Abstract
G-protein coupled receptor 4 (GPR4) belongs to a protein family comprised of 3 closely related G protein-coupled receptors. Recent studies have shown that GPR4 plays important roles in angiogenesis, proton sensing, and regulating tumor cells as an oncogenic gene. How GPR4 conducts its functions? Rare has been known. In order to detect the genes related to GPR4, microarray technology was employed. GPR4 is highly expressed in human vascular endothelial cell HMEC-1. Small interfering RNA against GPR4 was used to knockdown GPR4 expression in HMEC-1. Then RNA from the GPR4 knockdown cells and control cells were analyzed through genome microarray. Microarray results shown that among the whole genes and expressed sequence tags, 447 differentially expressed genes were identified, containing 318 up-regulated genes and 129 down-regulated genes. These genes whose expression dramatically changed may be involved in the GPR4 functions. These genes were related to cell apoptosis, cytoskeleton and signal transduction, cell proliferation, differentiation and cell-cycle regulation, gene transcription and translation and cell material and energy metabolism.
Keywords: GPR4, RNAi, microarray
Introduction
G-protein coupled receptor 4 (GPR4) belongs to a protein family comprised of 3 closely related G protein-coupled receptors (GPCRs): GPR4, OGR1/GPR68 and TDAG8/GPR65 [1,2]. GPR4 is highly conserved during evolution, with more than 90% homology in the amino acid sequences among mammalian orthologs, and 70% homology between human and zebrafish. GPR4 is expressed in many tissues including ovary, liver, heart, kidney, placenta and skeletal muscle [2-4]. G protein coupled receptor 4 (GPR4) is previously identified as a receptor for sphingosylphosphorylcholine (SPC) and lysophosphatidylcholine (LPC). The Kd values for SPC and LPC were estimated to be 36 and 159 nM, respectively, in the binding experiments of GPR4-expressing cells [5]. GPR4 is response to SPC and LPC, resulting in the stimulation of DNA synthesis and migration of Swiss 3T3 cells [5]. But later on, this ligand-receptor relationship has not been always confirmed [2]. Later studies have further showed that GPR4 function as a proton sensor. GPR4 senses extracellular protons and responds to acidic pH changes, leading to increased intracellular levels of cyclic AMP [3,4]. GPR4 is fully activated in the acidic pH 6.8. and partially activated in the physiological range (pH 7.4 to pH 6.8) [6-9]. The overexpression of GPR4 has been shown to stimulate serum response element (SRE)-, nuclear factor of activated T-cell (NFAT), and cAMP response element (CRE)-driven transcription [6,10].
Our previous studies and some other studies suggest that GPR4 may participate in the angiogenesis and the endothelial barrier function [11-14]. Our previous study indicated that GPR4 is capable of mediating the tube formation of blood vessels by regulating the function of endothelial cells [11]. We found GPR4 is highly expressed in vascular endothelial cells of both arteries and veins [11,13,15]. When GPR4 was knocked down in endothelial cells, the growth, migration, and tube formation of endothelial cells were significantly inhibited. In vivo studies have provided more evidence in support of the GPR4 functions in angiogenesis [16]. Higher perinatal mortality rate was found in GPR4 null neonates [3,16]. Dilated and tortuous subcutaneous blood vessels, spontaneous hemorrhages, and defective vascular smooth muscle cell coverage were found in a certain percentage of GPR4-null embryos and neonates. These observations indicated that GPR4 is required for the normal vascular development of multiple tissues/organs.
Besides, other available data suggested GPR4 may be oncogenic GPCRs with increased expression levels in different types of malignancies. GPR4 overexpression has been found in kidney tumors, ovarian tumors, colon tumors, breast tumors, and liver tumors [10]. Our previous study also suggested that GPR4 may play roles in the development of EOC, and its overexpression might be required for the angiogenesis of EOC [17]. Our study also shown that, after GPR4 was knockdown in ovarian cancer cells, cell colony formation, migration, invasion, and survivals were significantly inhibited, which suggested GPR4 behaves as an oncogenic gene.
But how GPR4 conducts its functions including proton sensor, angiogenic activity, and as an oncogene? Rare has been known. In order to detect the genes related to GPR4, microarray technology was employed. GPR4 is highly expressed in human vascular endothelial cell HMEC-1. Small interfering RNA against GPR4 was used to knockdown GPR4 expression in HMEC-1. Then RNA from the GPR4 knockdown cells and control cells were analyzed through genome microarray.
Expression profile microarray technology should provide a useful experimental strategy to define cellular target genes by identifying transcriptionally altered genes upon silencing of endogenous gene expression. Combining microarray with bioinformatics technology to analyze the gene expression patterns of various tumors is an important method in functional genomic studies. It surpasses the sole gene research pattern obviously and may illuminate the gene expression and regulation network of tumor cells in the whole.
Small interfering RNA (siRNA), sometimes known as short interfering RNA or silencing RNA, is a class of double-stranded RNA molecules, 20-25 base pairs in length. siRNA plays many roles, but its most notable is in the RNA interference (RNAi) pathway, where it interferes with the expression of specific genes with complementary nucleotide sequence [18]. siRNA also acts in RNAi-related pathways, e.g., as an antiviral mechanism or in shaping the chromatin structure of a genome.
A small hairpin RNA or short hairpin RNA (shRNA) is a sequence of RNA that makes a tight hairpin turn that can be used to silence target gene expression via RNA interference (RNAi) [19]. Expression of shRNA in cells is typically accomplished by delivery of plasmids or through viral or bacterial vectors. The promoter choice is essential to achieve robust shRNA expression. At first, polymerase III promoters such as U6 and H1 were used; however, these promoters lack spatial and temporal control. As such, there has been a shift to using polymerase II promoters to regulate expression of shRNA. shRNA is an advantageous mediator of RNAi in that it has a relatively low rate of degradation and turnover.
In order to detect the genes related to GPR4, microarray technology was employed. GPR4 is highly expressed in human vascular endothelial cell HMEC-1. Small interfering RNA against GPR4 was used to knockdown GPR4 expression in HMEC-1. Then RNA from the GPR4 knockdown cells and control cells were analyzed through genome microarray. Microarray results shown that among the whole genes and expressed sequence tags, 447 differentially expressed genes were identified, containing 318 up-regulated genes and 129 down-regulated genes. These genes whose expression dramatically changed may be involved in the GPR4 functions. These genes were related to cell apoptosis, cytoskeleton and signal transduction, cell proliferation, differentiation and cell-cycle regulation, gene transcription and translation and cell material and energy metabolism.
Materials and methods
Cell culture
The HMEC-1 cell line was from the Centers for Disease Control and Prevention (Atlanta, GA) and were maintained in RPMI 1640 Medium (Invitrogen) supplemented with 5 mM glucose, 10% foetal bovine serum (FBS; HyClone, Australia) and 0.48% (w/v) HEPES, pH 7.4, in a CO2 humid incubation chamber at 37°C.
Lentivirus vectors for GPR4 RNAi
The GPR4-specific target sequences were designed using the GPR4 reference sequence (Gene Bank Accession No NM_005282). Double-stranded DNA were synthesized according to the structure of a GV115 viral vector (GeneChem) and then inserted into a linearized vector. The shuttle vector and viral packaging system were cotransfected into HEK293T cells to replicate competent lentivirus. The lentivirus containing the GPR4 shRNA (short hairpin RNA) expressing cassette was used for lentivirus production and denoted as LV-shGPR4 in the next experiments. The GV115 mock vector was also packaged and used as a negative control, denoted as LV-eGFP, which has no significant homology to gene sequences. The titers averaged 1×108 TU/mL.
The sequences for the hairpin structures were chosen using the “RNAi oligo retriever” program (www.cshl.org/public/SCEINCE/hannon.html) provided by Dr. Greg Hannon. The U6 promoter followed by a sequence encoding a 29 nt short hairpin RNA (shRNA) targeting LPA3 or GPR4 was amplified by PCR with the primers AGATCTGATTTAGGTGACACTATAG and AAAAAAGAGACAATTCCAGCCCAGCGTAGGAACCGCAAGCTTCCGGTCCCCACACTGGGCTGGAATTGCCTCGGTGTTTCGTCCTTTCCACAA (for LPA3-RNAi) or AAAAAAGTGCTGGCGACAGCACCTTCAACTACACCCAAGCTTCGGTGCAGCTGAAGATGCTGCCGCCAGCACGGTGTTTCGTCCTTTCCACAA (for GPR4-RNAi).
Real-time PCR
Real-time PCR analysis was performed in a total volume of 20 μL containing template DNA, 100 nM of sense and antisense primers, 10 μL of SYBR® Green master mix (TaKaRa). After denatured at 95°C for 2 min, the mixtures were subjected to 40 amplification cycles (10 seconds at 98°C for denaturation and 20 sec for annealing and extension at 60°C). Incorporation of SYBR® Green dye into PCR products was monitored in real time by using a Light Cycler480 Detection System (Roche) and allowing determination of the threshold cycle (CT) at which exponential amplification of PCR products begins. After PCR, dissociation curves were generated with one peak, indicating the specificity of the amplification. A CT value was obtained from each amplification curve by using the software provided by the manufacturer (Roche).
Western blot
Cell lysates of lentivirus infected HMEC-1 cells and controls were harvested and subjected to SDS-PAGE in 10% polyacrylamide gel. Bands were probed for GPR4 protein expression using mouse monoclonal antibody and secondary goat anti-mouse antibody (Santa Cruz). GAPDH detection was performed using monoclonal anti-GAPDH (Santa Cruz). Western blot products on images captured by the Fusion FX6 system (Vilber Lourmat).
Gene expression analysis
LV-eGFP and LV-shGPR4 infected HMEC-1 cells were harvested and total RNA was extracted using the RNeasy Mini kit (Qiagen) according to the manufacturer’s instructions. RNA quantity and purity were assessed by measurement of OD260/280 using a NanoDrop® ND-1000 spectrophotometer. The quality was verified by the integrity of 28S and 18S rRNA using the Agilent Total RNA Nano chip assay on a Model 2100 Bioanalyzer (Agilent Technologies). The intensity of the 28S band should be twice the intensity of the 18S band. Gene expression profiling was conducted using the Human OneArray™ (Phalanx Biotech) containing over 30,000 sixty-mer polynucleotide probes with each probe mapped to the latest draft of the human genome (GoldenPath) printed on standard 1-in. x 3-in. glass slides. Analysis was performed according to the manufacturer’s recommendations. After hybridization, arrays were scanned using GenePix 4000B (Axon Instruments) and analyzed with GenePix Pro 6.0 (Axon Instruments) to obtain gene expression ratios. Transformed data were normalized using the Lowes procedure [1]. The normalized data were used for clustering analysis. Clustering analysis was performed using Genespring GX 10.0 software (Agilent Technologies) to provide a graphical display of the expression patterns.
Quantification of target RNA expression
Target gene expression was quantified using qPCR in an Light Cycler480 System (Roche) to confirm the upregulation of expression of target gene that were detected in the human OneArray™ from two cell lines. The expression level of each gene in LV-eGFP or LV-shGPR4 sample was represented relative to the expression of GAPDH, which was used as an internal control. We evaluated the expression levels by calculating the relative expression values in the two cell lines. Analysis of variance (ANOVA) was used for intergroup comparisons, and an appropriate post-hoc test was used to compensate for multiple comparisons (SigmaStat). P values, 0.05 were considered significant.
Results
Construction and analysis of the LV-shGPR4
The most efficient shRNA expression cassette was selected and constructed into the lentiviral vector, named LV-shGPR4. To determine the effect of LV-shGPR4 on the expression of GPR4, GFP expression was observed under a fluorescence microscope in the HMEC-1 cells 48 h after infection with LV-shGPR4 and LV-eGFP (Figure 1). Next, real-time PCR and Western blot were performed to determine the mRNA and protein levels of GPR4 in the LV-shGPR4 and LV-eGFP cell groups. As shown in Figure 2A and 2B, LV-shGPR4 significantly inhibited expression of GPR4 protein when compared with the levels in the HMEC-1 cells with LV-CON infection.
Figure 1.
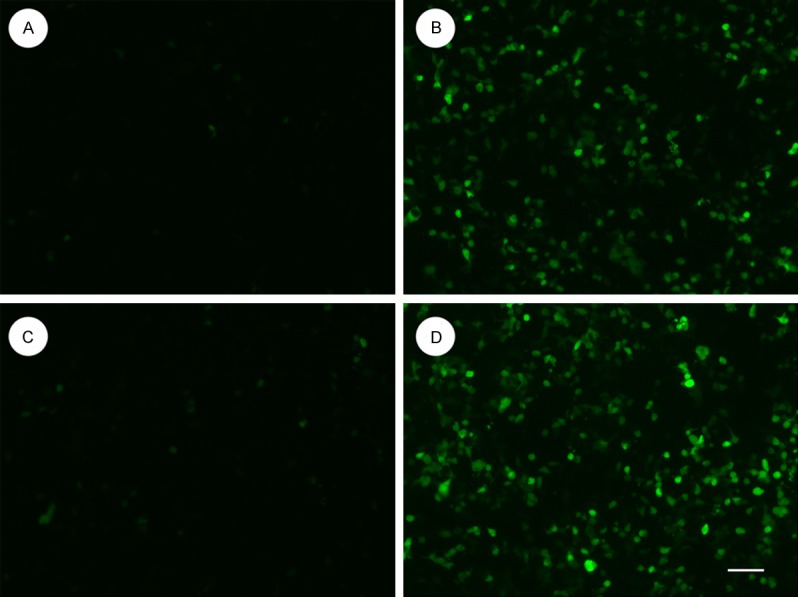
Verification of GPR4 knockdown in HMEC-1 cells by lentiviral-mediated RNA interference. Images of GFP expression showing shRNA delivery efficiency. Bar=50 μM. A: LV-CON-infected HMEC-1 cells for 24 h; B: LV-CON-infected HMEC-1 cells for 48 h; C: LV-shGPR4-infected HMEC-1 cells for 24 h; D: LV-shGPR4-infected HMEC-1 cells for 48 h.
Figure 2.
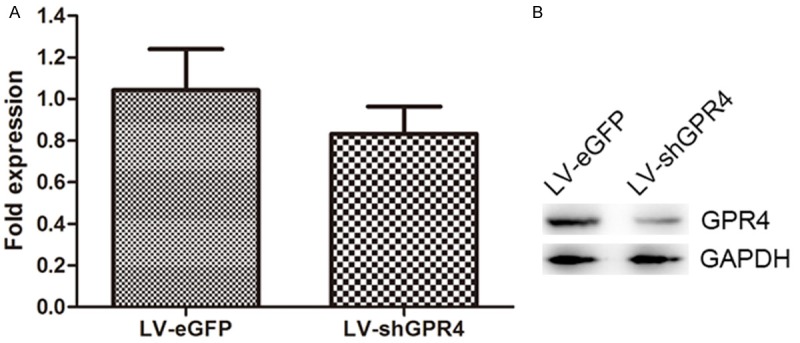
Lentiviral vector-mediated delivery of siRNA targeting GPR4 results in specific knockdown of expression. The indicated lentiviral siRNA expression constructs were cotransfected into HMEC-1 cells with an expression construct for the GPR4 target. A: Real-time PCR analyzed the expression of target gene expression in the RNA level. B: Immunoblotting of whole cell extracts was performed with anti-GPR4 antibody to detect GPR4 (upper panel). An anti-GAPDH antibody was used to confirm equal protein loading (bottom panel).
Gene array analysis of mrna levels in cells interfered by LV-shGPR4
To identify genes up-regulated or down-regulated by LV-shGPR4, we used human OneArray™ and defined the differential expression genes with a criterion (log ratio, P ≤ 0.01 and P ≥ 1.5-fold change in mRNA levels). Among the whole genes and expressed sequence tags, 447 differentially expressed genes were identified, containing 318 up-regulated genes and 129 down-regulated genes (Figure 3). These genes were principally classified into several biological process-related functions using the Panther analytical system, including (1) cell cycle; (2) apoptosis; (3) cell proliferation and differentiation; (4) protein biosynthesis, metabolism, and modification; (5) nucleobases, nucleoside, nucleotide, and nucleic acid metabolism; (6) signal transduction; (7) immune and defense; (8) transcription regulation; and so on.
Figure 3.

Cluster analysis of the gene expression. Clustering of differentially expressed genes in the HMEC-1 cells infected with LV-Con or LV-shGPR4.
Validation of gene expression changes with real-time PCR
Five genes including PPP1CC, ETS2, EIF4EBP2 and PCDH20 were testified with real-time PCR (Table 1). After detecting the expression of GPR4, the target 4 genes were analyzed by real-time PCR. The results of the 4 genes were in concord with microarray, signifying the high reliability of the microarray results (Figure 4).
Table 1.
Primers for real-time PC
| Primer No. | Sequence of primers (5’-3’) | Length (bp) |
|---|---|---|
| PPP1CC-F | TTCTGCTGTCATGGAGGTTTATC | 124 |
| PPP1CC-R | TATCGGGGTCAGACCACAAAA | |
| ETS2-F | CTGGGCATTCCAAAGAACCC | 85 |
| ETS2-R | CCAGACTGAACTCATTGGTGG | |
| EIF4EBP2-F | TAGCCCTGGCACCTTAATTGA | 91 |
| EIF4EBP2-R | ATCCCCAACTGCATGTTTCCT | |
| PCDH20-F | AAAATGCACCTGTAAACACCCG | 86 |
| PCDH20-R | GCGATAGGTCTGTACCCCATTA |
Figure 4.
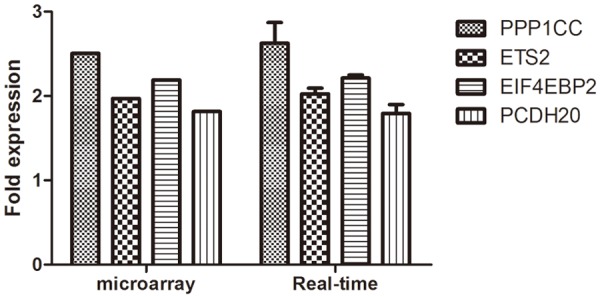
Comparison of 4 differential expression genes between the microarray and RT-PCR analyses.
Discussion
Silencing RNA is a highly specific tool for targeted gene knockdown, and it has advantages over the antisense oligo-DNA or ribozyme because it can be introduced into cells with a high efficiency and exerts its gene-silencing effect at a concentration several orders lower. Today, it is generally accepted that RNA interference is an effective, feasible, and stable approach for exploring gene function and identifying and validating new drug targets in functional genomic studies [20].
After GPR4 was knockdown in HMEC-1 cell, among the whole genes and expressed sequecnce tags, 447 differentially expressed genes were identified, containing 318 up-regulated genes and 129 down-regulated genes. These genes were principally classified into several biological process-related functions using the Panther analytical system, including 1) cell cycle; 2) apoptosis; 3) cell proliferation and differentiation; 4) protein biosynthesis, metabolism, and modification; 5) nucleobases, nucleoside, nucleotide, and nucleic acid metabolism; 6) signal transduction; 7) immune and defense; 8) transcription regulation and so on. Those genes whose expression dramatically changed may be involved in the GPR4 functions. These genes were related to cell apoptosis, cytoskeleton and signal transduction, cell proliferation, differentiation and cell-cycle regulation, gene transcription and translation and cell material and energy metabolism. This also confirm that GPR4 has a range of functions including 1) proton sensing; 2) regulating the growth, survive, migration, differentiation, tube formation of endothelial cells; 3) stimulating the cell proliferation, migration, invasion, colony formation, and survival of ovarian cancer cells.
In addition, 318 genes were upregulated in HMEC-1 cells treated with LV-shGPR4 compared with those treated with LV-CON, which were mainly involved in apoptosis, p53 signaling, and metastasis suppressor. Subsequently, RT-PCR was performed to confirm the GPR4-dependent expression of 4 genes identified in the microarray analysis. The expression of many genes is known to have important role in tumor progression.
The protein encoded by this gene belongs to the protein phosphatase family, PP1 subfamily. PP1 is an ubiquitous serine/threonine phosphatase that regulates many cellular processes, including cell division [21]. PPP1CC (protein phosphatase 1, catalytic subunit, gamma isozyme) is a protein-coding gene. Diseases associated with PPP1CC include ossifying fibroma, and lipoma, and among its related super-pathways are Regulation of lipid metabolism Insulin signaling-generic cascades and Mitotic Anaphase [22,23]. ETS2 (v-ets avian erythroblastosis virus E26 oncogene homolog 2) is a protein-coding gene. Diseases associated with ETS2 include frey syndrome, and Horner’s syndrome, and among its related super-pathways are Rho Family GTPases and OSM Pathway [24,25]. EIF4EBP2 (eukaryotic translation initiation factor 4E binding protein 2) encodes a member of the eukaryotic translation initiation factor 4E binding protein family. Diseases associated with EIF4EBP2 include sleep apnea, and Alzheimer’s disease, and among its related super-pathways are TGF-Beta Pathway and Erythropoietin Pathway [26]. PCDH20 (protocadherin 20) belongs to the protocadherin gene family, a subfamily of the cadherin superfamily and was associated with the WNT/β-catenin pathway [27,28]. Epigenetic silencing by hypermethylation of the CpG-rich promoter region of PCDH20 leads to loss of PCDH20 function, which may be a factor in the carcinogenesis of NSCLC [27].
GPR4 mediated an extremely complex and changing process. This process involves a variety of ways and multiple genes. Gene chip technology can provide comprehensive, reliable technical support for the screening of GPR4 related genes. This study may provide new evidence for GPR4 as a promising gene therapeutic target for cancer.
Acknowledgements
This manuscript is supported by the National Natural Science Foundations of China (Juan Ren, 31201060/C0709; Juan Ren, 30973175/H1621; and Juan Ren, 81172490/H1621); Supported by Program for New Century Excellent Talents in University (Juan Ren, NCET-12-0440); Scientific and Technological Research Foundation of Shaanxi Province (Juan Ren, 2012K13-01-06; Juan Ren, 2007K09-09); Project sponsored by Scientific Research Foundation for the Returned overseas Chinese Scholars of State Education Ministry, (Juan Ren, 0601-18920006); Research Foundation of Health Department of Shaan’xi Province (Juan Ren, 2010D41); Qing Nian Jiao Shi Gen Zong Ji Hua of Xi’an Jiaotong University (“The Fundamental Research Funds for the Central Universities”) (Juan Ren, 2012); Supported by Program for Changjiang Scholars and Innovative Research Team in University (Zongfang Li, PCSIRT:1171); Research Fundation of Xi’an Jiao Tong University of China, (Juan Ren).
Disclosure of conflict of interest
None.
References
- 1.Yang YH, Dudoit S, Luu P, Lin DM, Peng V, Ngai J, Speed TP. Normalization for cDNA microarray data: a robust composite method addressing single and multiple slide systematic variation. Nucleic Acids Res. 2002;30:e15. doi: 10.1093/nar/30.4.e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Seuwen K, Ludwig MG, Wolf RM. Receptors for protons or lipid messengers or both? J Recept Signal Transduct Res. 2006;26:599–610. doi: 10.1080/10799890600932220. [DOI] [PubMed] [Google Scholar]
- 3.Yang LV, Radu CG, Roy M, Lee S, McLaughlin J, Teitell MA, Iruela-Arispe ML, Witte ON. Vascular abnormalities in mice deficient for the G protein-coupled receptor GPR4 that functions as a pH sensor. Mol Cell Biol. 2007;27:1334–1347. doi: 10.1128/MCB.01909-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.An S, Tsai C, Goetzl EJ. Cloning, sequencing and tissue distribution of two related G protein-coupled receptor candidates expressed prominently in human lung tissue. FEBS Lett. 1995;375:121–124. doi: 10.1016/0014-5793(95)01196-l. [DOI] [PubMed] [Google Scholar]
- 5.Zhu K, Baudhuin LM, Hong G, Williams FS, Cristina KL, Kabarowski JH, Witte ON, Xu Y. Sphingosylphosphorylcholine and lysophosphatidylcholine are ligands for the G protein-coupled receptor GPR4. J Biol Chem. 2001;276:41325–41335. doi: 10.1074/jbc.M008057200. [DOI] [PubMed] [Google Scholar]
- 6.Tobo M, Tomura H, Mogi C, Wang JQ, Liu JP, Komachi M, Damirin A, Kimura T, Murata N, Kurose H, Sato K, Okajima F. Previously postulated “ligand-independent” signaling of GPR4 is mediated through proton-sensing mechanisms. Cell Signal. 2007;19:1745–1753. doi: 10.1016/j.cellsig.2007.03.009. [DOI] [PubMed] [Google Scholar]
- 7.Bektas M, Barak LS, Jolly PS, Liu H, Lynch KR, Lacana E, Suhr KB, Milstien S, Spiegel S. The G protein-coupled receptor GPR4 suppresses ERK activation in a ligand-independent manner. Biochemistry. 2003;42:12181–12191. doi: 10.1021/bi035051y. [DOI] [PubMed] [Google Scholar]
- 8.Im DS. Two ligands for a GPCR, proton vs lysolipid. Acta Pharmacol Sin. 2005;26:1435–1441. doi: 10.1111/j.1745-7254.2005.00237.x. [DOI] [PubMed] [Google Scholar]
- 9.Tomura H, Mogi C, Sato K, Okajima F. Proton-sensing and lysolipid-sensitive G-protein-coupled receptors: a novel type of multi-functional receptors. Cell Signal. 2005;17:1466–1476. doi: 10.1016/j.cellsig.2005.06.002. [DOI] [PubMed] [Google Scholar]
- 10.Sin WC, Zhang Y, Zhong W, Adhikarakunnathu S, Powers S, Hoey T, An S, Yang J. G protein-coupled receptors GPR4 and TDAG8 are oncogenic and overexpressed in human cancers. Oncogene. 2004;23:6299–6303. doi: 10.1038/sj.onc.1207838. [DOI] [PubMed] [Google Scholar]
- 11.Kim KS, Ren J, Jiang Y, Ebrahem Q, Tipps R, Cristina K, Xiao YJ, Qiao J, Taylor KL, Lum H, Anand-Apte B, Xu Y. GPR4 plays a critical role in endothelial cell function and mediates the effects of sphingosylphosphorylcholine. FASEB J. 2005;19:819–821. doi: 10.1096/fj.04-2988fje. [DOI] [PubMed] [Google Scholar]
- 12.Hashiya N, Jo N, Aoki M, Matsumoto K, Nakamura T, Sato Y, Ogata N, Ogihara T, Kaneda Y, Morishita R. In vivo evidence of angiogenesis induced by transcription factor Ets-1: Ets-1 is located upstream of angiogenesis cascade. Circulation. 2004;109:3035–3041. doi: 10.1161/01.CIR.0000130643.41587.DB. [DOI] [PubMed] [Google Scholar]
- 13.Qiao J, Huang F, Naikawadi RP, Kim KS, Said T, Lum H. Lysophosphatidylcholine impairs endothelial barrier function through the G protein-coupled receptor GPR4. Am J Physiol Lung Cell Mol Physiol. 2006;291:L91–101. doi: 10.1152/ajplung.00508.2005. [DOI] [PubMed] [Google Scholar]
- 14.Zou Y, Kim CH, Chung JH, Kim JY, Chung SW, Kim MK, Im DS, Lee J, Yu BP, Chung HY. Upregulation of endothelial adhesion molecules by lysophosphatidylcholine. Involvement of G protein-coupled receptor GPR4. FEBS J. 2007;274:2573–2584. doi: 10.1111/j.1742-4658.2007.05792.x. [DOI] [PubMed] [Google Scholar]
- 15.Xu Y. Sphingosylphosphorylcholine and lysophosphatidylcholine: G protein-coupled receptors and receptor-mediated signal transduction. Biochim Biophys Acta. 2002;1582:81–88. doi: 10.1016/s1388-1981(02)00140-3. [DOI] [PubMed] [Google Scholar]
- 16.Wyder L, Suply T, Ricoux B, Billy E, Schnell C, Baumgarten BU, Maira SM, Koelbing C, Ferretti M, Kinzel B, Muller M, Seuwen K, Ludwig MG. Reduced pathological angiogenesis and tumor growth in mice lacking GPR4, a proton sensing receptor. Angiogenesis. 2011;14:533–544. doi: 10.1007/s10456-011-9238-9. [DOI] [PubMed] [Google Scholar]
- 17.Ren J, Jin W, Gao YE, Xia L, Zhang Y, Li W, Zhang X, Zhao D, Li Z, Ma H, Wang J, Liu R, Chen Y, Qian J, Shi X, Liu Y. Relations between GPR4 Expression, microvascular density (MVD) and Clinical Pathological Characteristics of Patients with Epithelial Ovarian Carcinoma (EOC) Curr Pharm Des. 2013 doi: 10.2174/13816128113199990530. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 18.Matranga C, Tomari Y, Shin C, Bartel DP, Zamore PD. Passenger-strand cleavage facilitates assembly of siRNA into Ago2-containing RNAi enzyme complexes. Cell. 2005;123:607–620. doi: 10.1016/j.cell.2005.08.044. [DOI] [PubMed] [Google Scholar]
- 19.Davidson BL, McCray PB Jr. Current prospects for RNA interference-based therapies. Nat Rev Genet. 2011;12:329–340. doi: 10.1038/nrg2968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Martin SE, Caplen NJ. Applications of RNA interference in mammalian systems. Annu Rev Genomics Hum Genet. 2007;8:81–108. doi: 10.1146/annurev.genom.8.080706.092424. [DOI] [PubMed] [Google Scholar]
- 21.Cohen PT. Protein phosphatase 1--targeted in many directions. J Cell Sci. 2002;115:241–256. doi: 10.1242/jcs.115.2.241. [DOI] [PubMed] [Google Scholar]
- 22.Djouder N, Metzler SC, Schmidt A, Wirbelauer C, Gstaiger M, Aebersold R, Hess D, Krek W. S6K1-mediated disassembly of mitochondrial URI/PP1gamma complexes activates a negative feedback program that counters S6K1 survival signaling. Mol Cell. 2007;28:28–40. doi: 10.1016/j.molcel.2007.08.010. [DOI] [PubMed] [Google Scholar]
- 23.Windelinckx A, De Mars G, Huygens W, Peeters MW, Vincent B, Wijmenga C, Lambrechts D, Aerssens J, Vlietinck R, Beunen G, Thomis MA. Identification and prioritization of NUAK1 and PPP1CC as positional candidate loci for skeletal muscle strength phenotypes. Physiol Genomics. 2011;43:981–992. doi: 10.1152/physiolgenomics.00200.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chatterjee A, Dutta S, Mukherjee S, Mukherjee N, Chandra S, Mukherjee A, Sinha S, Panda CK, Chaudhuri K, Mukhopadyay K. Differential allelic distribution of V-ets erythroblastosis virus E26 oncogene homolog2 (ETS2) functional polymorphisms in different group of patients. Gene Expr. 2010;15:61–73. doi: 10.3727/105221611x12973615737541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dwyer J, Li H, Xu D, Liu JP. Transcriptional regulation of telomerase activity: roles of the the Ets transcription factor family. Ann N Y Acad Sci. 2007;1114:36–47. doi: 10.1196/annals.1396.022. [DOI] [PubMed] [Google Scholar]
- 26.Bailey SD, Xie C, Do R, Montpetit A, Diaz R, Mohan V, Keavney B, Yusuf S, Gerstein HC, Engert JC, Anand S. Variation at the NFATC2 locus increases the risk of thiazolidinedione-induced edema in the Diabetes REduction Assessment with ramipril and rosiglitazone Medication (DREAM) study. Diabetes Care. 2010;33:2250–2253. doi: 10.2337/dc10-0452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Imoto I, Izumi H, Yokoi S, Hosoda H, Shibata T, Hosoda F, Ohki M, Hirohashi S, Inazawa J. Frequent silencing of the candidate tumor suppressor PCDH20 by epigenetic mechanism in non-small-cell lung cancers. Cancer Res. 2006;66:4617–4626. doi: 10.1158/0008-5472.CAN-05-4437. [DOI] [PubMed] [Google Scholar]
- 28.Berndt JD, Aoyagi A, Yang P, Anastas JN, Tang L, Moon RT. Mindbomb 1, an E3 ubiquitin ligase, forms a complex with RYK to activate Wnt/beta-catenin signaling. J Cell Biol. 2011;194:737–750. doi: 10.1083/jcb.201107021. [DOI] [PMC free article] [PubMed] [Google Scholar]


