Abstract
MicroRNAs (miRNAs) have emerged as critical regulators of gene expression within cells. One particular miRNA, miR-155, is highly expressed within lymphocytes (both B and T cells) and mediates a number of important roles. These include shaping the transcriptome of lymphoid cells that control diverse biological functions vital in adaptive immunity. The use of mice engineered to be deficient in miR-155, as well as the identification of endogenous targets of miR-155 in T cells by transcriptome-wide analysis, has helped to unravel the crucial role that this miRNA plays in fine tuning the regulation of lymphocyte subsets such as B cells, CD8+ and CD4+ T cells ranging from T helper type 1 (Th1), Th2, Th17 and regulatory T cells. In this review, we summarize what we have learned about miR-155 in the regulation of lymphocyte responses at the cellular and molecular levels and in particular, we focus on the recent findings showing that miR-155 shapes the balance between tolerance and immunity.
Keywords: experimental autoimmune encephalomyelitis/multiple sclerosis, regulatory T cells, T cells, T helper type 1/type2 cells
Introduction
MicroRNAs (miRNAs) are small RNAs (19–22 nucleotides in length) that have vitally important roles in fine-tuning gene expression within cells. Within the immune system, they have also been shown to have critical roles in the developmental fate of lymphocytes and also in innate and adaptive immunity in general.1,2 One miRNA with an emerging role in regulating immune responses is miR-155. miR-155 is derived from the non-coding transcript of the proto-oncogene bic (B-cell integration cluster) gene and represents the only evolutionarily conserved sequence of this gene, suggesting that miR-155 has an important role in bic function. miR-155, like other miRNAs, works by binding to the 3′ untranslated region of mRNA via a protein complex called the RNA-induced silencing complex to effect either mRNA degradation or translational repression.3 A shorter seven nucleotide ‘seed’ region (nucleotides 2–8 from the 5′ end) within the miRNA determines which 3′ untranslated region of mRNAs can be targeted. Like other miRNAs, miR-155 has hundreds of potential target genes that it can regulate. Interestingly, using a technique that measured transcriptome-wide identification of endogenous targets of miR-155 in T cells revealed that up to 40% of miR-155 targets occurred at sites that did not have a perfect seed match.4 Although this approach together with the use of mice deficient in bic/miR155 have been very useful to unravel the important role of miR-155 in the function of specific lymphocyte populations, we need to unequivocally demonstrate whether miR-155 has a role in lymphocyte development.
Several studies showed that miR-155 controls differentiation of CD4+ T cells into the T helper type 1 (Th1), Th2 and Th17 subsets of helper T cells2,5,6 and that it affects the development of regulatory T (Treg) cells.7,8 miR-155 also regulates CD8+ T cells9,10 and is essential for normal B-cell differentiation and antibody production.5,6,11 As miRNAs are important regulators of T-cell immunity, and miR-155 particularly has received a great deal of attention in many ways due to its role in controlling protective immune responses and inflammation, we review here the literature that is available about miR-155-mediated regulation of lymphocyte subsets. We will report the role of miR-155 in adaptive immunity and summarize its role in various diseases ranging from haematological malignancies, cancers and viral infections to autoimmune diseases (Table 1).
Table 1.
Expression of miR-155 in different diseases. Main references are indicated
| Disease | Mouse | Human |
|---|---|---|
| Haematological malignancies | B-cell lymphoma39,40 | B-cell lymphoma (Hodgkin's,18,36,37 Burkitt's,37 DLBCL34,36 CLL34 |
| Cancers | Melanoma,47 breast cancer,48 ovarian cancer49 | |
| Viral infections | LCMV,47 influenza virus21 | HIV,50 HBV,53 HCV54 |
| Autoimmune disease | CIA,27 EAE2 | RA,55 MS,59 SLE,57 UC57 |
CIA, collagen-induced arthritis; CLL, chronic lymphocytic leukaemia; DLBCL, diffuse large cell B-cell lymphoma; EAE, experimental autoimmune encephalomyelitis; HBV, hepatitis B virus; HCV, hepatitis C virus; HIV, human immunodeficiency virus; LCMV, lymphocytic choriomeningitis virus; MS, multiple sclerosis; RA, rheumatoid arthritis; SLE, systemic lupus erythematosus; UC, ulcerative colitis.
The effect of miR-155 on lymphocyte biology
The miR-155 signature is specific to bone-marrow-derived haematopoietic cells in humans.12 In addition, the thymus exhibits the highest levels of miR-155 expression compared with other human tissues.13 miR-155 is also expressed in CD34+ cells isolated from peripheral blood or bone marrow and it was therefore hypothesized that miR-155 may inhibit their differentiation into a more mature cell type.14
Studies using mice deficient in bic/miR155 have revealed invaluable data on the role of miR-155 during haematopoiesis. miR-155 regulates both myelopoiesis14 and erythropoiesis.15,16 Mice deficient for bic show normal T-and B-lymphocyte subpopulation numbers and distribution.5 Natural killer cell numbers were however strikingly lower in the spleen as compared to wild-type mice.5
Importantly, multiple studies have demonstrated that miR-155 is consistently up-regulated following activation by CD3/CD28 co-stimulation in the case of lymphocytes, or by Toll-like receptor agonists in B cells and macrophages. These results suggest that bic/miR-155 may regulate the function of lymphocytes and myeloid cells.11,17
Microarray analysis of bic-deficient B and T cells under different conditions has revealed a wide spectrum of targets regulated by miR-155 and suggested mechanisms for the regulation of lymphocyte differentiation. miR-155 appears to be crucial during immune responses, inflammation and tumour development as further discussed below. However, the role of miR-155 in lymphocyte development, with the exception of Treg cells (as mentioned below), needs to be further explored.
miR-155 and adaptive immunity
miR-155 activity in B cells
Early studies have shown that miR-155 levels are up-regulated rapidly in both activated mature T and B cells18,19 but can also be up-regulated in germinal centre B cells,18 macrophages and dendritic cells. Mice lacking the bic/miR-155 gene were shown to produce significantly lower levels of specific antibody compared with control mice and also had germinal centres that were smaller than control mice.11 Other studies have confirmed that B-cell and T-cell responses are diminished in bic-deficient mice. Immunized mice produced significantly lower IgM and switched antigen-specific antibodies whereas T-cell responses were also shown to be defective in bic/miR-155-deficient mice. This was well demonstrated by failure to produce significant interferon-γ (IFN-γ) and interleukin-2 (IL-2) in immunized splenocytes from mutant mice (bic deficient) compared with wild-type mice.5 miR-155 has been shown to play important roles in immunoglobulin class switching via repression of activation-induced cytidine deaminases (AID) and the transcription factor PU.1.5,6,11,20
The role of miR-155 in CD8+ T cells
The role of miR-155 in CD8+ T cells has also been examined but a lot less is known about their functional relevance in this subset of lymphocytes. Like CD4+ T cells, in vitro stimulation of CD8+ T cells with anti-CD3 and anti-CD28 co-stimulation leads to a significant boost to miR-155 expression levels. Mice lacking miR-155 were found to have greatly reduced numbers of antigen-specific CD8+ T cells following a sub-lethal infection with an influenza virus compared with wild-type mice with normal levels of miR-155.21 Along with the reduced numbers of antigen-specific CD8+ T cells, there was also decreased viral clearance observed in the miR-155-deficient mice. Interestingly, over-expression of miR-155 led to improved antigen-specific CD8+ T-cell responses compared with the mice with normal levels of miR-155. Other studies have shown that mice lacking miR-155 have impaired primary responses to lymphocytic choriomeningitis virus and also to Listeria monocytogenes.9 In particular, there was a marked reduction in antigen-specific CD8+ T cells, as well as a functional defect with reduced ability to lyse targets using a cytotoxic T-lymphocyte assay. miR-155 also appears to be differentially expressed in CD8+ T-cell subsets, in particular sustained miR-155 levels are seen with effector/effector memory cells whereas lower levels of expression were seen with central memory CD8+ T cells.10 In the same study, mice engineered to lack miR-155 in the CD8+ T-cell compartment, showed much higher levels of viraemia (on exposure to a murine herpes virus). As with the CD4+ T cells discussed below, CD8+ T cells lacking miR-155 also had a lower frequency of cells that could produce IFN-γ.
Role of miR-155 in CD4+ T-cell subset differentiation and cytokine signalling
One important part of CD4+ T-cell differentiation is cytokine signal transduction and using a computational approach, targets of miR-155 that affected cytokine signal transduction were sought. The IFN-γRα chain was identified as a potential target of miR-155 and previous papers have shown that IFN-γ signalling through the IFN-γR is important in the differentiation of naive cells into Th1 cells.22,23 It was found that in Th1-inducing conditions, miR-155 was able to target IFN-γRα mRNA and therefore potentially affect T-cell differentiation through cytokine signalling.24
However, the interplay between miR-155 and cytokine/cytokine receptor signalling in CD4+ T cells remains complex. This is illustrated by papers showing that miR-155 is able to target Src homology-2 domain-containing inositol 5-phosphatase 1 (Ship1) in myeloid cells25 but also in T cells.26 In mice lacking miR-155, there were decreased numbers of IFN-γ-producing CD4+ T cells in the knockout mice and also Ship1 knockdown using small hairpin RNAs showed an increase in IFN-γ at the protein level in CD4+ T cells.26 This suggests that miR-155 can modulate IFN-γ levels through repression of Ship1.
Rodriguez et al. demonstrated that bic-deficient CD4+ T cells were intrinsically biased towards Th2 differentiation in view of higher numbers of IL-4-producing cells and enhanced levels of Th2 cytokines.5 The authors identified the transcription factor c-Maf (a potent transactivator of the IL-4 promoter) as a direct target of miR-155. However, the data suggested that bic/miR-155 was not required for Th1 differentiation even if Th1 cells exhibited an altered phenotype.5 These findings were confirmed by another group who found that there was a trend towards increased IL-4 levels but no significant difference in IFN-γ production in miR-155-deficient mice.27 Interestingly, Banerjee et al. showed that over-expression of miR-155 in activated CD4+ T cells promoted Th1 differentiation, although lacking miR-155 also led to a bias towards Th2 differentiation in CD4+ T cells.24 In addition, mice lacking bic/miR-155 also produced lower levels of tumour necrosis factor and lymphotoxin-α compared with control mice, with T cells from bic/miR-155-deficient mice skewing towards a Th2 phenotype.11
Role of miR-155 in Th1/Th17-mediated inflammation
A mouse model using an miR-155 knockout, demonstrated that following immunization with myelin oligodendrocyte glycoprotein (MOG), these mice were resistant to the development of experimental autoimmune encephalomyelitis (EAE), a murine model of multiple sclerosis (MS), compared with wild-type mice which expressed miR-155 and developed EAE following immunization with MOG.2 To further investigate the response to MOG, lymph node samples and splenocytes from these knockout mice were examined for the presence of IL-17 or IFN-γ (Th17 or Th1)-producing CD4+ T cells. Th17 cells were significantly reduced in both spleen and lymph nodes from miR-155 knockout mice whereas IFN-γ-producing Th1 CD4+ T cells were reduced in spleens but not lymph nodes of MOG-immunized mice lacking miR-155. The reduction in these cells was shown to be due to a defect in inflammatory T-cell development in the knockout mice.2 Other studies have demonstrated that mice lacking miR-155 have lower levels of Treg cells under normal physiological conditions,7,8 which were also seen in this study. Delayed type hypersensitivity responses were also shown to be impaired in mice lacking miR-155. Overall these studies have implicated miR-155 as having an important role in driving chronic inflammatory responses seen in a model of autoimmunity (EAE).
In another recent study, Hu et al. have investigated the role of miR-155 in differentiated Th17 cells.28 The authors have generated a mouse strain that allows for the isolation and analysis of viable miR-155−/− IL-17F-expressing CD4+ T cells that specifically recognize MOG35–55 antigen. Interestingly, they found that purified miR-155−/− Th17 cells were extremely defective in causing EAE following adoptive transfer when compared with wild-type controls. Furthermore, the authors showed that miR-155−/− Th17 cells are hypo-responsive to IL-23, revealing a new link between miR-155 and the highly relevant IL-23–IL-23R pathway.28 In accordance with this study, three others papers reported that the secretion of IL-17A by Th17 cells was positively regulated by miR-155.2,29,30
The role of miR-155 has also been studied in the mucosa and interestingly Das et al. revealed a transforming growth factor-β (TGF-β)-dependent role for miR-155 in modulating cytokine and T-cell immune responses in the gut.31 Consistent with the bioinformatics prediction that miR-155 targets the inducible T-cell kinase (itk) the authors demonstrated that over-expression of miR-155 in CD4+ T cells from lamina propria causes a decrease in itk mRNA, leading to a modest decline in IL-2 expression.31 Hence, they suggested that an excess of miR-155 in the presence of TGF-β might contribute to mucosal tolerance.
miR-155 and Treg cells
Although miR-155 is up-regulated in CD4+ T cells, its functional relevance in Treg cells still requires further work. Foxp3, the transcription factor that is a defining feature of Treg cells, appears to identify miR-155 (highly expressed in Treg cells) as a direct target of Foxp3. Using established protocols,32 we separated out naive Treg cells (CD25+ CD127low CD62L+ CD45RO−), effector/memory Treg cells (CD25+ CD127low CD62L+/− CD45RO+) and conventional naive CD4+ T cells (CD25− CD127high CD62L+ CD45RO−) from healthy donors and measured miR-155 levels in these subsets. We found miR-155 was strongly expressed in effector/memory Treg cells compared with lower expression levels in both naive Treg cells and naive CD4+ subsets.33 These data paralleled studies realized in miR-155-deficient mice showing reduced numbers of Treg cells in both the thymus and in the periphery, which was shown to be due to a defect in development.7 However, the suppressive function of Treg cells was not diminished in miR-155 knockout mice, nor was there any problem with bic/miR-155-deficient T cells to up-regulate Foxp3 under the right conditions. In line with the fact that miR-155 does not seem to play a role in Treg cell function, three independent studies reported that no significant differences were found in the production of IL-10 and TGF-β1 by Treg cells when miR-155 levels were modulated.2,29,30
Interestingly, there is evidence that miR-155 inhibits expression of the suppressor of cytokine signalling molecule (SOCS1).8 miR-155-deficient Treg cells resulted in increased SOCS1 expression, accompanied by impaired activation of signal transducer and activator of transcription 5 (STAT5) transcription factor in response to limiting amounts of IL-2, so leading to decreased Treg cell proliferation. Yao et al. reported that miR-155 contributes to the activation of IL-2/STAT5 and IL-6/STAT3 signalling pathways and the induction of Treg/Th17 cell differentiation.30 The authors suggested that miR-155 might be involved in modulating inflammatory diseases at least in part by regulating SOCS1.
miR-155 modulation in disease
The pivotal role of miR-155 for normal immune function was underlined by the studies of miR-155-deficient mice.5 These mice showed major disturbances in the function of B and T lymphocytes as has been outlined above. Moreover, these findings led to other important observations that described the role of miR-155 in various diseases, ranging from haematological malignancies, cancers and viral infections to autoimmune diseases.
Haematological malignancies
Over-expression of miR-155 is described in various B-cell lymphomas, such as Hodgkin's, Burkitt's and diffuse large cell B-cell lymphoma.18,34–38 Costinean et al. studied the role of miR-155 by producing the transgenic mice in which miR-155 is over expressed in B cells. These mice exhibited initially a preleukemic pre-B cell proliferation evident in spleen and bone marrow, followed by full-blown B-cell lymphomas.39 This suggested that miR-155 plays a major role in the early development of B cell lymphomas. More recently, Babar et al. produced an inducible knock-in model. As expected, induction of miR-155 in the lymphoid tissue led to disseminated lymphoma. Reduction of miR-155 led to the decrease in tumour size and other disease manifestations.40 In humans, Calin et al.41 studied extensively a microRNA signature associated with prognosis and progression in chronic lymphocytic leukaemia and found that miR-155 up-regulation is associated with poor prognosis. Interestingly, several lymphoma-associated viruses, including Epstein–Barr virus, Kaposi sarcoma-associated herpes virus and Marek's disease virus are associated with dysregulation of miR-155.42
Cancers
The oncogenic role of miR-155 is well established and described in several cancers including pancreatic, thyroid carcinoma, breast cancer, colon cancer and lung cancer where over-expression of miR-155 is generally linked with a poor prognosis.43–46 miR-155 regulates several pro-oncogenic pathways that are well described elsewhere43 and are beyond the scope of this review. From the immunological standpoint, the influence of miR-155 expression on anti-tumour response is underlined by several studies. Dudda et al. investigated the role of miR-155 in CD8+ T-cell anti-tumour response, using a mouse melanoma model.47 When compared with wild-type CD8+ T cells, mir-155−/− CD8+ T cells were not as effective in controlling the tumour growth and ensuring the longer survival of challenged mice. When melanoma antigen-specific CD8+ T cells were transduced with retrovirus encoding miRNA-155, resulting in increased miR-155 expression, they were much more effective in controlling the tumour growth than were cells transduced with scrambled RNA. Enhancing miR-155 expression within CD8+ T cells significantly prolonged the survival of tumour-challenged mice.47 miR-155 is also implicated in innate immune responses to tumours. Zonari et al. used a spontaneous breast cancer mouse model to assess the role of miR-155 primarily on innate immune responses.48 They found that knocking down miR-155 in the myeloid compartment accelerated tumour growth. Probable mechanisms included a decrease in the proportion of CD11c+ tumour-associated macrophages as well as their activation status. They also described a change of the cytokine milieu of the tumour toward an M2/Th2 phenotype.48 Cubillos-Ruiz et al. increased the activity of miR-155 in ovarian cancer-associated dendritic cells.49 They ‘fed’ them with nanoparticles loaded with Dicer substrate RNA duplexes that mimic the structure of endogenous precursor miR-155 hairpin molecules. This led to a complete shift in the dendritic cell phenotype that enabled a potent anti-tumour immune response.49 This study underlines a role of miR-155 in tumour immune dynamics and at the same time it proposes new strategies for safe induction of productive anti-tumour response.
Viral infections
Along with studying the effect of miR-155 on tumour-specific CD8+ T-cell responses, Dudda et al. extended their study on virally induced CD8+ T-cell responses employing both acute and chronic lymphocytic choriomeningitis virus infection models.47 Six to eight days after infecting the mice with lymphocytic choriomeningitis virus, they found an increase in effector CD8+ T cells expressing high levels of miR-155. This increase was highly dependent on the strength of T-cell receptor stimulation as high-affinity T-cell recptors induced higher levels of miR-155. Although mir155−/− CD8+ T cells were able to clear the virus, there was a major impairment of the robustness of the response, as well as the formation of central memory response.47 As noted earlier, Gracias et al. studied the role of miR-155 in the context of infection with influenza virus or Listeria monocytogenes.21 They found that miR-155 is highly expressed in effector and effector/memory CD8+ T cells and not in naive and central memory CD8+ T cells. Expression of miR-155 affected both optimal effector responses as well as development of a memory response.21 The mechanism by which miR-155 contributes to the establishment of the memory response is still not clear but it is of major interest, especially in vaccination and chronic infections. In the context of chronic infections, the study of the role of miR-155 in HIV-1 infection is coming into the limelight. When the expression of 377 miRNAs was evaluated in CD4+ T cells from HIV+ long-term non-progressors (LTNPs), naive patients and multiply exposed uninfected patients, several miRNAs differentiated multiply exposed uninfected patients from the naive patients and LTNPs. Only miR-155 expression discriminated between naive patients and LTNPs as it was up regulated in naive patients.50 This suggested that miR-155 might play a role in pathogenesis. Although this study had its critics51 because of the different conclusions from previous studies, it underlines the importance of miR-155 in HIV-1 infection as well as a potential drug target. As it was shown that miR-155 plays a role in Treg cell development,7 we investigated the levels of expression in different subsets of Treg cells. We found that miR-155 was strongly expressed in effector/memory Treg cells compared with lower expression levels in both naive Treg cells and naive CD4+ subsets. Further, we found that levels of effector/memory Treg cells are significantly increased in HIV-infected individuals with progressive infection versus LTNPs.52 The higher level of activated Tregs with presumably increased miR-155 levels may explain why these levels were different between the two HIV-1-infected groups reported by Bignami et al.52 We stressed the importance of analysing the expression of miR-155 in different T-cell subsets to better understand its role in HIV pathogenesis. HIV infection is not the only viral infection in which the role of miR-155 has been examined. It was shown in both hepatitis B virus53 and hepatitis C virus54 that miR-155 is implicated in several immune processes (Fig. 1).
Figure 1.
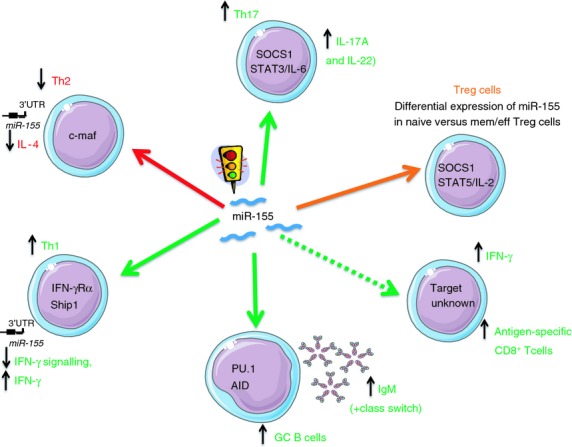
Role of miR-155 in immune cell function.
Autoimmune disease
miR-155 has also been implicated in many autoimmune disorders. Peripheral blood mononuclear cells from rheumatoid arthritis patients showed an increase in miR-155 expression compared with healthy controls.55 This is perhaps not surprising given that miR-155 has an important part in establishing and amplifying inflammatory responses through several mechanisms as reviewed elsewhere.56–58 Even though the role of miR-155 in rheumatoid arthritis as well as in other autoimmune disorders is considered detrimental, the role that its expression has on Treg cells leads to more ambiguous conclusions. Since miR-155 is involved in Treg cell differentiation and the maintenance as discussed before, the modulation of miR-155 expression for therapeutic purposes invites more caution. Animal models were very useful in defining the role of miR-155 in autoimmunity. miR-155-deficient mice do not develop collagen-induced arthritis (CIA) and they show the lower levels of IL-6 and IL-17 as well as the lower level of pathogenic IgG and auto-reactive T cells.27 As mentioned earlier, EAE is an animal model for study into MS. As in the case of arthritis, mir-155−/− mice are highly resistant to EAE because of aberrant differentiation of auto-reactive T cells, mostly of Th1 and Th17 phenotype. Furthermore, these mice showed impaired DC function needed for the development of these T-cell subsets.2 Paraboschi et al. monitored the differential expression of 22 immunity-related miRNAs in peripheral blood mononuclear cells to address their role in MS pathogenesis. The most up-regulated was miR-155, which suggest that it plays an important role in MS development and this calls for more investigations.59 Interestingly, increased expression of miR-155 was also described in brain lesions of MS patients.60 miR-155 has also a pro-inflammatory role in microglia and is necessary for the progression of the immune response through the modulation of SOCS-1, suggesting that, in a chronic inflammatory context, miR-155 inhibition can have a neuroprotective effect.61 Besides MS and rheumatoid arthritis, miR-155 is recognized in other autoimmune disorders, namely systemic lupus erythematosus and ulcerative colitis.57
Conclusions
miR-155 is a highly expressed and important miRNA with functional relevance in the biology of lymphocytes. Murine models that lack miR-155 in various lymphocyte compartments have highlighted the important role that miR-155 plays in particular with T-cell differentiation. Dysregulation of miR-155 has been shown to have relevance in a number of human tumours, autoimmune diseases and responses to viral infections by T cells and B cells. There is a paucity of information, however, regarding the role of miR-155 in certain T-cell subsets (such as T follicular helper cells) and research into the role of miR-155 in some of these T-cell subsets will help better define their functional relevance. The ability to manipulate miR-155 expression levels to see if disease outcomes can be altered (such as in autoimmune diseases such as MS), remains a lofty goal due to the difficulties of delivering these molecules precisely and the high likelihood of off-target effects. However, the altered expression of miR-155 in lymphocytes in various disease states, may lead to the identification of new protein targets that lead to novel therapies for these conditions. However, there remains a lot more to be understood about how miR-155, in concert with other miRNAs, regulate gene function in lymphocytes and also their precise role in disease pathogenesis.
Disclosures
The authors declare no conflict of interest.
References
- 1.Baltimore D, Boldin MP, O'Connell RM, Rao DS, Taganov KD. MicroRNAs: new regulators of immune cell development and function. Nat Immunol. 2008;9:839–45. doi: 10.1038/ni.f.209. [DOI] [PubMed] [Google Scholar]
- 2.O'Connell RM, Kahn D, Gibson WS, et al. MicroRNA-155 promotes autoimmune inflammation by enhancing inflammatory T cell development. Immunity. 2010;33:607–19. doi: 10.1016/j.immuni.2010.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–33. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Loeb GB, Khan AA, Canner D, Hiatt JB, Shendure J, Darnell RB, Leslie CS, Rudensky AY. Transcriptome-wide miR-155 binding map reveals widespread noncanonical microRNA targeting. Mol Cell. 2012;48:760–70. doi: 10.1016/j.molcel.2012.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rodriguez A, Vigorito E, Clare S, et al. Requirement of bic/microRNA-155 for normal immune function. Science. 2007;316:608–11. doi: 10.1126/science.1139253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vigorito E, Perks KL, Abreu-Goodger C, et al. microRNA-155 regulates the generation of immunoglobulin class-switched plasma cells. Immunity. 2007;27:847–59. doi: 10.1016/j.immuni.2007.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kohlhaas S, Garden OA, Scudamore C, Turner M, Okkenhaug K, Vigorito E. Cutting edge: the Foxp3 target miR-155 contributes to the development of regulatory T cells. J Immunol. 2009;182:2578–82. doi: 10.4049/jimmunol.0803162. [DOI] [PubMed] [Google Scholar]
- 8.Lu LF, Thai TH, Calado DP, et al. Foxp3-dependent microRNA155 confers competitive fitness to regulatory T cells by targeting SOCS1 protein. Immunity. 2009;30:80–91. doi: 10.1016/j.immuni.2008.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lind EF, Elford AR, Ohashi PS. Micro-RNA 155 is required for optimal CD8+ T cell responses to acute viral and intracellular bacterial challenges. J Immunol. 2013;190:1210–6. doi: 10.4049/jimmunol.1202700. [DOI] [PubMed] [Google Scholar]
- 10.Tsai CY, Allie SR, Zhang W, Usherwood EJ. MicroRNA miR-155 affects antiviral effector and effector Memory CD8 T cell differentiation. J Virol. 2013;87:2348–51. doi: 10.1128/JVI.01742-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Thai TH, Calado DP, Casola S, et al. Regulation of the germinal center response by microRNA-155. Science. 2007;316:604–8. doi: 10.1126/science.1141229. [DOI] [PubMed] [Google Scholar]
- 12.Landgraf P, Rusu M, Sheridan R, et al. A mammalian microRNA expression atlas based on small RNA library sequencing. Cell. 2007;129:1401–14. doi: 10.1016/j.cell.2007.04.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Martin MM, Lee EJ, Buckenberger JA, Schmittgen TD, Elton TS. MicroRNA-155 regulates human angiotensin II type 1 receptor expression in fibroblasts. J Biol Chem. 2006;281:18277–84. doi: 10.1074/jbc.M601496200. [DOI] [PubMed] [Google Scholar]
- 14.Georgantas RW, 3rd, Hildreth R, Morisot S, et al. CD34+ hematopoietic stem-progenitor cell microRNA expression and function: a circuit diagram of differentiation control. Proc Natl Acad Sci U S A. 2007;104:2750–5. doi: 10.1073/pnas.0610983104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Masaki S, Ohtsuka R, Abe Y, Muta K, Umemura T. Expression patterns of microRNAs 155 and 451 during normal human erythropoiesis. Biochem Biophys Res Commun. 2007;364:509–14. doi: 10.1016/j.bbrc.2007.10.077. [DOI] [PubMed] [Google Scholar]
- 16.O'Connell RM, Rao DS, Chaudhuri AA, Boldin MP, Taganov KD, Nicoll J, Paquette RL, Baltimore D. Sustained expression of microRNA-155 in hematopoietic stem cells causes a myeloproliferative disorder. J Exp Med. 2008;205:585–94. doi: 10.1084/jem.20072108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Grigoryev YA, Kurian SM, Hart T, et al. MicroRNA regulation of molecular networks mapped by global microRNA, mRNA, and protein expression in activated T lymphocytes. J Immunol. 2011;187:2233–43. doi: 10.4049/jimmunol.1101233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Berg van den A, Kroesen BJ, Kooistra K, et al. High expression of B-cell receptor inducible gene BIC in all subtypes of Hodgkin lymphoma. Genes Chromosom Cancer. 2003;37:20–8. doi: 10.1002/gcc.10186. [DOI] [PubMed] [Google Scholar]
- 19.Haasch D, Chen YW, Reilly RM, et al. T cell activation induces a noncoding RNA transcript sensitive to inhibition by immunosuppressant drugs and encoded by the proto-oncogene BIC. Cell Immunol. 2002;217:78–86. doi: 10.1016/s0008-8749(02)00506-3. [DOI] [PubMed] [Google Scholar]
- 20.Dorsett Y, McBride KM, Jankovic M, et al. MicroRNA-155 suppresses activation-induced cytidine deaminase-mediated Myc-Igh translocation. Immunity. 2008;28:630–8. doi: 10.1016/j.immuni.2008.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gracias DT, Stelekati E, Hope JL, et al. The microRNA miR-155 controls CD8+ T cell responses by regulating interferon signaling. Nat Immunol. 2013;14:593–602. doi: 10.1038/ni.2576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bach EA, Szabo SJ, Dighe AS, Ashkenazi A, Aguet M, Murphy KM, Schreiber RD. Ligand-induced autoregulation of IFN-γ receptor β chain expression in T helper cell subsets. Science. 1995;270:1215–8. doi: 10.1126/science.270.5239.1215. [DOI] [PubMed] [Google Scholar]
- 23.Tau GZ, Weid von der T, Lu B, et al. Interferon γ signaling alters the function of T helper type 1 cells. J Exp Med. 2000;192:977–86. doi: 10.1084/jem.192.7.977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Banerjee A, Schambach F, DeJong CS, Hammond SM, Reiner SL. Micro-RNA-155 inhibits IFN-γ signaling in CD4+ T cells. Eur J Immunol. 2010;40:225–31. doi: 10.1002/eji.200939381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.O'Connell RM, Chaudhuri AA, Rao DS, Baltimore D. Inositol phosphatase SHIP1 is a primary target of miR-155. Proc Natl Acad Sci U S A. 2009;106:7113–8. doi: 10.1073/pnas.0902636106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Huffaker TB, Hu R, Runtsch MC, et al. Epistasis between microRNAs 155 and 146a during T cell-mediated antitumor immunity. Cell Rep. 2012;2:1697–709. doi: 10.1016/j.celrep.2012.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bluml S, Bonelli M, Niederreiter B, et al. Essential role of microRNA-155 in the pathogenesis of autoimmune arthritis in mice. Arthritis Rheum. 2011;63:1281–8. doi: 10.1002/art.30281. [DOI] [PubMed] [Google Scholar]
- 28.Hu R, Huffaker TB, Kagele DA, Runtsch MC, Bake E, Chaudhuri AA, Round JL, O'Connell RM. MicroRNA-155 confers encephalogenic potential to Th17 cells by promoting effector gene expression. J Immunol. 2013;190:5972–80. doi: 10.4049/jimmunol.1300351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Murugaiyan G, Beynon V, Mittal A, Joller N, Weiner HL. Silencing microRNA-155 ameliorates experimental autoimmune encephalomyelitis. J Immunol. 2011;187:2213–21. doi: 10.4049/jimmunol.1003952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yao R, Ma YL, Liang W, Li HH, Ma ZJ, Yu X, Liao YH. MicroRNA-155 modulates Treg and Th17 cells differentiation and Th17 cell function by targeting SOCS1. PLoS ONE. 2012;7:e46082. doi: 10.1371/journal.pone.0046082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Das LM, Torres-Castillo MD, Gill T, Levine AD. TGF-β conditions intestinal T cells to express increased levels of miR-155, associated with down-regulation of IL-2 and itk mRNA. Mucosal Immunol. 2013;6:167–76. doi: 10.1038/mi.2012.60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Seddiki N, Santner-Nanan B, Martinson J, et al. Expression of interleukin (IL)-2 and IL-7 receptors discriminates between human regulatory and activated T cells. J Exp Med. 2006;203:1693–700. doi: 10.1084/jem.20060468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Seddiki N, Santner-Nanan B, Tangye SG, Alexander SI, Solomon M, Lee S, Nanan R, Fazekas de Saint Groth B. Persistence of naive CD45RA+ regulatory T cells in adult life. Blood. 2006;107:2830–8. doi: 10.1182/blood-2005-06-2403. [DOI] [PubMed] [Google Scholar]
- 34.Eis PS, Tam W, Sun L, Chadburn A, Li Z, Gomez MF, Lund E, Dahlberg JE. Accumulation of miR-155 and BIC RNA in human B cell lymphomas. Proc Natl Acad Sci USA. 2005;102:3627–32. doi: 10.1073/pnas.0500613102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Garzon R, Croce CM. MicroRNAs in normal and malignant hematopoiesis. Curr Opin Hematol. 2008;15:352–8. doi: 10.1097/MOH.0b013e328303e15d. [DOI] [PubMed] [Google Scholar]
- 36.Kluiver J, Poppema S, Jong de D, Blokzijl T, Harms G, Jacobs S, Kroesen BJ, Berg van den A. BIC and miR-155 are highly expressed in Hodgkin, primary mediastinal and diffuse large B cell lymphomas. J Pathol. 2005;207:243–9. doi: 10.1002/path.1825. [DOI] [PubMed] [Google Scholar]
- 37.Metzler M, Wilda M, Busch K, Viehmann S, Borkhardt A. High expression of precursor microRNA-155/BIC RNA in children with Burkitt lymphoma. Genes Chromosom Cancer. 2004;39:167–9. doi: 10.1002/gcc.10316. [DOI] [PubMed] [Google Scholar]
- 38.Vigorito E, Kohlhaas S, Lu D, Leyland R. miR-155: an ancient regulator of the immune system. Immunol Rev. 2013;253:146–57. doi: 10.1111/imr.12057. [DOI] [PubMed] [Google Scholar]
- 39.Costinean S, Zanesi N, Pekarsky Y, Tili E, Volinia S, Heerema N, Croce CM. Pre-B cell proliferation and lymphoblastic leukemia/high-grade lymphoma in E(mu)-miR155 transgenic mice. Proc Natl Acad Sci USA. 2006;103:7024–9. doi: 10.1073/pnas.0602266103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Babar IA, Cheng CJ, Booth CJ, Liang X, Weidhaas JB, Saltzman WM, Slack FJ. Nanoparticle-based therapy in an in vivo microRNA-155 (miR-155)-dependent mouse model of lymphoma. Proc Natl Acad Sci USA. 2012;109:E1695–704. doi: 10.1073/pnas.1201516109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Calin GA, Ferracin M, Cimmino A, et al. A MicroRNA signature associated with prognosis and progression in chronic lymphocytic leukemia. N Engl J Med. 2005;353:1793–801. doi: 10.1056/NEJMoa050995. [DOI] [PubMed] [Google Scholar]
- 42.Lawrie CH. MicroRNAs and lymphomagenesis: a functional review. Br J Haematol. 2013;160:571–81. doi: 10.1111/bjh.12157. [DOI] [PubMed] [Google Scholar]
- 43.Czyzyk-Krzeska MF, Zhang X. MiR-155 at the heart of oncogenic pathways. Oncogene. 2013 doi: 10.1038/onc.2013.26. doi: 10.1038/onc.2013.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gironella M, Seux M, Xie MJ, et al. Tumor protein 53-induced nuclear protein 1 expression is repressed by miR-155, and its restoration inhibits pancreatic tumor development. Proc Natl Acad Sci USA. 2007;104:16170–5. doi: 10.1073/pnas.0703942104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nikiforova MN, Tseng GC, Steward D, Diorio D, Nikiforov YE. MicroRNA expression profiling of thyroid tumors: biological significance and diagnostic utility. J Clin Endocrinol Metab. 2008;93:1600–8. doi: 10.1210/jc.2007-2696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Volinia S, Calin GA, Liu CG, et al. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc Natl Acad Sci USA. 2006;103:2257–61. doi: 10.1073/pnas.0510565103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dudda JC, Salaun B, Ji Y, et al. MicroRNA-155 is required for effector CD8+ T cell responses to virus infection and cancer. Immunity. 2013;38:742–53. doi: 10.1016/j.immuni.2012.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zonari E, Pucci F, Saini M, Mazzieri R, Politi LS, Gentner B, Naldini L. A role for miR-155 in enabling tumor-infiltrating innate immune cells to mount effective antitumor responses in mice. Blood. 2013;122:243–52. doi: 10.1182/blood-2012-08-449306. [DOI] [PubMed] [Google Scholar]
- 49.Cubillos-Ruiz JR, Baird JR, Tesone AJ, et al. Reprogramming tumor-associated dendritic cells in vivo using miRNA mimetics triggers protective immunity against ovarian cancer. Cancer Res. 2012;72:1683–93. doi: 10.1158/0008-5472.CAN-11-3160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bignami F, Pilotti E, Bertoncelli L, et al. Stable changes in CD4+ T lymphocyte miRNA expression after exposure to HIV-1. Blood. 2012;119:6259–67. doi: 10.1182/blood-2011-09-379503. [DOI] [PubMed] [Google Scholar]
- 51.Witwer KW, Clements JE. Evidence for miRNA expression differences of HIV-1-positive, treatment-naive patients and elite suppressors: a re-analysis. Blood. 2012;119:6395–6. doi: 10.1182/blood-2012-02-412742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Seddiki N, Swaminathan S, Phetsouphanh C, Kelleher AD. miR-155 is differentially expressed in Treg subsets, which may explain expression level differences of miR-155 in HIV-1 infected patients. Blood. 2012;119:6396–7. doi: 10.1182/blood-2012-02-412874. [DOI] [PubMed] [Google Scholar]
- 53.Su C, Hou Z, Zhang C, Tian Z, Zhang J. Ectopic expression of microRNA-155 enhances innate antiviral immunity against HBV infection in human hepatoma cells. Virol J. 2011;8:354. doi: 10.1186/1743-422X-8-354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bala S, Tilahun Y, Taha O, Alao H, Kodys K, Catalano D, Szabo G. Increased microRNA-155 expression in the serum and peripheral monocytes in chronic HCV infection. J Transl Med. 2012;10:151. doi: 10.1186/1479-5876-10-151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Pauley KM, Satoh M, Chan AL, Bubb MR, Reeves WH, Chan EK. Upregulated miR-146a expression in peripheral blood mononuclear cells from rheumatoid arthritis patients. Arthritis Res Ther. 2008;10:R101. doi: 10.1186/ar2493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Leah E. Rheumatoid arthritis: miR-155 mediates inflammation. Nat Rev Rheumatol. 2011;7:437. doi: 10.1038/nrrheum.2011.101. [DOI] [PubMed] [Google Scholar]
- 57.Leng RX, Pan HF, Qin WZ, Chen GM, Ye DQ. Role of microRNA-155 in autoimmunity. Cytokine Growth Factor Rev. 2011;22:141–7. doi: 10.1016/j.cytogfr.2011.05.002. [DOI] [PubMed] [Google Scholar]
- 58.Spoerl D, Duroux-Richard I, Louis-Plence P, Jorgensen C. The role of miR-155 in regulatory T cells and rheumatoid arthritis. Clin Immunol. 2013;148:56–65. doi: 10.1016/j.clim.2013.03.010. [DOI] [PubMed] [Google Scholar]
- 59.Paraboschi EM, Solda G, Gemmati D, et al. Genetic association and altered gene expression of mir-155 in multiple sclerosis patients. Int J Mol Sci. 2011;12:8695–712. doi: 10.3390/ijms12128695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Junker A, Krumbholz M, Eisele S, et al. MicroRNA profiling of multiple sclerosis lesions identifies modulators of the regulatory protein CD47. Brain. 2009;132(Pt 12):3342–52. doi: 10.1093/brain/awp300. [DOI] [PubMed] [Google Scholar]
- 61.Cardoso AL, Guedes JR, Pereira de Almeida L, Pedroso de Lima MC. miR-155 modulates microglia-mediated immune response by down-regulating SOCS-1 and promoting cytokine and nitric oxide production. Immunology. 2012;135:73–88. doi: 10.1111/j.1365-2567.2011.03514.x. [DOI] [PMC free article] [PubMed] [Google Scholar]


