Abstract
The gastrointestinal tract is a mucosal surface constantly exposed to foreign antigens and microbes, and is protected by a vast array of immunologically active structures and cells. Epithelial cells directly participate in immunological surveillance and direction of host responses in the gut and can express numerous pattern recognition receptors, including Toll-like receptor 5 (TLR5), TLR1, TLR2, TLR3, TLR9, and nucleotide oligomerization domain 2, as well as produce chemotactic factors for both myeloid and lymphoid cells following inflammatory stimulation. Within the epithelium and in the underlying lamina propria resides a population of innate lymphoid cells that, following stimulation, can become activated and produce effector cytokines and exert both protective and pathogenic roles during inflammation. Lamina propria dendritic cells play a large role in determining whether the response to a particular antigen will be inflammatory or anti-inflammatory. It is becoming clear that the composition and metabolic activity of the intestinal microbiome, as a whole community, exerts a profound influence on mucosal immune regulation. The microbiome produces short-chain fatty acids, polysaccharide A, α-galactosylceramide and tryptophan metabolites, which can induce interleukin-22, Reg3γ, IgA and interleukin-17 responses. However, much of what is known about microbiome–host immune interactions has come from the study of single bacterial members of the gastrointestinal microbiome and their impact on intestinal mucosal immunity. Additionally, evidence continues to accumulate that alterations of the intestinal microbiome can impact not only gastrointestinal immunity but also immune regulation at distal mucosal sites.
Keywords: immunity, inflammation, intestinal, microbiome, mucosal
Introduction
The gastrointestinal tract is the largest environment-exposed surface area in the body, and is in direct contact with a large and varied microbial community.1 Fortunately, the gastrointestinal tract is also home to a large variety of immune cells and structures that help maintain intestinal homeostasis in the face of microbial challenge.2–4 Intestinal epithelial cells physically separate underlying tissues from the intestinal lumen,5,6 while goblet cells maintain a mucus layer to prevent microbial contact with epithelial cells.7,8 Leucocytes beneath the epithelial cell layer can both promote and inhibit inflammatory responses,9–12 and are efficiently organized into effector and inductive sites.13–15 This organization largely prevents unwanted inflammation while retaining the ability to respond rapidly to a wide array of perturbations.
The gastrointestinal tract is also the home of the intestinal microbiome, defined as all of the microbial inhabitants (microbial community) and their collective genomes.16 While the microbiome provides numerous nutritional benefits to the host, including synthesizing vitamins17 and short chain-fatty acids (SCFAs),18 the presence of the microbiome is also vitally important for the development and functionality of the intestinal immune system.19,20 Animals devoid of intestinal microbial stimulation exhibit large defects in the organization and activity of immune structures in the gut, and proper activity can be restored via microbial stimulation.19,20 Individually, members of the microbiome can also have profound effects on host mucosal homeostasis, and specific microbes have been demonstrated to promote inflammatory21,22 or anti-inflammatory23,24 responses in the gut. Hence, cross-talk between the microbiome and the intestinal immune system is critical in the maintenance of mucosal homeostasis.
The small intestine and the large intestine are physiologically distinct sites. While nutrient absorption occurs in the small intestine, absorption of water occurs in the large intestine.1,25 Consistent with their varied physiological roles, the structure and organization of the small and large intestines are different. For example, Peyer's patches and isolated lymphoid follicles are both found within the small intestine,26 but only isolated lymphoid follicles have been described in the large intestine.27 Hence, we will first discuss the structural and cellular composition of the small and large intestines, establishing the proper context for our subsequent discussion of microbiome modulation of mucosal immunity (Fig. 1a,b).
Figure 1.
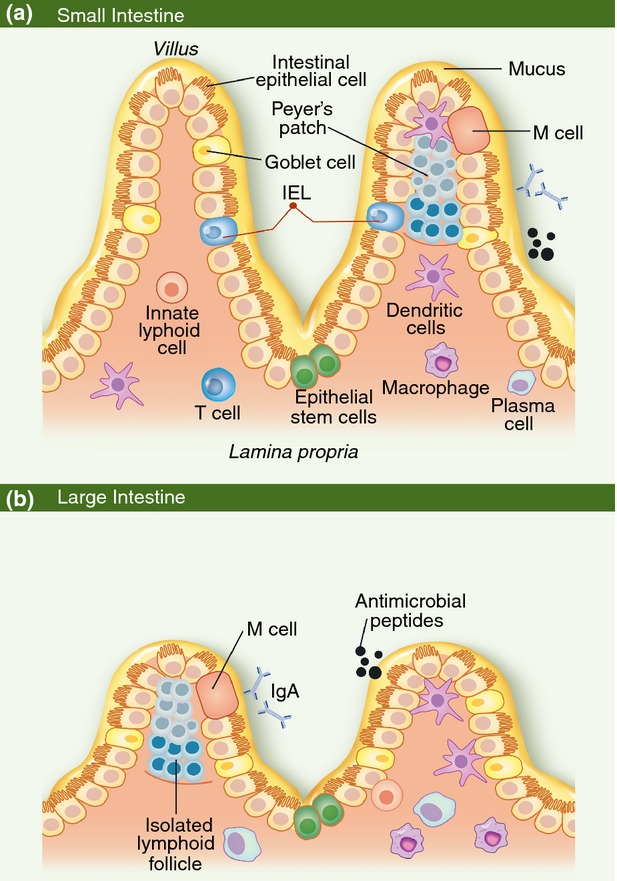
The cellular and structural composition of the small and large intestinal epithelium. (a) Organization of the small intestinal epithelium. Intestinal epithelial cells and a mucus layer separate the intestinal lumen from the underlying tissue. Lymphocytes beneath the intestinal epithelium are found in either inductive and effector sites. Inductive sites, such as Peyer's patches, generate mature lymphocytes that then migrate to effector sites, such as the lamina propria, to respond to microbial stimulation. (b) Organization of the large intestinal epithelium. The organization of the large intestinal epithelium is very similar to that of the small intestine, excepting the lack of Peyer's patches and a predominance of B cells instead of T cells in the underlying lamina propria. IEL, intraepithelial lymphocyte.
Structure and cellular composition of the small intestine
The small intestinal epithelium is actually a single layer of cells, all of which are derived from multipotent stem cells located within the intestinal crypts.2,6 Collectively, these cells are responsible for nutrient absorption, physical exclusion of luminal contents from underlying tissues, antimicrobial peptide production and maintenance of the intestinal mucus layer.2,6
Columnar epithelial cells constitute the majority of cells present in the intestinal epithelium.6,28 Enterocytes provide a physical barrier separating the luminal contents of the gastrointestinal tract from underlying tissues, as well as participating in the absorption of materials from lumen.5,6 Epithelial cells directly participate in immunological surveillance and direction of host responses in the gut. Epithelial cells can express numerous pattern recognition receptors, including Toll-like receptor 5 (TLR5),29 TLR1, TLR2, TLR3, TLR9,2 and nucleotide oligomerization domain 2,5 and can produce chemotactic factors for both myeloid and lymphoid cells following inflammatory stimulation.30 Interleukin-17 (IL-17) stimulation of intestinal epithelial cells can drive the expression of neutrophil chemokines.31 Epithelial cells can produce anti-microbial peptides, such as cathelicidin-related antimicrobial peptide, to directly influence microbial populations in the lumen of the gut.32 Additionally, epithelial cells can interact with leucocyte populations through the expression of both MHCII33 and MHCI.34 Therefore, enterocytes play a key role in not only preventing microbes and microbial products from penetrating to underlying tissues, but also initiating and directing inflammatory responses.
Within the epithelium resides a population of lymphocytes referred to as intraepithelial lymphocytes (IELs).35 Almost all IELs are T cells, with both αβ+ and γδ+ populations represented.35,36 Adherence of IELs to epithelial cells is mediated by interactions between CD103 expressed on IELS, and E-cadherin expressed on epithelial cells.37 Many IELs at baseline display a mixed phenotype, with expression of some activation markers but not others.38 However, following stimulation, IELs become activated and express effector cytokines including interferon-γ (IFN-γ) and keratinocyte growth factor.38–40 The IELs can exert both protective and pathogenic roles during inflammation: whereas IEL-derived keratinocyte growth factor is believed to protect the epithelium from damage during chemically induced colitis,41 IELs producing IFN-γ and tumour necrosis factor-α have been associated with the development of inflammatory bowel disease.42 The proximity of IELs to the lumen of the gut, and their ability to rapidly produce both inflammatory and epithelial-protective signals, make them key “first-line” defenders in the intestinal tract.
Underlying the intestinal epithelium is the lamina propria, an area rich in B and T lymphocytes.2 In contrast to Peyer's patches, which are inductive sites for the priming of lymphocytes, the lamina propria is an effector site where activated lymphocytes respond to appropriate stimulation.13–15 αβ T-cell receptor-positive T cells are the most common lymphocyte within the small intestinal lamina propria.36 In keeping with the effector function of the lamina propria, T cells found within the lamina propria express markers indicative of activation, including high levels of CD69 and CD25,43 as well as spontaneously secreting IL-4 and IFN-γ.44 Subsets within this population have drastically different activities: while CD4+ CD25+ regulatory T cells in the lamina propria can inhibit T-cell proliferation, cytokine production and the development of colitis,10,11 lamina propria CD4+ T cells can secrete both IL-17 and IL-22 and are associated with the development of intestinal inflammation.9,12 Therefore, lamina propria T cells have the ability to rapidly react to signals received from the luminal environment and initiate both inflammatory and anti-inflammatory responses.
Lamina propria dendritic cells (LPDCs) play a large role in determining whether the response to a particular antigen will be inflammatory or anti-inflammatory. LPDCs capture luminal antigen by extending their processes through the epithelial cell layer, a process dependent on CX3CR1.45 There are two broad classifications of LPDCs to consider: CD103+ and CD103−. The CD103+ LPDCS promote the generation of Foxp3+ regulatory T cells through the secretion of retinoic acid and in combination with transforming growth factor-β.3,4 In contrast, CD103− LPDCs support the development of inflammation, and increase expression of inflammatory mediators such as tumour necrosis factor-α and IL-6 following stimulation with TLR ligands.46 The presence of CD103+ LPDCs is particularly important in preventing unnecessary inflammation, as the absence of CD103+ CX3CR1− LPDCs enhances epithelial damage during colitis.47
Innate lymphoid cells (ILCs) are another cellular population found in the lamina propria.48,49 ILCs morphologically resemble lymphocytes, but do not possess recombination activating gene-dependent antigen receptors.50 They can be broken down into three broad groups.50 The defining characteristic of group 1 ILCs, such as natural killer cells, is the production of IFN-γ.50 Many group 1 ILCs are also T-bet+,50,51 and group 1 ILCs can be found at sites of mucosal inflammation.51 In contrast, generation of group 2 ILCs requires GATA3 and RORα,50 and IL-5 and IL-13 are the signature cytokines of this group.50 Group 2 ILCs are important in the response to nematode infections,50 and will be discussed no further in this review.
Particularly relevant to the intestinal tract are group 3 ILCs, which are primarily defined by their ability to produce IL-22 and IL-17.50 Additionally, the generation and activity of group 3 ILCs is dependent on RORγt.50 Recent evidence has strongly suggested that IL-17+ group 3 ILCs drive colonic inflammation during Helicobacter hepaticus infection.49 In contrast, during Citrobacter rodentium colitis, group 3 ILCs are known to produce IL-22.48 The IL-22 drives antimicrobial peptide expression and is required to prevent severe intestinal pathology and mortality during C. rodentium colitis.52 Hence, group 3 ILCs are important intestinal sources of IL-17 and IL-22, and can both promote and protect against intestinal pathology during insult.49,50,52
Peyer's patches are one of the most recognizable immune structures present in the small intestine (Fig. 1a). They are primarily a lymphoid structure, containing both germinal centres and a T-cell zone, as well as a subepithelial dome containing dendritic cells separated from the lumen of the gut by the follicle-associated epithelium (FAE).26,53 The FAE is functionally distinct from other sites in the epithelium: it contains fewer secretory cells, and IgA cannot be secreted across the FAE.54 A feature of the FAE is the presence of M cells, specialized epithelial cells that facilitate the uptake of antigen and microbes from the lumen of the gut and its delivery to underlying lymphoid tissue.54,55 Luminal antigens collected through the FAE are the primary antigens available in Peyer's patches because Peyer's patches have no afferent lymphatics.26 IgA+ B cells are prevalent in the Peyer's patch germinal centres,56 and the Peyer's patch dendritic cells promote IgA production from B cells.57 Additionally, isolated lymphoid follicles are structurally and functionally similar to Peyer's patches, but are smaller in size and can be found in both the small and large intestine.26,27,58–60 The presence of germinal centres within Peyer's patches and isolated lymphoid follicles, combined with their constant exposure to luminal antigen, make them an ideal site for the induction of adaptive responses along the intestinal tract.
Structure and cellular composition of the large intestine
In contrast to the small intestine, B cells are the predominant lymphocyte present in the lamina propria of the large intestine.36 Lamina propria B cells secrete dimeric IgA, which is transcytosed through epithelial cells to the lumen of the gut through the action of the polymeric immunoglobulin receptor.61,62 Although antigen-specific IgA can be generated during intestinal infection,63 intestinal IgA secretion also plays a key role at baseline by inhibiting the penetration of commensal microbes through the epithelium and enhancing the uptake of luminal bacterial by M cells.61 Intestinal IgA can also directly modulate the composition of the intestinal microbiome,64 highlighting the key role of IgA and lamina propria B cells in shaping both the membership and location of the microbiome.
Goblet cells are another class of specialized epithelial cells found in the intestinal epithelium.28,65 Goblet cells can be found in both the small and large intestines, but they represent approximately 15% of the cells found in the large intestinal epithelium.28,65 Goblet cells contain large mucus-laden vacuoles,65 and express high levels of the MUC2 gene.7 MUC2 is the major structural component of both intestinal mucus layers.66 The lower mucus layer makes direct contact with the intestinal epithelium and is rarely contaminated with bacteria, whereas the outer layer contacts the intestinal lumen and the intestinal microbiome.8 Goblet cells have also recently been found to produce the antimicrobial peptides Ang4, RegIIIγ and RegIIIβ.67,68 RegIIIγ activity is especially important in preventing microbial contact with the underlying epithelium.69 Goblet cells may also transfer antigens acquired in the intestinal lumen to dendritic cells in the lamina propria.70 These studies have demonstrated a potential role for goblet cells beyond mucus production by participating directly in the uptake of antigen and modulating the intestinal microbiome.
The influence of the microbiome on intestinal mucosal homeostasis
The mammalian gastrointestinal tract is home to a large community of bacteria, reaching a density of 1011 colony forming units/ml of colonic content in the large intestine, that provide an array of benefits to the host.71 The importance of the gastrointestinal tract microbiome in the generation of mucosal immune responses has been demonstrated using germ-free (GF) mice.19,20 The intestinal immune system is largely underdeveloped in the absence of microbial stimulation.19,20 Germ-free animals produce lower levels of antimicrobial peptides and have smaller numbers of IELs present than conventional animals.19,72 Additionally, the Peyer's patches of GF animals are less active and contain small germinal zones,20 and IgA+ plasma cell levels are also greatly reduced in these animals.73 The induction of oral tolerance is also deficient in GF mice.74–76 However, intestinal microbial stimulation in GF animals can restore the proper organization of the intestinal immune system.19,77
It is becoming clear that the composition and metabolic activity of the intestinal microbiome, as a whole community, exerts the greatest influence on mucosal immune regulation. However, much of what is known about microbiome–host immune interactions has come from the study of single bacterial members of the host microbiome. For example, Bacteroides fragilis produces a polysaccharide (polysaccharide A) with anti-inflammatory properties.23 Polysaccharide A, in a TLR2-dependent manner, mediates the conversion of CD4+ T cells into Foxp3+ regulatory T cells that produce IL-10, suppress IL-17 production and protect against numerous inflammatory insults.23,24,78,79 Bacteroides fragilis releases polysaccharide A in outer membrane vesicles that are detected by dendritic cells,79 whereas purified polysaccharide A can also prevent inflammation in vivo.23,24 Additionally, B. fragilis can also produce α-galactosylceramide (α-GalCerBf), a glycosphingolipid which is capable of binding CD1d and activating invariant natural killer T cells.80
In another example, monocolonization of GF mice with Bacteroides thetaiotamicron can cause changes in the expression of genes involved in intestinal nutrient absorption, mucosal barrier function and angiogenesis.81 Interestingly, while colonization by a complex microbiome is associated with high-level epithelial expression of RegIIIγ, a secreted C-type lectin that limits microbial contact with the epithelium,69,82 monocolonization of mice with B. thetaiotamicron is not.82 This failure to induce RegIIIγ expression is probably dependent on IgA, which presumably limits bacterial adhesion to and stimulation of the epithelium, as GF animals deficient in IgA express high levels of RegIIIγ following exposure to B. thetaiotamicron.82 Similarly, in mice lacking RegIIIγ there is increased bacterial colonization of intestinal epithelial surfaces and activation of intestinal adaptive immune responses, including increased levels of IgA+ cells.69 Recent work has also demonstrated that the microbiome produces signals that preferentially promote IL-22 transcription, which is required for RegIIIγ expression.52,83 Therefore, spatial separation of the microbiome and intestinal epithelium is maintained by a complex interplay between both microbiome and host-derived factors.
Another widely studied example is the role of segmented filamentous bacteria (SFB) in promoting intestinal T helper type 17 (Th17) responses.21,22 SFB associate closely with epithelial cells of the small intestine, and the presence of SFB in the terminal ileum is associated with an increase in the number of Th17 cells capable of expressing both IL-17 and IL-22 in the intestinal lamina propria.21,22 This enhanced inflammatory state appears to be protective for the host, as animals colonized with SFB are resistant to infection by the large intestine pathogen, C. rodentium.21 Monoassociation of GF mice with SFB promotes high levels of IgA, though only a small fraction of the total IgA produced is SFB-specific.84 IgA production, however, is critical for containing the SFB population; mice with deficient IgA levels (due to deficiency of activation-induced cytidine deaminase) have a marked expansion of SFB within the small intestine, which is reversed upon restoration of lamina propria IgA production.64 Hence, colonization of the small intestines of mice with SFB is a potent immunomodulatory signal for the mucosa, which in turn modulates the intestinal microbiome.
Recent studies have identified numerous species of Clostridia capable of inducing the development Foxp3+ regulatory T cells in the large intestine.85,86 Large intestine colonization with Clostridia from clusters IV and XIVa enhances transforming growth factor-β1 levels and promotes the development of IL-10-expressing Foxp3+ regulatory T cells in GF mice to levels comparable to those seen in conventionally reared animals.85,86 Colonization of conventionally reared animals with these Clostridia strains is also capable of reducing the severity of intestinal inflammation during chemically induced colitis.85,86 Since clostridial species are a major producer of SCFAs,87 one likely mechanism is the production of SCFA.
Short-chain fatty acids, such as butyric acid/butyrate, are by-products of fermentation by the microbiome and are detectable in the gastrointestinal tract.18 Butyrate also possesses potent anti-inflammatory activity on myeloid and lymphoid cells in a variety of in vitro culture systems.88–91 Butyrate has also been used to treat colitis and can reverse the increased mucosal permeability and intestinal ulceration seen in dextran sodium sulphate colitis.92,93 Conversely, in the absence of G-protein coupled receptor 43, one of the host receptors for SCFAs, mice are more susceptible to experimentally induced intestinal inflammation.94 Butyrate can also act directly on leucocytes, and inhibits IL-12 production, decreases co-stimulatory molecule expression, and blocks nuclear factor-κB translocation in human monocyte marrow-derived dendritic cells and macrophages.91,95
An anti-inflammatory role has been ascribed to members of the genus Lactobacillus.96,97 There are many reviews that discuss this field, so we will not discuss this topic in great detail. However, relevant to our previous point, though lactobacilli are poor producers of butyrate, they can produce ample quantities of lactic acid, which in turn can be rapidly converted to butyrate by other members of the microbiome,98,99 potentially accounting for one mechanism of their immunomodulatory activity. Taken together, these data clearly demonstrate that individual, non-pathogenic members of the intestinal microbiome can markedly alter the inflammatory state of the intestinal immune system to the benefit of the host (Fig. 2).
Figure 2.
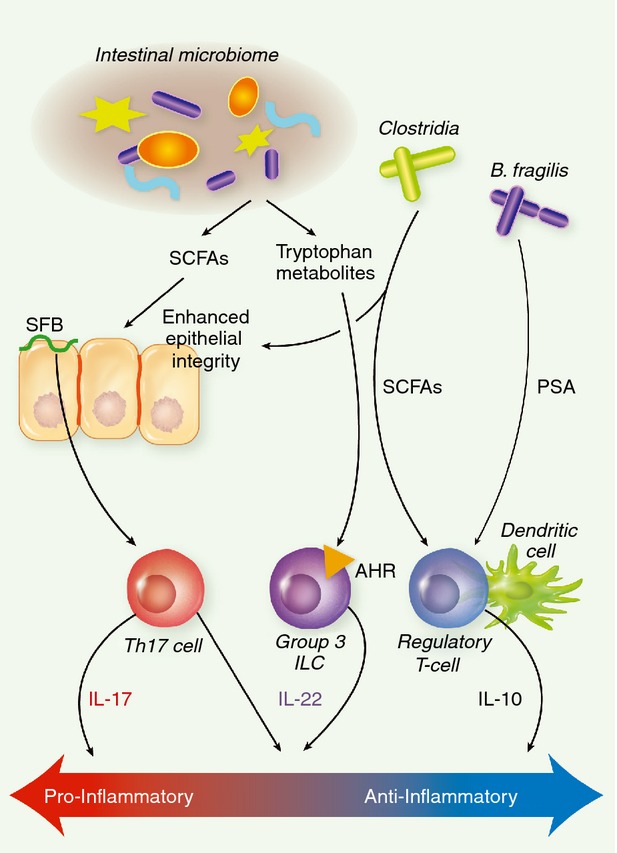
Microbial modulation of mucosal immunity. The intestinal microbiome generates numerous signals, which impact the regulation of intestinal mucosal immunity. The microbiome produces metabolic by-products, such as butyrate and tryptophan catabolites, which can enhance intestinal integrity and stimulate IL-22 production by group 3 ILCs, respectively. Certain members of the microbiome are known to activate specific arms of intestinal immunity. SFB colonization of the small bowel enhances Th17-mediated immunity, while colonization by Clostridia from clusters IV and XIVa promotes the development of regulatory T cells. Polysaccharide A, generated by Bacteroides fragilis, is also capable of enhancing regulatory T-cell activity in the gut. SFB, segmented filamentous bacteria; PSA, polysaccharide A; SCFAs, short-chain fatty acids; AHR, aryl hydrocarbon receptor; ILC, innate lymphoid cell; IL-17, interleukin-17.
Exogenous tryptophan metabolites play an important role in mammalian gut immune homeostasis via aryl hydrocarbon receptor signalling.83 The aryl hydrocarbon receptor promotes Th17 cell differentiation in vitro,100 as well as the homeostasis and function of Group 3 ILCs in vivo.101,102 Mice that are deficient for aryl hydrocarbon receptor have a significant deficiency in Group 3 ILCs, thereby resulting in much lower IL-22 production and increased susceptibility to intestinal infection.101 However, intestinal Th17 cells are increased in these mice, rather than decreased, because the reduction in IL-22 permits the expansion of SFB, thereby promoting Th17 cells.102 As recently demonstrated, the microbiome is one source of tryptophan metabolites, and tryptophan metabolism by Lactobacillus populations in mice produces indole-3-aldehyde, an aryl hydrocarbon receptor ligand that can drive IL-22 expression.83 This is yet another proposed mechanism of cross-talk between the microbiome and the host that promotes a balance between inflammatory and anti-inflammatory signalling and the overall maintenance of mucosal homeostasis.
Gastrointestinal microbiome and regulation of distal mucosal immunity
There is also increasing evidence that the gastrointestinal mucosa, the predominant site of microbiome–host interaction, can also play a role in the development of immune responses at distal mucosal sites. How might the gastrointestinal tract regulate responses to inhaled allergens or other antigens? The mucociliary architecture of the nasopharyngeal cavity and upper airways naturally sweeps all inhaled micro-particulates that stick to the mucus lining into the gastrointestinal tract. Shortly after intranasal inoculation or aerosol delivery, fluids, particles and microbes introduced into the nasal cavity are largely found in the gastrointestinal tract.103–105 In mice, intranasal inoculation of a volume as small as 2·5 ml still largely ends up in the gastrointestinal tract.104 Therefore, inhaled micro-particulates and aerosols (which comprise the vast majority of aeroallergens) are also swallowed. Using an animal model of allergic airway disease, it has been reported that 2 days after intranasal administration of antigenic peptide, corresponding antigen-specific CD4 T-cell division had not only occurred in the lymph nodes draining the lungs and nasopharyngeal cavity, but also in the mesenteric lymph nodes.106 No division was seen in peripheral non-draining nodes. In studies from our laboratory, we have been able to demonstrate that perturbation of the normal microbiome in mice can promote the development of allergic airway disease following allergen challenge.107,108 In other studies, oral delivery of various Lactobacillus strains can modulate pulmonary inflammation in murine systems.109–111 Other studies have shown that the microbiome composition can regulate the generation of virus-specific CD4 and CD8 T cells and antibody responses following respiratory influenza virus infection.112 These observations, combined with our knowledge of the mechanisms underlying the gastrointestinal afferent and systemic efferent mechanisms of oral tolerance, support the concept that the gastrointestinal and pulmonary mucosa both also respond to inhaled antigens and generate cross-regulatory immunity. It remains to be determined how these distal mucosal sites interact in generating mucosal immunity.
Concluding remarks
The maintenance of mucosal homeostasis is a delicate balance between the host and the intestinal microbiome. The host employs numerous mechanisms to contain the intestinal microbiome and prevent the development of inappropriate inflammation. At the same time, however, the intestinal immune system requires microbial stimulation for its proper development. Conversely, while certain members of the microbiome can activate specific arms of host mucosal immunity, these host responses often prevent inappropriate overgrowth or translocation of members of the microbiome. Additionally, microbiome-driven host responses can also prevent the development of inappropriate inflammation. The end result is an equilibrium state, where microbial stimulation promotes normal immune function in the intestine, which in turn allows the intestinal microbiome to flourish in the absence of unnecessary inflammation. Further investigation of these complex host–microbiome interactions will undoubtedly reveal new mechanisms underlying the maintenance of mucosal homeostasis and the development of inflammation in the intestines and distal mucosal sites.
Acknowledgments
The authors would like to acknowledge Dr Merritt Gillilland III for critical review of this manuscript.
Glossary
- FAE
follicle-associated epithelium
- GF
germ-free
- IELs
intraepithelial lymphocytes
- IFN-γ
interferon-γ
- IL-17
interleukin-17
- ILCs
innate lymphoid cells
- LPDC
lamina propria dendritic cell
- SCFA
short chain fatty acids
- SFB
segmented filamentous bacteria
- Th17
T helper type 17
- TLR
toll-like receptor
Disclosure
The authors declare no conflict of interest.
References
- 1.Vereecke L, Beyaert R, Loo van G. Enterocyte death and intestinal barrier maintenance in homeostasis and disease. Trends Mol Med. 2011;17:584–93. doi: 10.1016/j.molmed.2011.05.011. [DOI] [PubMed] [Google Scholar]
- 2.Abreu MT. Toll-like receptor signalling in the intestinal epithelium: how bacterial recognition shapes intestinal function. Nat Rev Immunol. 2010;10:131–44. doi: 10.1038/nri2707. [DOI] [PubMed] [Google Scholar]
- 3.Rio del ML, Bernhardt G, Rodriguez-Barbosa JI, Forster R. Development and functional specialization of CD103+ dendritic cells. Immunol Rev. 2010;234:268–81. doi: 10.1111/j.0105-2896.2009.00874.x. [DOI] [PubMed] [Google Scholar]
- 4.Sun CM, Hall JA, Blank RB, Bouladoux N, Oukka M, Mora JR, Belkaid Y. Small intestine lamina propria dendritic cells promote de novo generation of Foxp3 Treg cells via retinoic acid. J Exp Med. 2007;204:1775–85. doi: 10.1084/jem.20070602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Artis D. Epithelial-cell recognition of commensal bacteria and maintenance of immune homeostasis in the gut. Nat Rev Immunol. 2008;8:411–20. doi: 10.1038/nri2316. [DOI] [PubMed] [Google Scholar]
- 6.Yen TH, Wright NA. The gastrointestinal tract stem cell niche. Stem Cell Rev. 2006;2:203–12. doi: 10.1007/s12015-006-0048-1. [DOI] [PubMed] [Google Scholar]
- 7.Audie JP, Janin A, Porchet N, Copin MC, Gosselin B, Aubert JP. Expression of human mucin genes in respiratory, digestive, and reproductive tracts ascertained by in situ hybridization. J Histochem Cytochem. 1993;41:1479–85. doi: 10.1177/41.10.8245407. [DOI] [PubMed] [Google Scholar]
- 8.Matsuo K, Ota H, Akamatsu T, Sugiyama A, Katsuyama T. Histochemistry of the surface mucous gel layer of the human colon. Gut. 1997;40:782–9. doi: 10.1136/gut.40.6.782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kleinschek MA, Boniface K, Sadekova S, et al. Circulating and gut-resident human Th17 cells express CD161 and promote intestinal inflammation. J Exp Med. 2009;206:525–34. doi: 10.1084/jem.20081712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Makita S, Kanai T, Nemoto Y, et al. Intestinal lamina propria retaining CD4+ CD25+ regulatory T cells is a suppressive site of intestinal inflammation. J Immunol. 2007;178:4937–46. doi: 10.4049/jimmunol.178.8.4937. [DOI] [PubMed] [Google Scholar]
- 11.Makita S, Kanai T, Oshima S, et al. CD4+ CD25bright T cells in human intestinal lamina propria as regulatory cells. J Immunol. 2004;173:3119–30. doi: 10.4049/jimmunol.173.5.3119. [DOI] [PubMed] [Google Scholar]
- 12.Munoz M, Heimesaat MM, Danker K, et al. Interleukin (IL)-23 mediates Toxoplasma gondii-induced immunopathology in the gut via matrixmetalloproteinase-2 and IL-22 but independent of IL-17. J Exp Med. 2009;206:3047–59. doi: 10.1084/jem.20090900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bailey M, Plunkett FJ, Rothkotter HJ, Vega-Lopez MA, Haverson K, Stokes CR. Regulation of mucosal immune responses in effector sites. Proc Nutr Soc. 2001;60:427–35. doi: 10.1079/pns2001118. [DOI] [PubMed] [Google Scholar]
- 14.Brandtzaeg P, Pabst R. Let's go mucosal: communication on slippery ground. Trends Immunol. 2004;25:570–7. doi: 10.1016/j.it.2004.09.005. [DOI] [PubMed] [Google Scholar]
- 15.Lycke NY, Bemark M. The role of Peyer's patches in synchronizing gut IgA responses. Front Immunol. 2012;3:329. doi: 10.3389/fimmu.2012.00329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Turnbaugh PJ, Ley RE, Hamady M, Fraser-Liggett CM, Knight R, Gordon JI. The human microbiome project. Nature. 2007;449:804–10. doi: 10.1038/nature06244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kau AL, Ahern PP, Griffin NW, Goodman AL, Gordon JI. Human nutrition, the gut microbiome and the immune system. Nature. 2011;474:327–36. doi: 10.1038/nature10213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Topping DL, Clifton PM. Short-chain fatty acids and human colonic function: roles of resistant starch and nonstarch polysaccharides. Physiol Rev. 2001;81:1031–64. doi: 10.1152/physrev.2001.81.3.1031. [DOI] [PubMed] [Google Scholar]
- 19.Hooper LV. Bacterial contributions to mammalian gut development. Trends Microbiol. 2004;12:129–34. doi: 10.1016/j.tim.2004.01.001. [DOI] [PubMed] [Google Scholar]
- 20.Pollard M, Sharon N. Responses of the Peyer's patches in germ-free mice to antigenic stimulation. Infect Immun. 1970;2:96–100. doi: 10.1128/iai.2.1.96-100.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ivanov II, Atarashi K, Manel N, et al. Induction of intestinal Th17 cells by segmented filamentous bacteria. Cell. 2009;139:485–98. doi: 10.1016/j.cell.2009.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ivanov II, Frutos Rde L, Manel N, Yoshinaga K, Rifkin DB, Sartor RB, Finlay BB, Littman DR. Specific microbiota direct the differentiation of IL-17-producing T-helper cells in the mucosa of the small intestine. Cell Host Microbe. 2008;4:337–49. doi: 10.1016/j.chom.2008.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mazmanian SK, Round JL, Kasper DL. A microbial symbiosis factor prevents intestinal inflammatory disease. Nature. 2008;453:620–5. doi: 10.1038/nature07008. [DOI] [PubMed] [Google Scholar]
- 24.Round JL, Mazmanian SK. Inducible Foxp3+ regulatory T-cell development by a commensal bacterium of the intestinal microbiota. Proc Natl Acad Sci U S A. 2010;107:12204–9. doi: 10.1073/pnas.0909122107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Farre R, Tack J. Food and symptom generation in functional gastrointestinal disorders: physiological aspects. Am J Gastroenterol. 2013;108:698–706. doi: 10.1038/ajg.2013.24. [DOI] [PubMed] [Google Scholar]
- 26.Newberry RD, Lorenz RG. Organizing a mucosal defense. Immunol Rev. 2005;206:6–21. doi: 10.1111/j.0105-2896.2005.00282.x. [DOI] [PubMed] [Google Scholar]
- 27.Sipos F, Muzes G. Isolated lymphoid follicles in colon: switch points between inflammation and colorectal cancer? World J Gastroenterol. 2011;17:1666–73. doi: 10.3748/wjg.v17.i13.1666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chang WW, Leblond CP. Renewal of the epithelium in the descending colon of the mouse. I. Presence of three cell populations: vacuolated-columnar, mucous and argentaffin. Am J Anat. 1971;131:73–99. doi: 10.1002/aja.1001310105. [DOI] [PubMed] [Google Scholar]
- 29.Gewirtz AT, Navas TA, Lyons S, Godowski PJ, Madara JL. Cutting edge: bacterial flagellin activates basolaterally expressed TLR5 to induce epithelial proinflammatory gene expression. J Immunol. 2001;167:1882–5. doi: 10.4049/jimmunol.167.4.1882. [DOI] [PubMed] [Google Scholar]
- 30.Yang SK, Eckmann L, Panja A, Kagnoff MF. Differential and regulated expression of C-X-C, C-C, and C-chemokines by human colon epithelial cells. Gastroenterology. 1997;113:1214–23. doi: 10.1053/gast.1997.v113.pm9322516. [DOI] [PubMed] [Google Scholar]
- 31.Awane M, Andres PG, Li DJ, Reinecker HC. NF-κB-inducing kinase is a common mediator of IL-17-, TNF-α-, and IL-1β-induced chemokine promoter activation in intestinal epithelial cells. J Immunol. 1999;162:5337–44. [PubMed] [Google Scholar]
- 32.Iimura M, Gallo RL, Hase K, Miyamoto Y, Eckmann L, Kagnoff MF. Cathelicidin mediates innate intestinal defense against colonization with epithelial adherent bacterial pathogens. J Immunol. 2005;174:4901–7. doi: 10.4049/jimmunol.174.8.4901. [DOI] [PubMed] [Google Scholar]
- 33.Mayer L, Shlien R. Evidence for function of Ia molecules on gut epithelial cells in man. J Exp Med. 1987;166:1471–83. doi: 10.1084/jem.166.5.1471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Parr EL, Kirby WN. An immunoferritin labeling study of H-2 antigens on dissociated epithelial cells. J Histochem Cytochem. 1979;27:1327–36. doi: 10.1177/27.10.390033. [DOI] [PubMed] [Google Scholar]
- 35.Cheroutre H, Lambolez F, Mucida D. The light and dark sides of intestinal intraepithelial lymphocytes. Nat Rev Immunol. 2011;11:445–56. doi: 10.1038/nri3007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Resendiz-Albor AA, Esquivel R, Lopez-Revilla R, Verdin L, Moreno-Fierros L. Striking phenotypic and functional differences in lamina propria lymphocytes from the large and small intestine of mice. Life Sci. 2005;76:2783–803. doi: 10.1016/j.lfs.2004.08.042. [DOI] [PubMed] [Google Scholar]
- 37.Cepek KL, Shaw SK, Parker CM, Russell GJ, Morrow JS, Rimm DL, Brenner MB. Adhesion between epithelial cells and T lymphocytes mediated by E-cadherin and the αEβ7 integrin. Nature. 1994;372:190–3. doi: 10.1038/372190a0. [DOI] [PubMed] [Google Scholar]
- 38.Wang HC, Zhou Q, Dragoo J, Klein JR. Most murine CD8+ intestinal intraepithelial lymphocytes are partially but not fully activated T cells. J Immunol. 2002;169:4717–22. doi: 10.4049/jimmunol.169.9.4717. [DOI] [PubMed] [Google Scholar]
- 39.Moretto M, Weiss LM, Khan IA. Induction of a rapid and strong antigen-specific intraepithelial lymphocyte response during oral Encephalitozoon cuniculi infection. J Immunol. 2004;172:4402–9. doi: 10.4049/jimmunol.172.7.4402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Boismenu R, Havran WL. Modulation of epithelial cell growth by intraepithelial γδ T cells. Science. 1994;266:1253–5. doi: 10.1126/science.7973709. [DOI] [PubMed] [Google Scholar]
- 41.Chen Y, Chou K, Fuchs E, Havran WL, Boismenu R. Protection of the intestinal mucosa by intraepithelial γδ T cells. Proc Natl Acad Sci U S A. 2002;99:14338–43. doi: 10.1073/pnas.212290499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Simpson SJ, Hollander GA, Mizoguchi E, Allen D, Bhan AK, Wang B, Terhorst C. Expression of pro-inflammatory cytokines by TCR αβ+ and TCR γδ+ T cells in an experimental model of colitis. Eur J Immunol. 1997;27:17–25. doi: 10.1002/eji.1830270104. [DOI] [PubMed] [Google Scholar]
- 43.MacDonald TT, Pender SL. Lamina propria T cells. Chem Immunol. 1998;71:103–17. doi: 10.1159/000058721. [DOI] [PubMed] [Google Scholar]
- 44.Carol M, Lambrechts A, Gossum Van A, Libin M, Goldman M, Mascart-Lemone F. Spontaneous secretion of interferon γ and interleukin 4 by human intraepithelial and lamina propria gut lymphocytes. Gut. 1998;42:643–9. doi: 10.1136/gut.42.5.643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Niess JH, Brand S, Gu X, et al. CX3CR1-mediated dendritic cell access to the intestinal lumen and bacterial clearance. Science. 2005;307:254–8. doi: 10.1126/science.1102901. [DOI] [PubMed] [Google Scholar]
- 46.Rio del ML, Rodriguez-Barbosa JI, Bolter J, Ballmaier M, Dittrich-Breiholz O, Kracht M, Jung S, Forster R. CX3CR1+ c-kit+ bone marrow cells give rise to CD103+ and CD103– dendritic cells with distinct functional properties. J Immunol. 2008;181:6178–88. doi: 10.4049/jimmunol.181.9.6178. [DOI] [PubMed] [Google Scholar]
- 47.Varol C, Vallon-Eberhard A, Elinav E, et al. Intestinal lamina propria dendritic cell subsets have different origin and functions. Immunity. 2009;31:502–12. doi: 10.1016/j.immuni.2009.06.025. [DOI] [PubMed] [Google Scholar]
- 48.Satoh-Takayama N, Vosshenrich CA, Lesjean-Pottier S, et al. Microbial flora drives interleukin 22 production in intestinal NKp46+ cells that provide innate mucosal immune defense. Immunity. 2008;29:958–70. doi: 10.1016/j.immuni.2008.11.001. [DOI] [PubMed] [Google Scholar]
- 49.Buonocore S, Ahern PP, Uhlig HH, Ivanov II, Littman DR, Maloy KJ, Powrie F. Innate lymphoid cells drive interleukin-23-dependent innate intestinal pathology. Nature. 2010;464:1371–5. doi: 10.1038/nature08949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Spits H, Artis D, Colonna M, et al. Innate lymphoid cells – a proposal for uniform nomenclature. Nat Rev Immunol. 2013;13:145–9. doi: 10.1038/nri3365. [DOI] [PubMed] [Google Scholar]
- 51.Bernink JH, Peters CP, Munneke M, et al. Human type 1 innate lymphoid cells accumulate in inflamed mucosal tissues. Nat Immunol. 2013;14:221–9. doi: 10.1038/ni.2534. [DOI] [PubMed] [Google Scholar]
- 52.Zheng Y, Valdez PA, Danilenko DM, et al. Interleukin-22 mediates early host defense against attaching and effacing bacterial pathogens. Nat Med. 2008;14:282–9. doi: 10.1038/nm1720. [DOI] [PubMed] [Google Scholar]
- 53.Iwasaki A, Kelsall BL. Localization of distinct Peyer's patch dendritic cell subsets and their recruitment by chemokines macrophage inflammatory protein (MIP)-3α, MIP-3β, and secondary lymphoid organ chemokine. J Exp Med. 2000;191:1381–94. doi: 10.1084/jem.191.8.1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kraehenbuhl JP, Neutra MR. Epithelial M cells: differentiation and function. Annu Rev Cell Dev Biol. 2000;16:301–32. doi: 10.1146/annurev.cellbio.16.1.301. [DOI] [PubMed] [Google Scholar]
- 55.Owen RL, Pierce NF, Apple RT, Cray WC., Jr M cell transport of Vibrio cholerae from the intestinal lumen into Peyer's patches: a mechanism for antigen sampling and for microbial transepithelial migration. J Infect Dis. 1986;153:1108–18. doi: 10.1093/infdis/153.6.1108. [DOI] [PubMed] [Google Scholar]
- 56.Butcher EC, Rouse RV, Coffman RL, Nottenburg CN, Hardy RR, Weissman IL. Surface phenotype of Peyer's patch germinal center cells: implications for the role of germinal centers in B cell differentiation. J Immunol. 1982;129:2698–707. [PubMed] [Google Scholar]
- 57.Massacand JC, Kaiser P, Ernst B, Tardivel A, Burki K, Schneider P, Harris NL. Intestinal bacteria condition dendritic cells to promote IgA production. PLoS ONE. 2008;3:e2588. doi: 10.1371/journal.pone.0002588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hamada H, Hiroi T, Nishiyama Y, et al. Identification of multiple isolated lymphoid follicles on the antimesenteric wall of the mouse small intestine. J Immunol. 2002;168:57–64. doi: 10.4049/jimmunol.168.1.57. [DOI] [PubMed] [Google Scholar]
- 59.Lorenz RG, Chaplin DD, McDonald KG, McDonough JS, Newberry RD. Isolated lymphoid follicle formation is inducible and dependent upon lymphotoxin-sufficient B lymphocytes, lymphotoxin beta receptor, and TNF receptor I function. J Immunol. 2003;170:5475–82. doi: 10.4049/jimmunol.170.11.5475. [DOI] [PubMed] [Google Scholar]
- 60.Lorenz RG, Newberry RD. Isolated lymphoid follicles can function as sites for induction of mucosal immune responses. Ann N Y Acad Sci. 2004;1029:44–57. doi: 10.1196/annals.1309.006. [DOI] [PubMed] [Google Scholar]
- 61.Macpherson AJ, Slack E. The functional interactions of commensal bacteria with intestinal secretory IgA. Curr Opin Gastroenterol. 2007;23:673–8. doi: 10.1097/MOG.0b013e3282f0d012. [DOI] [PubMed] [Google Scholar]
- 62.Cerutti A. Location, location, location: B-cell differentiation in the gut lamina propria. Mucosal Immunol. 2008;1:8–10. doi: 10.1038/mi.2007.8. [DOI] [PubMed] [Google Scholar]
- 63.Frankel G, Phillips AD, Novakova M, Field H, Candy DC, Schauer DB, Douce G, Dougan G. Intimin from enteropathogenic Escherichia coli restores murine virulence to a Citrobacter rodentium eaeA mutant: induction of an immunoglobulin A response to intimin and EspB. Infect Immun. 1996;64:5315–25. doi: 10.1128/iai.64.12.5315-5325.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Suzuki K, Meek B, Doi Y, Muramatsu M, Chiba T, Honjo T, Fagarasan S. Aberrant expansion of segmented filamentous bacteria in IgA-deficient gut. Proc Natl Acad Sci U S A. 2004;101:1981–6. doi: 10.1073/pnas.0307317101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Karam SM. Lineage commitment and maturation of epithelial cells in the gut. Front Biosci. 1999;4:D286–98. doi: 10.2741/karam. [DOI] [PubMed] [Google Scholar]
- 66.Johansson ME, Phillipson M, Petersson J, Velcich A, Holm L, Hansson GC. The inner of the two Muc2 mucin-dependent mucus layers in colon is devoid of bacteria. Proc Natl Acad Sci U S A. 2008;105:15064–9. doi: 10.1073/pnas.0803124105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Forman RA, deSchoolmeester ML, Hurst RJ, Wright SH, Pemberton AD, Else KJ. The goblet cell is the cellular source of the anti-microbial angiogenin 4 in the large intestine post Trichuris muris infection. PLoS ONE. 2012;7:e42248. doi: 10.1371/journal.pone.0042248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Burger-van Paassen N, Loonen LM, Witte-Bouma J, et al. Mucin Muc2 deficiency and weaning influences the expression of the innate defense genes Reg3β, Reg3γ and angiogenin-4. PLoS ONE. 2012;7:e38798. doi: 10.1371/journal.pone.0038798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Vaishnava S, Yamamoto M, Severson KM, et al. The antibacterial lectin RegIIIγ promotes the spatial segregation of microbiota and host in the intestine. Science. 2011;334:255–8. doi: 10.1126/science.1209791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.McDole JR, Wheeler LW, McDonald KG, Wang B, Konjufca V, Knoop KA, Newberry RD, Miller MJ. Goblet cells deliver luminal antigen to CD103+ dendritic cells in the small intestine. Nature. 2012;483:345–9. doi: 10.1038/nature10863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.O'Hara AM, Shanahan F. The gut flora as a forgotten organ. EMBO Rep. 2006;7:688–93. doi: 10.1038/sj.embor.7400731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Bandeira A, Mota-Santos T, Itohara S, Degermann S, Heusser C, Tonegawa S, Coutinho A. Localization of γ/δ T cells to the intestinal epithelium is independent of normal microbial colonization. J Exp Med. 1990;172:239–44. doi: 10.1084/jem.172.1.239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Crabbe PA, Bazin H, Eyssen H, Heremans JF. The normal microbial flora as a major stimulus for proliferation of plasma cells synthesizing IgA in the gut. The germ-free intestinal tract. Int Arch Allergy Appl Immunol. 1968;34:362–75. doi: 10.1159/000230130. [DOI] [PubMed] [Google Scholar]
- 74.Moreau MC, Corthier G. Effect of the gastrointestinal microflora on induction and maintenance of oral tolerance to ovalbumin in C3H/HeJ mice. Infect Immun. 1988;56:2766–8. doi: 10.1128/iai.56.10.2766-2768.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Sudo N, Sawamura S, Tanaka K, Aiba Y, Kubo C, Koga Y. The requirement of intestinal bacterial flora for the development of an IgE production system fully susceptible to oral tolerance induction. J Immunol. 1997;159:1739–45. [PubMed] [Google Scholar]
- 76.Ostman S, Rask C, Wold AE, Hultkrantz S, Telemo E. Impaired regulatory T cell function in germ-free mice. Eur J Immunol. 2006;36:2336–46. doi: 10.1002/eji.200535244. [DOI] [PubMed] [Google Scholar]
- 77.Umesaki Y, Setoyama H, Matsumoto S, Okada Y. Expansion of αβ T-cell receptor-bearing intestinal intraepithelial lymphocytes after microbial colonization in germ-free mice and its independence from thymus. Immunology. 1993;79:32–7. [PMC free article] [PubMed] [Google Scholar]
- 78.Round JL, Lee SM, Li J, Tran G, Jabri B, Chatila TA, Mazmanian SK. The Toll-like receptor 2 pathway establishes colonization by a commensal of the human microbiota. Science. 2011;332:974–7. doi: 10.1126/science.1206095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Shen Y, Giardino Torchia ML, Lawson GW, Karp CL, Ashwell JD, Mazmanian SK. Outer membrane vesicles of a human commensal mediate immune regulation and disease protection. Cell Host Microbe. 2012;12:509–20. doi: 10.1016/j.chom.2012.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Wieland Brown LC, Penaranda C, Kashyap PC, et al. Production of α-galactosylceramide by a prominent member of the human gut microbiota. PLoS Biol. 2013;11:e1001610. doi: 10.1371/journal.pbio.1001610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Hooper LV, Wong MH, Thelin A, Hansson L, Falk PG, Gordon JI. Molecular analysis of commensal host–microbial relationships in the intestine. Science. 2001;291:881–4. doi: 10.1126/science.291.5505.881. [DOI] [PubMed] [Google Scholar]
- 82.Cash HL, Whitham CV, Behrendt CL, Hooper LV. Symbiotic bacteria direct expression of an intestinal bactericidal lectin. Science. 2006;313:1126–30. doi: 10.1126/science.1127119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Zelante T, Iannitti RG, Cunha C, et al. Tryptophan catabolites from microbiota engage aryl hydrocarbon receptor and balance mucosal reactivity via interleukin-22. Immunity. 2013;39:372–85. doi: 10.1016/j.immuni.2013.08.003. [DOI] [PubMed] [Google Scholar]
- 84.Talham GL, Jiang HQ, Bos NA, Cebra JJ. Segmented filamentous bacteria are potent stimuli of a physiologically normal state of the murine gut mucosal immune system. Infect Immun. 1999;67:1992–2000. doi: 10.1128/iai.67.4.1992-2000.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Atarashi K, Tanoue T, Shima T, et al. Induction of colonic regulatory T cells by indigenous Clostridium species. Science. 2011;331:337–41. doi: 10.1126/science.1198469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Atarashi K, Tanoue T, Oshima K, et al. Treg induction by a rationally selected mixture of Clostridia strains from the human microbiota. Nature. 2013;500:232–6. doi: 10.1038/nature12331. [DOI] [PubMed] [Google Scholar]
- 87.Louis P, Duncan SH, McCrae SI, Millar J, Jackson MS, Flint HJ. Restricted distribution of the butyrate kinase pathway among butyrate-producing bacteria from the human colon. J Bacteriol. 2004;186:2099–106. doi: 10.1128/JB.186.7.2099-2106.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Bohmig GA, Krieger PM, Saemann MD, Wenhardt C, Pohanka E, Zlabinger GJ. n-Butyrate downregulates the stimulatory function of peripheral blood-derived antigen-presenting cells: a potential mechanism for modulating T-cell responses by short-chain fatty acids. Immunology. 1997;92:234–43. doi: 10.1046/j.1365-2567.1997.00337.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Saemann MD, Bohmig GA, Osterreicher CH, et al. Anti-inflammatory effects of sodium butyrate on human monocytes: potent inhibition of IL-12 and up-regulation of IL-10 production. FASEB J. 2000;14:2380–2. doi: 10.1096/fj.00-0359fje. [DOI] [PubMed] [Google Scholar]
- 90.Cavaglieri CR, Nishiyama A, Fernandes LC, Curi R, Miles EA, Calder PC. Differential effects of short-chain fatty acids on proliferation and production of pro-and anti-inflammatory cytokines by cultured lymphocytes. Life Sci. 2003;73:1683–90. doi: 10.1016/s0024-3205(03)00490-9. [DOI] [PubMed] [Google Scholar]
- 91.Saemann MD, Parolini O, Bohmig GA, et al. Bacterial metabolite interference with maturation of human monocyte-derived dendritic cells. J Leukoc Biol. 2002;71:238–46. [PubMed] [Google Scholar]
- 92.Venkatraman A, Ramakrishna BS, Pulimood AB, Patra S, Murthy S. Increased permeability in dextran sulphate colitis in rats: time course of development and effect of butyrate. Scand J Gastroenterol. 2000;35:1053–9. doi: 10.1080/003655200451171. [DOI] [PubMed] [Google Scholar]
- 93.Okamoto T, Sasaki M, Tsujikawa T, Fujiyama Y, Bamba T, Kusunoki M. Preventive efficacy of butyrate enemas and oral administration of Clostridium butyricum M588 in dextran sodium sulfate-induced colitis in rats. J Gastroenterol. 2000;35:341–6. doi: 10.1007/s005350050358. [DOI] [PubMed] [Google Scholar]
- 94.Maslowski KM, Vieira AT, Ng A, et al. Regulation of inflammatory responses by gut microbiota and chemoattractant receptor GPR43. Nature. 2009;461:1282–6. doi: 10.1038/nature08530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Millard AL, Mertes PM, Ittelet D, Villard F, Jeannesson P, Bernard J. Butyrate affects differentiation, maturation and function of human monocyte-derived dendritic cells and macrophages. Clin Exp Immunol. 2002;130:245–55. doi: 10.1046/j.0009-9104.2002.01977.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Macho Fernandez E, Valenti V, Rockel C, Hermann C, Pot B, Boneca IG, Grangette C. Anti-inflammatory capacity of selected lactobacilli in experimental colitis is driven by NOD2-mediated recognition of a specific peptidoglycan-derived muropeptide. Gut. 2011;60:1050–9. doi: 10.1136/gut.2010.232918. [DOI] [PubMed] [Google Scholar]
- 97.Tien MT, Girardin SE, Regnault B, et al. Anti-inflammatory effect of Lactobacillus casei on Shigella-infected human intestinal epithelial cells. J Immunol. 2006;176:1228–37. doi: 10.4049/jimmunol.176.2.1228. [DOI] [PubMed] [Google Scholar]
- 98.Wullt M, Johansson Hagslatt ML, Odenholt I, Berggren A. Lactobacillus plantarum 299v enhances the concentrations of fecal short-chain fatty acids in patients with recurrent Clostridium difficile-associated diarrhea. Dig Dis Sci. 2007;52:2082–6. doi: 10.1007/s10620-006-9123-3. [DOI] [PubMed] [Google Scholar]
- 99.Tsukahara T, Koyama H, Okada M, Ushida K. Stimulation of butyrate production by gluconic acid in batch culture of pig cecal digesta and identification of butyrate-producing bacteria. J Nutr. 2002;132:2229–34. doi: 10.1093/jn/132.8.2229. [DOI] [PubMed] [Google Scholar]
- 100.Kimura A, Naka T, Nohara K, Fujii-Kuriyama Y, Kishimoto T. Aryl hydrocarbon receptor regulates Stat1 activation and participates in the development of Th17 cells. Proc Natl Acad Sci U S A. 2008;105:9721–6. doi: 10.1073/pnas.0804231105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Lee JS, Cella M, McDonald KG, et al. AHR drives the development of gut ILC22 cells and postnatal lymphoid tissues via pathways dependent on and independent of Notch. Nat Immunol. 2012;13:144–51. doi: 10.1038/ni.2187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Qiu J, Guo X, Chen ZM, He L, Sonnenberg GF, Artis D, Fu YX, Zhou L. Group 3 innate lymphoid cells inhibit T-cell-mediated intestinal inflammation through aryl hydrocarbon receptor signaling and regulation of microflora. Immunity. 2013;39:386–99. doi: 10.1016/j.immuni.2013.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Eyles JE, Spiers ID, Williamson ED, Alpar HO. Tissue distribution of radioactivity following intranasal administration of radioactive microspheres. J Pharm Pharmacol. 2001;53:601–7. doi: 10.1211/0022357011775929. [DOI] [PubMed] [Google Scholar]
- 104.Pickett TE, Pasetti MF, Galen JE, Sztein MB, Levine MM. In vivo characterization of the murine intranasal model for assessing the immunogenicity of attenuated Salmonella enterica serovar Typhi strains as live mucosal vaccines and as live vectors. Infect Immun. 2000;68:205–13. doi: 10.1128/iai.68.1.205-213.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Southam DS, Dolovich M, O'Byrne PM, Inman MD. Distribution of intranasal instillations in mice: effects of volume, time, body position, and anesthesia. Am J Physiol Lung Cell Mol Physiol. 2002;282:L833–9. doi: 10.1152/ajplung.00173.2001. [DOI] [PubMed] [Google Scholar]
- 106.Lambrecht BN, Pauwels RA, Fazekas De St Groth B. Induction of rapid T cell activation, division, and recirculation by intratracheal injection of dendritic cells in a TCR transgenic model. J Immunol. 2000;164:2937–46. doi: 10.4049/jimmunol.164.6.2937. [DOI] [PubMed] [Google Scholar]
- 107.Noverr MC, Falkowski NR, McDonald RA, McKenzie AN, Huffnagle GB. Development of allergic airway disease in mice following antibiotic therapy and fungal microbiota increase: role of host genetics, antigen, and interleukin-13. Infect Immun. 2005;73:30–8. doi: 10.1128/IAI.73.1.30-38.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Noverr MC, Noggle RM, Toews GB, Huffnagle GB. Role of antibiotics and fungal microbiota in driving pulmonary allergic responses. Infect Immun. 2004;72:4996–5003. doi: 10.1128/IAI.72.9.4996-5003.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Hisbergues M, Magi M, Rigaux P, et al. In vivo and in vitro immunomodulation of Der p 1 allergen-specific response by Lactobacillus plantarum bacteria. Clin Exp Allergy. 2007;37:1286–95. doi: 10.1111/j.1365-2222.2007.02792.x. [DOI] [PubMed] [Google Scholar]
- 110.Ezendam J, Loveren van H. Lactobacillus casei Shirota administered during lactation increases the duration of autoimmunity in rats and enhances lung inflammation in mice. Br J Nutr. 2008;99:83–90. doi: 10.1017/S0007114507803412. [DOI] [PubMed] [Google Scholar]
- 111.Forsythe P, Inman MD, Bienenstock J. Oral treatment with live Lactobacillus reuteri inhibits the allergic airway response in mice. Am J Respir Crit Care Med. 2007;175:561–9. doi: 10.1164/rccm.200606-821OC. [DOI] [PubMed] [Google Scholar]
- 112.Ichinohe T, Pang IK, Kumamoto Y, Peaper DR, Ho JH, Murray TS, Iwasaki A. Microbiota regulates immune defense against respiratory tract influenza A virus infection. Proc Natl Acad Sci U S A. 2011;108:5354–9. doi: 10.1073/pnas.1019378108. [DOI] [PMC free article] [PubMed] [Google Scholar]


