Abstract
Background
Autosomal dominant polycystic kidney disease (ADPKD) is the most common hereditary renal disorder caused by mutation in either one of two genes, PKD1 and PKD2. High structural and sequence complexity of PKD genes makes the mutational diagnostics of ADPKD challenging. The present study is the first detailed analysis of both PKD genes in a cohort of Czech patients with ADPKD using High Resolution Melting analysis (HRM) and Multiplex Ligation-dependent Probe Amplification (MLPA).
Methods
The mutational analysis of PKD genes was performed in a set of 56 unrelated patients. For mutational screening of the PKD1 gene, the long-range PCR (LR-PCR) strategy followed by nested PCR was used. Resulting PCR fragments were analyzed by HRM; the positive cases were reanalyzed and confirmed by direct sequencing. Negative samples were further examined for sequence changes in the PKD2 gene by the method of HRM and for large rearrangements of both PKD1 and PKD2 genes by MLPA.
Results
Screening of the PKD1 gene revealed 36 different likely pathogenic germline sequence changes in 37 unrelated families/individuals. Twenty-five of these sequence changes were described for the first time. Moreover, a novel large deletion was found within the PKD1 gene in one patient. Via the mutational analysis of the PKD2 gene, two additional likely pathogenic mutations were detected.
Conclusions
Probable pathogenic mutation was detected in 71% of screened patients. Determination of PKD mutations and their type and localization within corresponding genes could help to assess clinical prognosis of ADPKD patients and has major benefit for prenatal and/or presymptomatic or preimplantational diagnostics in affected families as well.
Keywords: ADPKD, PKD gene, Mutational analysis, Mutation, HRM, MLPA, Polycystic kidney disease
Background
ADPKD is a hereditary renal disorder with the incidence of 1:400 to 1:1000 live births, characterized by focal development and progressive enlargement of cysts in kidneys leading to the decline of renal function. End-stage renal disease (ESRD) occurs during the late phase of the illness and the majority of patients have to start dialysis and/or undergo kidney transplantation. The disease is frequently accompanied by many extra-renal symptoms, such as hypertension, formation of cysts in liver or pancreas, abnormalities of cardiovascular system or development of brain-aneurysms [1].
ADPKD is genetically determined by two genes, PKD1 (16p13.3) [2] and PKD2 (4q21-22) [3]. In clinical identified populations, mutations of the PKD1 gene account for approximately 85% of all ADPKD cases and are responsible for more serious form of the disease (the average age at the onset of ESRD is 58) [4]. Fifteen percent of ADPKD patients with milder clinical presentation (the average age at the onset of ESRD is 79) carry mutation in the PKD2 gene [5,6]. The hypothetical existence of the third PKD locus has not been supported by new reports [7,8].
The PKD1 gene (46 exons) encodes a large, transmembrane protein called polycystin-1 (4303 amino acids). The PKD2 gene product (15 exons) is an integral membrane protein polycystin-2 (968 amino acids) that acts as a transient receptor potential (TRP) ion channel involved in the regulation of intracellular Ca2+ concentration. Both proteins interact with each other in kidney primary cilia forming complex that hypothetically functions as a flow-dependent mechanosensor with a crucial role in the adhesion, proliferation and differentiation of tubular epithelial cells [9,10].
Both genes are highly polymorphic and sequence changes are spread over their whole length [11]. The majority of mutations are unique to a single family, recurrent mutations account only for 30% of the total [12]. Currently, 978 sequence variants of the PKD1 gene and 193 sequence variants of the PKD2 gene have been published in the Human Gene Mutation Database (HGMD) (http://www.hgmd.cf.ac.uk). The specialized Autosomal Dominant Polycystic Kidney Disease Mutation Database (PKDB) presents even 1,923 different sequence variants of PKD1 and 241 sequence variants of the PKD2 gene (http://pkdb.mayo.edu; [13]).
The high variability of sequence changes is not the only fact that makes the analysis of PKD genes difficult. First 33 exons of the PKD1 gene are duplicated six times with 97% sequential identity. These pseudo genes are located in the close surroundings of the original gene and significantly complicate the mutational screening of the PKD1 gene [14].
The clinical course of ADPKD is highly variable not only among families with different mutations but within families with a defined mutation as well. The large phenotypic variability of ADPKD within affected families highlights a role for the genetic background in disease presentation. Hypomorphic or incompletely penetrant PKD1 or PKD2 alleles have been described, causing mild cystic disease [15,16]. Coinheritance of hypomorphic allele in combination with an inactivating PKD allele can be associated with early-onset disease. About 2% of ADPKD patients show such an early and severe phenotype (renal enlargement in utero or in infancy) that can be clinically indistinguishable from autosomal recessive polycystic kidney disease (ARPKD). These severe cases have been explained by the presence of the expected familial germ-line mutation and, in addition, other supplementary mutations in PKD genes (including PKD1, PKD2, PKHD1 and also HNF-1β gene), which act as modifying genes and likely aggravate the final phenotype [17]. Moreover, mosaicism can also modulate the course of ADPKD in some families [16,18,19].
Diagnosis of ADPKD is usually based on an age-specific phenotype of affected kidneys (number and size of renal cysts on the ultrasound, computed tomography or magnetic resonance imaging), accompanying factors (for example hypertension) and positive family history [19-21]. Molecular diagnostics (traditional linkage analysis or direct mutational screening) is a useful tool to determine and confirm the definite diagnosis, especially for evaluation of potential living related kidney donors, for individuals with negative family history and for patients with early onset of ADPKD [22]. Molecular analysis of PKD genes is quite complicated and time-consuming, therefore new methods are being developed to simplify the whole process [23-25].
In the present study, the mutational analysis of both PKD genes in a cohort of 56 Czech patients with ADPKD was performed using HRM and MLPA techniques.
Methods
Patients
The mutational screening of PKD genes was performed in a set of 56 unrelated Czech patients with ADPKD diagnosed by renal ultrasound in accordance with previously described criteria [26]. The study was approved by the Ethics Committee of General University Hospital in Prague on May 23, 2006. All patients provided written informed consent. According to the linkage analysis, the disease was linked to the PKD1 gene in 20 individuals. In 26 patients a relation of the disease to the PKD1 gene was not proved and 10 patients had an isolated occurrence of the disease within a family.
Linkage analysis
Linkage analysis was carried out using a panel of CA-repeat flanking markers. The combination and number of markers used for analysis were selected to achieve at least two informative markers, one on the 5′end and the second on the 3′end of PKD gene to rule out possible recombination. The markers used for PKD1 analysis were: D16S521, D16S3024, KG8, CW4, CW3, CW2, and for PKD2 analysis: D4S231, D4S1534, JSTG3, JV108, AICA1, D4S1563 and D4S414.
Mutational screening
The genomic DNA was isolated from the peripheral blood lymphocytes using QIAGEN spin columns on a QIAcube (QIAGEN GmbH).
At first, the screening of the PKD1 gene was carried out in the selected set of 56 patients. For mutational analysis of the PKD1 gene, the technique of a long-range PCR (LR-PCR) followed by nested PCR was used [27]. Nested PCR amplicons were produced and analyzed by the method of High Resolution Melting (HRM); positive clones were reanalyzed and confirmed by sequencing in both directions. The protocol of the whole strategy (including primers and PCR reaction conditions) was previously described in detail by Reiterova et al. [16].
The DNA of patients where no definite pathogenic mutation was found within the PKD1 gene (29 samples) were subsequently analyzed via mutational screening of the PKD2 gene by HRM and via MLPA for large rearrangements of both PKD1 and PKD2 genes. This subset of 29 individuals included patients without any sequence changes found by mutational analysis of the PKD1 gene as well as patients with unpublished putative missense mutations, putative splice-site mutations and in-frame deletions/insertions detected in the PKD1 gene.
The coding region and intron-exon boundaries of the PKD2 gene were screened via 17 PCR fragments. The sequences of PCR primers (some primers were designed on the base of Tan et al.) and the conditions of PCR reactions are available upon request [28]. PCR fragments were produced and analyzed by HRM using the LightCycler® 480 (Roche Applied Science). The reaction mixture (total volume of 10 μl) contained 1 μl of diluted template DNA, 1 × LightCycler® 480 High Resolution Melting Master Mix (Roche Applied Science) and 5 pmol of primers. The LightCycler® 480 Gene Scanning Software (Roche Applied Science) analyses the melting profile of samples to identify changes in the shape of melting curves, which indicate both homozygous and heterozygous allelic variants in a sample (Figure 1) [29]. All PCR fragments showing aberrant melting curves by HRM were sequenced. The sequencing reaction was performed according to the manufacturer’s instructions using the BigDye® Terminator v1.1 Cycle Sequencing Kits (Applied Biosystems) and ran on an ABI PRISM® 310 Genetic Analyser (Applied Biosystems).
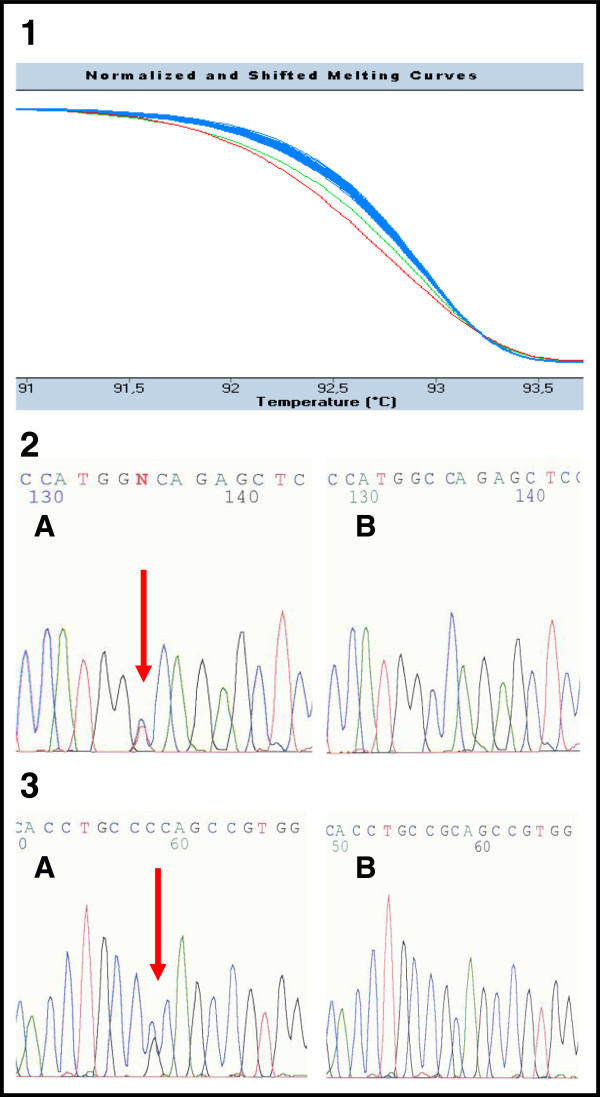
Figure 1.
The example of HRM analysis in patients 385 and 419 (exon 39). 1 Detection of mutations by HRM. The green curve in the diagram corresponds to the PCR fragment carrying the nonsense mutation c.11172G > A (p.Trp3724X) in patient 385. The red curve refers to the PCR fragment carrying the likely pathogenic substitution c.11248C > G (p.Arg3750Gly) in patient 419. The rest of fragments with blue curves are wild type samples (patients without sequence changes). 2, 3 Confirmation of mutations by direct sequencing in patient 385 (number 2) and patient 419 (number 3). Both sequences are in reverse direction. The letter A corresponds to the mutated sequence; B is the wild type.
MLPA analysis (MRC-Holland) was carried out with SALSA MLPA KIT P351-B1/P352-B1 PKD1-PKD2, which together covers 37 from 46 exons of the PKD1 gene, its upstream regulatory sequences, entire PKD2 gene and three exons of the TSC2 gene adjacent to the PKD1 gene [30]. The MLPA reaction was performed with 50–100 ng of genomic DNA according to the manufacturer’s instructions. The result of MLPA was carried out by fragmentation analysis with the use of ABI PRISM® 310 Genetic Analyser (Applied Biosystems). To obtain the final results, the generated raw data were normalized according to the manufacturer’s instructions. At first intra-sample normalization was performed by dividing the peak area of each probe’s amplification product by the total area of only the reference probes in the probemix. Subsequently, the normalization between individual samples was done by dividing the intra-normalized probe ratio of each sample by the average intra-normalized probe ratio of reference samples (negative controls). In-house developed method was used for the entry of all these calculations.
To prove the segregation of the potential mutation in PKD genes with the disease in the family, the DNAs of all available members of the respective family (including healthy individuals) were tested as well.
Data analysis and sequence changes classification
All sequenced samples were compared to the following NCBI reference sequences: [GeneBank: NM_001009944.2] for the PKD1 gene and [GeneBank: NM_000297.3] for the PKD2 gene. The sequence changes found within this study were checked with the list of PKD sequence variants currently published in the Human Gene Mutation Database (HGMD) (http://www.hgmd.cf.ac.uk) and the Autosomal Dominant Polycystic Kidney Disease Mutation Database (PKDB) (http://pkdb.mayo.edu). In case of novel mutations, their pathogenic potential was assessed. Nonsense or frameshift variants leading to a premature STOP codon were considered as definitely pathogenic.
The pathogenic potential of missense variants was evaluated computationally by examining interspecies sequence variations using PolyPhen2 (http://genetics.bwh.harvard.edu/pph2/; [4,31,32]) and Sorting Intolerant from Tolerant (SIFT) (http://sift.bii.a-star.edu.sg/; [4,32,33]) software. The sequence variant was considered as likely pathogenic, if both scores assessed its pathogenity. When only one test predicted the pathogenic potential, the variation was reported as indeterminate. The pathogenic relevance of putative splice site mutations was evaluated in the same way using NetGene2 (http://www.cbs.dtu.dk/services/NetGene2/; [34]) and Human Splicing Finder (HSF) (http://www.umd.be/HSF/; [35]) software. Further criteria for classification of missense or splice site variant as likely pathogenic mutation were its segregation within the affected family and absence of additional, more probable mutation in a concrete patient (family).
Possible influence of sex and type of mutations on the age of renal failure was evaluated. For this purpose, Shapiro-Wilk normality test for each group was performed and subsequently, according to results of normality test, unpaired two-sample t-test was used to compare the groups. All computations were carried out using in-house developed scripts in R language.
Results
We performed mutational analysis in PKD genes in a set of 56 unrelated Czech patients with ADPKD diagnosed by renal ultrasound. Twenty selected individuals came from families where ADPKD was linked unequivocally to the PKD1 gene according to the linkage analysis. Other 26 cases were the families with more serious manifestation of the disease (ESRD before 60 years of age). In this group of patients, a direct relation of the disease to the PKD1 gene has not been proven by the method of linkage analysis. At last, 10 unrelated individuals with an isolated occurrence of the disease within their families were examined (an assumption of de novo mutation).
Mutational screening of the PKD1 gene revealed 38 (36 different) likely pathogenic sequence changes in 37 from the total set of 56 patients (Table 1). In one individual, two different likely pathogenic mutations were found.
Table 1.
Likely pathogenic sequence changes of the PKD1 gene identified by HRM and direct sequencing in a set of patients from the Czech population
| Family | Exon/intron | Nucleotide change | Amino acid change/predicted effect | Predicted location within PKD1 domains | Reference |
|---|---|---|---|---|---|
|
394
ISOL.
|
4 |
c.450_459del10 |
p.Glu151ArgfsX135 |
LRRNT |
Novel |
|
338
PKD1
|
7 |
c.1564 T > C |
p.Cys522Arg |
C-type lectin domain |
Novel |
| 39
INDET.
|
15 |
c.3312_3313dupC |
p.Val1105ArgfsX4 |
PKD repeats |
[16] |
|
429
PKD1
|
15 |
c.4551_4581del31 |
p.Tyr1517X |
PKD repeats |
Novel |
|
236
PKD1
|
15 |
c.5650G > T |
p.Glu1884X |
PKD repeats |
Novel |
| 478
INDET.
|
15 |
c.5824_5825dupC |
p.Arg1942ProfsX47 |
PKD repeats |
[36] |
| 301
PKD1
|
15 |
c.5995G > A |
p.Gly1999Ser |
PKD repeats |
[12] |
| 496
ISOL.
| |||||
|
315
PKD1
|
15 |
c.6130A > G |
p.Asn2044Asp |
PKD repeats |
Novel |
|
249
INDET.
|
15 |
c.6586C > T |
p.Gln2196X |
REJ domain |
Novel |
|
427
ISOL.
|
15 |
c.6895delG |
p.Ala2299ProfsX14 |
REJ domain |
Novel |
|
237
PKD1
|
16 |
c.6960_6961insG |
p.Ser2321GlufsX98 |
REJ domain |
Novel |
| 253
PKD1
|
16 |
c.7000_7001dupGCTGGCG |
p.Val2334GlyfsX87 |
REJ domain |
[37] |
|
246
PKD1
|
17 |
c.7144_7156del13insTAC |
p.Ser2382TyrfsX234 |
REJ domain |
Novel |
|
112
INDET.
|
18 |
c.7484G > A |
p.Cys2495Tyr |
REJ domain |
Novel |
|
403
PKD1
|
20 |
c.7758_7761del4 |
p.Trp2587CysfsX31 |
REJ domain |
Novel |
|
357
INDET.
|
20 |
c.7856 T > G |
p.Leu2619Arg |
REJ domain |
Novel |
| 438
ISOL.
|
23 |
c.8309A > T |
p.Asn2770Ile |
REJ domain |
[36] |
| 336
INDET.
|
23 |
c.8311G > A |
p.Glu2771Lys |
REJ domain |
[37] |
| 389
PKD1
| |||||
| 275
ISOL.
|
23 |
c.8428G > T |
p.Glu2810X |
REJ domain |
[38] |
| c.8614delA |
p.Ile2872SerfsX3 |
|
[39] |
||
|
482
INDET.
|
IVS23 |
c.8792-2A > T |
Probable splice defect |
N/A |
Novel |
|
479
INDET.
|
27 |
c.9416_9417dupACGTGGG |
p.Ile3140ArgfsX40 |
PLAT domain |
Novel |
|
430
PKD1
|
28 |
c.9584G > A |
p.Trp3195X |
PLAT domain |
Novel |
|
391
PKD1
|
29 |
c.9904G > C9906_9907delTG |
p.Val3302LeufsX86 |
TM domain |
Novel |
|
395
ISOL.
|
33 |
c.10321C > T |
p.Gln3441X |
Not defined |
Novel |
|
413
INDET.
|
IVS36 |
c.10821 + 4_10821 + 6delAGG |
Probable splice defect |
N/A |
Novel |
| 330
INDET.
|
37 |
c.10951G > A |
p.Gly3651Ser |
Not defined |
[40] |
|
297
PKD1
|
37 |
c.10980_10982delAGC |
p.Glu3660_Ala3661delinsAsp |
Not defined |
Novel |
|
385
INDET.
|
39 |
c.11172G > A |
p.Trp3724X |
PKD channel domain |
Novel |
|
419
INDET.
|
39 |
c.11248C > G |
p.Arg3750Gly |
PKD channel domain |
Novel |
| 379
INDET.
|
41 |
c.11482_11484delGAG |
p.Glu3828del |
PKD channel domain |
[40] |
|
420
PKD1
|
43 |
c.11884 C > T |
p.Gln3962X |
PKD channel domain |
Novel |
|
161
INDET.
|
43 |
c.11993_11994dup9 |
p.Leu3998_Leu3999insPheLeuLeu |
PKD channel domain |
Novel |
|
425
INDET.
|
IVS 43-44 |
c.12004-15_12015del27 |
Probable splice defect |
N/A |
Novel |
| 215
PKD1
|
44 |
c.12061 C > T |
p.Arg4021X |
PKD channel domain |
[41] |
| 454 INDET. | 45 | c.12439_12441delAAG | p.Lys4147del | Not defined | Novel |
cDNA numbering is based on the reference database: Autosomal Dominant Polycystic Kidney Disease Mutation Database (PKDB) (http://pkdb.mayo.edu). Novel probable mutations are in boldface type. ISOL. patients with isolated occurrence of ADPKD in a family; PKD1 patients with proved linkage of ADPKD to the PKD1 gene; INDET. indeterminate patients (the linkage of ADPKD to PKD1 gene has not been proved); IVS – the intronic sequence; Novel mutation was not described in the Autosomal Dominant Polycystic Kidney Disease Mutation Database (PKDB) (http://pkdb.mayo.edu) and/or in the Human Gene Mutation Database (HGMD) (http://www.hgmd.cf.ac.uk). The potential location of mutations has been established on the basis of theoretical model of polycystin-1 by UniProtKB/Swiss-Prot database [P98161]. LRRNT – leucine rich repeat N-terminal domain; REJ – receptor for Egg Jelly; PLAT – Polycystin-1, Lipoxygenase, Alpha-Toxin domain; TM – transmembrane domain. As not defined are called sequences within PKD1 protein with unknown domain structure.
In total, eleven frameshift mutations (29%), nine nonsense mutations (24%), eleven missense mutations (29%), four small in-frame deletions/insertions (10%) and three putative splice-site mutations (8%) were detected in our set of patients. Via MLPA, one additional new large deletion was found within the PKD1 gene.
Fifty-seven percent (21) of likely pathogenic mutations found matched the definitions of being definitely pathogenic. Twenty-six of the mutations found in this study were novel. Moreover, three additional indeterminate sequence changes (two missense and one splice-site) were described (Table 2).
Table 2.
Indeterminate sequence changes of PKD genes identified by HRM and direct sequencing in a set of patients from the Czech population
| Family | Gene/exon/intron | Nucleotide change | Amino acid change/predicted effect | Predicted location within PKD domains | Reference |
|---|---|---|---|---|---|
| 308
INDET.
|
PKD1/15 |
c.3602C > T |
p.Ala1201Val |
PKD repeats |
Novel |
| 412
INDET.
|
PKD1/29 |
c.9718G > A |
p.Ala3240Thr |
Not defined |
Novel |
| 409
INDET.
|
PKD1/IVS42 |
c.11712 + 8C > A |
Probable splice defect |
N/A |
Novel |
| 466 INDET. | PKD2/IVS 9 | c.2019 + 9A > C | Probable splice defect | N/A | Novel |
cDNA numbering is based on the reference database: Autosomal Dominant Polycystic Kidney Disease Mutation Database (PKDB) (http://pkdb.mayo.edu). INDET. indeterminate patients (the linkage of ADPKD to PKD1 gene has not been proved); IVS the intronic sequence; Current paper mutation was not described in the Autosomal Dominant Polycystic Kidney Disease Mutation Database (PKDB) (http://pkdb.mayo.edu) and/or in the Human Gene Mutation Database (HGMD) (http://www.hgmd.cf.ac.uk). The potential location of mutations has been established on the basis of theoretical models of polycystins by UniProtKB/Swiss-Prot database (PKD1: P98161, PKD2:Q13563). As not defined are called sequences within PKD proteins with unknown domain structure.
A great deal of sequence variants of the PKD1 gene detected in our study were changes which do not alter the amino acid composition or have been already published as polymorphisms (or sequence changes with neutral effect on the function of polycystin 1). Complementary mutational analysis of the PKD2 gene revealed two already described likely pathogenic mutations and one indeterminate splice-site variant. All three sequence changes were found within patients without proven linkage of ADPKD to the PKD1 gene (Table 2 and 3). Screening of the PKD2 gene thus increased the final detection level of likely pathogenic mutations in this study to 71% (40 patients with a putative mutation from the set of 56 individuals in total).
Table 3.
Likely pathogenic sequence changes of the PKD2 gene identified by HRM and direct sequencing in a set of patients from the Czech population
| Family | Exon/intron | Nucleotide change | Amino acid change | Predicted location within PKD2 domains | Reference |
|---|---|---|---|---|---|
| 421
INDET.
|
6 |
c.1345_1346insGCAACAG |
Gly450AsnfsX22 |
Polycystin cation channel domain |
[42] |
| 467 INDET. | IVS 11 | c.2240 + 1G > T | Splice | N/A | [43] |
cDNA numbering is based on the reference database: Autosomal Dominant Polycystic Kidney Disease Mutation Database (PKDB) (http://pkdb.mayo.edu). INDET. indeterminate patients (the linkage of ADPKD to PKD1 gene has not been proved); IVS the intronic sequence. The potential location of mutations has been established on the basis of theoretical model of polycystin-2 by UniProtKB/Swiss-Prot database [Q13563].
Regarding the distribution of revealed mutations among the patients with distinctive phenotype and a family history, 16 likely pathogenic mutations (42%) were found in a cohort with more serious manifestation of ADPKD (ESRD reached before the age of 60 years). Fifteen putative mutations (approximately 40%) were detected among families with clear linkage to the PKD1 gene. The rest (18% of likely pathogenic mutations) referred to the set of patients without positive family history (isolated occurrence in a family).
From all 56 patients and their analyzed affected relatives, 20 males and 17 females have already reached ESRD (Table 4). Computation of dependency of sex and type of mutation (missense and small in-frame indels vs. truncating mutations) on the age of ESRD was performed in a group of 37 individuals. According to the Shapiro-Wilk test of normality, all tested groups had normal distribution (p > 0.1). Two-sample t-test did not reject null hypothesis in either sex (t = −0.9, p = 0.4), or type of mutation (t = 0.8, p = 0.4) dependency on age of renal failure.
Table 4.
Ages of end stage renal disease in families with likely pathogenic mutation found in the PKD1 gene
| Family | Nucleotide change | Amino acid change/predicted effect | Ages of end stage renal disease in the family in years, M-male, F-female |
|---|---|---|---|
| 429 |
c.4551_4581del31 |
p.Tyr1517X |
M 39, M 26 |
| 236 |
c.5650G > T |
p.Glu1884X |
M 52 |
| 478 |
c.5824_5825dupC |
p.Arg1942ProfsX47 |
M 40 |
| 301 |
c.5995G > A |
p.Gly1999Ser |
M 54 |
| 237 |
c.6960_6961insG |
p.Ser2321GlufsX98 |
M 48, F 45 |
| 253 |
c.7000_7001dupGCTGGCG |
p.Val2334GlyfsX87 |
M 61, M 60, M 36 |
| 403 |
c.7758_7761del4 |
p.Trp2587CysfsX31 |
F 60 |
| 357 |
c.7856 T > G |
p.Leu2619Arg |
M 41, F 57 |
| 482 |
c.8792-2A > T |
Probable splice defect |
M 43 |
| 479 |
c.9416_9417dupACGTGGG |
p.Ile3140ArgfsX40 |
F 79, M 55, M 51 |
| 430 |
c.9584G > A |
p.Trp3195X |
M 57 |
| 391 |
c.9904G > C9906_9907delTG |
p.Val3302LeufsX86 |
M 61 |
| 413 |
c.10821 + 4_10821 + 6delAGG |
Probable splice defect |
F43, F 49 |
| 330 |
c.10951G > A |
p.Gly3651Ser |
F73, F71, M 73 |
| 297 |
c.10980_10982delAGC |
p.Glu3660_Ala3661delinsAsp |
M 41, F 44, F 35 |
| 385 |
c.11172G > A |
p.Trp3724X |
F 49 |
| 419 |
c.11248C > G |
p.Arg3750Gly |
F 44 |
| 420 |
c.11884 C > T |
p.Gln3962X |
F 45, F 46 |
| 425 |
c.12004-15_12015del27 |
Probable splice defect |
F 65, F 51 |
| 215 |
c.12061 C > T |
p.Arg4021X |
F 58, M 50, M 50 |
| 454 | c.12439_12441delAAG | p.Lys4147del | M 67 |
Nonsense and frameshift mutations
One patient (275) from our study was found to have two truncating pathogenic mutations (one nonsense mutation and one frameshift mutation), both already published and both located within the exon 23 probably in cis regarding his mild phenotype of ADPKD without progression. Similar cases have been already described in the literature, but their existence is rare [11]. The patients 394, 395 and 427 are the representatives of the group with isolated occurrence of ADPKD within their families.
Missense mutations
Via our mutational analysis of both PKD genes, 9 likely pathogenic missense mutations have been identified; all within the PKD1 gene. Five mutations have been determined as novel and two already published substitutions (p.Gly1999Ser and p.Glu2771Lys) were recurrent (both present in two different individuals). Eight substitutions segregated with the disease in affected families (Table 5) and were confirmed as pathogenic by both predictor programs (PolyPhen2 and SIFT), one substitution occurred de novo.
Table 5.
Segregation of likely pathogenic missense and small in-frame indel mutations of PKD1 with polycystic kidney disease in affected families
| Amino acid change | Nucleotide change | Family | Number of affected/healthy family members |
|---|---|---|---|
| p.Cys522Arg |
c.1564 T > C |
338 |
5/2 |
| p.Gly1999Ser |
c.5995G > A |
301 |
3/2 |
| p.Asn2044Asp |
c.6130A > G |
315 |
6/4 |
| p.Cys2495Tyr |
c.7484G > A |
112 |
3/3 |
| p.Leu2619Arg |
c.7856 T > G |
357 |
2/3 |
| p.Glu2771Lys |
c.8311G > A |
336 |
2/1 |
| 389 |
6/5 |
||
| p.Gly3651Ser |
c.10951G > A |
330 |
2/3 |
| p.Arg3750Gly |
c.11248C > G |
419 |
2/2 |
| p.Glu3660_Ala3661delinsAsp |
c.10980_10982delAGC |
297 |
2/4 |
| p.Leu3998_Leu3999insPheLeuLeu |
c.11993_11994dup9 |
161 |
3/3 |
| p.Lys4147del | c.12439_12441delAAG | 454 | 3/2 |
Splice-site mutations
The probable splice-site mutations c.8792-2A > T (patient 482) and c.12004-15_12015del27 (patient 425) cover the consensus sequence of conserved splice-site and both predictor programs suggested their pathogenity. Although the probable splice-site mutation c.10821 + 4_10821 + 6del AGG (patient 413) is also situated within the conserved splice-site, its pathogenic impact is milder. According to NetGene2 and HSF, this mutation does not abolish the splice-site completely but decreases its efficiency significantly. Therefore this mutation was classified as pathogenic as well.
Small in-frame deletions/insertions
Within our set of 56 Czech individuals (families), four small in-frame deletions/insertions were detected; three of them were described as novel. The common molecular mechanisms of origin of such genomic events are non-homologous end joining (NHEJ) or microhomology-mediated replication-dependent recombination (MMRDR) [44]. All three novel small in-frame deletions/insertions characterized in the present study show high evolutionary conservation of involved amino acid residues; all three are the only probably pathogenic mutations identified in the patient and segregate with the disease in affected families (Table 5). Their pathogenic potential is therefore highly probable.
Large rearrangements
By the method of MLPA, one novel large deletion of the PKD1 gene was revealed in patient 105 with proven linkage of ADPKD to the PKD1 gene. The deletion was spread from the exon 3 to the first half of exon 15. The exact beginning of the deletion is arguable, because there is no probe for exon 2 in the MLPA kit used within our study. The deletion was identified in all available affected members of the family but was not characterized at the nucleotide level due to the sequence complexity of the PKD1 locus.
Indeterminate sequence changes
Four sequence changes found within our set of patients were categorized as indeterminate (Table 2). The possible splice-site mutations PKD1/c.11712 + 8C > A and PKD2/c.2019 + 9A > C segregate with the disease in affected families and are the only possible mutations found in patients 409 and 466, respectively. However, their location is out of the conventional range of the consensus splice-site sequence and both predictor programs (NetGene2 and HSF) evaluated them as non-pathogenic. Despite its segregation within affected family, also the substitution PKD1/p.Ala1201Val (patient 308) was determined as benign by both software programs used (PolyPhen2 and SIFT). Although the substitution PKD1/p.Ala3240Thr (patient 412) was predicted as possibly damaging by PolyPhen2, SIFT defined it as benign as well.
Discussion
The highest portion of mutations found within our study (54%) was predicted to truncate the protein. These truncating changes include frameshift mutations, nonsense mutations and one large deletion. Our findings seem to be in concordance with the recent publication by Cornec-Le Gall et al., where approximately two thirds of PKD1 mutation-positive pedigrees carried truncating mutations [6]. In addition, the most frequent definitely pathogenic mutation type of the PKD1 gene in our study is a frameshift mutation (29%), which is consistent with detailed current reports [6,12,36].
Characterization of mutations
In the present study, only 5.3% of mutations were recurrent (2 different already published mutations, both found in two different unrelated families). Both these mutations occur within CpG dinucleotides (G > A transitions), which are mutational hotspots typical for recurrent sequence changes [45]. The substitution p.Glu2771Lys is even published for the sixth time (always from a different location). This fact strongly supports its recurrent origin instead of the existence of a common ancestor.
Such a low percentage of recurrent mutations is in contrast with studies by other groups. Up to 30% of recurrent mutations were mentioned regarding PKD genes in the study by Rossetti et al.[12]. Also, the latest comprehensive mutational analysis of PKD genes in 700 unrelated patients revealed 20.8% of recurrent mutations [36]. The difference can be caused by a smaller number of patients analyzed within our study (56) compared to extensive studies by Rossetti et al. (202) and Audrézet et al. (700).
According to the recent data from literature, ADPKD without family history is present in about 10% of adult patients [46]. The clinical characteristics of patients with and without family history are very similar but only about a half of patients without positive family history show extrarenal abnormalities. In our case, 18% of found likely pathogenic mutations fall into the group of patients with isolated occurrence of ADPKD (the so called de novo mutations).
Large DNA rearrangements account for a maximum of 4% of all PKD1 mutations and even fewer of all PKD2 mutations [18]. Finding of only one large deletion of the PKD1 gene within our set of patients (approximately 2.5%) is thus in accordance with the data from literature.
Location of mutations
Sequence changes of all types were spread throughout the PKD1 gene (Table 1). The only evidence of a cluster of mutations (especially truncations) was at the end of exon 15. This region corresponds to the junction of the PKD repeats and the REJ domain of the resulted protein. The reference about this sort of clustering has been already mentioned in literature by Rossetti et al.[12]. No determination of mutational distribution in the PKD2 gene can be done as only one exonic mutation was found. Both PKD genes are characterized by a high level of allelic heterogeneity (a great number of polymorphisms), relatively high GC content and no mutation hotspots. The mutations are usually unique, highly variable and spread throughout the entire gene which fully corresponds with our observations [13]. However, the latest comprehensive study of PKD genes suggests the existence of theoretical hotspots for the three major types of mutations within the PKD1 gene [36]. Our results could not confirm this hypothesis because of the limited extent of the study that did not allow us such a specific computational analysis.
Genotype/phenotype correlation
Generally, the ADPKD phenotype is very variable and the correlation between genotype and phenotype is usually not apparent, even within one family. One of the explanations could be a different genetic background and the effect of heritable modifying factors.
The influence of gender, mutation type and mutation location on phenotype of ADPKD is limited and not completely clear. A slight correlation between gender and median age at ESRD onset has been described in literature (56.1 years for men versus 59.5 years for women) [6]. Although the influence of gender has been demonstrated in case of the PKD2 gene too, the most recent report has not proved this dependency [4,6]. Our data also suggested a slightly higher mean age of ESRD in ADPKD females than in males (53.8 ± 12.3 years versus 50.3 ± 11.4 years). The ages of ESRD in Czech patients compared with those described by Cornec-Le Gall et al. were slightly lower, but in concordance with observations made by Torra et al. (49.6 years for men versus 53.1 years for women) [47].
Another dependency examined within our study was the effect of distinctive type of mutation on the phenotype of ADPKD. We observed that the mean age of ESRD is slightly higher in case of Czech ADPKD patients with missense mutations and in-frame amino acids deletions (54.6 ± 14.4 years) in comparison with patients carrying protein truncating mutations (50.9 ± 11.1 years). This observation corresponds with a strong correlation between the mutation type and median age at ESRD onset that has been recently presented in literature (55.6 years in case of truncating mutations versus 67.9 years in case of mutations with no truncating effect) [6]. For further examination of this dependency a bigger data set would be necessary as only information from 11 patients with missense mutations and in-frame amino acids deletions was available.
Two PKD1 mutations were confirmed in a 24-year-old patient with normal renal function (patient 275 with two mutations within exon 23). The ultrasound of his parents was normal, without cysts. It seems probable that both mutations were de novo however DNA samples from parents were not available. Moreover, a family with large PKD1 deletion was described. The age of ESRD in two males (40 years and 53 years) was known in this family. Intrafamiliar variability has been repeatedly described in PKD families, and was noted in this family, as well. The clinical course in the male with ESRD at the age of 40 years was influenced by arterial hypertension. There were three more affected members in this family (35–50 years) with normal renal function or only mild renal insufficiency. The large PKD1 deletion does not lead to severe clinical course.
The more severe manifestation of the disease in patients with mutations in the 5′ region of the PKD1 gene has been described in comparison with patients carrying mutations in the 3′ region [11]. Nevertheless, a recent study with 741 patients has not shown the significant difference between the 5′ and 3′ end regions [6]. In the case of the PKD2 gene, no clear relationship between mutation location and ADPKD severity has been found, either [48]. Within our study, no effect of location of PKD1 mutations on the mean age of ESRD of Czech ADPKD patients has been observed.
Detection techniques
Mutation analysis of highly polymorphic PKD genes is challenging because of the length of genes (especially PKD1 gene), multi-exon structures, high GC content, marked allelic heterogeneity and existence of highly homologous pseudogenes of the PKD1 gene. On average, as much as 10.1 different sequence changes per patient are reported in case of the PKD1 gene [12,14]. Few complete screens of both genes have been already published with the use of different molecular techniques, but all described methods have still a lot of limitations, including time-demand, high price or insufficient sensitivity. The detection level of the present study is 71% (40 patients/families with likely pathogenic mutations in PKD genes from 56 patients in total) which is in accordance with the 75% efficiency reported by Bataille et al., where the same method of high resolution melting (HRM) was used [23]. The cases with no probable mutation found may correspond to allele dropout, different category of sequence changes that have not been analyzed within this study (such as deep intronic variants or missed mutations in promoter regions) or can be caused by incorrect original diagnosis. Hypomorphic variants or polygenic inheritance may also play an important role [15,17]. By denaturing high-performance liquid chromatography (DHPLC), approximately 64% efficiency for definitely pathogenic mutations has been achieved [11]. A current publication concerning DHPLC presented the total detection rate of 52.3% [39]. The combination of DHPLC with direct sequencing has increased the level of detection up to 89% [12]. Similarly, 86% efficiency has been reported with Transgenomic’s SURVEYOR Nuclease and WAVE Nucleic Acid High Sensitivity Fragment Analysis System [28]. Using direct sequencing alone, different results have been achieved ranging from 64.5% of mutations detected by Hoefele et al., 78% of probable pathogenic mutations detected by Garcia-Gonzales et al. to the highest efficiency of 93.8% reported by Cornec-Le Gall et al.[6,36,49,50]. In the most recent studies, the new high-throughput next generation sequencing (NGS) techniques have been tested for mutational analysis of PKD genes with the maximal detection rate of 63% and 69.4% [24,25]. The NGS deep sequencing helps with identification of missed or atypical sequence variants, which are hardly detectable through the conventional Sanger sequencing (especially allele dropout, gene conversion or deep intronic variants). Promising is also the new LR-PCR Sequencing Method developed by Tan et al. with very high sensitivity, improved intronic coverage and marked saving of time and costs [51].
In comparison with other detection techniques, HRM analysis has good and comparable efficacy. Ninety-six or even 394 samples could be analyzed at once through the simple strategy of PCR followed automatically by HRM analysis. The most important benefits of this method are thus the savings of time and costs and general convenience of the working strategy, decreasing the probability of mistakes within the whole process.
Conclusion
The methods for mutational identification are constantly improving and enable us to screen even such a complex gene as PKD1. The HRM and MLPA are relatively fast and convenient methods that together reached the detection level of 71%. The question is how to interpret the data from genetic analysis. The differences in severity of the disease within affected relatives suggest that the phenotype of ADPKD is influenced not only by the type or location of the mutation but also by genetic background and environmental factors. The assessment of patient’s prognosis on the basis of molecular diagnostics is therefore still impossible, but allows us a screening of presymptomatic relatives and kidney donors. Full characterization of molecular etiology of ADPKD should facilitate possible therapeutic intervention.
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
VE participated to carry out the molecular genetic studies of PKD genes and drafted the manuscript. JS participated in the conception and design of the study and carrying out of molecular genetic studies. LO carried out the mutational screening of PKD2 gene and revised final manuscript. JR participated in the conception and design of the study and collected the clinical data. MM and FL participated in recruitment of patients and collecting of clinical data. VT and MK conceived the study. All authors read and approved the final manuscript.
Pre-publication history
The pre-publication history for this paper can be accessed here:
Contributor Information
Lena Obeidova, Email: lena.obeidova@gmail.com.
Veronika Elisakova, Email: velisakova@post.cz.
Jitka Stekrova, Email: jstek@lf1.cuni.cz.
Jana Reiterova, Email: jreiterova@seznam.cz.
Miroslav Merta, Email: miroslav.merta@lf1.cuni.cz.
Vladimir Tesar, Email: vladimir.tesar@lf1.cuni.cz.
Frantisek Losan, Email: genetika@genetika-plzen.cz.
Milada Kohoutova, Email: mkoho@lf1.cuni.cz.
Acknowledgements
The study was funded by grants from: research projects IGA MZCR NR 9427-4/2007 and PRVOUK – P25/LF1/2. We would like to thank Stanislava Svobodova for the support with developing of methods, Anastassiya Zidkova for statistical support and Zdenka Hruskova for language corrections. We are also grateful for cooperation to patients, their families and clinical geneticists.
References
- Gabow PA. Autosomal dominant polycystic kidney disease. N Engl J Med. 1993;15:332–342. doi: 10.1056/NEJM199307293290508. [DOI] [PubMed] [Google Scholar]
- European Polycystic Kidney Disease Consortium. The polycystic kidney disease I gene encodes a 14 kb transcript and lies within a duplicated region on chromosome 16. Cell. 1994;15:881–894. doi: 10.1016/0092-8674(94)90137-6. [DOI] [PubMed] [Google Scholar]
- Kimberling WJ, Kumar S, Gabow PA, Kenyon JB, Connolly CJ, Somlo S. Autosomal dominant polycystic kidney disease: localization of the second gene to chromosome 4q13-q23. Genomics. 1993;15:467–472. doi: 10.1016/S0888-7543(11)80001-7. [DOI] [PubMed] [Google Scholar]
- Tan YC, Blumenfeld J, Rennert H. Autosomal dominant polycystic kidney disease: genetics, mutations and microRNAs. Biochim Biophys Acta. 1812;15:1202–1212. doi: 10.1016/j.bbadis.2011.03.002. [DOI] [PubMed] [Google Scholar]
- Hateboer N, van Dijk MA, Bogdanova N, Coto E, Saggar-Malik AK, San Millan JL, Torra R, Breuning M, Ravine D. Comparison of phenotypes of polycystic kidney disease types 1 and 2: European PKD1-PKD2 study group. Lancet. 1999;15:103–107. doi: 10.1016/S0140-6736(98)03495-3. [DOI] [PubMed] [Google Scholar]
- Cornec-Le Gall E, Audrézet MP, Chen JM, Hourmant M, Morin MP, Perrichot R, Charasse C, Whebe B, Renaudineau E, Jousset P, Guillodo MP, Grall-Jezequel A, Saliou P, Férec C, Le Meur Y. Type of PKD1 mutation influences renal outcome in ADPKD. J Am Soc Nephrol. 2013;15:1006–1013. doi: 10.1681/ASN.2012070650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daoust MC, Reynolds DM, Bichet DG, Somlo S. Evidence for a third genetic locus for autosomal dominant polycystic kidney disease. Genomics. 1995;15:733–736. doi: 10.1016/0888-7543(95)80020-M. [DOI] [PubMed] [Google Scholar]
- Paul BM, Consugar MB, Ryan Lee M, Sundsbak JL, Heyer CM, Rossetti S, Kubly VJ, Hopp K, Torres VE, Coto E, Clementi M, Bogdanova N, de Almeida E, Bichet DG, Harris PC. Evidence of a third ADPKD locus is not supported by re-analysis of designated PKD3 families. Kidney Int. 2013;15:383–392. doi: 10.1038/ki.2013.227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nauli SM, Alenghat FJ, Luo Y, Williams E, Vassilev P, Li X, Elia AE, Lu W, Brown EM, Quinn SJ, Ingberg DE, Zhou J. Polycystins 1 and 2 mediate mechanosensation in the primary cilium of kidney cells. Nat Genet. 2003;15:129–137. doi: 10.1038/ng1076. [DOI] [PubMed] [Google Scholar]
- Torres VE, Harris PC. Autosomal dominant polycystic kidney disease: the last 3 years. Kidney Int. 2009;15:149–168. doi: 10.1038/ki.2009.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossetti S, Burton S, Strmecki L, Pond GR, San Millán JL, Zerres K, Barratt TM, Ozen S, Torres VE, Bergstralh EJ, Winearls CG, Harris PC. The position of the polycystic kidney disease 1 (PKD1) gene mutation correlates with the severity of renal disease. J Am Soc Nephrol. 2002;15:1230–1237. doi: 10.1097/01.ASN.0000013300.11876.37. [DOI] [PubMed] [Google Scholar]
- Rossetti S, Consugar MB, Chapman AB, Torres VE, Guay-Woodford LM, Grantham JJ, Bennett WM, Meyers CM, Walker DL, Bae K, Zhang QJ, Thompson PA, Miller JP, Harris PC. CRISP Consortium. Comprehensive molecular diagnostics in autosomal dominant polycystic kidney disease. J Am Soc Nephrol. 2007;15:2143–2160. doi: 10.1681/ASN.2006121387. [DOI] [PubMed] [Google Scholar]
- Gout AM, Martin NC, Brown AF, Ravine D. PKDB: polycystic kidney disease mutation database–a gene variant database for autosomal dominant polycystic kidney disease. Hum Mutat. 2007;15:654–659. doi: 10.1002/humu.20474. [DOI] [PubMed] [Google Scholar]
- Bogdanova N, Markoff A, Gerke V, McCluskey M, Horst J, Dworniczak B. Homologues to the first gene for autosomal dominant polycystic kidney disease are pseudogenes. Genomics. 2001;15:333–341. doi: 10.1006/geno.2001.6568. [DOI] [PubMed] [Google Scholar]
- Rossetti S, Kubly VJ, Consugar MB, Hopp K, Roy S, Horsley SW, Chauveau D, Rees L, Barratt TM, van’t Hoff WG, Niaudet P, Torres VE, Harris PC. Incompletely penetrant PKD1 alleles suggest a role for gene dosage in cyst initiation in polycystic kidney disease. Kidney Int. 2009;15:848–855. doi: 10.1038/ki.2008.686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiterová J, Štekrová J, Merta M, Kotlas J, Elišáková V, Lněnička P, Korabečná M, Kohoutová M, Tesař V. Autosomal dominant polycystic kidney disease in a family with mosaicism and hypomorphic allele. BMC Nephrol. 2013;15:59. doi: 10.1186/1471-2369-14-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergmann C, van Bothmer J, Ortiz Brüchle N, Venghaus A, Frank V, Fehrenbach H, Hampel T, Pape L, Buske A, Jonsson J, Sarioglu N, Santos A, Ferreira JC, Becker JU, Cremer R, Hoefele J, Benz MR, Weber LT, Buettner R, Zerres K. Mutations in multiple PKD genes may explain early and severe polycystic kidney disease. J Am Soc Nephrol. 2011;15:2047–2056. doi: 10.1681/ASN.2010101080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Consugar MB, Wong WC, Lundquist PA, Rossetti S, Kubly VJ, Walker DL, Rangel LJ, Aspinwall R, Niaudet WP, Ozen S, David A, Velinov M, Bergstralh EJ, Bae KT, Chapman AB, Guay-Woodford LM, Grantham JJ, Torres VE, Sampson JR, Dawson BD, Harris PC. CRISP Consortium. Characterization of large rearrangements in autosomal dominant polycystic kidney disease and the PKD1/TSC2 contiguous gene syndrome. Kidney Int. 2008;15:1468–1479. doi: 10.1038/ki.2008.485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connor A, Lunt PW, Dolling C, Patel Y, Meredith AL, Gardner A, Hamilton NK, Dudley CR. Mosaicism in autosomal dominant polycystic kidney disease revealed by genetic testing to enable living related renal transplantation. Am J Transplant. 2008;15:232–237. doi: 10.1111/j.1600-6143.2007.02030.x. [DOI] [PubMed] [Google Scholar]
- Chapman AB, Guay-Woodford LM, Grantham JJ, Torres VE, Bae KT, Baumgarten DA, Kenney PJ, King BF Jr, Glockner JF, Wetzel LH, Brummer ME, O’Neill WC, Robbin ML, Bennett WM, Klahr S, Hirschman GH, Kimmel PL, Thompson PA, Miller JP. Consortium for Radiologic Imaging Studies of Polycystic Kidney Disease cohort. Renal structure in early autosomal-dominant polycystic kidney disease (ADPKD): the consortium for radiologic imaging studies of polycystic kidney disease (CRISP) cohort. Kidney Int. 2003;15:1035–1045. doi: 10.1046/j.1523-1755.2003.00185.x. [DOI] [PubMed] [Google Scholar]
- Pei Y. Diagnostic approach in autosomal dominant polycystic kidney disease. Clin J Am Soc Nephrol. 2006;15:1108–1114. doi: 10.2215/CJN.02190606. [DOI] [PubMed] [Google Scholar]
- Harris PC, Rossetti S. Molecular diagnostics for autosomal dominant polycystic kidney disease. Nat Rev Nephrol. 2010;15:197–206. doi: 10.1038/nrneph.2010.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bataille S, Berland Y, Fontes M, Burtey S. High resolution melt analysis for mutation screening in PKD1 and PKD2. BMC Nephrol. 2011;15:57. doi: 10.1186/1471-2369-12-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossetti S, Hopp K, Sikkink RA, Sundsbak JL, Lee YK, Kubly VJ, Eckloff BW, Ward CJ, Winearls CG, Torres VE, Harris PC. Identification of gene mutations in autosomal dominant polycystic kidney disease through targeted resequencing. J Am Soc Nephrol. 2012;15:915–933. doi: 10.1681/ASN.2011101032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi XP, Du ZF, Ma JM, Chen XL, Zhang Q, Fei J, Wei XM, Chen D, Ke HP, Liu XZ, Li F, Chen ZG, Su Z, Jin HY, Liu WT, Zhao Y, Jiang HL, Lan ZZ, Li PF, Fang MY, Dong W, Zhang XN. Genetic diagnosis of autosomal dominant polycystic kidney disease by targeted capture and next-generation sequencing: utility and limitations. Gene. 2013;15:93–100. doi: 10.1016/j.gene.2012.12.060. [DOI] [PubMed] [Google Scholar]
- Pei Y, Obaji J, Dupuis A, Paterson AD, Magistroni R, Dicks E, Parfrey P, Cramer B, Coto E, Torra R, San Millan JL, Gibson R, Breuning M, Peters D, Ravine D. Unified criteria for ultrasonographic diagnosis of ADPKD. J Am Soc Nephrol. 2009;15:205–212. doi: 10.1681/ASN.2008050507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watnick TJ, Piontek KB, Cordal TM, Weber H, Gandolph MA, Qian F, Lens XM, Neumann HP, Germino GG. An unusual pattern of mutation in the duplicated portion of PKD1 is revealed by use of a novel strategy for mutation detection. Hum Mol Genet. 1997;15:1473–1481. doi: 10.1093/hmg/6.9.1473. [DOI] [PubMed] [Google Scholar]
- Tan YC, Blumenfeld JD, Anghel R, Donahue S, Belenkaya R, Balina M, Parker T, Levine D, Leonard DG, Rennert H. Novel method for genomic analysis of PKD1 and PKD2 mutations in autosomal dominant polycystic kidney disease. Hum Mutat. 2009;15:264–273. doi: 10.1002/humu.20842. [DOI] [PubMed] [Google Scholar]
- Chateigner-Boutin AL, Small I. A rapid high-throughput method for the detection and quantification of RNA editing based on high-resolution melting of amplicons. Nucleic Acids Res. 2007;15:e114. doi: 10.1093/nar/gkm640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schouten JP, McElgunn CJ, Waaijer R, Zwijnenburg D, Diepvens F, Pals G. Relative quantification of 40 nucleic acid sequences by multiplex ligation-dependent probe amplification. Nucleic Acids Res. 2002;15:e57. doi: 10.1093/nar/gnf056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramensky V, Bork P, Sunyaev S. Human non-synonymous SNPs: server and survey. Nucleic Acids Res. 2002;15:3894–3900. doi: 10.1093/nar/gkf493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thusberg J, Olatubosun A, Vihinen M. Performance of mutation pathogenicity prediction methods on missense variants. Hum Mutat. 2011;15:358–368. doi: 10.1002/humu.21445. [DOI] [PubMed] [Google Scholar]
- Ng PC, Henikoff S. Accounting for human polymorphisms predicted to affect protein function. Genome Res. 2002;15:436–446. doi: 10.1101/gr.212802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunak S, Engelbrecht J, Knudsen S. Prediction of human mRNA donor and acceptor sites from the DNA sequence. J Mol Biol. 1991;15:49–65. doi: 10.1016/0022-2836(91)90380-O. [DOI] [PubMed] [Google Scholar]
- Desmet FO, Hamroun D, Lalande M, Collod-Béroud G, Claustres M, Béroud C. Human splicing finder: an online bioinformatics tool to predict splicing signals. Nucleic Acids Res. 2009;15:e67. doi: 10.1093/nar/gkp215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Audrézet MP, Cornec-Le Gall E, Chen JM, Redon S, Quéré I, Creff J, Bénech C, Maestri S, Le Meur Y, Férec C. Autosomal dominant polycystic kidney disease: comprehensive mutation analysis of PKD1 and PKD2 in 700 unrelated patients. Hum Mutat. 2012;15:1239–1250. doi: 10.1002/humu.22103. [DOI] [PubMed] [Google Scholar]
- Rossetti S, Strmecki L, Gamble V, Burton S, Sneddon V, Peral B, Roy S, Bakkaloglu A, Komel R, Winearls CG, Harris PC. Mutation analysis of the entire PKD1 gene: genetic and diagnostic implications. Am J Hum Genet. 2001;15:46–63. doi: 10.1086/316939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue S, Inoue K, Utsunomiya M, Nozaki J, Yamada Y, Iwasa T, Mori E, Yoshinaga T, Koizumi A. Mutation analysis in PKD1 of Japanese autosomal dominant polycystic kidney disease patients. Hum Mutat. 2002;15:622–628. doi: 10.1002/humu.10080. [DOI] [PubMed] [Google Scholar]
- Yu C, Yang Y, Zou L, Hu Z, Li J, Liu Y, Ma Y, Ma M, Su D, Zhang S. Identification of novel mutations in Chinese Hans with autosomal dominant polycystic kidney disease. BMC Med Genet. 2011;15:164. doi: 10.1186/1471-2350-12-164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S, Mei C, Zhang D, Dai B, Tang B, Sun T, Zhao H, Zhou Y, Li L, Wu Y, Wang W, Shen X, Song J. Mutation analysis of autosomal dominant polycystic kidney disease genes in Han Chinese. Nephron Exp Nephrol. 2005;15:e63–e76. doi: 10.1159/000084572. [DOI] [PubMed] [Google Scholar]
- Turco AE, Rossetti S, Bresin E, Corrà S, Restagno G, Carbonara A, De Prisco O, Gammaro L, Maschio G, Pignatti PF. Detection of two different nonsense mutations in exon 44 of the PKD1 gene in two unrelated Italian families with severe autosomal dominant polycystic kidney disease. Nephrol Dial Transplant. 1996;15:10–12. doi: 10.1093/ndt/11.supp6.10. [DOI] [PubMed] [Google Scholar]
- Reiterová J, Stekrová J, Peters DJ, Kapras J, Kohoutová M, Merta M, Zidovská J. Four novel mutations of the PKD2 gene in Czech families with autosomal dominant polycystic kidney disease. Hum Mutat. 2002;15:573. doi: 10.1002/humu.9035. [DOI] [PubMed] [Google Scholar]
- Chung W, Kim H, Hwang YH, Kim SY, Ko AR, Ro H, Lee KB, Lee JS, Oh KH, Ahn C. PKD2 gene mutation analysis in Korean autosomal dominant polycystic kidney disease patients using two-dimensional gene scanning. Clin Genet. 2006;15:502–508. doi: 10.1111/j.1399-0004.2006.00721.x. [DOI] [PubMed] [Google Scholar]
- Chen JM, Cooper DN, Férec C, Kehrer-Sawatzki H, Patrinos GP. Genomic rearrangements in inherited disease and cancer. Semin Cancer Biol. 2010;15:222–233. doi: 10.1016/j.semcancer.2010.05.007. [DOI] [PubMed] [Google Scholar]
- Cooper DN, Bacolla A, Férec C, Vasquez KM, Kehrer-Sawatzki H, Chen JM. On the sequence-directed nature of human gene mutation: the role of genomic architecture and the local DNA sequence environment in mediating gene mutations underlying human inherited disease. Hum Mutat. 2011;15:1075–1099. doi: 10.1002/humu.21557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neumann HPH, Bacher J, Nabulsi Z, Ortiz Brüchle N, Hoffmann MM, Schaeffner E, Nürnberger J, Cybulla M, Wilpert J, Riegler P, Corradini R, Kraemer-Guth A, Azurmendi P, Nunez M, Gläsker S, Zerres K, Jilg C. Adult patients with sporadic polycystic kidney disease: the importance of screening for mutations in the PKD1 and PKD2 genes. Int Urol Nephrol. 2012;15:1753–1762. doi: 10.1007/s11255-012-0125-0. [DOI] [PubMed] [Google Scholar]
- Torra R, Badenas C, Darnell A, Nicolau C, Volpini V, Revert L, Estivill X. Linkage, clinical features, and prognosis of autosomal dominant polycystic kidney disease types 1 and 2. J Am Soc Nephrol. 1996;15:2142–2151. doi: 10.1681/ASN.V7102142. [DOI] [PubMed] [Google Scholar]
- Hateboer N, Veldhuisen B, Peters D, Breuning MH, San Millán JL, Bogdanova N, Coto E, van Dijk MA, Afzal AR, Jeffery S, Saggar-Malik AK, Torra R, Dimitrakov D, Martinez I, de Castro SS, Krawczak M, Ravine D. Location of mutations within the PKD2 gene influences clinical outcome. Kidney Int. 2000;15:1444–1451. doi: 10.1046/j.1523-1755.2000.00989.x. [DOI] [PubMed] [Google Scholar]
- Garcia-Gonzalez MA, Jones JG, Allen SK, Palatucci CM, Batish SD, Seltzer WK, Lan Z, Allen E, Qian F, Lens XM, Pei Y, Germino GG, Watnick TJ. Evaluating the clinical utility of a molecular genetic test for polycystic kidney disease. Mol Genet Metab. 2007;15:160–167. doi: 10.1016/j.ymgme.2007.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoefele J, Mayer K, Scholz M, Klein HG. Novel PKD1 and PKD2 mutations in autosomal dominant polycystic kidney disease (ADPKD) Nephrol Dial Transplant. 2011;15:2181–2188. doi: 10.1093/ndt/gfq720. [DOI] [PubMed] [Google Scholar]
- Tan YC, Michaeel A, Blumenfeld J, Donahue S, Parker T, Levine D, Rennert H. A novel long-range PCR sequencing method for genetic analysis of the entire PKD1 gene. J Mol Diagn. 2012;15:305–313. doi: 10.1016/j.jmoldx.2012.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]