Abstract
Thymulin has been shown to present anti-inflammatory and anti-fibrotic properties in experimental lung diseases. We hypothesized that a biologically active thymulin analog gene, methionine serum thymus factor, delivered by highly compacted DNA nanoparticles composed of single molecule of plasmid DNA compacted with block copolymers of poly-l-lysine and polyethylene glycol (CK30PEG) that have been found safe in a human phase I clinical trial, may prevent lung inflammation and remodeling in a mouse model of allergic asthma. Thymulin plasmids were detected in the lungs of ovalbumin-challenged asthmatic mice up to 27 days after administration of DNA nanoparticles carrying thymulin plasmids. A single dose of DNA nanoparticles carrying thymulin plasmids prevented lung inflammation, collagen deposition and smooth muscle hypertrophy in the lungs of a murine model of ovalbumin-challenged allergic asthma, leading to improved lung mechanics. In the present model of chronic allergic asthma, highly compacted DNA nanoparticles using thymulin analog gene modulated the inflammatory and remodeling processes improving lung mechanics.
Keywords: allergy gene therapy, thymulin, DNA nanoparticles, lung remodeling
1. Introduction
Allergic asthma is a global health problem and is caused by unregulated production of cytokines secreted by allergen-specific helper T type 2 cells, which leads to airway inflammation and hyperresponsiveness [1]. Anti-inflammatory medication is the primary therapeutic option for patients with asthma to date [2, 3]. However, accumulated evidence suggests that airway remodeling results in the progressive loss of lung function [4], and that an intervention of asthma-associated structural changes in the lung, including smooth muscle hypertrophy, wall thickening and collagen deposition, would prevent the loss of lung function. The most widely employed asthma therapies, long-acting β2-agonist and/or inhaled corticosteroid, reduce inflammation [5], but no effective therapy has been established to reduce remodeling [6]. Therefore, therapeutic approaches to attenuate both inflammatory and remodeling processes are sorely needed [7, 8].
Thymulin, FTS (serum thymus factor), is a biologically inactive nonapeptide that becomes active when the peptide is coupled with zinc ions in vivo [9]. FTS has been shown to mediate anti-inflammatory and anti-fibrotic effects in several disease models by modulating intra- and extra-thymic T-cell differentiation [10]. Thymulin has been shown to reduce lung inflammation and fibrosis in a bleomycin-induced pulmonary fibrosis model [9]. Thymulin also inhibited pulmonary inflammation and hypertension caused by monocrotaline in the mouse lung through the suppression of pro-inflammatory p38 pathway [11]. However, the role of thymulin in allergic asthma, which shares similar pathophysiological properties with the aforementioned diseases, has never been evaluated, nor has a potentially long-lasting gene therapy approach for thymulin been attempted. Unlike conventional therapies for asthma, thymulin therapy may provide not only symptomatic therapy but also functional recovery of the lungs.
Local administration of thymulin genes to the lungs via an efficient gene delivery vehicle may provide sustainable thymulin protein expression and subsequent therapeutic effects. A majority of lung gene therapy trials have used viral gene vectors, but viral vectors may possess one or more of the following intrinsic shortcomings, including limited plasmid size, complicated manufacturing, and immunological inactivation upon repeated dosing [12]. Highly compacted DNA nanoparticles, composed of single molecules of plasmid DNA compacted with block copolymers of poly-L-lysine and polyethylene glycol linked by a cysteine residue (CK30PEG) [13–15] have been shown to be non-toxic and non-immunogenic in the lungs of mice and humans [15, 16], and provide functional gene expression in the lung epithelium of cystic fibrosis patients [15]. More recently, these DNA nanoparticles also mediated efficient transgene expression in the brain [17] and eyes [18] of rodents. Here, we tested our hypothesis that local administration of DNA nanoparticles carrying a thymulin analog gene encoding a biologically active form of thymulin, methionine serum thymus factor (metFTS), may prevent inflammatory and remodeling processes, thus improving lung mechanics in a murine model of ovalbumin (OVA)-challenged allergic asthma.
2. Methods
2.1. Preparation and physicochemical characterization of CK30PEG-DNA
A 31-mer peptide composed of 30 lysine residues and 1 cysteine residue (CK30) was synthesized by Fmoc-mediated solid-phase peptide synthesis using an automated peptide synthesizer (Symphony Quartet, Protein Technologies, Tucson, AZ). A block co-polymer of poly-L-lysine (i.e. CK30) and polyethylene glycol (PEG) (CK30PEG) was prepared as previously described [19]. The plasmid DNA pDC515-(T7)metFTS, was generously provided by Dr. Rodolpho G. Goya [20]. CK30PEG-DNA (metFTS and GFP) was manufactured by the drop-wise addition of 9 volumes of plasmid DNA solution (0.222 mg/ml in water) to 1 volume of CK30PEG polymer solution at a rate of 1 ml/min. The final molar ratio of side chain primary amines of CK30PEG to negatively charged plasmid DNA phosphates was 2:1. After incubating at room temperature for 30 min to form DNA nanoparticles, aggregates were removed by syringe filtration (0.2 µm). DNA nanoparticles were then washed twice with 10 volumes of isotonic saline to remove free polymers, and re-concentrated to 2 mg/ml using Amicon® Ultra Centrifugal Filters (100,000 MWCO, Millipore Corp., Billerica, MA)
The formulation of DNA nanoparticles was confirmed by transmission electron microscopy (TEM, Hitachi H7600, Japan). Hydrodynamic diameter and surface charge, as indicated by ζ-potential, were measured in 10 mM NaCl (pH ~7) by dynamic light scattering and laser Doppler anemometry, respectively, using a Zetasizer Nano ZS90 (Malvern Instruments, Southborough, MA). The ζ-potential values were calculated using the Smoluchowski equation. Turbidity and sedimentation assays were conducted as previously described [19, 21]; details are provided in the online supplementary method section.
2.2. Animal preparation and experimental protocol
This study was approved by the Animal Ethics Committee of the Health Sciences Centre, UFRJ (IBCCF019). Male BALB/c mice (20 – 25 g) were randomly assigned to be sensitized and challenged with sterile ovalbumin (OVA; Sigma-Aldrich, St. Louis, MO) or saline. In the allergic asthma groups, mice were sensitized by intraperitoneal (i.p.) injection of ovalbumin (OVA, 10 µg in 0.1 ml of saline) every other day up to 7 injections. Forty days after the beginning of sensitization, intratracheal OVA challenges were performed (20 µg in 20 µl saline) 3 times with 3-day intervals between the applications [22]. The control groups (C) received saline instead of OVA during both sensitization and challenge. Twenty days before the initial challenge, the C and OVA groups were randomly assigned to intratracheally receive 50 µl of CK30PEG-metFTS (THY-NANO), naked plasmid DNA (pDC515-(T7)metFTS, THY-pDNA), or saline (SAL) using a microsprayer (Penn Century-Micro Sprayer, Model IA-1C, Philadelphia, PA, USA). Twenty seven days after the administration (i.e. 24 hours after the last challenge with saline or OVA), lungs were harvested and all efficacy data was collected and analyzed as described below.
For the short-term in vivo gene transfer study, normal male BALB/c mice were intratracheally dosed with 50 µl of DNA nanoparticles carrying GFP plasmid (CK30PEG-GFP), naked GFP plasmid (GFP-pDNA), or SAL.
2.3. Mechanical parameters
Twenty-four hours after the last intratracheal challenge with saline or OVA, animals were sedated (diazepam 1 mg i.p.), anaesthetized (thiopental sodium 20 mg/kg i.p.), tracheotomized, paralyzed (vecuronium bromide 0.005 mg/kg intravenous (i.v.)), and ventilated with a constant flow ventilator (Samay VR15; Universidad de la Republica, Montevideo, Uruguay). Lung mechanics and airway responsiveness were analyzed using ANADAT software (RHT-InfoData, Inc., Montreal, Quebec, Canada) [22, 23]. For details, see the online supplementary method section.
2.4. Lung histology
After the animal's death, heparin (1,000 IU) was i.v. injected and lungs were removed. The lungs were fixed and then embedded in paraffin [3, 24]. Slices were cut and subjected to hematoxylin and eosin staining. Morphometric analysis was performed using a grid with 100 points and 50 lines (known length) coupled with a light microscope (Axioplan, Zeiss, Oberkochen, Germany), by a point-counting technique across 10 random non-coincident microscopic fields [3, 25].
Collagen fibers were quantified in the alveolar septa and the airways using a digital analysis system and an image processing software (Image-Pro Plus 5.1 for Windows; Media Cybernetics, Silver Spring, MD). For details, see the online supplementary method section.
2.5. Immunohistochemistry
Strips (2 × 2 × 10 mm) from the right lung were fixed and embedded in paraffin for immunohistochemistry using a monoclonal antibody against α-smooth muscle actin (Dako, Carpenteria, CA) at a 1:500 dilution. The analysis was performed on the slides stained for α- smooth muscle actin with application of the point-counting technique using a 121-point grid [3, 25]. For details, see the online supplementary method section.
2.6. Transmission electron microscopy
To obtain a stratified random sample, three 2 × 2 × 2-mm slices were cut from different segments of the left lung and fixed. Ultrathin sections from selected areas were examined and micrographed in a JEOL electron microscope (JSM-6100F; Tokyo, Japan). Submicroscopic analysis of lung tissue and pathological findings were graded according to a 5-point semi- quantitative severity-based scoring system [26]. For details, see the online supplementary method section.
2.7. Confocal microscopy
For the short-term in vivo gene transfer study, mice intratracheally dosed with DNA nanoparticles carrying GFP plasmid DNA were euthanized two days after the administration. Lungs were harvested, cyrosectioned and treated with 4′,6-diamidino-2-phenylindole dihydrochloride (DAPI)-supplemented mounting medium (Vectashield, Vector Labs, Burlingame, CA) to visualize cell nuclei. Fluorescent images of GFP expression in the lung were obtained using a laser scanning confocal microscope (Leica Microsystems Ltd., Heidelberg, Germany).
2.8. Evaluation of bronchoalveolar lavage fluid
To analyze the bronchoalveolar lavage fluid (BALF), a polyethylene cannula was inserted into the trachea and a total volume of 1.0 ml of phosphate-buffered saline (PBS) containing 10 mM EDTA was instilled and aspirated. Samples were centrifuged at 300 g for 10 minutes. The supernatant was removed, and the pellet was resuspended in 0.25 ml of PBS. Total leukocyte counts in the BALF were quantified in Neubauer chambers under light microscopy after dilution of the samples in Türk solution (2% acetic acid). Differential leukocyte counts were performed in cytocentrifuged smears stained by the May-Grünwald-Giemsa method as previously described [26].
2.9. Quantification of cytokines and growth factors
Interleukin-13 (IL-13), vascular endothelium growth factor (VEGF), interferon-γ (IFN-γ), eotaxin (Peprotech, New Jersey, NJ, USA) and transforming growth factor-β (TGF-β) (RND, Minnesota, MN, USA) were quantified in the lung tissues from various treatment groups by ELISA as per manufacturer’s protocol.
2.10. Detection of thymulin plasmid
DNA was isolated from lung tissues using QIAamp® DNA Mini and Blood Mini Kit (Qiagen, GmbH, Hilden, Germany) as per manufacturer’s protocol. Primers specific for thymulin and housekeeping (glyceraldehyde-3-phosphate dehydrogenase (GAPDH)) genes were designed (Invitrogen, Carlsbad, CA): thymulin (5-TAA TAC GAC TCA CTA TAG GG-3 and 5-TCG ATG CTA GAC GAT CCA GA-3) and GAPDH (5-AAC TTT GGC ATT GTG GAA GG-3 and 5-GTC TTC TGG GTG GCA GTG AT-3). Real-time polymerase chain reaction (PCR) (7500 Real-time PCR, Applied Biosystems, Foster City, CA) was conducted to amplify and detect thymulin plasmid DNA and GAPDH gene in the lung tissues. Relative plasmid DNA levels were quantified by measuring the intensity of SYBR green fluorescence using ABI 7500 real-time PCR (Applied Biosystems, Foster City, CA) and the Ct method where the amount of thymulin plasmid DNA is normalized by that of GAPDH gene.
2.11. Statistical analysis
Data were tested for normality using the Kolmogorov-Smirnov test with Lilliefors correction and the homogeneity of variances was assessed with the Levene median test. If both conditions were satisfied, two-way ANOVA followed by Tukey's test was used for the comparison among the groups. Parametric data were expressed as mean ± SD. All tests were performed using the SigmaPlot 11 software package (SYSTAT, Chicago, IL, USA), and statistical significance was established as p < 0.05.
3. Results
3.1. Physicochemical characterization of compacted DNA nanoparticles
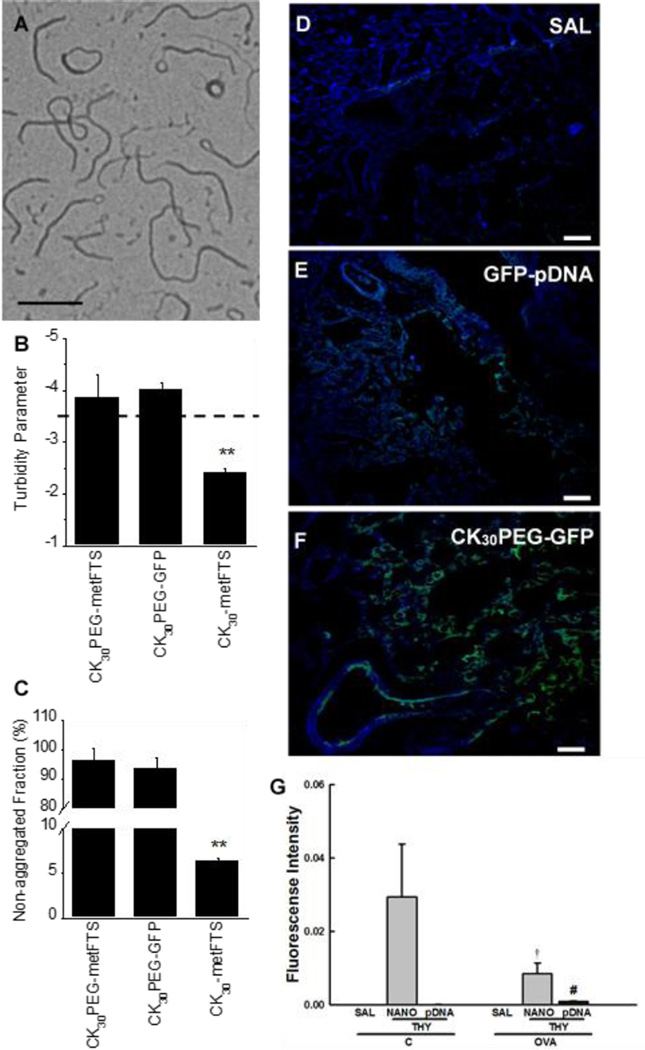
The formation of compacted DNA was confirmed by morphological examination via TEM, which revealed flexible rod-like nanostructures (Fig. 1A), in agreement with previous reports for CK30PEG-DNA nanoparticles [19, 27]. DNA nanoparticles exhibited a near neutral surface charge (−1.5 ± 3.9 mV), indicating the presence of PEG coating on the particle surface, and hydrodynamic diameter determined by dynamic light scattering was 89.0 ± 0.4 nm (PDI ~0.10). The colloidal stability of DNA nanoparticles carrying plasmid DNA encoding reporter green fluorescence protein (GFP) or metFTS (CK30PEG-GFP or CK30PEG-metFTS) in saline was confirmed by turbidity and sedimentation assays. A turbidity parameter less than −3.5 (above the dashed line) indicates DNA nanoparticles that are compacted and non-aggregated, whereas values greater than −3.5 (below the dashed line) indicate increasing aggregation of DNA nanoparticles (Fig. 1B). CK30PEG-metFTS and CK30PEG-GFP exhibited turbidity parameters indicative of negligible aggregation (Fig. 1B), in good agreement with sedimentation assays that showed that < 5% of compacted DNA aggregated and precipitated during this period (Fig. 1C).
Fig. 1.
Physicochemical characterization of DNA nanoparticles. (A) TEM image of compacted DNA nanoparticles (CK30PEG-metFTS). Scale bar represents 200 nm. (B) Colloidal stability of compacted DNA nanoparticles in saline. Turbidity parameter is an indicator of colloidal stability. The parameter less than -3.5 (above the dashed line) indicates that particles are compacted and non-aggregated. (C) Non-aggregated fraction of compacted DNA nanoparticles as determined by the fraction of particle sedimentation. Data represents the mean ± SD. ** denotes statistical significant differences (P < 0.01) compared to other treatment groups. Confocal images of lung parenchyma of at 2 days post-administration of (D) saline (SAL), (E) naked GFP plasmid DNA (GFP-pDNA), and (F) DNA nanoparticles carrying GFP plasmid DNA (CK30PEGGFP) (100 µg plasmid DNA). Scale bars represent 100 µm. Note the presence of GFP expression (green) in lung parenchyma. (G) Real-time PCR-based quantification of thymulin plasmid in the lung tissues of saline sensitized/challenged control (C) and ovalbumin-sensitized/challenged (OVA) mice at 27 days post-administration of saline (SAL), thymulin plasmid DNA (THY; 100 µg plasmid DNA) as naked plasmid DNA (pDNA) or DNA nanoparticles (NANO). Higher SYBR green fluorescence intensity indicates larger plasmid DNA content. Data represents the mean ± SD (n = 5 mice per group). Differences are statistically significant (p < 0.01) from C-THY-NANO (†) or OVA-THY-NANO (#).
3.2. In vivo gene transfer
To confirm whether CK30PEG-DNA nanoparticles mediate airway gene transfer in vivo, we intratracheally dosed mice with DNA nanoparticles carrying GFP plasmid DNA (CK30PEGGFP), and assessed transgene expression using confocal microscopy. As shown by the microscopic image without green fluorescence, no autofluorescence was detected in the lungs of normal mice treated with saline (Fig. 1D). Using the identical microscopic settings, we observed a sparse GFP expression in the lungs of normal mice dosed with naked GFP plasmid (GFP-pDNA) (Fig. 1E), whereas the distribution of GFP-positive cells was broader in the lungs of normal mice received CK30PEG-GFP (Fig. 1F) two days after the administration, in good agreement with previous findings [19, 21, 28].
We next investigated whether the DNA nanoparticles improve the retention of DNA in the mouse lungs (Fig. 1G). To do this, we intracheally dosed normal and OVA-challenged asthmatic mice with either saline (SAL), naked metFTS plasmid (metFTS-pDNA; pDNA), or DNA nanoparticles carrying the metFTS plasmid (CK30PEG-metFTS; NANO). The lung retention of metFTS plasmid was quantified by real-time polymerase chain reaction (PCR) at 27 days after the administration when the anti-inflammatory and anti-fibrotic efficacies were assessed in the subsequent studies. Surprisingly, metFTS plasmid was retained in the lungs of mice treated with metFTS-pDNA, but significantly larger quantities of metFTS plasmid were found in the lungs of both normal and asthmatic mice dosed with DNA nanoparticles (CK30PEG-metFTS) (p < 0.01). A significantly less amount of the plasmid was detected following the administration of CK30PEG-metFTS in the lungs of OVA-challenged mice compared to normal mice (p < 0.01).
3.3. Lung morphometry
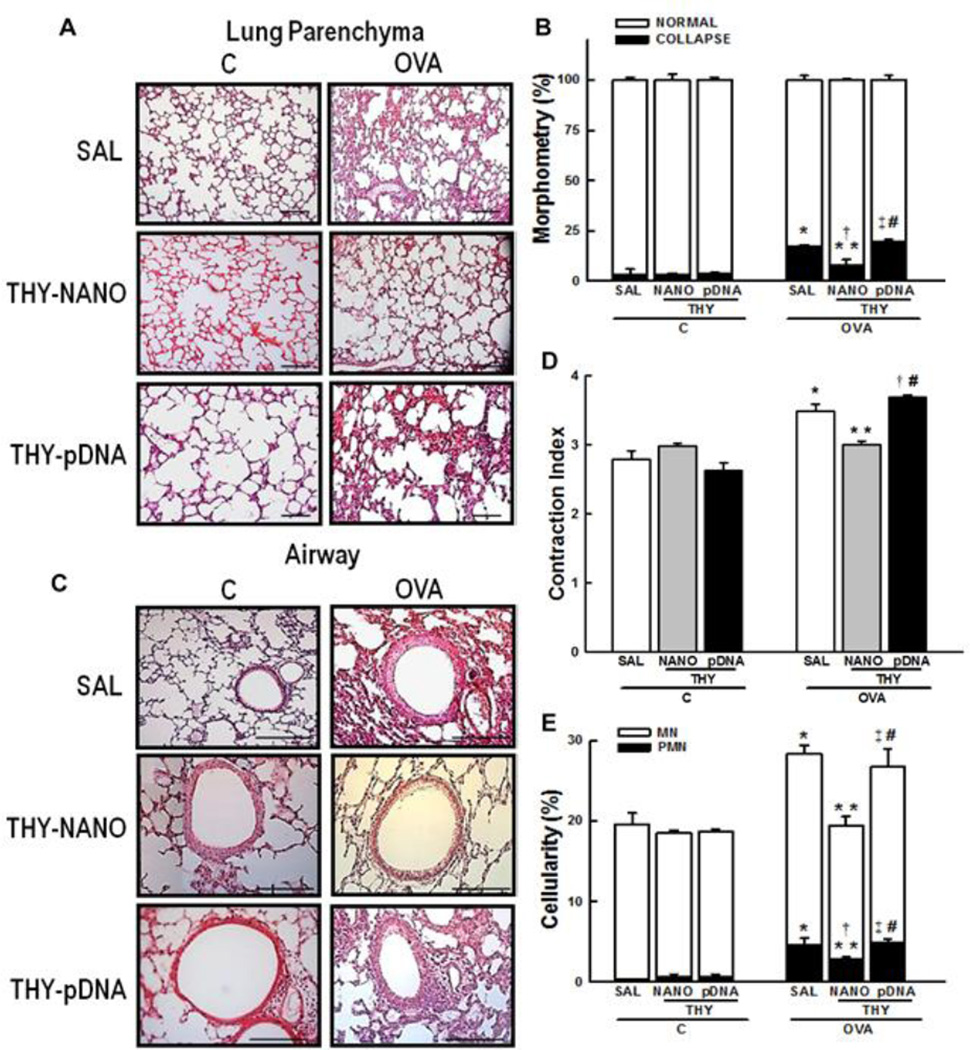
Morphometric examination of the lungs of OVA-challenged mice treated with saline (OVASAL group) demonstrated a significant increase in the fractional area of alveolar collapse (Figs. 2A, B), in a good agreement with previous observations [20, 24]. The alveoloar collapse induced by OVA was significantly mitigated in the lungs of mice treated with CK30PEG-metFTS (OVA-THY-NANO group) (p < 0.05). In contrast, the naked metFTS plasmid (OVA-THY-pDNA group) was not effective in reducing alveolar collapse (Figs. 2A, B). In addition, the diameter of the central airway was less constricted in mice in the OVA-THY-NANO group compared to OVA-SAL group (p < 0.05); this was not observed in OVA-THY-pDNA group (Figs. 2C, D).
Fig. 2.
Lung morphometry. Representative histological images of (A) lung parenchyma and (C) airways. Scale bars denote 100 µm (lung parenchyma) or 200 µm (airways). (B) The fractional area of normal and collapsed alveoli, (D) contraction index, and (E) tissue cellularity. Data represents the mean ± SD (n = 7 mice per group). C, mice sensitized/ challenged with saline; OVA, mice sensitized/challenged with ovalbumin; SAL, mice treated with saline; THY-NANO, mice treated with DNA nanoparticles (100 µg of plasmid DNA); THY-pDNA, mice treated with naked plasmid DNA (100 µg of plasmid DNA). Differences are statistically different from C-SAL (*), C-THY-NANO (†), C-THY-pDNA (‡), OVASAL (**), or OVA-THY-NANO (#).
3.4 Inflammatory process
As expected by well-reported OVA-induced inflammation [20,24], the number of mononuclear and polymorphonuclear cells in lung tissue was higher in the OVA-SAL group compared to control mice that were challenged and treated with saline only (p < 0.05) (C-SAL group). Mononuclear and polymorphonuclear cell infiltration was minimized in the OVA-THY-NANO group, but not in the OVA-THY-pDNA group (p < 0.05) (Fig. 2E).
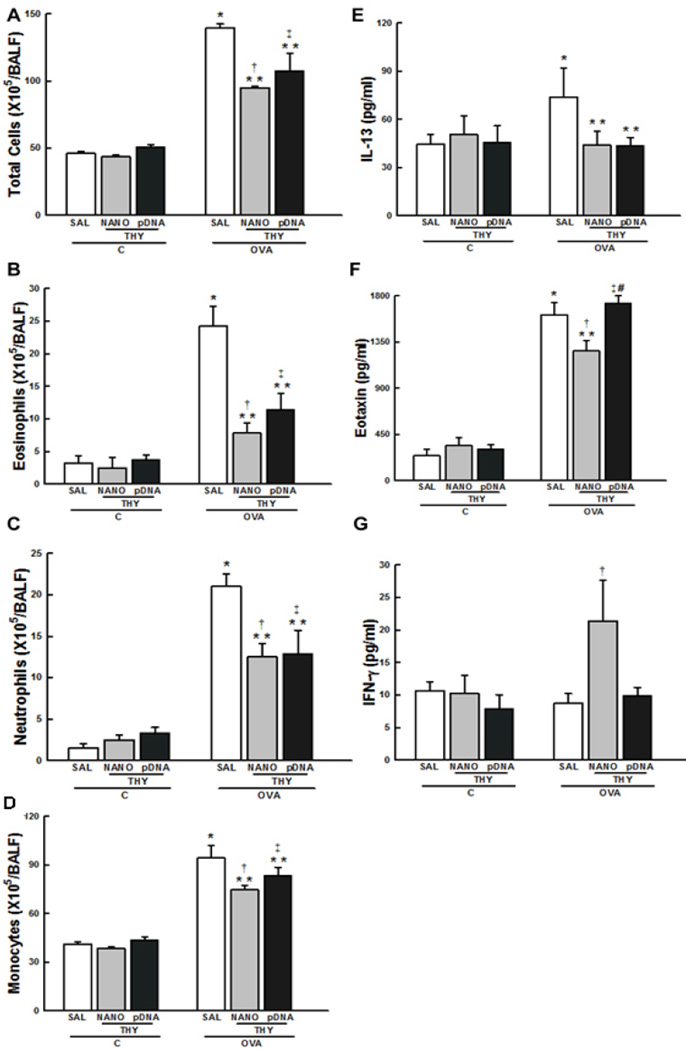
The total number of cells and the numbers of eosinophils, neutrophils, and monocytes in the BALF were higher in the OVA-SAL group than in the C-SAL group (p < 0.05) (Figs. 3A-D). These parameters were attenuated in both the OVA-THY-NANO and OVA-THY-pDNA treatment groups (Figs. 3A-D).
Fig. 3.
Lung inflammation. (A) Total number of cells and numbers of (B) eosinophils, (C) neutrophils, and (D) monocytes in BALF. Data represents the mean ± SD (n = 5 mice per group). The concentrations of (E) IL-13, (F) eotaxin, and (G) IFN-γ in the lung tissues, as determined by ELISA. Data represents the mean ± SD (n = 7 mice per group). C, mice sensitized/challenged with saline; OVA, mice sensitized/challenged with ovalbumin; SAL, mice treated with saline; THY-NANO, mice treated with DNA nanoparticles with (100 µg plasmid DNA); THY-pDNA, mice treated with naked plasmid DNA (100 µg plasmid DNA). Differences are statistically different from C-SAL (*), C-THY-NANO (†), C-THY-pDNA (‡), OVA-SAL (**), or OVA-THY-NANO (#).
The level of an asthmatic mediator, interleukin-13 (IL-13) [24], in the lung tissue was significantly higher in the OVA-SAL group (Fig. 3E) compared to the C-SAL group (p < 0.05). IL-13 was decreased in both the OVA-THY-NANO and OVA-THY-pDNA groups compared to OVA-SAL group (p < 0.05). The expression of an eosinophil chemotactic factor, eotaxin [29], was reduced in the OVA-THY-NANO group (p < 0.05), but not in the OVA-THY-pDNA group (Fig. 3F). Interferon- γ (IFN-γ), which has been shown to reduce allergic responses [30], was increased in the OVA-THY-NANO group compared to OVA-SAL and OVA-THY-pDNA groups (p < 0.05) (Fig. 3G).
3.5 Lung remodeling
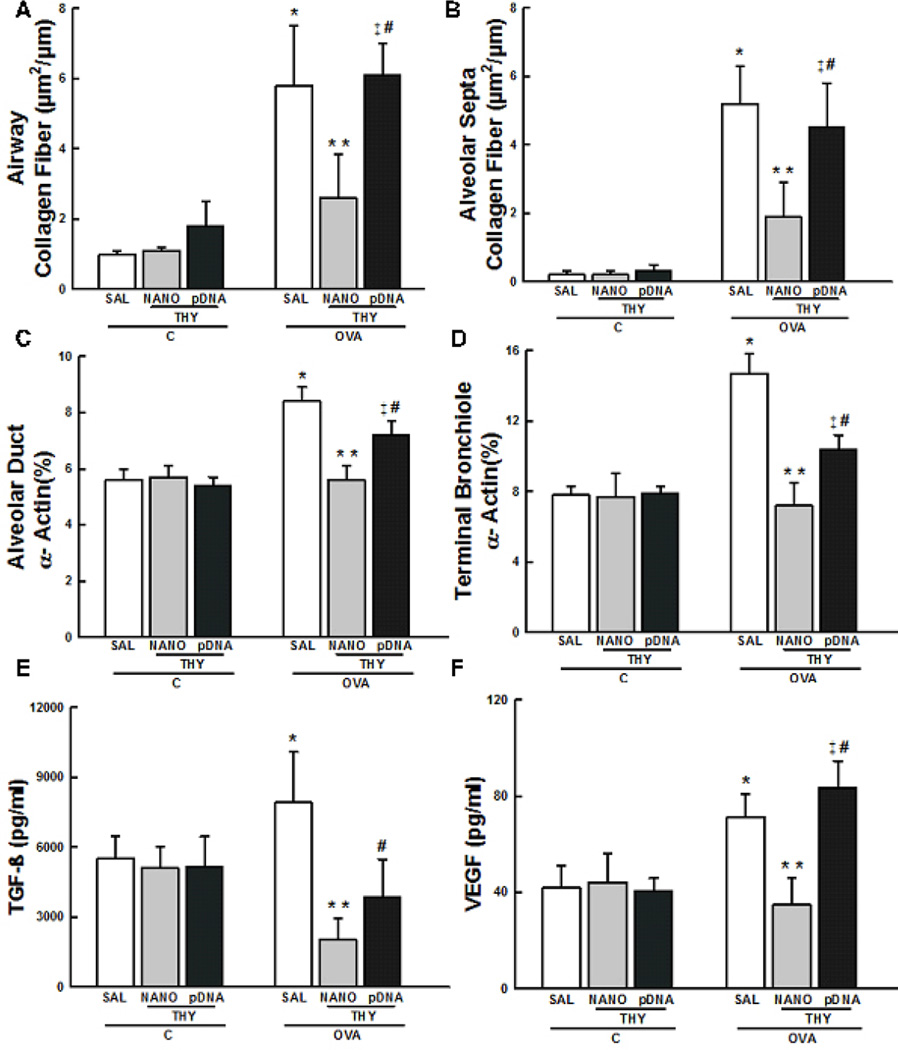
The collagen fiber content in the airways and alveolar septa, as well as the volume proportion of smooth muscle-specific actin (α-SMA) in terminal bronchioles and alveolar ducts, were higher in the OVA-SAL group compared to C-SAL group (p < 0.05) (Figs. 4A-D). The increase in α-SMA is a hallmark of smooth muscle hypertrophy associated with lung remodeling [31]. These changes were significantly reduced in the OVA-THY-NANO group (p < 0.05), but not in the OVA-THY-pDNA group (Figs. 4C, D).
Fig. 4.
Lung remodeling. Collagen fiber content in (A) airways and (B) alveolar septa. Volume proportion of smooth muscle-specific actin in (C) alveolar ducts and (D) terminal bronchioles. All values were computed in 10 random non-coincident fields per mice. The concentrations of (E) TGF-β and (F) VEGF in the lung tissues. Data represents the mean ± SD (n = 7 mice per group). C, mice sensitized/challenged with saline; OVA, mice sensitized/challenged with ovalbumin; SAL, mice treated with saline; THY-NANO, mice treated with DNA nanoparticles (100 µg plasmid DNA); THY-pDNA, mice treated with naked plasmid DNA (100 µg plasmid DNA). Differences are statistically different (p < 0.05) from CSAL (*), C-THY-NANO (†), C-THY-pDNA (‡), OVA-SAL (**), or OVA-THY-NANO (#).
We also quantified the critical chemical mediators of lung remodeling, including transforming growth factor-β (TGF-β) and vascular endothelial growth factor (VEGF) [32]. TGF-β expression was decreased in the OVA-THY-NANO and OVA-THY-pDNA groups compared to the OVA-SAL group (p < 0.05) (Fig. 4E). VEGF expression was reduced in the OVA-THY-NANO group compared to OVA-SAL group (p < 0.05), whereas no difference in VEGF levels were observed in the OVA-THY-pDNA group compared to OVA-SAL group (Fig. 4F).
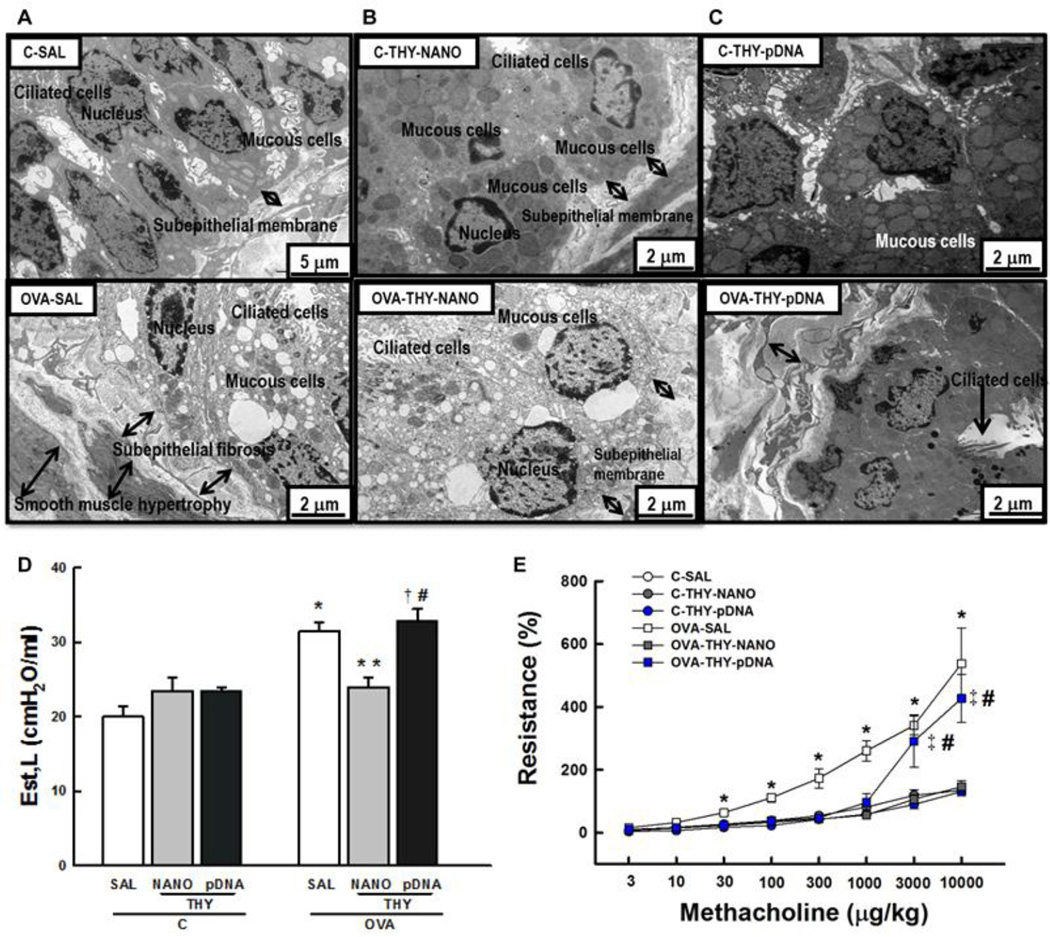
3.6. Ultrastructural changes and lung mechanics
We next performed TEM on airway sections from each animal group (Figs. 5A-C). Mice in the OVA-SAL group exhibited considerable subepithelial fibrosis and smooth muscle hypertrophy (Fig. 5A), hallmarks of the lung remodeling presented in patients afflicted with asthma [8]. Based on a semi-quantitative analysis of TEM images, mice in the OVA-SAL group showed degenerative changes in ciliated airway epithelial cells, epithelial detachment, edema formation, eosinophil and neutrophil infiltration and myofibroblast and mucous cell hyperplasia (Table 1). Remarkably, nanoparticle-mediated thymulin gene therapy (OVA-THY-NANO group) attenuated all of these ultrastructural changes resulted from lung remodeling; however, mice in the OVA-THY-pDNA group failed to show similar benefits (Table 1). Lung static elastance (Est, L) is expected to elevate in asthmatic lungs undergoing fibrotic process [34]. We found that Est, L was 55% higher in mice in the OVA-SAL group than in the C-SAL group (p < 0.05) (Fig. 5D). However, Est, L in the OVA-challenged mice treated with DNA nanoparticles carrying metFTS plasmid (OVATHY- NANO group) was not elevated (similar to the C-SAL group) (p < 0.05) (Fig. 5D). Furthermore, increase in airway resistance evoked by methacholine was significantly augmented in the OVA-SAL group; however, these changes were mitigated in OVA-THY-NANO group. The naked (uncompacted) metFTS was not efficient in minimizing these parameters (OVA-THY-pDNA group; Fig. 5E). It should be noted that OVA-THY-NANO group exhibited virtually identical airway resistance to control groups where there were no OVA challenges.
Fig. 5.
Lung ultrastructure and lung function. Electron microscopy of the lung of saline sensitized/challenged control (upper panel) and ovalbumin-sensitized/challenged mice (lower panel). Mice were intratracheally dosed with (A) saline, (B) DNA nanoparticles carrying thymulin plasmid DNA or (C) free thymulin plasmid DNA. (D) Lung static elastance (Est,L) and (E) airway hyperresponsiveness. Data represents the mean ± SD (n = 7 mice per group). C, mice sensitized/challenged with saline; OVA, mice sensitized/challenged with ovalbumin; SAL, mice treated with saline; THY-NANO, mice treated with DNA nanoparticles (100 µg plasmid DNA); THY-pDNA, mice treated with naked plasmid DNA (100 µg plasmid DNA). Differences are statistically significant (p < 0.05) from C-SAL (*), C-THY-pDNA (‡), OVA-SAL (**), or OVATHY- NANO (#).
Table 1.
Semi-quantitative analysis of TEM images of the lungs from various treatment groups.
| Pathologic finding |
C group | OVA group | ||||
|---|---|---|---|---|---|---|
| SAL | THY-NANO | THY-pDNA | SAL | THY-NANO | THY-pDNA | |
| Epithelial detachment |
0 (0–0) | 0 (0–0) | 0 (0–0) | 3 (2.75–3)* | 1 (0.75–1)*# | 2 (1.75–2)* |
| Edema formation |
0 (0–0) | 0 (0–0) | 0 (0–0) | 3 (2.75–3.25)* | 1(0–1)# | 2 (1–2.25)* |
| Eosinophil infiltration |
0 (0–0) | 0 (0–0) | 0 (0–0) | 3 (2–3.25)* | 1 (1–1.25)*# | 2 (1.75–2)* |
| Neutrophil infiltration |
0 (0–0) | 0 (0–0) | 0 (0–0) | 2 (1–2)* | 0 (0–1)# | 1 (1–1.25)* |
| Ciliated airway epithelial cell degeneration |
0 (0–0) | 0 (0–0) | 0 (0–0) | 3 (2.75–3)* | 0 (0–1)# | 2 (1–2)* |
| Subepithelial fibrosis |
0 (0–0) | 0 (0–0) | 0 (0–0) | 3 (3–4)* | 1 (0.75–1)*# | 3 (2.75–4)*† |
| Smooth muscle hypertrophy |
0 (0–0) | 0 (0–0) | 0 (0–0) | 4 (4–4)* | 1 (0.75–1)*# | 3 (2.75–3.25)* |
| Myofibroblast hyperplasia |
0 (0–0) | 0 (0–0) | 0 (0–0) | 4 (3–4)* | 1 (0.75–1)*# | 3 (2.75–3.25)* |
| Mucous cells hyperplasia |
0 (0–0) | 0 (0–0) | 0 (0–0) | 3 (2.75–4)* | 1 (0.75–1)*# | 3 (3–3.25)*† |
Values are the median (25th percentile–75th percentile) (n = 5 mice per group). Pathologic findings were graded according to a 5-point semi-quantitative severity-based scoring system: 0, normal lung parenchyma; 1, changes in 1–25%; 2, 26–50%; 3, 51–75%; 4, 76–100% of the examined tissues. C, mice sensitized/challenged with saline; OVA, mice sensitized/challenged with ovalbumin; SAL, mice treated with saline; THY-NANO, mice treated with DNA nanoparticles (100 µg of plasmid DNA); THY-pDNA, mice treated with naked plasmid DNA (100 µg plasmid DNA). Differences are statistically significant (p < 0.05)
from C-SAL (*),
OVA-SAL (#),
or OVA-THY-NANO (†).
4. Discussion
Current therapies for allergic airway diseases are limited to alleviating disease symptoms and exacerbations, and are unable to prevent or revert lung damage that occurs during the natural history of disease [33, 34]. In this study, we show that pulmonary delivery of a biologically active form of a thymulin analogue gene in highly compacted DNA nanoparticles mitigated airway inflammation and lung remodeling, leading to improved pulmonary mechanics, in the lungs of an asthmatic mouse model. To our knowledge, this is the first demonstration of airway gene therapy that improved lung function in an asthmatic animal model. We also confirm the previous finding that CK30PEG-DNA nanoparticles do not induce inflammation in the lungs of normal inbred mice [16]. This is likely due to the surface PEG coatings which shielded otherwise positive charged DNA nanoparticles [35–37]. Outcomes in the present study may be translated in human clinical trials with a relatively low regulatory burden, since the highly compacted DNA nanoparticles used in the present study have been safely administered to airways of human subjects previously [15].
Intratracheal administration of thymulin plasmid alone was able to reduce some aspects the inflammation in the lungs of asthmatic mice compared to saline-treated control. However, pathological lung remodeling was significantly intervened only if the thymulin plasmid was delivered via DNA nanoparticles, as shown by the significant reduction of both collagen deposition and smooth muscle hypertrophy in the lungs of asthmatic mice. Our findings of improvement on lung remodeling with nanoparticle-mediated thymulin gene therapy are consistent with a decrease in the secretion of key mediators, including VEGF and TGF-β. In addition, the anti-inflammatory effect was significantly greater with DNA nanoparticles compared to thymulin plasmid alone. This is likely due to improved gene transfer to the lung epithelium by DNA nanoparticles, as evident by the broad distribution of transgene expression throughout the conducting airways and alveolar region. The prolonged retention (at least 27 days post-administration) of thymulin plasmid achieved in the lungs of asthmatic mice with pulmonary administration of DNA nanoparticles provides further evidence of improved gene transfer mediated by DNA nanoparticles. This may be at least partially due to the stability of DNA nanoparticles in physiological conditions. It should be also noted that we have recently shown that DNA nanoparticles are highly stable in airway secretions produced by CF patients [19, 21]. In comparison, significantly lower amount of thymulin plasmid was retained at the same time post-administration when administered as naked DNA. We have previously reported that the DNA nanoparticles studied here enter human bronchial epithelial cells via a caveolin-mediated pathway, thereby bypassing degradative trafficking, and leading to rapid accumulation in the perinuclear region [38]. These characteristics of DNA nanoparticles may also have facilitated intracellular delivery of DNA payloads to the cell nucleus, leading to enhanced trangene expression compared to naked DNA.
The reduced airway inflammation observed following the intratracheal administration of DNA nanoparticles carrying thymulin plasmids corroborates the efficient thymulin gene expression demonstrated in the lungs of asthmatic mice. Allergic asthma is characterized by imbalance in the Th1/Th2 immune response where Th2 is highly upregulated compared to Th1 [39]. In this study, we found that a single administration of DNA nanoparticles carrying thymulin plasmid, but not plasmid alone, significantly increased IFN-γ, which is widely known to inhibit Th2-induced effector functions by stimulating Th1 cells [30]. This is in good agreement with previous findings that showed positive correlation between Th1 cytokine production and thymulin secretion [40]. The secretion of Th2 mediators involved in eosinophil recruitment, including IL-13 and eotaxin, was also significantly reduced in the lungs of asthmatic mice by DNA nanoparticles carrying thymulin plasmid.
The murine model of OVA-challenged allergic asthma has been widely utilized to investigate the mechanisms of airway inflammation and immunologic responses mediated by allergic asthma [23, 41]. We have customized the model by modifying the OVA sensitizing/challenging scheme to evaluate the inhibitory effect of biologically active thymulin analogue on the progression of lung inflammation and remodeling. Although the DNA nanoparticles carrying thymulin plasmid was dosed 20 days prior to the OVA challenges, we confirmed that initial OVA sensitizations readily established an increase in the inflammatory mediators even after the DNA nanoparticles were administered (Supplementary Fig. 1). The anti-inflammatory effect was evident at 27 days post-administration (i.e. 7 days after the initial OVA challenge), which is in good agreement with a previous observation that showed the peak activity by thymulin in thymectomized animal models 20 days after the intramuscular administration of adenoviral vectors carrying the thymulin analogue gene [42]. These observations suggest that a single dose of DNA nanoparticles therapeutically reduced inflammation in the lungs OVA-sensitized mice and retained the effect even after the OVA challenges. The anti-fibrotic effect of thymulin was likely preventive in this study, since lung remodeling took place only after the OVA challenges. Thus, asthmatic patients at relatively early stages, when lung fibrosis is not completely established, may benefit the most from a thymulin based gene therapy [43].
In summary, our results suggest that thymulin gene transfection mediated by highly compacted DNA nanoparticles effectively prevents the inflammatory and remodeling process in the asthmatic lungs, which enhances airway repair and thus improves lung mechanics in a leading mouse model of allergic inflammation. These findings motivate the further development of nanoparticle-based gene therapies for safe and effective delivery of therapeutic genes for asthma treatment.
Supplementary Material
Acknowledgments
The authors express their gratitude to Andre Benedito da Silva for animal care and Ana Lucia Neves da Silva for her help with microscopy. This work was supported by the following: Centers of Excellence Program (PRONEX-FAPERJ), Brazilian Council for Scientific and Technological Development (CNPq), Carlos Chagas Filho, Rio de Janeiro State Research Supporting Foundation (FAPERJ), Coordination for the Improvement of Higher Education Personnel (CAPES), São Paulo State Research Supporting Foundation (FAPESP), INCTINOFAR, the European Community's Seventh Framework Programme (TARKINAID, FP7- 2007-2013) and the National Institutes of Health (R01EB003558 and P01HL51811).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Holloway JW, Yang IA, Holgate ST. Genetics of allergic disease. J Allergy Clin Immunol. 2010;125(2 Suppl 2):S81–S94. doi: 10.1016/j.jaci.2009.10.071. [DOI] [PubMed] [Google Scholar]
- 2.Cates C. Inhaled corticosteroids in COPD: quantifying risks and benefits. Thorax. 2013;68(6):499–500. doi: 10.1136/thoraxjnl-2012-202959. [DOI] [PubMed] [Google Scholar]
- 3.Raissy HH, et al. Inhaled corticosteroids in lung diseases. Am J Respir Crit Care Med. 2013;187(8):798–803. doi: 10.1164/rccm.201210-1853PP. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pascual RM, Peters SP. Airway remodeling contributes to the progressive loss of lung function in asthma: an overview. J Allergy Clin Immunol. 2005;116(3):477–486. doi: 10.1016/j.jaci.2005.07.011. quiz 487. [DOI] [PubMed] [Google Scholar]
- 5.Kim SH, et al. Combined pharmacogenetic effect of ADCY9 and ADRB2 gene polymorphisms on the bronchodilator response to inhaled combination therapy. J Clin Pharm Ther. 2011;36(3):399–405. doi: 10.1111/j.1365-2710.2010.01196.x. [DOI] [PubMed] [Google Scholar]
- 6.Barnes PJ. Mechanisms and resistance in glucocorticoid control of inflammation. J Steroid Biochem Mol Biol. 2010;120(2–3):76–85. doi: 10.1016/j.jsbmb.2010.02.018. [DOI] [PubMed] [Google Scholar]
- 7.Durrani SR, Viswanathan RK, Busse WW. What effect does asthma treatment have on airway remodeling? Current perspectives. J Allergy Clin Immunol. 2011;128(3):439–48. doi: 10.1016/j.jaci.2011.06.002. quiz 449-50. [DOI] [PubMed] [Google Scholar]
- 8.O'Byrne PM. Global guidelines for asthma management: summary of the current status and future challenges. Pol Arch Med Wewn. 2010;120(12):511–517. [PubMed] [Google Scholar]
- 9.Yara S, et al. FTS reduces bleomycin-induced cytokine and chemokine production and inhibits pulmonary fibrosis in mice. Clin Exp Immunol. 2001;124(1):77–85. doi: 10.1046/j.1365-2249.2001.01493.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Savino W, Dardenne M. Neuroendocrine control of thymus physiology. Endocr Rev. 2000;21(4):412–443. doi: 10.1210/edrv.21.4.0402. [DOI] [PubMed] [Google Scholar]
- 11.Henriques-Coelho T, et al. Thymulin inhibits monocrotaline-induced pulmonary hypertension modulating interleukin-6 expression and suppressing p38 pathway. Endocrinology. 2008;149(9):4367–4373. doi: 10.1210/en.2008-0018. [DOI] [PubMed] [Google Scholar]
- 12.Lavigne MD, Gorecki DC. Emerging vectors and targeting methods for nonviral gene therapy. Expert Opin Emerg Drugs. 2006;11(3):541–557. doi: 10.1517/14728214.11.3.541. [DOI] [PubMed] [Google Scholar]
- 13.Liu G, et al. Nanoparticles of compacted DNA transfect postmitotic cells. J Biol Chem. 2003;278(35):32578–32586. doi: 10.1074/jbc.M305776200. [DOI] [PubMed] [Google Scholar]
- 14.Fink TL, et al. Plasmid size up to 20 kbp does not limit effective in vivo lung gene transfer using compacted DNA nanoparticles. Gene Ther. 2006;13(13):1048–1051. doi: 10.1038/sj.gt.3302761. [DOI] [PubMed] [Google Scholar]
- 15.Konstan MW, et al. Compacted DNA nanoparticles administered to the nasal mucosa of cystic fibrosis subjects are safe and demonstrate partial to complete cystic fibrosis transmembrane regulator reconstitution. Hum Gene Ther. 2004;15(12):1255–1269. doi: 10.1089/hum.2004.15.1255. [DOI] [PubMed] [Google Scholar]
- 16.Ziady AG, et al. Minimal toxicity of stabilized compacted DNA nanoparticles in the murine lung. Mol Ther. 2003;8(6):948–956. doi: 10.1016/j.ymthe.2003.09.002. [DOI] [PubMed] [Google Scholar]
- 17.Yurek DM, et al. Long-term transgene expression in the central nervous system using DNA nanoparticles. Mol Ther. 2009;17(4):641–650. doi: 10.1038/mt.2009.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Farjo R, et al. Efficient non-viral ocular gene transfer with compacted DNA nanoparticles. PLoS One. 2006;1:e38. doi: 10.1371/journal.pone.0000038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Boylan NJ, et al. Highly compacted DNA nanoparticles with low MW PEG coatings: in vitro, ex vivo and in vivo evaluation. J Control Release. 2012;157(1):72–79. doi: 10.1016/j.jconrel.2011.08.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Goya RG, et al. Thymulin gene therapy prevents the reduction in circulating gonadotropins induced by thymulin deficiency in mice. Am J Physiol Endocrinol Metab. 2007;293(1):E182–E187. doi: 10.1152/ajpendo.00085.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Suk JS, et al. N-acetylcysteine enhances cystic fibrosis sputum penetration and airway gene transfer by highly compacted DNA nanoparticles. Mol Ther. 2011;19(11):1981–1989. doi: 10.1038/mt.2011.160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xisto DG, et al. Lung parenchyma remodeling in a murine model of chronic allergic inflammation. Am J Respir Crit Care Med. 2005;171(8):829–837. doi: 10.1164/rccm.200408-997OC. [DOI] [PubMed] [Google Scholar]
- 23.Vieira RP, et al. Purinergic receptor type 6 contributes to airway inflammation and remodeling in experimental allergic airway inflammation. Am J Respir Crit Care Med. 2011;184(2):215–223. doi: 10.1164/rccm.201011-1762OC. [DOI] [PubMed] [Google Scholar]
- 24.Rocco PR, et al. Lung tissue mechanics and extracellular matrix remodeling in acute lung injury. Am J Respir Crit Care Med. 2001;164(6):1067–1071. doi: 10.1164/ajrccm.164.6.2007062. [DOI] [PubMed] [Google Scholar]
- 25.Weibel ER. Models of Lung Disease-Microscopy and Structural Methods. New York, NY, USA: 1990. Morphometry: stereological theory and practical methods; p. 48. [Google Scholar]
- 26.Abreu SC, et al. Effects of bone marrow-derived mononuclear cells on airway and lung parenchyma remodeling in a murine model of chronic allergic inflammation. Respir Physiol Neurobiol. 2011;175(1):153–163. doi: 10.1016/j.resp.2010.10.006. [DOI] [PubMed] [Google Scholar]
- 27.Ziady AG, et al. Bioluminescent imaging of reporter gene expression in the lungs of wildtype and model mice following the administration of PEG-stabilized DNA nanoparticles. Microsc Res Tech. 2010;73(9):918–928. doi: 10.1002/jemt.20855. [DOI] [PubMed] [Google Scholar]
- 28.Ziady AG, et al. Transfection of airway epithelium by stable PEGylated poly-L-lysine DNA nanoparticles in vivo. Mol Ther. 2003;8(6):936–947. doi: 10.1016/j.ymthe.2003.07.007. [DOI] [PubMed] [Google Scholar]
- 29.Coleman JM, et al. Epithelial eotaxin-2 and eotaxin-3 expression: relation to asthma severity, luminal eosinophilia and age at onset. Thorax. 2012;67(12):1061–1066. doi: 10.1136/thoraxjnl-2012-201634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mitchell C, et al. IFN-gamma acts on the airway epithelium to inhibit local and systemic pathology in allergic airway disease. J Immunol. 2011;187(7):3815–3820. doi: 10.4049/jimmunol.1100436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bentley JK, et al. Airway smooth muscle hyperplasia and hypertrophy correlate with glycogen synthase kinase-3(beta) phosphorylation in a mouse model of asthma. Am J Physiol Lung Cell Mol Physiol. 2009;296(2):L176–L184. doi: 10.1152/ajplung.90376.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Burgess JK. The role of the extracellular matrix and specific growth factors in the regulation of inflammation and remodelling in asthma. Pharmacol Ther. 2009;122(1):19–29. doi: 10.1016/j.pharmthera.2008.12.002. [DOI] [PubMed] [Google Scholar]
- 33.Manuyakorn W, Howarth PH, Holgate ST. Airway remodelling in asthma and novel therapy. Asian Pac J Allergy Immunol. 2013;31(1):3–10. [PubMed] [Google Scholar]
- 34.Verhoeven GT, et al. Effects of fluticasone propionate in COPD patients with bronchial hyperresponsiveness. Thorax. 2002;57(8):694–700. doi: 10.1136/thorax.57.8.694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vergaro V, et al. Drug-loaded polyelectrolyte microcapsules for sustained targeting of cancer cells. Adv Drug Deliv Rev. 2011;63(9):847–864. doi: 10.1016/j.addr.2011.05.007. [DOI] [PubMed] [Google Scholar]
- 36.Hatakeyama H, Akita H, Harashima H. A multifunctional envelope type nano device (MEND) for gene delivery to tumours based on the EPR effect: a strategy for overcoming the PEG dilemma. Adv Drug Deliv Rev. 2011;63(3):152–160. doi: 10.1016/j.addr.2010.09.001. [DOI] [PubMed] [Google Scholar]
- 37.Shim MS, Kwon YJ. Stimuli-responsive polymers and nanomaterials for gene delivery and imaging applications. Adv Drug Deliv Rev. 2012;64(11):1046–1059. doi: 10.1016/j.addr.2012.01.018. [DOI] [PubMed] [Google Scholar]
- 38.Kim AJ, et al. Non-degradative intracellular trafficking of highly compacted polymeric DNA nanoparticles. J Control Release. 2012;158(1):102–107. doi: 10.1016/j.jconrel.2011.10.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Doherty T, Broide D. Cytokines and growth factors in airway remodeling in asthma. Curr Opin Immunol. 2007;19(6):676–680. doi: 10.1016/j.coi.2007.07.017. [DOI] [PubMed] [Google Scholar]
- 40.Santarelli L, Bracci M, Mocchegiani E. In vitro and in vivo effects of mercuric chloride on thymic endocrine activity, NK and NKT cell cytotoxicity, cytokine profiles (IL-2, IFN-gamma, IL-6): role of the nitric oxide-L-arginine pathway. Int Immunopharmacol. 2006;6(3):376–389. doi: 10.1016/j.intimp.2005.08.028. [DOI] [PubMed] [Google Scholar]
- 41.Kudo M, et al. IL-17A produced by alphabeta T cells drives airway hyperresponsiveness in mice and enhances mouse and human airway smooth muscle contraction. Nat Med. 2012;18(4):547–554. doi: 10.1038/nm.2684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Reggiani PC, et al. Gene therapy for long-term restoration of circulating thymulin in thymectomized mice and rats. Gene Ther. 2006;13(16):1214–1221. doi: 10.1038/sj.gt.3302775. [DOI] [PubMed] [Google Scholar]
- 43.Holgate ST. A brief history of asthma and its mechanisms to modern concepts of disease pathogenesis. Allergy Asthma Immunol Res. 2010;2(3):165–171. doi: 10.4168/aair.2010.2.3.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.