Abstract
Amphotericin has remained the powerful but highly toxic last line of defense in treating life-threatening fungal infections in humans for over 50 years with minimal development of microbial resistance. Understanding how this small molecule kills yeast is thus critical for guiding development of derivatives with an improved therapeutic index and other resistance-refractory antimicrobial agents. In the widely accepted ion channel model for its mechanism of cytocidal action, amphotericin forms aggregates inside lipid bilayers that permeabilize and kill cells. In contrast, we report that amphotericin exists primarily in the form of large, extramembranous aggregates that kill yeast by extracting ergosterol from lipid bilayers. These findings reveal that extraction of a polyfunctional lipid underlies the resistance-refractory antimicrobial action of amphotericin and suggests a roadmap for separating its cytocidal and membrane-permeabilizing activities. This new mechanistic understanding is also guiding development of the first derivatives of amphotericin that kill yeast but not human cells.
The incidence of life-threatening systemic fungal infections continues to rise in parallel with expanding populations of immunocompromised patients.1 Substantially exacerbating this problem is the concomitant rise in pathogen resistance to almost all clinically approved antifungal agents. In contrast, amphotericin B (AmB) (Fig. 1a) has served as the gold standard treatment for systemic fungal infections for over five decades with minimal development of clinically significant microbial resistance.2 This exceptional track record reveals that resistance-refractory modes of antimicrobial action exist, and the mechanism by which AmB kills yeast is one of them. However, because of the often dose-limiting toxicity of this natural product, mortality rates for systemic fungal infections persist near 50%.3 Improving the notoriously poor therapeutic index of this drug and the development of other resistance-refractory antimicrobial agents thus represent two critically important objectives that stand to benefit from a clarified molecular description of the biological activities of AmB. Moreover, an advanced understanding of the biophysical interactions of this natural product within living systems would enable more effective utilization of its remarkable capacity to perform ion channel-like functions.
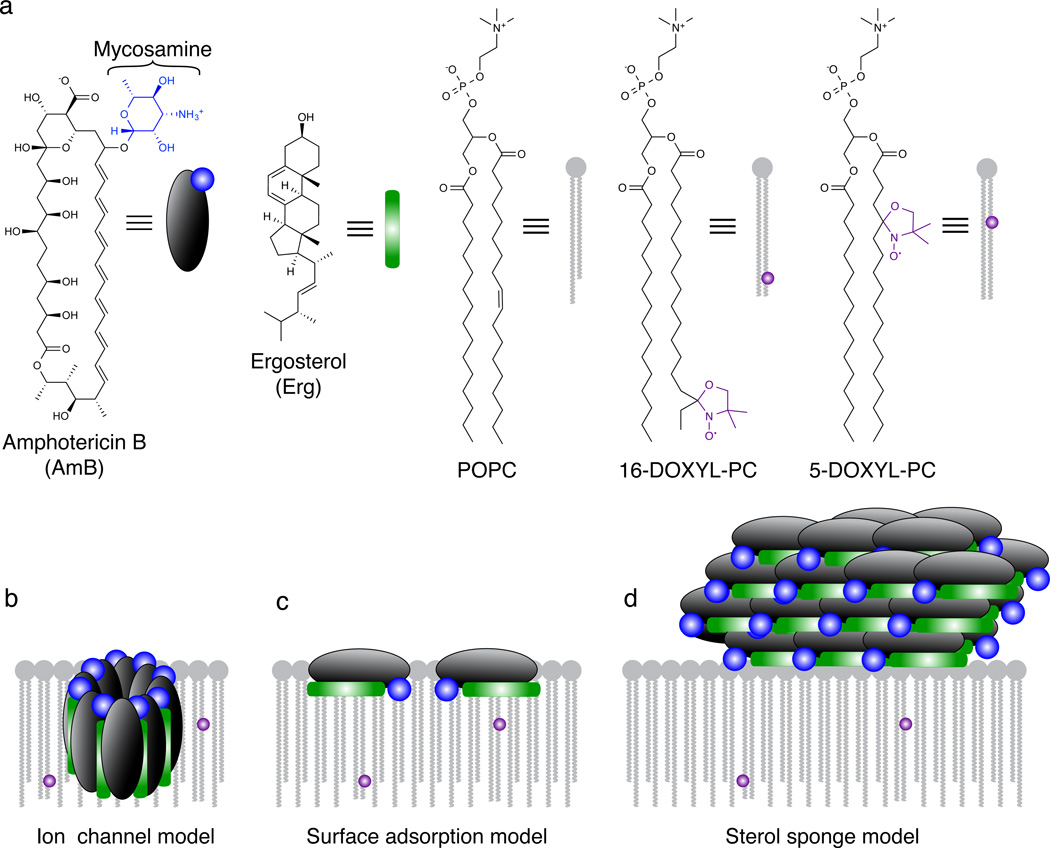
Figure 1. Models for the structure and function of AmB in the presence of lipid bilayers.
a, Structures of AmB, Erg, POPC, and paramagnetic probes 5-DOXYL-PC and 16-DOXYL-PC. 5-DOXYL and 16-DOXYL position a paramagnetic functional group at depths of ~12 and ~25 Å within the lipid bilayer, respectively. b, The classic ion channel model for the structure and function of AmB. c, Surface adsorption model. d, A new sterol sponge model, in which AmB primarily exists in the form of large extramembranous aggregates that extract Erg from lipid bilayers.
For decades, the prevailing theory has been that AmB primarily exists in the form of small ion channel aggregates that are inserted into lipid bilayers and thereby permeabilize and kill yeast cells (Fig. 1b).4–23 An extensive series of structural and biophysical studies, including those employing planar lipid bilayers,4–10 liposome permeability,9–13,17 Corey-Pauling-Kulton (CPK) modeling,7–9 UV/Vis spectroscopy,9–11,13,21 circular dichroism,10,11,13,21 fluorescence spectroscopy,9,11 Raman spectroscopy,10 differential scanning calorimetry,9,10,21 chemical modifications,11–14,17 atomic force microscopy,21 transmission electron microscopy,20 computer modeling,11,15 electron paramagnetic resonance,10 surface plasmon resonance,22 solution NMR spectroscopy,11 and solid-state NMR (SSNMR)16–19 spectroscopy have been interpreted through the lens of this ion channel model. Importantly, this model suggests that the path to an improved therapeutic index requires selective formation of ion channels in yeast versus human cells,10–20 that the search for other resistance-refractory antimicrobials should focus on membrane-permeabilizing compounds,24 and that the ion channel-forming and cytotoxic activities of AmB cannot be separated.
Recent studies show that the channel forming capacity of AmB is not required for fungicidal activity, whereas binding ergosterol (Erg) (Fig. 1a) is essential.25–27 However, the structural and biophysical underpinnings of this rare type of small molecule-small molecule interaction and its connection to cell killing all remained unclear. Sterols, including Erg in yeast, play many essential roles in eukaryotic cell physiology, including functional regulation of membrane proteins, microdomain formation, endocytosis, vacuole fusion, cell division, and cell signaling.28–31 We thus hypothesized that sequestering Erg and thereby concomitantly precluding its participation in multiple cellular functions may underlie the fungicidal action of AmB.
Guided by this hypothesis, we considered three possible models for the primary structure and function of AmB in the presence of Erg-containing phospholipid membranes (Fig. 1b–d): (i) In the classic channel model, AmB primarily exists in the form of small (~1 nm) ion channel aggregates inserted into the membrane, perpendicular to the membrane surface, with Erg molecules interdigitated between AmB molecules (Fig. 1b).7–9,11,12,15–19,22,23 (ii) In an alternative surface adsorption model, AmB is primarily positioned in the intermediate/headgroup region, oriented parallel to the plane of the membrane, sequestering Erg to the membrane surface (Fig. 1c).9,22 (iii) In a new sterol sponge model, AmB primarily exists as large extramembranous aggregates that extract Erg from lipid bilayers (Fig. 1d). In the latter two models, we envisioned that membrane-permeabilizing ion channels represent relatively minor contributors to both the structure and cytocidal activity of AmB. Here we report an extensive series of SSNMR, transmission electron microscopy (TEM), and cell-based experiments that all support the new sterol sponge model (Fig. 1d).
RESULTS
SSNMR paramagnetic relaxation enhancement experiments
Distinguishing among the aforementioned structural and functional models (Fig. 1b-d) required determining the location of AmB relative to lipid bilayers and the corresponding location of Erg in the absence and presence of AmB. Making these determinations turned out to be exceptionally challenging due to the lack of high-resolution methods for probing small molecule/membrane interactions.9–13,15,17–21 We thus developed an experiment based on the NMR paramagnetic relaxation enhancement (PRE) of 13C nuclei caused by lipid-appended spin labels.32–34 13C nuclei proximal to a stable radical, such as 4,4-dimethyloxazolidine-N-oxyl (DOXYL), experience large enhancements of their longitudinal relaxation rates (R1 = 1/T1). Due to the high gyromagnetic ratio of the electron spin, the PRE is detectable for distances up to ~20 Å. Harnessing this phenomenon, we designed a magic-angle spinning (MAS) SSNMR PRE experiment based on 16-DOXYL-PC and 5-DOXYL-PC to interrogate proximity to the hydrophobic core and intermediate/headgroup region, respectively (Fig. 1a). Importantly, the three models under consideration (Fig. 1b-d) predict distinct PRE effects for AmB. The ion channel model predicts large PREs with both spin labels; the surface adsorption model predicts large PREs only with 5-DOXYL-PC; and the sterol sponge model predicts little or no PRE effects with either spin label.
To execute this experiment with maximized signal-to-noise ratio, we prepared highly enriched uniformly 13C-labeled AmB (U-13C-AmB) via development of an improved biosynthetic protocol.18 By using uniformly 13C-labeled glucose (U-13C-glucose) as the primary carbon source, we developed a robust procedure for attaining >80% 13C incorporation, the highest 13C enrichment yet reported for this natural product (Supplementary Results, Supplementary Note). This highly enriched U-13C-AmB enabled confident assignment of 13C resonances and high sensitivity PRE measurements even at high lipid-to-AmB ratios.
We next identified a physiologically relevant lipid bilayer system in which we could execute the targeted experiments. Recent structure-function studies with AmB reveal that, in contrast to liposomes comprised of fully saturated lipids, liposomes derived from monounsaturated 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (POPC) and Erg yield results that mirror those obtained with live yeast cells.25,27 Also, unlike membranes derived from fully saturated lipids, the plasma membranes of yeast and bilayers derived from POPC:Erg are both in the liquid crystalline state at 23 °C.35,36 In yeast, POPC is highly abundant, and Erg is the most common sterol.37 Moreover, at the minimum inhibitory concentration for AmB against both Saccharomyces cerevisiae and Candida albicans, there is at least as much AmB as there is Erg.25 We thus prepared lipid bilayer samples throughout this study with molar ratios of at least 10:1 POPC:Erg; for those samples containing AmB, we used at least 1 equivalent of AmB (relative to Erg). These bilayers exhibited proper phase behavior and retained this behavior upon addition of DOXYL spin labels (Supplementary Fig. 1).
To first confirm the position of each spin label in the bilayer, we determined the respective PREs on 13C resonances in natural abundance POPC lipids with 5 mol% of each DOXYL spin label (Fig. 2a). Consistent with the structure of POPC membranes,35 bilayers doped with 5% 16-DOXYL-PC demonstrated a maximal PRE at the termini of the POPC fatty acid chains, and the PRE decreased as the distance from the center of the membrane increased (Fig. 2a). The incorporation of 5-DOXYL-PC alternatively yielded maximal PREs in the intermediate and headgroup regions of the lipid bilayer and much smaller, yet still readily observable, effects at the membrane interior. Thus, these two spin label probes enabled confident and complementary interrogation of the innermost and outermost regions of a lipid bilayer in a straightforward SSNMR experiment.
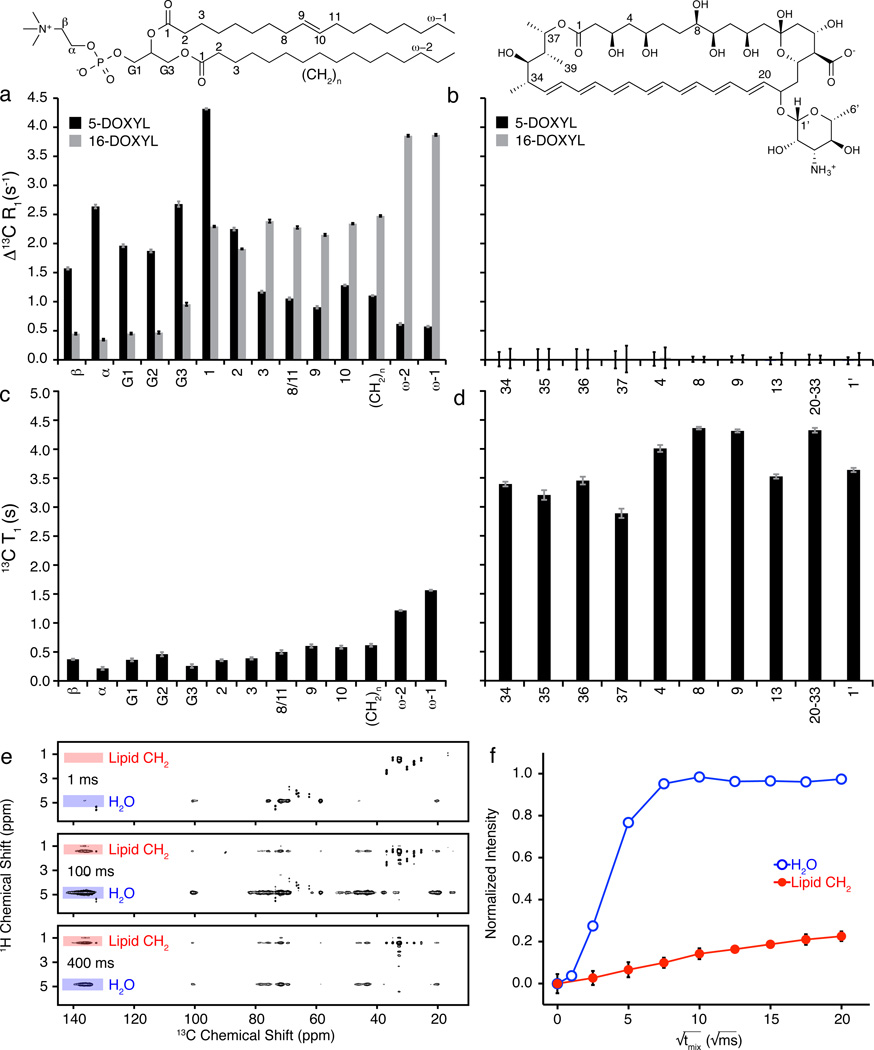
Figure 2. AmB primarily exists as large extramembranous aggregates.
a, Paramagnetic relaxation enhancements (PREs) have magnitude proportional to 1/r6 where r is the distance from spin label to NMR-detected nucleus. POPC controls demonstrated this proportionality in the presence of 5 mol% 5-DOXYL-PC (black) or 16-DOXYL-PC (gray). b, U-13C-AmB demonstrated no significant PRE effects (> 0.1 s−1) in the presence of either 5-DOXYL-PC (black) or 16-DOXYL-PC (gray) paramagnetic probes. c,d, Substantial differences were observed between longitudinal (T1) 13C relaxation times for sites in (c) POPC and (d) AmB. e, Selected 1H-13C 2D spectra were collected with 1 ms T2 filter, and 1H-1H spin diffusion times of 1 ms, 100 ms, and 400 ms; cross peaks from lipid acyl chains (red) and water (blue) to U-13C-AmB polyene region. f, The polarization transfer was quantified as a function of spin diffusion time from water and lipid to U-13C-AmB polyene. PRE values were derived from the difference between 13C R1 relaxation rates measured via inversion recovery for diamagnetic samples and samples containing 5- and 16-DOXYL-PC. Error bars were determined by chi-squared analysis. 1H-13C spin diffusion data were normalized relative to maximum intensity observed for both lipid and water cross peaks for a given 13C site after correcting for 1H T1 relaxation. Error bars were derived from signal-to-noise of the observed cross peak. Spectra were acquired at 14.1 T (600 MHz 1H frequency) at 20 °C, 10 kHz MAS.
We next prepared samples with U-13C-AmB in POPC/Erg bilayers and used a series of 13C chemical shift correlation experiments (including CTUC-COSY,38 DARR,39 and SPC5 recoupling40) to assign de novo the 13C resonances of AmB (Online Methods Section II, Supplementary Fig. 2 and 3, Supplementary Table 1, and Supplementary Note). We then performed PRE measurements in the presence of 16-DOXYL-PC or 5-DOXYL-PC spin probes. The results were striking and unambiguous. As predicted by the extramembranous sterol sponge model, and inconsistent with both the membrane-inserted ion channel and surface adsorption models, we observed no significant PREs to any 13C resonances of AmB with either probe (Fig. 2b). Thus, the majority of AmB in these samples was >20 Å away from the membrane-embedded spin labels.
AmB primarily exists as large extramembranous aggregates
A series of additional SSNMR experiments further revealed that AmB exists in the form of large aggregates that are more closely associated with water than lipids. The longitudinal relaxation times (T1 values) for AmB were substantially longer than those of the lipids, consistent with large and relatively immobile aggregates of AmB (Fig. 2c, 2d, Supplementary Table 2). SSNMR spin-diffusion experiments, designed for the purpose of probing membrane protein topology,41 revealed that lipid-AmB correlations reached maximum intensity only at very long mixing times (~400 ms) for all resolvable carbons on AmB (Fig. 2e, 2f, Supplementary Fig. 4), indicating that the majority of the lipids were >15 Å away from the AmB. In contrast, we observed strong correlations between water and AmB within just 25 ms, consistent with intimate proximity of the AmB aggregates to water.
To further probe these aggregates and distinguish between an intramembranous vs. extramembranous location, we also performed transmission electron microscopy analysis of large unilamellar vesicles (LUVs) comprised of the same ratio of POPC:Erg ± AmB. In the absence of added AmB, we observed well-formed LUVs (Fig. 3a, Supplementary Fig. 5a). When AmB was added, we observed large extramembranous aggregates (Fig. 3b, Supplementary Fig. 5b). These aggregates were associated with one or more LUVs, suggesting an interaction between the surfaces of the aggregate and the lipid bilayer. When we added the same amount of AmB to the same volume of buffer devoid of LUVs, similar aggregates of AmB were observed (Fig. 3c, Supplementary Fig. 5c). These observations are consistent with the spontaneous formation in aqueous buffer of large AmB aggregates that externally associate with the surface of lipid bilayers.
Figure 3. Direct visualization of large extramembranous aggregates of AmB by transmission election microscopy.
a, (supplementary Fig 5a), POPC:Erg 10:1 liposomes. b (supplementary Fig 5b), POPC:Erg 10:1 liposomes with 1 equivalent (relative to Erg) of added AmB. c (supplementary Fig 5c), AmB only.
Importantly, parallel potassium efflux experiments revealed readily observable membrane permeabilization upon adding the same concentration of AmB to suspensions of the same POPC:Erg LUVs (Supplementary Fig. 6). This observation was consistent with a minor fraction of AmB existing in the form of membrane-permeabilizing ion channels that are too small to be visualized by TEM. This analysis was also consistent with all of our SSNMR data, in which the limits of detection permit up to 5% of the AmB existing in the membrane (Online Methods Section II).
Extramembranous AmB aggregates extract Erg from bilayers
With the structural aspects of the sterol sponge model confirmed, we aimed to test the functional prediction that these large extramembranous aggregates of AmB extract Erg from lipid bilayers. We first performed a modified SSNMR PRE experiment in which we analyzed 13C-skip-labeled Erg (13C-Erg, Fig. 4a)19 in spin label-containing bilayers as a function of AmB:13C-Erg ratio (Fig. 4a). This labeling pattern provided sufficient sensitivity that the ratio of POPC to Erg was increased to 40:1, readily enabling titrations of the AmB:Erg molar ratio while retaining the biophysical properties of the lipid bilayer. Thus, we prepared bilayers comprised of POPC:13C-Erg 40:1 ± 5 mol% 16-DOXYL without or with increasing amounts of natural abundance AmB. AmB had minimal effect on the POPC PRE (Supplementary Fig. 7). In contrast, we observed a progressive decrease in the 13C-Erg PRE as the amount of AmB increased, indicating that Erg increasingly occupied a position outside the lipid bilayer (Fig. 4a, Supplementary Fig. 7a). In the absence of AmB (AmB:13C-Erg 0:1), we observed substantial PREs for the resolved 13C signals of 13C-Erg; for several sites, such as Erg-18, Erg-21, Erg-22, Erg-24 and Erg-26/27, the PRE was ~1.5 s−1 or greater, and the 13C T1 values were relatively short (<1.5 s) (Supplementary Fig. 7b). These findings are consistent with the structure of Erg-containing membranes in which the Erg was inserted into the hydrophobic core of the bilayer,35 with the isopropyl tail most deeply inserted and therefore most proximate to the 16-DOXYL label. These conformation-specific PREs for 13C-Erg decreased markedly upon the addition of AmB (Fig. 4a, Supplementary Fig. 7a). Specifically, with increasing amounts of natural abundance AmB (AmB:13C-Erg ratios of 1:1, 4:1, 8:1), we observed a progressive decrease, with at least a three-fold reduction in observed PRE in the AmB:13C-Erg 8:1 sample. These results support the interpretation that, in the presence of increasing amounts of AmB, Erg increasingly occupied a position outside the lipid bilayer membrane.
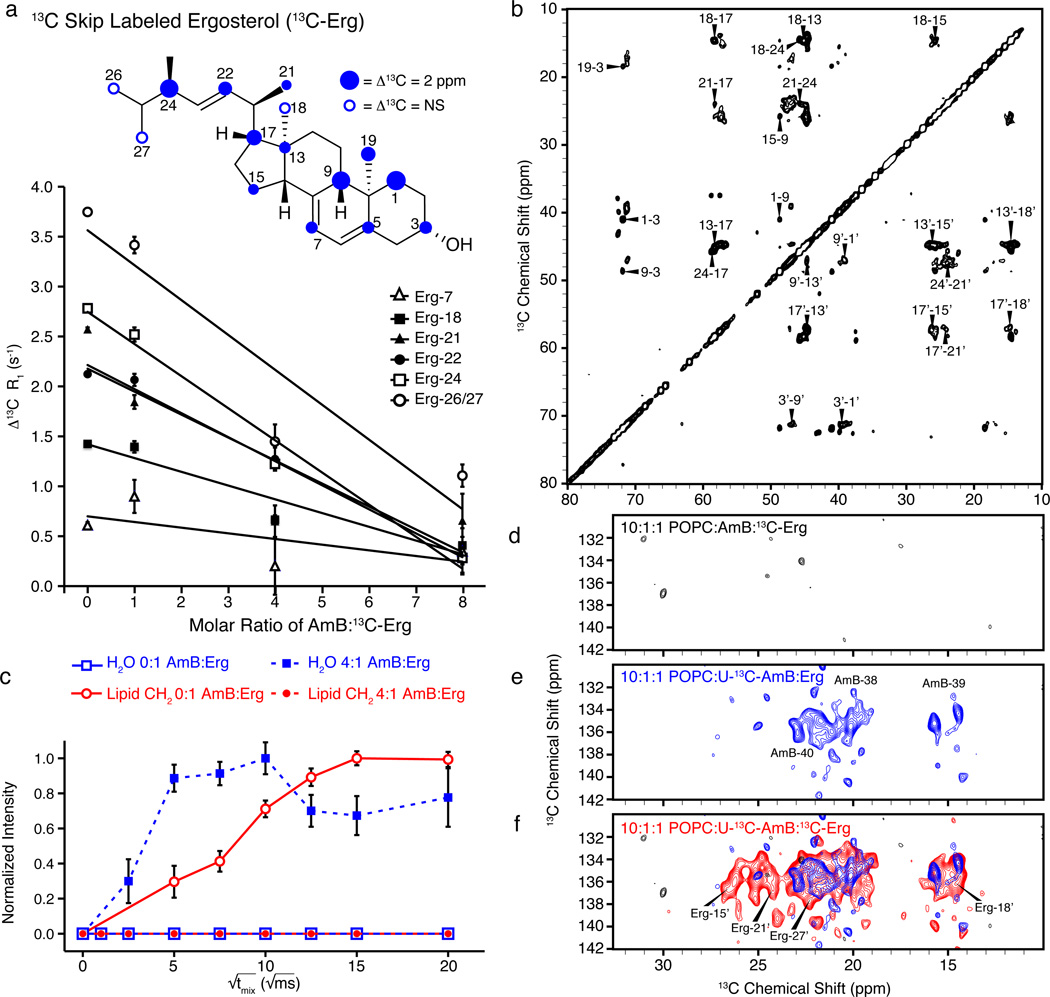
Figure 4. AmB extracts Erg from lipid bilayers.
a, Samples prepared using 40:1 POPC:13C-skip-labeled Erg (13C-Erg) ± 5 mol% 16-DOXYL-PC displayed progressive decrease in PRE effects of resolved Erg resonances as the ratio of AmB:13C-Erg increased. b, The 2D 13C-13C DARR spectrum of 13C-Erg (250 ms mixing, 10:1:1 POPC:AmB:13C-Erg) changed upon addition of AmB, exhibiting new cross peaks. c, The 1H-13C polarization transfers from water (blue) and lipid (red) to Erg-7 were substantially different in the absence (closed circles/squares, dashed line) and presence (open circles/squares, solid line) of AmB. d–f, Expansions of the olefin-to-methyl spectral region for 2D (1H)-13C-(1H-1H)-13C spectra42,43 of (d) only 13C-Erg (328 h signal averaging time), (e) only U-13C-AmB (187 hr), and (f) both 13C-Erg and U-13C-AmB (187 hr). Error bars in (a) were derived from chi-squared analysis of inversion recovery trajectories. The 1H-13C spin diffusion data in (c) were normalized relative to maximum intensity observed for both lipid and water cross peaks for a given 13C site after correcting for 1H T1 relaxation, and error bars were derived from signal-to-noise of the observed cross peak. Spectra were acquired at 14.1 T (600 MHz 1H frequency). Panels (a, b and c) were acquired at 10 kHz MAS, at 20 °C and 10° C, respectively. The (1H)-13C-(1H-1H)-13C spectra (d-f) were acquired at 10 °C, 11.628 kHz MAS, processed with 40 and 75 Hz linebroadening in the direct and indirect dimensions, respectively, and were drawn with contour threshold set to four times the root mean square noise.
Additional SSNMR experiments also supported this conclusion and further demonstrated that the extracted Erg is physically bound to the extramembranous aggregates of AmB. As the ratio of AmB:13C-Erg increased, Erg resonances, but not those of POPC, demonstrated inhomogeneous broadening,19 consistent with a transition from a mobile state to an immobile state (Supplementary Fig. 8). The average 13C T1 relaxation values for 13C-Erg also followed the expected trend, increasing with the AmB:13C-Erg ratio (Supplementary Fig. 7b). 2D 13C-13C correlation spectra further revealed several 13C-Erg resonances that shifted significantly upon the addition of AmB (Fig. 4b, and Supplementary Table 3), and resolved bound state resonances exhibited significantly higher linewidth and T1 values than those of the corresponding unbound state (Supplementary Fig. 9). In the absence of AmB, we observed very strong lipid-Erg correlations and no water-Erg correlations (Fig. 4c, Supplementary Fig. 10),41 whereas in the presence of AmB we observed strong water correlations to all resolved Erg sites, with polarization transfer rates similar to those observed for AmB (Fig. 4c, Supplementary Fig. 11). We also repeated 1D and 2D chemical shift, linewidth, and T1 analyses of 13C-Erg in the presence of amphoteronolide B (AmdeB), a synthesized derivative of AmB that lacks the mycosamine appendage and does not bind Erg,25,27 and observed no 13C-Erg chemical shift perturbations and only very small changes in linewidths and T1 values (Supplementary Fig. 12).
To definitively probe whether the extracted Erg is bound to the AmB aggregate, we prepared an additional series of samples in which 13C labels were placed on (i) only Erg (Fig. 4d), (ii) only AmB (Fig. 4e), and (iii) both AmB and Erg (Fig. 4f). (1H)-13C-(1H-1H)-13C spectra42,43 for the first two samples showed only the anticipated intramolecular correlations (Fig. 4d, 4e), while the sample containing labels on both AmB and Erg revealed many new intermolecular AmB-Erg cross peaks (Fig. 4f), consistent with Erg aligned parallel to the polyene region of AmB and directly confirming the formation of a small molecule-small molecule complex. We also measured the 1H-13C dipolar couplings for resolved sites in both AmB and Erg using the T-MREV recoupling sequence44 (Online Methods Section II, Supplementary Fig. 13) and Erg (Supplementary. Fig 14) to determine the relative mobility of these sites. In the absence of AmB, Erg was mobile as evidenced by the low order parameters, but in the presence of AmB, the order parameters shifted to the same rigid lattice limit observed for AmB (Supplementary Table 2). Furthermore, we observed line widths of >110 Hz for both AmB and Erg in the sterol sponge (Supplementary Table 2). Thus, AmB extracts Erg from lipid bilayers into large, extramembranous aggregates.
AmB extracts Erg from and thereby kills yeast cells
Finally, we tested the validity of the sterol sponge model in cells. First, we probed whether AmB extracts Erg from the cell membrane of yeast by adapting an ultracentrifugation-based membrane isolation assay45 to quantify the amount of Erg in the membranes of live Saccharomyces cerevisiae cells in the absence and presence of AmB (Online Methods Section V). As shown in Fig. 5a, AmB very effectively extracted Erg in a time-dependent fashion. In contrast, we observed no Erg extracting effects with the non-Erg-binding derivative AmdeB.
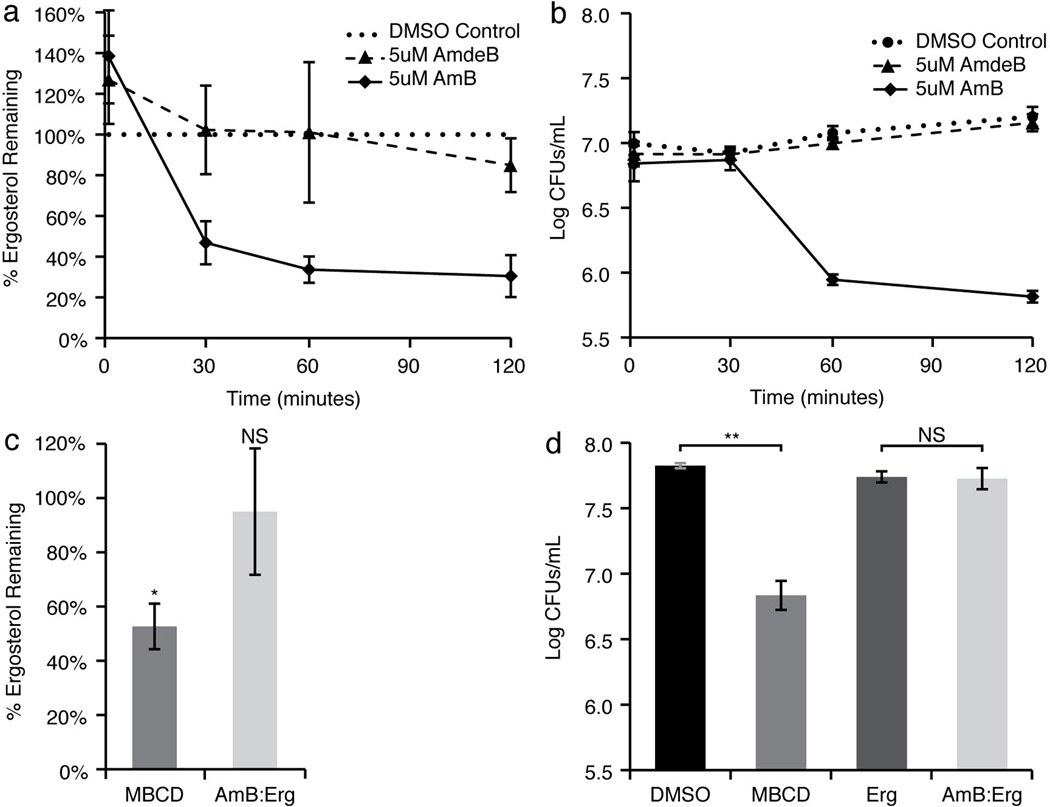
Figure 5. AmB extracts Erg from and thereby kills yeast cells.
a, AmB extracted Erg from the membranes of S. cerevisiae cells in a time-dependent manner, while the non-Erg-binding derivative AmdeB showed no Erg-extracting activity. The percentage of Erg remaining in the cell membranes was normalized to DMSO-only treated controls. b, Cell killing paralleled Erg extraction in AmB-treated cells. The non-Erg-extracting derivative, AmdeB showed no cell killing effects. c, Erg extraction after 120 minutes of incubation. 500 mM MBCD extracted Erg from the membranes of S. cerevisiae cells, whereas a pre-formed 5 µM AmB:25 µM Erg complex did not. The percentage of Erg remaining in the cell membranes was normalized to DMSO-only and 25 µM Erg in DMSO-only treated controls, respectively. d, Cell killing after 120 minutes incubation was observed for yeast treated 500 mM MBCD, but not for yeast treated with 5 µM AmB:25 µM Erg complex. Averages ± s.e.m. for at least 3 independent experiments. * P < 0.02, ** P < 0.002, NS not significant.
Further experiments demonstrated that the Erg-extracting activity of AmB was responsible for its cell killing effects. As shown in Fig. 5b, we observed no cell killing with DMSO or AmdeB, whereas AmB promoted robust cell killing with a time course that paralleled Erg extraction. In addition, methyl-beta-cyclodextrin (MBCD), a cyclic oligosaccharide known to extract sterols from membranes,46 similarly demonstrated both Erg extracting and cell killing activities (Fig. 5c and 5d). Finally, the sterol sponge model predicts that AmB aggregates pre-saturated with Erg will lose the ability to extract Erg from membranes and kill yeast. Enabling this hypothesis to be tested, we found conditions that promoted the formation of stable and soluble aggregates of AmB and Erg (Online Methods Section VI). As predicted, treating cells with this pre-formed AmB/Erg complex resulted in no Erg extraction (Fig. 5c), and no cell killing (Fig. 5d).
DISCUSSION
For decades, scientists have widely accepted that membrane-spanning ion channels primarily contribute to the structure and antifungal activity of AmB (Fig. 1b).4–23 In contrast, we found that AmB primarily forms large extramembranous aggregates that extract Erg from lipid bilayers and thereby kill yeast. Membrane-inserted ion channels are relatively minor contributors, both structurally and functionally, to the antifungal action of this natural product. While previous studies have reported large aggregates of AmB or its derivatives,17,21 the interpretation of these findings has been in terms of the ion channel model. Here we described PRE (Fig. 2b and 2d), 1H spin diffusion trajectory (Fig 2f and 4c, Supplementary Fig. 4, 10, 11), and TEM studies (Fig. 3a-c, Supplementary Fig. 5) that collectively demonstrated that AmB primarily exists in the form of large extramembranous aggregates. Moreover, changes in PREs, 1H spin diffusion trajectories, T1 relaxation, order parameters, line widths, and chemical shift perturbations, as well as the observation of direct intermolecular cross peaks and the results of cell-based ergosterol extraction experiments demonstrated that extramembranous aggregates of AmB directly bind Erg. We further confirmed that the AmB aggregates we observed in our SSNMR, TEM, and cell-based experiments were similar (Supplementary Fig 15). Collectively, these results strongly support the proposed sterol sponge model in which extramembranous aggregates of AmB extract ergosterol from phospholipid bilayers and thereby kill yeast.
The sterol sponge model provides a new foundation for better understanding and more effectively harnessing the unique biophysical, biological, and medicinal properties of this small molecule natural product. Based on the classic ion channel model, many efforts over the past several decades to improve the therapeutic index of AmB focused on selectively permeabilizing yeast versus human cells.11,13 This approach has not yielded a clinically viable derivative of the natural product. The sterol sponge model suggests that an alternative approach will be more effective. Specifically, analogous to the now clarified mechanism of antifungal activity, the extraction of cholesterol by large extramembranous aggregates of AmB may be primarily responsible for toxicity to human cells. This, in turn, suggests that the goal should be to maximize the relative binding affinity of AmB aggregates for Erg versus cholesterol. This insight is already guiding development of the first derivatives of AmB that are toxic to yeast cells but not human cells and thus hold exceptional promise for yielding an improved therapeutic index.47
A high-resolution structure of the large, extramembranous AmB aggregate, with and without bound ergosterol and cholesterol, would powerfully enable the discovery and/or further development of such derivatives. Importantly, the results described herein provide a strong platform for determining such a structure. Specifically, the large extramembranous aggregate of AmB, confirmed to reproducibly and stably form in the presence of POPC bilayers (Supplementary Fig 2, 15), represents an excellent substrate for SSNMR analysis, and the common relaxation properties of AmB and Erg are consistent with the existence of a stable complex. Moreover, the 2D (1H)-13C-(1H-1H)-13C spectra of the complex derived from U-13C-AmB and 13C-Erg (Fig. 4f) exhibited intermolecular AmB-Erg correlations with intensities indicating internuclear distances of ~6 Å or less. We further note that comparison of 13C-13C 2D spectra of 10:1:0 POPC:U-13C-AmB:Erg and 10:1:1 POPC:U-13C-AmB:Erg (Supplementary Fig. 2) showed that the structures of the AmB aggregates in the absence and presence of Erg were very similar. There were, however, some intriguing changes in the AmB resonances corresponding to the mycosamine appendage upon the binding of ergosterol (Supplementary Fig. 3), which will be the subject of future investigations.
We anticipate that further SSNMR studies, including those applied to derivatives of AmB and/or Erg/cholesterol with site-specific or skip-pattern isotopic labels, will enable us to define in high resolution the structure of this extramembranous aggregate and the interface between these small molecules. Such information may reveal the structural underpinnings of the small preference of AmB to bind Erg over cholesterol and further guide the development of derivatives of AmB that maximize this binding preference and thus the therapeutic index.47 In this vein, we note that the pattern of chemical shift perturbations observed for Erg in the absence and presence of AmB are consistent with tight association between AmB and the A and B rings of the sterol. Interestingly, the B ring of cholesterol, to which AmB binds but less strongly than Erg,27,47 is more sterically bulky than that of Erg, because it possesses an extra degree of saturation. Moreover, lanosterol, to which AmB does not bind,27 possesses both the same extra degree of saturation in the B ring and a sterically bulky gem dimethyl group on the A ring. While further studies are required to provide a detailed picture, our current data begin to support a structural rationale for the differential binding of AmB to Erg (strong), cholesterol (weak), and lanosterol (no binding). More broadly, relative to small molecules that bind proteins, small molecules that bind other small molecules in a biologically relevant fashion are very rare. A high-resolution structure of this prototypical AmB-Erg complex may enable rational pursuit and study of other biologically important small molecule-small molecule interactions.
The sterol sponge model also offers a new rationale for the paucity of clinically relevant microbial resistance that is a hallmark of AmB as a therapeutic. Because the extraction of Erg renders yeast membranes Erg-deficient, AmB may simultaneously perturb all cellular processes that depend on membrane Erg.28–31,48 This likely includes many different membrane proteins that directly bind Erg,28–31 and simultaneous mutation of all such proteins in a manner that alleviates this Erg dependence is highly improbable. It has also remained unclear why, in contrast to the rarity with which AmB resistant mutants are found in patients, it is relatively easy to generate AmB-resistant yeast mutants in cell culture experiments.49 The sterol sponge model provides a rationale for this dichotomy. AmB-resistant mutants generated in cell culture generally possess modified sterols in their membranes, e.g., lanosterol50 (and/or other biosynthetic precursors to Erg) to which AmB does not bind (see above).27 It was previously assumed that such changes in sterol content minimize antifungal potency by minimizing membrane-permeabilizing activity.9,10,13,49 The sterol sponge model alternatively suggests that, because AmB does not bind or extract lanosterol, this modified sterol remains in the membrane to serve as a surrogate binding partner for sterol-dependent proteins. Due to the structural differences between lanosterol and Erg described above, however, the former is likely only a minimally effective substitute, resulting in reduced activity of many proteins that require specific interactions with Erg to function properly. This, in turn, may translate into substantially reduced pathogenicity of the resulting yeast mutants. Consistent with this notion, strains of yeast with modified sterol content have markedly reduced pathogenicity in animal models.49 Such strains may routinely emerge in patients treated with AmB, but, due to their reduced pathogenicity, cannot thrive and/or are rapidly cleared by the immune system of the host. A recently reported alternative series of studies provide complementary support for these conclusions.49
The clarified picture of the structural and functional underpinnings of AmB activity provided by the sterol sponge model also illuminates a rational roadmap for separating the ion channel forming and cytotoxic activities of AmB. Small, membrane-spanning ion channel aggregates likely exist as minor components in equilibrium with the large extramembranous assemblies of AmB characterized herein. This proposal is consistent with the weak AmB-lipid correlations observed in the SSNMR spin diffusion experiments, and the limits of detection of the SSNMR PRE and TEM studies. As we have previously demonstrated, binding ergosterol in the absence of channel activity is sufficient for cell killing.25 Specifically, the capacity for channel formation can be selectively eliminated while preserving the capacity for both Erg binding and cell killing via deleting the C35 hydroxyl group appended to AmB.25 In the sterol sponge model, this result can be rationalized by invoking a selectively destabilizing influence of this functional group deletion on the smaller membrane-inserted channel aggregates. Future studies will aim to determine whether this putative equilibrium between large extramembranous and small membrane-spanning aggregates can be alternatively shifted to favor ion channel formation, thereby maximizing potentially useful membrane-permeabilizing functions25 while minimizing cytotoxic sterol extracting activity.
In summary, for more than half a century, the classic ion channel model has dominated the conceptual framework through which scientists have perceived and studied the structure and function of AmB in lipid bilayers. In contrast to this classic model, AmB primarily exists in the form of large, extramembranous aggregates that physically extract Erg from lipid bilayers and thereby kill yeast. This new sterol sponge model stands to more effectively guide the understanding, optimization, and clinical utilization of this prototypical small molecule natural product, as well as other small molecules that similarly interface with living systems.
Online Methods
I. General Methods
Materials
Commercially available materials were purchased from Sigma-Aldrich, Alfa Aesar, Avanti Polar Lipids, Cambridge Isotope Laboratories, or Fisher Scientific and were used without further purification unless stated otherwise. Natural abundance amphotericin (AmB) was purchased from Sigma-Aldrich or a gift from Bristol-Myers Squibb Company. Unless stated otherwise, all solvents were dispensed from a solvent purification system that passes solvents through packed columns according to the method of Pangborn and coworkers52 (THF, Et2O, CH2Cl2, toluene, dioxane, hexanes: dry neutral alumina; DMSO, DMF, CH3OH: activated molecular sieves). Water was dispensed from a MilliQ water purification system (Millipore Corporation, Billerica, MA).
Purification and Analysis
Preparative scale HPLC purification was performed using an Agilent 1260 series instrument equipped with a multiple-wavelength detector and a Waters SunFire Prep C18 OBD 5 µm 30×150 mm column at a flow rate of 25 mL/min. All HPLC solvents were filtered through 0.2 µm Millipore filters prior to use. UV/Vis analyses were performed on a Shimadzu PharmaSpec UV-1700 spectrophotometer. Electrospray ionization mass spectra (ESI-MS) were obtained at the University of Illinois mass spectrometry facility.
Amphotericin and Amphoteronolide B
Due to light and air sensitivity of polyenes, all manipulations of AmB and amphoteronolide B (AmdeB) were carried out under low-light conditions and compounds were stored under a dry argon atmosphere at −20 °C. AmdeB was prepared synthetically from natural abundance AmB as previously described.25–27 All AmB and AmdeB used for current experiments were purified by preparative scale HPLC. All manipulations of HPLC-purified AmB and AmdeB were performed using either Optima MeOH, 0.2 µm-filtered HPLC grade solvents, or solvents dispensed from a solvent purification system.52
For purification, solid AmB was dissolved in DMSO (10 mg/mL), filtered through Celite 545 and purified (100 µL injections) with gradient of 5% to 65% MeCN / 5 mM ammonium acetate (NH4OAc) over 12 minutes with detection at 406 nm. The column was subsequently flushed with isocratic 95% MeCN / 5 mM NH4OAc for 2 min and re-equilibrated to 5% MeCN / 5 mM NH4OAc prior to the next injection. The combined AmB solution was concentrated in vacuo, with filtered (0.2 µm) MeCN added back to the flask as needed for azeotropic removal of water. The resulting yellow solid was suspended via bath sonication in 1:1 MeCN:toluene and again concentrated in vacuo for azeotropic removal of residual NH4OAc. Residual solvent was removed under high vacuum for ≥ 8 h to furnish a pale yellow solid, which was stored under argon at −20 °C.
AmdeB was dissolved in DMF, filtered (Celite 545), injected, and eluted with a mobile phase gradient of 5% to 95% MeCN / 5 mM NH4OAc over 25 min.
Biosynthesis of U-13C-AmB
U-13C-AmB was prepared using a modified version of the method previously reported,18 with U-13C-glucose replacing natural abundance fructose in the culture medium. All simple carbon sources were thus uniformly 13C-labeled, resulting in unprecedented isotopic enrichment of >80%, as measured by mass spectrometry. After work up and precipitation, U-13C-AmB was purified by gradient C18 chromatography followed by HPLC. (Supplementary Note)
Ergosterol
Natural abundance ergosterol (Erg) was purchased from Sigma-Aldrich and recrystallized from EtOH before use. Stock solutions of 4 mg/mL Erg in CHCl3 were stored under argon at −20 °C for up to one month. 13C-skip-labeled Erg (13C-Erg) was prepared biosynthetically using the method previously described.19,51
II. Solid-state NMR spectroscopy
SSNMR experiments were performed using a 600 MHz InfinityPlus spectrometer (Varian, now a subsidiary of Agilent Technologies, Inc.) equipped with a 3.2 mm T3 HXY MAS probe tuned to 1H-31P-13C mode. Pulse widths (π/2) for 1H, 13C, and 31P were 2–2.5 µs, 3.2 µs, and 3.2 µs, respectively. Spinning was controlled with a Varian MAS controller to 10,000 ± 2 Hz. SPINAL-64 decoupling (~75 to 80 kHz) was used during evolution and acquisition periods.53 The flow rate of sample cooling gas was maintained at 100 scfh at 20 °C, resulting in a calibrated sample temperature of 19.2±1 °C. Chemical shifts were referenced externally with adamantane, with the downfield 13C resonance referenced to 40.48 ppm.54
13C T1 and PRE Experiments
T1 values were measured using standard T1 inversion recovery pulse sequence with a 5 second pulse delay. Data were processed and fit with Varian Spinsight software version 4.3.2. For each of the resolved methine and methylene in U-13C-labeled amphotericin (U-13C-AmB) and 13C skip labeled ergosterol (13C-Erg) the longitudinal 13C PRE was obtained by calculating the difference between the 13C R1 values for sample with and without 5 mol% of the DOXYL lipids, determined by modeling the individual relaxation trajectories as single exponential decays. T1 trajectories were fit using the integrated volume of a given peak as a function of delay time (tau_1); integration boundaries were set to the linewidth at half height. The average line widths were ~40–60 Hz for POPC, ~50 Hz for Erg with no AmB present, 127 Hz with AmB present (Supplementary Table 3), and ~187 Hz for AmB alone.
1H-13C Spin-Diffusion Experiments
We performed 1H-13C spin-diffusion correlation experiments as previously described41{Huster, 2002 #330} using a 1 ms T2 filter, to detect interactions between the mobile 1H signals of lipid acyl chains (1.35 ppm) and/or water (4.7 ppm) with the U-13C-AmB, and 13C-Erg in the presence and absence of AmB. 1H-13C polarization transfer trajectories were extracted from 1H-13C 2D spectra collected with 1H-13C mixing times ranging from 1 ms to 625 ms, by fitting peaks with a minimum signal to noise of 5, using a box integration method in Sparky. Trajectories were then normalized based on maximum observed intensity for a single resolved water or lipid 1H-13C cross peak after correction for 1H T1 relaxation, which was measured in a separate T1 inversion recovery experiment. Error bars are derived from the signal-to-noise ratios observed for each crosspeak.
Order Parameters from 1H-13C Dipolar Couplings
Dipolar order parameters (S) were measured using the T-MREV pulse sequence44 at an MAS rate of 8.333 kHz (N=4 condition, 100 kHz 1H decoupling nutation frequency, 2.5 µs 1H π/2 pulse length). The T-MREV 13C-1H dephasing was incremented by 30 µs and a total of 25 increments were recorded in t1. Fortran fitting routines55 were used to determine the 13C-1H dipolar coupling, taking into account the effects of relaxation and contributions from weaker couplings from neighboring protons. We calibrated the scaling factor of the T-MREV sequence by measuring the 13C-1H dipolar coupling for crystalline N-acetyl-L-valine under the identical experimental conditions.
(1H)-13C-(1H-1H)-13C Correlation Spectra
(1H)-13C-(1H-1H)-13C SSNMR experiments to yield performed at 10 °C, at an MAS rate 11.628 kHz, with the heteronuclear contact time (tHC) set to 400 µs, and 1H-1H mixing time of 400 µs. These conditions reveal cross peaks for internuclear 13C-13C distances of ~4–6 Å. In order to properly identify new intermolecular AmB-Erg cross peaks the (1H)-13C-(1H-1H)-13C spectra were acquired back-to-back under identical conditions, including and signal averaging, adjusting the total measurement time based on the amount of material. The rotors of POPC:U-13C-AmB:Erg (10:1:1 molar ratio) and POPC:U-13C -AmB:13C-Erg (10:1:1 molar ratio) were packed with ~25 mg and the spectra signal averaged for 7.8 days each. The 10:1:1 POPC:AmB:13C-Erg sample was ~16 mg and therefore signal averaged for 13.6 days. The three spectra were all processed identically, with 40 and 75 Hz 13C line broadening applied in the direct and indirect dimensions, respectively.
III. Preparation of samples for SSNMR
Preparation of stock solutions
A fresh stock solution of HPLC-purified AmB (natural abundance or U-13C-AmB) was prepared for each experiment by dissolving AmB in a large volume of Optima methanol, typically 75–100 mL for 10 mg of AmB. Stock solution concentration was measured in triplicate by dilution in MeOH and measuring absorbance at 406 nm (ε406 = 146000 M−1 cm−1).26
Stock solutions of Erg were prepared by dissolving recrystallized (commercial) or HPLC-purified (biosynthetic) Erg in a minimum volume of CHCl3 and the concentration determined by UV/Vis spectroscopy (ε282 = 10,400 M−1 cm−1).27 Erg stock solutions were stored in I-Chem vials under a dry argon atmosphere at −20 °C for up to 1 month.
Phospholipids were purchased as stock solutions in CHCl3 and these solutions were used directly for liposome preparation. Unused phospholipid solutions were stored in vials/bottles under a dry argon atmosphere at −20 °C, and discarded after 1 month.
Preparation of liposome vesicles for SSNMR
Liposomes were prepared using a modified version of the protocol previously reported.18 A suspension of POPC/Erg/AmB in 1:1 CHCl3/MeOH was prepared as follows: The desired amount of AmB stock solution (typically 30–40 mL) was concentrated in vacuo to 2–3 mL and transferred to a 7 mL Wheaton vial, with three Optima MeOH washes to ensure complete transfer. This resulting AmB suspension was concentrated in vacuo. The desired amounts of stock solutions of phospholipid and Erg were then added via Hamilton gastight syringe, and an equivalent volume of Optima MeOH was added to resuspend the AmB. The vial was capped and this suspension was briefly vortexed and bath-sonicated until no AmB remained adherent to the sides of the vial (2–3 cycles). Solvent was removed under a gentle stream of nitrogen gas. Residual solvent was removed under high vacuum for ≥8 h.
To the dried solid was added filter-sterilized 0.3 mM HEPES buffer, pH 7.0 to yield a final phospholipid concentration of 40 mM. This aqueous suspension was vortexed and sonicated 3 times or until a homogeneous suspension was observed. Samples were then submitted to 5 freeze/thaw cycles (liquid nitrogen, lukewarm tap water). Samples were again frozen in liquid nitrogen and lyophilized for ≥8 h. The lyophilization chamber was then back-filled with dry Ar to prevent samples from absorbing ambient water. Samples were immediately capped and packed into rotors for SSNMR as soon as possible.
Dry samples were packed in 3.2 mm diameter limited speed SSNMR rotors (Agilent Technologies, Inc.) and hydrated with 8–10 µL of MilliQ H2O. Rubber discs were used in the rotors to maintain hydration levels by creating a seal. Samples were placed at 4 °C for at least 24 hours to allow water to equilibrate.
IV. Electron Microscopy
General Information
LUVs were prepared by the method reported previously,25,27 and AmB was added to the LUV suspension as a freshly-prepared DMSO stock solution. Microscopy was performed using a 120-keV FEI Spirit Transmission Electron Microscope. Images were recorded using a bottom mount TVIPS CMOS based camera system at nominal magnifications of 23,000–49,000x at the specimen level. Measurements were taken in ImageJ32 (v 1.47).
Sample Preparation
AmB was prepared as a stock DMSO solution (8.82 mM). 5 µL of the stock AmB solution was added to 95 µL of the 50x-diluted LUV solutions. For AmB-free samples, 5 µL of DMSO was added to 95 µL of the 50x-diluted LUV solutions. Samples were vortexed gently for 5 seconds then incubated at 37 °C for 1 hour. EM samples were prepared as previously described56 with the following modifications. A 4 µL drop of the sample was applied to a negatively charged carbon-coated copper grid (Gilder 200 mesh, Ted Pella, Inc., Redding CA) for 30 seconds. Subsequently, two drops of freshly prepared 2% uranyl acetate were added to the sample and incubated for 1 minute before drying via aspiration. Samples were then screened on the electron microscope.
In vivo sterol extraction and membrane isolation
Growth Conditions for S. cerevisiae
S. cerevisiae was grown in autoclave-sterilized yeast peptone dextrose (YPD) media consisting of 10 g/L yeast extract, 20 g/L peptone and 20 g/L of filter-sterilized dextrose added as a sterile 40% w/v solution in water. Solid media was prepared by pouring sterile media containing agar (20 g/L) onto Corning (Corning, NY) 100×20 mm polystyrene plates. Liquid cultures were incubated at 30 °C on a rotary shaker and solid cultures were maintained at 30 °C in an incubator.
Sample Preparation
750 mL overnight cultures of S. cerevisiae were grown to stationary phase (OD600 of ~1.7 as measured with a Shimadzu PharmaSpec UV-1700 UV/Vis spectrophotometer). This culture was divided equally into 50 mL Falcon centrifuge tubes.
Stock solutions of AmdeB, AmB, and Erg were prepared in DMSO. Methyl-beta-cyclodextrin (MBCD) was added directly to the liquid culture. Cells were treated with either a DMSO only control, 5 µM AmdeB, or 5 µM AmB for 1, 30, 60, or 120 minutes. Cells were treated with DMSO control, 500 mM MBCD, 25 µM Erg control, and the 5 µM AmB:25 µM Erg complex (Section VII) for 120 minutes. Treated tubes were incubated on the rotary shaker (200 rpm) at 30°C for the time of exposure.
For the quantification of colony forming units (CFUs), at the end of exposure, aliquots were taken from the samples, diluted, and plated on YPD agar plates. The plates were then incubated for 48 hours at 30°C and colony-forming units were counted.
For the quantification of percent ergosterol remaining, yeast membranes were isolated using a modified version of Haas’ spheroplasting and isosmotic cell lysis protocol and simple differential ultracentrifugation.45 At the end of the exposure time, tubes were removed from the shaker and centrifuged for 5 minutes at 3000 µg at room temperature. The supernatant was decanted and 5 mL of wash buffer (dH2O, 1M DTT, 1M Tris-HCl, pH 9.4) was added. The tubes were vortexed to resuspend and incubated in a 30°C water bath for 10 minutes. Tubes were then centrifuged again for 5 minutes at 3000 µg and the supernatant decanted.
1 mL of spheroplasting buffer (1M KPi, YPD media, 4M Sorbitol) and 100 µL of a 5 mg/mL solution of lyticase from Arthrobacter luteus (L2524 Sigma-Aldrich) was added to each tube, and each tube was then vortexed to resuspend. Tubes were incubated in a 30°C water bath for 30 minutes, with occasional swirling. After incubation, tubes were centrifuged for 10 minutes at 1080 µg at 4 °C and the supernatant decanted.
1 mL of PBS buffer and 20 µL of a 0.4 mg/ml dextran in 8% Ficoll solution was added to each tube, mixed very gently to resuspend. This suspension was placed on ice for 4 minutes and then heat-shocked in a 30°C water bath for 3 minutes.
The suspensions were then transferred to Eppendorf tubes, vortexed to ensure complete lysis, and centrifuged at 15000 µg at 4 °C for 15 minutes to remove un-lysed cells and cell debris. The resulting supernatants were transferred to thick-wall polycarbonate ultracentrifuge tubes (3.5 mL, 13×51 mm, 349622 Beckman Coulter) and spun for 1 hour at 100,000 µg at 4°C in a Beckman Coulter TLA-100.3 fixed-angle rotor in a Beckman TL-100 Ultracentrifuge. The supernatant was poured off. The remaining membrane pellet was resuspended in 1 mL PBS buffer and stored at −80 °C until further analysis.
Gas chromatography quantification of sterols
750 µL of each membrane pellet sample and 20 µL of internal standard (4 mg/mL cholesterol in chloroform) were dissolved in 3 mL 2.5% ethanolic KOH in a 7 mL vial, which was then vortexed gently, capped, and heated in a heat block on a hot plate at 90°C for 1 hour. The vials were then removed from the heat source and allowed to cool to room temperature. 1 mL of brine was added to the contents of each vial. Extraction was performed twice, each with 3 mL of hexane. Organic layers were removed in both extractions, dried over magnesium sulfate, filtered through Celite® 545 (Sigma-Aldrich), and transferred to another 7 mL vial. The contents of the vial were then concentrated in vacuo in a 30°C water bath.
The resulting sterol films were resuspended in 100 µL pyridine and 100 µL N,O-Bis-(trimethylsilyl)-trifluoroacetamide with 1% trimethylchlorosilane (T6381-10AMP Sigma-Aldrich) by vortexing gently.57 This solution was heated at 60 °C for 1 hour. The vials were placed on ice and the solvent was evaporated off by nitrogen stream. Vials must be kept at a low temperature to prevent evaporation of the sterol TMS ethers along with the solvent. The resulting films were resuspended in 100 µL of decane, filtered and transferred to a GC vial insert for analysis.
Gas chromatography analysis was carried out on an Agilent 7890A gas chromatograph equipped with a FID, an Agilent GC 7693 Autosampler, and a Dell computer running Microsoft XP that utilizes ChemStation v.B.04.02 SP1. Samples were separated on a 30 m, 0.320 mm ID, 0.25 um film HP-5 capillary column (19091J-413 Agilent) using hydrogen as a carrier gas with an average velocity of 84.8 cm/s. Nitrogen make-up gas, hydrogen and compressed air were used for the FID. A split/splitless injector was used in a 20:1 split. The injector volume was 2 µL. The column temperature was initially held at 250 °C for 0.5 min, then ramped to 265 °C at a rate of 10 °C /min with a final hold time of 12.5 min. The injector and detector temperature were maintained at 270 °C and 290 °C, respectively. The value reported for each time point was calculated by dividing the value for the treatment group by the value for the DMSO control at the same time point, and then normalizing the DMSO control to 100%.
VI. Preparation of an Amphotericin/Ergosterol complex
Erg was prepared as a stock solution, 4 mg/mL in CHCl3, and the solvent removed under a gentle stream of nitrogen gas. Residual solvent was removed under high vacuum for at least 8 h. A DMSO solution of 5 µM AmB was then added to this solid Erg (25 µM final Erg concentration, 5:1 mole ratio Erg:AmB). The resulting suspension was gently vortexed and then heated to 80 °C for one hour in an aluminum heating block to allow Erg to fully dissolve. The resulting AmB/Erg solution was then allowed to cool to room temperature. This solution was left to complex at room temperature for another hour before use.
The absorbance spectra of the two types of aggregate, (1) 5 µM AmB only in PBS buffer, (2) 5 µM AmB:25 µM Erg complex in PBS buffer, and the monomeric form of AmB (AmB in 25% PBS buffer, 75% methanol) were investigated using a Shimadzu PharmaSpec UV-1700 UV/Vis spectrophotometer.58 Supplementary Fig. 15 shows the distinct shift in UV spectra between the different forms of AmB and AmB bound to Erg in a complex.
Supplementary Material
Acknowledgements
Paul J. Hergenrother and Eric Oldfield are gratefully acknowledged for helpful discussions, and Dr. Jakob J. Lopez is thanked for preliminary spin diffusion SSNMR experiments. Portions of this work were supported by the NIH (R01GM080436, F30DK081272), the University of Illinois at Urbana-Champaign (Centennial Scholar Award to C.M.R.). M.D.B. is an HHMI Early Career Scientist. M.C.C. is an American Heart Association Predoctoral Fellow. T.M.A. is a Ruth L. Kirchstein NIH NRSA Predoctoral Fellow. The Gonen lab is funded by the Howard Hughes Medical Institute.
Footnotes
Supplementary Information is available in the online version of the paper.
Author Contributions. T.M.A., M.C.C., A.G.C., K.A.D., A.J.N., G.C., T.G., C.M.R., and M.D.B. designed research. T.M.A., N.M., and A.G.C. prepared U-13C-AmB and 13C-Erg. T.M.A., M.C.C., A.G.C., G.S.H., A.J.N., G.C., and B.E.U. prepared samples for SSNMR. M.C.C., A.J.N., G.C., G.S.H., M.D.T., and C.M.R. acquired SSNMR data. A.G.C. and T.G. performed microscopy. K.A.D. performed cell-based assays. T.M.A., M.C.C., A.G.C., K.A.D., G.S.H., M.D.T., A.J.N., G.C., S.W., B.E.U., E.L.W., T.G., C.M.R., and M.D.B. analyzed data. T.M.A., M.C.C., A.G.C., K.A.D., C.M.R., and M.D.B. wrote the paper.
C.M.R. and M.D.B. declare no competing financial interests.
REFERENCES
- 1.Cannon RD, et al. Candida albicans drug resistance - another way to cope with stress. Microbiology. 2007;153:3211–3217. doi: 10.1099/mic.0.2007/010405-0. [DOI] [PubMed] [Google Scholar]
- 2.Mora-Duarte J, et al. Comparison of caspofungin and amphotericin B for invasive candidiasis. New. Engl. J. Med. 2002;347:2020–2029. doi: 10.1056/NEJMoa021585. [DOI] [PubMed] [Google Scholar]
- 3.Monk BC, Goffeau A. Outwitting multidrug resistance to antifungals. Science. 2008;321:367–369. doi: 10.1126/science.1159746. [DOI] [PubMed] [Google Scholar]
- 4.Ermishkin LN, Kasumov KM, Potzeluyev VM. Single ionic channels induced in lipid bilayers by polyene antibiotics amphotericin-B and nystatine. Nature. 1976;262:698–699. doi: 10.1038/262698a0. [DOI] [PubMed] [Google Scholar]
- 5.Andreoli TE, Monahan M. The interaction of polyene antibiotics with thin lipid membranes. J. Gen. Physiol. 1968;52:300–325. doi: 10.1085/jgp.52.2.300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cass A, Finkelstein A, Krespi V. The ion permeability induced in thin lipid membranes by the polyene antibiotics nystatin and amphoterici B. J. Gen. Physiol. 1970;56:100–124. doi: 10.1085/jgp.56.1.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Finkelstein A, Holz R. Aqueous pores created in thin lipid membranes by the polyene antibiotics nystatin and amphotericin B. Membranes. 1973;2:377–408. [PubMed] [Google Scholar]
- 8.Andreoli TE. Structure and function of amphotericin B-cholesterol pores in lipid bilayer membranes. Ann N.Y. Acad. Sci. 1974;235:448–468. doi: 10.1111/j.1749-6632.1974.tb43283.x. [DOI] [PubMed] [Google Scholar]
- 9.de Kruijf B, Demel RA. Polyene antibiotic-sterol interactions in membranes of acholeplasma-laidlawii cells and lecithin liposomes. 3. molecular-structure of polyene sntibiotic-cholesterol complexes. Biochim. Biophys. Acta. 1974;339:57–70. doi: 10.1016/0005-2736(74)90332-0. [DOI] [PubMed] [Google Scholar]
- 10.Bolard J. How do the polyene macrolide antibiotics affect the cellular membraneproperties. Biochim. Biophys. Acta. 1986;864:257–304. doi: 10.1016/0304-4157(86)90002-x. [DOI] [PubMed] [Google Scholar]
- 11.Volmer AA, Szpilman AM, Carreira EM. Synthesis and biological evaluation of amphotericin B derivatives. Nat. Prod. Rep. 2010;27:1329–1349. doi: 10.1039/b820743g. [DOI] [PubMed] [Google Scholar]
- 12.Cereghetti DM, Carreira EM. Amphotericin B: 50 years of chemistry and biochemistry. Synthesis-Stuttgart. 2006:914–942. [Google Scholar]
- 13.Cheron M, et al. Quantitative structure-activity relationships in amphotericin B derivatives. Biochem. Pharmacol. 1988;37:827–836. doi: 10.1016/0006-2952(88)90168-2. [DOI] [PubMed] [Google Scholar]
- 14.Croatt MP, Carreira EM. Probing the role of the mycosamine C2'-OH on the activity of amphotericin B. Org. Lett. 2011;13:1390–1393. doi: 10.1021/ol2000765. [DOI] [PubMed] [Google Scholar]
- 15.Baginski M, Resat H, Borowski E. Comparative molecular dynamics simulations of amphotericin B-cholesterol/ergosterol membrane channels. Biochim. Biophys. Acta. 2002;1567:63–78. doi: 10.1016/s0005-2736(02)00581-3. [DOI] [PubMed] [Google Scholar]
- 16.Murata M, et al. Ion channel complex of antibiotics as viewed by NMR. Pure Appl. Chem. 2009;81:1123–1129. [Google Scholar]
- 17.Matsumori N, Sawada Y, Murata M. Large molecular assembly of amphotericin B formed in ergosterol-containing membrane evidenced by solidstate NMR of intramolecular bridged derivative. J. Am. Chem. Soc. 2006;128:11977–11984. doi: 10.1021/ja063433w. [DOI] [PubMed] [Google Scholar]
- 18.Matsuoka S, et al. Membrane interaction of amphotericin B as single-length assembly examined by solid state NMR for uniformly 13C-enriched agent. Bioorg. Med. Chem. 2006;14:6608–6614. doi: 10.1016/j.bmc.2006.06.001. [DOI] [PubMed] [Google Scholar]
- 19.Umegawa Y, et al. Head-to-tail interaction between amphotericin B and ergosterol occurs in hydrated phospholipid membrane. Biochemistry. 2012;51:83–89. doi: 10.1021/bi2012542. [DOI] [PubMed] [Google Scholar]
- 20.Verkleij AJ, et al. Freeze-Etch Electron-Microscopy of erythrocytes, acholeplasma-laidlawii cells and liposomal membranes after action of filipin and amphotericin-B. Biochim. Biophys. Acta. 1973;291:577–581. doi: 10.1016/0005-2736(73)90509-9. [DOI] [PubMed] [Google Scholar]
- 21.Milhaud J, Ponsinet V, Takashi M, Michels B. Interactions of the drug amphotericin B with phospholipid membranes containing or not ergosterol: New insight, into them role of ergosterol. Biochim. Biophys. Acta. 2002;1558:95–108. doi: 10.1016/s0005-2736(01)00416-3. [DOI] [PubMed] [Google Scholar]
- 22.Mouri R, et al. Complex formation of amphotericin B in sterol-containing membranes as evidenced by surface plasmon resonance. Biochemistry. 2008;47:7807–7815. doi: 10.1021/bi800334p. [DOI] [PubMed] [Google Scholar]
- 23.Katzung BG, Masters SB, Trevor AJ. Basic and Clinical Pharmacology. 12th edn. Europe: McGraw-Hill Education; 2012. [Google Scholar]
- 24.Fernandez-Lopez S, et al. Antibacterial agents based on the cyclic D,L-alpha-peptide architecture. Nature. 2001;412:452–455. doi: 10.1038/35086601. [DOI] [PubMed] [Google Scholar]
- 25.Gray KC, et al. Amphotericin primarily kills yeast by simply binding ergosterol. Proc. Natl. Acad. Sci. USA. 2012;109:2234–2239. doi: 10.1073/pnas.1117280109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Palacios DS, Anderson TM, Burke MD. A post-PKS oxidation of the amphotericin B skeleton predicted to be critical for channel formation is not required for potent antifungal activity. J. Am. Chem. Soc. 2007;129:13804–13805. doi: 10.1021/ja075739o. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Palacios DS, et al. Synthesis-enabled functional group deletions reveal key underpinnings of amphotericin B ion channel and antifungal activities. Proc. Natl. Acad. Sci. USA. 2011;108:6733–6738. doi: 10.1073/pnas.1015023108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jin H, McCaffery JM, Grote E. Ergosterol promotes pheromone signaling and plasma membrane fusion in mating yeast. J. Cell Biol. 2008;180:813–826. doi: 10.1083/jcb.200705076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kato M, Wickner W. Ergosterol is required for the Sec18/ATP-dependent priming step of homotypic vacuole fusion. EMBO J. 2001;20:4035–4040. doi: 10.1093/emboj/20.15.4035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Heese-Peck A, et al. Multiple functions of sterols in yeast endocytosis. Mol. Biol. Cell. 2002;13:2664–2680. doi: 10.1091/mbc.E02-04-0186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.te Welscher YM, et al. Polyene antibiotic that inhibits membrane transport proteins. Proc. Natl. Acad. Sci. USA. 2012;109:11156–11159. doi: 10.1073/pnas.1203375109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Solomon I. Relaxation processes in a system of 2 spins. Phys. Rev. 1955;99:559–565. [Google Scholar]
- 33.Nadaud PS, Helmus JJ, Hofer N, Jaroniec CP. Long-range structural restraints in spin-labeled proteins probed by solid-state nuclear magnetic resonance spectroscopy. J. Am. Chem. Soc. 2007;129:7502–7503. doi: 10.1021/ja072349t. [DOI] [PubMed] [Google Scholar]
- 34.Sankaram MB, Thompson TE. Cholesterol-induced fluid-phase immiscibility in membranes. Proc. Natl. Acad. Sci. USA. 1991;88:8686–8690. doi: 10.1073/pnas.88.19.8686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hsueh YW, et al. Ergosterol in POPC membranes: Physical properties and comparison with structurally similar sterols. Biophys. J. 2007;92:1606–1615. doi: 10.1529/biophysj.106.097345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Watson K, Bertoli E, Griffiths DE. Phase-transitions in yeast mitochondrial-membranes - effect of temperature on energies of activation of respiratory enzymes ofsaccharomyces-cerevisiae . Biochem. J. 1975;146:401–407. doi: 10.1042/bj1460401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ejsing CS, et al. Global analysis of the yeast lipidome by quantitative shotgun mass spectrometry. Proc. Natl. Acad. Sci. USA. 2009;106:2136–2141. doi: 10.1073/pnas.0811700106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chen LL, et al. Constant-time through-bond 13C correlation spectroscopy for assigning protein resonances with solid-state NMR spectroscopy. J. Am. Chem. Soc. 2006;128:9992–9993. doi: 10.1021/ja062347t. [DOI] [PubMed] [Google Scholar]
- 39.Takegoshi K, Nakamura S, Terao T. 13C-1H dipolar-assisted rotational resonance in magic-angle spinning NMR. Chem. Phys. Lett. 2001;344:631–637. [Google Scholar]
- 40.Hohwy M, Rienstra CM, Jaroniec CP, Griffin RG. Fivefold symmetric homonuclear dipolar recoupling in rotating solids: Application to double quantum spectroscopy. J. Chem. Phys. 1999;110:7983–7992. [Google Scholar]
- 41.Huster D, Yao XL, Hong M. Membrane protein topology probed by 1H spin diffusion from lipids using solid-state NMR spectroscopy. J. Am. Chem. Soc. 2002;124:874–883. doi: 10.1021/ja017001r. [DOI] [PubMed] [Google Scholar]
- 42.Lange A, Luca S, Baldus M. Structural constraints from proton-mediated rare-spin correlation spectroscopy in rotating solids. J. Am. Chem. Soc. 2002;124:9704–9705. doi: 10.1021/ja026691b. [DOI] [PubMed] [Google Scholar]
- 43.Tang M, Waring AJ, Hong M. Intermolecular packing and alignment in an ordered beta-hairpin antimicrobial peptide aggregate from 2D solid-state NMR. J. Am. Chem. Soc. 2005;127:13919–13927. doi: 10.1021/ja0526665. [DOI] [PubMed] [Google Scholar]
- 44.Hohwy M, et al. Local structure and relaxation in solid-state NMR: Accurate measurement of amide N-H bond lengths and H-N-H bond angles. J. Am. Chem. Soc. 2000;122:3218–3219. [Google Scholar]
- 45.Haas A. A quantitative assay to measure homotypic vacuole fusion in vitro. Methods Cell Sci. 1995;17:283–294. [Google Scholar]
- 46.Yancey PG, et al. Cellular cholesterol effect mediated by cyclodextrins - Demonstration of kinetic pools and mechanism of efflux. J. Biol. Chem. 1996;271:16026–16034. doi: 10.1074/jbc.271.27.16026. [DOI] [PubMed] [Google Scholar]
- 47.Wilcock BC, Endo MM, Uno BE, Burke MD. C2′-OH of amphotericin B plays an important role in binding the primary sterol of human cells but not yeast cells. J. Am. Chem. Soc. 2013;135:8488–8491. doi: 10.1021/ja403255s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kang CK, et al. Visualization analysis of the vacuole-targeting fungicidal activity of amphotericin B against the parent strain and an ergosterol-less mutant of Saccharomyces cerevisiae. Microbiology. 2013;159:939–947. doi: 10.1099/mic.0.065714-0. [DOI] [PubMed] [Google Scholar]
- 49.Vincent BM, et al. Fitness trade-offs restrict the evolution of resistance to amphotericin B. PLoS Biol. 2013;11:e1001692. doi: 10.1371/journal.pbio.1001692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sanglard D, et al. Candida albicans mutations in the ergosterol biosynthetic pathway and resistance to several antifungal agents. Antimicrob. Agents Chemother. 2003;47:2404–2412. doi: 10.1128/AAC.47.8.2404-2412.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Seo S, et al. Biosynthesis of sitosterol, cycloartenol, and 24-methylenecycloartanol in tissue-cultures of higher-plants and of ergosterol in yeast from [1,2-13C2]-Acetate and [2-13C-2H3]-Acetate and [5-13C-2H2] MVA. J. Chem. Soc.Perkin Trans. 1. 1988:2407–2414. [Google Scholar]
- 52.Pangborn AB, et al. Safe and convenient procedure for solvent purification. Organometallics. 1996;15:1518–1520. [Google Scholar]
- 53.Comellas G, Lopez JJ, Nieuwkoop AJ, Lemkau LR, Rienstra CM. Straightforward, effective calibration of SPINAL-64 decoupling results in the enhancement of sensitivity and resolution of biomolecular solid-state NMR. J. Magn. Reson. 2011;209:131–135. doi: 10.1016/j.jmr.2010.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Morcombe CR, Zilm KW. Chemical shift referencing in MAS solid state NMR. J. Magn. Reson. 2003;162:479–486. doi: 10.1016/s1090-7807(03)00082-x. [DOI] [PubMed] [Google Scholar]
- 55.Rienstra CM, et al. Determination of multiple torsion-angle constraints in U-13C,15N-labeled peptides: 3D 1H-15N-13C-1H dipolar chemical shift NMR spectroscopy in rotating solids. J. Am. Chem. Soc. 2002;124:11908–11922. doi: 10.1021/ja020802p. [DOI] [PubMed] [Google Scholar]
- 56.Ohi M, Li Y, Cheng Y, Walz T. Negative staining and image classification - powerful tools in modern electron microscopy. Biol. Proced. Online. 2004;6:23–34. doi: 10.1251/bpo70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Winkler JK, Rennick KA, Eller FJ, Vaughn SF. Phytosterol and tocopherol components in extracts of corn distiller's dried grain. J. Agric. Food. Chem. 2007;55:6482–6486. doi: 10.1021/jf070594q. [DOI] [PubMed] [Google Scholar]
- 58.Shervani Z, et al. Aggregation of polyene antibiotics as studied by electronic absorption and circular dichroism spectroscopies. Colloid. Surface. B. 1996;7:31–38. [Google Scholar]
- 59.Fung BM, Khitrin AK, Ermolaev K. An improved broadband decoupling sequence for liquid crystals and solids. J. Magn. Reson. 2000;142:97–101. doi: 10.1006/jmre.1999.1896. [DOI] [PubMed] [Google Scholar]
- 60.Shervani Z, Etori H, Taga K, Yoshida T, Okabayashi H. Aggregation of polyene antibiotics as studied by electronic absorption and circular dichroism spectroscopies. Colloid. Surface. B. 1996;7:31–38. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.