Abstract
Streptomyces albus J1074 is a streptomycete strain widely used as a host for expression of secondary metabolite gene clusters. Bioinformatic analysis of the genome of this organism predicts the presence of 27 gene clusters for secondary metabolites. We have used three different strategies for the activation of some of these silent/cryptic gene clusters in S. albus J1074: two hybrid polyketide-non-ribosomal peptides (PK-NRP) (antimycin and 6-epi-alteramides), a type I PK (candicidin), a non-ribosomal peptides (NRP) (indigoidine) and glycosylated compounds (paulomycins). By insertion of a strong and constitutive promoter in front of selected genes of two clusters, production of the blue pigment indigoidine and of two novel members of the polycyclic tetramate macrolactam family (6-epi-alteramides A and B) was activated. Overexpression of positive regulatory genes from the same organism also activated the biosynthesis of 6-epi-alteramides and heterologous expression of the regulatory gene pimM of the pimaricin cluster activated the simultaneous production of candicidins and antimycins, suggesting some kind of cross-regulation between both clusters. A cluster for glycosylated compounds (paulomycins) was also identified by comparison of the high-performance liquid chromatography profiles of the wild-type strain with that of a mutant in which two key enzymes of the cluster were simultaneously deleted.
Introduction
Actinomycetes are Gram-positive bacteria belonging to the phylum Actinobacteria, one of the largest taxonomic groups within the domain Bacteria (Gao and Gupta, 2012). Filamentous actinomycetes are characterized by a complex life cycle of morphological differentiation that starts with the germination of a spore to give a substrate mycelium and later an aerial mycelium, followed then by a sporulation stage (Chater, 1998). Actinobacteria are widely distributed in terrestrial, especially soil, and aquatic ecosystems (McCarthy and Williams, 1992; Stach and Bull, 2005). They are important for soil formation by decomposing and recycling complex mixtures of polymers in dead plant, animal and fungal materials using extracellular enzymes (McCarthy and Williams, 1992). They also exhibit diverse physiological and metabolic properties, such as the production of volatile substances (Gust et al., 2003) and a wide variety of secondary metabolites, many of which are potent antibiotic, antifungal, antitumor, immunosuppressant, antiviral or antiparasitic agents. Among these compounds, some structural types such as polyene macrolides, large-membered macrolides, anthracyclines, polyethers, cyclopolylactones, aminoglycosides, streptothricins, actinomycins and quinoxaline peptides are produced almost exclusively by actinomycetes. In addition, glycopeptides and orthosomycins are mainly produced by non-Streptomyces species (Bérdy, 2012). This has turned actinomycetes into the primary bioactive metabolite-producing organisms exploited by the pharmaceutical industry and has prompted the study of these microorganisms at all levels: taxonomy, genetics and physiology (Hopwood, 1999).
Since the development of recombinant DNA technology, an increasing number of biosynthesis gene clusters for bioactive metabolites have been isolated and characterized from actinomycetes, leading to the development of genetic engineering approaches to develop new bioactive compounds (Olano et al., 2011). In the last decade, the increasing efficiency and reduced cost of DNA sequencing has prompted researchers to join whole prokaryotic genome-sequencing projects (Lasken, 2012; Loman et al., 2012). At the present time, at least 5611 bacterial genomes are available, 779 corresponding to actinobacterias and among them 214 to Streptomyces species (http://www.ncbi.nlm.nih.gov/genomes/MICROBES/microbial_taxtree.html). This, together with the improvement of bioinformatics annotation (Torrieri et al., 2012; Wood et al., 2012) and biosynthesis gene clusters search tools (Fedorova et al., 2012) has shown that the metabolic capabilities of microorganisms known to produce bioactive compounds have been clearly underestimated (Nett et al., 2009). Some of the first sequenced actinomycete genomes corresponded to Streptomyces avermitilis, Streptomyces coelicolor, Saccharopolyspora erythraea, Salinispora tropica or S. griseus, producers of avermectin, actinorhodin, erythromycin, salinosporamide and streptomycin respectively (Nett et al., 2009). Analysis of these genome sequences has revealed the presence of numerous biosynthetic gene clusters (18 to 37) that might be involved in the biosynthesis of additional secondary metabolites belonging to different structural classes (Nett et al., 2009). These gene clusters are defined as cryptic or orphan because the identity of the natural product of the encoded pathway is unknown. The putative structure of these metabolites can be predicted, in some cases, using bioinformatics tools that take into consideration the function of the genes present in the cluster (Ziemert and Jensen, 2012). Various approaches have been described through genome mining to confirm the function of these pathways and to identify the products, including the activation of low-or non-expressed pathways, known as silent pathways (Zerikly and Challis, 2009; Winter et al., 2011).
Streptomyces albus J1074 is a derivative of S. albus G, defective in both restriction and modification enzymes of the SalI system (Chater and Wilde, 1976). Under normal growth conditions, this strain is not known to produce any bioactive natural product and it is widely used as a host for expression of Streptomyces secondary metabolite gene clusters (Baltz, 2010). In this work, we describe the identification of several compounds produced by S. albus J1074 using genome-mining approaches and activation of the expression of the corresponding gene clusters.
Results
Bioinformatic analysis of S. albus J1074 genome and identification of secondary metabolite gene clusters
The S. albus J1074 genome was sequenced and annotated by the Broad Institute (Cambridge, MA, USA), the sequence being available since 2008. It contains 6 823 670 bp [73.2% guanine-cytosine (GC) content] of which supercont3.1 genomic scaffold shotgun sequence represents 6 813 830 bp (accession number NZ_DS999645.1). This sequence comprises 5968 genes coding for 5902 predicted proteins. At the time the genome sequence was released, no production of secondary metabolites had been reported in S. albus J1074. However, S. albus G, parental strain of S. albus J1074 (Chater and Wilde, 1976), was shown (after treatment with N-methyl-N′-nitro-N-nitrosoguanidine and isolation of mutant J1670) to produce paulomycins A and B (Majer and Chater, 1987) and other S. albus strains were shown to produce the polyether salinomycin (Izumikawa et al., 2003a; Knirschová et al., 2007; Jiang et al., 2012; Yurkovich et al., 2012). We searched the genome sequence of S. albus J1074 chromosome using the bioinformatic tool antibiotics and Secondary Metabolite Analysis Shell (antismash) (Medema et al., 2011; Blin et al., 2013), and we found 26 secondary metabolite biosynthesis gene clusters predicted (Table 1). One additional cluster (cluster 23, Table 1), containing genes directing the biosynthesis and attachment of deoxyhexoses, was not identified by antismash but rather by specific search in the S. albus J1074 chromosome for genes encoding Nucleotide DiPhosphate-stereochemistry of sugar (NDP-D)-glucose synthase and NDP-D-glucose 4,6-dehydratase, both required for the biosynthesis of 6-deoxyhexoses.
Table 1.
Secondary metabolite biosynthesis gene clusters identified in S. albus J1074 by genome mining
| Cluster | Locationa | Type | Predicted producta |
|---|---|---|---|
| 1 | SSHG_00001–00036 | Type I PKS-NRPS | Unknown |
| 2 | SSHG_00040–00050 | Lantipeptide | Class II lantibiotic |
| 3 | SSHG_00051–00053 | Type I PKS-NRPS | Unknown |
| 4 | SSHG_00054–00069 | Type I PKS-NRPS | Antimycins |
| 5 | SSHG_00076–00101 | Type I PKS | Heptaene macrolide |
| 6 | SSHG_00144–00145 | Type III PKS | THN, flaviolin |
| 7 | SSHG_00244–00268 | Terpene | Isorenieratene |
| 8 | SSHG_00271 | Bacteriocin | Linocin M18-family |
| 9 | SSHG_00311–00315 | NRPS | Blue pigment |
| 10 | SSHG_00980–00990 | Ectoine synthase | 5-Hydroxyectoine |
| 11 | SSHG_01746–01756 | Siderophore | Desferrioxamine |
| 12 | SSHG_01964–02004 | NRPS | Unknown |
| 13 | SSHG_02468–02510 | NRPS | Lipopeptide |
| 14 | SSHG_02783–02815 | NRPS | Unknown |
| 15 | SSHG_03580–03598 | Lantipeptide | Class III lantibiotic |
| 16 | SSHG_03731 | Bacteriocin | Lactococcin 972-family |
| 17 | SSHG_03888–03912 | Lantipeptide | Class I lantibiotic |
| 18 | SSHG_04343–04344 | Terpene | Albaflavenone |
| 19 | SSHG_04630 | Terpene | Geosmin |
| 20 | SSHG_04857–04868 | Siderophore | Unknown |
| 21 | SSHG_04926–04963 | NRPS | Lipopeptide |
| 22 | SSHG_05166–05175 | Bacteriocin | Unknown |
| 23b | SSHG_05313–5354 | Deoxysugars | Glycosylated compound |
| 24 | SSHG_05575 | Bacteriocin | Unknown |
| 25 | SSHG_05647–05659 | Terpene | Hopene |
| 26 | SSHG_05699–05729 | Type I PKS-NRPS | Polycyclic tetramate macrolactam |
| 27 | SSHG_05882–05890 | NRPS | Unknown |
Location and predicted products based on antismash analysis of S. albus J1074 genome sequence (accession number NZ_DS999645.1) and individual blast analysis of selected genes.
Cluster 23 was not located by antismash analysis but only by Blast analysis of individual genes.
Twelve clusters containing modular enzyme-coding genes [polyketide synthase (PKS) or non-ribosomal peptide synthetase (NRPS)] have been identified in S. albus J1074. Two of them (clusters 5 and 6) contain PKS genes belonging to type I and type III PKS respectively. The first one (cluster 5) could correspond to a type I PKS involved in the biosynthesis of a heptaene macrolide of the polyene family because of its high degree of similarity with the FR-008/candicidin cluster identified in several streptomycetes such as S. griseus IMRU 3570, Streptomyces sp. FR-008 or Streptomyces sp. S4 (Campelo and Gil, 2002; Chen et al., 2003; Seipke et al., 2011). The other PKS cluster (cluster 6) contains a type III PKS possibly involved in the production of 1,3,6,8-tetrahydroxynaphthalene and its auto-oxidation product flaviolin (Izumikawa et al., 2003b) because this cluster is well conserved in several actinomycetes, particularly S. coelicolor A3(2) and S. avermitilis MA-4680 (Nett et al., 2009; Craney et al., 2013; Ikeda et al., 2013). In addition, four clusters (clusters 1, 3, 4 and 26) contain hybrid type I PKS-NRPS genes. In the case of clusters 1 and 3, despite the fact that most of the amino acids incorporated by each adenylation domain can be predicted by several bioinformatic tools (Weber et al., 2009), it is quite hazardous to propose even a presumptive structure for the product of these clusters. On the other hand, cluster 4 and 26 might be involved in the biosynthesis of antifungal compounds. Cluster 4 is conserved among different Streptomyces in particular in Streptomyces sp. S4 (Seipke et al., 2011) and cluster 26 shows high similarity to Lysobacter enzymogenes strain C3 cluster involved in the production of a heat-stable antifungal factor (Yu et al., 2007). Six additional clusters are comprised of NRPS genes, five of them not conserved in other actinomycetes (clusters 12, 13, 14, 21 and 27). The NRPSs of two of them show high similarity to lipopeptide biosynthesis gene clusters (clusters 13 and 21), but for the same reasons mentioned above, no presumptive structure can be proposed. In addition, the NRPS-encoding gene sshg_00313 from cluster 9 shows a high degree of similarity to genes involved in the biosynthesis of a blue pigment produced by different bacteria such as Erwinia chrysanthemi or several Streptomyces species (Reverchon et al., 2002; Takahashi et al., 2007; Novakova et al., 2010; Yu et al., 2013).
There are ten additional clusters involved in the biosynthesis of other peptides or amino acidic compounds such as siderophores (clusters 11 and 20), ribosomally derived lantipeptides (clusters 2, 15 and 17), bacteriocins (clusters 8, 16, 22 and 24) belonging to different families and ectoine (cluster 10, Table 1). Cluster 11 might be involved in the biosynthesis of desferrioxamine, a cluster well conserved among different streptomycetes (Barona-Gómez et al., 2004; 2006; Nett et al., 2009). Cluster 10, present in most actinomycetes (Nett et al., 2009), might determine the biosynthesis of osmoprotectant ectoine. The production of ectoine and 5-hydroxyectoine have been shown in S. coelicolor A3(2) to be triggered upon exposure to high salinity or elevated temperature (Bursy et al., 2008). Four clusters identified by antiSMASH determine the production of known terpenes (clusters 7, 18, 19 and 25) all of them also produced by both S. coelicolor A3(2) and S. avermitilis MA-4680 (Craney et al., 2013; Ikeda et al., 2013). Cluster 23 is the only one from S. albus J1074 identified by the presence of genes determining the biosynthesis of 2,6-deoxyhexoses. This cluster contains in addition two genes encoding glycosyltransferases (sshg_05324 and sshg_05335).
Activation of a small NRPS cluster for the biosynthesis of the blue pigment indigoidine
In order to activate some cryptic/silent gene clusters in S. albus J1074, we decided to use a strategy based on the insertion of a strong and constitutive promoter in front of a selected gene of the cluster. We used the promoter of the erythromycin resistance gene (ermE*p) of Sac. erythraea (erythromycin producer). As a proof of concept, we decided to apply this strategy to a small NRPS cluster. In cluster 9, the sshg_00313 gene codes for a single module NRPS that resembles the bpsA genes from S. lavendulae (Takahashi et al., 2007) and from S. aureofaciens (Novakova et al., 2010) and the indC genes from S. chromofuscus (Yu et al., 2013) and from Erwinia chrysanthemi (Reverchon et al., 2002). Analysis of the amino acid sequence of this gene indicates that would code for a NRPS containing an oxidation domain that is embedded in an adenylation domain with the signature sequence DAWQFGLINK for recognition of L-glutamine. In addition, SSHG_00313 contains a thiolation and thioesterase domains, structural organization shared with the BpsA and IndC homologues previously mentioned. Surrounding the sshg_00313, there are other genes for which homologues are also found in the vicinity of bpsA and indC in Erwinia and Streptomyces species such as: SSHG_00311 IndA-like, SSHG_00314 IndB-like, transmembrane transporter SSHG_00315 and phosphoribosyl transferase SSHG_00316. On the other hand, SSHG_00312 encodes a 4-oxalocrotonate tautomerase that is fused to S. chromofuscus IndC NRPS. The indC-like bpsA gene is involved in the biosynthesis of the blue pigment indigoidine by condensation of two L-glutamines, this being the only activity required to generate the blue pigment (Müller et al., 2012). Under normal laboratory cultivation conditions, cultures of S. albus J1074 on agar plates do not show any blue pigmentation at all (Fig. 1A). Therefore, we considered that cluster 9 of S. albus J1074 might also be involved in the biosynthesis of this pigment, its expression being silent under normal cultivation conditions. To activate this small cluster, we inserted the ermE*p in front of the sshg_00313 gene through homologous recombination using plasmid pOJ313 (Fig. 1A), and the result was that the resulting recombinant strain produced a blue colour (Fig. 1B), indicating that expression of the cluster was activated.
Figure 1.
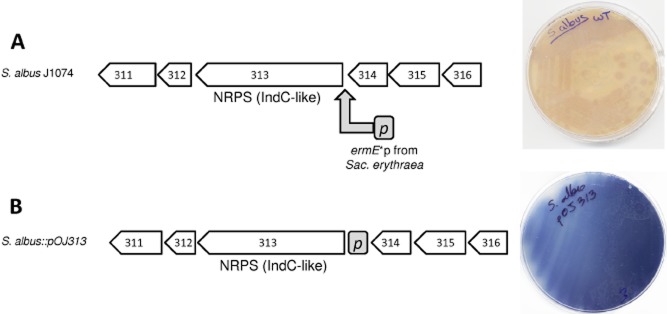
A. Genetic organization of cluster 9 and S. albus J1074 phenotype when grown in R5A solid medium. The grey arrow shows the location where the ermE*p was introduced using pOJ313. B. Genetic organization of activated cluster 9 and S. albus::pOJ313 phenotype when grown in R5A solid medium.
Activation and identification of a cluster for the hybrid PK-NRP 6-epi-alteramides A and B
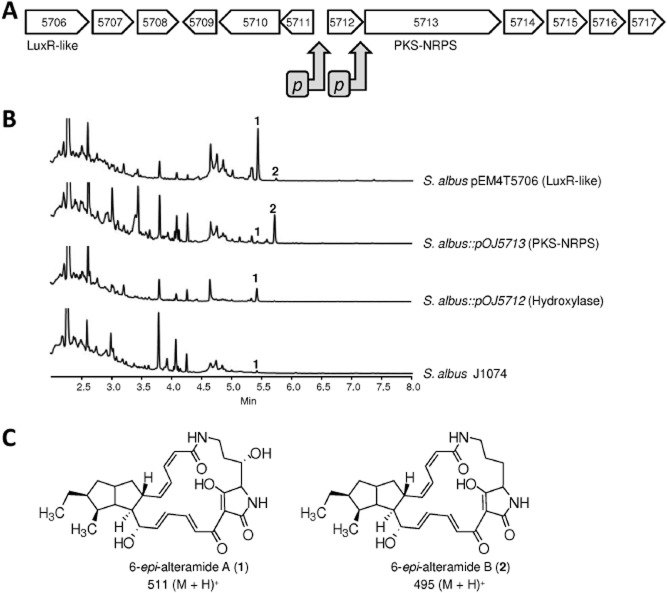
One of the clusters in S. albus (cluster 26) contains a gene coding for a hybrid type I PKS-NRPS (sshg_05713). This protein contains nine domains organized in two modules, one PKS module containing ketosynthase, acyltransferase, dehydratase, ketoreductase and acyl carrier domains, and an NRPS module composed by condensation, adenylation, peptidyl carrier protein and thioesterase domains. The PKS acyltransferase might incorporate malonate because it contains the characteristic GHSxGE signature while the NRPS adenylation domain would incorporate ornithine based in its conserved motif and characteristic-binding pocket signature DVGEIGSIDK. Surrounding the hybrid PKS-NRPS gene, there are several other genes coding for enzymatic activities possibly participating in the biosynthesis of a hybrid polyketide-peptide compound (cyclase/dehydrase SSHG_05711; flavin adenine dinucleotide-dependent oxidoreductases SSHG_05714 and SSHG_05715; oxidoreductase SSHG_05716; cytochrome P450 SSHG_05717 and hydroxylase SSHG_05712). This cluster showed similarity with other clusters from Streptomyces sp. SPB74 and Streptomyces sp. SPB78 that have been shown to be involved in the biosynthesis of the polycyclic tetramate macrolactams frontalamides A and B (Blodgett et al., 2010). To identify the compound synthesized by this cluster in S. albus, we inserted independently the ermE*p in front of sshg_05712 and of sshg_05713 using pOJ5712 and pOJ5713 respectively (Fig. 2A). Analysis by high-performance liquid chromatography-mass spectrometry (HPLC-MS) of ethyl acetate extracts of the corresponding recombinant strains showed the appearance in the case of the sshg_05712 gene of a peak with a mass of m/z 511 [M + H]+, and in the case of the sshg_05713 gene, the same peak was observed and an additional peak with a mass of m/z 495 [M + H]+ was also present (Fig. 2B). Mass analysis allowed us to discard the possibility that they were coincident with frontalamides A and B because these compounds have a different mass [m/z 525 (M + H)+ and m/z 509 (M + H)+, respectively] and slightly different absorption spectrum, and also to exclude other members of the family such as ikarugamycin [mass m/z 481 (M + H)+], cylindramide [mass m/z 467 (M + H)+] or dihydromaltophilin [mass m/z 513 (M + H)+]. The first peak was detected as a very tiny peak in extracts of the wild-type strain and the second one was undetected. The two compounds were isolated, purified and their structures elucidated by nuclear magnetic resonance (NMR) and mass spectrometry (MS; see Supporting information) and found to correspond to novel compounds 6-epi-alteramide A and 6-epi-alteramide B (Fig. 2C). They differ in the presence of a hydroxyl group at C26 of 6-epi-alteramide A, which is absent in 6-epi-alteramide B.
Figure 2.
A. Genetic organization of S. albus J1074 cluster 26. The grey arrows show the location where the ermE*p was introduced using pOJ5712 and pOJ5713. B. UPLC chromatograms, monitored at 272 nm, of S. albus J1074, S. albus::pOJ5712, S. albus::pOJ5713 and S. albus pEM4T5706 grown during 7 days in R5A liquid medium. C. Chemical structure of novel compounds 6-epi-alteramide A (1) and 6-epi-alteramide B (2).
In the vicinity of sshg_05712, 7.6 kb upstream, we found sshg_05706 that would code for a transcriptional regulator of the LuxR-family. The region comprising from sshg_05706 to sshg_05711 is not conserved in other clusters involved in the biosynthesis of polycyclic tetramate macrolactams (Blodgett et al., 2010). However, to assess the possible involvement of SSHG_05706 in the regulation of the biosynthesis of 6-epi-alteramides, we expressed sshg_05706 under the control of ermE*p using a high copy number plasmid (pEM4T5706). Streptomyces albus J1704 expressing sshg_05706 overproduced 6-epi-alteramide A and small amounts of 6-epi-alteramide B were also detected (Fig. 2B).
Simultaneous activation of candicidins and antimycins gene clusters
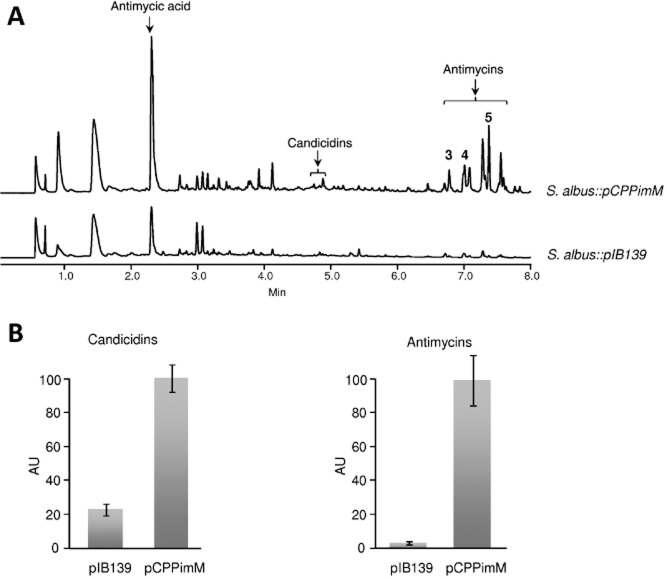
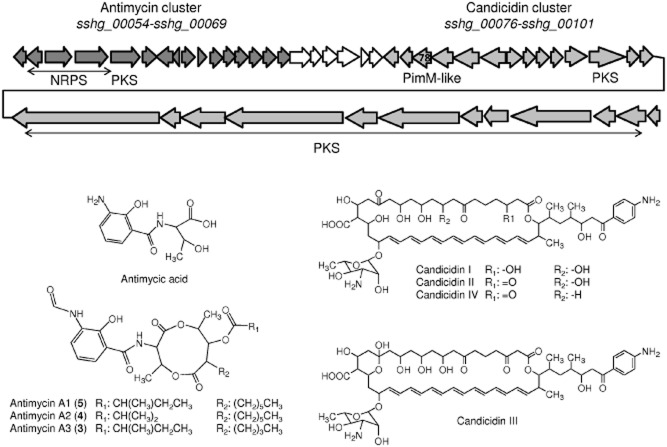
One of the largest clusters in S. albus J1074 genome is that of a type I polyketide (sshg_00076 to sshg_00101) that could code for a heptaene macrolide. It comprises a region of approximately 137 kb, and it contains a large number of type I PKS-coding genes. Most of the genes of this cluster shows high similarity with genes involved in the biosynthesis of the macrolide polyene FR-008/candicidin (Campelo and Gil, 2002; Chen et al., 2003; Seipke et al., 2011). Polyenes possess a characteristic absorption spectrum with maxima at 230, 360, 380 and 400 nm. However, under different incubation conditions, no clear peaks were detected in extracts of S. albus that could indicate the presence of a polyene. To attempt the activation of this cluster and to identify the compound it codes, we used a regulatory gene from another cluster in another organism. This was the pimM gene from the pimaricin cluster in S. natalensis that codes for a positive activator of the LuxR-family in the biosynthesis of pimaricin (Antón et al., 2007). This gene was expressed in S. albus using an integrative vector under the control of the ermE*p promoter (pCPPimM). Upon integration of the vector into the S. albus chromosome, we detected the appearance of small peaks with the characteristic absorption spectra of heptaenes (Fig. 3A). These peaks, although small, were reproducible and clearly showed a significant increase in the production yields (fourfold) (Fig. 3B). Mass analysis showed ions of m/z 1111, 1109, 1109 and 1093 [M + H]+ and co-migration with pure candicidins I, II, III and IV respectively, used as standard (Fig. 4). Interestingly, we also observed several peaks in the chromatogram with a later chromatographic retention and another peak with an earlier retention time (Fig. 3A). Mass analyses of these peaks showed ions of m/z 255, 549, 535 and 521 [M + H]+ respectively, and absorption spectra with maxima at 229 and 319 nm. Co-migration with pure samples used as standards revealed that they corresponded to different members of the antimycins family (antimycins A1, A2 and A3) and its biosynthetic precursor antimycic acid (Fig. 4). Antimycins showed a magnified production yield upon expression of PimM (29-fold) (Fig. 3B). A careful reanalysis of the S. albus genome prompted us to identify one of the PKS-NRPS clusters located in the vicinity of the candicidin cluster (genes sshg_00054 to sshg_00069) as the putative antimycin cluster (Fig. 4). This cluster showed similarity with other antimycin clusters described from Streptomyces S4, Streptomyces ambofaciens and other Streptomyces strains (Seipke et al., 2011; Yan et al., 2012; Aigle et al., 2013). In the S. albus candicidin cluster, there is a pimM homologous gene (sshg_00078). We also expressed this gene (pIB00078) in S. albus using the same integrative vector and we obtained similar results as with PimM (data not shown).
Figure 3.
A. UPLC chromatograms, monitored at 244 nm, of S. albus::pCPPimM expressing regulatory gene pimM cloned in pIB139 and S. albus::pIB139 used as a control, both grown during 7 days in R5A liquid medium. The arrows indicate the mobility of candicidins and antimycins used as standards. B. Production of candicidins and antimycins by S. albus J1074 [wild-type (WT)], S. albus::pIB139 (pIB139) and S. albus::pCPPimM (pCPPimM) grown during 7 days in R5A liquid medium.
Figure 4.
Genetic organization of S. albus J1074 clusters 4 and 5 involved in the biosynthesis of antimycins and candicidins, respectively, and chemical structure of compounds of these families.
Identification of the paulomycin gene cluster
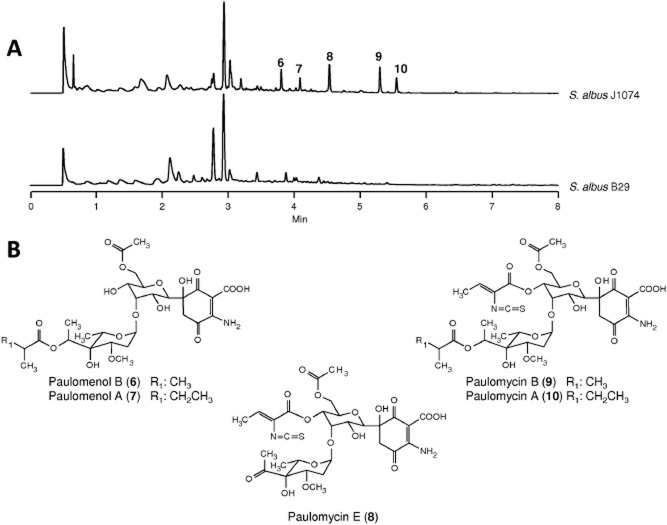
Analysis of the genome sequence of S. albus J1074 for the presence of gene clusters coding for glycosylated secondary metabolites only revealed the presence of one cluster containing genes involved in the biosynthesis of 6-deoxyhexoses (cluster 23). This cluster would comprise a region of approximately 60 kb and, in addition to several genes involved in 6-deoxysugar biosynthesis, two glycosyltransferases and three putative acyltransferases, we also observed the presence of genes coding for enzymes acting on chorismate to generate 3-hydroxyanthranilic acid or other chorismate-derived compounds. We therefore anticipated that the compound coded by this cluster could contain a central core derived from chorismate, which would be glycosylated with two deoxysugars and it should contain three acylations. To identify the compound produced by this cluster, we generated a mutant (S. albus B29) by the simultaneous deletion of two consecutive genes involved in chorismate modification, sshg_05327 and sshg_05328 that would code for an anthranilate synthase and an isochorismatase respectively. Inactivation of these genes produced the disappearance of five peaks from the chromatogram (Fig. 5A). Two of them (peaks 6 and 7) shared a similar absorption spectrum with maxima at 244 and 321 nm and mass analysis revealed ions of m/z 648 and 662 [M + H]+ respectively. The other three peaks (peaks 8, 9 and 10) shared a different spectrum with maxima at 234, 275 and 323 nm and masses of m/z 701, 773 and 787 [M + H]+ respectively. At this stage, it is important to mention that the presence of these five peaks in ethyl acetate extracts of S. albus J1074 was very variable and we could not find a reproducible pattern for production of these compounds using different batches of media or changing the culture media. The five compounds were purified and found to correspond to paulomycin A (peak 10), paulomycin B (peak 9), paulomycin E (peak 8), paulomenol A (peak 7) and paulomenol B (peak 6) by comparing their chromatographic mobilities, absorption spectra, masses and co-migration with authentic samples of these compounds (Braña et al., 2014). These compounds were originally isolated from S. paulus (Argoudelis et al., 1982; 1988a,b). The main difference between the two groups of compounds is the presence in the paulomycins of an acylation with a paulic acid residue containing an isothiocyanate residue that is not present in paulomenols and that is responsible of the absorbance peak at 275 nm observed in paulomycins (Argoudelis et al., 1988b). Paulomenols have been proposed to be true intermediates in the biosynthesis of paulomycins (Argoudelis et al., 1988b). However, some experiments prompted us to speculate that perhaps paulomenols were not real intermediates but rather degradation products of paulomycins. To answer this question, we carried out feeding experiments using as biotransformation host the non-producing deletion mutant described before (S. albus B29) blocked in early steps of the biosynthesis of paulomycins (Fig. 6). We fed paulomenol A to this mutant after 48 h of growth, and after a further 48 h, we could not observe any conversion of paulomenols into paulomycins (Fig. 6A). In contrast, when we fed the same mutant with paulomycin A, there was a progressive appearance of paulomenol A in parallel to a decrease in paulomycin A (Fig. 6B). These experiments demonstrated that paulomycin A was quite unstable and paulomenols arise as degradation products upon incubation. Therefore, the conclusion is that paulomenols are not real biosynthetic intermediates in the biosynthesis of paulomycins but rather degradation products.
Figure 5.
A. UPLC chromatograms, monitored at 244 nm, of S. albus J1074 and mutant S. albus B29 in which sshg_05727 and sshg_05728 have been deleted. Peaks absent in the mutant strain are labelled as compounds 6 to 10. B. Chemical structures of paulomenols B (6) and A (7) and paulomycins E (8), B (9) and A (10).
Figure 6.
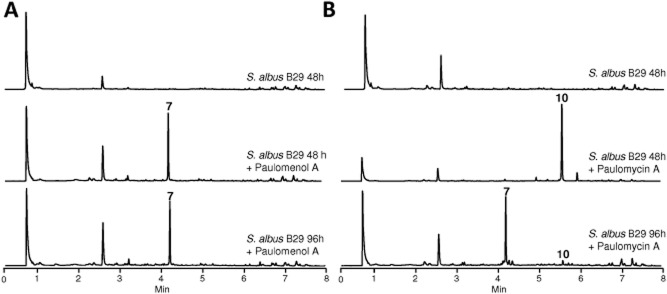
A. UPLC chromatograms, monitored at 244 nm, of mutant S. albus B29 grown in R5A liquid medium and fed with 50 μg ml−1 of paulomenol A. B. UPLC chromatograms, monitored at 244 nm, of mutant S. albus B29 grown in R5A liquid medium and fed with 50 μg ml−1 of paulomycin A.
Discussion
Filamentous bacteria of the actinomycetes family are amazingly prolific in the number of bioactive they can produce. About 75% of the antibiotics are produced by actinomycetes and most of them are synthesized by the genus Streptomyces (Demain, 2013). Nevertheless, in the last decade, the number of novel chemical entities from actinomycetes has declined concomitantly with a decrease of new drug discovery by the pharmaceutical industry. However, the development of whole genome-sequencing technology and its application to actinomycetes has revealed that these microorganisms have a greater potential for the production of bioactive natural products that was anticipated (Challis, 2008; Corre and Challis, 2009; Nett et al., 2009). Many Streptomyces species, previously known to produce one or two bioactive compounds, have been shown to possess gene clusters for a larger number of secondary metabolites. However, most of them are not expressed during standard fermentations (Olano et al., 2008; Baltz, 2011). Therefore, there is a big potential in actinomycetes for the discovery of novel bioactive natural products that can be useful for clinical or pharmaceutical applications. A key issue for the success in finding novel compounds is to find strategies to turn on (or turn up) the expression of these cryptic or low (or non) expressed biosynthesis pathways to identify the corresponding compounds and to test their biological activities. We have applied three different strategies for the activation and identification of silent/cryptic biosynthesis pathways in S. albus J1074: insertion of a strong promoter, overexpression of a positive regulatory gene and generation of a non-producing mutant. Streptomyces albus J1704 is a Streptomyces strain not known for its capability to produce a number of bioactive compounds but widely used by researchers in the actinomycetes field as a host for gene expression. Bioinformatic analysis of the genome of S. albus J1074 showed that in agreement with what has been found in other actinomycetes, it possesses a large number of gene clusters for secondary metabolites. Assuming that the lack of expression of some gene clusters could be due to the absence of some signals to be recognized by specific promoter regions of the cluster, we selected as one of the strategies the insertion of a strong and constitutive promoter in front of some key genes of selected clusters. This was the promoter of the erythromycin resistance genes, ermE*p. The proof of concept indicating that this strategy could function was successfully obtained through the activation of the expression of a small cluster coding for the blue pigment indigoidine. This strategy was also used to activate the expression of a cluster coding for a hybrid PK-NRP compound of the polycyclic tetramate macrolactams family. Insertion of the ermE*p in front of two genes of the cluster induced the biosynthesis of two novel compounds of this family, 6-epi-alteramides A and B, epimers at position 6 of alteramides A and B, compounds isolated from the bacterium Alteromonas sp. living in symbiosis with a marine sponge (Shigemori et al., 1992). This approach bypasses the natural induction of the expression of the cluster by the corresponding intracellular signal.
A second strategy we have used for cluster activation was the overexpression of positive regulatory genes. This is not a novel strategy for yield enhancement and has been used in a considerable number of systems (Bibb, 2005; Olano et al., 2008). Such approach can use regulatory genes from the same cluster or heterologously expressed regulatory genes from other clusters. We have used both approaches in two different clusters. The 6-epi-alteramides gene cluster was also activated by the overexpression in S. albus of a regulatory gene (sshg_05706) of the same organism belonging to the LuxR-family and located 7.6 kb upstream of the hybrid PKS-NRPS gene. Gene clusters for the biosynthesis of different members of this macrolactam family have been shown to be present in different actinomycetes (Blodgett et al., 2010). However, when we searched for the presence of homologous genes to sshg_05706 in several of these organisms, we could not find any homologous gene neither in the same position nor in the vicinity of these clusters. At present, it is not clear if this LuxR-like gene of S. albus is a pathway-specific regulator or a pleiotropic regulatory gene positively affecting the expression of 6-epi-alteramides among other functions. However, the high similarity among these clusters both at the gene organization level and at the DNA and protein sequence level and the absence of LuxR-like genes in these organisms are more in favour to support the view that the LuxR-like SSHG_05706 of S. albus is a pleiotropic regulator. Expression of a LuxR-like gene from S. ambofaciens has been also used to trigger the expression of stambomycins A-D, 51-membered glycosylated macrolides (Laureti et al., 2011).
Activation of clusters by the heterologous expression of a regulatory gene gave us surprising results. From the bioinformatic analysis, we suspected the presence of a cluster coding for the polyene candicidins. PimM is a regulatory gene of the pimaricin pathway in S. natalensis that combines an N-terminal PAS sensory domain with a C-terminal helix–turn–helix motif of the LuxR type for DNA binding (Santos-Aberturas et al., 2010). Insertion of this gene in the chromosome of S. albus allowed the identification of candicidins as products of the corresponding gene cluster. Surprisingly, a cluster for the biosynthesis of antimycins was also activated, this cluster being located in the vicinity of the candicidins cluster. The simultaneous activation of both clusters suggests that either the two of them use a common regulatory gene or that there could be some kind of cross-talk between them. Cross-regulation between different biosynthesis gene clusters has been reported in several Streptomyces (Fuente et al., 2002; Huang et al., 2005). It is interesting to mention that gene clusters for the biosynthesis of candicidins are quite frequent in the genomes of actinomycetes (Haeder et al., 2009; Jørgensen et al., 2009) and quite frequently an antimycins cluster is also present closely linked to the candicidins cluster (Seipke et al., 2011; Yan et al., 2012; this manuscript). Both compounds show antifungal activity, and it is possible that through evolution actinomycetes have preserved these biosynthesis pathways to combat or to protect themselves or associated organisms in which they live in symbiosis (i.e. ants) of fungal infections.
Glycosylated compounds are quite frequent within bioactive natural products (Weymouth-Wilson, 1997; Méndez and Salas, 2001; Salas and Méndez, 2007). However, searching in the genomes of actinomycetes reveals that the presence of gene clusters for glycosylated compounds is rare in this group of organisms. In the genome of S. albus only one cluster for a glycosylated compound is present, and it was not identified with antismash. We identified the compounds produced by this cluster using a third strategy, the generation of a non-producing mutant by deleting two key genes in the cluster. We cannot consider this cluster as silent/cryptic because the peaks corresponding to paulomycins and paulomenols were present in ethyl acetate extracts of the wild-type strain. However, regulation of the expression of this cluster seems to be very loose because there were clear qualitative and quantitative variations in the presence and levels of production of the five compounds. It is worth mentioning that two of these compounds, paulomenols A and B, were initially supposed to be intermediates in the biosynthesis of paulomycins (Argoudelis et al., 1988b). However, they were not converted into paulomycins when fed to a mutant blocked in early stages of the biosynthesis while feeding paulomycins gave rise to the appearance of paulomenols. This is a clear indication of the instability of paulomycins that could easily lose the paulic acid residue, thus generating paulomenols. According to this experimental evidence, paulomenols would not be real biosynthetic intermediates but rather degradation products.
In conclusion, we have demonstrated the presence in S. albus J1074 of gene clusters for the biosynthesis of two hybrid PK-NRP (antimycin and 6-epi-alteramides), a type I PK (candicidin), an NRP (indigoidine) and a glycosylated family of compounds (paulomycins). Activation of the expression of these clusters was achieved by insertion of a strong and constitutive promoter and through the overexpression of positive regulatory genes. In the case of paulomycins, the identification came from comparison of the HPLC profiles between the wild-type strain and a non-producing mutant. Several other clusters are also present in S. albus, and we are now proceeding using these and other strategies to attempt the identification of all bioactive compounds produced by this organism.
Experimental procedures
Strains, culture conditions and plasmids
Bacterial strains used in this work were S. albus J1074 (Chater and Wilde, 1976), Escherichia coli DH10B (Invitrogen, Grand Island, NY, USA) and ET12567 (pUB307) (Kieser et al., 2000) used for subcloning and intergeneric conjugation respectively. Growth medium for S. albus and mutants was tryptone soya broth; A medium was used for sporulation (Fernández et al., 1998) and R5A as production medium (Fernández et al., 1998). Escherichia coli media were those described in the literature (Sambrook et al., 1989). When plasmid-containing clones were grown, the medium was supplemented with the appropriate antibiotics: 100 μg ml−1 ampicillin, 20 μg ml−1 tobramycin, 25 μg ml−1 apramycin, 50 μg ml−1 thiostrepton, 10 μg ml−1 tetracycline, 25 μg ml−1 chloramphenicol or 50 μg ml−1 nalidixic acid.
Plasmids used in this work were pOJ260P (Olano et al., 2004) and pEFBAoriT, (Horna et al., 2011) for gene disruption and gene replacement respectively. pEM4T (Menéndez et al., 2006) and pIB139 (Wilkinson et al., 2002) were used for gene expression. pLHyg (Olano et al., 2004) was the source of the hygromycin resistance gene hyg. pCR-BLUNT (Invitrogen) was used for cloning polymerase chain reaction (PCR) products. pCPPimM (Antón et al., 2007) was used to expresses pimM under the control of ermE*p.
DNA analysis of S. albus J1074 chromosome
Computer-aided database searching and sequence analysis were carried out using the bioinformatic tool antismash (Medema et al., 2011; Blin et al., 2013) and the blast program (Altschul et al., 1997). Analysis of PKS-and NRPS-predicted proteins were carried out using programs asmpks (Tae et al., 2007) and NRPS predictor (Rausch et al., 2005).
DNA manipulation
Deoxyribonucleic acid manipulations were performed according to standard procedures for E. coli (Sambrook et al., 1989) and Streptomyces (Kieser et al., 2000). PCR conditions used for all amplifications were 97°C, 5 min.; 30 cycles of 95°C, 30 s, 50°C, 45 s and 68°C, 1 min and a final extension cycle at 68°C, 10 min. Pfx DNA polymerase (Invitrogen) and 2.5% dimethyl sulfoxide (DMSO) were used for all amplifications. To introduce the ermE*p promoter in front of the sshg_00313 gene through homologous recombination, a 2 kb fragment upstream of sshg_00313 containing its start codon was amplified using oligoprimers SA313A and SA313B (Supporting Information Table S1 in Supplementary Information). The corresponding PCR product was cloned in pCR-BLUNT (Invitrogen) and confirmed by DNA sequencing. Afterwards the fragment was subcloned into the BamHI-EcoRI-digested pOJ260P, where sshg_00313 is located downstream of ermE*p promoter leading to plasmid pOJ313.
pOJ5712 and pOJ5713 were generated to clone the ermE*p promoter in front of the sshg_05712 and sshg_05713 genes through homologous recombination. A 1.53 kb PCR fragment containing the start codon of sshg_05712 was amplified using oligoprimers SA5712A and SA5712B (Supporting Information Table S1). The PCR product was cloned in pCR-BLUNT and then subcloned into the PstI-XbaI-digested pOJ260P, leading to pOJ5712. pOJ5713 was generated using the same approach by amplification of a 1.58 kb PCR fragment, containing the start codon of sshg_05712, using oligoprimers SA5713A and SA5713B (Supporting Information Table S1).
For the expression of ssh_05706 and sshg_00078 (under the control of ermE*p) in S. albus J1074, the corresponding genes were amplified by PCR. The sshg_05706 gene was amplified as a 2628 bp fragment using oligoprimers 11R1A1 and 11R1B1 (Supporting Information Table S1). Oligoprimers 078FW and 078RV (Supporting Information Table S1) were used to amplify an 800 bp fragment containing sshg-00078. After verification by sequencing of each PCR fragment, these were subcloned into the BamHI-EcoRI digested pEM4T (for ssh_05706) or pIB139 (for sshg_00078) leading to constructs pEM4T5796 and pIB00078 respectively.
Deletion of genes sshg_05327 and sshg_05328 was accomplished by amplification of two DNA fragments of 1.5 kb. Fragment A, amplified using the oligoprimers SA5238FW and SA5328RV (Supporting Information Table S1), was cloned into SpeI-SphI-digested pEFBAoriT leading to pT5328A. Fragment B, amplified using the oligoprimers SA5327FW and SA5327RV (Supporting Information Table S1), was cloned into NdeI-BamHI-digested pT5328A, leading to pT5328/27 where fragments A and B are flanking the apramycin resistance gene aac(3)IV. Finally, pT5328/27 was digested with XbaI, and the gene hyg from pLHyg was subcloned as a SpeI-NheI fragment to obtain plasmid p5328/27Hyg used for the generation of S. albus mutant B29.
Analysis of metabolites by ultra performance liquid chromatography (UPLC) and liquid chromatography–mass spectrometry (LC-MS)
Whole cultures were extracted with ethyl acetate containing 1% formic acid (to enhance the extraction of compounds containing ionizing groups) and analysed by reversed phase chromatography in an Acquity UPLC instrument fitted with a BEH C18 column (1.7 μm, 2.1 × 100 mm; Waters), with acetonitrile and 0.1% trifluoroacetic acid (TFA) as solvents. Samples were eluted with 10% acetonitrile for 1 min, followed by a linear gradient from 10% to 100% acetonitrile over 7 min, at a flow rate of 0.5 ml min−1 and a column temperature of 35°C. For HPLC-MS analysis, an Alliance chromatographic system coupled to a ZQ4000 mass spectrometer and a SunFire C18 column (3.5 μm, 2.1 × 150 mm; Waters) was used. Solvents were the same as above and elution was performed with an initial isocratic hold with 10% acetonitrile during 4 min followed by a linear gradient from 10% to 88% acetonitrile over 26 min, at 0.25 ml min−1. MS analysis were done by electrospray ionization in the positive mode, with a capillary voltage of 3 kV and a cone voltage of 20 V. Detection and spectral characterization of peaks was performed in both cases by photodiode array detection in the range from 200 nm to 500 nm, using empower software (Waters) to extract bidimensional chromatograms at different wavelengths, depending on the spectral characteristics of the desired compound.
Isolation of 6-epi-alteramides
Strains S. albus::pOJ5712 and S. albus::pOJ5713 were grown in R5A medium without sucrose for the purification of 6-epi-alteramide A (1) and B (2) respectively. In each case, 40 Erlenmeyer flasks (250 ml), each containing 50 ml of medium, were inoculated with spores and incubated in an orbital shaker (Climo-Shaker ISF4-X, Adolf Kühner AG, Basel, Switzerland) at 30°C and 250 r.p.m. during 3 days. The cultures were centrifuged, the supernatants were discarded and the pellets were extracted twice with ethyl acetate acidified with 1% formic acid. The organic extracts were evaporated in vacuo, and the resulting dry extract was re-dissolved in 3 ml of a mixture of DMSO and methanol (1:1). The compounds of interest were purified by preparative HPLC using a SunFire C18 column (10 μm, 10 × 250 mm; Waters). Compounds were chromatographed with mixtures of acetonitrile and 0.05% TFA in water in isocratic conditions optimized for each peak at 7 ml min−1. After every purification step, the collected compounds were diluted fourfold with water and were desalted and concentrated by solid-phase extraction (Sep-Pak C18, Waters). Finally, the compounds were dissolved in tert-butanol and lyophilized. The resulting yields were 2.7 mg of 6-epi-alteramide A (1) and 13.8 mg of 6-epi-alteramide B (2).
Structural characterization of 6-epi-alteramides
6-epi-alteramide A (1) and B (2) were purified from cultures of S. albus::pOJ5712 and of S. albus::pOJ5713 respectively. Their structural elucidation was carried out using 1D 1H, 1D 13C, 2D 1H COSY, 2D 1H TOCSY, 1H, 13C heteronuclear multiple-quantum correlation-edited and heteronuclear multiple-bond correlation NMR experiments using deuterated methanol (CD3OD) as solvent (Supporting Information Figs. S1–S15 in Supplementary Information). NMR spectra were recorded at 24°C on a Bruker AVANCE III-600 MHz using a 1.7 mm microcryoprobe (Bruker). 6-epi-alteramide A and B NMR data are shown in Tables S2–S4.
Acknowledgments
We like to thank Professor Jesús Aparicio for providing construct pCPPimM and Dr. Fernando Reyes from Fundación Medina for technical support in the structural elucidation of 6-epi-alteramides.
Conflict of interest
None declared.
Funding Information
This research was supported by the Spanish Ministry of Economy and Competitiveness grants (BIO2012–33596 to J.A.S and PIM2010EEI-00752 to C.M.). A.G. was the recipient of a fellowship of FICYT (Asturias, Spain). We thank Obra Social Cajastur for financial support to C.O.
Supporting Information
Additional Supporting Information may be found in the online version of this article at the publisher's web-site:
Fig. S1. Key NOESY correlations observed in the structure of 6-epi-alteramide A.
Fig. S2. 1H NMR spectrum of 6-epi-alteramide A (CD3OD, 500 MHz).
Fig. S3. 1H NMR spectrum of 6-epi-alteramide A (1:1 CD3OD/CDCl3, 500 MHz).
Fig. S4. COSY spectrum of 6-epi-alteramide A (1:1 CD3OD/CDCl3).
Fig. S5. NOESY spectrum of 6-epi-alteramide A (1:1 vCD3OD/CDCl3, mixing time 600 msec).
Fig. S6. HSQC spectrum of 6-epi-alteramide A (1:1 CD3OD/CDCl3).
Fig. S7. HSQC spectrum of 6-epi-alteramide B (1:1 CD3OD/CDCl3).
Fig. S8. 1H NMR spectrum of 6-epi-alteramide B (CD3OD, 500 MHz).
Fig. S9. Comparison 1H NMR spectra of 6-epi-alteramide A (blue) and 6-epi-alteramide B (red) (1:1 CD3OD/CDCl3, 500 MHz).
Fig. S10. COSY spectrum of 6-epi-alteramide B (CD3OD).
Fig. S11. TOCSY spectrum of 6-epi-alteramide B (CD3OD, mixing time 90 msec).
Fig. S12. NOESY spectrum of 6-epi-alteramide B (CD3OD, mixing time 600 msec).
Fig. S13. HSQC spectrum of 6-epi-alteramide B (CD3OD).
Fig. S14. Expansion (olefinic region) of the HSQC spectrum of 6-epi-alteramide B (CD3OD). Assignments are indicated by numbers. Circled signals correspond to the cycloaddition product.
Fig. S15. Expansion (aliphatic region) of the HSQC spectrum of 6-epi-alteramide B (CD3OD). Assignments are indicated by numbers.
Table S1. Primer sets used in this work for the amplification of DNA regions used in gene inactivation and gene expression.
Table S2. 6-epi-alteramide A 1H and 13C NMR data acquired in 1:1 CD3OD/CDCl3 (500 MHz, 24 °C).
Table S3. 6-epi-alteramide A 1H and 13C NMR data acquired in CD3OD (500 MHz, 24 °C).
Table S4. 6-epi-alteramide B 1H and 13C NMR data acquired in CD3OD (500 MHz, 24 °C).
References
- Aigle B, Lautru S, Spiteller D, Dickschat JS, Challis GL, Leblond P, Pernodet JL. Genome mining of Streptomyces ambofaciens. J Ind Microbiol Biotechnol. 2013;41:251–263. doi: 10.1007/s10295-013-1379-y. [DOI] [PubMed] [Google Scholar]
- Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antón N, Santos-Aberturas J, Mendes MV, Guerra SM, Martín JF, Aparicio JF. PimM, a PAS domain positive regulator of pimaricin biosynthesis in Streptomyces natalensis. Microbiology. 2007;153:3174–3183. doi: 10.1099/mic.0.2007/009126-0. [DOI] [PubMed] [Google Scholar]
- Argoudelis AD, Brinkley TA, Brodasky TF, Buege JA, Meyer HF, Mizsak SA. Paulomycins A and B. Isolation and characterization. J Antibiot. 1982;35:285–294. doi: 10.7164/antibiotics.35.285. [DOI] [PubMed] [Google Scholar]
- Argoudelis AD, Baczynskyj L, Haak WJ, Knoll WM, Mizsak SA, Shilliday FB. New paulomycins produced by Streptomyces paulus. J Antibiot. 1988a;41:157–169. doi: 10.7164/antibiotics.41.157. [DOI] [PubMed] [Google Scholar]
- Argoudelis AD, Baczynskyj L, Mizsak SA, Shilliday FB. O-Demethylpaulomycins A and B, U-77 802 and U-77 803, paulomenols A and B, new metabolites produced by Streptomyces paulus. J Antibiot. 1988b;41:1316–1330. doi: 10.7164/antibiotics.41.1316. [DOI] [PubMed] [Google Scholar]
- Baltz RH. Streptomyces and Saccharopolyspora hosts for heterologous expression of secondary metabolite gene clusters. J Ind Microbiol Biotechnol. 2010;37:759–772. doi: 10.1007/s10295-010-0730-9. [DOI] [PubMed] [Google Scholar]
- Baltz RH. Strain improvement in actinomycetes in the postgenomic era. J Ind Microbiol Biotechnol. 2011;38:657–666. doi: 10.1007/s10295-010-0934-z. [DOI] [PubMed] [Google Scholar]
- Barona-Gómez F, Wong U, Giannakopulos AE, Derrick PJ, Challis GL. Identification of a cluster of genes that directs desferrioxamine biosynthesis in Streptomyces coelicolor M145. J Am Chem Soc. 2004;126:16282–16283. doi: 10.1021/ja045774k. [DOI] [PubMed] [Google Scholar]
- Barona-Gómez F, Lautru S, Francou FX, Leblond P, Pernodet JL, Challis GL. Multiple biosynthetic and uptake systems mediate siderophore-dependent iron acquisition in Streptomyces coelicolor A3(2) and Streptomyces ambofaciens ATCC 23877. Microbiology. 2006;152:3355–3366. doi: 10.1099/mic.0.29161-0. [DOI] [PubMed] [Google Scholar]
- Bérdy J. Thoughts and facts about antibiotics: Where we are now and where we are heading. J Antibiot. 2012;65:385–395. doi: 10.1038/ja.2012.27. [DOI] [PubMed] [Google Scholar]
- Bibb MJ. Regulation of secondary metabolism in streptomycetes. Curr Opin Microbiol. 2005;8:208–215. doi: 10.1016/j.mib.2005.02.016. [DOI] [PubMed] [Google Scholar]
- Blin K, Medema MH, Kazempour D, Fischbach MA, Breitling R, Takano E, Weber T. antiSMASH 2.0-a versatile platform for genome mining of secondary metabolite producers. Nucleic Acids Res. 2013;41:W204–W212. doi: 10.1093/nar/gkt449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blodgett JA, Oh DC, Cao S, Currie CR, Kolter R, Clardy J. Common biosynthetic origins for polycyclic tetramate macrolactams from phylogenetically diverse bacteria. Proc Natl Acad Sci USA. 2010;107:11692–11697. doi: 10.1073/pnas.1001513107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braña AF, Rodríguez M, Pahari P, Rohr J, García LA, Blanco G. Activation and silencing of secondary metabolites in Streptomyces albus and Streptomyces lividans after transformation with cosmids containing the thienamycin gene cluster form Streptomyces cattleya. 2014. Arch Microbiol (in press) [DOI] [PubMed]
- Bursy J, Kuhlmann AU, Pittelkow M, Hartmann H, Jebbar M, Pierik AJ, Bremer E. Synthesis and uptake of the compatible solutes ectoine and 5-hydroxyectoine by Streptomyces coelicolor A3(2) in response to salt and heat stresses. Appl Environ Microbiol. 2008;74:7286–7296. doi: 10.1128/AEM.00768-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campelo AB, Gil JA. The candicidin gene cluster from Streptomyces griseus IMRU 3570. Microbiology. 2002;148:51–59. doi: 10.1099/00221287-148-1-51. [DOI] [PubMed] [Google Scholar]
- Challis GL. Mining microbial genomes for new natural products and biosynthetic pathways. Microbiology. 2008;154:1555–1569. doi: 10.1099/mic.0.2008/018523-0. [DOI] [PubMed] [Google Scholar]
- Chater KF. Taking a genetic scalpel to the Streptomyces colony. Microbiology. 1998;144:1465–1478. doi: 10.1099/00221287-144-6-1465. [DOI] [PubMed] [Google Scholar]
- Chater KF, Wilde LC. Restriction of a bacteriophage of Streptomyces albus G involving endonuclease SalI. J Bacteriol. 1976;128:644–650. doi: 10.1128/jb.128.2.644-650.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S, Huang X, Zhou X, Bai L, He J, Jeong KJ, et al. Organizational and mutational analysis of a complete FR-008/candicidin gene cluster encoding a structurally related polyene complex. Chem Biol. 2003;10:1065–1076. doi: 10.1016/j.chembiol.2003.10.007. [DOI] [PubMed] [Google Scholar]
- Corre C, Challis GL. New natural product biosynthetic chemistry discovered by genome mining. Nat Prod Rep. 2009;26:977–986. doi: 10.1039/b713024b. [DOI] [PubMed] [Google Scholar]
- Craney A, Ahmed S, Nodwell J. Towards a new science of secondary metabolism. J Antibiot. 2013;66:387–400. doi: 10.1038/ja.2013.25. [DOI] [PubMed] [Google Scholar]
- Demain AL. Importance of microbial natural products and the need to revitalize their discovery. J Ind Microbiol Biotechnol. 2013;41:185–201. doi: 10.1007/s10295-013-1325-z. [DOI] [PubMed] [Google Scholar]
- Fedorova ND, Moktali V, Medema MH. Bioinformatics approaches and software for detection of secondary metabolic gene clusters. Methods Mol Biol. 2012;944:23–45. doi: 10.1007/978-1-62703-122-6_2. [DOI] [PubMed] [Google Scholar]
- Fernández E, Weissbach U, Sánchez Reillo C, Braña AF, Méndez C, Rohr J, Salas JA. Identification of two genes from Streptomyces argillaceus encoding glycosyltransferases involved in transfer of a disaccharide during biosynthesis of the antitumor drug mithramycin. J Bacteriol. 1998;180:4929–4937. doi: 10.1128/jb.180.18.4929-4937.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuente A, Lorenzana LM, Martín JF, Liras P. Mutants of Streptomyces clavuligerus with disruptions in different genes for clavulanic acid biosynthesis produce large amounts of holomycin: possible cross-regulation of two unrelated secondary metabolic pathways. J Bacteriol. 2002;184:6559–6565. doi: 10.1128/JB.184.23.6559-6565.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao B, Gupta RS. Phylogenetic framework and molecular signatures for the main clades of the phylum Actinobacteria. Microbiol Mol Biol Rev. 2012;76:66–112. doi: 10.1128/MMBR.05011-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gust B, Challis GL, Fowler K, Kieser T, Chater KF. PCR-targeted Streptomyces gene replacement identifies a protein domain needed for biosynthesis of the sesquiterpene soil odor geosmin. Proc Natl Acad Sci USA. 2003;100:1541–1546. doi: 10.1073/pnas.0337542100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haeder S, Wirth R, Herz H, Spiteller D. Candicidin-producing Streptomyces support leaf-cutting ants to protect their fungus garden against the pathogenic fungus Escovopsis. Proc Natl Acad Sci USA. 2009;106:4742–4746. doi: 10.1073/pnas.0812082106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopwood DA. Forty years of genetics with Streptomyces: from in vivo through in vitro to in silico. Microbiology. 1999;145:2183–2202. doi: 10.1099/00221287-145-9-2183. [DOI] [PubMed] [Google Scholar]
- Horna DH, Gómez C, Olano C, Palomino-Schätzlein M, Pineda-Lucena A, Carbajo RJ, et al. Biosynthesis of the RNA polymerase inhibitor streptolydigin in Streptomyces lydicus: tailoring modification of 3-methyl-aspartate. J Bacteriol. 2011;193:2647–2651. doi: 10.1128/JB.00108-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J, Shi J, Molle V, Sohlberg B, Weaver D, Bibb MJ, et al. Cross-regulation among disparate antibiotic biosynthetic pathways of Streptomyces coelicolor. Mol Microbiol. 2005;58:1276–1287. doi: 10.1111/j.1365-2958.2005.04879.x. [DOI] [PubMed] [Google Scholar]
- Ikeda H, Shin-Ya K, Omura S. Genome mining of the Streptomyces avermitilis genome and development of genome-minimized hosts for heterologous expression of biosynthetic gene clusters. J Ind Microbiol Biotechnol. 41:233–250. doi: 10.1007/s10295-013-1327-x. [DOI] [PubMed] [Google Scholar]
- Izumikawa M, Murata M, Tachibana K, Ebizuka Y, Fujii I. Cloning of modular type I polyketide synthase genes from salinomycin producing strain of Streptomyces albus. Bioorg Med Chem. 2003a;11:3401–3405. doi: 10.1016/s0968-0896(03)00337-7. [DOI] [PubMed] [Google Scholar]
- Izumikawa M, Shipley PR, Hopke JN, O'Hare T, Xiang L, Noel JP, Moore BS. Expression and characterization of the type III polyketide synthase 1,3,6,8-tetrahydroxynaphthalene synthase from Streptomyces coelicolor A3(2) J Ind Microbiol Biotechnol. 2003b;30:510–515. doi: 10.1007/s10295-003-0075-8. [DOI] [PubMed] [Google Scholar]
- Jiang C, Wang H, Kang Q, Liu J, Bai L. Cloning and characterization of the polyether salinomycin biosynthesis gene cluster of Streptomyces albus XM211. Appl Environ Microbiol. 2012;78:994–1003. doi: 10.1128/AEM.06701-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jørgensen H, Fjaervik E, Hakvåg S, Bruheim P, Bredholt H, Klinkenberg G, et al. Candicidin biosynthesis gene cluster is widely distributed among Streptomyces spp. isolated from the sediments and the neuston layer of the Trondheim fjord, Norway. Appl Environ Microbiol. 2009;75:3296–3303. doi: 10.1128/AEM.02730-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kieser T, Bibb MJ, Buttner MJ, Chater KF, Hopwood DA. Practical Streptomyces Genetics. Norwich, England: The John Innes Foundation; 2000. [Google Scholar]
- Knirschová R, Nováková R, Fecková L, Timko J, Turna J, Bistáková J, Kormanec J. Multiple regulatory genes in the salinomycin biosynthetic gene cluster of Streptomyces albus CCM 4719. Folia Microbiol. 2007;52:359–365. doi: 10.1007/BF02932090. [DOI] [PubMed] [Google Scholar]
- Lasken RS. Genomic sequencing of uncultured microorganisms from single cells. Nat Rev Microbiol. 2012;10:631–640. doi: 10.1038/nrmicro2857. [DOI] [PubMed] [Google Scholar]
- Laureti L, Song L, Huang S, Corre C, Leblond P, Challis GL, Aigle B. Identification of a bioactive 51-membered macrolide complex by activation of a silent polyketide synthase in Streptomyces ambofaciens. Proc Natl Acad Sci USA. 2011;108:6258–6263. doi: 10.1073/pnas.1019077108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loman NJ, Constantinidou C, Chan JZ, Halachev M, Sergeant M, Penn CW, et al. High-throughput bacterial genome sequencing: an embarrassment of choice, a world of opportunity. Nat Rev Microbiol. 2012;10:599–606. doi: 10.1038/nrmicro2850. [DOI] [PubMed] [Google Scholar]
- McCarthy AJ, Williams ST. Actinomycetes as agents of biodegradation in the environment-a review. Gene. 1992;115:189–192. doi: 10.1016/0378-1119(92)90558-7. [DOI] [PubMed] [Google Scholar]
- Majer J, Chater KF. Streptomyces albus G produces an antibiotic complex identical to paulomycins A and B. J Gen Microbiol. 1987;133:2503–2507. doi: 10.1099/00221287-133-9-2503. [DOI] [PubMed] [Google Scholar]
- Méndez C, Salas JA. Altering the glycosylation pattern of bioactive compounds. Trends Biotechnol. 2001;19:449–456. doi: 10.1016/s0167-7799(01)01765-6. [DOI] [PubMed] [Google Scholar]
- Medema MH, Blin K, Cimermancic P, Jager de V, Zakrzewski P, Fischbach MA, et al. antiSMASH: rapid identification, annotation and analysis of secondary metabolite biosynthesis gene clusters in bacterial and fungal genome sequences. Nucleic Acids Res. 2011;39:W339–W346. doi: 10.1093/nar/gkr466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menéndez N, Nur-e-Alam M, Fischer C, Braña AF, Salas JA, Rohr J, Méndez C. Deoxysugar transfer during chromomycin A3 biosynthesis in Streptomyces griseus subsp. griseus: new derivatives with antitumor activity. Appl Environ Microbiol. 2006;72:167–177. doi: 10.1128/AEM.72.1.167-177.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller M, Ausländer S, Ausländer D, Kemmer C, Fussenegger M. A novel reporter system for bacterial and mammalian cells based on the non-ribosomal peptide indigoidine. Metab Eng. 2012;14:325–335. doi: 10.1016/j.ymben.2012.04.002. [DOI] [PubMed] [Google Scholar]
- Nett M, Ikeda H, Moore BS. Genomic basis for natural product biosynthetic diversity in the actinomycetes. Nat Prod Rep. 2009;26:1362–1384. doi: 10.1039/b817069j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novakova R, Odnogova Z, Kutas P, Feckova L, Kormanec J. Identification and characterization of an indigoidine-like gene for a blue pigment biosynthesis in Streptomyces aureofaciens CCM 3239. Folia Microbiol. 2010;55:119–125. doi: 10.1007/s12223-010-0018-5. [DOI] [PubMed] [Google Scholar]
- Olano C, Wilkinson B, Sánchez C, Moss SJ, Sheridan R, Math V, et al. Biosynthesis of the angiogenesis inhibitor borrelidin by Streptomyces parvulus Tü4055: cluster analysis and assignment of functions. Chem Biol. 2004;11:87–97. doi: 10.1016/j.chembiol.2003.12.018. [DOI] [PubMed] [Google Scholar]
- Olano C, Lombo F, Mendez C, Salas JA. Improving production of bioactive secondary metabolites in actinomycetes by metabolic engineering. Metab Eng. 2008;10:281–292. doi: 10.1016/j.ymben.2008.07.001. [DOI] [PubMed] [Google Scholar]
- Olano C, Méndez C, Salas JA. Gene clusters for bioactive natural products in actinomycetes and their use in combinatorial biosynthesis. In: Dyson P, editor. Streptomyces Molecular Biology and Biotechnology. Norfolk, England: Caiser Academic Press; 2011. pp. 195–232. [Google Scholar]
- Rausch C, Weber T, Kohlbacher O, Wohlleben W, Huson DH. Specificity prediction of adenylation domains in nonribosomal peptide synthetases (NRPS) using transductive support vector machines (TSVM) Nucleic Acids Res. 2005;33:5799–5808. doi: 10.1093/nar/gki885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reverchon S, Rouanet C, Expert D, Nasser W. Characterization of indigoidine biosynthetic genes in Erwinia chrysanthemi and role of this blue pigment in pathogenicity. J Bacteriol. 2002;184:654–665. doi: 10.1128/JB.184.3.654-665.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salas JA, Méndez C. Engineering the glycosylation of natural products in actinomycetes. Trends Microbiol. 2007;15:219–232. doi: 10.1016/j.tim.2007.03.004. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. New York: Cold Spring Harbour Laboratory Press; 1989. [Google Scholar]
- Santos-Aberturas J, Vicente CM, Guerra SM, Payero T, Martín JF, Aparicio JF. Molecular control of polyene macrolide biosynthesis. Direct binding of the regulator PimM to eight promoters of pimaricin genes and identification of binding boxes. J Biol Chem. 2010;286:9150–9161. doi: 10.1074/jbc.M110.182428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seipke RF, Barke J, Brearley C, Hill L, Yu DW, Goss RJ, Hutchings MI. A single Streptomyces symbiont makes multiple antifungals to support the fungus farming ant Acromyrmex octospinosus. PLoS ONE. 2011;6:e22028. doi: 10.1371/journal.pone.0022028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shigemori H, Bae MA, Yazawa K, Sasaki T, Kobayashi J. Alteramide A a new tetracyclic alkaloid from a bacterium Alteromonas sp. associated with the marine sponge Halichondria okadai. J Org Chem. 1992;57:4317–4320. [Google Scholar]
- Stach JE, Bull AT. Estimating and comparing the diversity of marine actinobacteria. Antonie Van Leeuwenhoek. 2005;87:3–9. doi: 10.1007/s10482-004-6524-1. [DOI] [PubMed] [Google Scholar]
- Tae H, Kong EB, Park K. ASMPKS: an analysis system for modular polyketide synthases. BMC Bioinformatics. 2007;8:327–335. doi: 10.1186/1471-2105-8-327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi H, Kumagai T, Kitani K, Mori M, Matoba Y, Sugiyama M. Cloning and characterization of a Streptomyces single module type non-ribosomal peptide synthetase catalyzing a blue pigment synthesis. J Biol Chem. 2007;282:9073–9081. doi: 10.1074/jbc.M611319200. [DOI] [PubMed] [Google Scholar]
- Torrieri R, Oliveira FS, Oliveira G, Coimbra RS. Automatic assignment of prokaryotic genes to functional categories using literature profiling. PLoS ONE. 2012;7:e47436. doi: 10.1371/journal.pone.0047436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber T, Rausch C, Lopez P, Hoof I, Gaykova V, Huson DH, Wohlleben W. CLUSEAN: a computer-based framework for the automated analysis of bacterial secondary metabolite biosynthetic gene clusters. J Biotechnol. 2009;140:13–17. doi: 10.1016/j.jbiotec.2009.01.007. [DOI] [PubMed] [Google Scholar]
- Weymouth-Wilson AC. The role of carbohydrates in biologically active natural products. Nat Prod Rep. 1997;14:99–110. doi: 10.1039/np9971400099. [DOI] [PubMed] [Google Scholar]
- Wilkinson CJ, Hughes-Thomas ZA, Martin CJ, Böhm I, Mironenko T, Deacon M, et al. Increasing the efficiency of heterologous promoters in actinomycetes. J Mol Microbiol Biotechnol. 2002;4:417–426. [PubMed] [Google Scholar]
- Winter JM, Behnken S, Hertweck C. Genomics-inspired discovery of natural products. Curr Opin Chem Biol. 2011;15:22–31. doi: 10.1016/j.cbpa.2010.10.020. [DOI] [PubMed] [Google Scholar]
- Wood DE, Lin H, Levy-Moonshine A, Swaminathan R, Chang YC, Anton BP, et al. Thousands of missed genes found in bacterial genomes and their analysis with COMBREX. Biol Direct. 2012;7:37. doi: 10.1186/1745-6150-7-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan Y, Zhang L, Ito T, Qu X, Asakawa Y, Awakawa T, et al. Biosynthetic pathway for high structural diversity of a common dilactone core in antimycin production. Org Lett. 2012;14:4142–4145. doi: 10.1021/ol301785x. [DOI] [PubMed] [Google Scholar]
- Yu D, Xu F, Valiente J, Wang S, Zhan J. An indigoidine biosynthetic gene cluster from Streptomyces chromofuscus ATCC 49982 contains an unusual IndB homologue. J Ind Microbiol Biotechnol. 2013;40:159–168. doi: 10.1007/s10295-012-1207-9. [DOI] [PubMed] [Google Scholar]
- Yu F, Zaleta-Rivera K, Zhu X, Huffman J, Millet JC, Harris SD, et al. Structure and biosynthesis of heat-stable antifungal factor (HSAF), a broad-spectrum antimycotic with a novel mode of action. Antimicrob Agents Chemother. 2007;51:64–72. doi: 10.1128/AAC.00931-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yurkovich ME, Tyrakis PA, Hong H, Sun Y, Samborskyy M, Kamiya K, Leadlay PF. A late-stage intermediate in salinomycin biosynthesis is revealed by specific mutation in the biosynthetic gene cluster. Chembiochem. 2012;13:66–71. doi: 10.1002/cbic.201100590. [DOI] [PubMed] [Google Scholar]
- Zerikly M, Challis GL. Strategies for the discovery of new natural products by genome mining. Chembiochem. 2009;10:625–633. doi: 10.1002/cbic.200800389. [DOI] [PubMed] [Google Scholar]
- Ziemert N, Jensen PR. Phylogenetic approaches to natural product structure prediction. Methods Enzymol. 2012;517:161–182. doi: 10.1016/B978-0-12-404634-4.00008-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. Key NOESY correlations observed in the structure of 6-epi-alteramide A.
Fig. S2. 1H NMR spectrum of 6-epi-alteramide A (CD3OD, 500 MHz).
Fig. S3. 1H NMR spectrum of 6-epi-alteramide A (1:1 CD3OD/CDCl3, 500 MHz).
Fig. S4. COSY spectrum of 6-epi-alteramide A (1:1 CD3OD/CDCl3).
Fig. S5. NOESY spectrum of 6-epi-alteramide A (1:1 vCD3OD/CDCl3, mixing time 600 msec).
Fig. S6. HSQC spectrum of 6-epi-alteramide A (1:1 CD3OD/CDCl3).
Fig. S7. HSQC spectrum of 6-epi-alteramide B (1:1 CD3OD/CDCl3).
Fig. S8. 1H NMR spectrum of 6-epi-alteramide B (CD3OD, 500 MHz).
Fig. S9. Comparison 1H NMR spectra of 6-epi-alteramide A (blue) and 6-epi-alteramide B (red) (1:1 CD3OD/CDCl3, 500 MHz).
Fig. S10. COSY spectrum of 6-epi-alteramide B (CD3OD).
Fig. S11. TOCSY spectrum of 6-epi-alteramide B (CD3OD, mixing time 90 msec).
Fig. S12. NOESY spectrum of 6-epi-alteramide B (CD3OD, mixing time 600 msec).
Fig. S13. HSQC spectrum of 6-epi-alteramide B (CD3OD).
Fig. S14. Expansion (olefinic region) of the HSQC spectrum of 6-epi-alteramide B (CD3OD). Assignments are indicated by numbers. Circled signals correspond to the cycloaddition product.
Fig. S15. Expansion (aliphatic region) of the HSQC spectrum of 6-epi-alteramide B (CD3OD). Assignments are indicated by numbers.
Table S1. Primer sets used in this work for the amplification of DNA regions used in gene inactivation and gene expression.
Table S2. 6-epi-alteramide A 1H and 13C NMR data acquired in 1:1 CD3OD/CDCl3 (500 MHz, 24 °C).
Table S3. 6-epi-alteramide A 1H and 13C NMR data acquired in CD3OD (500 MHz, 24 °C).
Table S4. 6-epi-alteramide B 1H and 13C NMR data acquired in CD3OD (500 MHz, 24 °C).