Abstract
Streptomyces clavuligerus ATCC 27064 and S. clavuligerus ΔccaR::tsr cultures were grown in asparagine-starch medium, and samples were taken in the exponential and stationary growth phases. Transcriptomic analysis showed that the expression of 186 genes was altered in the ccaR-deleted mutant. These genes belong to the cephamycin C gene cluster, clavulanic acid gene cluster, clavams, holomycin, differentiation, carbon, nitrogen, amino acids or phosphate metabolism and energy production. All the clavulanic acid biosynthesis genes showed Mc values in the order of −4.23. The blip gene-encoding a β-lactamase inhibitory protein was also controlled by the cephamycin C-clavulanic acid cluster regulator (Mc −2.54). The expression of the cephamycin C biosynthesis genes was greatly reduced in the mutant (Mc values up to −7.1), while the genes involved in putative β-lactam resistance were less affected (Mc average −0.88). Genes for holomycin biosynthesis were upregulated. In addition, the lack of clavulanic acid and cephamycin production negatively affected the expression of genes for the clavulanic acid precursor arginine and of miscellaneous genes involved in nitrogen metabolism (amtB, glnB, glnA3, glnA2, glnA1). The transcriptomic results were validated by quantative reverse transcription polymerase chain reaction and luciferase assay of luxAB-coupled promoters. Transcriptomic analysis of the homologous genes of S. coelicolor validated the results obtained for S. clavuligerus primary metabolism genes.
Introduction
Streptomyces species are the largest group of antibiotic-producing micro-organisms. Antibiotic biosynthesis genes are usually clustered and their expression is regulated in response to nutritional environment, culture time and cell density. Most of the gene clusters for antibiotic biosynthesis contain regulators responding to these parameters (Martín and Liras, 2010). Regulators of LysR-(Pérez-Redondo et al., 1998) or large ATP-binding regulators of the LuxR family (LAL)-type (Wilson et al., 2001; Antón et al., 2004; Tahlan et al., 2007) are present in several clusters, but the most common ones are the activator proteins belonging to the SARP (Streptomyces antibiotic regulatory proteins) family. They are present in many antibiotic clusters (Bate et al., 2002; Garg and Parry, 2010; He et al., 2010) and bind specific deoxyribonucleic acid sequences activating expression of antibiotic biosynthesis genes.
Cephamycin C biosynthesis in Streptomyces cattleya is controlled by a SARP-type protein encoded by thnU, a gene that is not located in the cephamycin C cluster (Rodríguez et al., 2008). In Streptomyces clavuligerus, a similar protein, cephamycin C-clavulanic acid cluster regulator (CcaR), encoded by a gene located in the cephamycin C cluster, controls both cephamycin and clavulanic acid (CA) biosynthesis (Pérez-Llarena et al., 1997; Santamarta et al., 2011). The two clusters are located side by side in the chromosome.
Specific sequences for the binding of SARP proteins have been described in antibiotic biosynthesis genes (Wietzorrek and Bibb, 1997). In S. clavuligerus, triple heptameric conserved sequences are responsible for CcaR binding and controls the expression of the lat, cefF, cefD and ccaR genes in the cephamycin C cluster. In the CA cluster, CcaR binds sequences upstream of ceaS2, which encodes the first enzyme of the CA pathway, and of claR, for the LysR-type regulator controlling late steps in CA biosynthesis (Santamarta et al., 2011).
At the beginning of this study, the S. clavuligerus genome was not published, but many S. clavuligerus genes had been deposited in the database and additional genes were available through the DSM (Delft, The Netherlands) S. clavuligerus sequencing project. Therefore, we constructed microarrays containing probes for 800 genes of the S. clavuligerus genome as well as for 7728 genes of the Streptomyces coelicolor genome in order to compare the housekeeping genes of both strains. This microarray has been used to detect gene expression in different S. clavuligerus mutants. In this article, we report the results observed for S. clavuligerus ΔccaR::tsr, a strain lacking the SARP regulator that controls cephamycin C and CA biosynthesis.
Results
Transcriptomic analysis of antibiotic biosynthesis genes in S. clavuligerus ATCC 27064 and S. clavuligerus ΔccaR::tsr
Streptomyces clavuligerus ATCC 27064 and S. clavuligerus ΔccaR::tsr cultures were grown in asparagine-starch (SA) medium, and samples were taken in the exponential and stationary growth phases (Fig. 1) as indicated in Experimental Procedures. The microarrays analysis showed differences in expression in 186 genes of the ccaR-negative mutant at one specific sampling time at least (Table 1).
Figure 1.
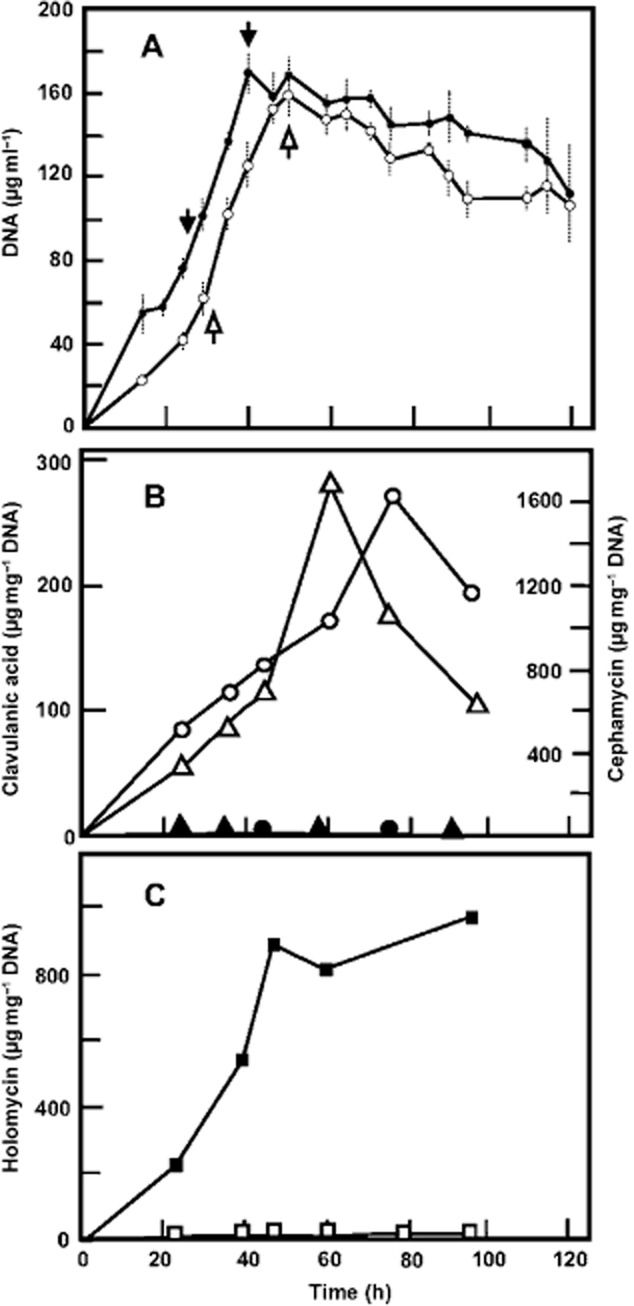
Growth and antibiotic production in Asparagine-starch medium. Strains used: S. clavuligerus ATCC 27064 (open symbols) and S. clavuligerus ΔccaR::tsr (closed symbols). A. Growth (circles) as measured by the DNA content. B. Cephamycin C (circles) or clavulanic acid (triangles) production. C. Holomycin production (squares).
Table 1.
Streptomyces clavuligerus ΔccaR::tsr gene expression as compared with S. clavuligerus ATCC 27064
| Exponential phase |
Stationary phase |
|||||
|---|---|---|---|---|---|---|
| Code | Gene | Product | Mc | FDR | Mc | FDR |
| Clavulanic acid biosynthesis | ||||||
| SCLAV_4189 | cyp | Cytochrome P450 | −5.34 | < 1E−06 | −5.19 | < 1E−06 |
| SCLAV_4191 | claR | Transcriptional regulator | −4.61 | < 1E−06 | −3.99 | < 1E−06 |
| SCLAV_4187 | orf12 | Beta-lactamase protein-like | −5.09 | < 1E−06 | −4.51 | < 1E−06 |
| SCLAV_4181 | gcas | Carboxylase | −5.44 | < 1E−06 | −4.98 | < 1E−06 |
| SCLAV_4186 | orf13 | Export pump | −3.73 | < 1E−06 | −3.25 | < 1E−06 |
| SCLAV_4185 | orf14 | Acetyl transferase | −4.77 | < 1E−06 | −3.84 | < 1E−06 |
| SCLAV_4183 | oppA2 | Oligopeptide-binding protein | −5.28 | < 1E−06 | −4.72 | < 1E−06 |
| SCLAV_4182 | orf16 | DUF482 domain-containing protein | −5.03 | < 1E−06 | −4.40 | < 1E−06 |
| SCLAV_4180 | orf18 | Penicillin-binding protein | −0.49 | 7.48E−04 | −0.58 | 2.02E−05 |
| SCLAV_4179 | orf19 | Penicillin-binding protein | −0.61 | 9.36E−03 | −0.65 | 1.99E−03 |
| SCLAV_4178 | orf20 | Cytochrome P450 | −0.95 | 3.93E−05 | −1.68 | < 1E−06 |
| SCLAV_4196 | bls2 | Beta-lactam synthetase 2 | −6.38 | < 1E−06 | −5.36 | < 1E−06 |
| SCLAV_4190 | car | Clavaldehyde dehydrogenase | −3.94 | < 1E−06 | −3.03 | < 1E−06 |
| SCLAV_4194 | cas2 | Clavaminato sintase 2 | −6.55 | < 1E−06 | −6.00 | < 1E−06 |
| SCLAV_4197 | ceaS2 | Carboxyethyl arginine synthetase 1 | −5.78 | < 1E−06 | −5.33 | < 1E−06 |
| SCLAV_4193 | oat2 | Similar to ornithine acetyltransferase | −2.67 | < 1E−06 | −2.81 | < 1E−06 |
| SCLAV_4192 | oppA1 | Oligopeptide-binding protein | −4.97 | < 1E−06 | −4.44 | < 1E−06 |
| SCLAV_4195 | pah2 | Proclavaminate amidinohydrolase 2 | −6.66 | < 1E−06 | −6.01 | < 1E−06 |
| Cephamycin C biosynthesis | ||||||
| SCLAV_4198 | pcbR | Beta-lactam antibiotics resistance | −1.02 | 4.88E−05 | −0.92 | 9.76E−05 |
| SCLAV_4199 | pcbC | Isopenicillin N synthetase | −3.25 | < 1E−06 | −2.82 | < 1E−06 |
| SCLAV_4200 | pcbAB | ACV Synthetase | −5.21 | < 1E−06 | −3.85 | < 1E−06 |
| SCLAV_4201 | lat | L-lysine epsilon amino transferase | −7.19 | < 1E−06 | −5.87 | < 1E−06 |
| SCLAV_4202 | blp | Similar to beta-lactamase inhibitory protein | −6.23 | < 1E−06 | −4.84 | < 1E−06 |
| SCLAV_4203 | orf10 | Secreted protein | −3.40 | < 1E−06 | −3.20 | < 1E−06 |
| SCLAV_4204 | ccaR | Transcriptional regulator | −7.12 | < 1E−06 | −7.48 | < 1E−06 |
| SCLAV_4205 | cmcH | Cabamoyl transferase | −5.96 | < 1E−06 | −4.16 | < 1E−06 |
| SCLAV_4206 | cefF | Deacetyl cephalosporin C synthetase | −6.83 | < 1E−06 | −5.48 | < 1E−06 |
| SCLAV_4207 | cmcJ | Methyl transferase | −7.23 | < 1E−06 | −5.32 | < 1E−06 |
| SCLAV_4208 | cmcI | Cepahalosporin hydroxylase | −7.48 | < 1E−06 | −6.25 | < 1E−06 |
| SCLAV_4210 | cefD | Isopenicillin N epimerase | −6.15 | < 1E−06 | −5.18 | < 1E−06 |
| SCLAV_4211 | cefE | Deacetoxycephalosporin C synthetase (DAOCS) | −5.12 | < 1E−06 | −4.45 | < 1E−06 |
| SCLAV_4212 | pcd | Piperideine carboxylate dehydrogenase | −3.40 | < 1E−06 | −2.08 | < 1E−06 |
| SCLAV_4213 | cmcT | Efflux protein | −3.74 | < 1E−06 | −2.29 | < 1E−06 |
| SCLAV_4214 | pbp | Penicillin-binding protein | −0.51 | 2.25E−03 | −0.63 | 4.27E−05 |
| Proteins blip | ||||||
| SCLAV_4456 | atpA | Similar to ABC transporter ATP-binding domain | −1.08 | 3.95E−02 | −1.62 | 2.88E−04 |
| SCLAV_4457 | atpA2 | Similar to ABC transporter ATP-binding domain | −1.07 | < 1E−06 | −1.65 | < 1E−06 |
| SCLAV_4455 | blip | Beta-lactamase inhibitory protein | −2.54 | < 1E−06 | −2.19 | < 1E−06 |
| SCLAV_4452 | Putative regulatory protein | −0.71 | 2.32E−01 | −0.99 | 2.71E−02 | |
| SCLAV_4453 | Hypothetical protein | −0.76 | 1.10E−03 | −0.98 | < 1E−06 | |
| Clavams biosynthesis | ||||||
| SCLAV_p1072 | orf8 | Serine hydroxymethyltransferase | −2.59 | < 1E−06 | −3.18 | < 1E−06 |
| SCLAV_p1076 | pah1 | Proclavaminate amidinohydrolase 1 | −0.98 | 3.99E−02 | −3.23 | < 1E−06 |
| SCLAV_p1077 | oat1 | Ornithine acetyltransferase isoenzyme | −1.58 | 6.48E−04 | −2.60 | < 1E−06 |
| SCLAV_p1078 | cvm6P | Pyridoxal phosphate dependent aminotransferase | −1.04 | 9.64E−03 | −2.28 | < 1E−06 |
| SCLAV_p1079 | cvm7P | Transcriptional regulator | −0.90 | 1.23E−03 | −1.97 | < 1E−06 |
| SCLAV_2922 | cvm6 | Pyridoxal phosphate dependent aminotransferase | −0.61 | 1.61E−02 | −1.98 | < 1E−06 |
| SCLAV_2923 | cvm5 | Flavin-dependent oxidoreductase | −1.51 | 2.27E−05 | −2.88 | < 1E−06 |
| SCLAV_2924 | cvm4 | Deacetylcephalosporin C acetyltransferase | −1.69 | 7.88E−03 | −2.56 | < 1E−06 |
| SCLAV_2925 | cas1 | Clavaminate synthase 1 | −3.32 | 2.79E−02 | −4.29 | 1.01E−03 |
| SCLAV_2926 | cvm1 | Aldo/keto reductase family 2 | −1.17 | 1.80E−04 | −1.99 | < 1E−06 |
| SCLAV_2927 | cvm2 | Ribulose 5 phosphate epimerase | −1.14 | 2.09E−05 | −2.40 | < 1E−06 |
| SCLAV_2928 | cvm3 | Oxidoreductase | −0.99 | 1.30E−03 | −2.18 | < 1E−06 |
| SCLAV_2932 | cvm13 | Asparaginase | −0.95 | 7.30E−04 | −1.45 | < 1E−06 |
| Holomycin biosynthesis | ||||||
| SCLAV_5267 | hlmA | Acetyl transferase | 5.95 | < 1E−06 | 4.41 | < 1E−06 |
| SCLAV_5268 | hlmB | Acyl-CoA dehydrogenase | 6.03 | < 1E−06 | 4.60 | < 1E−06 |
| SCLAV_5269 | hlmC | Thioesterase | 6.11 | < 1E−06 | 4.39 | < 1E−06 |
| SCLAV_5270 | hlmD | Probable dehydrogenase | 6.11 | < 1E−06 | 4.07 | < 1E−06 |
| SCLAV_5272 | hlmF | DNA/pantothenate metabolism flavoprotein | 5.47 | < 1E−06 | 3.83 | < 1E−06 |
| SCLAV_5273 | hlmG | Hypothetical protein – Frankia sp. EAN1pec | 5.41 | < 1E−06 | 3.69 | < 1E−06 |
| SCLAV_5275 | hlmI | Putative reductase – S. coelicolor | 3.96 | < 1E−06 | 4.23 | < 1E−06 |
| SCLAV_5278 | hlmM | Putative transcriptional regulator | 2.01 | 2.59E−04 | 1.36 | 8.01E−03 |
| Arginine biosynthesis | ||||||
| SCLAV_0799 | argB | Acetylglutamate kinase | −2.15 | < 1E−06 | −2.98 | < 1E−06 |
| SCLAV_0801 | argC | N-acetyl-gamma-glutamyl-phosphate reductase | −1.70 | 3.58E−02 | −2.49 | 3.43E−04 |
| SCLAV_0798 | argD | Acetonitrile aminotransferase | −1.67 | 3.19E−03 | −2.60 | < 1E−06 |
| SCLAV_0796 | argG | Argininosuccinate synthase | −2.39 | < 1E−06 | −2.91 | < 1E−06 |
| SCLAV_0795 | argH | Argininosuccinate lyase | −2.37 | 1.29E−05 | −2.81 | < 1E−06 |
| SCLAV_0800 | argJ | Glutamate N-acetyltransferase | −2.32 | 2.99E−05 | −3.32 | < 1E−06 |
| SCLAV_0797 | argR | Transcriptional regulator | −1.45 | 1.30E−04 | −2.34 | < 1E−06 |
| Drugs resistance | ||||||
| SCLAV_0793 | epeA | Transmembrane-transport protein | −0.19 | 8.42E−01 | −1.59 | 2.42E−03 |
| SCLAV_0794 | epeR | TetR-family transcriptional regulator | −0.48 | 2.76E−01 | −1.93 | < 1E−06 |
| Strict response | ||||||
| SCLAV_0744 | relA | ppGpp synthetase | 1.19 | 5.69E−04 | 1.22 | 1.36E−04 |
| Cell differentiation | ||||||
| SCLAV_5713 | rarE | Putative cytochrome P450 | −0.72 | 5.18E−03 | −1.22 | < 1E−06 |
| SCLAV_1816 | rarB | RarB Roadblock/LC7 protein | −0.91 | 2.60E−05 | −0.56 | 6.25E−03 |
| SCLAV_1817 | rarC | RarC protein | −0.90 | < 1E−06 | −0.51 | 4.58E−03 |
| SCLAV_1818 | rarD | RarD ATP/GTP-binding protein | −0.82 | 2.70E−03 | −0.74 | 2.61E−03 |
| Energy | ||||||
| SCLAV_0790 | Putative glycerophosphoryl diester phosphodiesterase | −0.91 | 1.16E−03 | −1.29 | < 1E−06 | |
| SCLAV_1370 | Probable cytochrome c oxidase polypeptide IV | −0.82 | 9.14E−05 | −0.15 | 4.78E−01 | |
| SCLAV_1372 | Probable cytochrome c oxidase polypeptide II | −0.53 | 3.40E−02 | −0.05 | 8.30E−01 | |
| SCLAV_1613 | aceE | Pyruvate dehydrogenase E1 component | −0.54 | 1.32E−04 | −0.95 | 9.70E−12 |
| SCLAV_3564 | nuoA1 | NADH-quinone oxidoreductase chain | −0.96 | < 1E−06 | −0.48 | 8.03E−03 |
| SCLAV_3970 | Putative succinate dehydrogenase flavoprotein subunit | −1.83 | < 1E−06 | −0.95 | 4.30E−04 | |
| SCLAV_3969 | Fumarate reductase iron-sulfur subunit | −1.67 | 1.82E−05 | −1.15 | 1.66E−03 | |
| SCLAV_4767 | Monophosphatase | −1.86 | < 1E−06 | −0.34 | 2.16E−01 | |
| Carbon metabolism | ||||||
| SCLAV_0631 | glpF2 | Putative glycerol uptake facilitator protein | −1.49 | 1.12E−05 | −1.95 | < 1E−06 |
| SCLAV_0632 | glpK2 | Putative glycerol kinase | −1.13 | 9.59E−03 | −1.82 | < 1E−06 |
| SCLAV_0876 | gylR | Glycerol operon regulatory protein | −0.31 | 3.62E−01 | −1.34 | < 1E−06 |
| SCLAV_0877 | glpF1 | Putative glycerol uptake facilitator protein | 0.30 | 6.19E−01 | −0.42 | 2.76E−01 |
| SCLAV_0878 | glpK1 | Glycerol kinase | −0.07 | 9.08E−01 | −1.19 | 1.99E−05 |
| SCLAV_0879 | glpD | Glycerol-3-phosphate dehydrogenase | −0.02 | 9.35E−01 | −0.94 | < 1E−06 |
| SCLAV_5509 | gap2 | Glyceraldehyde-3-phosphate dehydrogenase 2 | −2.61 | < 1E−06 | −1.75 | < 1E−06 |
| SCLAV_4529 | glcP | Glucose permease | 1.79 | < 1E−06 | −0.60 | 7.97E−02 |
| SCLAV_p0975 | Ribulose-phosphate 3-epimerase | −1.25 | < 1E−06 | −1.36 | < 1E−06 | |
| Nitrogen metabolism | ||||||
| SCLAV_4534 | amtB | Ammonium transporter | −4.10 | < 1E−06 | −5.04 | < 1E−06 |
| SCLAV_4535 | glnB | Putative nitrogen regulatory protein P-II | −4.39 | < 1E−06 | −5.09 | < 1E−06 |
| SCLAV_p1452 | glnIII | Glutamine synthetase III | −0.60 | 3.33E−02 | −1.54 | < 1E−06 |
| SCLAV_1431 | glnA3 | Glutamine synthetase (Glutamate-ammonia ligase) | −4.67 | < 1E−06 | −5.32 | < 1E−06 |
| SCLAV_1416 | glnA2 | Glutamine synthetase I (Glutamate-ammonia ligase I) | −3.56 | < 1E−06 | −2.95 | < 1E−06 |
| SCLAV_1473 | Glutamine synthetase | −0.88 | 6.84E−03 | −2.35 | < 1E−06 | |
| SCLAV_4660 | gluD1 | Glutamate transporter permease | −0.84 | 1.62E−02 | −1.28 | 4.14E−05 |
| SCLAV_0834 | glnA1 | Putative glutamine synthetase | −1.77 | < 1E−06 | −2.41 | < 1E−06 |
| Phosphate metabolism | ||||||
| SCLAV_1719 | phoH | Phosphate starvation-induced protein | 1.25 | 1.37E−04 | −0.06 | 8.67E−01 |
| SCLAV_3166 | pstB | PstB protein – Phosphate import ATP-binding | −0.93 | 1.41E−02 | −1.50 | 1.14E−05 |
| SCLAV_3167 | pstA | PstA protein – Permease component | −1.12 | 4.84E−05 | −1.61 | < 1E−06 |
| SCLAV_3168 | pstC | PstC protein – Permease component | −1.11 | 2.41E−02 | −1.77 | 4.12E−05 |
| SCLAV_3169 | pstS | PstS protein precursor – Periplasmic component | −0.93 | 3.10E−02 | −1.41 | 1.66E−04 |
| SCLAV_3220 | phoU | Putative phosphate transport system regulatory protein | −1.01 | 3.52E−04 | −1.23 | < 1E−06 |
| Lipid metabolism | ||||||
| SCLAV_4986 | Putative acetyl-coenzyme A synthetase | −0.55 | 4.01E−02 | −2.18 | < 1E−06 | |
| SCLAV_3406 | Putative acetyl/propionyl CoA carboxylase alpha | −1.49 | 2.87E−03 | −2.15 | < 1E−06 | |
| SCLAV_3405 | Putative acetyl/propionyl CoA carboxylase beta | −1.64 | < 1E−06 | −2.41 | < 1E−06 | |
| Transcriptional and regulatory proteins | ||||||
| SCLAV_p0826 | Putative AraC-family transcriptional regulator | 0.88 | 2.87E−05 | −0.05 | 8.17E−01 | |
| SCLAV_p0894 | brp | Gamma-butyrolactone receptor protein | 0.86 | 7.97E−04 | −0.55 | 1.98E−02 |
| SCLAV_p1319 | Putative transcriptional regulator AraC family | −1.72 | 1.75E−04 | −1.10 | 1.03E−02 | |
| SCLAV_1096 | Transcriptional regulator, GntR-family protein | −0.66 | 1.20E−04 | −0.09 | 6.16E−01 | |
| SCLAV_1433 | Putative regulatory protein | −1.18 | 2.33E−02 | −0.60 | 1.94E−01 | |
| SCLAV_1621 | Putative MerR-family transcriptional regulator | 0.67 | 2.33E−02 | −0.93 | 2.92E−04 | |
| SCLAV_1957 | adpA | AraC-family transcriptional regulator | −1.22 | 5.42E−04 | −1.43 | 1.52E−05 |
| SCLAV_1958 | ornA | Oligoribonuclease | −1.00 | < 1E−06 | −0.97 | < 1E−06 |
| SCLAV_2732 | Two-component transcriptional regulator | 1.31 | 2.20E−03 | −0.23 | 6.00E−01 | |
| SCLAV_3001 | Putative gntR-family transcriptional regulator | −0.68 | 8.78E−04 | −1.51 | < 1E−06 | |
| SCLAV_4054 | WhiB-family transcriptional regulator | 2.62 | < 1E−06 | 0.32 | 3.13E−01 | |
| SCLAV_4937 | Putative regulatory protein | 1.05 | 9.79E−06 | −1.06 | < 1E−06 | |
| SCLAV_5278 | AmphRI-like transcriptional regulator | 2.01 | 2.59E−04 | 1.36 | 8.01E−03 | |
| Unknown function | ||||||
| SCLAV_0018 | Cytochrome P450 monooxygenase | −1.16 | 8.47E−03 | −1.47 | 1.77E−04 | |
| SCLAV_0633 | ATP–GTP-binding protein | −0.78 | 4.44E−02 | −0.52 | 1.19E−01 | |
| SCLAV_0636 | Putative large secreted protein | −0.76 | 1.59E−02 | −0.17 | 5.91E−01 | |
| SCLAV_0646 | Putative inhibitor of KinA | −0.93 | 3.99E−02 | 0.08 | 8.53E−01 | |
| SCLAV_0743 | Peroxidase | 1.46 | < 1E−06 | 1.40 | < 1E−06 | |
| SCLAV_1335 | Two-component system sensor kinase | 0.51 | 3.31E−02 | −0.37 | 7.66E−02 | |
| SCLAV_1344 | Conserved phosphoesterase | −0.78 | 1.36E−02 | −1.11 | 8.41E−05 | |
| SCLAV_1564 | Acetyl-CoA acetyltransferase | −2.06 | < 1E−06 | −2.68 | < 1E−06 | |
| SCLAV_1565 | Cytochrome P450 hydroxylase | −1.26 | < 1E−06 | −2.67 | < 1E−06 | |
| SCLAV_1617 | Hypothetical protein | 1.02 | < 1E−06 | 0.03 | 8.67E−01 | |
| SCLAV_1748 | DUF143 domain-containing protein | 0.45 | 1.28E−02 | 0.14 | 4.26E−01 | |
| SCLAV_1959 | Sensor protein | −0.91 | < 1E−06 | −1.36 | < 1E−06 | |
| SCLAV_2623 | SclavP3 predicted orf | 1.03 | 1.10E−02 | 0.05 | 9.07E−01 | |
| SCLAV_2625 | Subtilase-type protease inhibitor precursor | 0.96 | 4.44E−02 | −0.04 | 9.33E−01 | |
| SCLAV_3194 | DUF1416 domain-containing protein | −0.77 | 2.64E−02 | 0.55 | 7.31E−02 | |
| SCLAV_4131 | Metallophosphoesterase | −0.80 | 3.06E−02 | −1.40 | 1.54E−05 | |
| SCLAV_4308 | Methylmalonyl-CoA epimerase | −0.68 | 1.77E−05 | −1.32 | 1.31E−15 | |
| SCLAV_4352 | Integrin-like protein | 1.15 | 7.63E−03 | 0.42 | 2.98E−01 | |
| SCLAV_4355 | Hypothetical protein | −0.64 | 2.99E−02 | −1.35 | < 1E−06 | |
| SCLAV_4359 | Metalloendopeptidase | −2.95 | 1.92E−04 | −3.36 | < 1E−06 | |
| SCLAV_4530 | Acetiltransferase | 2.52 | < 1E−06 | −0.21 | 6.30E−01 | |
| SCLAV_4717 | Putative hydroxylase | −1.20 | 6.98E−03 | −1.71 | 2.05E−05 | |
| SCLAV_5249 | Membrane protein | 1.31 | 6.40E−04 | 0.20 | 6.10E−01 | |
| SCLAV_p0763 | Amidohydrolase:Amidohydrolase-like precursor | −0.84 | 3.63E−02 | −0.10 | 7.94E−01 | |
| SCLAV_p1123 | Putative methyltransferase | 3.26 | < 1E−06 | 3.44 | < 1E−06 | |
| SCLAV_p1142 | Rhs family protein | −1.01 | < 1E−06 | −1.77 | < 1E−06 | |
| pSCL2 Plasmid | ||||||
| SclaA2_010100027605 | Helicase | 1.02 | 3.06E−03 | −0.11 | 7.59E−01 | |
| SclaA2_010100027610 | Hypothetical protein | 0.91 | 3.35E−04 | 0.14 | 6.04E−01 | |
| SclaA2_010100027625 | Hypothetical protein | 0.91 | 4.42E−06 | 1.27 | < 1E−06 | |
| SclaA2_010100027920 | Transposase | 1.00 | 7.48E−04 | 0.11 | 7.16E−01 | |
| SclaA2_010100027930 | Hypothetical protein | 2.15 | < 1E−06 | 1.27 | 1.21E−05 | |
| SclaA2_010100027935 | Hypothetical protein | 1.07 | 4.80E−03 | 0.86 | 1.27E−02 | |
| SclaA2_010100027955 | GntR-family regulatory protein | 0.85 | 3.89E−02 | 0.69 | 5.42E−02 | |
| SclaA2_010100027975 | Hypothetical protein | 0.50 | 2.02E−02 | 0.96 | < 1E−06 | |
| SclaA2_010100027990 | Hypothetical protein | 0.70 | 6.76E−03 | 0.11 | 6.74E−01 | |
| SclaA2_010100028020 | Hypothetical protein | 1.64 | 8.91E−08 | 1.18 | 5.15E−05 | |
| SclaA2_010100028015 | Phosphatase | 0.88 | 8.96E−03 | −0.16 | 6.20E−01 | |
| SclaA2_010100028325 | Hypothetical protein | 1.85 | < 1E−06 | 0.42 | 1.64E−01 | |
| SclaA2_010100028330 | Hypothetical protein | 2.71 | < 1E−06 | 0.98 | 4.07E−02 | |
| SclaA2_010100028335 | Transferase | 2.97 | < 1E−06 | 1.82 | 3.19E−05 | |
| SclaA2_010100028340 | Telomere-associated protein | 1.15 | 6.58E−03 | 0.27 | 5.12E−01 | |
| SclaA2_010100028350 | Hypothetical protein | 0.73 | 6.58E−03 | −0.29 | 2.42E−01 | |
| SclaA2_010100028360 | Hypothetical protein | 0.75 | 3.28E−02 | 0.10 | 7.66E−01 | |
| SclaA2_010100028340 | Telomere-associated protein | 0.79 | 1.61E−02 | 0.02 | 9.56E−01 | |
| pSCL1 Plasmid | ||||||
| SclaA2_010100027570 | Hypothetical protein | 1.69 | 4.80E−03 | 0.24 | 6.85E−01 | |
| SclaA2_010100027560 | Hypothetical protein | 1.95 | 4.33E−13 | 1.07 | 1.82E−05 | |
| SclaA2_010100027550 | Hypothetical protein | 1.11 | 1.81E−02 | −0.22 | 6.21E−01 | |
| SclaA2_010100027545 | Hypothetical protein | 1.11 | 6.58E−03 | −0.51 | 1.73E−01 | |
ABC, ATP-binding cassette transporters; ATP, adenosine triphosphate; BLIP, β-lactamase-inhibitory protein; GTP, guanosine triphosphate; NADH, nicotinamide adenine dinucleotide.
These genes belong to the cephamycin C gene cluster, CA gene cluster, clavams, holomycin, cellular differentiation, carbon, nitrogen, amino acids or phosphate metabolism and energy production.
CA biosynthesis genes
Transcriptomic studies showed that expression of the four genes of the ceaS2 to cas2 operon, encoding enzymes for the early steps of CA biosynthesis, dropped in the mutant to an average Mc value of −6.34 in the exponential phase. Expression of claR, encoding the regulatory protein ClaR, is under CcaR control (Pérez-Redondo et al., 1998; Santamarta et al., 2011). This gene was underexpressed in the ccaR-deleted mutant (Mc −4.61) and concomitantly transcription of the genes for the late steps of the pathway car, gcaS as well as cyp, orf12, orf13, orf14, oppA2, oppA1 and orf16, of unknown function, dropped an average Mc value of −4.23, being oat2, orf13 and car above this value.
Expression of orf18 and orf19, which encode penicillin-binding proteins, orf20 for a cytochrome P450 and pcbR, tentatively involved in β-lactam resistance, was lower in the mutant than in the wild-type strain. All these genes were expressed at higher Mc values (average Mc −0.88) than the biosynthetic genes in the ccaR-mutant.
An interesting finding was made in relation to genes encoding β-lactamase proteins. The blp gene, encoding a putative β-lactamase inhibitory protein, which is located downstream of ccaR, was clearly underexpressed in the mutant (Mc −6.2 and −4.8 in the exponential and stationary phases), which might be due to the absence of the ccaR promoter in the ccaR-mutant. The second gene, blip, encoding a well-characterized β-lactamase inhibitory protein (Doran et al., 1990), was also underexpressed in the mutant. This gene, located outside the CA gene cluster, presented Mc values of −2.54 and −2.19 in the exponential and stationary phases, respectively.
A gene external to the CA cluster, adpA, encoding a regulatory protein involved in antibiotic biosynthesis (López-García et al., 2010) was also slightly downregulated.
Genes for CA precursors
Clavulanic acid biosynthesis occurs through condensation of arginine and glyceraldehyde-3-phosphate by the carboxyethylargininesynthase, the first enzyme of the CA pathway. Streptomyces clavuligerus and S. coelicolor probes of genes for arginine biosynthesis and glycerol utilization were present in the microarrays.
The arginine biosynthesis genes were downregulated in the ccaR-deleted mutant. Six genes for arginine biosynthesis, located in three separated locations in the S. clavuligerus genome, showed average Mc values of −2.01 and −2.85 in the exponential and stationary phases respectively. These genes are controlled by the argR-encoded regulator ArgR, whose expression decreased to −1.45 and −2.34, at both sampling times.
The co-transcribed genes for glycerol utilization, glpF1-glpK1-glpD1, were weakly but significantly affected by the lack of the CcaR regulator, and the same occurred to the gylR gene, encoding the glycerol utilization regulatory protein (Baños et al., 2009).
However, a positive effect of CcaR on the second cluster for glycerol utilization might occur, because glpF2 and glpK2 were underexpressed in the ccaR-deleted strain. The most affected gene in this group was gap2, encoding glyceraldehyde-3-phosphate dehydrogenase (Mc −2.61 in the exponential growth phase).
Cephamycin C biosynthesis genes
In relation to cephamycin C biosynthesis, the transcriptomic data fitted well with those previously obtained by quantative reverse transcription polymerase chain reaction (Santamarta et al., 2011). The Mc values for lat, cmcI, cmcJ, cefD and cefF genes, either starting transcriptional units or close to the first gene of the operon, showed values ranging from −6.15 to −7.48 in the exponential phase, close to the −7.12 value of ccaR (which is deleted in the mutant), indicating a complete lack of transcription of these units. All other genes related to cephamycin biosynthesis showed Mc values between −3.2 and −7.12 in the exponential phase and their expression dropped in the ccaR-mutant by an average of 18% in the stationary phase.
The pbp74 and bla genes were barely affected in the exponential phase but were less expressed in the mutant in the stationary phase.
Genes for other antibiotics produced by S. clavuligerus
All the holomycin biosynthesis genes tested were overexpressed in the ccaR-deleted mutant, with an average Mc value of 5.57 in the exponential phase, while the regulatory gene hlmM had a Mc value of 2.01. These results coincide with the formation of holomycin by the S. clavuligerus ccaR::aph disrupted mutant detected by Fuente and colleagues (2002). A downregulation was observed in the clavams and CA paralogous clusters, especially in the stationary phase. Production of clavams is relatively variable and medium-dependent. However, cas1 encoding the second clavaminate synthase isoenzyme (Mc −3.32 and −4.29) and cvm8, encoding a serine hydroxymethyltransferase (Mc −2.59 and −3.18) were strongly downregulated in the ccaR-deleted mutant.
Nitrogen and phosphate metabolism
The lack of CcaR and, therefore, of CA and cephamycin C formation, affected nitrogen metabolism. A strong downregulation was observed in the exponential phase in the expression of the ammonium transporter-encoding gene amtB (Mc −4.1) and the downstream gene, glnB (Mc −4.39), which encodes the uridilyltransferase regulatory protein PII. This effect was stronger in the stationary phase (Mc −5.04 and −5.09, for amtB and glnB respectively). Also genes for glutamine synthetases (glnA3, glnA2, glnA1) were underexpressed in the ccaR-deleted mutant (Mc −4.67, −3.56 and −1.77) in the exponential phase. A weaker, but clear downregulation also occurred in gluD1, encoding a glutamate permease. The homologous probes of S. coelicolor (also present in the arrays) for glutamine synthetase (glnA, glnII, SCO1613) as well of genes for urease (ureAB) and an allantoinase (SCO6247) gave a lower signal when hybridized with S. clavuligerus ΔccaR::tsr messenger ribonucleic acid (mRNA) in relation to S. clavuligerus ATCC 27064 mRNA. This might indicate that cephamycin C and CA production requires a strong demand of nitrogen-derived precursors and, therefore, nitrogen metabolism slows down when antibiotic production is blocked.
Some genes involved in phosphate transport (pstA, pstC and pstS) showed a weak but clear underexpression in the ccaR-mutant, especially in the stationary phase sampling point.
Miscellaneous genes
Several genes involved in energy production or lipid metabolism were affected in the ccaR-deleted strain. This was the case for the genes encoding putative acetyl-CoA synthetases and acetyl-CoA carboxylases (SCLAV_4986, SCLAV_3405, SCLAV_3406) and the same occurred with the acetyltransferase-and methyltransferase-encoding genes (SCLAV_4530 and SCLAV_p1123). A set of genes whose function is unknown (SclaA2_010100027930, SclaA2_010100028330 and SclaA2_010100028335) all located in plasmid pSCL2, showed a clear overexpression in the mutant in the exponential phase of growth.
Effect on regulatory genes
The transcription of 13 genes encoding different types of regulators was affected in the mutant, including adpA (see previous discussion). A very strong downregulation was observed for the whiB-family transcriptional regulator (Mc 2.62) and for the gene encoding an AmphR1-like regulator (SCLAV_5278) with an Mc value of 2.0 in the exponential phase.
Expression of S. clavuligerus genes as detected by transcriptomic analysis of orthologous S. coelicolor genes
The whole S. coelicolor genome was represented in the microarrays. Therefore, we were able to compare expression of miscellaneous genes by heterologous hybridization of S. coelicolor probes present in the microarray with mRNA from S. clavuligerus. Heterologous transcriptomic studies confirmed the lower expression of the glnA, glnII, SCLAV_0834, ureAB and gap2 genes in the ΔccaR mutant. Other blocks of underexpressed genes were those encoding succinate dehydrogenases (SCLAV_3969 and SCLAV_3970) and the nuo genes (nuoEFJKLMN) for different subunits of the nicotinamide adenine dinucleotide (NADH) dehydrogenase. Genes for membrane proteins or for hypothetical proteins were also found to be underexpressed; but some were overexpressed, including the orthologous aminotransferase encoded by SCLAV_5663 (Mc 2.63) and the methyltransferase encoded by SCLAV_5654 (Mc 2.56) (Supporting Information Table S1).
Validation studies
Quantitative reverse transcription polymerase chain reaction (qRT-PCR)
The validation of many genes differentially expressed in this transcriptomic analysis has already been carried out using proteomics or qRT-PCR (Santamarta et al., 2011). Such is the case for all the downregulated genes of the CA pathway and for the lat, cmcI, cefD or cmcT genes in the cephamycin C gene cluster.
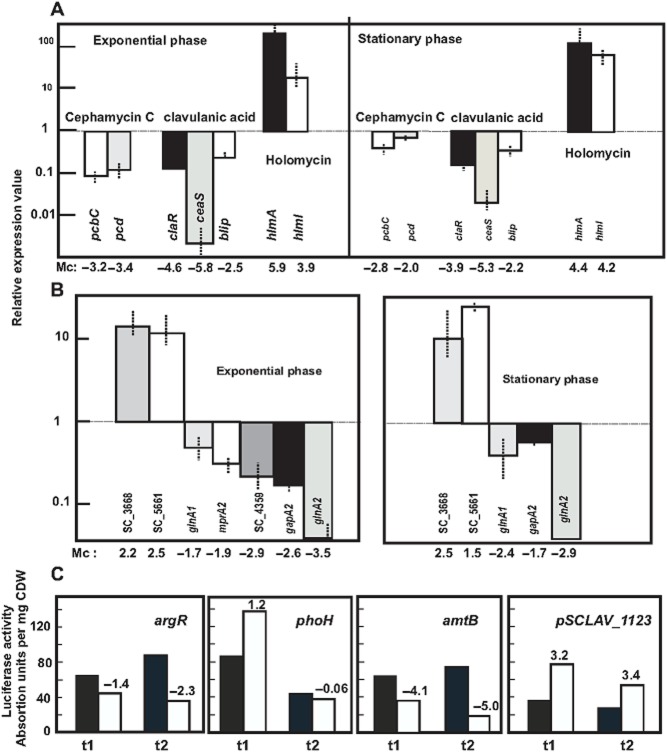
Other differentially regulated genes present in Table 1 and Supporting Information Table S1 were validated by qRT-PCR performed on reverse-transcribed RNA samples. A total of 15 genes (pcbC, pcd, claR, ceaS2, blip, hlmI, hlmA, gapA2, glnA1, glnA2, SCLAV_4359, SCLAV_3668, mprA2, SCLAV_5661 and, as control, hrdB) were chosen for analysis (Fig. 2). The Pearson's correlation coefficient (R2 = 0.9243) between Mc values and relative expression values of qRT-PCR indicates a good validation of the results.
Figure 2.
Validation of the microarray results by qRT-PCR. Quantitative RT-PCR of genes indicated below using the oligonucleotides shown in Supporting Information Table S2. The relative values are referred to 1, assigned relative value for the expression of each gene in S. clavuligerus ATCC 27064. Error bars were calculated by measuring the standard deviation among biological replicates of each sample. A. Antibiotic biosynthesis genes. B. Miscellaneous genes indicated below. C. Validation of the microarray data using the luciferase coupled method. Streptomyces clavuligerus ATCC 27064 (black bars); S. clavuligerus ΔccaR::tsr (white bars). The numbers indicate for each gene the relative Mc values of S. clavuligerus ΔccaR::tsr for each gene.
Luciferase activity
Promoters of the argR, phoH, amtB and SCLAV_p1123 genes were coupled to the luxAB genes and the constructions were introduced in S. clavuligerus ATCC 27064 and S. clavuligerus ΔccaR::tsr. Luciferase activity was lower in S. clavuligerus ΔccaR::tsr when the luxAB genes were expressed from the argR and phoH promoters (Fig. 2C) and higher when expression was carried out from the amtB and SCLAV_p1123 promoters in agreement with the results obtained in the transcriptomic experiments. The correlation index between the data obtained, measuring luciferase activity and the Mc values of the respective genes, was 0.796. Luciferase activity differed slightly from the expected data of the microarrays when the enzyme was formed in the stationary phase using the phoH promoter.
Discussion
A transcriptomic study reveals the complete gene expression in a micro-organism, allowing the comparison of overproducing strains and wild-type strains (Medema et al., 2010). However, while this method is convenient for understanding the metabolic flow in industrial strains, these strains have been obtained through heavy mutagenesis and selection programs; therefore, the final results observed might not be targeted at a particular mutation but derive from indirect effects of randomly mutated genes.
In this study, we compared the wild-type strain S. clavuligerus ATCC 27064 and a ccaR-deleted mutant obtained by molecular methods (Alexander and Jensen, 1998). As expected, the CA and cephamycin C biosynthesis genes are strongly downregulated in the mutant in agreement with the activator effect found previously for CcaR (Pérez-Llarena et al., 1997; Santamarta et al., 2011). These differences are very marked (a 37-fold decrease in the exponential phase) for all the CA biosynthesis genes, or the cephamycin biosynthesis genes (559-fold) but lower (average 1.6 fold) for genes located at the ends of the clusters (orf18, orf19, orf20, pcbR, pbpA), which are supposed to be involved in β-lactam antibiotic resistance. Interestingly, two genes encoding β-lactamase inhibitory proteins, blip and blp, are downregulated in the mutant. This is the first description of a relationship between CcaR and blip expression, a gene located outside the CA cluster. According to the transcriptomic data, blip is expressed as an independent gene. A putative heptameric sequence for CcaR binding is present 104 bp upstream of blip, suggesting a direct effect of CcaR on the expression of this gene; however, electrophoretic mobility shift assay analysis has to be performed to confirm this hypothesis. The function of the blp-encoded protein has not been clearly elucidated (Thai et al., 2001), but this gene is strongly downregulated in the ccaR-mutant, perhaps due to a co-transcription with the ccaR gene located upstream.
Medema and colleagues (2010) found several uncharacterized gene clusters, including one for secondary metabolism, that were overexpressed in the high-producing strain. The opposite is found in S. clavuligerus ΔccaR::tsr, in which all the genes for holomycin biosynthesis (hlmA to hlmM) are overexpressed (average values of 32-fold) confirming that the ccaR-knockout mutant produces more holomycin than the wild-type strain (Fuente et al., 2002; Robles-Reglero et al., 2013). These results agree with those of Chen and colleagues (2009) who reported an overexpression of the filipin biosynthesis genes in an ave1-disrupted mutant unable to produce avermectin and suggest that precursor and energy flow might be directed to the production of a different secondary metabolite once the most abundant metabolite pathway has been disrupted.
Expression of the clavams biosynthesis cluster or the ‘paralogous’ CA cluster, including the cvm7 gene, encoding a LAL regulator, was weakly downregulated except for the cas1 and orf8 genes, encoding the clavaminate synthetase1 isoenzyme and a serine hydroxymethyltransferase, which were strongly downregulated. This result contrasts with that of Medema and colleagues (2010) who described a significant overexpression of all these genes in the CA overproducing mutant that overexpresses ccaR.
The effects of CcaR absence on the transcription of S. clavuligerus primary metabolism genes are validated by the parallel transcription results for S. coelicolor genes present in the microarray.
Clavulanic acid is formed from arginine and a C3 compound derived from glycerol. All genes for arginine biosynthesis are strongly downregulated in the mutant suggesting that cells detect the lack of arginine requirement for CA biosynthesis in the ccaR-deleted mutant and shut down the arginine pathway. Genes for glycerol utilization (glpF2, glpK2) are expressed in the CA non-producing mutants in the order of 0.4-fold in relation to the control strain, which is a small reduction if compared with the two-fold expression increase observed for these genes by Medema and colleagues (2010) in the high CA-producing strain; however, our result in the gap2 gene, for glyceraldehyde-3-phosphate dehydrogenase 2, indicates that this gene is downregulated (0.16-fold), confirming what was observed by Medema and colleagues (2010) in the high CA-producing strain and the relevance of gap2-disruption in increasing the glyceraldehyde-3-phoshate required for CA production (Li and Townsend, 2006).
In summary, our results confirm most of those previously obtained for a CcaR overproducing strain. CcaR binds heptameric sequences in many CA and cephamycin C genes. However, no clear heptameric sequences have been found in nitrogen metabolism genes, genes controlling energy flow or genes for the antibiotic precursors suggesting that the lack of CA and cephamycin C production directly affects the flow of these pathways.
Experimental procedures
Culture conditions
Streptomyces clavuligerus ATCC 27064 and the mutants S. clavuligerus ΔccaR::tsr (Alexander and Jensen, 1998) and S. clavuligerus ccaR::aph (Pérez-Llarena et al., 1997) were used in this work. Strain S. clavuligerus ΔccaR::tsr was chosen for the transcriptomic experiments because the ccaR was deleted. To determine the more homogeneous and repetitive conditions for RNA sampling, the wild-type strain and S. clavuligerus ΔccaR::tsr were grown in SA medium and DNA, cell dry weight and antibiotics production were analyzed. Trypticase soy broth medium (100 ml) was inoculated with 1 ml of frozen mycelia, and the culture was grown to an OD600nm of 6.5. This culture was used to inoculate (5% v/v) 500 ml baffled flasks containing 100 ml of semidefined SA medium (Aidoo et al., 1994). The cultures were maintained at 28°C with 220 r.p.m. shaking. Exponential phase sampling was done when the cultures reached a DNA content of 75 and 80 μg ml−1 in the wild type and the mutant (24 and 32 h respectively) and 160 and 170 μg DNA ml−1 in the wild type and the mutant for the early stationary phase (40 and 50 h respectively) (Fig. 1).
RNA isolation and purification
Samples (2 ml) from S. clavuligerus ATCC 27064 and S. clavuligerus ΔccaR::tsr in the exponential and stationary growth phase in SA medium were stabilized with two volumes of RNA Protect Bacteria Reagent (Qiagen) for 5 min, then 1% β-mercaptoethanol was added. The samples were treated as indicated by Álvarez-Álvarez and colleagues (2013).
Labelling and microarray hybridizations
Streptomyces clavuligerus microarrays were obtained from Agilent Technologies (Santa Clara, CA, USA), in the Agilent 8 × 15K format. They include S. clavuligerus quadruple probes for about 800 genes and intergenic regions of some clusters involved in secondary metabolism, and also duplicated probes for 7728 chromosomal genes (out of 7825) of the S. coelicolor genome. Four biological replicates were made for each condition (two strains and two growth times). Labelling reactions were performed according to the recommendations described by BioPrime Array CGH Genomic Labelling Systems (Life Technologies, Carlsbad, CA, USA). Total RNA was labelled with Cy3-dCTP (Amersham, Freiburg, Germany) using random primers and Superscript II reverse transcriptase (Invitrogen, Carlsbad, CA, USA). Genomic deoxyribonucleic acid (gDNA) was labelled with Cy5-dCTP (Amersham) from random primers extended with the Klenow fragment of DNA polymerase (Roche, Basel, Switzerland). The final products were purified with MinElute columns (Qiagen, Venlo, the Netherlands) and labelling efficiencies were quantified spectrophotometrically. Cy3-cDNA (300 ng) and Cy5-labelled gDNA (10 pmol) were mixed, vacuum dried, resuspended in 32 μL of hybridization solution (Agilent) and applied on the microarray surface. Hybridizations were carried out at 55°C and extended to 60 h to improve the quality of the results (Sartor et al., 2004). Washing, scanning with an Agilent DNA Microarray Scanner G2565BA and image quantification were carried out as indicated previously (Rodríguez-García et al., 2007).
Identification of differentially transcribed genes and transcription profile classification
Microarray data were normalized and analyzed with the Bioconductor package limma (Smyth and Speed, 2003; Smyth, 2004). Weighted median was applied within arrays. Weights were assigned as follows: 1, probes corresponding to S. coelicolor genes showing a raw Cy3 intensity value higher than 2000; 0.25, probes corresponding to S. clavuligerus genes; and 0, the remaining probes. The normalized log2 of Cy3/Cy5 intensities is referred to as the Mg value, which is proportional to the abundance of transcripts for a particular gene (Mehra et al., 2006). The procedure by Smyth and colleagues (2005) to include the information from within-array replicates was applied to the set of quadruple probes. The Mg transcription values of the four experimental conditions were compared using two contrasts, mutant versus wild type, corresponding to the two studied growth phases (exponential and stationary). For each gene, the Mc value is the binary log of the differential transcription between the mutant and the wild strain. A positive Mc value indicates upregulation, and a negative one, downregulation. False discovery rate (FDR) correction for multiple testing was applied. A result was considered as statistically significant if the FDR-corrected P-value < 0.05. The microarray data have been deposited in National Center for Biotechnology Information-Gene Expression Omnibus under accession number GSE51435.
qRT-PCR
The oligonucleotide primers used in this work are shown in Table S2 (Supporting Information). All PCR reactions were performed using total DNA of S. clavuligerus strains as a template in a T-gradient (Biometra, Goettingen, Germany) thermocycler. The PCR reaction (30 μl) was performed as described by Kieser and colleagues (2000), and contained 300 ng DNA template, 0.5 mM each oligonucleotide, 28 mM each dGTP and dCTP, 12 mM each dATP and dTTP, 1 mM MgCl2, dimethylsulfoxide 5%, and 0.8 U Taq DNA polymerase. The amplification programme was as follows: after a step of 95°C for 30 s, the annealing temperature was reduced in a touchdown of 1°C from 60°C to 55°C in one cycle, with an annealing time of 30 s; an annealing temperature of 55°C was used in the next 25 cycles with an extension step of 1 min at 72°C. Quantification and purity analysis of all PCR products was determined using a NanoDrop ND-1000 Spectrophotometer (Thermo Scientific, Waltham, MA, USA).
Gene expression analysis by qRT-PCR was performed as previously described (López-García et al., 2010). Streptomyces clavuligerus RNA was obtained in the same way as that used for microarray experiments. RNA samples were prepared using RNeasy mini-spin columns. The samples were treated with DNase I (Qiagen) and Turbo DNase (Ambion, Carlsbad, CA, USA) to eliminate DNA. Negative controls on qRT-PCR amplification (to confirm the absence of contaminating DNA) were carried out with each set of primers. The efficiency of the primers used was measured by serial dilution of genomic DNA as template. Relative quantification of gene expression was performed by the ΔΔCt method (Livak and Schmittgen, 2001), using the constitutive housekeeping gene hrdB as reference (Buttner et al., 1990).
Luciferase assay
For luciferase reporter analysis, promoter regions were amplified with primers containing NdeI and BamHI restriction sites (Supporting Information Table S2) to clone the promoters in the ATG codon of the luxA gene in pLUXAR-neo. Cultures of S. clavuligerus exconjugants harbouring the promoter–probe constructs were carried out in SA medium. Sample treatment and the luciferase assays were done as described by Pérez-Redondo and colleagues (2012). At least two different cultures from the same strain were analyzed for luminescence production and measured in triplicate.
Acknowledgments
We acknowledge the reception of S. clavuligerus ΔccaR::tsr from Prof. S. E. Jensen. Streptomyces clavuligerus gene sequences were obtained from DSM (The Netherlands). R. Álvarez-Álvarez and Y. Martínez-Burgo received Formación del Profesorado Universitario fellowships from the Ministry of Science and Innovation.
Conflict of interest
There no conflict of interest in this article.
Funding Information
This work was financed by Grants BIO2012–34723 from the Ministry of Science and Innovation and LE046-A11-2 from the Junta de Castilla y León.
Supporting information
Additional Supporting Information may be found in the online version of this article at the publisher's web-site:
Expression of S. clavuligerus genes as detected by hybridization in microarrays with S. coelicolor probes
Oligonucleotides used in this work
References
- Aidoo KA, Wong A, Alexander DC, Rittammer RA, Jensen SE. Cloning, sequencing and disruption of a gene from Streptomyces clavuligerus involved in clavulanic acid biosynthesis. Gene. 1994;147:41–46. doi: 10.1016/0378-1119(94)90036-1. [DOI] [PubMed] [Google Scholar]
- Alexander DC, Jensen SE. Investigation of the Streptomyces clavuligerus cephamycin C gene cluster and its regulation by the CcaR protein. J Bacteriol. 1998;180:4068–4079. doi: 10.1128/jb.180.16.4068-4079.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Álvarez-Álvarez R, Martínez-Burgo Y, Pérez-Redondo R, Braña AF, Martín JF, Liras P. Expression of the endogenous and heterologous clavulanic acid cluster in Streptomyces flavogriseus: why a silent cluster is sleeping. Appl Microbiol Biotechnol. 2013;97:9451–9463. doi: 10.1007/s00253-013-5148-7. [DOI] [PubMed] [Google Scholar]
- Antón N, Mendes MV, Martín JF, Aparicio JF. Identification of PimR as a positive regulator of pimaricin biosynthesis in Streptomyces natalensis. J Bacteriol. 2004;186:2567–2575. doi: 10.1128/JB.186.9.2567-2575.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baños S, Pérez-Redondo R, Koekman B, Liras P. Glycerol utilization gene cluster in Streptomyces clavuligerus. Appl Environ Microbiol. 2009;75:2991–2995. doi: 10.1128/AEM.00181-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bate N, Stratigopoulos G, Cundliffe E. Differential roles of two SARP-encoding regulatory genes during tylosin biosynthesis. Mol Microbiol. 2002;43:449–458. doi: 10.1046/j.1365-2958.2002.02756.x. [DOI] [PubMed] [Google Scholar]
- Buttner MJ, Chater KF, Bibb MJ. Cloning, disruption and transcriptional analysis of three RNA polymerase sigma factor genes of Streptomyces coelicolorA3(2) J Bacteriol. 1990;172:3367–3378. doi: 10.1128/jb.172.6.3367-3378.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Chen J, Jiang Y, Zhang W, Jiang W, Lu Y. Transcriptomics analyses reveal global roles of the regulator AveI in Streptomyces avermitilis. FEMS Microbiol Lett. 2009;298:199–207. doi: 10.1111/j.1574-6968.2009.01721.x. [DOI] [PubMed] [Google Scholar]
- Doran JL, Leskiw BK, Aippersbach S, Jensen SE. Isolation and characterization of a β-lactamase-inhibitory protein from Streptomyces clavuligerus and cloning and analysis of the corresponding gene. J Bacteriol. 1990;172:4909–4918. doi: 10.1128/jb.172.9.4909-4918.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuente A, Lorenzana de la LM, Martín JF, Liras P. Mutants of Streptomyces clavuligerus with disruptions in different genes for clavulanic acid biosynthesis produce large amounts of holomycin: possible cross-regulation of two unrelated secondary metabolic pathways. J Bacteriol. 2002;184:6559–6565. doi: 10.1128/JB.184.23.6559-6565.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garg RP, Parry RJ. Regulation of valanimycin biosynthesis in Streptomyces viridifaciens: characterization of VlmI as a Streptomyces antibiotic regulatory protein (SARP) Microbiology. 2010;156:472–483. doi: 10.1099/mic.0.033167-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He X, Li R, Pan Y, Liu G, Tan H. SanG, a transcriptional activator, controls nikkomycin biosynthesis through binding to the sanN-sanO intergenic region in Streptomyces ansochromogenes. Microbiology. 2010;156:828–837. doi: 10.1099/mic.0.033605-0. [DOI] [PubMed] [Google Scholar]
- Kieser T, Bibb MJ, Buttner MJ, Chater KF, Hopwood DA. Practical Streptomyces Genetics. Norwich, UK: John Innes Foundation; 2000. [Google Scholar]
- Li R, Townsend CA. Rational strain improvement for enhanced clavulanic acid production by genetic engineering of the glycolytic pathway in Streptomyces clavuligerus. Metabol Eng. 2006;8:240–252. doi: 10.1016/j.ymben.2006.01.003. [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2–ΔΔCT. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- López-García MT, Santamarta I, Liras P. Morphological differentiation and clavulanic acid formation are affected in an S. clavuligerus ΔadpA-deleted mutant. Microbiology. 2010;156:2354–2365. doi: 10.1099/mic.0.035956-0. [DOI] [PubMed] [Google Scholar]
- Martín JF, Liras P. Engineering of regulatory cascades and networks controlling antibiotic biosynthesis in Streptomyces. Curr Opin Microbiol. 2010;13:263–273. doi: 10.1016/j.mib.2010.02.008. [DOI] [PubMed] [Google Scholar]
- Medema MH, Alam MT, Heijne WHM, Berg van der MA, Müller U, Trefzer A, et al. Genome wide gene expression changes in an industrial clavulanic acid overproduction strain of Streptomyces clavuligerus. Microbial Biotechnol. 2010;4:300–305. doi: 10.1111/j.1751-7915.2010.00226.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehra S, Lian W, Jayapal KP, Charaniya SP, Sherman DH, Hu W-S. A framework to analyze multiple time series data: a case study with Streptomyces coelicolor. J Ind Microbiol Biotechnol. 2006;33:159–172. doi: 10.1007/s10295-005-0034-7. [DOI] [PubMed] [Google Scholar]
- Pérez-Llarena FJ, Liras P, Rodríguez-García A, Martín JF. A regulatory gene (ccaR) required for cephamycin and clavulanic acid production in Streptomyces clavuligerus: amplification results in overproduction of both beta-lactam compounds. J Bacteriol. 1997;170:2053–2059. doi: 10.1128/jb.179.6.2053-2059.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez-Redondo R, Rodríguez-García A, Martín JF, Liras P. The claR gene of Streptomyces clavuligerus, encoding a LysR-type regulatory protein controlling clavulanic acid biosynthesis, is linked to the clavulanate-9-aldehyde reductase (car) gene. Gene. 1998;211:311–321. doi: 10.1016/s0378-1119(98)00106-1. [DOI] [PubMed] [Google Scholar]
- Pérez-Redondo R, Rodríguez-García A, Botas A, Santamarta I, Martín JF, Liras P. ArgR of Streptomyces coelicolor is a versatile regulator. PLoS ONE. 2012;7:e32697. doi: 10.1371/journal.pone.0032697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robles-Reglero V, Santamarta I, Álvarez-Álvarez R, Martín JF, Liras P. Transcriptional analysis and proteomics of the holomycin gene cluster in overproducer mutants of Streptomyces clavuligerus. J Biotechnol. 2013;163:69–76. doi: 10.1016/j.jbiotec.2012.09.017. [DOI] [PubMed] [Google Scholar]
- Rodríguez M, Núñez LE, Braña AF, Méndez C, Salas JA, Blanco G. Identification of transcriptional activators for thienamycin and cephamycin C biosynthetic genes within the thienamycin gene cluster from Streptomyces cattleya. Mol Microbiol. 2008;69:633–645. doi: 10.1111/j.1365-2958.2008.06312.x. [DOI] [PubMed] [Google Scholar]
- Rodríguez-García A, Barreiro C, Santos-Beneit F, Sola-Landa A, Martín JF. Genome-wide transcriptomic and proteomic analysis of the primary response to phosphate limitation in Streptomyces coelicolor M145 and in a ΔphoP mutant. Proteomics. 2007;7:2410–2429. doi: 10.1002/pmic.200600883. [DOI] [PubMed] [Google Scholar]
- Santamarta I, López-García MT, Kurt A, Nárdiz N, Alvarez-Álvarez R, Pérez-Redondo R, et al. Characterization of DNA-binding sequences for CcaR in the cephamycin-clavulanic acid supercluster of Streptomyces clavuligerus. Mol Microbiol. 2011;81:968–981. doi: 10.1111/j.1365-2958.2011.07743.x. [DOI] [PubMed] [Google Scholar]
- Sartor M, Schwanekamp J, Halbleib D, Mohamed I, Karyala S, Medvedovic M, et al. Microarray results improve significantly as hybridization approaches equilibrium. Biotechniques. 2004;36:790–796. doi: 10.2144/04365ST02. [DOI] [PubMed] [Google Scholar]
- Smyth GK. Linear models and empirical Bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol. 2004;3:1544–6115. doi: 10.2202/1544-6115.1027. [DOI] [PubMed] [Google Scholar]
- Smyth GK, Speed TP. Normalization of cDNA microarray data. Methods. 2003;31:265–273. doi: 10.1016/s1046-2023(03)00155-5. [DOI] [PubMed] [Google Scholar]
- Smyth GK, Michaud J, Scott HS. Use of within-array replicate spots for assessing differential expression in microarray experiments. Bioinformatics. 2005;21:2067–2075. doi: 10.1093/bioinformatics/bti270. [DOI] [PubMed] [Google Scholar]
- Tahlan K, Anders C, Wong A, Mosher RH, Beatty PH, Brumlik MJ, et al. 5S clavam biosynthetic genes are located in both the clavam and paralog gene clusters in Streptomyces clavuligerus. Chem Biol. 2007;14:131–142. doi: 10.1016/j.chembiol.2006.11.012. [DOI] [PubMed] [Google Scholar]
- Thai W, Paradkar AS, Jensen SE. Construction and analysis of ss-lactamase-inhibitory protein (BLIP) non-producer mutants of Streptomyces clavuligerus. Microbiology. 2001;147:325–335. doi: 10.1099/00221287-147-2-325. [DOI] [PubMed] [Google Scholar]
- Wietzorrek A, Bibb M. A novel family of proteins that regulate antibiotic production in streptomycetes appears to contain an OmpR-like DNA-binding fold. Mol Microbiol. 1997;25:1177–1184. doi: 10.1046/j.1365-2958.1997.5421903.x. [DOI] [PubMed] [Google Scholar]
- Wilson DJ, Xue Y, Reynolds KE, Sherman DH. Characterization and analysis of the PikD regulatory factor in the pikromycin biosynthetic pathway of Streptomyces venezuelae. J Bacteriol. 2001;183:3468–3475. doi: 10.1128/JB.183.11.3468-3475.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Expression of S. clavuligerus genes as detected by hybridization in microarrays with S. coelicolor probes
Oligonucleotides used in this work