Recent advances in genomics technology have led to the explosive discovery of pervasive transcription activity in most of the eukaryotic genomic regions that were once considered “junk DNA” or “dark matter” (Birney et al., 2007; Carninci et al., 2005; Djebali et al., 2012; Kapranov et al., 2007; Orgel and Crick, 1980). The latest ENCODE data collection from 15 cell lines shows that more than 70% of the human genome generates primary transcripts with protein-coding genes only accounting for 2.94% (Djebali et al., 2012). While a portion of the novel transcripts are ascribed to novel isoforms of previously known protein-coding genes, the most prominent finding in the latest human genome annotation is the catalog of a large number of novel long non-coding RNAs (lncRNAs) comprising 9,277 manually annotated genes producing 14,880 transcripts (Derrien et al., 2012). Accordingly, lncRNAs have recently drawn intense research efforts and one bright perspective is that the lncRNAs may represent a new regulatory layer in the complexity of mammalian gene regulatory networks underlying a wide range of pathophysiology of human diseases. However, the vast majority of lncRNAs exhibit much lower abundance compared with typical protein-coding mRNAs, which raises a precaution that some of them might be the product of transcriptional noise without any biological function (Natoli and Andrau, 2012; Struhl, 2007). Moreover the functional assignments of individual lncRNAs have often been solely based on their expression correlations with neighboring protein-coding genes without any mechanistic understanding and/or functional characterization (Dinger et al., 2008; Mercer et al., 2008; Rinn and Chang, 2012). Therefore, additional work would certainly be necessary before we can accurately appreciate the nature of the lncRNA regulatory networks and their roles in various biological processes as well as human diseases. Despite the limited knowledge of this newly emerging class of RNA molecules, a growing number of studies, especially in the last several years, have begun to elucidate the functional mechanisms of lncRNAs. In most cases where the functions of lncRNAs have been relatively well characterized, their prime roles lie at the regulation of gene expression and epigenetic processes in the nucleus. Consistently, analyses of subcellular RNA fractions have shown that the lncRNAs are highly enriched in the nucleus, many of which are tightly associated with the chromatin fraction (Djebali et al., 2012; Khalil et al., 2009; Ng et al., 2012). Gene expression in neurons is dynamically controlled by both an intrinsic genetic program and sensory stimulation, which is critical for brain function and development (Lyons and West, 2011). The brain is one of the richest sources for lncRNAs, which are expressed in regional, cellular, and temporal patterns in developing and adult brains (Mercer et al., 2008; Qureshi et al., 2010). However very little is known for the role of lncRNAs in brain development and function through the regulation of nuclear gene expression programs. This review will focus on recently emerging mechanistic principles that underlie the nuclear functions of lncRNAs in order to provide neuroscientists with molecular insights that will help future research on lncRNAs in the brain.
Genomic organization and expression feature of lncRNAs
The lncRNAs are conventionally defined as a transcript longer than 200 nucleotides in length with lack of protein-coding capability (Rinn and Chang, 2012). The idea that RNA itself might be a functional regulatory entity was originally developed from early studies investigating the epigenetic mechanisms of genomic imprinting and X-chromosome inactivation (XCI) (Lee, 2011; Lyon, 1961). In therian mammals, XCI is the mechanism of dosage compensation in which one of two X chromosomes in females is epigenetically silenced to account for the difference in X-linked gene dosage between XX females and XY males. One of the first prototype lncRNAs, Xist is highly expressed from one of the X chromosomes during the onset of XCI (Brown et al., 1992; Clemson et al., 1996). This 17-kb long transcript does not encode any protein but instead coats the inactive X chromosome (Xi) in cis and recruits Polycomb repressive complex 2 (PRC2) to the Xi through a conserved repeat motif to induce heterochromatin formation, thereby silencing associated genes (Zhao et al., 2008). Later it turned out that Xist action is controlled by two additional lncRNAs, Tsix and Jpx. The negative regulator Tsix is transcribed in an antisense orientation from its own promoter, located downstream of the Xist gene, and represses Xist transcription on one allele (therefore determining the active X chromosome) by means of several mechanisms including the recruitment of DNA methyltransferase 3a (Dnmt3a) to the Xist promoter region (Bacher et al., 2006; Lee et al., 1999; Xu et al., 2006). The positive regulator Jpx appears to be the Xist activator as its deletion or knock-down blocks XCI (Chureau et al., 2002; Tian et al., 2010).
The advent of high throughput genomic technologies such as DNA microarray and next generation sequencing (NGS) then enabled systematic interrogation of the lncRNAs in various cell types across developmental stages. One way to classify lncRNAs is based on their relative locations to nearby protein-coding genes. LncRNAs can be intergenic or intragenic, and depending on their origin and orientation, they can also be categorized as divergent or antisense (Kung et al., 2013). However, compared to protein-coding RNAs, lncRNAs are expressed at much lower levels, and their sequences have been subject to weak evolutionary constraints (Cabili et al., 2011; Clark et al., 2012; Derrien et al., 2012; Djebali et al., 2012; Pang et al., 2006; Ponting et al., 2009; Tani et al., 2012; Wang et al., 2004). These properties of lncRNAs not only imposed a difficulty in reliable detection and accurate assembly of transcript structure, but also raised a concern that a large portion of the novel noncoding transcripts might be the consequence of transcriptional noise (Natoli and Andrau, 2012; Struhl, 2007). In an effort to identify the most likely functional lncRNAs across the genome, chromatin-state maps were used to characterize the genomic origins of lncRNAs. Functional regulatory regions, such as active promoters, enhancers and coding regions, are uniquely decorated by a combination of post-translational modifications occurring at the N-terminal tails of individual histone subunits (Heintzman et al., 2007; Hon et al., 2009; Visel et al., 2009; Wang et al., 2009). Actively transcribing genes are marked by trimethylation of the lysine 4 residue of histone H3 (H3K4me3) at their promoter and trimethylation of lysine 36 of histone H3 (H3K36me3) along the transcribed region. Enrichment levels of these marks are well correlated with the levels of RNA expression. An integrative analysis of the transcriptomes and the K4-K36 domain profiles in four mouse cell types revealed ~1,600 intergenic regions that produce multi-exonic lncRNAs with relatively strong purifying selection in their genomic loci [originally termed as “lincRNAs” (large intervening non-coding RNAs), but later on more preferably referred to as “lncRNAs”] (Guttman et al., 2009). Attributed to the selection criteria, the genomic origin and structural properties of the lncRNAs identified by this approach are virtually indistinguishable from those of protein-coding mRNAs (Figure 1). Both classes of RNAs are transcribed by RNA polymerase II (RNAPII) from highly conserved promoters and undergo maturation processes such as 5′ capping, splicing and 3′ polyadenylation. This is also true for well-characterized lncRNAs such as HOTAIR, NRON, and Xist (Brown et al., 1992; Clemson et al., 1996; Rinn et al., 2007; Willingham et al., 2005). Subsequently, the GENCODE consortium within the framework of the ENCODE project performed the most comprehensive analysis of human lncRNAs and identified a total of 9,277 lncRNAs (Derrien et al., 2012). The analyses confirmed that lncRNAs show a high degree of similarity with protein-coding genes with regard to the chromatin architecture surrounding their origins, splicing signals, and exon/intron lengths. However, lncRNAs do have some distinguishable features. Although their promoter regions are conserved, human lncRNAs are under weaker selective constraints than protein-coding genes, and one-third seem to have arisen within the primate lineage (Ponting et al., 2009; Qureshi and Mehler, 2012). They display more tissue-specific expression patterns and tend to be shorter in length, containing fewer exons (Derrien et al., 2012). Co-expression clustering analysis between lncRNAs and protein-coding mRNAs offers an effective bioinformatic method to infer the function of lncRNAs on a global scale based on the known functions of co-expressed protein-coding genes. Such an analysis discovered that lncRNAs might be associated with a wide range of biological processes including cell proliferation, neuronal processes, and embryonic stem cell (ESC) pluripotency (Dinger et al., 2008; Guttman et al., 2009; Mercer et al., 2010; Ng et al., 2012).
Figure 1.
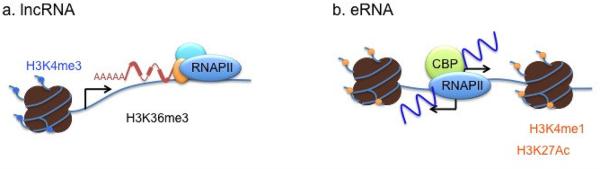
Genomic organization of stereotypical IncRNAs and eRNAs
Another class of long non-coding transcripts was recently identified from genome-wide characterization studies of enhancers in several mammalian cell types. Enhancer regions show high levels of mono-methylation at histone H3 lysine 4 (H3K4me1) without noticeable promoter-specific mark H3K4me3, and are typically bound by general transcription coactivators, such as CBP or p300 (Heintzman et al., 2009; Heintzman et al., 2007; Kim et al., 2010; Visel et al., 2009). A growing number of studies are showing that active enhancers directly recruit RNAPII to produce RNA transcripts, which are now collectively known as enhancer RNAs (eRNAs) (Creyghton et al., 2010; De Santa et al., 2010; Djebali et al., 2012; Hah et al., 2011; Kim et al., 2010; Koch and Andrau, 2011; Lam et al., 2013; Li et al., 2013; Melo et al., 2012; Mousavi et al., 2013; Rada-Iglesias et al., 2011; Wang et al., 2011a). Beside the unique chromatin architecture surrounding their genomic origin, eRNAs differ from previously characterized lncRNAs in that eRNAs are even shorter (< 2kb) than typical lncRNAs and show little evidence of maturation (Figure 1). Moreover, the majority of eRNAs are transcribed bi-directionally from the center of the enhancer domain and mainly reside inside the nucleus. The latest characterization of eRNAs in 15 human cell types performed by the ENCODE consortium, largely concurred with earlier findings but further revealed that eRNA-producing enhancers display stronger signals for H3K4me1, H3K27ac and H3K79me2 along with higher levels of RNA polymerase II compared to non-transcribed enhancers, and that eRNAs exhibit a great diversity in the levels of polyadenylation, subcellular localization, and transcriptional direction (Djebali et al., 2012). Notably, there is a strong concordance in the levels of expression between eRNAs and nearby mRNAs, suggesting a possible functional relationship between eRNAs and nearby mRNAs. Taken together, these studies imply that enhancers might play a more complex role in gene expression than previously appreciated (Creyghton et al., 2010; Djebali et al., 2012; Hah et al., 2011; Kim et al., 2010; Rada-Iglesias et al., 2011; Wang et al., 2011a).
LncRNAs in the brain
Brain development and function are tightly regulated by epigenetic mechanisms that modulate gene expression in response to intrinsic and extrinsic signals (Qureshi et al., 2010; Qureshi and Mehler, 2012). A growing body of evidence suggests that the lncRNAs may represent an important epigenetic network that plays integral roles in virtually every aspect of the gene expression process. LncRNAs are highly enriched in the brain, many of which show developmental stage- or region-specific expression patterns, derived from imprinted gene loci or key neural protein-coding genes, in cis-antisense, intronic, or bi-directional configurations. The collection of in situ hybridization data generated from the Alan Brain Atlas showed that over 800 lncRNAs expressed in the adult mouse brain are associated with specific neuroanatomical regions, cell types, or subcellular compartments (Mercer et al., 2008). The analysis of 659 evolutionary conserved intergenic lncRNAs revealed that those expressed in the brain are more frequently conserved and preferentially located adjacent to protein-coding genes (Ponjavic et al., 2009). The lncRNAs are also dynamically expressed during pluripotency and differentiation into neural or glial cells (Lin et al., 2011; Ng et al., 2012). Knock-down of four lncRNAs that are associated with neuronal differentiation alters cellular differentiation fate from a neurogenic to a gliogenic program, suggesting the functional role of the lncRNAs in neural cell fate specification (Ng et al., 2012). Evf2 is another lncRNA involved in neural development. It is transcribed from the ultraconserved enhancer regions critical for expression of the homeodomain transcription factors DLX5 and DLX6 in the developing mouse forebrain (Feng et al., 2006). The Evf2 transcript was shown to form a stable complex with another homeodomain transcription factor DLX2 to activate the Dlx5/6 enhancer in a target and homeodomain-specific manner (Bond et al., 2009). Evf2 mouse mutants generated by the insertion of transcription termination sites into exon 1 exhibit reduced numbers of GABAergic interneurons in early postnatal hippocampus and dentate gyrus, providing the in vivo evidence for its functional significance. However disruption of Evf2 specifically increased Dlx5/6 expression, which is opposite of the expected result based on its proposed function. In a rescue experiment, a low level of ectopic Evf2 expression decreased (rescued) Dlx5 expression but not Dlx6, whereas a higher level of Evf2 increased both, suggesting a dose-dependent effect of Evf2. Further analysis showed that the perplexing result might result from the involvement of the negative transcription factor MeCP2, which also binds to the Dlx5/6 enhancer region in an Evf2-dependent manner (Bond et al., 2009). The result of ectopic Evf2 expression suggests that the Evf2 transcript might be involved only in Dlx5 transcriptional control. As the Evf2 transcribing region encompasses the Dlx6 gene in an anti-sense direction, it was also proposed that Evf2 mediated transcriptional control of the Dlx6 gene would involve anti-sense inhibition in cis.
A few examples also illustrate the regulatory roles of lncRNAs in synaptic development, maintenance, and plasticity (Bernard et al., 2010:,Mercer, 2010 #1348:Ip, 2012 #1349; Eom et al., 2011; Ling et al., 2011; Wang et al., 2002; Zhong et al., 2009). For example, a lncRNA, Malat1 (metastasis-associated lung adenocarcinoma transcript 1; also known as NEAT2) is abundantly expressed in neurons and regulates synapse formation by modulating the expression of genes involved in synapse formation and/or maintenance. Malat1 is enriched in nuclear speckles together with the pre-mRNA- splicing factor SF2/ASF and modulates the recruitment of SR proteins to the transcription sites of genes involved in nuclear and synapse function (Bernard et al., 2010). Other lncRNAs that are potentially implicated in modulating synapse development and plasticity are eRNAs. The eRNAs were first identified as a class of novel lncRNAs that is specifically expressed from activity-induced enhancers in mouse cortical neurons. Neuronal activity triggers rapid induction of eRNAs from more than 2,000 neuronal enhancers genome-wide in a highly correlative manner with nearby protein-coding genes, suggesting a possible “activating” function of the eRNAs (Kim et al., 2010). Although the exact function and mechanism of eRNAs in neuronal gene expression are yet to be determined, recent functional studies of eRNAs expressed in other cell types provided evidence supporting their role in gene expression (Lam et al., 2013; Li et al., 2013; Melo et al., 2012; Mousavi et al., 2013).
Several genetic loci associated with a wide range of neurological and psychiatric disorders also express lncRNAs either bi-directionally or in anti-sense orientation (Knauss and Sun, 2013; Pastori and Wahlestedt, 2012; Ponjavic et al., 2007). FMR4 is a primate-specific lncRNA derived from upstream of the fragile X mental retardation gene (FMR1) in an anti-sense orientation and was shown to have a possible role in cell survival (Khalil et al., 2008). A triplet nucleotide repeat expansion within the 5′ UTR of FMR1 underlies the pathogenesis of the fragile X syndrome (FXS) and interestingly the same triplet repeat also silences the expression of FMR4, suggesting the possible involvement of FMR4 in aspects of the clinical presentation of fragile X syndrome and/or related disorders. However, as knock-down of FMR4 does not affect FMR1 expression, FMR4 might independently contribute to FXS. Spinocerebellar ataxia type 7 (SCA7) is a neuro-degenerative disorder caused by CAG/polyglutamine repeat expansions in the ataxin-7 gene. SCAANT1 (spino-cerebellar ataxia 7 antisense noncoding transcript 1) is an anti-sense lncRNA expressed from an alternative promoter located adjacent to the ataxin-7 repeat. Its knock-down causes derepression of the ataxin-7 gene, suggesting a possible role in the repression of ataxin-7 in cis (Sopher et al., 2011). Transcription of anti-sense lncRNAs have also been observed in genomic loci of genes implicated in Angelman syndrome (Ube3a-ATS) (Huang et al., 2012), neuro-degeneration (BACE1-AS) (Faghihi et al., 2008), and psychiatric diseases such as schizophrenia and bipolar disease (DISC2 and PSZA11q14) (Blackwood et al., 2001; Millar et al., 2000; Sokolov et al., 2003). Although our current knowledge on the lncRNA-mediated functional mechanisms in the brain is very limited and mostly inferred based on expression correlations between the lncRNAs and associated protein-coding genes, widespread expression of lncRNAs in a manner that tightly associates with key genes implicated in neuropsychiatric diseases illustrates pivotal regulatory potentials of the lncRNA network in modulating a wide range of aspects in neural functions and development.
Epigenetic role for lncRNAs in gene silencing/repression
The ENCODE transcriptome analysis of nuclear and cytoplasmic fractions from six different cell lines clearly demonstrated that lncRNAs are predominantly localized in the nucleus (Djebali et al., 2012). Moreover many lncRNAs are significantly enriched in the chromatin fraction. This would imply a possible role of lncRNAs in the regulation of epigenetic and/or gene expression programs. Consistently, several studies have demonstrated that a common function of the lncRNAs is to recruit the repressive chromatin modifying complexes to create a repressive chromatin state (Gupta et al., 2010; Khalil et al., 2009; Ng et al., 2012; Zhao et al., 2010). The Polycomb repressive complex 2 (PRC2) mediates the di- or tri-methylation of Lys 27 of histone H3 (H3K27me2/3) through its enzymatic subunits EZH1 and EZH2 and is shown to be responsible for gene silencing (Margueron and Reinberg, 2011; Simon and Kingston, 2009). Expressed from the X-inactivation center (Xic), Xist forms an “Xist cloud” and recruits PRC2 to mediate XCI (Zhao et al., 2008). HOTAIR, expressed in the mammalian HOXC locus, is required for PRC2-mediated silencing of the Hoxd cluster in trans (Rinn et al., 2007). Increased expression of HOTAIR is observed in various types of cancers, and forced expression of HOTAIR can lead to altered histone H3 lysine 27 methylation, gene expression, and increased cancer invasiveness and metastasis in a manner dependent on PRC2 (Gupta et al., 2010). The lncRNA-mediated PRC2 recruitment can also play a role in gene imprinting. Kcnq1ot1 is a 91kb-long ncRNA that is expressed from the paternal allele, reciprocal to the expression of several maternally expressed imprinted gene clusters (Kanduri et al., 2006). Kcnq1ot1 interacts with PRC2 as well as the H3K9-specific histone methyltransferase G9a in a lineage-specific manner to mediate the silencing of the Kcnq1 gene in the paternal allele in placenta (Pandey et al., 2008). Similarly, a lncRNA, Air (Antisense to Igf2r RNA; also known as Airn, which stands for Antisense to Igf2r RNA Noncoding) is paternally expressed from the second intron of the Igf2r gene and recruits G9a to mediate allele-specific silencing of the cis-linked Slc22a3, Slc22a2, and Igf2r genes in mouse placenta (Nagano et al., 2008). Subsequently, genome-scale analyses have demonstrated that a substantial proportion of lncRNAs do physically interact with repressive chromatin-modifying complexes such as PRC2 and coREST, suggesting that repressive chromatin modification through the recruitment of histone modifying complexes is a shared mechanism for many lncRNAs (Khalil et al., 2009; Ng et al., 2012; Zhao et al., 2010). Besides histone modifications, lncRNAs such as Tsix and Kcnq1ot1 have been shown to modulate the DNA methylation state at the promoter regions of their target genes (Xist and Kcnq1, respectively) by interacting with a DNA methyltransferase (Mohammad et al., 2010; Sado et al., 2006). Taken together, the generation of repressive chromatin architecture, and resulting gene silencing/repression are clearly a common shared mechanism of lncRNA function.
Transcriptional repression mediated by lncRNAs
Many lncRNAs have been shown to act as negative regulators of transcription. In quiescent cells, expression of the gene encoding dihydrofolate reductase (DHFR) is suppressed by a lncRNA initiated from the upstream minor promoter (Martianov et al., 2007). The defined biochemical mechanism involves the formation of a stable complex between the non-coding RNA and the major promoter through a direct interaction of the non-coding RNA with the general transcription factor IIB, leading to the dissociation of the preinitiation complex from the major promoter. Another example comes from two lncRNAs that are induced by p53 upon DNA damage. LncRNA-p21 is a direct target of p53 and serves as a repressor in p53-dependent transcriptional responses by negatively regulating the expression of hundreds of p53 target genes through the physical association with heterogeneous nuclear ribonucleoprotein K (hnRNP-K) (Huarte et al., 2010). PANDA is one of five lncRNAs induced by p53 from the CDK1A promoter and functions as a decoy for the transcription factor NF-YA to limit expression of pro-apoptotic genes and enable cell-cycle arrest (Hung et al., 2011). Another decoy mechanism for a transcription regulator is shown by Gas5. Upon growth-arrest, Gas5 is abundantly expressed to promote cellular apoptosis by suppressing glucocorticoid-dependent induction of several responsive genes (Kino et al., 2010). The region of the Gas5 sequence between nucleotides 400 and 598 can mimic the glucocorticoid response element (GRE), thus competing with cognate DNA GRE sites for binding to the glucocorticoid receptor (GR). These studies illustrate an emerging role of lncRNAs in negative regulation of the transcriptional process via diverse biochemical mechanisms.
Epigenetic role for lncRNAs in gene activation
Although the majority of known lncRNA functions are implicated in gene silencing or repression, several lncRNAs have been shown to function in gene activation by establishing transcriptionally competent chromatin structure at their target gene loci. Systematic loss-of-function (LOF) analysis of lncRNAs expressed in mouse ES cells found that comparable numbers of activated and repressed genes were observed upon knock-down of the lncRNAs, suggesting that lncRNAs regulate gene expression in either direction (Guttman et al., 2011). In Drosophila, Trithorax (TrxG) group genes are defined as suppressors of Polycomb (PcG) phenotypes and are critically required for homeotic gene expression throughout development (Schuettengruber et al., 2011). A member of the TrxG group, ASH1 is a histone methyltransferase that can methylate lysine residues 4 and 9 in H3 and 20 in H4. As an epigenetic activator, ASH1 maintains an activated transcription state for the expression of the homeotic gene Ultrabithorax (Ubx) in the third-leg and haltere imaginal discs. The specific targeting of ASH1 to the Ubx gene is mediated by noncoding RNAs that are expressed from three trithorax response elements (TREs) located 22kb upstream of the Ubx promoter (Sanchez-Elsner et al., 2006). HOTTIP mediates coordinated activation of Hoxa genes (Wang et al., 2011b). Its knock-down leads to a broad loss of H3K4me2/3 across the Hoxa locus and a decrease in distal Hox gene transcription. Although expressed from the distal end of the human Hoxa cluster, HOTTIP can be physically located in close proximity to Hoxa gene promoters via pre-configured chromatin looping. There it is able to recruit a member of the Mixed Lineage Leukemia (MLL) family of SET domain-containing lysine methyltransferases, MLL1 through an interaction with the adaptor protein WDR5. Therefore lncRNAs can play an epigenetic role in gene activation by serving as an intermediate in targeting a chromatin modifier to specific cis-regulatory regions.
Enhancer-like activity of lncRNAs
As seen from these examples, lncRNAs often regulate the expression of neighboring protein-coding genes, which is reminiscent of enhancer activity. In fact, a recent study characterizing the function of a set of new GENCODE annotated lncRNAs further demonstrated an enhancer-like mechanism of lncRNAs (Orom et al., 2010). Knock-down of a subset of the lncRNAs, termed ncRNA-activating (ncRNA-a) led to a decrease in neighboring protein-coding genes, and heterologous reporter assays revealed that the sequences corresponding to the lncRNA transcription unit can activate expression of a heterologous promoter in an orientation-independent manner, but cannot act as a promoter itself. Replacement of the lncRNA unit with irrelevant protein-coding genes, while keeping the endogenous lncRNA promoter, failed to display increased gene expression, suggesting that the lncRNA itself but not its transcription is important for the potentiation of gene expression. A subsequent study further revealed that the ncRNA-a specifically interacts with a transcriptional co-activator complex, Mediator to induce chromatin looping between the ncRNA-a loci and target gene promoters, and kinase activity towards histone H3 serine 10, a histone modification that is known to be associated with transcriptional activation (Lai et al., 2013). The ncRNA-a was shown to bind directly to the MED12 subunit and interestingly, the Mediator complex containing disease-causing mutations within the MED12 subunit (Opitz–Kaveggia syndrome) significantly diminishes its association with ncRNA-a, demonstrating the clinical significance of the ncRNA-a-dependent function. Another example of enhancer-like lncRNA is NeST (nettoie Salmonella pas Theiler’s [cleanup Salmonella not Theiler’s]), which is located adjacent to the interferon (IFN)-γ-encoding gene in both mice (Ifng) and humans (IFNG) (Gomez et al., 2013). This genomic region is part of the Tmevp3 (Theiler’s murine encephalitis virus persistence 3) locus, one of the loci responsible for strain-specific variations in the ability of inbred mice to clear Theiler’s infection. The SJL/J-derived alleles confer higher NeST expression, increased interferon-γ in activated CD8+ T cells, increased Theiler’s virus persistence, and decreased Salmonella enterica pathogenesis. Interestingly, transgenic expression of SJL/J-derived NeST alone was sufficient to confer all phenotypes of the SJL/J locus. Further analysis revealed that overexpressed NeST could increase the expression of interferon-γ by CD8+ T cells by promoting an active chromatin state at the interferon-γ locus. Just like the HOTTIP action at the Hoxa locus (Wang et al., 2011b), NeST recruits the MLL/SET1 H3K4 methyltransferase complex to the interferon-γ locus by interacting with the WDR subunit of the complex. One noticeable feature of NeST is the ability to regulate target genes in trans, which has not been shown by other activating lncRNAs (HOTTIP and ncRNA-a). Therefore, understanding the molecular nature of various lncRNAs underlying the target specificity will be an important subject of future study.
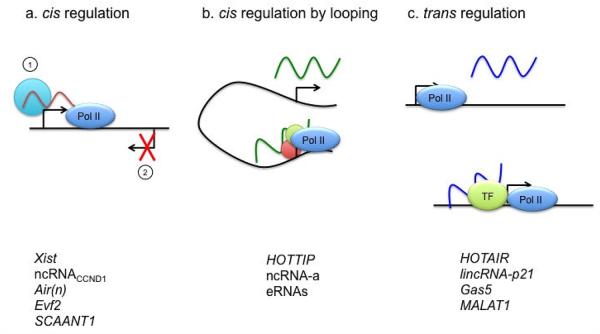
LncRNA action in cis
Several unique properties of lncRNAs make them ideal cellular machinery for regulating their targets that are located on or near the site of their synthesis (cis-regulation) (Figure 2). Conceptually, lncRNAs have great potential to function as tethering or guidance molecules in that unlike protein-coding mRNAs, they can act on sites where they are synthesized without needing to leave the nucleus. One proposed mechanism is that 5′ sides of long ncRNA transcripts might bind to protein partners as soon as they are synthesized, while transcriptionally lagging 3′ ends are still tethered to chromatin by transcriptionally engaged RNA polymerase II (Lee, 2012). Additional features of many lncRNAs suitable for cis-regulation are their relatively fast turnover rates and low copy numbers (Cabili et al., 2011; Clark et al., 2012; De Santa et al., 2010; Derrien et al., 2012; Sun et al., 2006a; Tani et al., 2012). Although these properties could also be used for favoring the transcriptional noise idea, the relatively short half-life of the lncRNAs and possible tethering mechanism would provide an effective way for locus- or allele-specific regulation by preventing lncRNAs from diffusing away from where they are generated. Specific PRC2 targeting to the X inactivation center (Xic) might be mediated by this type of lncRNA-dependent mechanism. Both Xist and a 1.6 kb-long ncRNA, RepA expressed within the Xist area harbor a common repeat A motif at the very 5′ end, which was shown to be responsible for PRC2 binding (Zhao et al., 2008). It’s been proposed that the interaction between PRC2 and RepA might occur co-transcriptionally to hold the RNA-protein complex in place for its cis action. An additional mechanism to confine the Xist activity in cis involves Polycomb group transcription factor, YY1, which can load the Xist-PRC2 complex onto the Xic through its bivalent capability of binding both Xist and DNA regions within the first exon of Xist (Jeon and Lee, 2011). Therefore the combinatorial action of co-transcriptional Xist tethering and YY1 acting as an adaptor between Xist and its chromatin target explains how PRC2-dependent XCI is mediated by Xist in cis. Another example of the RNA tethering mechanism could be seen by the action of the lncRNA (ncRNACCND1) expressed from the 5′ regulatory region of the cyclin D1 (CCND1) gene (Wang et al., 2008). Upon DNA damage signals, ncRNACCND1 is rapidly expressed, but in a low-copy number, and recruits an RNA binding protein, TLS (translocated in liposarcoma) to inhibit CBP/p300 activities on the CCND1 gene in cis, causing gene-specific repression. Subcellular fractionation studies found that ncRNACCND1 is mainly bound to chromatin. Although the exact mechanism of tethering remains to be determined, the observed cis-regulation could be mediated by the action of ncRNACCND1 tethered to transcribing RNA polymerase II.
Figure 2.
Targeting mechanisms of IncRNAs
Depending on the sequence complementarity, lncRNAs could be tethered to a particular chromosomal region through the formation of a DNA:RNA triplex, possibly via Hoogsteen base pairing. The pRNA (promoter-associated RNA) is an ncRNA derived from the RNA polymerase I-mediated transcription of ribosomal DNA (rDNA) repeat units, and its sequence is complementary to the ribosomal DNA promoter (Schmitz et al., 2010). The formation of this DNA:RNA triplex structure occurs at the binding site of TTF-1, which is the major transcription factor for ribosomal RNA transcription. The triplex structure causes silencing of the rRNA gene via two independent mechanisms. In addition to preventing TTF-1 binding, it can also recruit a de novo DNA methyltransferase, DNMT3b to facilitate DNA methylation at the promoter region. The transcriptional repression of the DHFR gene in quiescent cells is also mediated by a DNA:RNA hybrid that is formed between the major promoter region of the DHFR gene and the complimentary lncRNA expressed from the upstream minor promoter (Martianov et al., 2007).
Natural antisense transcripts (NATs)
A prominent feature of the lncRNA expression profile is that many of them are transcribed from the DNA strand opposite to a protein-coding gene in an overlapping fashion, therefore sharing at least some sequence complementarity with their counterpart sense transcripts. This group of lncRNAs is collectively defined as Natural Antisense Transcripts (NATs) (Lapidot and Pilpel, 2006; Magistri et al., 2012). Genome-wide analyses of several eukaryotic species reported that 5-29% of transcription units account for sense-antisense pairing (Chen et al., 2004; Katayama et al., 2005; Sun et al., 2006b; Yelin et al., 2003). Aforementioned lncRNAs, Xist and Kcnq1ot1 also belong to the NAT class. Intriguingly a growing list of validated sense-antisense pairs is implicated in brain development and various types of human diseases such as Alzheimer’s disease (BACE1 and BACE1-AS) and Fragile X mental retardation (FMR1 and ASFMR1) (Faghihi and Wahlestedt, 2009). Due to their transcription orientation and overlapping feature with corresponding sense transcripts, NATs are well suited for cis-regulation of sense strand transcription at multiple levels including imprinting, RNA processing and export, and transcriptional regulation. Consistently, a systematic loss-of-function RNAi approach targeting 797 antisense transcripts conserved between human and mouse provided evidence for a regulatory role for a number of natural antisense transcripts (Faghihi et al., 2010a).
The mechanisms for the antisense-mediated regulation of sense mRNA can largely be grouped into three different categories: mechanisms related to transcription (including epigenetic interactions), RNA–DNA interactions, and RNA–RNA interactions (Faghihi and Wahlestedt, 2009). Several examples illustrated that the act of transcription in the antisense direction could alter transcription of sense RNA (see more details in the following section). As seen from the action mechanisms of Kcnq1ot1 and Xist, a key mechanism of antisense transcript-mediated silencing of sense RNA transcription involves conversion of chromatin architecture to a repressive state through the interactions of locally accumulated NATs with DNA and/or chromatin modifying enzymes (Bernstein and Allis, 2005). ANRIL is a NAT expressed from the genomic locus encoding three tumor suppressor genes, p15INK4b, p16INK4a, and a regulator of the p53 pathway, ARF (Pasmant et al., 2007). The INK4a/ARF/INK4b locus is often found to be deleted or silenced in various types of human cancers (Kim and Sharpless, 2006; Popov and Gil, 2010). Expression levels of all three of the genes are low in young and normal cells but upregulated in aging cells or upon oncogenic stimuli. In concert with a repressive histone mark, H3K27me3, ANRIL mediates the silencing of the locus by physically interacting with a subunit of polycomb repressive complex 1 (PRC1), chromobox 7 (CBX7) (Yap et al., 2010). The polycomb repressive complex 2 (PRC2) was also shown to contribute to the silencing of the gene cluster through a similar interaction with ANRIL (Kotake et al., 2011). Both in human and mouse, expression of Brain-derived neurotrophic factor (Bdnf) is also regulated by a NAT, Bdnf-AS, which has 225 nucleotides of full complementarity to all 11 splice variants of Bdnf mRNA (Modarresi et al., 2012). Knockdown of Bdnf-AS increases Bdnf mRNA expression in vivo and is accompanied by a specific reduction in the levels of the H3K27me3 mark in both the sense-antisense overlapping region and in the upstream Bdnf promoter region. The binding level of a PRC2 component, EZH2 was also reduced near the Bdnf promoter region. NATs can also function as cis-acting epigenetic activators. During the primitive streak phase of embryoid bodies (EB) differentiation, two novel NATs, Evx1as and Hoxb5/6as are concordantly up-regulated with their associated homeotic genes, Evx1 and Hoxb5/6, respectively (Dinger et al., 2008). RNA-ChIP analysis showed that both NATs were associated with H3K4me3 chromatin fractions and the mammalian trithorax protein MLL, which can trimethylate H3K4. Therefore, the role of NATs in the alteration of chromatin structure in cis can influence the expression of neighboring genes positively and negatively. Due to the sequence complementarity with the sense transcript, a NAT can form duplex RNA, which then possibly influences several sense RNA processing steps such as splicing, editing, stability, and translation (Faghihi and Wahlestedt, 2009; Guil and Esteller, 2012). Supporting this notion, a large scale genome-wide analysis of RNA expression data from 176 lymphoblastoid cell lines revealed that the majority of expressed sense–antisense pairs exhibited alternative splicing events that were correlated to the expression of the antisense gene (Morrissy et al., 2011). During the Snail1-induced epithelial–mesenchymal transition (EMT), translational up-regulation of a transcriptional repressor of E-cadherin, Zeb2 is mediated by a NAT that overlaps the 5′-UTR intron region of Zeb2. This intron contains an internal ribosome entry site (IRES) necessary for the expression of Zeb2. Snail1-dependent induction or ectopic expression of the Zeb2 NAT correlates with the inclusion of the 5′ UTR intron, leading to a model where the Zeb2 NAT prevents binding of the spliceosome to the 5′ splice site possibly by forming an RNA duplex. Consequently, the intron present in the 5′-UTR is conserved. RNA duplex formation was also shown to increase mRNA stability by masking miRNA targeting sequences. BACE-AS is a NAT expressed from the β-secretase-1 (BACE1) locus and exhibits 104 nucleotides of full complementarity to exon 6 of human BACE1 mRNA, which contains a miR-485-5p binding site (Faghihi et al., 2008; Faghihi et al., 2010b). BACE-AS forms an RNA duplex with BACE mRNA and increases BACE mRNA stability by preventing miRNA- induced BACE1 mRNA silencing. As a central enzyme in the Alzheimer’s disease pathophysiology, BACE1 proteins are present at a higher level in brains of Alzheimer’s patients compared with unaffected controls. Importantly up-regulation of BACE1-AS was also observed in subjects with Alzheimer’s disease and in amyloid precursor protein transgenic mice, highlighting the functional significance of NAT regulation in human diseases (Faghihi et al., 2010b).
The role of ncRNA transcription
Despite many examples showing the biological roles for mature lncRNA transcripts, several studies have also reported that the act of transcription, but not the transcript itself, is functional. The most straightforward case for this type of regulation would be when a target gene is located nearby or is physically overlapping a lncRNA gene either in the sense or antisense direction. In this arrangement, transcription of lncRNA can block transcription of a neighboring gene in cis by the transcriptional interference (TI) mechanism, which was originally identified in prokaryotes and yeast (Mazo et al., 2007). The SER3 gene in S. cerevisiae is repressed during growth in rich medium through the expression of the upstream ncRNA, SRG1 (Martens et al., 2004). The TI occurs as SRG1 transcription extends past the SER3 promoter. Replacement of the SRG1 sequence with the sequence for the URA3 gene still showed SER3 repression. However, insertion of a transcription-termination sequence before the SER3 promoter led to the derepression of the SER3 gene, suggesting that the act of SRG1 transcription is important. A subsequent study revealed that SRG1 transcription causes repression of SER3 by directing a high level of nucleosomes over the SER3 promoter, suggesting that chromatin remodeling is the underlying mechanism of the TI (Hainer et al., 2011). Transcription of ncRNA can also do the opposite. In S. pombe, transcription of several species of ncRNAs initiated upstream of the fbp1+ gene progressively converts the chromatin region at the fbp1+ gene to an open configuration, which in turn leads to the translocation of RNA polymerase II to the transcription start site of the fbp1+ gene (Hirota et al., 2008). This effect was abolished when a transcription terminator is inserted into the upstream region. In erythroid cells, the HS2 enhancer within the β-globin locus control region initiates ncRNA transcription through the intervening DNA region into the cis-linked promoter and gene, but in a manner that is independent of a cis-linked globin promoter (Ashe et al., 1997; Johnson et al., 2003). Insertion of a transcription terminator or an insulator between HS2 and the ε-globin gene promoter in stably integrated reporter genes not only blocks the elongation of enhancer-initiated transcription but also reduces the level of mRNA synthesized from the ε-globin gene promoter (Ling et al., 2004; Ling et al., 2005; Zhao and Dean, 2004). In addition, blocking the enhancer initiated transcription results in a widespread decrease in histone acetylation at the intervening DNA region as well as the target gene. Although these findings cannot rule out the possibility of the HS2 initiated ncRNA transcript having a role in target gene expression, it was proposed that the tracking of the HS2 enhancer-assembled transcriptional machinery to the cis-linked promoter might be critical for enhancer-mediated gene activation. In some cases, the act of transcription and resulting lncRNA transcripts have independent biological functions. A paternally expressed lncRNA, Air(n) silences in cis the paternal alleles of imprinted genes, Igf2r, Slc22a3, and Slc22a2 (Sleutels et al., 2002). Interestingly, Igf2r is silenced in all developmental stages, whereas Slc22a3 and Slc22a2 are only silenced in some extraembryonic lineages, providing a hint that Air(n) might differentially regulate these genes (Hudson et al., 2011; Yamasaki et al., 2005). It turns out that the Air(n) transcript accumulates at the Slc22a3 promoter and recruits the H3K9 histone methyltransferase G9a to repress Slc22a3 expression (Nagano et al., 2008). However allele-specific silencing of the Igf2r gene neither requires the action of the Air(n) transcript nor is dependent on PcG proteins or DNA methylation, suggesting that the alteration in chromatin structure does not play a role in this process. A recent in vivo study showed that it is not the Air(n) transcript but instead transcription activity of the Air(n) gene that is sufficient to silence the Igf2r gene (Latos et al., 2012). As the Air(n) transcribing region overlaps with the Igf2r promoter, Air(n) transcription interferes with RNA polymerase II binding to the Igf2r gene promoter even in the absence of repressive chromatin.
Cis-regulation mediated by chromatin looping
Chromatin looping allows another layer of the cis regulatory mechanism by which lncRNAs expressed from a distant location can be physically in close proximity to their target genes (Figure 2). For example, HOTTIP, an activating lncRNA is synthesized at the 5′ tip of the Hoxa gene cluster and chromatin looping brings HOTTIP near target Hoxa genes to allow coordinate activation (Wang et al., 2011b). Chromatin looping is also likely to play a role in the lncRNA-dependent repression/silencing of long distance genes as 3C-on-chip (4C) analysis in Drosophila discovered large interaction networks between PcG protein target genes (Tolhuis et al., 2011). On the other hand, some lncRNAs were shown to facilitate chromatin looping. The Mediator-cohesion complex was previously shown to function in connecting the enhancers and core promoters of active genes by facilitating chromatin looping (Kagey et al., 2010). The enhancer-like lncRNAs, ncRNA-as recruit the Mediator complex to induce chromosomal pairing with specific target genes and promote their transcription. Supporting this model, depletion of Mediator subunits or ncRNA-a reduced the chromatin looping between the two loci (Lai et al., 2013).
eRNAs: functional RNA or transcription noise?
Although eRNAs exhibit concordant expression patterns with nearby protein-coding RNAs, the low abundance and little evidence of maturation shown by the majority of eRNAs initially casted doubt on the possibility of eRNAs having a regulatory function in gene expression. However, recent studies have provided multiple lines of evidence supporting regulatory roles of eRNAs in gene activation. Knock-down of several eRNAs caused reduction in the expression of nearby target genes in various cell types (Lam et al., 2013; Li et al., 2013; Melo et al., 2012; Mousavi et al., 2013). These eRNAs are non-polyadenylated and bi-directionally transcribed from the defined enhancer regions (high levels of H3K4me1 and H3K27ac, and low level of H3K4me3), therefore resembling initially discovered neuronal eRNAs (Kim et al., 2010). Characterization of eRNAs expressed from estrogen receptor α(ER-α)-bound enhancers in MCF-7 human breast cancer cells demonstrated that eRNAs increase the strength of specific enhancer-promoter looping in part by interacting with cohesin (Li et al., 2013). Consistently, the artificial tethering of an eRNA upstream of a minimal promoter in a plasmid-based reporter system enhances the reporter gene expression. A similar reporter plasmid-based study in mouse macrophages further suggested that the activating function of eRNAs appears to be sequence or strand-specific although the critical determinants of eRNA action have not been identified (Lam et al., 2013). Although chromatin looping appears to be a critical regulatory step on which distinct classes of activating lncRNAs commonly act, a recent study in mouse C2C12 skeletal muscle cells found that eRNAs could facilitate the transcription activation step after the enhancer-promoter looping has occurred (Mousavi et al., 2013). Knock-down of eRNA (CERNA) expressed from the core enhancer region (CE) upstream of the Myod1 gene did not alter the chromatin occupancies of the cohesin complex, which facilitates enhancer-promoter looping. Instead chromatin accessibility and subsequent assembly of RNAPII transcription machinery at the promoter region of the Myod1 gene was diminished. Therefore eRNAs appear to be functionally engaged in multiple stages of enhancer-mediated transcription activation.
LncRNA action in trans
Despite several prominent examples of lncRNA action in cis, a recent loss-of-function analysis on 226 lncRNAs present in mouse ESCs has shown that the majority of the lncRNAs affect gene expression in trans, affecting a large number of genes at great distances, including at other chromosomes (Guttman et al., 2011) (Figure 2). Initial analysis of HOTAIR expressed from the Hoxc locus showed that it mediates transcriptional repression of the Hoxd locus located on a different chromosome via PRC2 recruitment (Gupta et al., 2010; Rinn et al., 2007). Subsequent genome-wide analysis of HOTAIR occupancy revealed 832 HOTAIR occupancy sites across the genome in MDA-MB-231 breast cancer cells occurring on multiple chromosomes in addition to the Hoxd locus. Supporting its role in trans, over-expression of HOTAIR in epithelial cancer cells induced genome-wide re-targeting of PRC2, leading to altered H3K27me3, gene expression, and increased cancer invasiveness and metastasis in a manner dependent on PRC2. The lincRNA-p21 also acts as a trans-acting repressor regulating hundreds of p53-dependent gene targets through the physical association with hnRNP-K (Huarte et al., 2010). Although these studies provide clear functional evidence for lncRNA function in trans, the precise mechanism by which these lncRNAs can target specific loci remains to be determined. As seen from the cis-regulatory lncRNAs, targeting of the trans-acting lncRNAs could be mediated by forming DNA:RNA triple-helical structures. Alternatively, unknown transcription factors could play a role in the recruitment of the lncRNAs to their targets. In the case of Gas5, a spliced ~1kb-long lncRNA induced upon starvation and growth arrest, the transacting mechanism for repressing a large number of GR-regulated genes does not involve targeting of the Gas5 to specific loci (Kino et al., 2010). Instead, expressed Gas5 competes with DNA GREs for binding to the GR by the portion of its sequences that mimic the GRE. PANDA also utilizes the decoy mechanism to inhibit expression of apoptotic genes to cause cell-cycle arrest through sequestration of NF-YA, a transcription factor required for activating the apoptotic program upon DNA damage (Hung et al., 2011).
Perspectives
A great extent of the functional flexibility and specificity of the lncRNAs that can be created by their intrinsic structural properties and biogenesis mechanism can certainly offer some functional advantages over proteins in precise control over many biological processes. Recent functional studies performed in non-neuronal cell types have revealed that lncRNAs can function as a guide, decoy, or scaffold and such a versatile capacity of the lncRNA allows for their key epigenetic roles in diverse aspects of transcriptional, epigenetic and nuclear processes (Kung et al., 2013; Wang et al., 2011a). Nuclear gene expression in neurons represents a cell-wide adaptation mechanism that mediates the long-lasting form of synaptic and behavioral plasticity by allowing the stimulus-specific production and deployment of the effector proteins that stably alter neural function (Lyons and West, 2011). Sensory experience-evoked synaptic activity triggers various calcium-dependent signaling events that ultimately regulate the expression of a group of genes involved in various aspects of neuronal function (Flavell and Greenberg, 2008). Therefore recently emerged functions and mechanisms of the lncRNAs would also fit nicely into the gene regulatory mechanisms that are critically required for executing complex neurobiological processes.
In light of the lncRNA functionality in brain, evolutionary features of the lncRNA sequences provide interesting perspectives. Evolutionary studies initially casted doubt on the functionality of the lncRNAs as their sequences were much less conserved with many displaying rapid sequence evolution (Pang et al., 2006; Wang et al., 2004). However, further analyses have found that the lncRNAs show reduced nucleotide substitutions, insertions and deletions, both within their promoters and within their sequences, and higher conservation at their dinucleotide splice sites than expected by chance, suggesting that at least from an evolutionary point of view, some lncRNAs are likely to have a function that is moderately conserved among diverse ranges of species (Chodroff et al., 2010; Cogan et al., 2007; Guttman et al., 2010; Marques and Ponting, 2009; Ponjavic et al., 2009; Ponjavic et al., 2007; Ponting and Belgard, 2010; Ponting et al., 2009). Evf2 is an example for the case in which a lncRNA can be derived from evolutionarily conserved genomic elements. Evf2 is expressed from an ultra-conserved enhancer and functions to regulate expression of Dlx5/6 genes during forebrain development (Bond et al., 2009). Nonetheless, compared with protein-coding sequences, lncRNA sequences are less constrained in general. One interpretation could be that they might have been rapidly evolved in higher organisms to fulfill the demands for performing more complex and higher order functions such as human cognition. One example as evidence for such a positive selection is a lncRNA, HAR1F, which was identified as the region of the human genome that has undergone the most rapid sequence changes in the human lineage since divergence from the great apes (Pollard et al., 2006). HAR1F is co-expressed with Reelin in Cajal-Retzius neurons, suggesting its possible role in the establishment of regional forebrain organization. However, this exciting finding was later challenged by the arguments that the accelerated changes in HAR1F may not be the consequence of the activities of adaptive evolution, but instead is caused by biased gene conversion (BGC), a neutral process associated with recombination (Galtier and Duret, 2007; Ponting et al., 2009). While to what extent the HAR (Human accelerated regions) can be derived by pure positive selection is still an on-going subject of study (Duret and Galtier, 2009; Katzman et al., 2010; Kostka et al., 2012), it is also noteworthy that the relative amount of non-protein-coding sequence increases consistently with developmental complexity (Taft et al., 2007), which could in part explain the G value paradox–the inconsistent relationship between organismal complexity and the number of protein-coding genes (Hahn and Wray, 2002).
An increasing number of human genetic studies show that human disease-associated genetic mutations reside in noncoding and intergenic regions (Halvorsen et al., 2010; Simon-Sanchez and Singleton, 2008; Walsh et al., 2008). Given the pervasive nature of lncRNA transcription occurring throughout the mammalian genome, it is not difficult to predict that many mutations would be transmitted to the transcriptome, potentially affecting a large number of lncRNAs. As cumulative evidence supports a convergent view on the lncRNAs as a new layer of complexity in the molecular architecture of human diseases, further biochemical and functional characterizations will definitely be a worthwhile avenue of future neuroscience research to understand the lncRNA functions in fundamental biological processes and to establish their causal roles for human diseases, which should aid the development of novel diagnostic and therapeutic strategies.
Highlights.
Long non-coding RNAs (lncRNAs) are involved in various processes of gene regulation.
LncRNAs can be sub-divided based on their genomic organizations.
LncRNAs can be targeted in cis, cis by chromatin looping, and trans.
LncRNAs are enriched in the brain.
LncRNAs might modulate a wide range of aspects in neural functions and development.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errorsmaybe discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Ashe HL, Monks J, Wijgerde M, Fraser P, Proudfoot NJ. Intergenic transcription and transinduction of the human beta-globin locus. Genes & development. 1997;11:2494–2509. doi: 10.1101/gad.11.19.2494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bacher CP, Guggiari M, Brors B, Augui S, Clerc P, Avner P, Eils R, Heard E. Transient colocalization of X-inactivation centres accompanies the initiation of X inactivation. Nat Cell Biol. 2006;8:293–299. doi: 10.1038/ncb1365. [DOI] [PubMed] [Google Scholar]
- Bernard D, Prasanth KV, Tripathi V, Colasse S, Nakamura T, Xuan Z, Zhang MQ, Sedel F, Jourdren L, Coulpier F, et al. A long nuclear-retained non-coding RNA regulates synaptogenesis by modulating gene expression. EMBO J. 2010;29:3082–3093. doi: 10.1038/emboj.2010.199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein E, Allis CD. RNA meets chromatin. Genes & development. 2005;19:1635–1655. doi: 10.1101/gad.1324305. [DOI] [PubMed] [Google Scholar]
- Birney E, Stamatoyannopoulos JA, Dutta A, Guigo R, Gingeras TR, Margulies EH, Weng Z, Snyder M, Dermitzakis ET, Thurman RE, et al. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature. 2007;447:799–816. doi: 10.1038/nature05874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackwood DH, Fordyce A, Walker MT, St Clair DM, Porteous DJ, Muir WJ. Schizophrenia and affective disorders–cosegregation with a translocation at chromosome 1q42 that directly disrupts brain-expressed genes: clinical and P300 findings in a family. Am J Hum Genet. 2001;69:428–433. doi: 10.1086/321969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bond AM, Vangompel MJ, Sametsky EA, Clark MF, Savage JC, Disterhoft JF, Kohtz JD. Balanced gene regulation by an embryonic brain ncRNA is critical for adult hippocampal GABA circuitry. Nat Neurosci. 2009;12:1020–1027. doi: 10.1038/nn.2371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown CJ, Hendrich BD, Rupert JL, Lafreniere RG, Xing Y, Lawrence J, Willard HF. The human XIST gene: analysis of a 17 kb inactive X-specific RNA that contains conserved repeats and is highly localized within the nucleus. Cell. 1992;71:527–542. doi: 10.1016/0092-8674(92)90520-m. [DOI] [PubMed] [Google Scholar]
- Cabili MN, Trapnell C, Goff L, Koziol M, Tazon-Vega B, Regev A, Rinn JL. Integrative annotation of human large intergenic noncoding RNAs reveals global properties and specific subclasses. Genes & development. 2011;25:1915–1927. doi: 10.1101/gad.17446611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carninci P, Kasukawa T, Katayama S, Gough J, Frith MC, Maeda N, Oyama R, Ravasi T, Lenhard B, Wells C, et al. The transcriptional landscape of the mammalian genome. Science. 2005;309:1559–1563. doi: 10.1126/science.1112014. [DOI] [PubMed] [Google Scholar]
- Chen J, Sun M, Kent WJ, Huang X, Xie H, Wang W, Zhou G, Shi RZ, Rowley JD. Over 20% of human transcripts might form sense-antisense pairs. Nucleic Acids Res. 2004;32:4812–4820. doi: 10.1093/nar/gkh818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chodroff RA, Goodstadt L, Sirey TM, Oliver PL, Davies KE, Green ED, Molnar Z, Ponting CP. Long noncoding RNA genes: conservation of sequence and brain expression among diverse amniotes. Genome Biol. 2010;11:R72. doi: 10.1186/gb-2010-11-7-r72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chureau C, Prissette M, Bourdet A, Barbe V, Cattolico L, Jones L, Eggen A, Avner P, Duret L. Comparative sequence analysis of the X-inactivation center region in mouse, human, and bovine. Genome Res. 2002;12:894–908. doi: 10.1101/gr.152902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark MB, Johnston RL, Inostroza-Ponta M, Fox AH, Fortini E, Moscato P, Dinger ME, Mattick JS. Genome-wide analysis of long noncoding RNA stability. Genome Res. 2012;22:885–898. doi: 10.1101/gr.131037.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clemson CM, McNeil JA, Willard HF, Lawrence JB. XIST RNA paints the inactive X chromosome at interphase: evidence for a novel RNA involved in nuclear/chromosome structure. J Cell Biol. 1996;132:259–275. doi: 10.1083/jcb.132.3.259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cogan NO, Drayton MC, Ponting RC, Vecchies AC, Bannan NR, Sawbridge TI, Smith KF, Spangenberg GC, Forster JW. Validation of in silico-predicted genic SNPs in white clover (Trifolium repens L.), an outbreeding allopolyploid species. Molecular genetics and genomics : MGG. 2007;277:413–425. doi: 10.1007/s00438-006-0198-5. [DOI] [PubMed] [Google Scholar]
- Creyghton MP, Cheng AW, Welstead GG, Kooistra T, Carey BW, Steine EJ, Hanna J, Lodato MA, Frampton GM, Sharp PA, et al. Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proc Natl Acad Sci U S A. 2010;107:21931–21936. doi: 10.1073/pnas.1016071107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Santa F, Barozzi I, Mietton F, Ghisletti S, Polletti S, Tusi BK, Muller H, Ragoussis J, Wei CL, Natoli G. A large fraction of extragenic RNA pol II transcription sites overlap enhancers. PLoS Biol. 2010;8:e1000384. doi: 10.1371/journal.pbio.1000384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derrien T, Johnson R, Bussotti G, Tanzer A, Djebali S, Tilgner H, Guernec G, Martin D, Merkel A, Knowles DG, et al. The GENCODE v7 catalog of human long noncoding RNAs: analysis of their gene structure, evolution, and expression. Genome Res. 2012;22:1775–1789. doi: 10.1101/gr.132159.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinger ME, Amaral PP, Mercer TR, Pang KC, Bruce SJ, Gardiner BB, Askarian-Amiri ME, Ru K, Solda G, Simons C, et al. Long noncoding RNAs in mouse embryonic stem cell pluripotency and differentiation. Genome Res. 2008;18:1433–1445. doi: 10.1101/gr.078378.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Djebali S, Davis CA, Merkel A, Dobin A, Lassmann T, Mortazavi A, Tanzer A, Lagarde J, Lin W, Schlesinger F, et al. Landscape of transcription in human cells. Nature. 2012;489:101–108. doi: 10.1038/nature11233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duret L, Galtier N. Biased gene conversion and the evolution of mammalian genomic landscapes. Annual review of genomics and human genetics. 2009;10:285–311. doi: 10.1146/annurev-genom-082908-150001. [DOI] [PubMed] [Google Scholar]
- Eom T, Berardi V, Zhong J, Risuleo G, Tiedge H. Dual nature of translational control by regulatory BC RNAs. Mol Cell Biol. 2011;31:4538–4549. doi: 10.1128/MCB.05885-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faghihi MA, Kocerha J, Modarresi F, Engstrom PG, Chalk AM, Brothers SP, Koesema E, St Laurent G, Wahlestedt C. RNAi screen indicates widespread biological function for human natural antisense transcripts. PLoS One. 2010a;5 doi: 10.1371/journal.pone.0013177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faghihi MA, Modarresi F, Khalil AM, Wood DE, Sahagan BG, Morgan TE, Finch CE, St Laurent G, 3rd, Kenny PJ, Wahlestedt C. Expression of a noncoding RNA is elevated in Alzheimer’s disease and drives rapid feed-forward regulation of beta-secretase. Nat Med. 2008;14:723–730. doi: 10.1038/nm1784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faghihi MA, Wahlestedt C. Regulatory roles of natural antisense transcripts. Nature reviews Molecular cell biology. 2009;10:637–643. doi: 10.1038/nrm2738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faghihi MA, Zhang M, Huang J, Modarresi F, Van der Brug MP, Nalls MA, Cookson MR, St-Laurent G, 3rd, Wahlestedt C. Evidence for natural antisense transcript-mediated inhibition of microRNA function. Genome Biol. 2010b;11:R56. doi: 10.1186/gb-2010-11-5-r56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng J, Bi C, Clark BS, Mady R, Shah P, Kohtz JD. The Evf-2 noncoding RNA is transcribed from the Dlx-5/6 ultraconserved region and functions as a Dlx-2 transcriptional coactivator. Genes & development. 2006;20:1470–1484. doi: 10.1101/gad.1416106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flavell SW, Greenberg ME. Signaling mechanisms linking neuronal activity to gene expression and plasticity of the nervous system. Annu Rev Neurosci. 2008;31:563–590. doi: 10.1146/annurev.neuro.31.060407.125631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galtier N, Duret L. Adaptation or biased gene conversion? Extending the null hypothesis of molecular evolution. Trends in genetics : TIG. 2007;23:273–277. doi: 10.1016/j.tig.2007.03.011. [DOI] [PubMed] [Google Scholar]
- Gomez JA, Wapinski OL, Yang YW, Bureau JF, Gopinath S, Monack DM, Chang HY, Brahic M, Kirkegaard K. The NeST long ncRNA controls microbial susceptibility and epigenetic activation of the interferon-gamma locus. Cell. 2013;152:743–754. doi: 10.1016/j.cell.2013.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guil S, Esteller M. Cis-acting noncoding RNAs: friends and foes. Nature structural & molecular biology. 2012;19:1068–1075. doi: 10.1038/nsmb.2428. [DOI] [PubMed] [Google Scholar]
- Gupta RA, Shah N, Wang KC, Kim J, Horlings HM, Wong DJ, Tsai MC, Hung T, Argani P, Rinn JL, et al. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature. 2010;464:1071–1076. doi: 10.1038/nature08975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guttman M, Amit I, Garber M, French C, Lin MF, Feldser D, Huarte M, Zuk O, Carey BW, Cassady JP, et al. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature. 2009;458:223–227. doi: 10.1038/nature07672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guttman M, Donaghey J, Carey BW, Garber M, Grenier JK, Munson G, Young G, Lucas AB, Ach R, Bruhn L, et al. lincRNAs act in the circuitry controlling pluripotency and differentiation. Nature. 2011;477:295–300. doi: 10.1038/nature10398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guttman M, Garber M, Levin JZ, Donaghey J, Robinson J, Adiconis X, Fan L, Koziol MJ, Gnirke A, Nusbaum C, et al. Ab initio reconstruction of cell type-specific transcriptomes in mouse reveals the conserved multi-exonic structure of lincRNAs. Nat Biotechnol. 2010;28:503–510. doi: 10.1038/nbt.1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hah N, Danko CG, Core L, Waterfall JJ, Siepel A, Lis JT, Kraus WL. A rapid, extensive, and transient transcriptional response to estrogen signaling in breast cancer cells. Cell. 2011;145:622–634. doi: 10.1016/j.cell.2011.03.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn MW, Wray GA. The g-value paradox. Evolution & development. 2002;4:73–75. doi: 10.1046/j.1525-142x.2002.01069.x. [DOI] [PubMed] [Google Scholar]
- Hainer SJ, Pruneski JA, Mitchell RD, Monteverde RM, Martens JA. Intergenic transcription causes repression by directing nucleosome assembly. Genes & development. 2011;25:29–40. doi: 10.1101/gad.1975011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halvorsen M, Martin JS, Broadaway S, Laederach A. Disease-associated mutations that alter the RNA structural ensemble. PLoS Genet. 2010;6:e1001074. doi: 10.1371/journal.pgen.1001074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heintzman ND, Hon GC, Hawkins RD, Kheradpour P, Stark A, Harp LF, Ye Z, Lee LK, Stuart RK, Ching CW, et al. Histone modifications at human enhancers reflect global cell-type-specific gene expression. Nature. 2009;459:108–112. doi: 10.1038/nature07829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heintzman ND, Stuart RK, Hon G, Fu Y, Ching CW, Hawkins RD, Barrera LO, Van Calcar S, Qu C, Ching KA, et al. Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nat Genet. 2007;39:311–318. doi: 10.1038/ng1966. [DOI] [PubMed] [Google Scholar]
- Hirota K, Miyoshi T, Kugou K, Hoffman CS, Shibata T, Ohta K. Stepwise chromatin remodelling by a cascade of transcription initiation of non-coding RNAs. Nature. 2008;456:130–134. doi: 10.1038/nature07348. [DOI] [PubMed] [Google Scholar]
- Hon GC, Hawkins RD, Ren B. Predictive chromatin signatures in the mammalian genome. Human Molecular Genetics. 2009;18:R195–201. doi: 10.1093/hmg/ddp409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang HS, Allen JA, Mabb AM, King IF, Miriyala J, Taylor-Blake B, Sciaky N, Dutton JW, Jr., Lee HM, Chen X, et al. Topoisomerase inhibitors unsilence the dormant allele of Ube3a in neurons. Nature. 2012;481:185–189. doi: 10.1038/nature10726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huarte M, Guttman M, Feldser D, Garber M, Koziol MJ, Kenzelmann-Broz D, Khalil AM, Zuk O, Amit I, Rabani M, et al. A large intergenic noncoding RNA induced by p53 mediates global gene repression in the p53 response. Cell. 2010;142:409–419. doi: 10.1016/j.cell.2010.06.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson QJ, Seidl CI, Kulinski TM, Huang R, Warczok KE, Bittner R, Bartolomei MS, Barlow DP. Extra-embryonic-specific imprinted expression is restricted to defined lineages in the post-implantation embryo. Dev Biol. 2011;353:420–431. doi: 10.1016/j.ydbio.2011.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hung T, Wang Y, Lin MF, Koegel AK, Kotake Y, Grant GD, Horlings HM, Shah N, Umbricht C, Wang P, et al. Extensive and coordinated transcription of noncoding RNAs within cell-cycle promoters. Nat Genet. 2011;43:621–629. doi: 10.1038/ng.848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeon Y, Lee JT. YY1 tethers Xist RNA to the inactive X nucleation center. Cell. 2011;146:119–133. doi: 10.1016/j.cell.2011.06.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson KD, Grass JA, Park C, Im H, Choi K, Bresnick EH. Highly restricted localization of RNA polymerase II within a locus control region of a tissue-specific chromatin domain. Mol Cell Biol. 2003;23:6484–6493. doi: 10.1128/MCB.23.18.6484-6493.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kagey MH, Newman JJ, Bilodeau S, Zhan Y, Orlando DA, van Berkum NL, Ebmeier CC, Goossens J, Rahl PB, Levine SS, et al. Mediator and cohesin connect gene expression and chromatin architecture. Nature. 2010;467:430–435. doi: 10.1038/nature09380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanduri C, Thakur N, Pandey RR. The length of the transcript encoded from the Kcnq1ot1 antisense promoter determines the degree of silencing. EMBO J. 2006;25:2096–2106. doi: 10.1038/sj.emboj.7601090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapranov P, Cheng J, Dike S, Nix DA, Duttagupta R, Willingham AT, Stadler PF, Hertel J, Hackermuller J, Hofacker IL, et al. RNA maps reveal new RNA classes and a possible function for pervasive transcription. Science. 2007;316:1484–1488. doi: 10.1126/science.1138341. [DOI] [PubMed] [Google Scholar]
- Katayama S, Tomaru Y, Kasukawa T, Waki K, Nakanishi M, Nakamura M, Nishida H, Yap CC, Suzuki M, Kawai J, et al. Antisense transcription in the mammalian transcriptome. Science. 2005;309:1564–1566. doi: 10.1126/science.1112009. [DOI] [PubMed] [Google Scholar]
- Katzman S, Kern AD, Pollard KS, Salama SR, Haussler D. GC-biased evolution near human accelerated regions. PLoS Genet. 2010;6:e1000960. doi: 10.1371/journal.pgen.1000960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khalil AM, Faghihi MA, Modarresi F, Brothers SP, Wahlestedt C. A novel RNA transcript with antiapoptotic function is silenced in fragile X syndrome. PLoS One. 2008;3:e1486. doi: 10.1371/journal.pone.0001486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khalil AM, Guttman M, Huarte M, Garber M, Raj A, Rivea Morales D, Thomas K, Presser A, Bernstein BE, van Oudenaarden A, et al. Many human large intergenic noncoding RNAs associate with chromatin-modifying complexes and affect gene expression. Proc Natl Acad Sci U S A. 2009;106:11667–11672. doi: 10.1073/pnas.0904715106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim TK, Hemberg M, Gray JM, Costa AM, Bear DM, Wu J, Harmin DA, Laptewicz M, Barbara-Haley K, Kuersten S, et al. Widespread transcription at neuronal activity-regulated enhancers. Nature. 2010;465:182–187. doi: 10.1038/nature09033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim WY, Sharpless NE. The regulation of INK4/ARF in cancer and aging. Cell. 2006;127:265–275. doi: 10.1016/j.cell.2006.10.003. [DOI] [PubMed] [Google Scholar]
- Kino T, Hurt DE, Ichijo T, Nader N, Chrousos GP. Noncoding RNA gas5 is a growth arrest- and starvation-associated repressor of the glucocorticoid receptor. Sci Signal. 2010;3:ra8. doi: 10.1126/scisignal.2000568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knauss JL, Sun T. Regulatory mechanisms of long noncoding RNAs in vertebrate central nervous system development and function. Neuroscience. 2013;235:200–214. doi: 10.1016/j.neuroscience.2013.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch F, Andrau JC. Initiating RNA polymerase II and TIPs as hallmarks of enhancer activity and tissue-specificity. Transcription. 2011;2:263–268. doi: 10.4161/trns.2.6.18747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kostka D, Hubisz MJ, Siepel A, Pollard KS. The role of GC-biased gene conversion in shaping the fastest evolving regions of the human genome. Molecular biology and evolution. 2012;29:1047–1057. doi: 10.1093/molbev/msr279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotake Y, Nakagawa T, Kitagawa K, Suzuki S, Liu N, Kitagawa M, Xiong Y. Long non-coding RNA ANRIL is required for the PRC2 recruitment to and silencing of p15(INK4B) tumor suppressor gene. Oncogene. 2011;30:1956–1962. doi: 10.1038/onc.2010.568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kung JT, Colognori D, Lee JT. Long noncoding RNAs: past, present, and future. Genetics. 2013;193:651–669. doi: 10.1534/genetics.112.146704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai F, Orom UA, Cesaroni M, Beringer M, Taatjes DJ, Blobel GA, Shiekhattar R. Activating RNAs associate with Mediator to enhance chromatin architecture and transcription. Nature. 2013;494:497–501. doi: 10.1038/nature11884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam MT, Cho H, Lesch HP, Gosselin D, Heinz S, Tanaka-Oishi Y, Benner C, Kaikkonen MU, Kim AS, Kosaka M, et al. Rev-Erbs repress macrophage gene expression by inhibiting enhancer-directed transcription. Nature. 2013;498:511–515. doi: 10.1038/nature12209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lapidot M, Pilpel Y. Genome-wide natural antisense transcription: coupling its regulation to its different regulatory mechanisms. EMBO reports. 2006;7:1216–1222. doi: 10.1038/sj.embor.7400857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latos PA, Pauler FM, Koerner MV, Senergin HB, Hudson QJ, Stocsits RR, Allhoff W, Stricker SH, Klement RM, Warczok KE, et al. Airn transcriptional overlap, but not its lncRNA products, induces imprinted Igf2r silencing. Science. 2012;338:1469–1472. doi: 10.1126/science.1228110. [DOI] [PubMed] [Google Scholar]
- Lee JT. Gracefully ageing at 50, X-chromosome inactivation becomes a paradigm for RNA and chromatin control. Nature reviews Molecular cell biology. 2011;12:815–826. doi: 10.1038/nrm3231. [DOI] [PubMed] [Google Scholar]
- Lee JT. Epigenetic regulation by long noncoding RNAs. Science. 2012;338:1435–1439. doi: 10.1126/science.1231776. [DOI] [PubMed] [Google Scholar]
- Lee JT, Davidow LS, Warshawsky D. Tsix, a gene antisense to Xist at the X-inactivation centre. Nat Genet. 1999;21:400–404. doi: 10.1038/7734. [DOI] [PubMed] [Google Scholar]
- Li W, Notani D, Ma Q, Tanasa B, Nunez E, Chen AY, Merkurjev D, Zhang J, Ohgi K, Song X, et al. Functional roles of enhancer RNAs for oestrogen-dependent transcriptional activation. Nature. 2013;498:516–520. doi: 10.1038/nature12210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin M, Pedrosa E, Shah A, Hrabovsky A, Maqbool S, Zheng D, Lachman HM. RNA-Seq of human neurons derived from iPS cells reveals candidate long non-coding RNAs involved in neurogenesis and neuropsychiatric disorders. PLoS One. 2011;6:e23356. doi: 10.1371/journal.pone.0023356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ling J, Ainol L, Zhang L, Yu X, Pi W, Tuan D. HS2 enhancer function is blocked by a transcriptional terminator inserted between the enhancer and the promoter. J Biol Chem. 2004;279:51704–51713. doi: 10.1074/jbc.M404039200. [DOI] [PubMed] [Google Scholar]
- Ling J, Baibakov B, Pi W, Emerson BM, Tuan D. The HS2 enhancer of the beta-globin locus control region initiates synthesis of non-coding, polyadenylated RNAs independent of a cis-linked globin promoter. J Mol Biol. 2005;350:883–896. doi: 10.1016/j.jmb.2005.05.039. [DOI] [PubMed] [Google Scholar]
- Ling KH, Hewitt CA, Beissbarth T, Hyde L, Cheah PS, Smyth GK, Tan SS, Hahn CN, Thomas T, Thomas PQ, et al. Spatiotemporal regulation of multiple overlapping sense and novel natural antisense transcripts at the Nrgn and Camk2n1 gene loci during mouse cerebral corticogenesis. Cerebral cortex. 2011;21:683–697. doi: 10.1093/cercor/bhq141. [DOI] [PubMed] [Google Scholar]
- Lyon MF. Gene action in the X-chromosome of the mouse (Mus musculus L.) Nature. 1961;190:372–373. doi: 10.1038/190372a0. [DOI] [PubMed] [Google Scholar]
- Lyons MR, West AE. Mechanisms of specificity in neuronal activity-regulated gene transcription. Prog Neurobiol. 2011;94:259–295. doi: 10.1016/j.pneurobio.2011.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magistri M, Faghihi MA, St Laurent G, 3rd, Wahlestedt C. Regulation of chromatin structure by long noncoding RNAs: focus on natural antisense transcripts. Trends in genetics : TIG. 2012;28:389–396. doi: 10.1016/j.tig.2012.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margueron R, Reinberg D. The Polycomb complex PRC2 and its mark in life. Nature. 2011;469:343–349. doi: 10.1038/nature09784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marques AC, Ponting CP. Catalogues of mammalian long noncoding RNAs: modest conservation and incompleteness. Genome Biol. 2009;10:R124. doi: 10.1186/gb-2009-10-11-r124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martens JA, Laprade L, Winston F. Intergenic transcription is required to repress the Saccharomyces cerevisiae SER3 gene. Nature. 2004;429:571–574. doi: 10.1038/nature02538. [DOI] [PubMed] [Google Scholar]
- Martianov I, Ramadass A, Serra Barros A, Chow N, Akoulitchev A. Repression of the human dihydrofolate reductase gene by a non-coding interfering transcript. Nature. 2007;445:666–670. doi: 10.1038/nature05519. [DOI] [PubMed] [Google Scholar]
- Mazo A, Hodgson JW, Petruk S, Sedkov Y, Brock HW. Transcriptional interference: an unexpected layer of complexity in gene regulation. J Cell Sci. 2007;120:2755–2761. doi: 10.1242/jcs.007633. [DOI] [PubMed] [Google Scholar]
- Melo CA, Drost J, Wijchers PJ, van de Werken H, de Wit E, Vrielink JA, Elkon R, Melo SA, Leveille N, Kalluri R, et al. eRNAs Are Required for p53-Dependent Enhancer Activity and Gene Transcription. Mol Cell. 2012 doi: 10.1016/j.molcel.2012.11.021. [DOI] [PubMed] [Google Scholar]
- Mercer TR, Dinger ME, Sunkin SM, Mehler MF, Mattick JS. Specific expression of long noncoding RNAs in the mouse brain. Proc Natl Acad Sci U S A. 2008;105:716–721. doi: 10.1073/pnas.0706729105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercer TR, Qureshi IA, Gokhan S, Dinger ME, Li G, Mattick JS, Mehler MF. Long noncoding RNAs in neuronal-glial fate specification and oligodendrocyte lineage maturation. BMC Neurosci. 2010;11:14. doi: 10.1186/1471-2202-11-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millar JK, Wilson-Annan JC, Anderson S, Christie S, Taylor MS, Semple CA, Devon RS, St Clair DM, Muir WJ, Blackwood DH, et al. Disruption of two novel genes by a translocation co-segregating with schizophrenia. Hum Mol Genet. 2000;9:1415–1423. doi: 10.1093/hmg/9.9.1415. [DOI] [PubMed] [Google Scholar]
- Modarresi F, Faghihi MA, Lopez-Toledano MA, Fatemi RP, Magistri M, Brothers SP, van der Brug MP, Wahlestedt C. Inhibition of natural antisense transcripts in vivo results in gene-specific transcriptional upregulation. Nat Biotechnol. 2012;30:453–459. doi: 10.1038/nbt.2158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohammad F, Mondal T, Guseva N, Pandey GK, Kanduri C. Kcnq1ot1 noncoding RNA mediates transcriptional gene silencing by interacting with Dnmt1. Development. 2010;137:2493–2499. doi: 10.1242/dev.048181. [DOI] [PubMed] [Google Scholar]
- Morrissy AS, Griffith M, Marra MA. Extensive relationship between antisense transcription and alternative splicing in the human genome. Genome Res. 2011;21:1203–1212. doi: 10.1101/gr.113431.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mousavi K, Zare H, Dell’orso S, Grontved L, Gutierrez-Cruz G, Derfoul A, Hager GL, Sartorelli V. eRNAs promote transcription by establishing chromatin accessibility at defined genomic loci. Mol Cell. 2013;51:606–617. doi: 10.1016/j.molcel.2013.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagano T, Mitchell JA, Sanz LA, Pauler FM, Ferguson-Smith AC, Feil R, Fraser P. The Air noncoding RNA epigenetically silences transcription by targeting G9a to chromatin. Science. 2008;322:1717–1720. doi: 10.1126/science.1163802. [DOI] [PubMed] [Google Scholar]
- Natoli G, Andrau JC. Noncoding transcription at enhancers: general principles and functional models. Annu Rev Genet. 2012;46:1–19. doi: 10.1146/annurev-genet-110711-155459. [DOI] [PubMed] [Google Scholar]
- Ng SY, Johnson R, Stanton LW. Human long non-coding RNAs promote pluripotency and neuronal differentiation by association with chromatin modifiers and transcription factors. EMBO J. 2012;31:522–533. doi: 10.1038/emboj.2011.459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orgel LE, Crick FH. Selfish DNA: the ultimate parasite. Nature. 1980;284:604–607. doi: 10.1038/284604a0. [DOI] [PubMed] [Google Scholar]
- Orom UA, Derrien T, Beringer M, Gumireddy K, Gardini A, Bussotti G, Lai F, Zytnicki M, Notredame C, Huang Q, et al. Long noncoding RNAs with enhancer-like function in human cells. Cell. 2010;143:46–58. doi: 10.1016/j.cell.2010.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey RR, Mondal T, Mohammad F, Enroth S, Redrup L, Komorowski J, Nagano T, Mancini-Dinardo D, Kanduri C. Kcnq1ot1 antisense noncoding RNA mediates lineage-specific transcriptional silencing through chromatin-level regulation. Mol Cell. 2008;32:232–246. doi: 10.1016/j.molcel.2008.08.022. [DOI] [PubMed] [Google Scholar]
- Pang KC, Frith MC, Mattick JS. Rapid evolution of noncoding RNAs: lack of conservation does not mean lack of function. Trends in genetics : TIG. 2006;22:1–5. doi: 10.1016/j.tig.2005.10.003. [DOI] [PubMed] [Google Scholar]
- Pasmant E, Laurendeau I, Heron D, Vidaud M, Vidaud D, Bieche I. Characterization of a germ-line deletion, including the entire INK4/ARF locus, in a melanoma-neural system tumor family: identification of ANRIL, an antisense noncoding RNA whose expression coclusters with ARF. Cancer research. 2007;67:3963–3969. doi: 10.1158/0008-5472.CAN-06-2004. [DOI] [PubMed] [Google Scholar]
- Pastori C, Wahlestedt C. Involvement of long noncoding RNAs in diseases affecting the central nervous system. RNA Biol. 2012;9:860–870. doi: 10.4161/rna.20482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollard KS, Salama SR, Lambert N, Lambot MA, Coppens S, Pedersen JS, Katzman S, King B, Onodera C, Siepel A, et al. An RNA gene expressed during cortical development evolved rapidly in humans. Nature. 2006;443:167–172. doi: 10.1038/nature05113. [DOI] [PubMed] [Google Scholar]
- Ponjavic J, Oliver PL, Lunter G, Ponting CP. Genomic and transcriptional colocalization of protein-coding and long non-coding RNA pairs in the developing brain. PLoS Genet. 2009;5:e1000617. doi: 10.1371/journal.pgen.1000617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponjavic J, Ponting CP, Lunter G. Functionality or transcriptional noise? Evidence for selection within long noncoding RNAs. Genome Res. 2007;17:556–565. doi: 10.1101/gr.6036807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponting CP, Belgard TG. Transcribed dark matter: meaning or myth? Hum Mol Genet. 2010;19:R162–168. doi: 10.1093/hmg/ddq362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponting CP, Oliver PL, Reik W. Evolution and functions of long noncoding RNAs. Cell. 2009;136:629–641. doi: 10.1016/j.cell.2009.02.006. [DOI] [PubMed] [Google Scholar]
- Popov N, Gil J. Epigenetic regulation of the INK4b-ARF-INK4a locus: in sickness and in health. Epigenetics. 2010;5:685–690. doi: 10.4161/epi.5.8.12996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qureshi IA, Mattick JS, Mehler MF. Long non-coding RNAs in nervous system function and disease. Brain Res. 2010;1338:20–35. doi: 10.1016/j.brainres.2010.03.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qureshi IA, Mehler MF. Emerging roles of non-coding RNAs in brain evolution, development, plasticity and disease. Nature reviews Neuroscience. 2012;13:528–541. doi: 10.1038/nrn3234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rada-Iglesias A, Bajpai R, Swigut T, Brugmann SA, Flynn RA, Wysocka J. A unique chromatin signature uncovers early developmental enhancers in humans. Nature. 2011;470:279–283. doi: 10.1038/nature09692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinn JL, Chang HY. Genome regulation by long noncoding RNAs. Annual review of biochemistry. 2012;81:145–166. doi: 10.1146/annurev-biochem-051410-092902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinn JL, Kertesz M, Wang JK, Squazzo SL, Xu X, Brugmann SA, Goodnough LH, Helms JA, Farnham PJ, Segal E, et al. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell. 2007;129:1311–1323. doi: 10.1016/j.cell.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sado T, Hoki Y, Sasaki H. Tsix defective in splicing is competent to establish Xist silencing. Development. 2006;133:4925–4931. doi: 10.1242/dev.02670. [DOI] [PubMed] [Google Scholar]
- Sanchez-Elsner T, Gou D, Kremmer E, Sauer F. Noncoding RNAs of trithorax response elements recruit Drosophila Ash1 to Ultrabithorax. Science. 2006;311:1118–1123. doi: 10.1126/science.1117705. [DOI] [PubMed] [Google Scholar]
- Schmitz KM, Mayer C, Postepska A, Grummt I. Interaction of noncoding RNA with the rDNA promoter mediates recruitment of DNMT3b and silencing of rRNA genes. Genes & development. 2010;24:2264–2269. doi: 10.1101/gad.590910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuettengruber B, Martinez AM, Iovino N, Cavalli G. Trithorax group proteins: switching genes on and keeping them active. Nature reviews Molecular cell biology. 2011;12:799–814. doi: 10.1038/nrm3230. [DOI] [PubMed] [Google Scholar]
- Simon JA, Kingston RE. Mechanisms of polycomb gene silencing: knowns and unknowns. Nature reviews Molecular cell biology. 2009;10:697–708. doi: 10.1038/nrm2763. [DOI] [PubMed] [Google Scholar]
- Simon-Sanchez J, Singleton A. Genome-wide association studies in neurological disorders. Lancet neurology. 2008;7:1067–1072. doi: 10.1016/S1474-4422(08)70241-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sleutels F, Zwart R, Barlow DP. The non-coding Air RNA is required for silencing autosomal imprinted genes. Nature. 2002;415:810–813. doi: 10.1038/415810a. [DOI] [PubMed] [Google Scholar]
- Sokolov BP, Polesskaya OO, Uhl GR. Mouse brain gene expression changes after acute and chronic amphetamine. J Neurochem. 2003;84:244–252. doi: 10.1046/j.1471-4159.2003.01523.x. [DOI] [PubMed] [Google Scholar]
- Sopher BL, Ladd PD, Pineda VV, Libby RT, Sunkin SM, Hurley JB, Thienes CP, Gaasterland T, Filippova GN, La Spada AR. CTCF regulates ataxin-7 expression through promotion of a convergently transcribed, antisense noncoding RNA. Neuron. 2011;70:1071–1084. doi: 10.1016/j.neuron.2011.05.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Struhl K. Transcriptional noise and the fidelity of initiation by RNA polymerase II. Nature structural & molecular biology. 2007;14:103–105. doi: 10.1038/nsmb0207-103. [DOI] [PubMed] [Google Scholar]
- Sun BK, Deaton AM, Lee JT. A transient heterochromatic state in Xist preempts X inactivation choice without RNA stabilization. Mol Cell. 2006a;21:617–628. doi: 10.1016/j.molcel.2006.01.028. [DOI] [PubMed] [Google Scholar]
- Sun M, Hurst LD, Carmichael GG, Chen J. Evidence for variation in abundance of antisense transcripts between multicellular animals but no relationship between antisense transcriptionand organismic complexity. Genome Res. 2006b;16:922–933. doi: 10.1101/gr.5210006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taft RJ, Pheasant M, Mattick JS. The relationship between non-protein-coding DNA and eukaryotic complexity. Bioessays. 2007;29:288–299. doi: 10.1002/bies.20544. [DOI] [PubMed] [Google Scholar]
- Tani H, Mizutani R, Salam KA, Tano K, Ijiri K, Wakamatsu A, Isogai T, Suzuki Y, Akimitsu N. Genome-wide determination of RNA stability reveals hundreds of short-lived noncoding transcripts in mammals. Genome Res. 2012;22:947–956. doi: 10.1101/gr.130559.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian D, Sun S, Lee JT. The long noncoding RNA, Jpx, is a molecular switch for X chromosome inactivation. Cell. 2010;143:390–403. doi: 10.1016/j.cell.2010.09.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tolhuis B, Blom M, Kerkhoven RM, Pagie L, Teunissen H, Nieuwland M, Simonis M, de Laat W, van Lohuizen M, van Steensel B. Interactions among Polycomb domains are guided by chromosome architecture. PLoS Genet. 2011;7:e1001343. doi: 10.1371/journal.pgen.1001343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visel A, Blow MJ, Li Z, Zhang T, Akiyama JA, Holt A, Plajzer-Frick I, Shoukry M, Wright C, Chen F, et al. ChIP-seq accurately predicts tissue-specific activity of enhancers. Nature. 2009;457:854–858. doi: 10.1038/nature07730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh CA, Morrow EM, Rubenstein JL. Autism and brain development. Cell. 2008;135:396–400. doi: 10.1016/j.cell.2008.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D, Garcia-Bassets I, Benner C, Li W, Su X, Zhou Y, Qiu J, Liu W, Kaikkonen MU, Ohgi KA, et al. Reprogramming transcription by distinct classes of enhancers functionally defined by eRNA. Nature. 2011a;474:390–394. doi: 10.1038/nature10006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Iacoangeli A, Popp S, Muslimov IA, Imataka H, Sonenberg N, Lomakin IB, Tiedge H. Dendritic BC1 RNA: functional role in regulation of translation initiation. J Neurosci. 2002;22:10232–10241. doi: 10.1523/JNEUROSCI.22-23-10232.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Zhang J, Zheng H, Li J, Liu D, Li H, Samudrala R, Yu J, Wong GK. Mouse transcriptome: neutral evolution of ‘non-coding’ complementary DNAs. Nature. 2004;431 1 p following 757; discussion following 757. [PubMed] [Google Scholar]
- Wang KC, Yang YW, Liu B, Sanyal A, Corces-Zimmerman R, Chen Y, Lajoie BR, Protacio A, Flynn RA, Gupta RA, et al. A long noncoding RNA maintains active chromatin to coordinate homeotic gene expression. Nature. 2011b;472:120–124. doi: 10.1038/nature09819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Arai S, Song X, Reichart D, Du K, Pascual G, Tempst P, Rosenfeld MG, Glass CK, Kurokawa R. Induced ncRNAs allosterically modify RNA-binding proteins in cis to inhibit transcription. Nature. 2008;454:126–130. doi: 10.1038/nature06992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Schones DE, Zhao K. Characterization of human epigenomes. Current Opinion in Genetics & Development. 2009;19:127–134. doi: 10.1016/j.gde.2009.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willingham AT, Orth AP, Batalov S, Peters EC, Wen BG, Aza-Blanc P, Hogenesch JB, Schultz PG. A strategy for probing the function of noncoding RNAs finds a repressor of NFAT. Science. 2005;309:1570–1573. doi: 10.1126/science.1115901. [DOI] [PubMed] [Google Scholar]
- Xu N, Tsai CL, Lee JT. Transient homologous chromosome pairing marks the onset of X inactivation. Science. 2006;311:1149–1152. doi: 10.1126/science.1122984. [DOI] [PubMed] [Google Scholar]
- Yamasaki Y, Kayashima T, Soejima H, Kinoshita A, Yoshiura K, Matsumoto N, Ohta T, Urano T, Masuzaki H, Ishimaru T, et al. Neuron-specific relaxation of Igf2r imprinting is associated with neuron-specific histone modifications and lack of its antisense transcript Air. Hum Mol Genet. 2005;14:2511–2520. doi: 10.1093/hmg/ddi255. [DOI] [PubMed] [Google Scholar]
- Yap KL, Li S, Munoz-Cabello AM, Raguz S, Zeng L, Mujtaba S, Gil J, Walsh MJ, Zhou MM. Molecular interplay of the noncoding RNA ANRIL and methylated histone H3 lysine 27 by polycomb CBX7 in transcriptional silencing of INK4a. Mol Cell. 2010;38:662–674. doi: 10.1016/j.molcel.2010.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yelin R, Dahary D, Sorek R, Levanon EY, Goldstein O, Shoshan A, Diber A, Biton S, Tamir Y, Khosravi R, et al. Widespread occurrence of antisense transcription in the human genome. Nat Biotechnol. 2003;21:379–386. doi: 10.1038/nbt808. [DOI] [PubMed] [Google Scholar]
- Zhao H, Dean A. An insulator blocks spreading of histone acetylation and interferes with RNA polymerase II transfer between an enhancer and gene. Nucleic Acids Res. 2004;32:4903–4919. doi: 10.1093/nar/gkh832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J, Ohsumi TK, Kung JT, Ogawa Y, Grau DJ, Sarma K, Song JJ, Kingston RE, Borowsky M, Lee JT. Genome-wide identification of polycomb-associated RNAs by RIP-seq. Molecular Cell. 2010;40:939–953. doi: 10.1016/j.molcel.2010.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J, Sun BK, Erwin JA, Song JJ, Lee JT. Polycomb proteins targeted by a short repeat RNA to the mouse X chromosome. Science. 2008;322:750–756. doi: 10.1126/science.1163045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong J, Chuang SC, Bianchi R, Zhao W, Lee H, Fenton AA, Wong RK, Tiedge H. BC1 regulation of metabotropic glutamate receptor-mediated neuronal excitability. J Neurosci. 2009;29:9977–9986. doi: 10.1523/JNEUROSCI.3893-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]