Abstract
Poly(A) tails are important regulators of mRNA stability and translational efficiency. Cytoplasmic removal of poly(A) tails by 3′-to-5′ exonucleases (deadenylation) is the rate-limiting step in mRNA degradation. Two exonuclease complexes contribute the majority of the deadenylation activity in eukaryotes: Ccr4-Not and Pan2/Pan3. These can be specifically recruited to mRNA to regulate mRNA stability or translational efficiency, thereby fine-tuning gene expression. In the present review, we discuss the activities and roles of the Pan2/Pan3 deadenylation complex.
Keywords: mRNA, polyA tail, gene expression, exonuclease, mRNA decay
Introduction
Almost every eukaryotic mRNA is synthesised with a polyA tail of uniform length: yeast mRNAs have an ~70 nucleotide (nt) polyA tail whereas mRNAs in higher eukaryotes contain polyA tails of 200-250 nt. PolyA tails are added co-transcriptionally and are required for mRNA export from the nucleus [1]. They are bound by polyA binding protein or PABP (Pab1p in yeast, PABPC1 in mammals). The polyA tail (and PABP) protects mRNA 3′ ends from non-specific exonuclease activities [2]. In addition, the polyA tail and PABPs promote translational efficiency. The Pan2/Pan3 and Ccr4-Not multi-protein complexes comprise the main deadenylation activities in eukaryotic cells [3]. Both complexes play key roles in regulation of polyA tail length and gene expression. Here we review the function of Pan2/Pan3, with a focus on the yeast complex.
Discovery of PAN
Early experiments addressing the function of polyA tails showed that depletion or inactivation of the essential polyA binding protein Pab1p in yeast results in mRNAs with an increased average polyA tail length and reduced translation [4,5]. This led to the proposal that polyA tail removal and translation initiation are stimulated by Pab1p [5]. Indeed, later work showed that polyA binding proteins interact with the eukaryotic translation initiation factor 4F (eIF4F) to stimulate initiation of translation [6,7]. eIF4F also interacts with the 5′ cap and therefore circularises the mRNA. This “closed-loop” conformation of mRNA provides a quality control mechanism to prevent translation of partially degraded mRNA or aberrant mRNAs lacking a polyA tail. Longer polyA tails stimulate translation whereas deadenylation results in translational repression [8,9].
Although polyA binding proteins protect mRNA from degradation by non-specific nucleases, they also appeared to stimulate polyA-specific exonucleases. For example, addition of purified Pab1p to extracts made from a Pab1p-deficient yeast strain stimulates shortening of polyA tails [10]. Initial searches for the eukaryotic deadenylase, the PolyA Nuclease (PAN), were performed by fractionating yeast extracts [10]. A 3′-to-5′ exonuclease activity was identified: this PAN activity could degrade a 3′ polyA tail but not the mRNA body, and was dependent on two proteins - Pan2p and Pan3p [11,12]. Neither of these proteins are essential for viability in yeast but their disruption results in an increased average polyA tail length in vivo [11]. Deadenylation is completely eliminated only when the both CCR4 (one of the two deadenylase subunits of Ccr4-Not) and PAN2 are disrupted.
Pan2
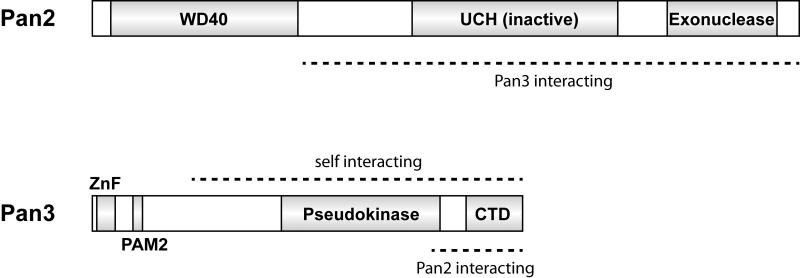
Pan2 is a 127 kDa protein (Figure 1), first identified in yeast by Boeck et al. [11] but conserved across all eukaryotes. It contains a nuclease domain of the DEDD/RNaseD family of 3′-to-5′ exonucleases, the same type of domain found in the Caf1/Pop2 exonuclease subunit of the Ccr4-Not complex [13,14]. The exonuclease domain of Pan2 is located at its C-terminus.
Figure 1. Domain structures of Pan2 and Pan3.
The domain structures of Pan2 and Pan3 are depicted showing the WD40, ubiquitin C-terminal hydrolase (UCH) and exonuclease domains of Pan2; and the zinc finger (ZnF), PAM2 PABP interacting motif, pseudokinase and C-terminal domain (CTD) of Pan3. Regions involved in protein protein interactions are indicated.
Pan2 also contains a WD40 repeat domain (at its N-terminus) and a ubiquitin C-terminal hydrolase domain (UCH) (Figure 1). WD40 domains often mediate protein-protein interactions but an interaction partner for Pan2’s WD40 domain has not been identified. Interestingly, the UCH domain is predicted to be inactive due to lack of active site residues [15]. It is unclear whether the UCH domain binds ubiquitin and what role this plays within the complex. Deletion of the first 332 amino acids of yeast Pan2p does not inhibit complex formation with Pan3p suggesting that their interaction is mediated through either the linker, UCH or exonuclease domain [16] (Figure 1).
Pan3
Pan3 is the 76 kDa interacting partner of Pan2 [12] (Figure 1). It contains an N-terminal zinc finger, a pseudokinase domain and a highly-conserved C-terminal domain (aa 560-679 in yeast Pan3p) of unknown function. The C-terminal domain likely interacts with Pan2, as shown by yeast-2-hybrid and pulldown experiments [16,17]. An unstructured linker that connects the zinc finger and pseudokinase domains contains a PABP-interacting motif 2 (PAM2) that binds to the C-terminal domain of polyA binding proteins [18]. Yeast-2-hybrid experiments also show that the C-terminal two-thirds of yeast Pan3p (aa 250-679) can self interact [16].
Deadenylation activity
The biochemical activity of the Pan2/Pan3 complex has been extensively characterised in vitro. Early work showed that Pan2/Pan3 binds to polyU- and polyA-sepharose suggesting a high affinity for RNA [10]. Pan2/Pan3 purified from yeast or human cells is dependent on magnesium, requires a 3′-OH group and the product of the reaction is 5′-AMP [17,19].
Pan2/Pan3 was initially purified from a Pab1p-deficient yeast strain as an exonuclease that requires Pab1p to remove a polyA tail from a model mRNA substrate [10]. Similarly, the human complex is also greatly stimulated by polyA binding protein [17]. Pan2/Pan3 is specific for polyA - it is unable to degrade the body of the mRNA. Although some conditions, e.g. addition of spermidine, allow PABP-independent deadenylation, it is generally assumed that PABP is required for recruitment to mRNA in vivo [17,19]. In agreement with this, Pan2/Pan3 does not efficiently remove the final 20-25 nt of the polyA tail [19]. PABP binding to polyA saturates at approximately 25 A residues per molecule [4,20] so this suggests that Pan2/Pan3 is only active on a polyA:PABP complex. In addition, Pab1p mutation in yeast results in longer polyA tails - the same phenotype observed in PAN2 and PAN3 deletion strains [10].
Pan2/Pan3 is predominantly cytoplasmic. However, it is unclear whether it acts solely in the cytoplasm, or whether it is also involved in maturation of the polyA tail in the nucleus [21-24]. A second polyA binding protein found in the nucleus in yeast, Nab2p, may protect polyA tails from premature deadenylation [22].
Roles in mRNA decay
Deadenylation of mRNA is the first and rate-limiting step of mRNA degradation [25]. Following the lag phase due to deadenylation, mRNA degradation proceeds rapidly either by decapping and 5′-to-3′ degradation by Xrn1, or by the 3′- to-5′ exonuclease activity of the cytoplasmic exosome [2,26]. Many of the mRNA decay factors localise to mammalian P-bodies (processing bodies), including Pan2 and Pan3 [27].
A two phase model of deadenylation has been proposed wherein Pan2/Pan3 removes polyA until PABP can no longer bind (12-25 As) [21]. Subsequently, Ccr4-Not removes the remaining As in the “terminal deadenylation” phase triggering rapid mRNA degradation [28,29]. Indeed deletion of any single exonuclease subunit (Pan2p, Ccr4p, Caf1p) in yeast results in longer polyA tails but not a severe growth phenotype [11,12] whereas a double PAN2/CCR4 deletion is very detrimental to cell growth with no residual deadenylation [28]. In agreement with this, deletion of CCR4 in yeast impairs terminal deadenylation (removal of the final ~20 As) whereas deletion of PAN2 impairs the removal of the distal portion of a polyA tail [28]. Similar conclusions have been made in mammalian cells where both complexes contribute to deadenylation, and overexpression of PAN2 promotes the first phase of deadenylation but overexpression of CCR4 promotes the second phase [29,30]. These data also suggest functional redundancy between the two complexes.
Different mRNAs have different deadenylation rates, contributing to differences in their relative stabilities in vivo [19,25]. For example, the normally distributive deadenylation activity of Pan2/Pan3 appears to be processive on certain mRNAs (e.g. MFA2) [19]. Interestingly, in trypanosomes where transcription is polycistronic and mRNA degradation plays a larger role in controlling gene expression, depletion of Pan2 causes stabilisation of mRNAs with intermediate half lives while depletion of Caf1 stabilises most mRNAs [31].
Importantly, deadenylation can signal not only mRNA degradation but can also fine-tune gene expression. Deadeylation represses translation, and readenylation by cytoplasmic polyA polymerases enhances translation [32]. This control mechanism of deadenylation/readenylation is used to control translation in oocytes, inflammation and neurons [32].
Recruitment to mRNA and regulation
A major mRNA recruitment mechanism for Pan2/Pan3 is via the Pan3-PABP interaction [18]. However, it is unclear whether Pan2/Pan3 is able to interact with every PABP-bound mRNA, or whether other signals prevent or promote this interaction. Recent studies by Huang et al show that phosphorylation of a cluster of serine and threonine residues adjacent to the PAM2 motif in Pan3 may regulate its interaction with PABP [33]. Ccr4-Not is recruited to mRNA through interactions with proteins that directly bind sequences in mRNA 3′ untranslated regions (UTRs), e.g. Puf/Pumilio, and Tristetraprolin/TTP proteins [32,34]. For instance, the Puf3p protein promotes mRNA deadenylation by both Pan2/Pan3 and Ccr4-Not, although a direct interaction between Puf3p and Pan2/Pan3 has not been demonstrated [35]. There are few examples of specific recruitment of Pan2/Pan3 to mRNAs.
Both Pan2/Pan3 and Ccr4-Not complexes are recruited to mRNAs via the microRNA-induced silencing complex (miRISC). Specifically Pan3, CNOT1 and PABP are reported to bind to the C-terminal silencing domain of the GW182/TNRC6 subunit of miRISC [36-38] and these interactions are highly conserved [39]. In this context, the deadenylation complexes mediate rapid biphasic deadenylation of miRNA-targeted mRNAs and as expected, this leads to either translational repression or mRNA decay [40]. Interestingly, there is some evidence that GW182 proteins not only recruit deadenylation complexes but may stimulate their deadenylation activity, at least for Ccr4-Not [36]. Overexpression of dominant negative mutants of Pan2 or Caf1 slows the first and second phases of miRNA-mediated deadenylation, respectively [40]. In addition, Ccr4-Not appears to mediate translational repression in response to miRNAs, independent of its deadenylation activity: tethering a catalytically inactive version of the deadenylase subunit Caf1 to a reporter mRNA represses translation, as does tethering other Ccr4-Not components to reporter mRNAs lacking polyA tails [41]. It is possible that Ccr4-Not actively inhibits translation, for example through inhibition of the activity of a translation initiation factor [42]. Alternatively, Ccr4-Not could disrupt PABP interaction or the closed-loop conformation of mRNA to inhibit translation initiation indirectly [43]. Further studies will be required to clarify this.
Pan3p interacts with the Dun1 kinase and dun1pan2 or dun1pan3 double mutants are hypersensitive to replication stress [44]. Thus, it has been proposed that Dun1 and Pan2/Pan3 function together to regulate Rad5 mRNA. Interestingly, stress inhibits deadenylation (both by Ccr4-Not and Pan2/Pan3), mRNA decay and translation, possibly to allow selective translation of mRNAs specific to the stress response while retaining other mRNAs to be used upon recover from the stress [45,46]. The mechanism of stress-induced inhibition of deadenylation is unknown.
Outlook
PolyA tails are required to regulate mRNA stability and efficient translation. Deadenylation is a key step of mRNA degradation, possibly because release of the closed-loop mRNA structure is required to expose the mRNA 5′-cap for decapping [47]. Studies of deadenylation of eukaryotic mRNAs suggest that transcription, mRNA processing, translation and mRNA decay are closely linked [48]. The enzymes involved in each of these key events communicate with each other to create an integrated control mechanism for gene expression. In support of this, genomic methods to monitor mRNA synthesis and decay demonstrate that yeast compensate for a reduction in transcription due to mutants in RNA Polymerase II by globally increasing mRNA stability, while cells compensate for deletion of Ccr4-Not subunits by decreasing mRNA synthesis [49]. In addition, there is evidence that the Pan2/Pan3 and Ccr4-Not complexes interact in larger assembly [27]. Future research directed towards understanding deadenylation will be required to reveal the molecular mechanisms of the individual components and their larger assemblies.
ACKNOWLEDGEMENTS
This work was supported by ERC starting grant no. 261151 to L.A.P. and Medical Research Council grant U105192715.
Abbreviations
- aa
amino acids
- miRNA
microRNA
- nt
nucleotides
- P-bodies
processing bodies
- PABP
polyA binding protein
- PAM2
PABP-interacting motif 2
- PAN
polyA nuclease
- RISC
RNA-induced silencing complex
- UCH
ubiquitin C-terminal hydrolase
- UTR
untranslated region
REFERENCES
- 1.Zhao J, Hyman L, Moore C. Formation of mRNA 3′ ends in eukaryotes: mechanism, regulation, and interrelationships with other steps in mRNA synthesis. Microbiol. Mol. Biol. Rev. 1999;63:405–445. doi: 10.1128/mmbr.63.2.405-445.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Garneau NL, Wilusz J, Wilusz CJ. The highways and byways of mRNA decay. Nat. Rev. Mol. Cell Biol. 2007;8:113–126. doi: 10.1038/nrm2104. [DOI] [PubMed] [Google Scholar]
- 3.Wiederhold K, Passmore LA. Cytoplasmic deadenylation: regulation of mRNA fate. Biochem. Soc. Trans. 2010;38:1531–1536. doi: 10.1042/BST0381531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sachs AB, Davis RW, Kornberg RD. A single domain of yeast poly(A)-binding protein is necessary and sufficient for RNA binding and cell viability. Mol. Cell. Biol. 1987;7:3268–3276. doi: 10.1128/mcb.7.9.3268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sachs AB, Davis RW. The poly(A) binding protein is required for poly(A) shortening and 60S ribosomal subunit-dependent translation initiation. Cell. 1989;58:857–867. doi: 10.1016/0092-8674(89)90938-0. [DOI] [PubMed] [Google Scholar]
- 6.Tarun SZ, Sachs AB. Association of the yeast poly(A) tail binding protein with translation initiation factor eIF-4G. EMBO J. 1996;15:7168–7177. [PMC free article] [PubMed] [Google Scholar]
- 7.Kapp LD, Lorsch JR. The molecular mechanics of eukaryotic translation. Annu. Rev. Biochem. 2004;73:657–704. doi: 10.1146/annurev.biochem.73.030403.080419. [DOI] [PubMed] [Google Scholar]
- 8.Novoa I, Gallego J, Ferreira PG, Mendez R. Mitotic cell-cycle progression is regulated by CPEB1 and CPEB4-dependent translational control. Nat. Cell Biol. 2010;12:447–456. doi: 10.1038/ncb2046. [DOI] [PubMed] [Google Scholar]
- 9.Beilharz TH, Preiss T. Widespread use of poly(A) tail length control to accentuate expression of the yeast transcriptome. RNA. 2007;13:982–997. doi: 10.1261/rna.569407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sachs AB, Deardorff JA. Translation initiation requires the PAB-dependent poly(A) ribonuclease in yeast. Cell. 1992;70:961–973. doi: 10.1016/0092-8674(92)90246-9. [DOI] [PubMed] [Google Scholar]
- 11.Boeck R, Tarun S, Rieger M, Deardorff JA, Müller-Auer S, Sachs AB. The yeast Pan2 protein is required for poly(A)-binding protein-stimulated poly(A)-nuclease activity. J. Biol. Chem. 1996;271:432–438. doi: 10.1074/jbc.271.1.432. [DOI] [PubMed] [Google Scholar]
- 12.Brown CE, Tarun SZ, Boeck R, Sachs AB. PAN3 encodes a subunit of the Pab1p-dependent poly(A) nuclease in Saccharomyces cerevisiae. Mol. Cell. Biol. 1996;16:5744–5753. doi: 10.1128/mcb.16.10.5744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Moser MJ, Holley WR, Chatterjee A, Mian IS. The proofreading domain of Escherichia coli DNA polymerase I and other DNA and/or RNA exonuclease domains. Nucleic Acids Res. 1997;25:5110–5118. doi: 10.1093/nar/25.24.5110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zuo Y, Deutscher MP. Exoribonuclease superfamilies: structural analysis and phylogenetic distribution. Nucleic Acids Res. 2001;29:1017–1026. doi: 10.1093/nar/29.5.1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Quesada V, Díaz-Perales A, Gutiérrez-Fernández A, Garabaya C, Cal S, López-Otín C. Cloning and enzymatic analysis of 22 novel human ubiquitin-specific proteases. Biochem. Biophys. Res. Commun. 2004;314:54–62. doi: 10.1016/j.bbrc.2003.12.050. [DOI] [PubMed] [Google Scholar]
- 16.Mangus DA, Evans MC, Agrin NS, Smith M, Gongidi P, Jacobson A. Positive and negative regulation of poly(A) nuclease. Mol. Cell. Biol. 2004;24:5521–5533. doi: 10.1128/MCB.24.12.5521-5533.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Uchida N, Hoshino S-I, Katada T. Identification of a human cytoplasmic poly(A) nuclease complex stimulated by poly(A)-binding protein. J. Biol. Chem. 2004;279:1383–1391. doi: 10.1074/jbc.M309125200. [DOI] [PubMed] [Google Scholar]
- 18.Siddiqui N, Mangus DA, Chang T-C, Palermino J-M, Shyu A-B, Gehring K. Poly(A) nuclease interacts with the C-terminal domain of polyadenylate-binding protein domain from poly(A)-binding protein. J. Biol. Chem. 2007;282:25067–25075. doi: 10.1074/jbc.M701256200. [DOI] [PubMed] [Google Scholar]
- 19.Lowell JE, Rudner DZ, Sachs AB. 3′-UTR-dependent deadenylation by the yeast poly(A) nuclease. Genes Dev. 1992;6:2088–2099. doi: 10.1101/gad.6.11.2088. [DOI] [PubMed] [Google Scholar]
- 20.Baer BW, Kornberg RD. Repeating structure of cytoplasmic poly(A)-ribonucleoprotein. Proc. Natl. Acad. Sci. U.S.A. 1980;77:1890–1892. doi: 10.1073/pnas.77.4.1890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Brown CE, Sachs AB. Poly(A) tail length control in Saccharomyces cerevisiae occurs by message-specific deadenylation. Mol. Cell. Biol. 1998;18:6548–6559. doi: 10.1128/mcb.18.11.6548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schmid M, Poulsen MB, Olszewski P, Pelechano V, Saguez C, Gupta I, Steinmetz LM, Moore C, Jensen TH. Rrp6p controls mRNA poly(A) tail length and its decoration with poly(A) binding proteins. Mol. Cell. 2012;47:267–280. doi: 10.1016/j.molcel.2012.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dunn EF, Hammell CM, Hodge CA, Cole CN. Yeast poly(A)-binding protein, Pab1, and PAN, a poly(A) nuclease complex recruited by Pab1, connect mRNA biogenesis to export. Genes Dev. 2005;19:90–103. doi: 10.1101/gad.1267005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dheur S, Nykamp KR, Viphakone N, Swanson MS, Minvielle-Sebastia L. Yeast mRNA Poly(A) tail length control can be reconstituted in vitro in the absence of Pab1p-dependent Poly(A) nuclease activity. J. Biol. Chem. 2005;280:24532–24538. doi: 10.1074/jbc.M504720200. [DOI] [PubMed] [Google Scholar]
- 25.Decker CJ, Parker R. A turnover pathway for both stable and unstable mRNAs in yeast: evidence for a requirement for deadenylation. Genes Dev. 1993;7:1632–1643. doi: 10.1101/gad.7.8.1632. [DOI] [PubMed] [Google Scholar]
- 26.Parker R, Song H. The enzymes and control of eukaryotic mRNA turnover. Nat. Struct. Mol. Biol. 2004;11:121–127. doi: 10.1038/nsmb724. [DOI] [PubMed] [Google Scholar]
- 27.Zheng D, Ezzeddine N, Chen C-Y, Zhu W, He X, Shyu AB. Deadenylation is prerequisite for P-body formation and mRNA decay in mammalian cells. J. Cell Biol. 2008;182:89–101. doi: 10.1083/jcb.200801196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tucker M, Valencia-Sanchez MA, Staples RR, Chen J, Denis CL, Parker R. The transcription factor associated Ccr4 and Caf1 proteins are components of the major cytoplasmic mRNA deadenylase in Saccharomyces cerevisiae. Cell. 2001;104:377–386. doi: 10.1016/s0092-8674(01)00225-2. [DOI] [PubMed] [Google Scholar]
- 29.Yamashita A, Chang T-C, Yamashita Y, Zhu W, Zhong Z, Chen C-YA, Shyu A-B. Concerted action of poly(A) nucleases and decapping enzyme in mammalian mRNA turnover. Nat. Struct. Mol. Biol. 2005;12:1054–1063. doi: 10.1038/nsmb1016. [DOI] [PubMed] [Google Scholar]
- 30.Bönisch C, Temme C, Moritz B, Wahle E. Degradation of hsp70 and other mRNAs in Drosophila via the 5″ 3″ pathway and its regulation by heat shock. J. Biol. Chem. 2007;282:21818–21828. doi: 10.1074/jbc.M702998200. [DOI] [PubMed] [Google Scholar]
- 31.Fadda A, Färber V, Droll D, Clayton C. The roles of 3′-exoribonucleases and the exosome in trypanosome mRNA degradation. RNA. 2013;19:937–947. doi: 10.1261/rna.038430.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Weill L, Belloc E, Bava F-A, Mendez R. Translational control by changes in poly(A) tail length: recycling mRNAs. Nat. Struct. Mol. Biol. 2012;19:577–585. doi: 10.1038/nsmb.2311. [DOI] [PubMed] [Google Scholar]
- 33.Huang K-L, Chadee AB, Chen C-YA, Zhang Y, Shyu A-B. Phosphorylation at intrinsically disordered regions of PAM2 motif-containing proteins modulates their interactions with PABPC1 and influences mRNA fate. RNA. 2013;19:295–305. doi: 10.1261/rna.037317.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wahle E, Winkler GS. RNA decay machines: Deadenylation by the Ccr4-Not and Pan2-Pan3 complexes. Biochim. Biophys. Acta. 2013 doi: 10.1016/j.bbagrm.2013.01.003. [DOI] [PubMed] [Google Scholar]
- 35.Lee D, Ohn T, Chiang Y-C, Quigley G, Yao G, Liu Y, Denis CL. PUF3 acceleration of deadenylation in vivo can operate independently of CCR4 activity, possibly involving effects on the PAB1-mRNP structure. J. Mol. Biol. 2010;399:562–575. doi: 10.1016/j.jmb.2010.04.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fabian MR, Cieplak MK, Frank F, Morita M, Green J, Srikumar T, Nagar B, Yamamoto T, Raught B, Duchaine TF, et al. miRNA-mediated deadenylation is orchestrated by GW182 through two conserved motifs that interact with CCR4-NOT. Nat. Struct. Mol. Biol. 2011;18:1211–1217. doi: 10.1038/nsmb.2149. [DOI] [PubMed] [Google Scholar]
- 37.Braun JE, Huntzinger E, Fauser M, Izaurralde E. GW182 proteins directly recruit cytoplasmic deadenylase complexes to miRNA targets. Mol. Cell. 2011;44:120–133. doi: 10.1016/j.molcel.2011.09.007. [DOI] [PubMed] [Google Scholar]
- 38.Chekulaeva M, Mathys H, Zipprich JT, Attig J, Colic M, Parker R, Filipowicz W. miRNA repression involves GW182-mediated recruitment of CCR4-NOT through conserved W-containing motifs. Nat. Struct. Mol. Biol. 2011;18:1218–1226. doi: 10.1038/nsmb.2166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kuzuoglu-Öztürk D, Huntzinger E, Schmidt S, Izaurralde E. The Caenorhabditis elegans GW182 protein AIN-1 interacts with PAB-1 and subunits of the PAN2-PAN3 and CCR4-NOT deadenylase complexes. Nucleic Acids Res. 2012;40:5651–5665. doi: 10.1093/nar/gks218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chen C-YA, Zheng D, Xia Z, Shyu A-B. Ago-TNRC6 triggers microRNA-mediated decay by promoting two deadenylation steps. Nat. Struct. Mol. Biol. 2009;16:1160–1166. doi: 10.1038/nsmb.1709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cooke A, Prigge A, Wickens M. Translational repression by deadenylases. J. Biol. Chem. 2010;285:28506–28513. doi: 10.1074/jbc.M110.150763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Meijer HA, Kong YW, Lu WT, Wilczynska A, Spriggs RV, Robinson SW, Godfrey JD, Willis AE, Bushell M. Translational repression and eIF4A2 activity are critical for microRNA-mediated gene regulation. Science. 2013;340:82–85. doi: 10.1126/science.1231197. [DOI] [PubMed] [Google Scholar]
- 43.Zekri L, Kuzuoglu-Öztürk D, Izaurralde E. GW182 proteins cause PABP dissociation from silenced miRNA targets in the absence of deadenylation. EMBO J. 2013;32:1052–1065. doi: 10.1038/emboj.2013.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hammet A, Pike BL, Heierhorst J. Posttranscriptional regulation of the RAD5 DNA repair gene by the Dun1 kinase and the Pan2-Pan3 poly(A)-nuclease complex contributes to survival of replication blocks. J. Biol. Chem. 2002;277:22469–22474. doi: 10.1074/jbc.M202473200. [DOI] [PubMed] [Google Scholar]
- 45.Hilgers V, Teixeira D, Parker R. Translation-independent inhibition of mRNA deadenylation during stress in Saccharomyces cerevisiae. RNA. 2006;12:1835–1845. doi: 10.1261/rna.241006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gowrishankar G, Winzen R, Dittrich-Breiholz O, Redich N, Kracht M, Holtmann H. Inhibition of mRNA deadenylation and degradation by different types of cell stress. Biol. Chem. 2006;387:323–327. doi: 10.1515/BC.2006.043. [DOI] [PubMed] [Google Scholar]
- 47.Caponigro G, Parker R. Multiple functions for the poly(A)-binding protein in mRNA decapping and deadenylation in yeast. Genes Dev. 1995;9:2421–2432. doi: 10.1101/gad.9.19.2421. [DOI] [PubMed] [Google Scholar]
- 48.Dori-Bachash M, Shema E, Tirosh I. Coupled evolution of transcription and mRNA degradation. PLoS Biol. 2011;9:e1001106. doi: 10.1371/journal.pbio.1001106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sun M, Schwalb B, Schulz D, Pirkl N, Etzold S, Larivière L, Maier KC, Seizl M, Tresch A, Cramer P. Comparative dynamic transcriptome analysis (cDTA) reveals mutual feedback between mRNA synthesis and degradation. Genome Res. 2012;22:1350–1359. doi: 10.1101/gr.130161.111. [DOI] [PMC free article] [PubMed] [Google Scholar]