Abstract
The lasso peptide microcin J25 is known to hijack the siderophore receptor FhuA for initiating internalization. Here, we provide the first structural evidence on the recognition mechanism and our biochemical data show that another closely related lasso peptide cannot interact with FhuA. Our work provides an explanation on the narrow activity spectrum of lasso peptides and opens the path to the development of new antibacterials.
Under iron starvation, Gram-negative bacteria are capable of acquiring iron in the form of ferric siderophore complexes that are taken up by high affinity outer membrane iron receptors, such as FhuA, FepA, FecA and Cir in Escherichia coli. These receptors provide an advantage for the cells to scavenge iron, but can also be exploited by antibacterial compounds produced by other bacteria, or in infection by bacteriophages. Microcins form a class of gene-encoded antibacterial peptides of low molecular weight (< 10 kDa) involved in the defence strategies of Enterobacteriaceae for colonisation and survival 1. Several post-translationally modified microcins exploit the iron uptake receptors of closely-related sensitive cells and kill them, through a TonB-dependent process 1. Microcin J25 (MccJ25) is a 21-amino acid peptide characterized by a lasso topology. It contains an N-terminal macrolactam ring (Gly1-Glu8) through which a C-terminal tail (Tyr9-Gly21) is threaded and sterically locked by the bulky residues Phe19 and Tyr20 located below and above the ring, respectively 2, 3 (Supplementary Results, Supplementary Fig. 1). MccJ25 exerts potent, narrow spectrum antibacterial activity against Escherichia and Salmonella species through inhibition of RNA polymerase 4. In E. coli, MccJ25 enters the target bacteria through the outer membrane siderophore receptor FhuA 5 and inner membrane protein SbmA 6. We have previously shown that MccJ25 can bind to FhuA with a KD of 1.2 μM 7.
The physiological role of FhuA is to transport Fe(III) chelated to the hydroxamate siderophore ferrichrome to ensure iron uptake for bacterial growth. Fe(III) is highly insoluble and poorly accessible in the environment and requires the formation of soluble iron siderophore complexes, to be taken up by bacteria. FhuA is also the receptor for the siderophore-conjugated antibiotic albomycin, a structural analogue of ferrichrome 8, the rifamycin CGP 4832, which has no structural similarity with ferrichrome 9, the bacterial toxin colicin M and phages T1, T5 and Φ80, which all hijack this receptor. Ferrichrome and MccJ25 lack structural similarity, raising questions as to how MccJ25 hijacks the FhuA receptor. Here, we provide the first structural evidence on the interaction of an antibacterial lasso peptide with an outer membrane receptor. Using competition experiments with phage T5 and ligand binding studies by non-denaturing mass spectrometry, we have identified a key residue in MccJ25 that is important for recognition by FhuA. These data, combined with modelling and biochemical experiments, lead to a proposal for how lasso peptides have acquired their narrow spectrum of antibacterial activity.
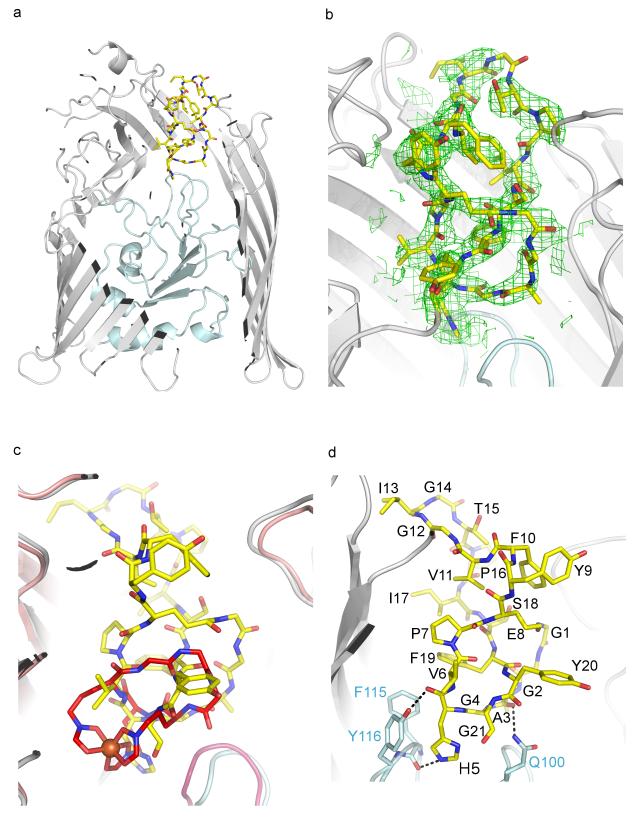
We first determined the crystal structure of FhuA in complex with MccJ25 at 2.3 Å resolution (Fig. 1a and Supplementary Table 1). The overall structure of FhuA is similar to previously published structures 10, 11. FhuA is a monomeric β-barrel protein consisting of 22 antiparallel β-strands with its N-terminus folded inside the β-barrel from the periplasmic side, thus forming the cork domain (residues 20-160) and a large extracellular ligand binding pocket open to the external medium. Translocation of the ferrichrome requires displacement of the ‘cork’. FhuA uses the proton motive force from the inner membrane through the TonB/ExbB/ExbD complex to transport its substrates. The N-terminus of the cork domain contains the switch helix, which harbours the TonB-box, a conserved seven-amino acid sequence that has been shown to be important for interaction with TonB 12.
Fig 1.
Structure of E. coli FhuA in complex with MccJ25. (a) MccJ25 (yellow sticks) binds to the extracellular pocket of the outer membrane ferrichrome receptor FhuA. FhuA is displayed as grey ribbons and the plug domain is shown in light blue. The front face of FhuA, detergent molecules and LPS have been omitted for clarity. (b) Unambiguous 2Fo-Fc electron density was observed for MccJ25 after molecular replacement and rigid body refinement. MccJ25 was not included in the refinement but is shown for clarity. The map is contoured at 1σ (see also Supplementary Fig. 2). (c) MccJ25 (yellow sticks) mimics the binding of the ferrichrome (red sticks; iron is shown as an orange sphere). (d) MccJ25 displays three hydrogen bonds with the plug domain. Other interactions are shown in Supplementary Fig. 4. MccJ25 carbons are shown as yellow sticks, oxygens in red and nitrogen in blue. The FhuA plug domain side chains carbons are in light blue. The rest of the atoms are as in MccJ25.
After molecular replacement and initial structure refinement, clear electron density was observed for MccJ25 bound at the extracellular pocket of FhuA, detergent molecules and a lipopolysaccharide (LPS) at the outer surface of the β-barrel (Fig. 1b and Supplementary Fig. 2). The electron density allowed identifying the macrolactam ring (residues 1-8) of MccJ25. Using the NMR structure of MccJ25 2, the macrolactam ring was placed in the density without any manipulation. It sits vertically inside FhuA, whereas the β-hairpin loop region (residues 9-18) shows significant conformational changes compared to the NMR structure (Supplementary Fig. 3). In the NMR structure, the loop region forms two short β-strands, whereas in complex with FhuA the loop has lost its β-strand structure and has adopted a wider and less structured conformation. The FhuA-bound MccJ25 can be superimposed with the NMR structure with an rmsd of 2.3 Å over 21 Ca atoms and an rmsd of 0.6 Å when aligned on the macrolactam ring. MccJ25 completely occupies and occludes the FhuA channel. Residues from the FhuA β-barrel, the cork domain and extracellular loops contribute to the binding of MccJ25 side chains and backbone (Supplementary Fig. 4).
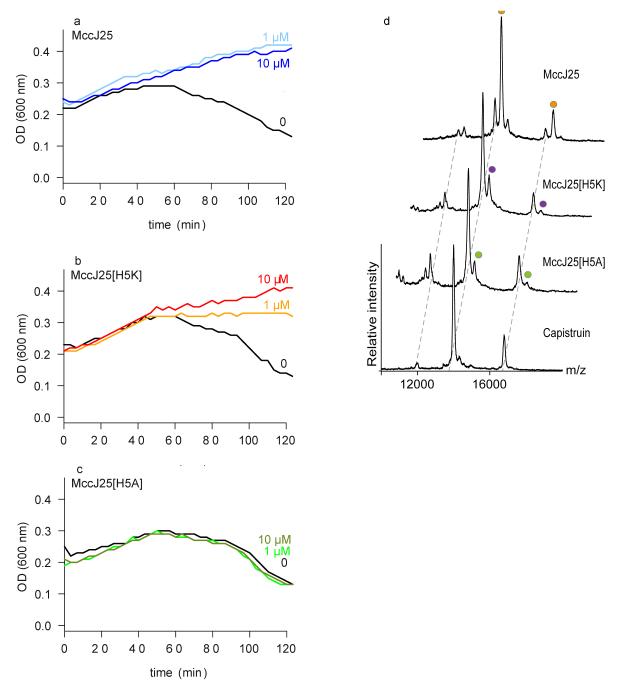
Comparison of the MccJ25- and ferrichrome-bound FhuA structures show MccJ25 binds at a very similar location to ferrichrome 10, 11 (Fig. 1c). Ferrichrome displays extensive hydrogen bonds with the cork domain, whereas only two MccJ25 residues have hydrogen bonds with this domain. The imidazole ring and backbone carbonyl group of His5MccJ25 make hydrogen bonds with the backbone carbonyl group of Phe115FhuA (2.8 Å) and side chain of Tyr116FhuA (3.2 Å) respectively, whereas the backbone carbonyl group of Ala3MccJ25 is within hydrogen bond distance from the side chain of Gln100FhuA (3.3 Å) (Fig. 1d and Supplementary Fig. 4). The lack of extensive bonding between MccJ25 and the cork domain indicates that minimal conserved interactions with the receptor are sufficient for MccJ25 to hijack the binding pocket of FhuA. It has been proposed that binding of the ligand to the extracellular pocket transmits allosteric changes along the cork domain 11. Indeed, the imidazole ring of His5MccJ25 is “buried” further inside the cork domain relative to the other residues. We investigated the role of the His5MccJ25 residue in competition experiments with infection of E. coli cells by phage T5 7. Phage infection involves a strong interaction of its receptor binding protein pb5 with FhuA 13 and release of its DNA. Phage T5 infection therefore provides a functional assay to assess the binding of MccJ25 variants to FhuA in vivo. In competition experiments with phage T5, the charge-conserved MccJ25H5K variant showed slightly altered protection against phage infection compared to MccJ25 (Fig. 2a, b), while the MccJ25H5A variant did not compete with phage (Fig. 2c). As a second line of evidence, we performed ligand-binding studies by non-denaturing mass spectrometry to calculate the affinity of MccJ25 and its variants for FhuA. This technique was used to characterise the interaction of OmpF with bacteriocin colicin E9 14. The affinity of MccJ25 for FhuA was calculated at 0.98 ± 0.02 μM, in agreement with our previously published value of 1.2 μM determined by ITC 7 (Fig. 2d, Supplementary Fig. 5). The affinity for the MccJ25H5K variant (16.9 ± 0.4 μM) is more than 10-fold decreased. Interestingly, despite the absence of protection from phage T5 infection, the MccJ25H5A variant can still bind to FhuA, although with much lower affinity (23.6 ± 0.8 μM). Different conformations in the loop region could be acquired by the variants upon interaction with FhuA, which could account for the lack of correlation between affinity of the variants and ability to compete with T5. In addition, the distinct mechanisms of the MccJ25 antibacterial activity and phage T5 infection could also account for the apparent inconsistency. However, taken together, our data suggest that His5MccJ25 is essential for recognition and binding to the cork domain of FhuA, and subsequently for transport. These results are further consistent with the minimal inhibitory concentrations (MICs) measured for the peptides on E. coli W3110 (Supplementary Table 2). Interestingly, FhuA cannot efficiently transport the antibiotic rifamycin, while it can transport the rifamycin derivative CGP 4832 15, which displays more contacts with the cork domain 9. Indeed, CGP 4832 displays hydrogen bonds with the plug domain residues Gly99FhuA (3.4 Å), Ser101FhuA (3.1 Å) and Tyr116FhuA (3.2 Å). Alignment of the FhuA-MccJ25 structure with that of CGP 4832 reveals that His5MccJ25 is in the same position as the methyl-piperidine group of CGP 4832 (Supplementary Fig. 6), providing further evidence on the ability of MccJ25 to contact the plug domain.
Fig 2.
Interaction studies of lasso peptides with FhuA by phage T5 infection and non-denaturing mass spectrometry. (a-c) E. coli W3110 was incubated with MccJ25 (a), MccJ25H5K (b), and MccJ25H5A (c) at concentrations of 1 or 10 μM, or with solvent only. After addition of phage T5, lysis was monitored by measuring culture turbidity (OD 600 nm) for 120 min. The data shown are the average of three measurements for each condition. (d) Non-denaturing mass spectra showing binding of MccJ25, MccJ25H5K and MccJ25H5A to FhuA. Increasing concentrations of the peptides were titrated to FhuA (Supplementary Fig. 5). Only the 7.5 μM peptide concentration of the MccJ25 and its variants are shown. Cap is shown at 20 μM peptide concentration. Each mass spectrum and mass-to-charge peak is assigned according to incubation condition. In the mass spectra we observed higher molecular weight species with low intensity that probably corresponded to non-specific binding of a second peptide at highest concentration; a second binding molecule was also observed from the ITC data 7.
Previous knockout studies of the fhuA gene have confirmed the importance of FhuA for internalization of MccJ25 inside the cell 6. Using a tonB minus strain we showed that MccJ25 internalization is dependent on the FhuA-TonB system (Supplementary Fig. 7), providing direct evidence that MccJ25 can induce displacement of the cork domain for transport via the TonB-dependent pathway. Therefore, MccJ25 and the MccJ25H5K variant could provide protection against T5 infection through the possible displacement of the cork domain, since T5 cannot infect a cork-less FhuA 16, allowing another explanation for the different activities of the MccJ25 variants. We have previously shown that the MccJ25 loop is also essential for recognition by detergent-purified FhuA 7. This is consistent with the absence of binding to FhuA (Supplementary Fig. 5, 8 and Supplementary Table 3) and competition with phage T5 observed for the thermolysin-cleaved MccJ25 (t-MccJ25). Finally, we investigated if recognition of MccJ25 by FhuA is independent on the lasso topology of the macrolactam ring by using the cyclic-branched topoisomer (MccJ25-lcm), which contains the macrolactam ring but not the lasso topology. We did not observe protection against T5 infection or binding (Supplementary Fig. 5, 8 and Supplementary Table 3). We conclude that the macrolactam ring itself cannot mediate binding of MccJ25 to FhuA and that the integrity of the loop is also essential for binding.
Lasso antibacterial peptides usually display a very narrow spectrum of activity against bacteria closely related to their producers. MccJ25 exerts its potent antibacterial activity exclusively against Escherichia and some Salmonella species 17. Capistruin (Cap), which is another lasso peptide produced by Burkholderia thailandensis, has a moderate antibacterial activity directed against Burkholderia species 18, though its internalization process is not known. Since both lasso peptides inhibit E. coli RNA polymerase with equal efficiency in vitro 4, 19, it is likely that the narrow spectrum of activity is due to specific interaction of the lasso peptides with the outer membrane receptors. The NMR structure of Cap, which has a shorter loop and longer tail than MccJ25, could be modelled in the FhuA extracellular pocket with some steric clashes (Supplementary Fig. 9) that could be relieved by conformational changes of the C-terminal tail 18. However, Cap did not provide protection against phage T5 (Supplementary Fig. 8), did not bind to E. coli FhuA (Fig. 2d) and did not show antibacterial activity on E. coli W3110 at concentrations up to 100 μM (Supplementary Table 2). Sequence analysis of FhuA receptors from Burkholderia and E. coli show they share weak similarity (35% sequence identity) and the residues aligning the extracellular pocket are not very well conserved (Supplementary Fig. 10). This suggests that bacterial antimicrobial peptides have acquired structural features to target a very narrow spectrum of related bacteria by complementing specific outer membrane receptor binding pockets.
In conclusion, we have shown that MccJ25 can hijack the outer membrane siderophore receptor FhuA by mimicking the binding mode of ferrichrome to induce TonB-dependent transport. In addition, its narrow spectrum of antibacterial activity can be attributed to its overall structure and specific interactions with the FhuA binding pocket and cork domain. The emergence of resistant bacteria remains a critical problem and in the context of poor development of novel antimicrobial drugs, the possibility of developing antibiotics that penetrate bacteria efficiently through high affinity iron-transport receptors is very attractive 20. Internalization of siderophore-antibiotic conjugates such as albomycin is strongly affected by modifications, while MccJ25 internalization and inhibitory properties have been shown to accept a wide range of sequence modifications 21, 22. These sequence variations could be mapped on the FhuA-MccJ25 complex to provide structural models for the enhancement of MccJ25 uptake and activity (Supplementary Fig. 11). The antibacterial activity of MccJ25 has been demonstrated both ex vivo, in complex fluid matrices, and in vivo in a mouse model of infection by Salmonella 23, and without haemolytic activity on human erythrocytes, thus suggesting a potential therapeutic role of this lasso peptide. The unique and very stable lasso structure of MccJ25 makes it an ideal platform to be considered for the design of novel molecules against bacterial infections and the structure of MccJ25 in complex with FhuA will prove a valuable and helpful tool for such developments.
Online Methods
Protein expression and purification
FhuA from E. coli MG1655 was cloned with an internal His-tag and overexpressed as previously described 24. Outer membranes from 10 L cell culture were prepared as in 25. FhuA was extracted using 1 % lauryldimethylamine-oxide (LDAO) for 1 h at room temperature in 1 × PBS. The solubilized suspension was spun at 120,000 g for 1 h to remove unsolubilized material. Subsequent purification steps were performed at 4°C. The supernatant was brought to 50 mM imidazole and was passed through a 5 mL His-Trap column (GE Healthcare) equilibrated in 1 × PBS, 30 mM imidazole and 0.1 % LDAO. The column was washed with 5 column volumes (CV) of equilibration buffer and with a further 5 CV of 1 × PBS, 50 mM imidazole and 0.1 % LDAO. FhuA was eluted in the same buffer containing 300 mM imidazole. The eluted protein was concentrated using a 50 kDa cut-off concentrator and injected in a Superdex S200 10/300 size exclusion column (GE Healthcare) equilibrated in 50 mM ammonium acetate pH 8.0, 10 mM NaCl and 0.06% LDAO. The fraction corresponding to FhuA was concentrated to 15 mg ml−1 for crystallization.
Peptides
MccJ25 was produced from a culture of E. coli K12 MC4100 harboring the plasmid pTUC202, as described previously 26. The variants MccJ25[His5Ala] and [His5Lys] were obtained by site-directed mutagenesis using the Quick-Change II XL kit (Agilent) 22. Cap was produced from a culture of Burkholderia thailandensis E264, as previously described 18. The peptide t-MccJ25 (MccJ25 hydrolyzed between Phe10 and Val11) was obtained by incubation of MccJ25 in the presence of thermolysin. The purification of the peptides was carried out by solid phase extraction followed by semi-preparative reverse-phase-HPLC 22 (HPLC only for t-MccJ25). The purity and lasso topology of the peptides were checked by liquid-chromatography – mass spectrometry, as previously reported 22 (Supplementary Fig. 12 and Supplementary Table 4). The synthetic cyclic branched peptide MccJ25-lcm was obtained from Genepep (Saint Jean de Védas, France), with a purity > 95%.
Inhibition of infection with phage T5
A 10 mL preculture of E. coli W3110 in LB medium was incubated for overnight at 37°C and 250 rpm. 10 mL LB medium were inoculated with 200 μL preculture, and incubated at 37°C and 250 rpm until reaching an optical density at 600 nm of 1, corresponding to a viable bacteria concentration of 5×108 c.f.u. (colony-forming units)/mL. 90 μL of the bacterial suspension were deposited in a 96-well microplate, and 10 μL peptide were added (MccJ25 or MccJ25 variants or Cap dissolved in methanol/water 1:1), with a final peptide concentration in the range 0.25-100 μM. A control was performed with 10 μL solvent added to the bacterial suspension. 10 μL phage T5 in phage buffer (Tris/HCl 10 mM, pH 7.5, NaCl 150 mM, CaCl2 1 mM, MgSO4 1 mM) was added to every well, to reach a multiplicity of infection (MOI) of 2.5. The microplate was incubated at 37°C under shaking in a POLARstar OMEGA microplate reader (BMG Labtech), and bacterial growth and lysis were monitored over 120 min by measurement of the optical density at 600 nm. A control for bacterial growth was performed in the presence of peptide and in the absence of phage T5 (Supplementary Fig. 8). Each experiment was made in triplicate.
Antibacterial activities
The MICs were measured using a liquid growth inhibition assay in poor broth (PB: 1% bactotryptone, 0.5% NaCl w/v). 10 μL of serial dilutions of peptides (MccJ25, MccJ25[His5Lys], MccJ25[His5Ala], Cap) were added to 90 μL of a mid-logarithmic growth phase culture of E. coli W3110 diluted in PB to an optical density of 0.001 at 600 nm. Each peptide dilution was tested in triplicate. The plates were incubated at 37°C under shaking in a POLARstar OMEGA microplate reader (BMG Labtech), and the bacterial growth was monitored over 120 min by measurement of the optical density at 600 nm. The MICs were determined as the lowest peptide concentrations showing complete inhibition of the bacterial growth.
tonB antibacterial assay
E. coli W3110 tonB− strain was a kind gift from Prof. Lucienne Letellier (IBBMC, Orsay, France). The antibacterial activity of MccJ25 towards E. coli W3110 wildtype and tonB− strains was tested by a spot-on-lawn assay. Briefly, 107 E. coli cells were mixed with 3 mL LB soft agar and plated onto a LB agar plate. After solidification, 10 μL MccJ25 in MeOH (1-100 μg/mL) were spotted. A positive control using kanamycin was set up in parallel. The plates were incubated at 37°C overnight.
Non-denaturing mass spectrometry
FhuA protein sample was buffer exchanged to 200 mM ammonium acetate supplemented with 0.03% (w/w) LDAO detergent using Bio-Spin 6 (Bio-Rad) columns prior to mass spectrometric analysis. The spectra were acquired on a QToF mass spectrometer (Waters) previously modified for transmission of high masses and also modified for high energy in collision cell 27, 28. Samples were introduced into the mass spectrometer from a gold coated capillary needle using following conditions: capillary voltage 1.7 kV, cone voltage 200 V and collision cell energy 320 V. Lowering the energy in collision cell gave rise to higher charge states of the protein possibly corresponding to unfolded population. Further decrease resulted in an unresolved hump.
MccJ25 and variant peptides binding to FhuA
Non-denaturing electrospray titration experiments were performed using the MccJ25 and variant peptides using FhuA at a constant concentration of 7.5 μM. Peptide were diluted to 100 μM concentration in 200 mM ammonium acetate in presence of 0.03% LDAO. For any particular point titration, peptide was diluted from the stock solution to a concentration double that required. The peptide was then added to FhuA solution at 1:1 / v:v ratio, to yield the required concentration of the peptide. For Cap, Mccj25-lcm and t-MccJ25 only 20 μM of each peptide was used (Supplementary Fig. 5).
Crystallization
Prior to crystallization, the protein (15 mg ml−1) was incubated with 1 mM MccJ25 at room temperature overnight. Crystals were grown at 20°C using the vapour diffusion method by mixing protein and reservoir solutions at 1:1. Crystals were grown from a precipitant solution containing 20 mM Tris pH 7.5, 120 mM lithium sulfate, 100 mM sodium citrate pH 5, 20 % PEG300. The crystals grew after seven days and achieved maximum size after 2 weeks. The crystals were directly frozen into liquid nitrogen and diffraction screening and data collection were performed at Diamond Light Source synchrotron. The crystals diffracted X-rays to a maximum of 2.3 Å resolution.
Data collection
Diffraction data to 2.3 Å resolution were collected on I04-1 at Diamond Light Source at a wavelength of 0.92 Å using a Pilatus 2M detector. Data were processed with xia2 29. The resolution of the data and anisotropy analysis were evaluated by half-dataset correlation coefficient in Aimless (cut-off less than 0.5) 30. The space group was determined to be C2221 with one copy of FhuA in the asymmetric unit. The data collection statistics are summarized in Supplementary Table 1.
Structure solution and refinement
The structure was solved by molecular replacement in Phaser 31 using the apo FhuA structure (PDB ID: 2FCP) 10 as a search model. All model building was performed in Coot. Initial rounds of refinement were carried out in Phenix 32 and later in Buster 33. After rigid body and restrained refinement extra electron density corresponding to MccJ25 (Fig. 1b and Supplementary Fig. 2), one molecule of lipopolysaccharide and several detergent molecules were identified. In the PDB, there are two MccJ25 NMR structures deposited (PDB ID: 1Q71 2 and 1PP5 3) that are identical in structure. The NMR structure of MccJ25 with PDB ID 1Q71 was used to build the microcin into the density. The high resolution of the electron density maps, allowed us to identify the macrolactam ring and side chains of MccJ25, which could fit in the density without any manipulation, whereas the loop region had undergone conformational changes upon binding to FhuA (loss of β-hairpin loop). Water molecules were added using ARP/wARP 34. The final FhuA structure contains residues 20-404 and 418-725. The electron density at the internal His-tag loop region was too weak/absent to allow building residues 405-417 reliably. The final model has an Rfactor of 21.2 % and an Rfree of 25.5 %. The refinement statistics are summarized in Supplementary Table 1. The FhuA structure has 96.3 % of the residues in the favoured Ramachandran region and 0.14 % outliers as judged by MolProbity. The outlier corresponds to Gly566 in a loop turn.
Capistruin modelling
The NMR structure of Cap (Biological Magnetic Resonance Data Bank ID: 20014) 18 was used for modelling into the FhuA receptor. The macrolactam ring of Cap was superposed onto the MccJ25 macrolactam ring using the program Superpose in CCP4; the two molecules aligned with an rmsd deviation of 1.74 Å over 8 Cα atoms. The loops could not be aligned against each other due to significant differences in their length and conformation. At the current orientation there are significant clashes between FhuA and the C-terminal tail of Cap that can be relieved with manipulation of the overall conformation of the tail. The current model satisfies some of the hydrophobic contacts and hydrogen bonds between FhuA and Cap. Rotation of Cap by 90 °, 180 ° or flipping it along its axis results to a worst model with clashes between the C-terminal tail and FhuA that cannot be relieved even after manipulation.
The FhuA-MccJ25 coordinates and structure factors have been deposited to the Protein Data Bank with PDB ID code 4cu4.
Supplementary Material
Acknowledgments
We are grateful to Dr. Pascale Boulanger (Institute of Biochemistry and Molecular and Cellular Biophysics, Orsay University Paris Sud-11, France) for her kind gift of phage T5. We would like to thank Diamond Light Source for beam time allocation and access. We thank the mass spectrometry platform at the MNHN for access to the spectrometers, together with Kok-Phen Yan and Zoë Falk for contributing to the site-directed mutagenesis and phage competition experiments, respectively. The Oxford University Mass Spectrometry facility is funded by Medical Research Council (DNRUBH0 to C.V.R.). MPL is funded by the Wellcome Trust (WT/099165/Z/12/Z to Professor So Iwata). I.M. is supported by a Ministry of Science, Technology and Innovation (MOSTI) postgraduate scholarship. Part of this work was supported by the Biotechnology and Biological Sciences Research Council (BB/H01778X/1 to K.B.).
Footnotes
Competing financial interests
The authors declare no competing financial interests.
References
- 1.Duquesne S, Destoumieux-Garzon D, Peduzzi J, Rebuffat S. Nat. Prod. Rep. 2007;24:708–734. doi: 10.1039/b516237h. [DOI] [PubMed] [Google Scholar]
- 2.Rosengren KJ, et al. J Am Chem Soc. 2003;125:12464–12474. doi: 10.1021/ja0367703. [DOI] [PubMed] [Google Scholar]
- 3.Bayro MJ, et al. J Am Chem Soc. 2003;125:12382–12383. doi: 10.1021/ja036677e. [DOI] [PubMed] [Google Scholar]
- 4.Delgado MA, Rintoul MR, Farias RN, Salomon RA. J. Bacteriol. 2001;183:4543–4550. doi: 10.1128/JB.183.15.4543-4550.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Salomon RA, Farias RN. J. Bacteriol. 1993;175:7741–7742. doi: 10.1128/jb.175.23.7741-7742.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Salomon RA, Farias RN. J. Bacteriol. 1995;177:3323–3325. doi: 10.1128/jb.177.11.3323-3325.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Destoumieux-Garzon D, et al. Biochem. J. 2005;389:869–876. doi: 10.1042/BJ20042107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ferguson AD, et al. Protein Sci. 2000;9:956–963. doi: 10.1110/ps.9.5.956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ferguson AD, et al. Structure. 2001;9:707–716. doi: 10.1016/s0969-2126(01)00631-1. [DOI] [PubMed] [Google Scholar]
- 10.Ferguson AD, Hofmann E, Coulton JW, Diederichs K, Welte W. Science. 1998;282:2215–2220. doi: 10.1126/science.282.5397.2215. [DOI] [PubMed] [Google Scholar]
- 11.Locher KP, et al. Cell. 1998;95:771–778. doi: 10.1016/s0092-8674(00)81700-6. [DOI] [PubMed] [Google Scholar]
- 12.Killmann H, Herrmann C, Torun A, Jung G, Braun V. Microbiology. 2002;148:3497–3509. doi: 10.1099/00221287-148-11-3497. [DOI] [PubMed] [Google Scholar]
- 13.Flayhan A, Wien F, Paternostre M, Boulanger P, Breyton C. Biochimie. 2012;94:1982–1989. doi: 10.1016/j.biochi.2012.05.021. [DOI] [PubMed] [Google Scholar]
- 14.Housden NG, et al. Science. 2013;340:1570–1574. doi: 10.1126/science.1237864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pugsley AP, Zimmerman W, Wehrli W. J. Gen. Microbiol. 1987;133:3505–3511. doi: 10.1099/00221287-133-12-3505. [DOI] [PubMed] [Google Scholar]
- 16.Braun M, Endriss F, Killmann H, Braun V. J. Bacteriol. 2003;185:5508–5518. doi: 10.1128/JB.185.18.5508-5518.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Salomon RA, Farias RN. J. Bacteriol. 1992;174:7428–7435. doi: 10.1128/jb.174.22.7428-7435.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Knappe TA, et al. J. Am. Chem. Soc. 2008;130:11446–11454. doi: 10.1021/ja802966g. [DOI] [PubMed] [Google Scholar]
- 19.Kuznedelov K, et al. J. Mol. Biol. 2011;412:842–848. doi: 10.1016/j.jmb.2011.02.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cotter PD, Ross RP, Hill C. Nat. Rev. Microbiol. 2013;11:95–105. doi: 10.1038/nrmicro2937. [DOI] [PubMed] [Google Scholar]
- 21.Pavlova O, Mukhopadhyay J, Sineva E, Ebright RH, Severinov K. J. Biol. Chem. 2008;283:25589–25595. doi: 10.1074/jbc.M803995200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ducasse R, et al. Chembiochem. 2012;13:371–380. doi: 10.1002/cbic.201100702. [DOI] [PubMed] [Google Scholar]
- 23.Lopez FE, Vincent PA, Zenoff AM, Salomon RA, Farias RN. J. Antimicrob. Chemother. 2007;59:676–680. doi: 10.1093/jac/dkm009. [DOI] [PubMed] [Google Scholar]
- 24.Ferguson AD, Breed J, Diederichs K, Welte W, Coulton JW. Protein Sci. 1998;7:1636–1638. doi: 10.1002/pro.5560070719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Beis K, Whitfield C, Booth I, Naismith JH. Int. J. Biol. Macromol. 2006;39:10–14. doi: 10.1016/j.ijbiomac.2005.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zirah S, et al. J. Am. Soc. Mass Spectrom. 2011;22:467–479. doi: 10.1007/s13361-010-0028-1. [DOI] [PubMed] [Google Scholar]
- 27.Sobott F, Hernandez H, McCammon MG, Tito MA, Robinson CV. Anal. Chem. 2002;74:1402–1407. doi: 10.1021/ac0110552. [DOI] [PubMed] [Google Scholar]
- 28.Benesch JL, et al. Anal. Chem. 2009;81:1270–1274. doi: 10.1021/ac801950u. [DOI] [PubMed] [Google Scholar]
- 29.Winter G. J. Appl. Cryst. 2010;43:186–190. [Google Scholar]
- 30.Evans P. Acta Crystallogr. D Biol. Crystallogr. 2006;62:72–82. doi: 10.1107/S0907444905036693. [DOI] [PubMed] [Google Scholar]
- 31.McCoy AJ, et al. J. Appl. Cryst. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Terwilliger TC, et al. Acta Crystallogr. D Biol. Crystallogr. 2008;64:61–69. doi: 10.1107/S090744490705024X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Blanc E, et al. Acta Crystallogr. D Biol. Crystallogr. 2004;60:2210–2221. doi: 10.1107/S0907444904016427. [DOI] [PubMed] [Google Scholar]
- 34.Langer G, Cohen SX, Lamzin VS, Perrakis A. Nat. Protoc. 2008;3:1171–1179. doi: 10.1038/nprot.2008.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.