Abstract
BACKGROUND & AIMS
Positive-sense RNA viruses remodel intracellular membranes to generate specialized membrane compartments for viral replication. Several RNA viruses, including poliovirus and hepatitis C virus (HCV), require phosphatidylinositol (PI) 4-kinases for their replication. However, it is not known how PI 4-kinases and their product, PI(4)P, facilitate host membrane reorganization and viral replication. Furthermore, although the HCV replication compartment, known as the membranous web, is believed to be cholesterol-enriched, the mechanisms by which this occurs have not been elucidated. We aimed to identify and characterize a PI 4-kinase effector in HCV replication.
METHODS
We used a combination of microscopic and biochemical methods to study HCV replication, web morphology, the distribution of intracellular protein and PI(4)P, along with cholesterol trafficking in HCV-infected cells. PI 4-kinase and oxysterol-binding protein (OSBP) were inhibited using RNA interference or small molecules in cells expressing a full-length genotype 1b replicon or infected with the JFH-1 strain of HCV.
RESULTS
OSBP was required for HCV replication and membranous web integrity. OSBP was recruited to membranous webs in a PI 4-kinase-dependent manner, and both these factors were found to regulate cholesterol trafficking to the web. We also found OSBP to be required for poliovirus infection but dispensable for dengue virus.
CONCLUSIONS
OSBP is a PI 4-kinase effector in HCV infection, and contributes to the integrity and cholesterol enrichment of the membranous web. OSBP might also be a PI 4-kinase effector in poliovirus infection and could be involved in replication of other viruses that require PI 4-kinases.
Keywords: lipid transfer proteins, phosphatidylinositol 4-phosphate, lipid kinase, viral replication
INTRODUCTION
Positive-sense single-stranded RNA viruses remodel intracellular membranes to generate specialized membrane compartments for viral replication1, which might facilitate genome replication and packaging by restricting diffusion, and may shield viral components from host innate immunity. Hepatitis C virus (HCV) is a positive-sense RNA virus in the family Flaviviridae. While its genome is initially translated at the host endoplasmic reticulum, RNA replication occurs in association with virus-induced intracellular membranes termed the “membranous web”2,3. Although the HCV membranous web is thought to be enriched in cholesterol4–6, the molecular mechanisms by which this occurs are unknown.
The host phosphatidylinositol 4-kinase PI4KA and its product phosphatidylinositol 4-phosphate (PI(4)P) are essential for HCV replication7–13 and membranous web integrity8,9,14–16. Although the related PI 4-kinase PI4KB is essential for the replication of several other positive-sense RNA viruses17,18, evidence for its importance in HCV infection is mixed10,11,15. PI4KA requires lipid kinase activity to support HCV replication, suggesting that PI(4)P is also essential for HCV replication9,15. PI4KA is recruited to the HCV membranous web by a direct interaction with NS5A, and HCV infection is associated with increased intracellular PI(4)P levels and enrichment of PI(4)P at the membranous web.
A crucial and as yet unanswered question is how PI 4-kinases and PI(4)P facilitate RNA virus replication and HCV web integrity. One possibility is that PI(4)P recruits phosphoinositide-binding proteins to viral replication compartments. In particular, oxysterol-binding protein (OSBP) is a plausible candidate PI4KA effector in the HCV life cycle for several reasons. First, OSBP interacts with HCV NS5A, and its silencing inhibits both infectious particle secretion and replication19,20. Second, OSBP binds to PI(4)P via a pleckstrin homology (PH) domain at its N-terminus21. Third, OSBP encodes a sterol-binding domain22 and can mediate PI(4)P-dependent cholesterol transfer between liposomes in vitro23. However, there has been controversy regarding the role of OSBP-related proteins (ORPs) in sterol trafficking in yeast24,25, and there has been little direct evidence that OSBP or ORPs regulate sterol trafficking in mammalian cells.
Herein, we focus on the role of OSBP as a possible effector of PI4KA and PI(4)P in supporting HCV replication. OSBP is essential not only for the replication of HCV, but also for the integrity of the HCV membranous web. OSBP is recruited to the membranous web in HCV-infected cells in a PI(4)P-dependent manner. In addition, we find that OSBP and PI4KA regulate the transport of cholesterol to the membranous web. Finally, the dependence of poliovirus and dengue virus replication on OSBP correlates with their PI 4-kinase dependence, suggesting that OSBP may be a PI 4-kinase effector for multiple RNA viruses. This study sheds light on how viruses modulate host lipid membrane compositions to their own benefit by exploiting host phosphoinositide signaling pathways.
Results
OSBP is relevant to HCV replication
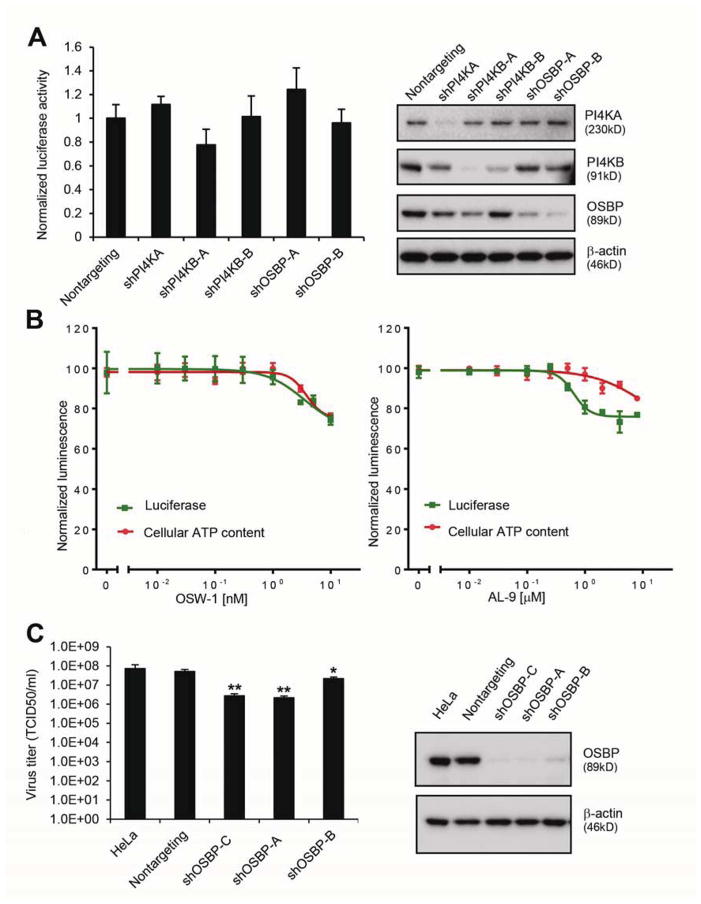
To test the role of OSBP in HCV replication, OR6 cells containing a full-length genotype 1b HCV replicon expressing a Renilla luciferase reporter gene26 were transduced with five independent shRNA lentiviral vectors targeting OSBP. The magnitude of HCV replication inhibition correlated with the degree of OSBP silencing (Supplementary Figure 1A). Two shRNAs that efficiently silenced OSBP significantly inhibited HCV replication and reduced HCV NS5A protein levels without detectable cytotoxicity in OR6 cells (Figure 1A) as well as in the genotype 2a JFH-1 infectious HCV system (Figure 1B), indicating that the dependency of HCV on OSBP is not genotype-specific.
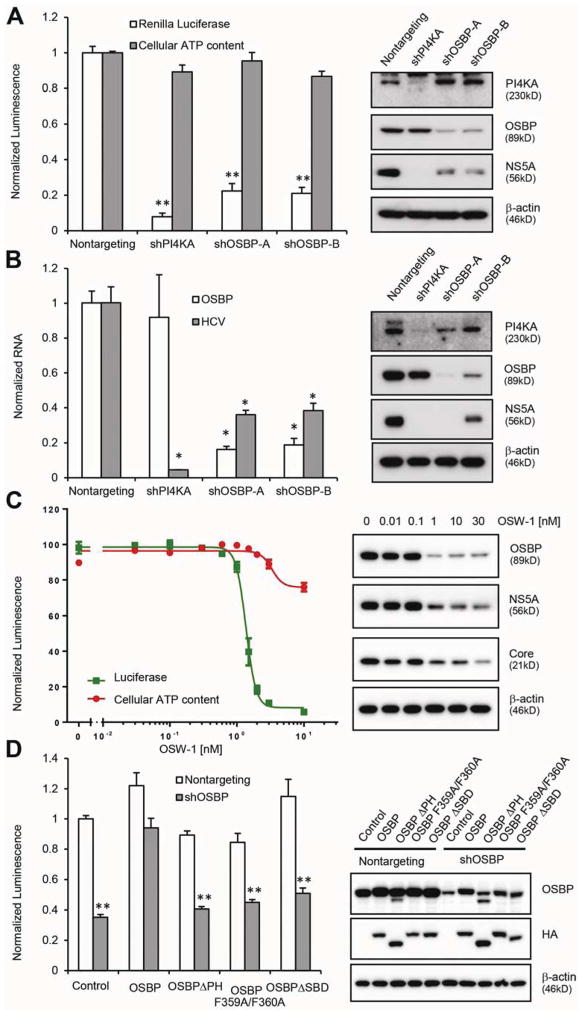
Figure 1. OSBP is relevant to HCV replication.
(A) OR6 replicon cells were transduced with the indicated shRNAs. 72 hr later, replication and cell viability were assessed by Renilla luciferase activity and cellular ATP content, respectively (left panel). Right panel, immunoblotting of cell lysates. Values (means ± SEM, three independent experiments) are normalized to a nontargeting shRNA. **, P<.001 versus nontargeting control.
(B) Huh7.5.1 cells were transduced with the indicated shRNAs 72 hr before infection with the JFH-1 strain of HCV (MOI = 0.1). HCV and OSBP mRNA were quantified by qRT-PCR at 48 hr postinfection (left panel). Right panel, immunoblotting of cell lysates. Values (means ± SD, three independent experiments) are normalized to nontargeting shRNA. *, P<.005 versus nontargeting control.
(C) OR6 cells were treated with OSW-1 for 24 hr prior to measurement of HCV replication and cell viability (left panel). Right panel, immunoblotting of cell lysates. Values are means ± SEM, three independent experiments.
(D) OR6 cells stably expressing a nontargeting shRNA or an OSBP shRNA targeting the 3′UTR were transduced with lentiviral vectors encoding the indicated HA-tagged OSBP constructs lacking the 3′UTR. Luciferase activity was measured 4 days later (left panel). Right panel, immunoblotting of cell lysates. Values are mean ± SEM of three independent experiments and are normalized to nontargeting control. **, P<.001 versus nontargeting control.
Using an orthogonal pharmacologic approach, we found that the compound OSW-1, which inhibits OSBP by binding as well as through proteasomal degradation27, inhibited OR6 replication and JFH-1 infection in a dose-dependent manner (IC50 1.37 ± 0.07 nM) with cytotoxicity only at the highest tested doses (Figure 1C and Supplementary Figure 1B).
To further exclude the possibility that OSBP shRNAs inhibit HCV replication by off-target effects and to assess the domains of OSBP necessary for HCV replication, we conducted a rescue assay. OR6 cells stably expressing an shRNA targeting the 3′UTR of the endogenous OSBP mRNA were transduced with lentiviral vectors encoding OSBP or OSBP mutant constructs lacking the OSBP 3′UTR. We generated three OSBP mutants: deletion of the PI(4)P-binding PH domain (ΔPH), a F359A/F360A substitution in the FFAT (two phenylalanines in an acidic tract) motif that interacts with ER proteins called Vesicle-Associated Membrane Protein-Associated Proteins (VAPs), and deletion of the steroid-binding domain (ΔSBD). The inhibition of HCV by knockdown of endogenous OSBP could be rescued by expression of wild-type OSBP but not by the mutant constructs (Figure 1D), indicating that the PH domain, FFAT motif, and SBD are all required for OSBP function.
OSBP localizes to NS5A-positive structures in HCV-infected cells
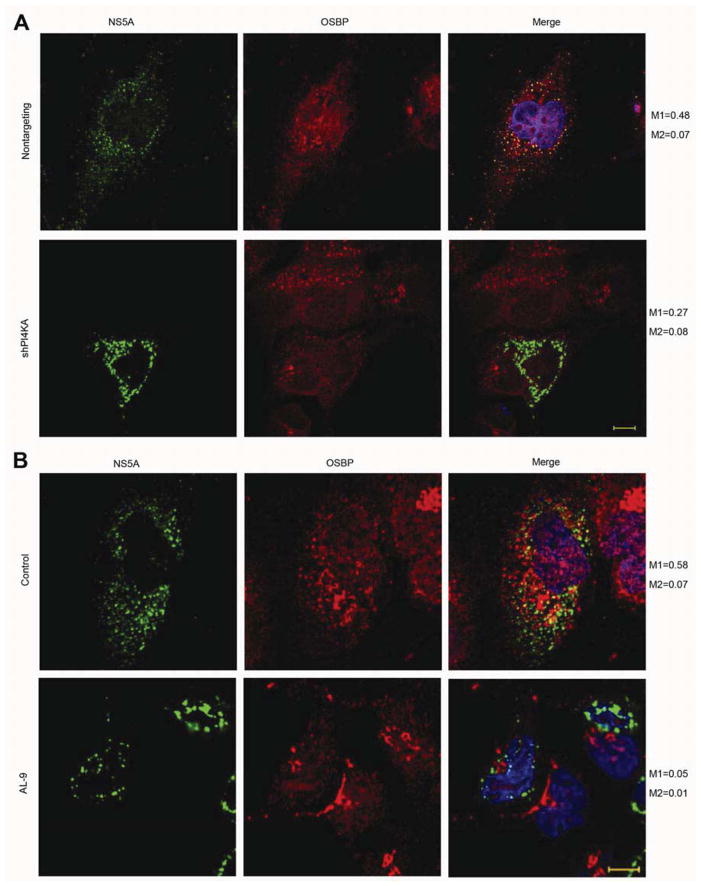
Since OSBP is essential for HCV replication, we hypothesized that OSBP might associate with HCV membranous webs. In HCV-infected cells, most of the endogenous OSBP was in a paranuclear distribution consistent with Golgi localization. However, we also observed colocalization of viral NS5A with some endogenous OSBP (Figure 2A, Manders’ coefficient M1=0.60 (fraction of NS5A overlapping OSBP); see also Supplementary Figure 2A), suggesting that some OSBP localizes to the membranous web. We used Manders’ coefficients here because they more accurately quantitate partial overlap between two channels. OSBP-ΔPH, however, no longer appeared to localize to Golgi or NS5A-positive membranes (Supplementary Figure 2B). In contrast, the OSBP F359A/F360A and OSBP ΔSBD mutants still showed partial colocalization with NS5A (Supplementary Figure 2B), suggesting that OSBP localization to NS5A-positive membranes requires the PH domain but not the FFAT motif or the SBD.
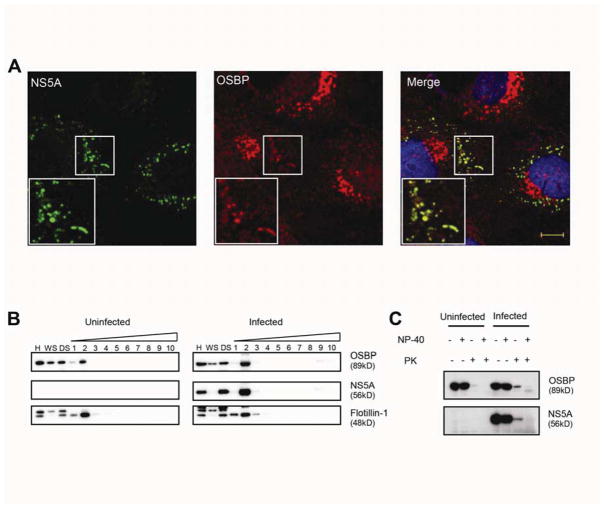
Figure 2. OSBP localizes to membranous webs.
(A) Immunostaining of JFH-1 infected cells for NS5A (green) and endogenous OSBP (red) with DAPI nuclear counterstaining (blue). Manders’ coefficients: M1=0.60 (fraction of NS5A overlapping OSBP); M2=0.07 (fraction of OSBP overlapping NS5A). Bar, 10 μm.
(B) Uninfected or HCV-infected Huh7.5.1 cell homogenates (H) were centrifuged to prepare a crude membrane pellet and a “water soluble” supernatant (WS). The pellet was treated with cold 1% NP-40 and spun again; the “detergent soluble” (DS) fraction was removed and detergent-resistant membranes were fractionated on a density gradient. Fractions (numbered in order from light to heavy) were analyzed by immunoblotting for the indicated proteins.
(C) Immunoblots of homogenates from uninfected or HCV-infected Huh7.5.1 cells, treated with or without cold 1% NP-40 followed by treatment with or without proteinase K (PK).
Membrane-associated HCV replicase components are resistant to extraction with cold NP-40 detergent and exhibit low buoyant densities on density gradient centrifugation4,5, which are properties of cholesterol-enriched “lipid raft” microdomains. As expected, NS5A cofractionated with the DRM marker flotilin-1 (Figure 2B). HCV infection was associated with an increase in DRM-associated OSBP, consistent with our hypothesis that OSBP associates with membranous webs in HCV-infected cells.
The membranous structure of the HCV replication complex can protect viral and cellular components from exogenous proteases and nucleases in vitro1,4. We found that a fraction of NS5A was resistant to proteinase K digestion in the absence of detergent, while NS5A was completely digested after detergent treatment (Figure 2C). Consistent with our model, HCV infection was associated with increased amounts of protease-resistant OSBP in the absence of detergent (Figure 2C).
OSBP is required for membranous web integrity
Silencing or pharmacologic inhibition of PI4KA induces the accumulation of membrane “clusters” containing HCV NS5A and other viral replication components8,9,14–16, indicating that PI4KA and its product PI(4)P are required for membranous web integrity. Either PI4KA or OSBP silencing induced the formation of NS5A-positive membrane clusters in HCV-infected cells (Figure 3A). Although the efficiency of HCV infection was markedly reduced by PI4KA or OSBP silencing, a low percentage of stably silenced cells could be infected with HCV, likely reflecting heterogeneous knockdown. The pharmacologic OSBP inhibitor OSW-1 also induced membrane cluster formation in JFH-1 infected cells (Figure 3B). To exclude the possibility that this effect was dependent on inhibition of HCV RNA replication, we expressed the NS3-5B polyprotein in Huh7.5.1 cells using T7 RNA polymerase that renders viral protein expression independent of viral replication15, and found similar membrane clusters in OSBP-silenced cells (Supplementary Figure 3A).
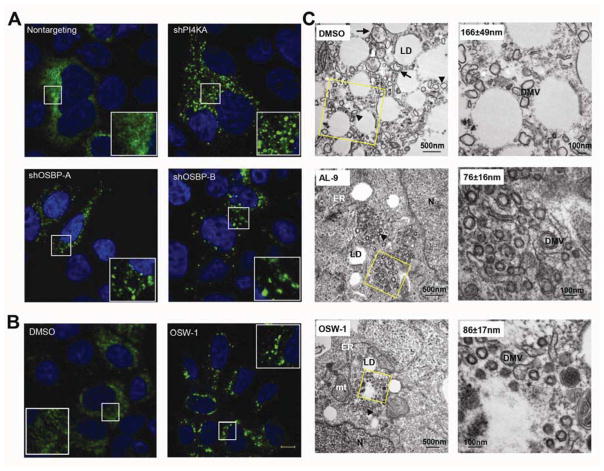
Figure 3. OSBP or PI4KA inhibition alters membranous web morphology.
(A) HCV-infected Huh7.5.1 cells stably transduced by nontargeting, PI4KA, or OSBP shRNA were immunostained for HCV NS5A (green) with DAPI nuclear counterstaining (blue).
(B) HCV-infected cells were treated with 0.1% DMSO or 30 nM OSW-1 for 24 hr before NS5A immunostaining. Bar, 10 μm.
(C) HCV-infected cells were treated with 0.1% DMSO, 4 μM AL-9, or 20 nM OSW-1 for 18 hr before preparation for EM. Enlargements of the boxed areas are shown in the right panels; values are DMV mean diameter ± SD. Arrowheads: double-membrane vesicles (DMVs); Arrows: multi-membrane vesicles (MMVs); LD: lipid droplets; ER: endoplasmic reticulum; mt: mitochondria; N, nucleus.
Next, we examined the ultrastructure of the altered membranous webs induced by OSBP and PI4KA inhibition using electron microscopy. In mock treated cells, JFH-1 infection induced the formation of double membrane vesicles (DMVs) of somewhat heterogeneous size (mean diameter 166 ± 49 nm) and morphology, as well as multi-membrane vesicles (MMVs) (Figure 3C, upper panels), consistent with previous reports28. These DMVs and MMVs were typically found in the vicinity of lipid droplets (LDs) and were not seen in uninfected cells (Supplementary Figure 3B). In contrast, DMVs were smaller and more homogeneous in size (average diameter 76 ± 16 nm) in HCV-infected cells treated with the PI4KA inhibitor AL-914. These small DMVs were often aggregated in clusters (Figure 3C and Supplementary Figure 3B) but were also occasionally dispersed (Supplementary Figure 3B). These clusters of small DMVs seen by TEM likely correspond to the NS5A-positive clusters seen by immunofluorescence (Supplementary Figure 3C and 9). Similarly, OSW-1 treatment was associated with clusters of small, homogenous DMVs with diameters (86±17nm) (Figure 3C, lower panels) similar to those of AL-9 treated cells, as well as individual small DMVs adjacent to LDs (Supplementary Figure 3B). In contrast, the NS5B polymerase inhibitor 2′CMeA did not reduce mean DMV diameter or induce DMV clustering (Supplementary Figure 3B), indicating that the observed effect of PI4KA/OSBP inhibition was not a nonspecific result of inhibiting HCV replication.
Collectively, these results indicate that OSBP, like PI4KA, is required for HCV membranous web integrity. The similar phenotypes of the membrane alterations induced by OSBP or PI4KA inhibition support the hypothesis that these two host factors function along the same pathway.
HCV induced PI(4)P increase does not require OSBP and is not sufficient for membranous web integrity
Intracellular PI(4)P levels are increased by HCV infection both in vitro and in vivo in a PI4KA-dependent manner9,14,16. We assessed the relative effects of PI4KA or OSBP silencing on PI(4)P levels in HCV-expressing cells. To avoid confounding effects of PI4KA and OSBP silencing on HCV replication, this experiment was conducted with Huh7/T7 cells expressing the NS3-5B/GFP polyprotein described above. PI(4)P was elevated in NS3-NS5B/GFP expressing cells transduced with a nontargeting shRNA by approximately 3.5-fold (Figure 4B) as compared to HCV-nonexpressing cells (Figure 4A). As expected, PI4KA silencing blocked the upregulation of intracellular PI(4)P levels by HCV polyprotein expression and was also associated with the appearance of NS5A-positive membrane clusters. In contrast, OSBP silencing did not block the upregulation of intracellular PI(4)P levels in HCV-expressing cells despite the formation of membrane clusters (Figures 4A and B), consistent with OSBP functioning downstream of PI4KA and PI(4)P.
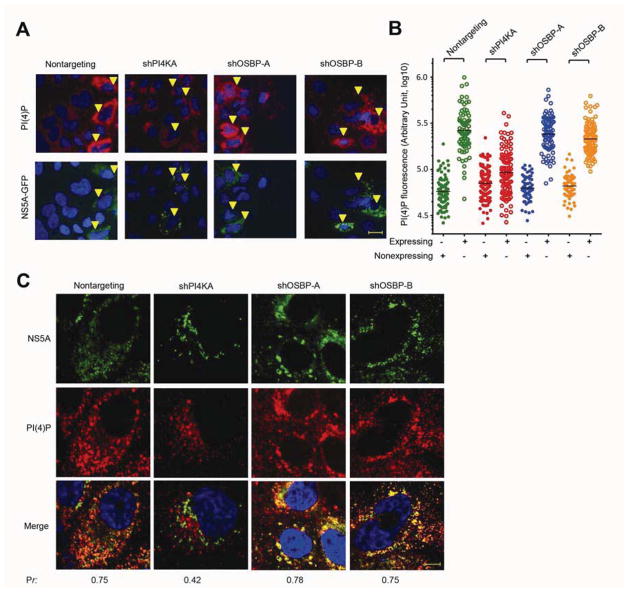
Figure 4. OSBP inhibition alters web morphology without blocking HCV-induced PI(4)P accumulation.
(A) Nonreplicative expression of the NS3-5B(NS5A/GFP) polyprotein in Huh7.5.1/T7 cells stably expressing T7 RNA polymerase and the indicated shRNAs. 48 hr post-transfection, cells were immunostained for PI(4)P (red) and GFP (green) with DAPI nuclear counterstaining (blue). Arrowheads indicate cells expressing the NS3-5B/GFP polyprotein. Bar, 20 μm.
(B) Quantitation of PI(4)P fluorescence from HCV-expressing cells (open symbols) versus HCV-nonexpressing cells (closed symbols). Each point denotes the integrated fluorescence signal from a single cell with the mean fluorescence indicated by the black lines.
(C) Immunostaining of JFH-1 infected Huh7.5.1 cells stably expressing the indicated shRNAs for PI(4)P (red) and GFP (green) with DAPI nuclear counterstaining (blue). Pr, Pearson’s correlation coefficients; bar, 10 μm.
HCV expression is associated not only with increased total levels of intracellular PI(4)P but also with PI(4)P enrichment at the membranous web15–17. In HCV-infected control cells expressing a nontargeting shRNA, there was substantial NS5A-PI(4)P colocalization (Figure 4C, Pearson’s correlation Pr=0.75). As expected, in PI4KA-silenced cells, we observed less total intracellular PI(4)P and little colocalization between PI(4)P and NS5A at membrane clusters (Pr=0.42). In contrast, in OSBP-silenced cells, there was prominent PI(4)P staining at membrane clusters (Figure 4C; Pr=0.78 and 0.75 for shOSBP-A and shOSBP-B, respectively).
We conclude from these results that although OSBP is required for membranous web integrity, PI4KA-mediated PI(4)P accumulation does not require OSBP, and that PI(4)P upregulation is not sufficient for normal membranous web morphology. These results suggest that OSBP functions downstream of PI4KA and PI(4)P in membranous web formation.
OSBP localization to NS5A-positive structures requires PI4KA activity
Targeting of OSBP to membranes requires its PI(4)P-binding PH domain (21 and Supplementary Figure 2B). We asked whether OSBP association with NS5A-positive structures requires PI4KA and PI(4)P. OSBP-NS5A colocalization could be observed in NS3-NS5B/GFP expressing Huh7/T7 cells transduced with a nontargeting shRNA (Figure 5A), as we had observed in HCV-infected cells (Figure 2A). However, PI4KA silencing (Figure 5A) or pharmacologic inhibition (Figure 5B) led to the loss of OSBP colocalization with membrane clusters, indicating that PI4KA and its lipid kinase activity are required for OSBP recruitment to the membranous web.
Figure 5. OSBP localization to membranous web requires PI4KA activity.
(A) The NS3-5B(NS5A/GFP) polyprotein was expressed in Huh7.5.1/T7 cells stably expressing a nontargeting or a PI4KA shRNA. 48 hr post-transfection, cells were immunostained for OSBP (red) and NS5A (green) with DAPI nuclear counterstaining (blue). M1 and M2, Manders’ coefficients; M1 represents the fraction of NS5A overlapping OSBP. Bar, 10 μm.
(B) The NS3-NS5B(NS5A/GFP) polyprotein was expressed in Huh7.5.1/T7 cells. 48 hr later, cells were treated with 0.1% DMSO or 4 μM AL-9 for 24 hr, and then immunostained for NS5A (green) and endogenous OSBP (red) with DAPI nuclear counterstaining (blue). M1 and M2, Manders’ coefficients; M1 represents the fraction of NS5A overlapping OSBP. Bar, 10 μm.
PI4KA and OSBP are required for cholesterol trafficking to NS5A-positive, detergent-resistant membranes
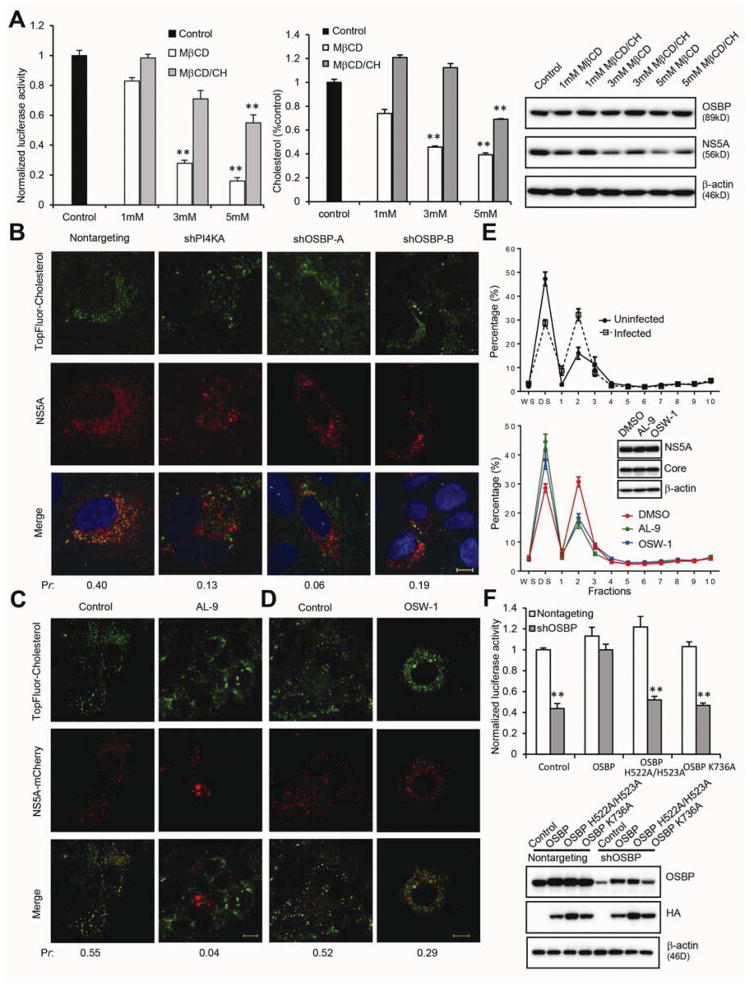
The HCV membranous web is cholesterol-enriched4–6, and cholesterol biosynthesis inhibitors hamper HCV replication4. To test directly whether cholesterol is required for HCV replication, OR6 replicon cells were treated with methyl-β-cyclodextrin (MβCD), which binds to and depletes cholesterol from membranes. MβCD caused a dose-dependent decrease in replication (Figure 6A, left panel) with little cytotoxicity (Supplementary Figure 4A). This decrease was partially reversed if MβCD was presaturated with cholesterol (MβCD/CH; Figure 6A, left panel), indicating that MβCD inhibits HCV replication primarily by cholesterol removal rather than by nonspecific effects. Immunoblotting also demonstrated a dose-dependent reduction of NS5A expression by MβCD treatment that was partially reversed by cholesterol presaturation (Figure 6A, right panel). MβCD and MβCD-cholesterol complexes had the expected effects on cellular cholesterol content (Figure 6A, middle panel).
Figure 6. Cholesterol depletion inhibits HCV replication, and cholesterol trafficking to membranous webs requires PI4KA and OSBP.
(A) OR6 cells were treated with the indicated concentrations of MβCD or MβCD-cholesterol complexes for 4 hr, washed, and then incubated for 24 hr prior to measurement of luciferase activity (left panel), cellular cholesterol content (middle panel), and immunoblotting (right panel). Values (means ± SEM, three independent experiments) are normalized to control. **, P<.001 versus control.
(B) HCV-infected Huh 7.5.1 cells stably expressing the indicated shRNAs were labeled with 2 μM TopFluor-cholesterol (green) for 2 hr followed by immunostaining for NS5A (red) with DAPI nuclear counterstaining (blue). Pr, Pearson’s correlation coefficients; bar, 10 μm.
(C, D) Live-cell imaging of cells stably expressing a subgenomic replicon with mCherry-tagged NS5A (red) treated with 0.1% DMSO (left panels), 4 μM AL-9 (C, right panels), or 30 nM OSW-1 (D, right panels) for 8 hr before labeling with 2 μM TF-cholesterol (green) for another 16 hr. Pr, Pearson’s correlation coefficients; bar, 10 μm.
(E) Upper panel: Uninfected (solid line) or HCV-infected (dotted line) cells were labeled with 2 μM TF-cholesterol for 4 hr; Lower panel: JFH-1 infected cells were treated with 0.1% DMSO (red), 5 μM AL-9 (green), or 50 nM OSW-1 (blue) for 4 hr in the presence of 2 μM TF-cholesterol. Cell homogenates were then fractionated as in Figure 2B. Fluorescence intensity of TF-cholesterol was measured from each fraction and represented as a percentage of the total TF-cholesterol fluorescence in all fractions. Values are mean ± SEM, four independent experiments. Inset, immunoblotting of total cell homogenates.
(F) OR6 cells stably expressing the indicated shRNAs were transduced with lentiviral vectors encoding HA-OSBP constructs lacking the 3′UTR, followed 4 days later by luciferase activity measurement (upper panel) and immunoblotting (lower panel). Values (mean ± SEM, four independent experiments) are normalized to nontargeting control. **, P<.001 versus nontargeting control.
OSBP binds to cholesterol in vitro22 and can catalyze the PI(4)P-dependent transfer of cholesterol between synthetic liposomes23. To test whether cholesterol is transported to NS5A-positive HCV membranous webs and if so, whether this process requires OSBP, we labeled JFH-1 infected Huh7.5.1 cells with TopFluor-cholesterol (TF-cholesterol), a fluorescent cholesterol analog that closely mimics the membrane partitioning and trafficking of native cholesterol29. TF-cholesterol colocalized extensively with NS5A in nontargeting shRNA-expressing cells (Figure 6B), suggesting that TF-cholesterol is transported to the membranous web. In contrast, there was little colocalization between TF-cholesterol and NS5A in cells silenced for either PI4KA or OSBP, indicating that cholesterol trafficking to NS5A-positive structures requires both PI4KA and OSBP.
To exclude possible artifacts from cell fixation, we also examined TF-cholesterol transport to membranous webs in living cells harboring a subgenomic HCV replicon with mCherry-tagged NS5A. In control cells, TF-cholesterol colocalized extensively with NS5A-positive membranes (Figure 6C, D). In contrast, treatment with the PI4K inhibitor AL-9 (Figure 6C) and the OSBP inhibitor OSW-1 (Figure 6D) inhibited transport of cholesterol to NS5A-positive membrane clusters.
The requirement of PI4KA and OSBP for cholesterol transport to the membranous web was further tested by a biochemical assay. HCV-infected cells labeled with TF-cholesterol were treated with AL-9 or OSW-1 to inhibit PI4KA or OSBP, respectively, and we quantitated the amount of incorporated TF-cholesterol fluorescence in isolated DRM fractions and in detergent-soluble membranes. In HCV-infected control cells, an increased proportion of TF-cholesterol was detected in DRM fractions compared to uninfected cells (Figure 6E, upper panel), while AL-9 or OSW-1 treatment reduced the proportion of TF-cholesterol found in DRMs (Figure 6E, lower panel). Brief AL-9 or OSW-1 treatment did not lower the total amount of NS5A protein (Figure 6E, lower panel inset), and silencing of PI4KA or OSBP did not significantly block TF-cholesterol cellular uptake (Supplementary Figure 4B). These results collectively suggest a specific requirement of PI4KA and OSBP for cholesterol trafficking to DRMs in HCV-infected cells.
OSBP and its yeast homolog, Osh4p, can exchange sterols for PI(4)P between lipid bilayers30,31. Although Osh4p lacks a PH domain, it interacts with PI(4)P at a binding site that overlaps with the sterol-binding domain (SBD)30. Several residues at the entrance of the SBD (including H143/H144 and K336 in Osh4p) form essential contacts with PI(4)P but are dispensable for sterol binding and transfer activity30, and we mapped the corresponding residues in OSBP to H522/H523 and K736 (Supplementary Figure 4C). To test whether these PI(4)P-interacting residues at the OSBP SBD are essential for its ability to support HCV replication, we conducted a rescue assay. HCV inhibition caused by shRNA targeting the OSBP 3′UTR could be rescued by exogenous expression of wild type OSBP but not by the OSBP mutants H522A/H523A or K736A (Figure 6F). Immunostaining showed that these two OSBP mutants colocalize with NS5A, but fail to transport TF-cholesterol to NS5A-positive structures (Supplementary Figure 4D and 4E), consistent with our previous finding that the SBD is not required for OSBP colocalization with NS5A.
The dependence of RNA viruses on OSBP correlates with their dependence on PI 4-kinases
Like HCV, Dengue virus (DENV) is also a flavivirus, but its replication does not require PI4KA32. Silencing of PI4KA, PI4KB, or OSBP failed to substantially inhibit replication of a DENV replicon (Figure 7A). Consistent with these results, the PI4KA inhibitor AL-9 and the OSBP inhibitor OSW-1 had only minor effects on DENV replication (Figure 7B) at concentrations that strongly inhibit HCV replication (Figure 1C and Supplementary Figure 5). These results suggest that DENV replication is not dependent on PI4KA, PI4KB, or OSBP.
Figure 7. The dependence of RNA viruses on OSBP correlates with their dependence on PI 4-kinases.
(A) Huh7.5.1 cells containing a subgenomic DENV replicon expressing a Renilla luciferase reporter were transduced with the indicated shRNAs for 72 hr prior to luciferase activity measurement (left panel) and immunoblotting (right panel). Values (means ± SD, two independent experiments) are normalized to control cells
(B) DENV replicon cells were treated with the indicated concentrations of OSW-1 (left panel) or AL-9 (right panel) for 24 hr before luciferase activity measurement. Values are means ± SD, two independent experiments.
(C) HeLa cells stably expressing a nontargeting shRNA or three independent OSBP shRNAs were infected with poliovirus (MOI 0.1) 24 hr before measurement of viral titers (left panel), and immunoblotting (right panel). *, P = .004 and **, P< .001 versus nontargeting shRNA.
Conversely, poliovirus (PV) is a positive-sense RNA virus that exploits the PI 4-kinase PI4KB for its replication17. Three independent OSBP shRNAs inhibited PV infection in HeLa cells (Figure 7C). We conclude that the dependence of three RNA viruses on OSBP correlates with their dependence on PI 4-kinases, supporting a link between PI 4-kinases and OSBP in RNA virus infection.
DISCUSSION
Although PI 4-kinases and PI(4)P have been implicated in positive sense RNA virus-induced remodeling of intracellular membranes8,9,14,16,17, their effectors in this process have not been defined. In this study, we identify OSBP as a PI4KA effector in the formation of the HCV membranous web. While HCV exploits PI4KA for its replication, several picornaviruses instead depend on the related PI 4-kinase PI4KB17,18,33. The utilization of two different PI 4-kinases by RNA viruses suggests a common requirement for PI(4)P, and possibly also for common cellular PI(4)P effectors. Consistent with this hypothesis, we find that OSBP appears to be a cofactor for poliovirus replication, even though PV exploits PI4KB rather than PI4KA. Conversely, DENV does not require PI4KA, PI4KB, or OSBP for its replication. If these findings are extended to other RNA viruses, then inhibition of PI 4-kinase effectors may be a potential broadly antiviral strategy.
Non-vesicular intracellular cholesterol trafficking in mammalian cells has been proposed to be regulated by lipid transfer proteins such as OSBP. However, there has been little evidence that OSBP can regulate cholesterol trafficking in mammalian cells. Here we find that cholesterol trafficking to the HCV membranous web is regulated by OSBP and that it functions downstream of PI4KA. We propose that recruitment of PI4KA to the membranous web and its activation by HCV NS5A9 leads to the local accumulation of PI(4)P. OSBP recruitment to the web by PI(4)P and possibly also by VAPs, which are also enriched at webs and interact with HCV NS5A34,35, then stimulates the delivery of cholesterol to the web, possibly by exchanging cholesterol with PI(4)P30,31. This is consistent with the requirement of the PH domain, the FFAT domain, the sterol-binding domain, and PI(4)P-interacting residues within the SBD for OSBP to support HCV replication. Although intact membranous webs have not been isolated from HCV-infected cells at sufficient purity to permit direct measurement of their lipid composition, a recent study has shown that affinity-purified NS4B-enriched membranes from HCV-infected cells are cholesterol-enriched, and also found that cholesterol depletion could inhibit HCV replication6.
In addition to cholesterol transport, OSBP regulates sphingomyelin synthesis at the Golgi by stimulating the recruitment of ceramide transfer protein36. Lipid raft membranes are also enriched in sphingomyelin and glycophingolipids, and inhibitors of sphingolipid synthesis suppress HCV replication37. Our results do not exclude the possibility that PI4KA and OSBP also regulate the accumulation of sphingolipids at the membranous web, although we have not observed efficient trafficking of fluorescent sphingolipids to webs (not shown).
We do not know why HCV replication might be dependent on cholesterol and/or sphingolipid enrichment at the membranous web. Possibilities include alteration of membrane fluidity or curvature, recruitment of cellular or viral factors necessary for viral replication, and direct stimulation of viral enzymatic activity by binding to sterols and/or sphingolipids38. The development of systems to reconstitute HCV genome replication in vitro from purified components would greatly facilitate our ability to discriminate among these alternatives.
Materials and Methods
Additional methods are described in Supplementary Material.
Viruses and viral replicons
The genotype 2a HCV strain JFH-1 was produced and propagated as described in 39. A subgenomic replicon expressing mCherry-tagged NS5A based on the JFH-1 genotype 2a sequence, termed here as pSGR-JFH1(NS5A/mCherry) and a dengue virus replicon based on the DENV-2 16681 strain with a Renilla luciferase reporter gene are described in Supplemental information. Poliovirus (PV) used in this study is type 1 Mahoney.
Immunofluorescence staining
Immunofluorescent staining was performed as described in 15. PI(4)P staining of intracellular membranes was performed exactly as described in 40. A Nikon A1 laser scanning confocal microscope was used with sequential scanning mode to limit crosstalk between fluorochromes. PI(4)P fluorescence intensity was quantified using ImageJ software from multiple random fields obtained with identical acquisition settings.
Subcellular fractionation
Huh7.5.1 cells were infected with JFH-1 (MOI = 0.1) and cells were harvested at 5 days postinfection. For AL-9 or OSW-1 treatment, Huh7.5.1 cells were infected 4 days prior to drug treatment for 4 hr before harvesting. DRM isolation was performed as described previously41, and described further in Supplementary Material.
TopFluor-cholesterol cell labeling
Huh7.5.1 cells stably transduced with nontargeting, PI4KA, or OSBP shRNA were infected with JFH-1 (MOI = 0.1). 4 days later, TopFluor-Cholesterol (TF-cholesterol; Avanti Polar Lipids, Alabaster, AL) was added to the culture medium to a final concentration of 2 μM for 2 hr before they were immunolabeled for NS5A. Drug treatment with OSW-1 or AL-9 was performed before the addition of TF-Cholesterol. TF-cholesterol labeled replicon cells underwent live-cell imaging, while HCV-infected cells underwent subcellular fractionation. The fluorescence intensity of TF-cholesterol in the collected fractions was measured with a Synergy2 plate reader equipped with 485±20 nm excitation and 520±25 nm emission filters.
Supplementary Material
Acknowledgments
Grant Support: This work was supported by grants AI083785 and DK097374 from the National Institutes of Health and the Greenview Foundation Hepatitis C Research Fund (to AWT)
We thank Dotty Sorenson of the Microscopy & Image Analysis Laboratory at the University of Michigan for technical assistance with electron microscopy. Huh7.5.1 cells, the OR6 cell line, the infectious JFH-1 clone, anti-NS5A antibody (clone 9E10), OSW-1, and an OSBP construct were generous gifts from Dr. Francis Chisari (Scripps Institute, La Jolla, CA), Dr. Nobuyuki Kato (Okayama University, Okayama, Japan), Dr. Takaji Wakita (National Institute of Infectious Diseases, Tokyo, Japan), Dr. Charles Rice (Rockefeller University, New York, NY), Dr. Matthew Shair (Harvard University, Cambridge, MA), and Dr. Marci Scidmore (Cornell University, Ithaca, NY), respectively.
Abbreviations used in this paper
- DMV
double-membrane vesicle
- DRM
detergent-resistant membrane
- FFAT
two phenylalanines in an acidic tract
- HCV
hepatitis C virus
- MMV
multi-membrane vesicle
- ORP
OSBP-related protein
- OSBP
oxysterol-binding protein
- PH
pleckstrin homology
- PI
phosphatidylinositol
- PI(4)P
phosphatidylinositol 4-phosphate
- SBD
sterol-binding domain
- TF
TopFluor
- VAP
vesicle-associated membrane protein-associated protein
Footnotes
Disclosures: The authors disclose no conflicts.
Writing assistance: none
Author Contributions: Study concept and design (HW and AWT), acquisition of data (HW, JWP, and ASL), analysis and interpretation of data (HW, JWP, ASL, AWT), drafting of the manuscript (HW and AWT), critical revision of the manuscript (HW, JWP, PN, RDF, AWT), reagents (ASL, PN, RDF)
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Miller S, Krijnse-Locker J. Modification of intracellular membrane structures for virus replication. Nature Reviews Microbiology. 2008;6:363–74. doi: 10.1038/nrmicro1890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Egger D, Wölk B, Gosert R, et al. Expression of hepatitis C virus proteins induces distinct membrane alterations including a candidate viral replication complex. Journal of Virology. 2002;76:5974–84. doi: 10.1128/JVI.76.12.5974-5984.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gosert R, Egger D, Lohmann V, et al. Identification of the hepatitis C virus RNA replication complex in Huh-7 cells harboring subgenomic replicons. Journal of Virology. 2003;77:5487–92. doi: 10.1128/JVI.77.9.5487-5492.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Aizaki H, Lee KJ, Sung VM, et al. Characterization of the hepatitis C virus RNA replication complex associated with lipid rafts. Virology. 2004;324:451–61. doi: 10.1016/j.virol.2004.03.034. [DOI] [PubMed] [Google Scholar]
- 5.Shi ST, Lee KJ, Aizaki H, et al. Hepatitis C virus RNA replication occurs on a detergent-resistant membrane that cofractionates with caveolin-2. Journal of Virology. 2003;77:4160–8. doi: 10.1128/JVI.77.7.4160-4168.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Paul D, Hoppe S, Saher G, et al. Morphological and biochemical characterization of the membranous hepatitis C virus replication compartment. Journal of Virology. 2013;87:10612–27. doi: 10.1128/JVI.01370-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Berger KL, Cooper JD, Heaton NS, et al. Roles for endocytic trafficking and phosphatidylinositol 4-kinase III alpha in hepatitis C virus replication. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:7577–82. doi: 10.1073/pnas.0902693106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tai AW, Benita Y, Peng LF, et al. A functional genomic screen identifies cellular cofactors of hepatitis C virus replication. Cell Host and Microbe. 2009;5:298–307. doi: 10.1016/j.chom.2009.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Reiss S, Rebhan I, Backes P, et al. Recruitment and activation of a lipid kinase by hepatitis C virus NS5A is essential for integrity of the membranous replication compartment. Cell Host and Microbe. 2011;9:32–45. doi: 10.1016/j.chom.2010.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Trotard M, Lepère-Douard C, Régeard M, et al. Kinases required in hepatitis C virus entry and replication highlighted by small interference RNA screening. FASEB Journal. 2009;23:3780–9. doi: 10.1096/fj.09-131920. [DOI] [PubMed] [Google Scholar]
- 11.Borawski J, Troke P, Puyang X, et al. Class III phosphatidylinositol 4-kinase alpha and beta are novel host factor regulators of hepatitis C virus replication. Journal of Virology. 2009;83:10058–74. doi: 10.1128/JVI.02418-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vaillancourt FH, Pilote L, Cartier M, et al. Identification of a lipid kinase as a host factor involved in hepatitis C virus RNA replication. Virology. 2009;387:5–10. doi: 10.1016/j.virol.2009.02.039. [DOI] [PubMed] [Google Scholar]
- 13.Li Q, Brass AL, Ng A, et al. A genome-wide genetic screen for host factors required for hepatitis C virus propagation. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:16410–5. doi: 10.1073/pnas.0907439106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bianco A, Reghellin V, Donnici L, et al. Metabolism of Phosphatidylinositol 4-Kinase IIIalpha-Dependent PI4P Is Subverted by HCV and Is Targeted by a 4-Anilino Quinazoline with Antiviral Activity. PLoS Pathogens. 2012;8:e1002576. doi: 10.1371/journal.ppat.1002576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tai AW, Salloum S. The Role of the Phosphatidylinositol 4-Kinase PI4KA in Hepatitis C Virus-Induced Host Membrane Rearrangement. PLoS ONE. 2011;6:e26300. doi: 10.1371/journal.pone.0026300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Berger KL, Kelly SM, Jordan TX, et al. Hepatitis C Virus Stimulates the Phosphatidylinositol 4-Kinase III Alpha-Dependent Phosphatidylinositol 4-Phosphate Production That Is Essential for Its Replication. Journal of Virology. 2011;85:8870–83. doi: 10.1128/JVI.00059-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hsu NY, Ilnytska O, Belov G, et al. Viral reorganization of the secretory pathway generates distinct organelles for RNA replication. Cell. 2010;141:799–811. doi: 10.1016/j.cell.2010.03.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Greninger AL, Knudsen GM, Betegon M, et al. The 3A protein from multiple picornaviruses utilizes the golgi adaptor protein ACBD3 to recruit PI4KIIIbeta. J Virol. 2012;86:3605–16. doi: 10.1128/JVI.06778-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Amako Y, Sarkeshik A, Hotta H, et al. Role of oxysterol binding protein in hepatitis C virus infection. Journal of Virology. 2009;83:9237–46. doi: 10.1128/JVI.00958-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Amako Y, Syed GH, Siddiqui A. Protein kinase D negatively regulates hepatitis C virus secretion through phosphorylation of oxysterol-binding protein and ceramide transfer protein. Journal of Biological Chemistry. 2011;286:11265–74. doi: 10.1074/jbc.M110.182097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Levine TP, Munro S. Targeting of Golgi-specific pleckstrin homology domains involves both PtdIns 4-kinase-dependent and -independent components. Current Biology. 2002;12:695–704. doi: 10.1016/s0960-9822(02)00779-0. [DOI] [PubMed] [Google Scholar]
- 22.Wang PY, Weng J, Lee S, et al. The N terminus controls sterol binding while the C terminus regulates the scaffolding function of OSBP. Journal of Biological Chemistry. 2008;283:8034–45. doi: 10.1074/jbc.M707631200. [DOI] [PubMed] [Google Scholar]
- 23.Ngo M, Ridgway ND. Oxysterol binding protein-related Protein 9 (ORP9) is a cholesterol transfer protein that regulates Golgi structure and function. Molecular Biology of the Cell. 2009;20:1388–99. doi: 10.1091/mbc.E08-09-0905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Raychaudhuri S, Im YJ, Hurley JH, et al. Nonvesicular sterol movement from plasma membrane to ER requires oxysterol-binding protein-related proteins and phosphoinositides. Journal of Cell Biology. 2006;173:107–19. doi: 10.1083/jcb.200510084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tariq H, Manzoor S, Parvaiz F, et al. An overview: In vitro models of HCV replication in different cell cultures. Infect Genet Evol. 2011 doi: 10.1016/j.meegid.2011.10.009. [DOI] [PubMed] [Google Scholar]
- 26.Ikeda M, Abe K, Dansako H, et al. Efficient replication of a full-length hepatitis C virus genome, strain O, in cell culture, and development of a luciferase reporter system. Biochemical and Biophysical Research Communications. 2005;329:1350–9. doi: 10.1016/j.bbrc.2005.02.138. [DOI] [PubMed] [Google Scholar]
- 27.Burgett AW, Poulsen TB, Wangkanont K, et al. Natural products reveal cancer cell dependence on oxysterol-binding proteins. Nature Chemical Biology. 2011;7:639–47. doi: 10.1038/nchembio.625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Romero-Brey I, Merz A, Chiramel A, et al. Three-dimensional architecture and biogenesis of membrane structures associated with hepatitis C virus replication. PLoS Pathogens. 2012;8:e1003056. doi: 10.1371/journal.ppat.1003056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Holtta-Vuori M, Uronen RL, Repakova J, et al. BODIPY-cholesterol: a new tool to visualize sterol trafficking in living cells and organisms. Traffic. 2008;9:1839–49. doi: 10.1111/j.1600-0854.2008.00801.x. [DOI] [PubMed] [Google Scholar]
- 30.de Saint-Jean M, Delfosse V, Douguet D, et al. Osh4p exchanges sterols for phosphatidylinositol 4-phosphate between lipid bilayers. Journal of Cell Biology. 2011;195:965–78. doi: 10.1083/jcb.201104062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mesmin B, Bigay J, Moser von Filseck J, et al. A Four-Step Cycle Driven by PI(4)P Hydrolysis Directs Sterol/PI(4)P Exchange by the ER-Golgi Tether OSBP. Cell. 2013;155:830–43. doi: 10.1016/j.cell.2013.09.056. [DOI] [PubMed] [Google Scholar]
- 32.Heaton NS, Perera R, Berger KL, et al. Dengue virus nonstructural protein 3 redistributes fatty acid synthase to sites of viral replication and increases cellular fatty acid synthesis. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:17345–50. doi: 10.1073/pnas.1010811107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sasaki J, Ishikawa K, Arita M, et al. ACBD3-mediated recruitment of PI4KB to picornavirus RNA replication sites. EMBO Journal. 2011 doi: 10.1038/emboj.2011.429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gao L, Aizaki H, He JW, et al. Interactions between viral nonstructural proteins and host protein hVAP-33 mediate the formation of hepatitis C virus RNA replication complex on lipid raft. Journal of Virology. 2004;78:3480–8. doi: 10.1128/JVI.78.7.3480-3488.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hamamoto I, Nishimura Y, Okamoto T, et al. Human VAP-B is involved in hepatitis C virus replication through interaction with NS5A and NS5B. Journal of Virology. 2005;79:13473–82. doi: 10.1128/JVI.79.21.13473-13482.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Banerji S, Ngo M, Lane CF, et al. Oxysterol binding protein-dependent activation of sphingomyelin synthesis in the golgi apparatus requires phosphatidylinositol 4-kinase IIα. Molecular Biology of the Cell. 2010;21:4141–50. doi: 10.1091/mbc.E10-05-0424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sakamoto H, Okamoto K, Aoki M, et al. Host sphingolipid biosynthesis as a target for hepatitis C virus therapy. Nature Chemical Biology. 2005;1:333–7. doi: 10.1038/nchembio742. [DOI] [PubMed] [Google Scholar]
- 38.Weng L, Hirata Y, Arai M, et al. Sphingomyelin activates hepatitis C virus RNA polymerase in a genotype specific manner. Journal of Virology. 2010;84:11761–70. doi: 10.1128/JVI.00638-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wakita T, Pietschmann T, Kato T, et al. Production of infectious hepatitis C virus in tissue culture from a cloned viral genome. Nature Medicine. 2005;11:791–6. doi: 10.1038/nm1268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hammond GR, Schiavo G, Irvine RF. Immunocytochemical techniques reveal multiple, distinct cellular pools of PtdIns4P and PtdIns(4,5)P(2) Biochemical Journal. 2009;422:23–35. doi: 10.1042/BJ20090428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Saxena V, Lai CK, Chao TC, et al. Annexin A2 is involved in the formation of hepatitis C virus replication complex on the lipid raft. Journal of Virology. 2012 doi: 10.1128/JVI.06327-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.