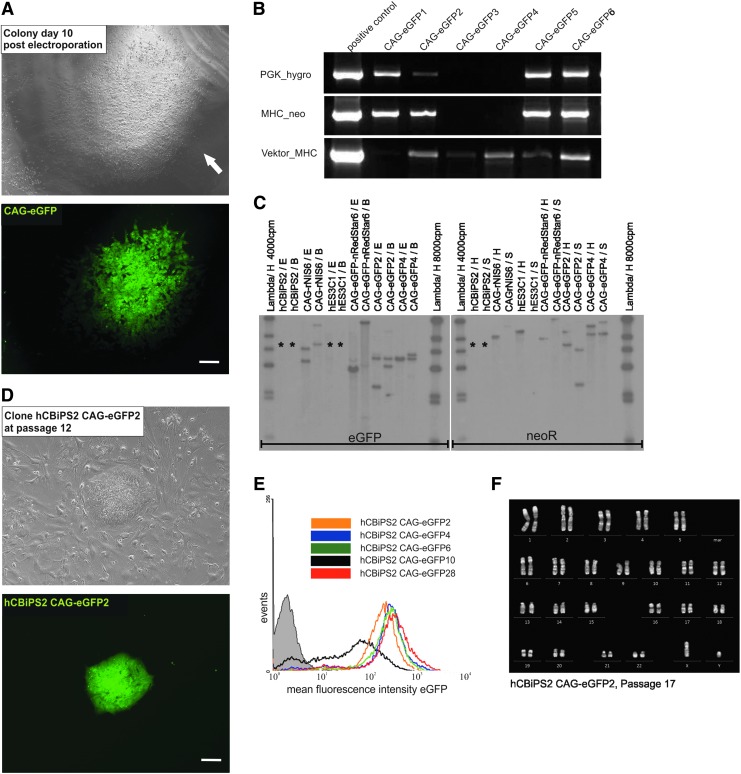
To establish stable multitransgenic hPSC lines, colonies were manually picked from Matrigel-coated dishes about 2 weeks after electroporation and subsequently transferred to feeder-based cultures (Fig. 2A and Fig. 3A and D), whereupon further hygromycin selection was omitted. Resulting clones were expanded for four or five passages before reanalysis of genomic transgene(s) integration and expression. The genomic integration of the selection vector αMHCneoPGKhygro was assessed by PCR (Fig. 3B). Clones from which all of the three tested vector-specific fragments (representing key functional elements of the vector; Supplementary Fig. S2) could be amplified (∼50% of all clones tested; n=59) were analyzed by Southern blotting (Fig. 3C), using respective restriction endonucleases and a NeoR gene-specific hybridization probe. Southern blot analysis confirmed and expanded the PCR results by demonstrating the presence of one or two copies of the transgene, respectively, in all expected clones (Fig. 3C). Applying alternative restriction enzymes and an eGFP gene-specific hybridization probe, we have further demonstrated the genomic integration of the cotransfected plasmid CAG-eGFP (Fig. 3C). The hybridization pattern suggests that three of these representative clones had two copies of the CAG-eGFP plasmid integrated, whereas one clone apparently exhibited a single integration event.
FIG. 3.
Characterization of multitransgenic colonies and hiPSC lines derived therefrom. (A) Light and fluorescence microscopic appearance of a primary eGFPpos hCBiPS2 cell colony (highlighted by white arrow) on day 10 after cotransfection with plasmids αMHCneoPGKhygro and CAG-eGFP (i.e., after 7 days of hygromycin selection). Scale bar: 100 μm. (B) Established (picked and expanded) eGFPpos clones were analyzed for the integrity of the selection vector αMHCneoPGKhygro by PCR using primers specific to three individual vector elements. As shown in this gel, ∼50% of all clones tested (n=59) showed only partial integration of the plasmid. Those were excluded from further analysis. (C) Southern blot analysis of several transgenic clones revealed differences in copy numbers of the transgenes eGFP (representative of CAG-eGFP, left) and neoR (representative of αMHCneoPGKhygro, right). Lanes representing DNA from nontransfected control cells are marked with an asterisk. (D) To establish stable clones (and subsequently transgenic cell lines), respective colonies were individually picked, transferred to feeder cells, and acquired typical hPSC morphology. Depicted is a representative eGFP-expressing colony of a transgenic cell line 12 passages after clone picking. (E) Analysis of five established hCBiPS2 αMHCneoPGKhygro_CAG-eGFP (CAG-eGFP) transgenic cell lines by flow cytometry. Four lines showed homogeneous eGFP expression with only slight shifts in their median expression intensity (peak of the bell curve) whereas one line (designated hCBiPS2 CAG-eGFP10; black line) showed a mosaic pattern, whereby a part of the cell population showed no expression (overlay with nontransgenic control shown in the gray histogram). (F) Exemplary karyotype analysis of two transgenic cell lines revealed no chromosomal abnormalities at passage 17 after isolation of the initial clones. The depicted karyogram represents the cell line hCBiPS2 CAG-eGFP2. Color images available online at www.liebertpub.com/hgtb
However, loss of fluorescence marker expression occurred in ∼20% of picked clones during the first ∼3 weeks of cultivation; eGFP expression remained more stable compared with nRedStar, tallying with the higher number of initially eGFPpos colonies recovered after hygromycin selection in our approach (Table 1). Remaining fluorescence-expressing clones were further passaged four or five times, stored, and termed transgenic cell lines thereafter.
Multitransgenic cell lines remain pluripotent and express typical pluripotency markers
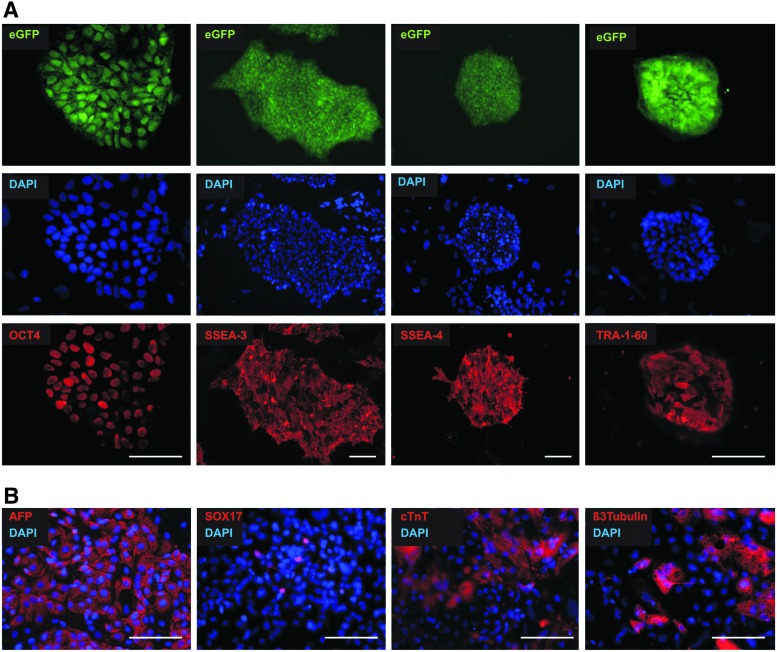
Transgenic hPSC lines showed characteristics of their parental cell lines (Supplementary Fig. S1) regarding their proliferation characteristics (equivalent splitting interval and ratio; data not shown) and colony morphology (Fig. 3D, Fig. 4A, and Fig. 5A). As demonstrated by flow cytometry the mean fluorescence intensity remained stable in most established cell lines tested for eGFP expression for up to 23 passages after clonal selection. In rare cases, however, homogeneous fluorescence expression turned into a mosaic-like expression pattern, as exemplified in the flow cytometry-based histogram in Fig. 3E. Notably, no apparent karyotypic abnormalities were identified when two representative lines were tested at passage 17 after clone isolation, as exemplified in Fig. 3F. Transgenic lines also showed the expected expression patterns of the pluripotency-associated genes OCT4, TRA-1-60, SSEA3, and SSEA4 as demonstrated by immunocytology (Fig. 4A). To show maintenance of the differentiation potential, we applied a standard embryoid body (EB)-based protocol to test the general multilineage differentiation. Immunocytology on EB outgrowth on day 21 of differentiation revealed the presence of derivatives of all three germ layers expressing endodermal (α-fetoprotein [AFP] and SOX17), mesodermal (cardiac troponin T), and ectodermal (β3-tubulin) marker proteins (Fig. 4B).
FIG. 4.
Multitransgenic cells display typical hPSC morphology, retain expression of pluripotency markers, and differentiate into derivatives of all three germ layers. (A) As exemplified for the cell line hCBiPS2 αMHCneoPGKhygro_CAG-eGFP2 (CAG-eGFP2) in passage 21 homogeneous staining of cell colonies for pluripotency associated markers OCT4, SSEA-3, SSEA-4, and TRA-1-60 (in red; eGFP expression in green; DAPI-stained nuclei in blue) was observed. Scale bars: 100 μm. (B) 21 days after induction of EB-based differentiation of this cell line immunocytological analysis revealed the expression of markers representative of endoderm (α-fetoprotein [AFP] and SOX17), mesoderm (cardiac troponin T [cTnT]), and ectoderm (β3 tubulin). Scale bars: 100 μm. Color images available online at www.liebertpub.com/hgtb
FIG. 5.
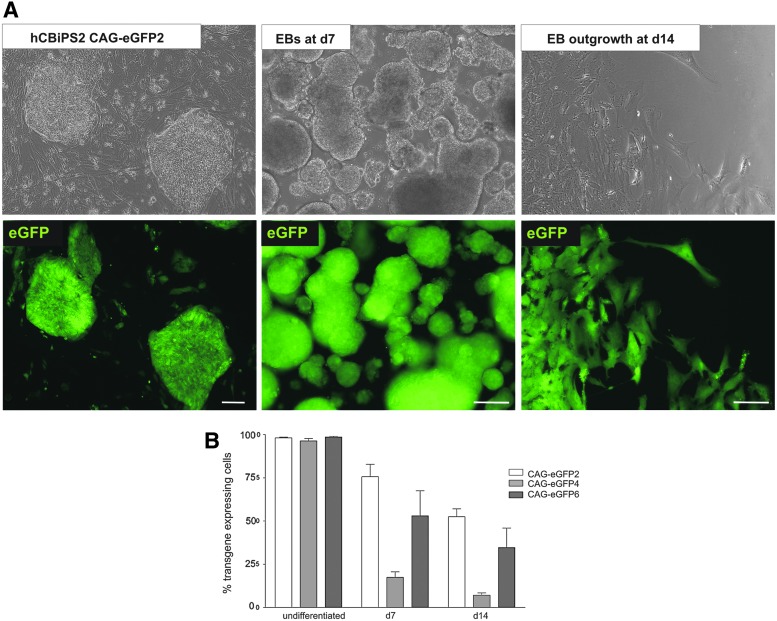
Maintenance of CAG promoter-controlled transgene expression on differentiation is transgene line dependent. (A) Phase-contrast and fluorescence microscopy images of eGFP expression in the transgenic cell line hCBiPS2 CAG-eGFP2 at the undifferentiated stage (left), after spontaneous differentiation induced by EB-formation on day 7 (middle), and in an EB outgrowth on day 14 (right). Scale bars: 100 μm. (B) Graph depicting the quantitative analysis of three independent cell lines at the same respective differentiation stages by flow cytometry, revealed a general but differential, cell line-dependent decrease in the proportion of eGFPpos cells (“silencing” of transgene expression). Color images available online at www.liebertpub.com/hgtb
Maintenance of transgene expression on differentiation
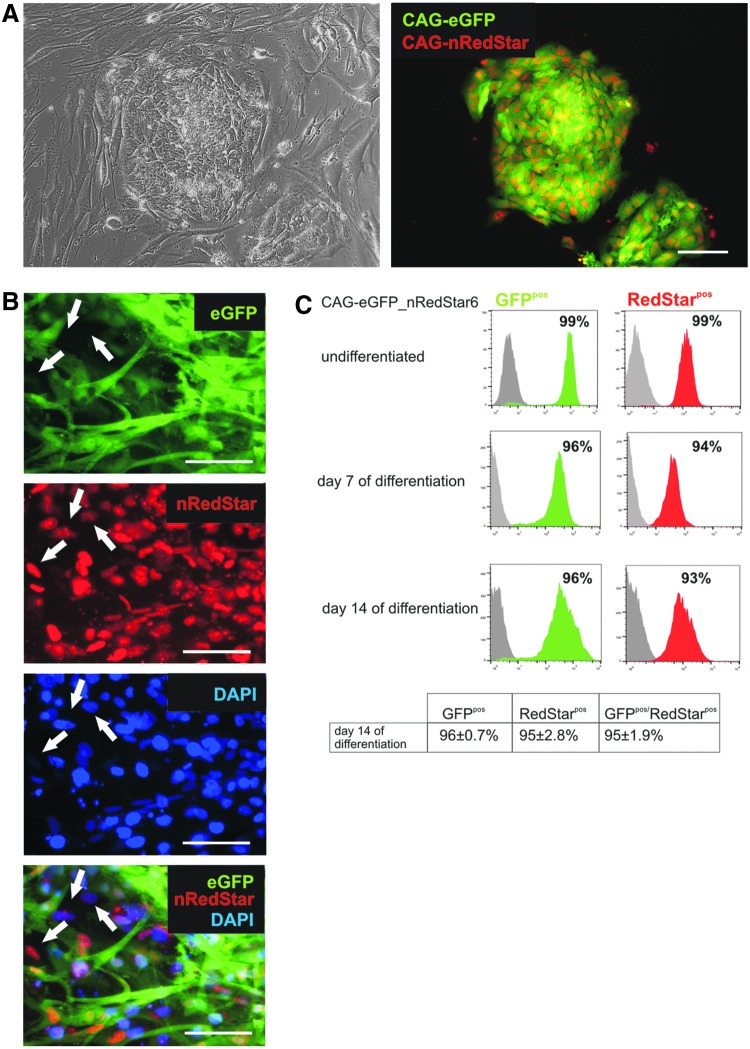
During EB-induced differentiation downregulation of CAG promoter-controlled fluorescence expression was observed to some extent. Whereas the amount of eGFPpos cells was in the same range in established lines in the pluripotent state (98±0.3%; Fig. 5), the proportion of transgene-expressing cells declined in a transgenic line-dependent manner, ranging from 53±4.5% (strongest preservation) to 7±1.3% (highest downregulation) on day 14 of differentiation (Fig. 5A and B). This phenomenon was generally observed for all fluorescence reporters tested. The comparative analyses of three double-transgenic lines, which all resulted from transfection with the same vector combination (CAG-eGFP plus αMHCneoPGKhygro), in Fig. 5B support the notion that the issue was transgenic line-related, likely reflecting the influence of different genomic integration sites and possibly varying copy numbers of the transgene(s) as well. Of note, differentiated derivatives of a triple-transgenic line (resulting from αMHCneoPGKhygro, CAG-eGFP, and CAG-nRedStar transfection; Fig. 6A) showed a highly stable nRedStar/eGFP expression level of 95±1.9% double-positive cells for at least 14 days of differentiation, with detection of some intermingled cells that were positive for either one or the other fluorescence marker only (Fig. 6B and C).
FIG. 6.
Fluorescence reporter expression is mainly preserved during differentiation in a triple-transgenic hiPSC line. (A) Stable expression of cytoplasmic eGFP and nuclear RedStar in undifferentiated cells of the triple-transgenic line hCBiPS2 αMHCneoPGKhygro_CAG-eGFP-nRedStar6 (CAG-eGFP-nRedStar6). Depicted is a phase-contrast (left) and fluorescence image (right) of a representative colony in passage 8 after clone picking. Scale bars: 100 μm. (B) Fluorescence images of differentiated cells from the same line on day 14 of differentiation. Although both fluorescence reporters were still coexpressed in the vast majority of the differentiated cells, a minority of cells lost either eGFP or nRedStar expression (white arrows). The graph in (C) depicts the flow cytometry-based quantification of single- and double-positive cells in the pluripotent state (top) and at different time points after induction of differentiation. Color images available online at www.liebertpub.com/hgtb
Efficient transgene-based cardiomyocyte enrichment
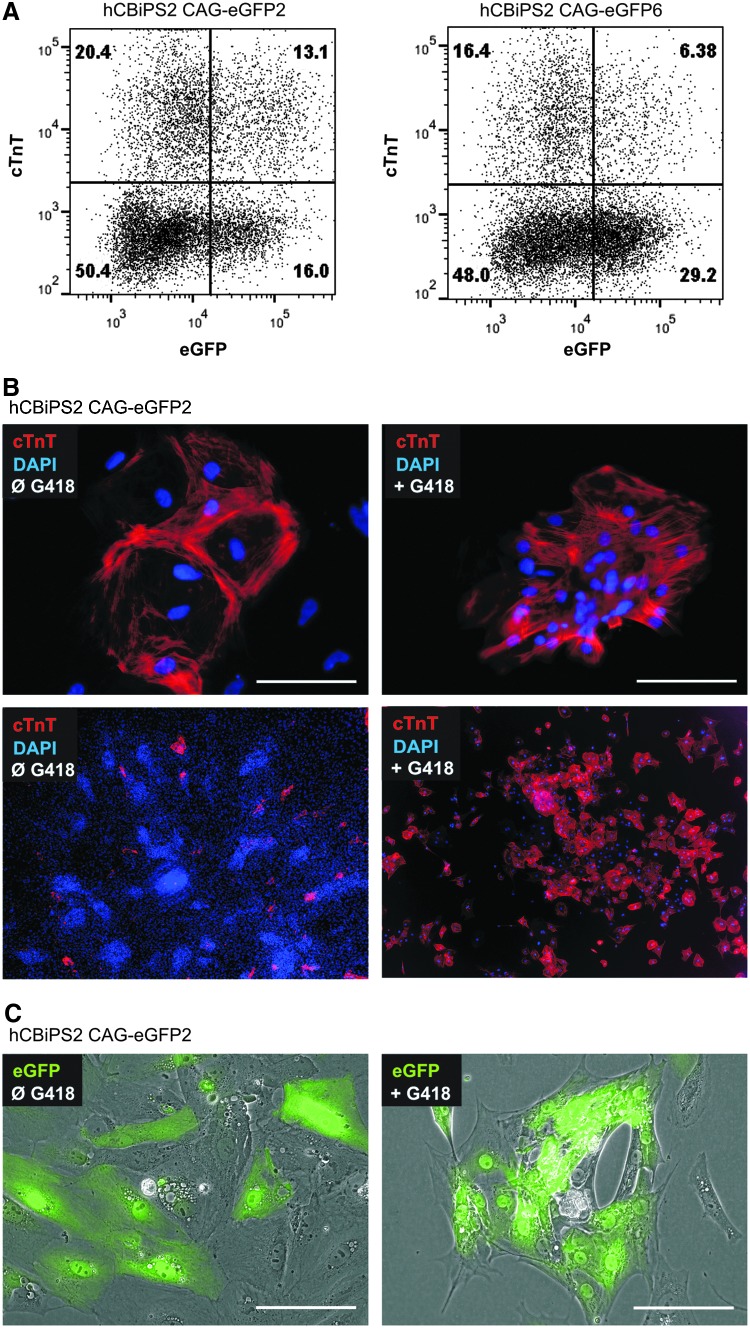
As an example of the practical application of the technique, three αMHCneo/eGFPpos double-transgenic lines were tested for the ability to enrich hiPSC-derived cardiomyocytes (CMs) based on the expression of αMHC promoter-dependent neomycin resistance. The cardiac differentiation efficiencies were equivalent between all transgenic lines tested (i.e., induction of ∼22–33% cardiac troponin T [cTnT]-positive CMs, shown in Fig. 7A) and to nontransgenic parental controls as well (data not shown). For CM enrichment addition of G418 was started after the appearance of beating foci (∼days 9–11 of differentiation). Quantification of enriched CM populations by immunocytological staining specific to cTnT (counterstained with DAPI to visualize cell nuclei; Fig. 7B) revealed >90% cTnTpos cells. Similar to nonselected, differentiated cells (∼6–13% of cTnTpos/eGFPpos CMs; Fig. 7A) a mixed pattern of eGFPhigh, eGFPlow, and GFPneg cells was observed in G418-selected CMs (Fig. 7C), suggesting that transgene silencing was apparently not cell type (i.e., cardiomyocyte) specific, but seems to occur somewhat stochastically in individual cells on differentiation, whereby the overall incidence of eGFP expression silencing apparently varies between individual transgenic cell lines, as outlined previously.
FIG. 7.
Genetic enrichment of hiPSC-derived cardiomyocytes (CMs) and maintenance of fluorescence reporter gene expression. (A) Dot plots depicting flow cytometric analysis of two differentiated but not CM-enriched cell lines (i.e., hCBiPS2 CAG-eGFP2 [left] and hCBiPS2 CAG-eGFP6 [right]) revealed the induction of ∼33.5% cardiac troponin T-positive (cTnTpos) CMs (top row in the left plot) whereby 29.1% of the cells expressed eGFP and 13.1% of the total cell population appeared to be double positive for cTnT and GFP, respectively. Slightly lower cardiomyogenic differentiation efficiency and thus a different proportion of cTnTpos versus cTnTneg cells, was observed for the line hCBiPS2 CAG-eGFP6 (right). (B) The line hCBiPS2 CAG-eGFP2 was differentiated in an EB-based approach in serum-free medium, leading to various cell lineages, among them beating CM clusters, as depicted by the cTnT (in red; DAPI stained nuclei, blue)-specific stain of fixed cells in the top and bottom left pictures. G418 (neomycin analog) selection resulted in a highly enriched cardiac cell population with almost all cells positively stained for cTnT (right). (C) Partial eGFP fluorescence reporter silencing was observed in unselected cells (left) and in G418-enriched CMs as well (right), resulting in mosaic-like expression patterns. Scale bars: 100 μm. Color images available online at www.liebertpub.com/hgtb
Noninvasive cell tracking
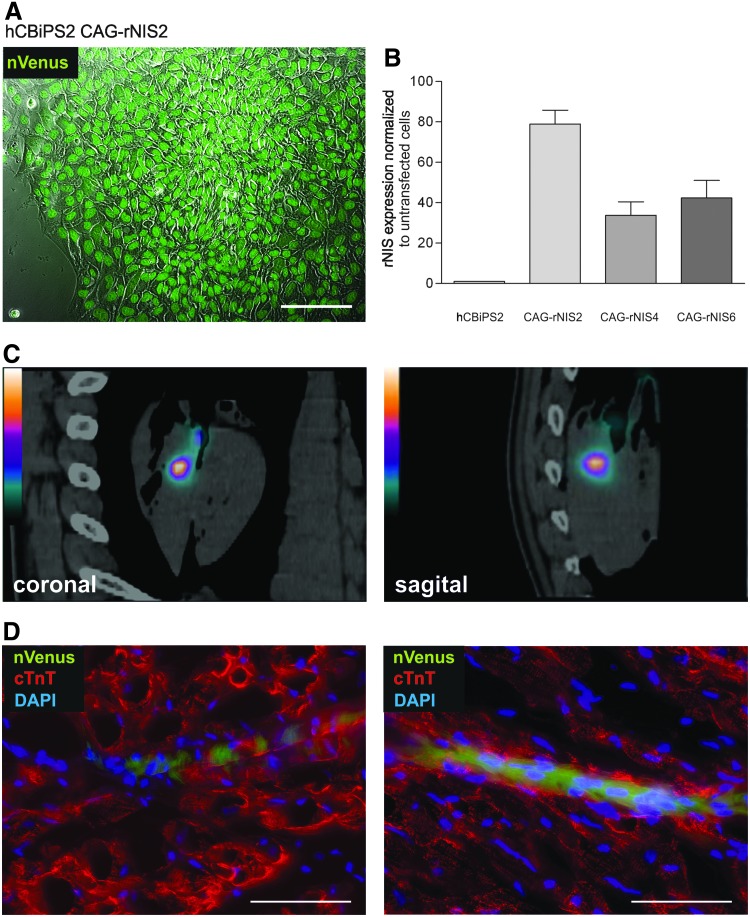
To provide a second practical approach for the application of our method, we assessed the expression of the rat sodium iodide symporter (rNIS) transgene as a tool for noninvasive cell tracking in large animal models in situ and potentially in vivo. Toward this end hCBiPS2 clones carrying two plasmids encoding four functional transgenes were generated. These included the plasmid αMHCneoPGKhygro enabling, first, clonal selection and, second, cardiomyogenic enrichment, as discussed previously (Fig. 7). The cointroduced bicistronic vector CAG-rNIS-nVenus encodes the transmembrane protein rNIS. Endogenous expression of the symporter is almost exclusively restricted to the thyroid gland; its expression results in the active uptake of iodide isotopes in transgenic cells, which can then be monitored via SPECT or positron emission tomography (PET) after transplantation (Templin et al., 2012). Finally, established cell lines expressed nVenus (Fig. 8A) as a cytological marker to facilitate the detection of injected donor cells in histological tissue sections after termination. Expression of rNIS was analyzed in three transgenic hCBiPS2 lines via qRT-PCR and the relative level of transgene expression was compared with a nontransfected hCBiPS2 control (Fig. 8B). For functional testing rNISpos-hCBiPS cells were labeled by incubation with the iodide isotope 123I followed by catheter-based intramyocardial cell injection into explanted pig hearts to simulate a clinically applicable route of cell administration. The cells were injected into the anterior wall of the left ventricle and monitored by SPECT-CT, allowing the detection of 123I-prelabeled cells at the site of transplantation (Fig. 8C). Notably, successful cell tracing was possible through a dissected pig chest that had been placed above the injected heart to mimic signal attenuation in vivo. For confirmation of the SPECT-CT imaging results tissue biopsies were collected at the sites of injection and analyzed by immunohistology. Indeed, nVenuspos hCBiPS cells were detected along the presumed injection canal within the myocardial tissue, which was demarcated by cTnT-specific staining (Fig. 8D).
FIG. 8.
Transgenic rNIS expression enables in situ imaging of multitransgenic hiPSCs in a pig heart. (A) Fluorescence microscopy of a colony of an established multitransgenic hCBiPS2 line (αMHCneoPGKhygro_CAG-rNIS-nVenus2; CAG-rNIS2) coexpressing the rat sodium iodide symporter (rNIS; see graph in B) and the nuclear yellow GFP variant Venus (nVenus, green). (B) Three independent transgenic hCBiPS2 CAG-rNIS lines were analyzed by qRT-PCR for rNIS transgene expression. rNIS expression was normalized to cellular β-Actin and respective expression levels are shown relative to non-transfected control cells (left column). (C) Iodine isotope 123I prelabeled hCBiPS2 CAG-rNIS6 cells were injected ex vivo into explanted pig hearts and placed under a dissected pig chest in order to simulate in vivo imaging after clinical cell transplantation. After the injection of 5×106 prelabeled cells, the heart was scanned with a Hybrid SPECT-CT. Depicted are SPECT-CT images in the coronal and sagital plane demonstrating tracer signal at the site of cell injection. (D) Detection of injected cells using an anti-GFP antibody (cross reacting with the yellow GFP variant Venus, green). On respective microtome sections the heart muscle tissue was visualized by anti-cTnT staining (in red) and cell nuclei were counter stained by DAPI (in blue). nVenuspos cells were detected along the presumed injection channel. Scale bars: 100 μm. Notably, fixation and immunhistological treatment of the tissue led to a more cytoplasmatic staining pattern of the otherwise nuclear Venus that is in contrast to the strictly nucleus-restricted pattern in viable cells as shown in A. Color images available online at www.liebertpub.com/hgtb
Discussion
The ability to genetically modify human PSCs is essential to fully exploit their potential for in vitro assay development, disease modeling, tissue engineering, and ultimately in regenerative medicine. For this reason substantial efforts have been invested to establish more efficient protocols for transgenic hPSC line generation (Costa et al., 2005, 2007; Braam et al., 2008a,b, 2010; Xu et al., 2008b). Development of improved electroporation devices has facilitated these efforts.
In this study, we have combined and optimized protocols for single-cell dissociation of hPSCs (Olmer et al., 2010; Zweigerdt et al., 2011) with prominent advantages provided by the Neon transfection system resulting in a fast and relative efficient protocol, which is directly applicable to conventional feeder-based cultures and does not require time-consuming preadaptation to feeder-free cultures. Another key element of progress in our work is the straightforward generation of double- and triple-transgenic clones, that is, stably expressing transgenes encoded on two or three independent plasmids, based on the antibiotic-based selection encoded on one of these vectors, only.
An efficient transfection protocol, which was successfully applicable to five independent hPSC lines including three hiPSC lines, all established from different somatic cell sources (see Materials and Methods for details), builds the basis of our approach. This is notable given the observed heterogeneity of hiPSC lines with respect to electroporation-based transfection (Cahan and Daley, 2013). Apart from the effects of vector structure (i.e., type and arrangement of promoter and reporter transgenes), a transient transfection efficiency of up to ∼45% for eGFP and ∼65% for nRedStar was observed for hiPSC lines whereas ∼40% eGFP positivity was observed for hES3. This is significantly higher compared with previous reports for electroporation (2±0.4%), nucleofection (16±3.6%), and lentiviral transduction (25±4.8%) (Cao et al., 2010) and equivalent to results observed for the Neon transfection system by others investigators (Moore et al., 2010).
Cell viability postelectroporation was also relatively high in our protocol and subsequently enabled the robust recovery of antibiotic-resistant colonies. For instance, an average number of ∼80 hygromycin-resistant colonies was observed after transfection of 1.5×106 hCBiPS2 cells, using the two-vector combination of CAG-nRedStar and αMHCneoPGKhygro, thus resulting in a total colony formation efficiency of ∼5.3×10−5 (Table 1). Approximately 25% of these colonies (∼19 on average; Table 1) also expressed nRedStar, resulting in an efficiency of 1.3×10−5 double-transgenic colonies; an equivalent value of 1.2×10−5 was also detected for eGFP-expressing colonies observed after cotransfection of CAG-eGFP and αMHCneoPGKhygro (Table 1).
Thus, our efficiency for the genomic integration and expression of two independent plasmids was equivalent or moderately lower compared with efficiencies achieved after single-vector transfection by Moore and colleagues (4.9×10−5; Moore et al., 2010) and Braam and colleagues (up to 1.9×10−4, also using the CAG promoter; Braam et al., 2008b). However, these published studies critically relied on the adaptation of hPSCs to feeder-free conditions before genetic manipulation, with the known disadvantages of increased time requirements and the enrichment of potentially abnormal cell clones that better tolerate these culture conditions. Our method is therefore also considerably faster and requires only ∼14 days until the appearance of multitransgenic colonies when starting with conventional feeder-based cultures. Another feature of our strategy is the fact that feeder cells were dispensable in the antibiotic selection phase. This makes the protocol adaptable to any type of antibiotic resistance transgene and independent of the availability of resistant feeder cells.
We noted, however, that the efficiency of fluorescence marker expression (encoded on separate vectors) of hygromycin-resistant colonies was ∼5- to 10-fold lower (range, 0.1–0.2×10−5) for the other cell lines tested (Table 1). These results correlate with the lower seeding efficiency and/or cell viability that was observed for these cell lines at 48 hr postelectroporation, thus resulting in lower cell numbers per well before the initiation of antibiotic-based colony selection (see Fig. 1D for hES3 cells). Following our strategy shown here, further cell line-specific adaptations of protocol parameters, that is, by modifying the dissociation conditions before electroporation, the DNA concentration, and the cell-seeding density after the treatment, should overcome this issue.
Using the hCBiPS2 line, we have further observed an efficiency of 0.4×10−5 for the formation of hygromycin-resistant colonies coexpressing eGFP and nRedStar, thus representing three-plasmid (triple)-transgenic cell lines (Table 1). To the best of our knowledge this is the first study demonstrating the possibility of achieving stable cointroduction of multiple plasmid vectors into hPSCs with only one of these plasmids carrying an antibiotic resistance marker. We acknowledge, however, that our coapplied vectors carried constitutively expressed fluorescence markers, which clearly facilitated the identification of (multi-) transgene-expressing colonies.
We have intentionally used common eukaryotic expression vectors without specific elements that might support their episomal replication and segregation such as scaffold/matrix attachment regions (S/MARS) (Argyros et al., 2011; Wade-Martins, 2011) or autonomous functional elements present on human artificial chromosomes (HACs; Mandegar et al., 2011). This fact makes the episomal persistence of plasmids in transgenic colonies and cell lines derived thereof extremely unlikely. In one study we have also applied PCR to monitor the persistence of plasmid vectors (which were of equivalent structure to vectors applied here) after transient hPSC transfection by electroporation (Hartung et al., 2013). We found that high plasmid levels persisted for ∼12 hr in hPSCs but that plasmids were almost undetectable 48 hr after cell transfection even by PCR, arguing against their prominent episomal replication, if any. Here, we have also performed Southern blot analysis of single or multitransgenic cell lines that were passaged up to 14 times after clone isolation (notably in the absence of any antibiotic-based selection pressure) before genomic DNA was isolated and analyzed. The respective hybridization patterns unequivocally confirmed the stable integration of the respective plasmid vectors in the genome.
Despite the discussed cell line-dependent variability of the method's efficiency, our approach generally worked with all five hPSC lines tested. However, by focusing on the stable transgene introduction into the parental cell line hCBiPS2, we have also observed a remarkable transgene dependency. Such disparities were apparently reporter gene dependent and, more specifically, seem to be related to the subcellular localization of respective fluorescence transgenes. Whereas 40% of cytoplasmic eGFP-expressing clones gave rise to stable fluorescent protein-expressing cell lines (i.e., 24 lines of 59 clones tested; Table 1), only 23% (3 of 13) of nVenus lines were generated and no stable nRedStar only-expressing lines could be established, despite serious attempts (0 of 33); notably 1 stable eGFP/nRedStar-coexpressing line (1 of 10) was successfully generated (Table 1 and Fig. 6). These results were unexpected, particularly in the light of the ∼2-fold higher transient expression levels found for nRedStar compared with eGFP as depicted for hCBiPS2 and hFF1-iPS4 in Fig. 1B. Whether the RedStar protein itself or its intended nuclear localization underlies these observations requires further side-by-side comparison of different reporters expressed in the same subcellular compartments.
Our results confirm previous findings (Liew et al., 2007) demonstrating that the CAG promoter-controlled reporter gene expression remained stable in the pluripotent state in established hiPSC lines during long-term cultivation, that is, for up to 23 passages, in contrast to retroviral and lentiviral vectors (Pannell and Ellis, 2001; Ellis and Yao, 2005). We further found that the expression of one or even two fluorescence reporters in transgenic lines did not interfere with their pluripotency and multilineage differentiation potential. This point is worth highlighting because culture adaptation, which is usually required during the generation of transgenic cell lines, has been reported to alter culture and differentiation characteristics (Enver et al., 2005; Catalina et al., 2008).
Besides stable transgene expression in undifferentiated hPSCs, maintenance of transgene expression in differentiated derivatives is of special interest for various applications. The undesired but frequently observed silencing of transgenes on differentiation is known to depend on the vector design and especially the promoter used.
Using the CAG promoter, we have analyzed several transgenic cell lines regarding the expression of eGFP or coexpression of eGFP/nRedStar after induction of differentiation. As previously reported for other plasmids or lentiviral vectors using this promoter (Hong et al., 2007; Xia et al., 2007; Chen et al., 2011), we found for three transgenic lines tested some level of silencing and subsequently a mosaic-like expression of eGFP in differentiated progenies (Figs. 5 and 7A). This was also true in αMHC promoter-neoR-enriched cardiomyocytes (Fig. 7C). Whether the presence of different cardiomyocyte subtypes (i.e., pacemaker-, atrial-, or ventricular-like cells), the heterogeneous myocyte maturation grade, or somewhat stochastic silencing (i.e., in some daughter cells but not in others) accounts for these results rather than cell type (i.e., cardiomyocyte)-specific events that occur during differentiation requires further investigation.
Although the CAG promoter offers a valuable tool for transgene expression in hPSCs in the pluripotent state and on differentiation, it is well established that promoter methylation, which might be integration site specific, is a main cause of transgene silencing (Alexopoulou et al., 2008). Interestingly, studies have shown that ubiquitous chromatin-opening elements (UCOEs) can strongly reduce promoter DNA methylation and thereby prevent epigenetic transgene silencing in mouse and human PSCs and their progenies (Pfaff et al., 2013; Ackermann et al., 2014). Thus, the strategy might support stable transgene expression controlled by ubiquitous and lineage-specific promoters independent of the (random) genomic integration site.
Finally, we have tested the overexpression of an rNIS symporter as a proof-of-concept for cell tracking in a clinically relevant animal model of heart damage, namely the pig. Constitutive nVenus/rNIS-expressing undifferentiated hiPS cells were injected intramyocardially into an isolated pig heart ex vivo. We showed that our transgenic cells can be detected by SPECT-CT imaging in situ, and subsequently the presence of injected cells was confirmed by means of a second transgene, that is, nVenus, on histological tissue sections. These are important steps for the efficient, unequivocal monitoring of donor cell survival and distribution in large animal models, which is a key issue in the field (Chang et al., 2006; Yang, 2012). First experiments applying respective cell lines generated by our method for longitudinal monitoring of hPSC engraftment in a pig myocardial infarct model in vivo have already been performed (Templin et al., 2012).
In summary, we provide a novel method enabling the fast and relative efficient generation of multitransgenic hPSC lines expressing transgenes located on independent plasmids with only one antibiotic resistance marker. Our improved technology also has potential to support the targeting of transgenes into specific loci via homologous recombination in future experiments.
Supplementary Material
Acknowledgments
This work was funded by: Cluster of Excellence REBIRTH (DFG EXC62/1), German Ministry for Education and Science (BMBF, 01GN0958, 315493 and 13N12606), BIOSCENT (FP7/2007-2013, grant 214539), StemBANCC (Support from the Innovative Medicines Initiative joint undertaking under grant agreement 115439-2, resources of which are composed of financial contribution from the European Union [FP7/2007-2013] and EFPIA companies' in-kind contribution), Swiss National Research Foundation Sonderprogramm Universitäre Medizin (33CM30-124112/1), Swiss Life Foundation and the Gottfried and Julia Bangerter-Rhvner-Foundation, and the CORTISS foundation (Hannover, Germany) providing a research grant to K. Schwanke. The authors thank T. Scheper for providing bFGF and A. Kirschning/G. Dräger for providing Y-27632. The vectors αMHCneoPGKhygro and CAG-eGFP (pCAGGS2) were kindly provided by Loren Field (Indianapolis, IN). The NIS transgene was kindly provided by Simon P. Hoerstrup (University Hospital Zurich, Zurich, Switzerland). The authors are grateful to Ennio Müller for technical assistance and to the Department of Nuclear Medicine at the University Hospital Zurich for providing the infrastructure for SPECT CT imaging.
Author Disclosure Statement
No competing financial interests exist.
References
- Ackermann M., Lachmann N., Hartung S., et al. (2014). Promoter and lineage independent anti-silencing activity of the A2 ubiquitous chromatin opening element for optimized human pluripotent stem cell-based gene therapy. Biomaterials 35, 1531–1542 [DOI] [PubMed] [Google Scholar]
- Acton P.D., and Kung H.F. (2003). Small animal imaging with high resolution single photon emission tomography. Nucl. Med. Biol. 30, 889–895 [DOI] [PubMed] [Google Scholar]
- Alexopoulou A.N., Couchman J.R., and Whiteford J.R. (2008). The CMV early enhancer/chicken β actin (CAG) promoter can be used to drive transgene expression during the differentiation of murine embryonic stem cells into vascular progenitors. BMC Cell Biol. 9, 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amit M., and Itskovitz-Eldor J. (2002). Derivation and spontaneous differentiation of human embryonic stem cells. J. Anat. 200, 252–232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Argyros O., Wong S.P., and Harbottle R.P. (2011). Non-viral episomal modification of cells using S/MAR elements. Expert Opin. Biol. Ther. 11, 1177–1191 [DOI] [PubMed] [Google Scholar]
- Boecker W., Bernecker O.Y., Wu J.C., et al. (2004). Cardiac-specific gene expression facilitated by an enhanced myosin light chain promoter. Mol. Imaging 3, 69–75 [DOI] [PubMed] [Google Scholar]
- Braam S.R., Denning C., Matsa E., et al. (2008a). Feeder-free culture of human embryonic stem cells in conditioned medium for efficient genetic modification. Nat. Protoc. 3, 1435–1443 [DOI] [PubMed] [Google Scholar]
- Braam S.R., Denning C., Van Den Brink S., et al. (2008b). Improved genetic manipulation of human embryonic stem cells. Nat. Methods 5, 389–392 [DOI] [PubMed] [Google Scholar]
- Braam S.R., Denning C., and Mummery C.L. (2010). Genetic manipulation of human embryonic stem cells in serum and feeder-free media. Methods Mol. Biol. 584, 413–423 [DOI] [PubMed] [Google Scholar]
- Burridge P.W., Thompson S., Millrod M.A., et al. (2011). A universal system for highly efficient cardiac differentiation of human induced pluripotent stem cells that eliminates interline variability. PLoS One 6, e18293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cahan P., and Daley G.Q. (2013). Origins and implications of pluripotent stem cell variability and heterogeneity. Nat. Rev. Mol. Cell. Biol. 14, 357–368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao F., Xie X., Gollan T., et al. (2010). Comparison of gene-transfer efficiency in human embryonic stem cells. Mol. Imaging Biol. 12, 15–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catalina P., Montes R., Ligero G., et al. (2008). Human ESCs predisposition to karyotypic instability: Is a matter of culture adaptation or differential vulnerability among hESC lines due to inherent properties? Mol. Cancer 7, 76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang G.Y., Xie X., and Wu J.C. (2006). Overview of stem cells and imaging modalities for cardiovascular diseases. J. Nucl. Cardiol. 13, 554–569 [DOI] [PubMed] [Google Scholar]
- Chen C.M., Krohn J., Bhattacharya S., and Davies B. (2011). A comparison of exogenous promoter activity at the ROSA26 locus using a PhiC31 integrase mediated cassette exchange approach in mouse ES cells. PLoS One 6, e23376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen R., John J., Lavrentieva A., et al. (2012). Cytokine production using membrane adsorbers: Human basic fibroblast growth factor produced by Escherichia coli. Eng. Life Sci. 12, 29–38 [Google Scholar]
- Cherry S.R., Biniszkiewicz D., Van Parijs L., et al. (2000). Retroviral expression in embryonic stem cells and hematopoietic stem cells. Mol. Cell. Biol. 20, 7419–7426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa M., Dottori M., Ng E., et al. (2005). The hESC line Envy expresses high levels of GFP in all differentiated progeny. Nat. Methods 2, 259–260 [DOI] [PubMed] [Google Scholar]
- Costa M., Dottori M., Sourris K., et al. (2007). A method for genetic modification of human embryonic stem cells using electroporation. Nat. Protoc. 2, 792–796 [DOI] [PubMed] [Google Scholar]
- Dai W., Field L.J., Rubart M., et al. (2007). Survival and maturation of human embryonic stem cell-derived cardiomyocytes in rat hearts. J. Mol. Cell. Cardiol. 43, 504–516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- David R., Stieber J., Fischer E., et al. (2009). Forward programming of pluripotent stem cells towards distinct cardiovascular cell types. Cardiovasc. Res. 84, 263–272 [DOI] [PubMed] [Google Scholar]
- Ellis J., and Yao S. (2005). Retrovirus silencing and vector design: Relevance to normal and cancer stem cells? Curr. Gene Ther. 5, 367–373 [DOI] [PubMed] [Google Scholar]
- Enver T., Soneji S., Joshi C., et al. (2005). Cellular differentiation hierarchies in normal and culture-adapted human embryonic stem cells. Hum. Mol. Genet. 14, 3129–3140 [DOI] [PubMed] [Google Scholar]
- Graichen R., Xu X., Braam S.R., et al. (2008). Enhanced cardiomyogenesis of human embryonic stem cells by a small molecular inhibitor of p38 MAPK. Differentiation 76, 357–370 [DOI] [PubMed] [Google Scholar]
- Haas D.L., Case S.S., Crooks G.M., and Kohn D.B. (2000). Critical factors influencing stable transduction of human CD34+ cells with HIV-1-derived lentiviral vectors. Mol. Ther. 2, 71–80 [DOI] [PubMed] [Google Scholar]
- Haase A., Olmer R., Schwanke K., et al. (2009). Generation of induced pluripotent stem cells from human cord blood. Cell Stem Cell 5, 434–441 [DOI] [PubMed] [Google Scholar]
- Hartung S., Schwanke K., Haase A., et al. (2013). Directing cardiomyogenic differentiation of human pluripotent stem cells by plasmid-based transient overexpression of cardiac transcription factors. Stem Cells Dev. 22, 1112–1125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong S., Hwang D.Y., Yoon S., et al. (2007). Functional analysis of various promoters in lentiviral vectors at different stages of in vitro differentiation of mouse embryonic stem cells. Mol. Ther. 15, 1630–1639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huber I., Itzhaki I., Caspi O., et al. (2007). Identification and selection of cardiomyocytes during human embryonic stem cell differentiation. FASEB J. 21, 2551–2563 [DOI] [PubMed] [Google Scholar]
- Jang J.E., Shaw K., Yu X.J., et al. (2006). Specific and stable gene transfer to human embryonic stem cells using pseudotyped lentiviral vectors. Stem Cells Dev. 15, 109–117 [DOI] [PubMed] [Google Scholar]
- Kita-Matsuo H., Barcova M., Prigozhina N., et al. (2009). Lentiviral vectors and protocols for creation of stable hESC lines for fluorescent tracking and drug resistance selection of cardiomyocytes. PLoS One 4, e5046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klug M.G., Soonpaa M.H., Koh G.Y., and Field L.J. (1996). Genetically selected cardiomyocytes from differentiating embryonic stem cells form stable intracardiac grafts. J. Clin. Invest. 98, 216–224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knop M., Barr F., Riedel C.G., et al. (2002). Improved version of the red fluorescent protein (drFP583/DsRed/RFP). Biotechniques 33, 592, 594, 596–598passim. [DOI] [PubMed] [Google Scholar]
- Kustikova O., Fehse B., Modlich U., et al. (2005). Clonal dominance of hematopoietic stem cells triggered by retroviral gene marking. Science 308, 1171–1174 [DOI] [PubMed] [Google Scholar]
- Liew C.G., Draper J.S., Walsh J., et al. (2007). Transient and stable transgene expression in human embryonic stem cells. Stem Cells 25, 1521–1528 [DOI] [PubMed] [Google Scholar]
- Liu J., Jones K.L., Sumer H., and Verma P.J. (2009). Stable transgene expression in human embryonic stem cells after simple chemical transfection. Mol. Reprod. Dev. 76, 580–586 [DOI] [PubMed] [Google Scholar]
- Livak K.J., and Schmittgen T.D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCt method. Methods 25, 402–408 [DOI] [PubMed] [Google Scholar]
- Ma Y., Ramezani A., Lewis R., et al. (2003). High-level sustained transgene expression in human embryonic stem cells using lentiviral vectors. Stem Cells 21, 111–117 [DOI] [PubMed] [Google Scholar]
- Mandegar M.A., Moralli D., Khoja S., et al. (2011). Functional human artificial chromosomes are generated and stably maintained in human embryonic stem cells. Hum. Mol. Genet. 20, 2905–2913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mauritz C., Martens A., Rojas S.V., et al. (2011). Induced pluripotent stem cell (iPSC)-derived Flk-1 progenitor cells engraft, differentiate, and improve heart function in a mouse model of acute myocardial infarction. Eur. Heart J. 32, 2634–2641 [DOI] [PubMed] [Google Scholar]
- Moore J.C., Van Laake L.W., Braam S.R., et al. (2005). Human embryonic stem cells: Genetic manipulation on the way to cardiac cell therapies. Reprod. Toxicol. 20, 377–391 [DOI] [PubMed] [Google Scholar]
- Moore J.C., Atze K., Yeung P.L., et al. (2010). Efficient, high-throughput transfection of human embryonic stem cells. Stem Cell Res. Ther. 1, 23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norrman K., Fischer Y., Bonnamy B., et al. (2010). Quantitative comparison of constitutive promoters in human ES cells. PLoS One 5, e12413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okita C., Sato M., and Schroeder T. (2004). Generation of optimized yellow and red fluorescent proteins with distinct subcellular localization. Biotechniques 36, 418–22, 424. [DOI] [PubMed] [Google Scholar]
- Olmer R., Haase A., Merkert S., et al. (2010). Long term expansion of undifferentiated human iPS and ES cells in suspension culture using a defined medium. Stem Cell Res. 5, 51–64 [DOI] [PubMed] [Google Scholar]
- Palecek J., Zweigerdt R., Olmer R., et al. (2011). A practical synthesis of Rho-Kinase inhibitor Y-27632 and fluoro derivatives and their evaluation in human pluripotent stem cells. Org. Biomol. Chem. 9, 5503–5510 [DOI] [PubMed] [Google Scholar]
- Pannell D., and Ellis J. (2001). Silencing of gene expression: Implications for design of retrovirus vectors. Rev. Med. Virol. 11, 205–217 [DOI] [PubMed] [Google Scholar]
- Pfaff N., Lachmann N., Ackermann M., et al. (2013). A ubiquitous chromatin opening element prevents transgene silencing in pluripotent stem cells and their differentiated progeny. Stem Cells 31, 488–499 [DOI] [PubMed] [Google Scholar]
- Philippe S., Sarkis C., Barkats M., et al. (2006). Lentiviral vectors with a defective integrase allow efficient and sustained transgene expression in vitro and in vivo. Proc. Natl. Acad. Sci. U.S.A. 103, 17684–17689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pichon C., Billiet L., and Midoux P. (2010). Chemical vectors for gene delivery: Uptake and intracellular trafficking. Curr. Opin. Biotechnol. 21, 640–645 [DOI] [PubMed] [Google Scholar]
- Reubinoff B.E., Pera M.F., Fong C.Y., et al. (2000). Embryonic stem cell lines from human blastocysts: Somatic differentiation in vitro. Nat. Biotechnol. 18, 399–404 [DOI] [PubMed] [Google Scholar]
- Ritner C., Wong S.S., King F.W., et al. (2011). An engineered cardiac reporter cell line identifies human embryonic stem cell-derived myocardial precursors. PLoS One 6, e16004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schinzel R.T., Ahfeldt T., Lau F.H., et al. (2011). Efficient culturing and genetic manipulation of human pluripotent stem cells. PLoS One 6, e27495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlegelberger B., Metzke S., Harder S., et al. (1999). Classical and molecular cytogenetics of tumor cells. In Diagnostic Cytogenetics. Wegner R.-D., ed. (Springer Verlag, New York: ), pp. 151–185 [Google Scholar]
- Sun N., Lee A., and Wu J.C. (2009). Long term non-invasive imaging of embryonic stem cells using reporter genes. Nat. Protoc. 4, 1192–1201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Templin C., Zweigerdt R., Schwanke K., et al. (2012). Transplantation and tracking of human-induced pluripotent stem cells in a pig model of myocardial infarction: Assessment of cell survival, engraftment, and distribution by hybrid single photon emission computed tomography/computed tomography of sodium iodide symporter transgene expression. Circulation 126, 430–439 [DOI] [PubMed] [Google Scholar]
- Wade-Martins R. (2011). Developing extrachromosomal gene expression vector technologies: An overview. Methods Mol. Biol. 738, 1–17 [DOI] [PubMed] [Google Scholar]
- Wurm M., Gross B., Sgodda M., et al. (2011). Improved lentiviral gene transfer into human embryonic stem cells grown in co-culture with murine feeder and stroma cells. Biol. Chem. 392, 887–895 [DOI] [PubMed] [Google Scholar]
- Xia X., Zhang Y., Zieth C.R., and Zhang S.C. (2007). Transgenes delivered by lentiviral vector are suppressed in human embryonic stem cells in a promoter-dependent manner. Stem Cells Dev. 16, 167–176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X.Q., Graichen R., Soo S.Y., et al. (2008a). Chemically defined medium supporting cardiomyocyte differentiation of human embryonic stem cells. Differentiation 76, 958–970 [DOI] [PubMed] [Google Scholar]
- Xu X.Q., Zweigerdt R., Soo S.Y., et al. (2008b). Highly enriched cardiomyocytes from human embryonic stem cells. Cytotherapy 10, 376–389 [DOI] [PubMed] [Google Scholar]
- Yang P.C. (2012). Is reliable in vivo detection of stem cell viability possible in a large animal model of myocardial injury? Circulation 126, 388–390 [DOI] [PubMed] [Google Scholar]
- Yao S., Sukonnik T., Kean T., et al. (2004). Retrovirus silencing, variegation, extinction, and memory are controlled by a dynamic interplay of multiple epigenetic modifications. Mol. Ther. 10, 27–36 [DOI] [PubMed] [Google Scholar]
- Ziello J.E., Huang Y., and Jovin I.S. (2010). Cellular endocytosis and gene delivery. Mol. Med. 16, 222–229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zwaka T.P., and Thomson J.A. (2003). Homologous recombination in human embryonic stem cells. Nat. Biotechnol. 21, 319–321 [DOI] [PubMed] [Google Scholar]
- Zweigerdt R., Olmer R., Singh H., et al. (2011). Scalable expansion of human pluripotent stem cells in suspension culture. Nat. Protoc. 6, 689–700 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.