Abstract
BACKGROUND & AIMS
Hyperactivation of the RAS-RAF signaling pathway in colorectal tumors is associated with metastasis and poor outcomes of patients. Little is known about how RAS–RAF signaling is turned off once activated. We investigated how the pH domain and leucine-rich repeat protein phosphatases (PHLPPs) control RAS–RAF signaling and colorectal cancer (CRC) development.
METHODS
We used co-immunoprecipitation assays to identify substrates of PHLPP1 and PHLPP2.We studied phosphorylation of RAF1 in CRC cells that express transgenic PHLPP1 or PHLPP2, or lentiviral-based small hairpin (sh)RNAs against their transcripts; we measured effects on cell motility, migration, and invasion in vitro. Tumor progression and survival were analyzed in Phlpp1−/− mice, ApcMin mice, and ApcMin/Phlpp1−/− mice. Microarray data sets of colorectal tumor and non-tumor tissues were analyzed for PHLPP gene expression.
RESULTS
PHLPP1 and 2 were found to dephosphorylate RAF1 at S338, inhibiting its kinase activity in vitro and in CRC cells. In cells, shRNA knockdown of PHLPP1 or PHLPP2 increased the amplitude and duration of RAF-MEK-ERK signaling downstream of EGFR and KRAS, whereas overexpression had the opposite effect. Knockdown of PHLPP1 or PHLPP2 caused CRC cells to express markers of the epithelial-mesenchymal transition (EMT), and increased migration and invasion in vitro. ApcMin/Phlpp1−/− mice had decreased survival and developed larger intestinal and colon tumors than ApcMin mice, which developed mostly low-grade adenomas; in contrast, 20% of the tumors that developed in ApcMin/Phlpp1−/− mice were invasive adenocarcinomas. Normal villi and adenomas of ApcMin/Phlpp1−/− mice had significantly fewer apoptotic cells than ApcMin mice. Human CRC patient microarray data revealed that the expression of PHLPP1 or PHLPP2 is positively correlated with CDH1.
CONCLUSIONS
PHLPP1 and 2 dephosphorylate RAF1 to reduce its signaling, increase the invasive and migratory activities of CRC cells, and activate the EMT. In ApcMin mice, loss of PHLPP1 promotes tumor progression.
Keywords: colorectal cancer, metastasis, tumor suppressor, phosphatase
INTRODUCTION
PHLPP (PH domain Leucine-rich-repeats Protein Phosphatase) belongs to a novel family of Ser/Thr protein phosphatases that play an important role in maintaining the balance in cell signaling1. Two isoforms of PHLPP, namely PHLPP1 and PHLPP2, are found in this phosphatase family1. Previous studies have identified Akt as a substrate of PHLPP, and PHLPP-mediated dephosphorylation of Akt is associated with an increase in apoptosis and a decrease in proliferation in cancer cells2-4. The PHLPP1 gene is located to a chromosome region (18q21.33) that has been shown to encounter the most frequent loss of heterozygosity (LOH) in CRC5, 6. This notion has recently been confirmed by the genome-scale sequencing analysis conducted by the Cancer Genome Atlas Network. More significantly, 18q deletion has been associated with tumor aggressiveness and advanced stage of CRC7. However, the role of PHLPP-loss in promoting tumor progression and metastasis has not been determined.
RAF1 is a proto-oncogene that serves as a pivotal member downstream of growth factors and RAS8. RAF1 transduces signals from the cell membrane to nucleus by initiating a cascade of phosphorylation events resulting in sequential activation of MEK and ERK8, 9. In addition to membrane translocation of RAF1 upon binding to active RAS, activation of RAF1 kinase requires phosphorylation of four key residues including S338/Y341 in the N-region (negatively-charge regulatory region) and T491/S494 in the activation loop of the kinase8, 10. Phosphorylation of the activation loop is necessary but not sufficient to activate the kinase, whereas phosphorylation of S338 and Y341 is absolutely required for RAF1 activity as loss of phosphorylation at either site blocks the activation8, 11, 12. Since the activity of RAF1 is tightly controlled by phosphorylation at S338, dephosphorylation of this site would effectively inactivate the kinase. However, the identity of a protein phosphatase that directly dephosphorylates RAF1 and suppresses its oncogenic function remains elusive.
In this study, we report the identification of RAF1 as a novel substrate of PHLPP. We show that the PHLPP negatively regulates RAF/MEK/ERK signaling by directly inhibiting RAF1 activity. Functionally, silencing PHLPP expression increases the invasiveness of CRC cells by inducing EMT in vitro, and deletion of Phlpp1 gene promotes intestinal tumor progression in ApcMin mice.
MATERIALS AND METHODS
Animals
All animal procedures were performed by following protocols approved by the University of Kentucky Institutional Animal Care and Use Committee. Phlpp1 null mice on 129 Sv/C57BL6 background as recently described13 were maintained by random inter-crossing to sustain a heterogeneous mixed genetic background. To produce animals used in the experiments, Phlpp1−/− mice were bred with ApcMin mice on a C57BL6 background. These mice were then inter-crossed to produce four cohorts of animals including wild-type, Phlpp1−/−, ApcMin/Phlpp1+/+ (referred to as ApcMin), and ApcMin/Phlpp1−/−. To monitor survival, these four cohorts of mice were followed for up to 35 weeks.
Intestinal histopathology and IHC staining
Tissue processing and analysis of tumor histopathology were described in details in the Supplementary Materials and Methods section.
Cell culture, antibodies, expression constructs and other standard experiments
Cell culture conditions and sources of antibodies as well as all plasmids and other standard experimental methods used were detailed in the Supplementary Materials and Methods section.
Statistical Analysis
Microarray and patient clinical data from two colorectal cancer studies14, 15 were downloaded from the Oncomine database. The TCGA14 data set contains 192 adenocarcinoma and 22 normal samples, within which 13 were matched pairs. The Skrzypczak et al15 data set contains 81 tumor and 24 normal samples. Two-sample t-tests or linear mixed models were used to compare PHLPP1 and PHLPP2 expression between adenocarcinoma and normal samples. Pearson’s correlation coefficient was calculated to quantify the correlation between the expression of PHLPP1 and PHLPP2 with CDH1.
RESULTS
PHLPP directly dephosphorylates RAF1 and inactivates RAF/MEK/ERK signaling
In our search for proteins that co-immunoprecipitated with PHLPP1, we identified RAF1 by mass spectrometry analysis. Subsequent co-immunoprecipitation experiments confirmed that both PHLPP isoforms were able to bind RAF1. Interestingly, RAF1 interacted with PHLPP1 with a higher affinity compared to PHLPP2 (Fig. 1A). Moreover, the interaction between endogenous PHLPP1 and RAF1 was detected in SW480 cells, and PHLPP1 interacted with RAF1 more strongly when treated with EGF (Fig. 1B). This confirmation-sensitive interaction between PHLPP and RAF1 was confirmed by using a constitutively active RAF1/DDED (S338D/Y341D/T491E/S494D) and a kinase dead RAF1/KD (T491A/S494A) mutant. More RAF1/DDED co-immunoprecipitated with PHLPP compared to WT RAF1 whereas no detectable amount of RAF1/KD was detected (Supplementary Fig. S1). Furthermore, we determined the domains in both PHLPP and RAF1 required for the interaction. Consistent with the notion that protein phosphatases interact with their substrates with relatively high affinity, our data showed that the phosphatase domain of PHLPP is involved in binding with the kinase domain of RAF1 (Supplementary Fig. S1).
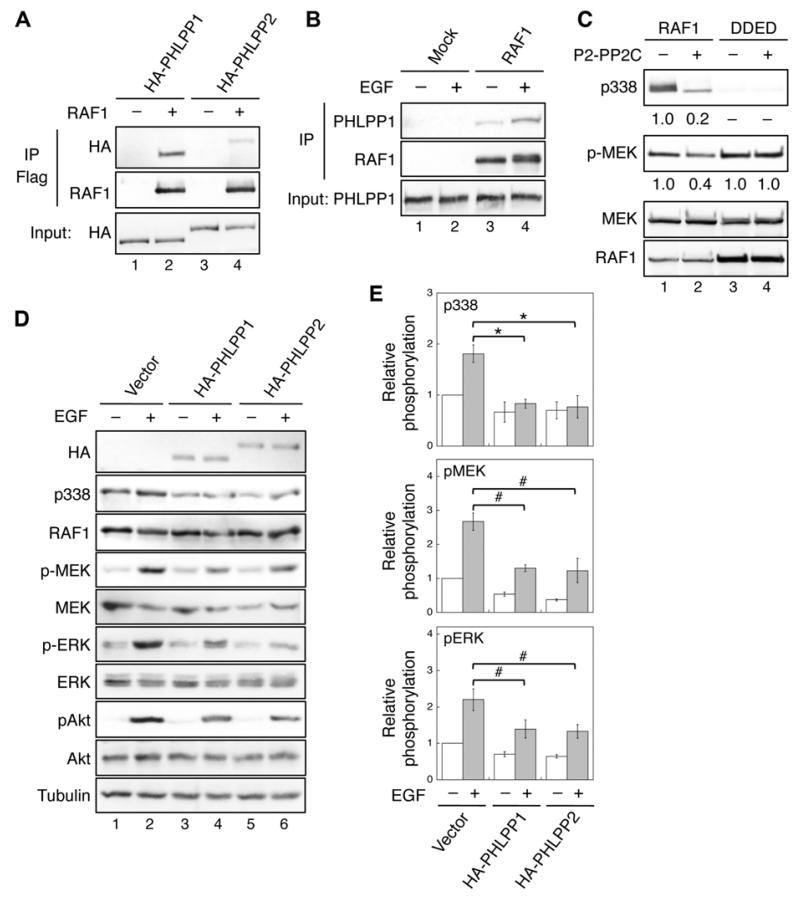
Figure 1. Identification of RAF1 as a substrate of PHLPP.
(A) 293T cells co-transfected with HA-PHLPP1 (lanes 1-2) or HA-PHLPP2 (lanes 3-4) together with vector or Flag-RAF1 were immunoprecipitated with the anti-Flag agarose and analyzed by immunoblotting. The expression of PHLPP1 and PHLPP2 was detected using the HA antibody. (B) 293T cells were serum starved overnight and subsequently treated with EGF (10 ng/ml) for 15 minutes. Cell lysates were incubated with protein A/G beads alone (Mock) or beads plus the RAF1 monoclonal antibody. The presence of PHLPP1 and RAF1 in the immunoprecipitates was detected using the PHLPP1 and RAF1 antibodies, respectively. (C) In vitro dephosphorylation of RAF1. Flag-RAF1 and Flag-RAF1/DDED were incubated with purified PP2C domain of PHLPP2 and subsequently used to phosphorylate recombinant MEK1. The relative phosphorylation of RAF1 and MEK1 were quantified by normalizing to total proteins and shown below the phospho-blots. (D) Stable vector, HA-PHLPP1, or HA-PHLPP2 overexpressing SW480 cells were serum starved overnight and treated with EGF for 15 minutes. Cell lysates were analyzed for phosphorylation and total protein expression. Note that since it was difficult to detect the phosphorylation of endogenous RAF1 at S338 in cell lysates directly, RAF1 was immunoprecipitated from cell lysates using the RAF1 antibody. Similar approach was taken to detect S338 phosphorylation in RAF1 in all subsequent experiments. (E) Relative phosphorylation levels of RAF1, MEK and ERK were obtained by normalizing ECL signals of p338, p-MEK and p-ERK antibodies to those of total RAF1, MEK and ERK, respectively. The level in untreated control cells was set to 1. Data represent the mean ± SEM (n=3, * p<0.01 and # p<0.05 by two-sample t-tests).
To determine whether RAF1 is a substrate of PHLPP, we performed in vitro dephosphorylation experiments using the purified PP2C domain of PHLPP2 (P2-PP2C) as the phosphatase. Incubating phosphorylated RAF1 isolated from transfected 293T cells with P2-PP2C resulted in an 80% decrease in S338 phosphorylation (Fig. 1C). Once dephosphorylated by PHLPP, the kinase activity of RAF1 towards its substrate, purified GST-MEK1, was largely reduced (Fig. 1C). Similarly, RAF1 was dephosphorylated by purified PP2C domain of PHLPP1 (data not shown). Serving as a negative control, the kinase activity of the phosphomimetic mutant RAF1/DDED was not affected by treating with P2-PP2C (Fig. 1C), suggesting that PHLPP-mediated dephosphorylation of RAF1 directly inactivates RAF1.
We next determined the effect of PHLPP-mediated dephosphorylation of RAF1 in SW480 cells stably expressing HA-PHLPP1 or HA-PHLPP2. Since SW480 cells harbor homozygous mutant KRASG12V alleles16, the phosphorylation of S338 in RAF1 was maintained in control cells after serum starvation; and this phosphorylation was increased in EGF treated cells. Overexpression of either PHLPP isoform resulted in a significant decrease in both basal and EGF-induced phosphorylation of RAF1, and this decrease in RAF1 phosphorylation coincided with reduced activation of downstream MEK and ERK (Fig. 1D-E). As a control, the phosphorylation of Akt at S473, a known substrate of PHLPP, was also decreased (Fig. 1D). Interestingly, although the basal constitutive phosphorylation of T308 was not readily affected by PHLPP overexpression, we found that EGF-induced phosphorylation of T308 was attenuated in stable SW480 cells (Supplementary Fig. S1). Collectively, our data indicated that overexpression of PHLPP inhibits RAF1 activity in CRC cells harboring mutant KRAS.
Knockdown of PHLPP activates RAF/MEK/ERK signaling in CRC cells
To determine the effect of endogenous PHLPP on RAF1 phosphorylation, PHLPP was silenced in CRC cells using lentiviral-mediated RNAi. Knocking down either PHLPP isoform resulted in a marked increase in RAF1 phosphorylation upon EGF treatment and a consequent increase in MEK and ERK phosphorylation (Fig. 2A-B). Similarly, the amplitude of RAF1, MEK, and ERK phosphorylation induced by EGF stimulation was also elevated in PHLPP knockdown HT29 cells (Supplementary Fig. S2). Moreover, we found that the amplitude of basal and EGF-induced phosphorylation of RAF1 was elevated in Phlpp1−/− MEF compared to WT cells, which led to increased activation of MEK and ERK downstream of RAF1 (Fig. 2C-D).
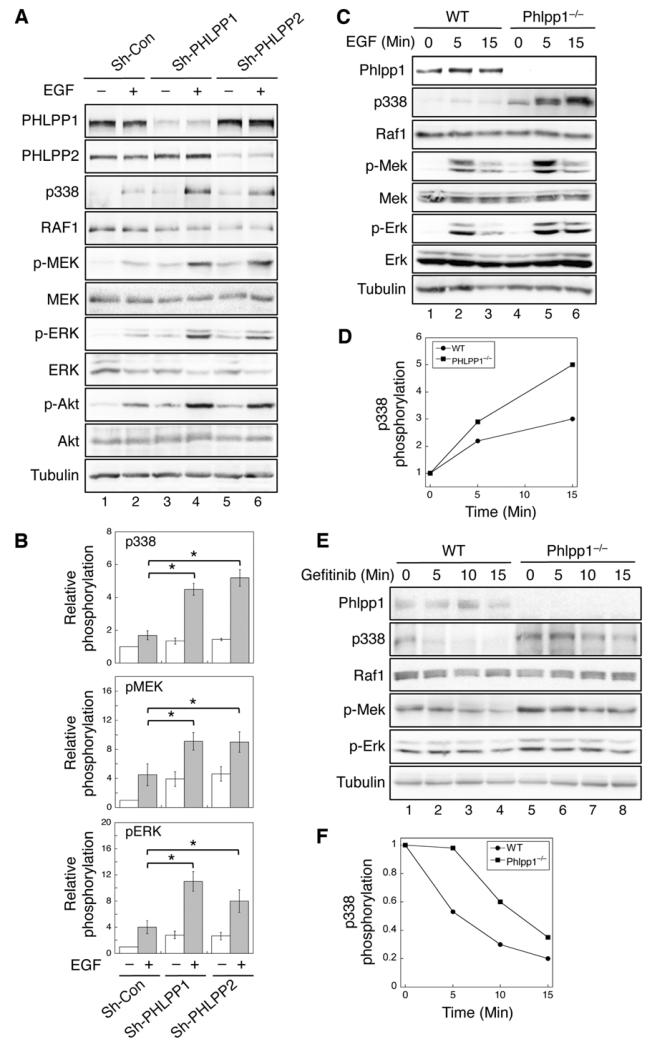
Figure 2. Loss of PHLPP expression enhances RAF/MEK/ERK signaling.
(A) Stable control (sh-Con), sh-PHLPP1, and sh-PHLPP2 SW480 cells were serum starved overnight and treated with EGF for 15 minutes, and cell lysates were analyzed for phosphorylation and total protein expression. (B) Relative phosphorylation levels of RAF1, MEK and ERK were obtained by normalizing ECL signals of p338, p-MEK and p-ERK antibodies to those of total RAF1, MEK and ERK, respectively. The level in untreated control cells was set to 1. Data represent the mean ± SEM (n=3, * p<0.05 by two-sample t-tests). (C) WT and Phlpp1−/− MEF cells were serum starved for 4 hours and subsequently treated with EGF for 0, 5, and 15 minutes. (D) Graphic representation of EGF-induced RAF1 phosphorylation at S338. The relative S338 phosphorylation was determined by normalizing ECL signals of the p338 antibody to that of total RAF1, and levels of S338 phosphorylation at time 0 in both cell lines were normalized to 1. (E) WT and Phlpp1−/− MEF cells grown in media containing 10% FBS were treated with gefitinib (5 μM) for 0, 5, 10, and 15 minutes. (F) Graphic representation of gefitinib-induced dephosphorylation of Raf1 at S338 in WT and Phlpp1−/− MEF cells. Western blots shown in (D) were quantified by normalizing ECL signals of the p338 antibody to that of total Raf1, and the levels of S338 phosphorylation at time 0 in both cell lines were normalized to 1.
We next determined the effect of PHLPP on inactivating RAF1 in cells treated with EGFR inhibitor gefitinib. Gefitinib induced a rapid dephosphorylation of RAF1 in WT MEF cells. However, the time course of RAF1 dephosphorylation was slowed down in Phlpp1−/− MEF cells (Fig. 2E-F). Interestingly, the time course of MEK and ERK dephosphorylation was not significantly altered. This is likely due to the fact that the dephosphorylation of MEK and ERK is controlled by PP2A and dual specific protein phosphatases17, respectively, rather than PHLPP. To confirm that PHLPP-mediated effect depends on its ability to dephosphorylate RAF1, we showed that EGF-induced phosphorylation of RAF1 and subsequent activation of MEK and ERK were inhibited by PHLPP even in cells overexpressing KRASG12V, whereas overexpression of RAF1/DDED rendered PHLPP incapable of inducing dephosphorylation of MEK and ERK. Moreover, gefitinib treatment largely blocked the EGF-induced activation of RAF/MEK/ERK signaling in both WT and Phlpp1−/− MEF cells (Supplementary Fig. S3). Collectively, we demonstrated that depleting PHLPP expression activates RAF1-dependent signaling and PHLPP functions at the level of RAF1 downstream of both EGFR and KRAS.
Knockdown of PHLPP promotes cell migration and invasion by inducing EMT in CRC cells
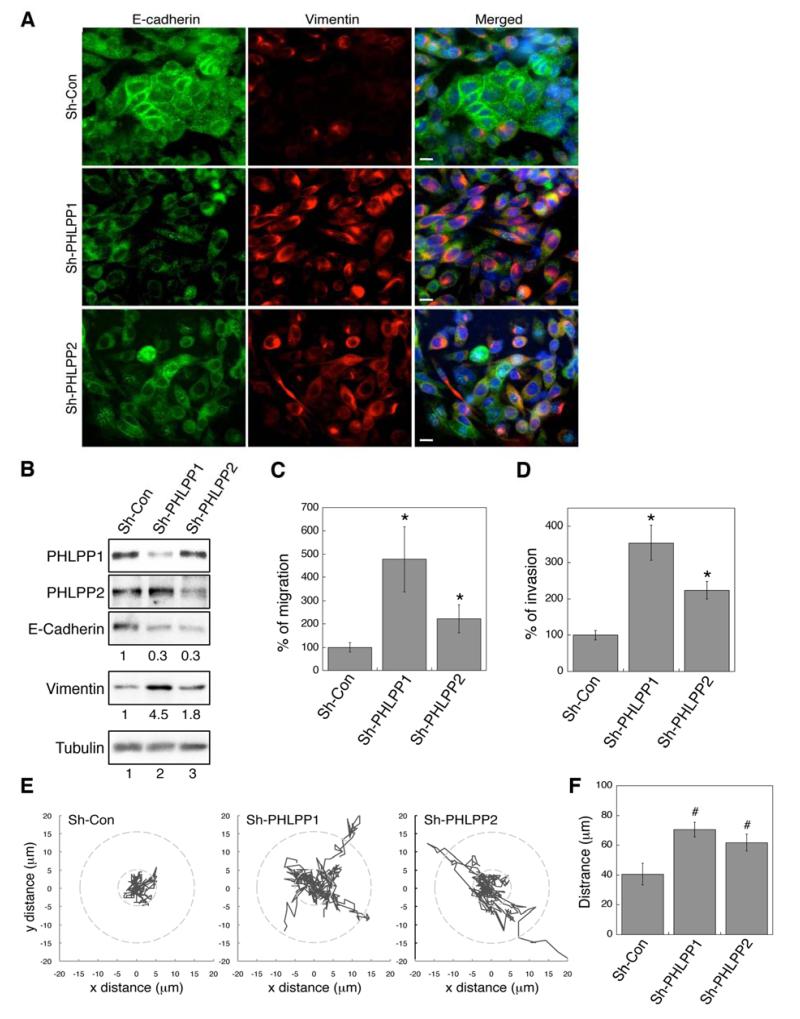
While working with PHLPP knockdown SW480 cells, we observed that the cells became elongated and adopted a spindle-like shape whereas control cells had a more cobblestone epithelial morphology (data not shown). To test whether this morphological change is associated with EMT, we examined the expression of E-cadherin (an epithelial cell marker) and vimentin (a fibroblast cell marker) using immunofluorescence staining. The expression of E-cadherin was readily detected along the plasma membrane and in the cytoplasm in control cells, whereas the expression of vimentin was low (Fig. 3A). In contrast, no plasma membrane staining of E-cadherin was detected in PHLPP knockdown cells while the expression of vimentin was largely increased (Fig. 3A). This decrease of E-cadherin and increase of vimentin expression were confirmed by Western blotting analysis (Fig. 3B). Furthermore, we found that PHLPP knockdown cells migrated significantly faster than control cells as determined by Transwell migration assays (Fig. 3C). Similarly, the ability of SW480 cells to invade through Matrigel was significantly increased in PHLPP knockdown cells (Fig. 3D). Knockdown of PHLPP1 resulted in a larger increase in the rate of migration and invasion compared to knocking down PHLPP2. This is likely due to that fact that PHLPP1 interacted with RAF1 with a higher affinity (Fig. 1A). In addition, the rate of cell migration was also increased in PHLPP knockdown HT29 cells (Supplementary Fig. S2). Conversely, overexpression of PHLPP promoted mesenchymalepithelial transition (MET), the reverse process of EMT, in SW480 cells. We found that re-expression of either PHLPP isoform was sufficient to elevate the level of E-cadherin, and decrease the level of vimentin and the rate of cell migration in cells overexpressing PHLPP1 or PHLPP2 (Supplementary Fig. S4). Moreover, PHLPP overexpressing cells failed to invade through Matrigel completely in the invasion assays. When tracking the cell movement in real time, we found that PHLPP knockdown cells were considerably more motile and average distances traveled by PHLPP knockdown cells were significantly increased compared to the control cells (Fig. 3E and 3F). Taken together, we have identified a novel role of PHLPP in negatively regulating EMT and cell motility in CRC cells.
Figure 3. Knockdown of PHLPP expression induces EMT and promotes migration and invasion in CRC cells.
(A) Stable control and PHLPP knockdown SW480 cells were co-stained with E-cadherin and vimentin antibodies and analyzed using fluorescence microscopy. Cell nuclei were stained with DAPI. Scale bars, 10 μm. (B) Cell lysates prepared from stable SW480 knockdown cells were analyzed using immunoblotting. The relative expression levels of E-cadherin and vimentin were quantified by normalizing to tubulin. (C-D) Stable SW480 knockdown cells were subjected to Transwell migration assays (C) using collagen and EGF as chemoattractants and Transwell invasion assays (D) using 5% FBS as the chemoattractant. Each experiment was done in duplicates and three independent experiments were averaged and expressed as mean ± SD (* p<0.01 by two-sample t-tests compared to the control cells). (E) Migration patterns of stable SW480 knockdown cells. The trajectories of 10 randomly chosen cells for each cell line were shown in the graphs. The radius of 5 and 10 μm from the origin is shown as dashed circles for reference. (F) The average distance traveled by stable SW480 knockdown cells during 6 hours of migration. Data represent mean ± SD (n = 10 cells/line, # p<0.05 by two-sample t-tests).
PHLPP inhibits EMT and cell migration by downregulating RAF/MEK/ERK signaling
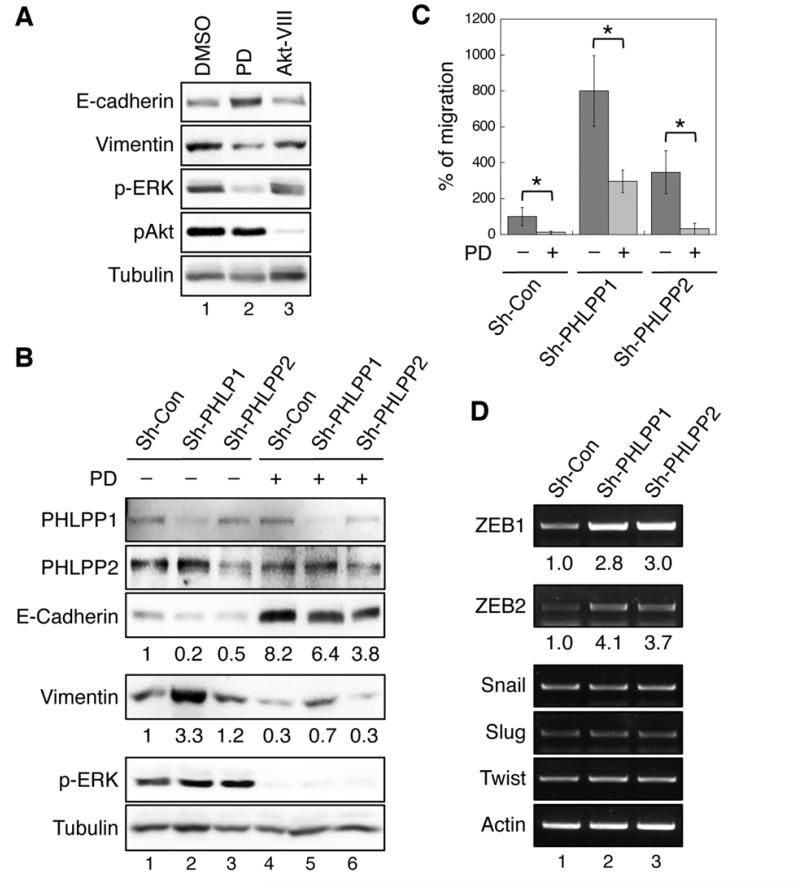
To determine whether PHLPP-mediated effect on EMT depends on its ability to suppress RAF/MEK/ERK signaling, SW480 cells were treated with either a MEK inhibitor (PD325901) or an Akt inhibitor (Akt VIII) for 36 hours. Both inhibitors functioned properly at inhibiting their targets as shown by decreased phosphorylation of ERK and Akt, respectively (Fig. 4A). However, inhibition of Akt had no effect on the expression of E-cadherin and vimentin, whereas MEK inhibitor readily reversed the EMT phenotype (Fig. 4A). Furthermore, inhibition of MEK largely blocked EMT induced by silencing PHLPP in SW480 cells, as the expression of E-cadherin increased and vimentin decreased (Fig. 4B). Consistently, the rate of cell migration was significantly decreased in MEK inhibitor treated cells, and PHLPP-depletion induced increased in cell migration was effectively blocked by MEK inhibitor (Fig. 4C). In addition, we found that PHLPP-mediated regulation of RAF1 and cell migration was independent of its ability to dephosphorylate Akt, as knockdown of Phlpp1 or Phlpp2 increased Raf1 phosphorylation and the rate of cell migration in Akt1/2 double knockout MEF cells (Supplementary Fig. S5). Finally, we showed that the levels of transcription factors that are known to regulate EMT were altered in PHLPP knockdown colon cancer cells. Semi-quantitative RT-PCR results revealed that ZEB1 and ZEB2 were specifically upregulated in PHLPP knockdown cells whereas other transcription factors including snail, slug, and twist were not affected (Fig. 4D). These results established a link between loss of PHLPP expression and reprogramming of cells by EMT-associated transcription factors. Although PHLPP negatively regulates both Akt and RAF1 signaling, our findings suggested that PHLPP-loss induced activation of RAF/MEK/ERK pathway is likely responsible for EMT and increased cell motility in CRC cells.
Figure 4. Loss of PHLPP promotes EMT by enhancing RAF/MEK/ERK signaling in CRC cells.
(A) SW480 cells were treated with DMSO, MEK inhibitor (PD0325901, 10 nM), or Akt-VIII (1 μM) for 30 hours, and cell lysates were analyzed by immunoblotting. (B) Stable SW480 knockdown cells were treated with DMSO or PD0325901 for 30 hours, and cell lysates were analyzed by immunoblotting. The relative expression of E-cadherine and vimentin were quantified by normalizing to tubulin. (C) Stable SW480 knockdown cells pretreated with DMSO or PD0325901 were subjected to Transwell migration assays in the presence or absence of PD0325901 using collagen and EGF as chemoattractants. Data shown in the graph represent the mean ± SD (n=3, * p<0.01 by two-sample t-tests). (F) The mRNA expression of ZEB1, ZEB2, Snail, Slug, Twist, and β-actin in control and PHLPP knockdown SW480 cells were assessed by semi-quantitative RT-PCR. The relative levels of ZEB1 and ZEB2 were quantified by normalizing to actin.
Deletion of Phlpp1 gene promotes invasive tumor growth in ApcMin mice
We next sought to determine whether loss of PHLPP promotes tumor progression to an invasive phenotype in vivo. Since knockdown of PHLPP1 had more pronounced effect on inducing EMT phenotype and increasing cell motility compared to PHLPP2 (see Figs. 3 and 4), and Phlpp1-null mice are currently available, we crossed Phlpp1-null mice onto ApcMin mouse model to investigate the susceptibility of mice deficient in Phlpp1 to colorectal adenomas. Deletion of Phlpp1 on its own produced microscopic hyperplasic lesions in moue intestine at ~80% penetrance within 12 months of age. The life span of Phlpp1−/− mice was slightly reduced compared to their wild-type littermates (Fig. 5A). However, normal physiological functions in Phlpp1−/− animals were generally not affected. In marked contrast, ApcMin/Phlpp1−/− compound mutant mice had significantly decreased survival compared to ApcMin mice (median survival time 17.5 weeks vs. 35 weeks) (Fig. 5A). Adenomas were detected in both intestine and colon regions of ApcMin/Phlpp1−/− mice, and the distribution of tumors was similar as that observed in ApcMin mice. Although the adenoma multiplicity of ApcMin/Phlpp1−/− mice was not increased compare to ApcMin mice, the tumor size was significantly increased (Supplementary Table S1). Consistent with previous reports, histological analysis indicated that almost all tumors developed in ApcMin mice were low-grade adenomas; however, over 20% of total tumors developed in ApcMin/Phlpp1−/− mice were invasive adenocarcinomas (Fig. 5B). Representative H&E images of adenomas and carcinomas found in ApcMin and ApcMin/Phlpp1−/− mice, as well as small adenoma lesions (<1mm) in Phlpp1−/− mice, are shown in Figure 5C. Moreover, the invasive adenocarcinomas found in ApcMin/Phlpp1−/− mice were associated with increased desmoplastic reactions in the stroma similar as those seen in human colon cancers (Fig. 5C insert). The epithelial origin of invasive carcinomas in ApcMin/Phlpp1−/− mice was confirmed as they stained positive for both β-catenin and colon epithelial cell marker cytokeratin 20 (CK20) (Fig. 5D). Interestingly, the CK20 staining not only detected an epithelial tumor invaded into muscularis propria, also revealed smaller clusters of tumor cells as they disseminated and started to invade into submucosal regions (Fig. 5D, marked by arrowheads in CK20 panel).
Figure 5. Phlpp1 deletion decreases survival and promotes tumor progression in ApcMin mice.
(A) Kaplan-Meier survival curve showing mice of WT, Phlpp1−/−, ApcMin, and ApcMin/Phlpp1−/−. Numbers of mice in the four cohorts are: WT (n=40), Phlpp1−/− (n=30), ApcMin (n=28), and ApcMin/Phlpp1−/− (n=34). Statistical significance (determined by Log Rank test) is given for comparison between WT and Phlpp1−/− (non-significant, ns), WT and ApcMin/Phlpp1−/− (p<0.0001), ApcMin and ApcMin/Phlpp1−/− (p<0.0001), and Phlpp1−/− and ApcMin/Phlpp1−/− (p<0.0001). (B) Graph showing the percentage of invasive adenocarcinomas observed in ApcMin and ApcMin/Phlpp1−/− mice. Mice were sacrificed at ages of 24 weeks and intestine and colon tissues were fixed and analyzed using H&E staining. Numbers of mice analyzed are: ApcMin (n=11) and ApcMin/Phlpp1−/− (n=13). Total numbers of tumors were counted and invasive adenocarcinomas were defined as tumors that invaded into submucosa or muscularis propria. (C) Histology analysis of intestine andenomas in 24-week old Phlpp1−/−, ApcMin, and ApcMin/Phlpp1−/− mice. The regions indicated by blue arrows in Phlpp1−/− and ApcMin/Phlpp1−/− panels were enlarged and shown as inserts. Scale bars, 500 μm. (D) Development of invasive adenocarcinomas in ApcMin/Phlpp1−/− mice. Tissue sections from ApcMin/Phlpp1−/− mice were analyzed using H&E staining and IHC staining for β-catenine and CK20. The H&E panel shows tumor invasion into muscularis propria (marked by a blue arrow). The blue arrowheads in the CK20 panel marked disseminating tumor cells of epithelial origin inside the submucosal layer. Scale bars, 100 μm.
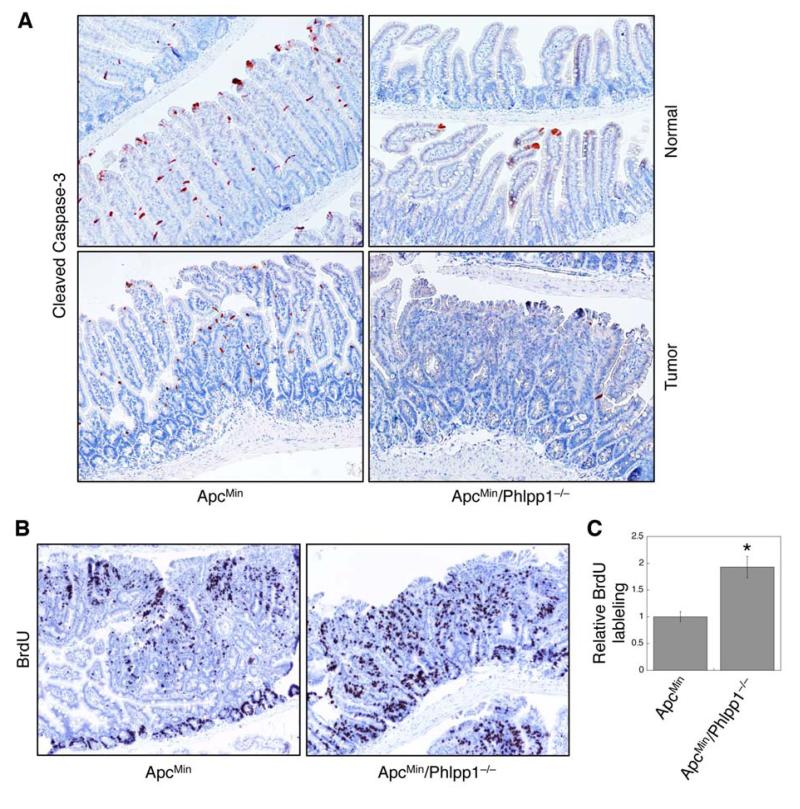
Furthermore, the effect of Phlpp1-loss on apoptosis and cell proliferation was analyzed in normal intestine/colon regions and in adenomas. The number of apoptotic cells as indicated by cleaved caspase-3 staining were drastically reduced in both normal villi and adenomas of ApcMin/Phlpp1−/− mice (Fig. 6A). In addition, a significantly higher number of proliferating tumor cells as measured by BrdU incorporation were observed in ApcMin/Phlpp1−/− mice (Fig. 6B-C). The proliferation of normal intestine epithelial cells along the crypt-villus axis was slightly increased in ApcMin/Phlpp1−/− mice compared to ApcMin mice, however, not statistically significant (data not shown). Taken together, these results indicate that loss of PHLPP expression enhances tumor growth in vivo by synergistically inhibiting apoptosis and promoting cell proliferation.
Figure 6. Phlpp1 deletion inhibits apoptosis and promotes cell proliferation in ApcMin mice.
(A) Intestine tissue sections from 24-week old ApcMin and ApcMin/Phlpp1−/− mice were stained for cleaved caspase 3. Representative images were taken from normal intestinal regions and adenomas. (B) Representative images showing BrdU labeled adenomas in ApcMin and ApcMin/Phlpp1−/− mice. (C) The numbers of BrdU positive cells in adenomas from ApcMin and ApcMin/Phlpp1−/− mice were quantified and analyzed using the Leica Application Suite EZ software. Ten randomly chosen tumors were averaged, and data in the graph represent the mean ± SD (* p<0.05).
Deletion of Phlpp1 gene downregulates E-cadherin and activates both Akt and Raf1 in ApcMin mice
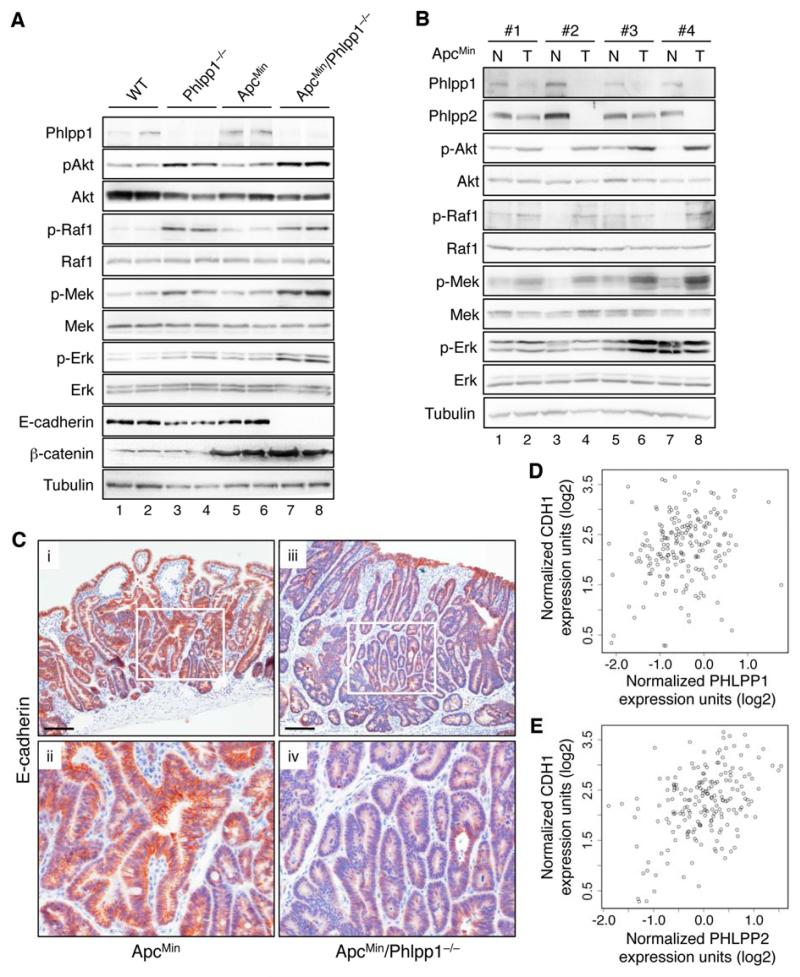
To begin determine the mechanism underlying the tumor suppressor function of PHLPP in ApcMin mice, we examined the activation status of RAF/MEK/ERK and Akt pathways in Phlpp1-/- mice. Knockout of Phlpp1 alone resulted in an increase in both Akt and Raf1 phosphorylation in mouse intestine tissues, whereas ApcMin mutation by itself had no effect on these two pathways (Fig. 7A). A modest increase in both Mek and Erk phosphorylation was observed in Phlpp1−/− mice, and Phlpp1-loss in ApcMin mice further enhanced the activation of Mek and Erk (Fig. 7A). In addition, the phosphorylation of Akt and Erk was increased in tumor tissues in ApcMin/Phlpp1−/− mice compared to ApcMin mice as detected by IHC staining; and Phlpp1-loss resulted in an increase in T308 phosphorylation of Akt in mouse normal intestinal tissues (Supplementary Fig. S6).
Figure 7. Phlpp1 deletion induces activation of both Akt and Raf1 signaling and downregulation of E-cadherin expression.
(A) Western blotting analysis of Akt and Raf1 activation in intestine tissues of 24-week old mice. Lysates were prepared from a mixture of normal and tumor tissues. Two samples obtained from two different mice were shown in each cohort. (B) Western blotting analysis of Phlpp expression and Akt and Raf1 activation in normal and tumor tissues prepared from ApcMin mice. Intestine isolated from 24-week old mice were opened longitudinally and inspected visually. The segments with visible tumor lesions (polyps) were separated from normal-looking tissues and used as tumor (T) and normal (N) tissues, respectively. (C) E-cadherin expression in adenomas of ApcMin (panels i-ii) and ApcMin/Phlpp1−/− (panels iii-iv) mice. Representative images were taken from intestine tissues of 24-week old mice analyzed by IHC staining. Scale bars, 100 μm. Panels ii and iv are enlarged images of areas enclosed by the white boxes shown in panels i and iii, respectively. (D-E) The correlation between PHLPP1 (D) and PHLPP2 (E) expression with CDH1 were quantified by Pearson’s correlation coefficient analysis using the TCGA CRC data set14. Pearson’s correlation coefficient for PHLPP1 and CDH1 was 0.20 (95% confidence interval 0.06 to 0.33, p=0.007), and for PHLPP2 and CDH1 was 0.42 (95% confidence interval 0.30 to 0.53, p<0.001)].
More significantly, we found that the tumorigenesis process in ApcMin mice was associated with decrease in Phlpp expression. As shown in Fig. 7B, the expression of both Phlpp isoforms was downregulated in tumors compared to matched normal intestine tissues, whereas the phosphorylation of Raf1/Mek/Erk and Akt were increased in ApcMin mice. Consistent with this finding, analysis of two large CRC patient microarray data sets revealed that expressions of PHLPP1 and PHLPP2 were significantly reduced in tumors compared to normal samples (Supplementary Fig. S7). Furthermore, E-cadherin levels were decreased in ApcMin/Phlpp1−/− mice compared to ApcMin mice as detected by Western blotting and IHC staining (Fig. 7A and 7C). The CRC patient microarray data analysis also revealed that expressions of PHLPP1 and PHLPP2 were positively correlated CDH1 (E-cadherin) expression (Fig. 7D-E and Supplementary Fig. S7), suggesting that loss of PHLPP expression is assoicated with loss of epithelial cell characteristics in CRC patients. In addition, point mutations, including nonsense mutations in the phosphatase domain of both PHLPP isoforms, have been identified in the genome-scale CRC sequencing project7 (Supplementary Table S2). Taken together, our data indicate that PHLPP-loss is a necessary and clinically relevant step that promotes EMT and tumor progression in vivo.
DISCUSSION
The funtional importance of PHLPP as a tumor suppressor in different types of cancer has been investigated in several recent studies2-4, 13, 18, 19. Evidence starts to emerge indicating that PHLPP not only plays an important role in inhibiting cell proliferation and promoting apoptosis but also negatively regulates tumor metatasis. However, the molecular mechanism underlying PHLPP-mediated regulation of cell invasion and metastasis remains elusive. In this study, we identified RAF1 as a novel substrate of PHLPP, and PHLPP-mediated dephosphorylation of S338 in RAF1 directly inhibits its kinase activity. Functionally, knockdown of PHLPP induces EMT and enhances cell motility in CRC cells, and Phlpp1-loss promotes invasive tumor growth in ApcMin mice.
Although knockdown of PHLPP results in an increase in both Akt and RAF1 activation in CRC cells examined in this study, the subsequent increase in cell migration and induction of EMT depends on RAF1 but not Akt signaling as inhibition of MEK/ERK activity reverses the EMT phenotype whereas inhibition of Akt had no effect. This finding is consistent with a previous report that stimulation of ERK activity is required for TGF-β1 induced EMT in CRC cells20. In addition, we show here that loss of PHLPP leads to upregulation of RAF/MEK/ERK signaling and pro-EMT transcription factors ZEB1 and ZEB2. Similarly, overexpression of a constitutively active ERK2 has been shown to increase ZEB1 and ZEB2 expression in breast cancer cells21. On the other hand, since different Akt isoforms may have opposite effect in regulating EMT and cell migration22, 23, and PHLPP is known to inhibit both Akt1 and Akt2 isoforms in CRC cells4, 24, the overall effect of PHLPP-mediated regulation of Akt may not directly correlate with the alteration in cell motility. Interestingly, we found that PHLPP is capable of inhibiting T308 phosphorylation in Akt upon growth factor stimulation. The preference of PHLPP for S473 is likely due to the fact that S473 is more surface accessible. In fact, PHLPP is capable of dephosphorylating T308 in vitro2. Conceivably, the confirmational change induced by kinase activation may allow a better access of PHLPP to T308. Future studies are needed to better understand how PHLPP regulates T308 phosphorylation. Here, since Phlpp1-loss in ApcMin mice promotes both Akt and Raf1 signaling, phenotypes observed in ApcMin/Phlpp1−/− mice are likely the result of combined activation of Akt and Raf1.
It has been shown previously that S338 of RAF1 can be dephosphorylated by protein phosphatase 5 (PP5) to inhibit neurite outgrowth in PC12 cells25. However, overexpression of PP5 has been associated with increased rate of tumor growth in breast cancer, thus suggesting an oncogenic role of PP526. Our study here identifies PHLPP as the first protein phosphatase that dephosphorylates RAF1 and suppresses its oncogenic function by directly inactivating its enzyme activity. Interestingly, the interaction between PHLPP and RAF1 is strengthened when RAF1 is in its active confirmation, suggesting that PHLPP-mediated dephosphorylation can be triggered upon activation of RAF1. This confirmation-dependent association between PHLPP and RAF1 may help to provide a tighter control of RAF1 activity. Furthermore, the notion that PHLPP functions at the RAF1 level downstream of EGFR and KRAS allows it to inhibit RAF/MEK/ERK signaling in CRC cells expressing either KRAS or BRAF mutant alleles (e.g. SW480 and HT29, respectively).
Since CRC patients with activated RAS/RAF signaling are known to have a high degree of liver metastases27, inhibition of this pathway represents a highly desirable approach in preventing tumor progression. In this study, we have provided several lines of evidence demonstrating the clinical relevance of PHLPP-loss in CRC patients: i) the expression of both PHLPP genes is downregulated in CRC patient samples; ii) the expression of PHLPP genes positively correlates with CDH1; and iii) point mutations in both PHLPP genes are identified in 2-3% of tumors. In conclusion, the identification of RAF1 as a novel substrate of PHLPP has uncovered a pivotal role of PHLPP in suppressing tumor progression and metastasis in CRC. Our finding may provide a mechanistic insight into the prevalence of LOH 18q in late stage colon cancer. Future studies on PHLPP-dependent inhibition of tumor progression will help to explore the potential application of using PHLPP as a translational target or diagnostic marker in the treatment of colon caner.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Dr. Doug Andres (University of Kentucky) for providing the expression constructs of RAF1. We also thank Garretson Epperly for his assistance with time-lapse live cell imaging, and Dana Napier for helping with tissue preparation and sectioning. The Biostatistics Shared Resource Facility at the Markey Cancer Center provided assistance with statistical analysis.
Grant Support: This work was supported by R01CA133429 (TG), American Cancer Society RSG0822001TBE (TG), and Kentucky Young Researchers Program (MY).
Abbreviations
- CRC
colorectal cancer
- EMT
epithelial-mesenchymal transition
- H&E
Hematoxylin and Eosin
- LOH
loss of heterozygosity
- MEF
mouse embryonic fibroblast
- MET
mesenchymal-epithelial transition
- WT
wild-type
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author Contributions: study concept and design: XL, PDS, JL, TG; data acquisition: XL, PDS, JL, HY, WW, MDS, MY, EYL, TG; critical revision for content: All authors; statistical analysis: XL, TG; study supervision: TG
Disclosures: The authors have no conflicts in association with publication of this manuscript.
REFERENCES
- 1.Brognard J, Newton AC. PHLiPPing the switch on Akt and protein kinase C signaling. Trends Endocrinol Metab. 2008;19:223–30. doi: 10.1016/j.tem.2008.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gao T, Furnari F, Newton AC. PHLPP: a phosphatase that directly dephosphorylates Akt, promotes apoptosis, and suppresses tumor growth. Mol Cell. 2005;18:13–24. doi: 10.1016/j.molcel.2005.03.008. [DOI] [PubMed] [Google Scholar]
- 3.Brognard J, Sierecki E, Gao T, Newton AC. PHLPP and a second isoform, PHLPP2, differentially attenuate the amplitude of Akt signaling by regulating distinct Akt isoforms. Mol Cell. 2007;25:917–31. doi: 10.1016/j.molcel.2007.02.017. [DOI] [PubMed] [Google Scholar]
- 4.Liu J, Weiss HL, Rychahou P, Jackson LN, Evers BM, Gao T. Loss of PHLPP expression in colon cancer: role in proliferation and tumorigenesis. Oncogene. 2009;28:994–1004. doi: 10.1038/onc.2008.450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kinzler KW, Vogelstein B. Lessons from hereditary colorectal cancer. Cell. 1996;87:159–70. doi: 10.1016/s0092-8674(00)81333-1. [DOI] [PubMed] [Google Scholar]
- 6.Pino MS, Chung DC. The chromosomal instability pathway in colon cancer. Gastroenterology. 2010;138:2059–72. doi: 10.1053/j.gastro.2009.12.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.The Cancer Genome Atlas Network Comprehensive molecular characterization of human colon and rectal cancer. Nature. 2012;487:330–7. doi: 10.1038/nature11252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wellbrock C, Karasarides M, Marais R. The RAF proteins take centre stage. Nat Rev Mol Cell Biol. 2004;5:875–85. doi: 10.1038/nrm1498. [DOI] [PubMed] [Google Scholar]
- 9.McKay MM, Morrison DK. Integrating signals from RTKs to ERK/MAPK. Oncogene. 2007;26:3113–21. doi: 10.1038/sj.onc.1210394. [DOI] [PubMed] [Google Scholar]
- 10.Maurer G, Tarkowski B, Baccarini M. Raf kinases in cancer-roles and therapeutic opportunities. Oncogene. 2011;30:3477–88. doi: 10.1038/onc.2011.160. [DOI] [PubMed] [Google Scholar]
- 11.Chong H, Lee J, Guan K-L. Positive and negative regulation of Raf kinase activity and function by phosphorylation. Embo J. 2001;20:3716–3727. doi: 10.1093/emboj/20.14.3716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mason CS, Springer CJ, Cooper RG, Superti-Furga G, Marshall CJ, Marais R. Serine and tyrosine phosphorylations cooperate in Raf-1, but not B-Raf activation. Embo J. 1999;18:2137–48. doi: 10.1093/emboj/18.8.2137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen M, Pratt CP, Zeeman ME, Schultz N, Taylor BS, O’Neill A, Castillo-Martin M, Nowak DG, Naguib A, Grace DM, Murn J, Navin N, Atwal GS, Sander C, Gerald WL, Cordon-Cardo C, Newton AC, Carver BS, Trotman LC. Identification of PHLPP1 as a Tumor Suppressor Reveals the Role of Feedback Activation in PTEN-Mutant Prostate Cancer Progression. Cancer Cell. 2011;20:173–86. doi: 10.1016/j.ccr.2011.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.TCGA The Cancer Genome Atlas. http://tcga-data.nci.nih.gov/tcga/
- 15.Skrzypczak M, Goryca K, Rubel T, Paziewska A, Mikula M, Jarosz D, Pachlewski J, Oledzki J, Ostrowski J. Modeling oncogenic signaling in colon tumors by multidirectional analyses of microarray data directed for maximization of analytical reliability. PLoS One. 2010:5. doi: 10.1371/journal.pone.0013091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Brink M, de Goeij AF, Weijenberg MP, Roemen GM, Lentjes MH, Pachen MM, Smits KM, de Bruine AP, Goldbohm RA, van den Brandt PA. K-ras oncogene mutations in sporadic colorectal cancer in The Netherlands Cohort Study. Carcinogenesis. 2003;24:703–10. doi: 10.1093/carcin/bgg009. [DOI] [PubMed] [Google Scholar]
- 17.Junttila MR, Li SP, Westermarck J. Phosphatase-mediated crosstalk between MAPK signaling pathways in the regulation of cell survival. FASEB J. 2008;22:954–65. doi: 10.1096/fj.06-7859rev. [DOI] [PubMed] [Google Scholar]
- 18.Molina JR, Agarwal NK, Morales FC, Hayashi Y, Aldape KD, Cote G, Georgescu MM. PTEN, NHERF1 and PHLPP form a tumor suppressor network that is disabled in glioblastoma. Oncogene. 2012;31:1264–74. doi: 10.1038/onc.2011.324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nitsche C, Edderkaoui M, Moore RM, Eibl G, Kasahara N, Treger J, Grippo PJ, Mayerle J, Lerch MM, Gukovskaya AS. The phosphatase PHLPP1 regulates Akt2, promotes pancreatic cancer cell death, and inhibits tumor formation. Gastroenterology. 2012;142:377–87. doi: 10.1053/j.gastro.2011.10.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pino MS, Kikuchi H, Zeng M, Herraiz MT, Sperduti I, Berger D, Park DY, Iafrate AJ, Zukerberg LR, Chung DC. Epithelial to mesenchymal transition is impaired in colon cancer cells with microsatellite instability. Gastroenterology. 2010;138:1406–17. doi: 10.1053/j.gastro.2009.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shin S, Dimitri CA, Yoon SO, Dowdle W, Blenis J. ERK2 but not ERK1 induces epithelial-to-mesenchymal transformation via DEF motif-dependent signaling events. Mol Cell. 2010;38:114–27. doi: 10.1016/j.molcel.2010.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Toker A, Yoeli-Lerner M. Akt signaling and cancer: surviving but not moving on. Cancer Research. 2006;66:3963–6. doi: 10.1158/0008-5472.CAN-06-0743. [DOI] [PubMed] [Google Scholar]
- 23.Irie HY, Pearline RV, Grueneberg D, Hsia M, Ravichandran P, Kothari N, Natesan S, Brugge JS. Distinct roles of Akt1 and Akt2 in regulating cell migration and epithelialmesenchymal transition. J Cell Biol. 2005;171:1023–34. doi: 10.1083/jcb.200505087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li X, Yang H, Liu J, Schmidt MD, Gao T. Scribble-mediated membrane targeting of PHLPP1 is required for its negative regulation of Akt. Embo Rep. 2011;12:818–24. doi: 10.1038/embor.2011.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.von Kriegsheim A, Pitt A, Grindlay GJ, Kolch W, Dhillon AS. Regulation of the Raf-MEK-ERK pathway by protein phosphatase 5. Nat Cell Biol. 2006;8:1011–6. doi: 10.1038/ncb1465. [DOI] [PubMed] [Google Scholar]
- 26.Golden T, Aragon IV, Rutland B, Tucker JA, Shevde LA, Samant RS, Zhou G, Amable L, Skarra D, Honkanen RE. Elevated levels of Ser/Thr protein phosphatase 5 (PP5) in human breast cancer. Biochim Biophys Acta. 2008;1782:259–70. doi: 10.1016/j.bbadis.2008.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Santini D, Loupakis F, Vincenzi B, Floriani I, Stasi I, Canestrari E, Rulli E, Maltese PE, Andreoni F, Masi G, Graziano F, Baldi GG, Salvatore L, Russo A, Perrone G, Tommasino MR, Magnani M, Falcone A, Tonini G, Ruzzo A. High concordance of KRAS status between primary colorectal tumors and related metastatic sites: implications for clinical practice. Oncologist. 2008;13:1270–5. doi: 10.1634/theoncologist.2008-0181. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.