Abstract
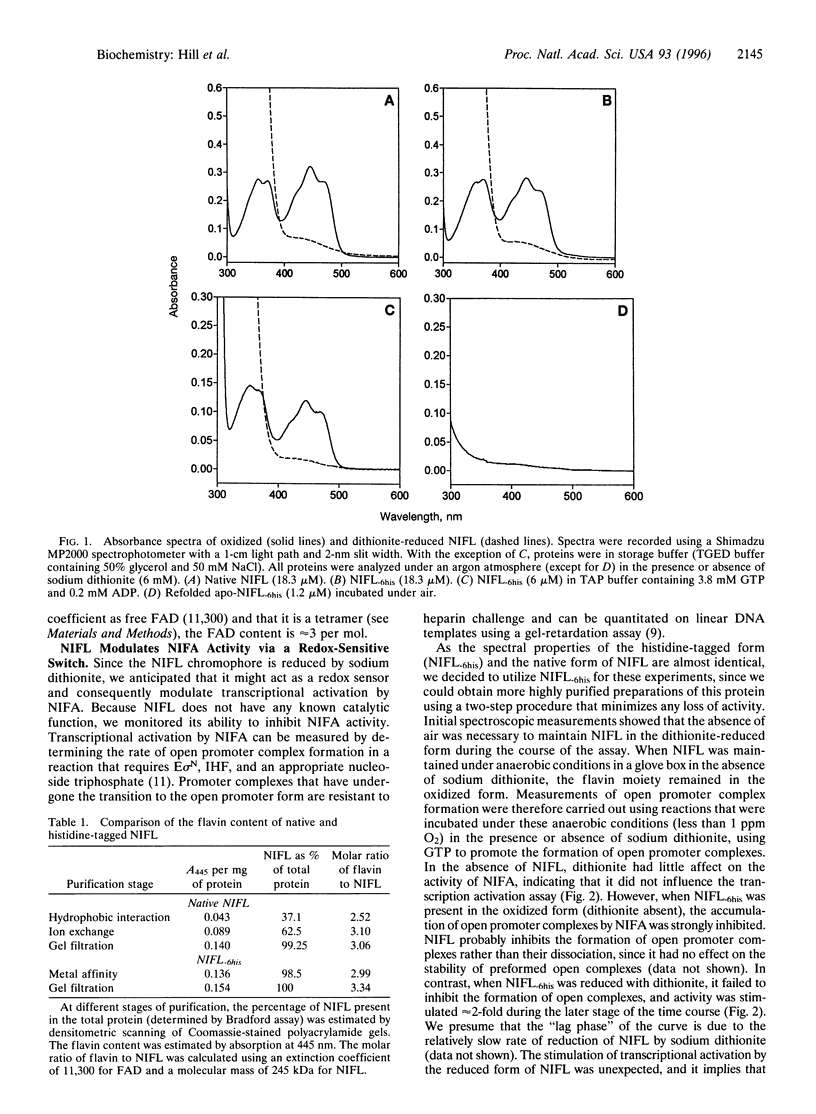
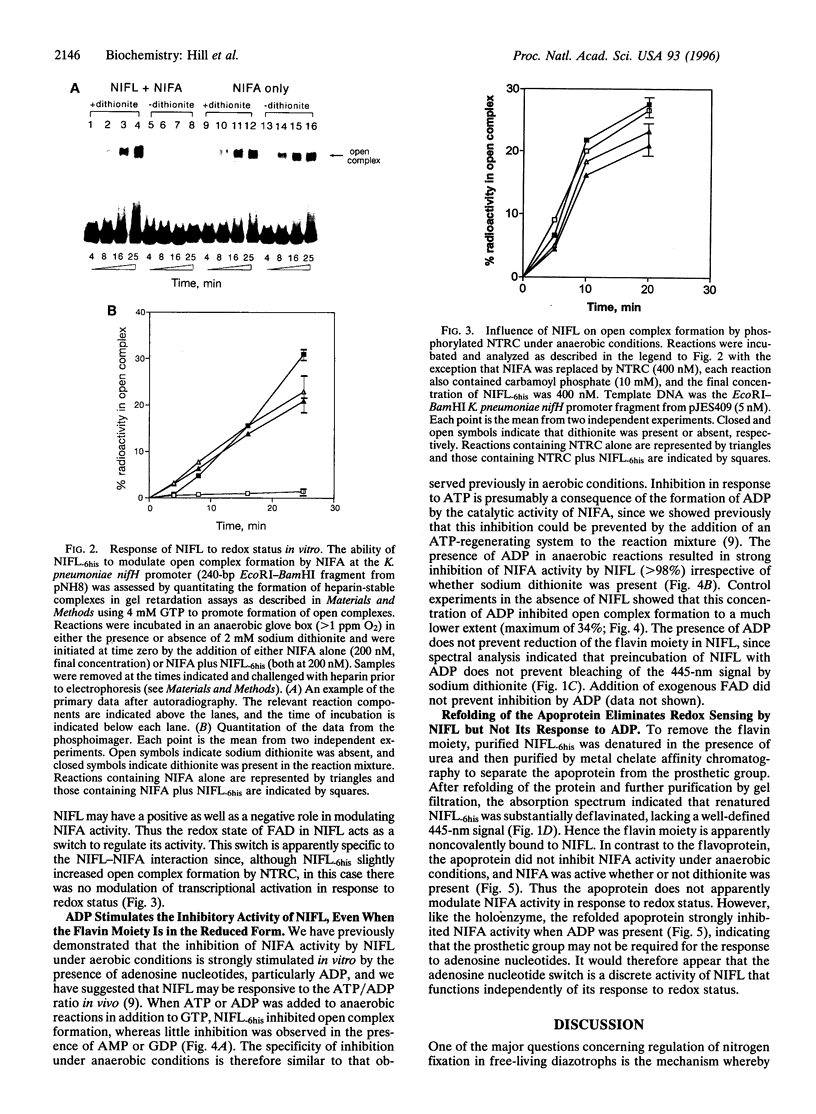
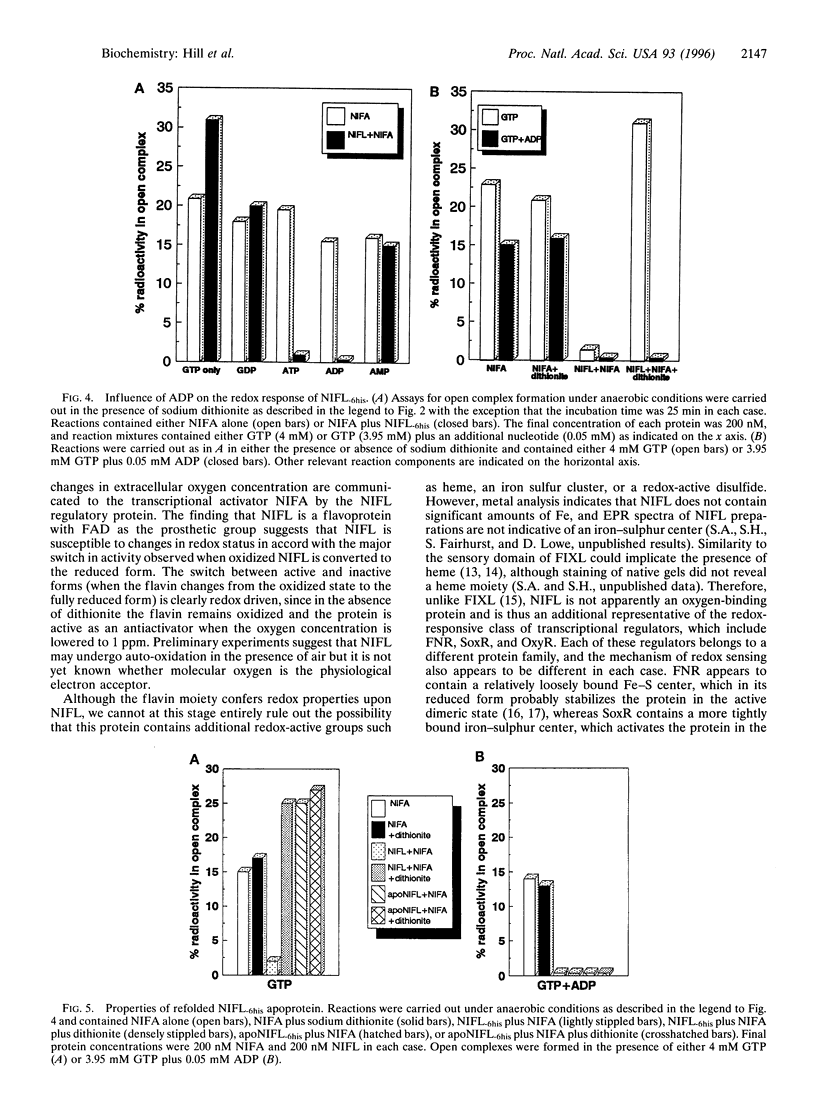
The NIFL regulatory protein controls transcriptional activation of nitrogen fixation (nif) genes in Azotobacter vinelandii by direct interaction with the enhancer binding protein NIFA. Modulation of NIFA activity by NIFL, in vivo occurs in response to external oxygen concentration or the level of fixed nitrogen. Spectral features of purified NIFL and chromatographic analysis indicate that it is a flavoprotein with FAD as the prosthetic group, which undergoes reduction in the presence of sodium dithionite. Under anaerobic conditions, the oxidized form of NIFL inhibits transcriptional activation by NIFA in vitro, and this inhibition is reversed when NIFL is in the reduced form. Hence NIFL is a redox-sensitive regulatory protein and may represent a type of flavoprotein in which electron transfer is not coupled to an obvious catalytic activity. In addition to its ability to act as a redox sensor, the activity of NIFL is also responsive to adenosine nucleotides, particularly ADP. This response overrides the influence of redox status on NIFL and is also observed with refolded NIFL apoprotein, which lacks the flavin moiety. These observations suggest that both energy and redox status are important determinants of nif gene regulation in vivo.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Agron P. G., Ditta G. S., Helinski D. R. Oxygen regulation of nifA transcription in vitro. Proc Natl Acad Sci U S A. 1993 Apr 15;90(8):3506–3510. doi: 10.1073/pnas.90.8.3506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Austin S., Buck M., Cannon W., Eydmann T., Dixon R. Purification and in vitro activities of the native nitrogen fixation control proteins NifA and NifL. J Bacteriol. 1994 Jun;176(12):3460–3465. doi: 10.1128/jb.176.12.3460-3465.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bali A., Blanco G., Hill S., Kennedy C. Excretion of ammonium by a nifL mutant of Azotobacter vinelandii fixing nitrogen. Appl Environ Microbiol. 1992 May;58(5):1711–1718. doi: 10.1128/aem.58.5.1711-1718.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger D. K., Narberhaus F., Kustu S. The isolated catalytic domain of NIFA, a bacterial enhancer-binding protein, activates transcription in vitro: activation is inhibited by NIFL. Proc Natl Acad Sci U S A. 1994 Jan 4;91(1):103–107. doi: 10.1073/pnas.91.1.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanco G., Drummond M., Woodley P., Kennedy C. Sequence and molecular analysis of the nifL gene of Azotobacter vinelandii. Mol Microbiol. 1993 Aug;9(4):869–879. doi: 10.1111/j.1365-2958.1993.tb01745.x. [DOI] [PubMed] [Google Scholar]
- Contreras A., Drummond M., Bali A., Blanco G., Garcia E., Bush G., Kennedy C., Merrick M. The product of the nitrogen fixation regulatory gene nfrX of Azotobacter vinelandii is functionally and structurally homologous to the uridylyltransferase encoded by glnD in enteric bacteria. J Bacteriol. 1991 Dec;173(24):7741–7749. doi: 10.1128/jb.173.24.7741-7749.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eydmann T., Söderbäck E., Jones T., Hill S., Austin S., Dixon R. Transcriptional activation of the nitrogenase promoter in vitro: adenosine nucleotides are required for inhibition of NIFA activity by NIFL. J Bacteriol. 1995 Mar;177(5):1186–1195. doi: 10.1128/jb.177.5.1186-1195.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilles-Gonzalez M. A., Ditta G. S., Helinski D. R. A haemoprotein with kinase activity encoded by the oxygen sensor of Rhizobium meliloti. Nature. 1991 Mar 14;350(6314):170–172. doi: 10.1038/350170a0. [DOI] [PubMed] [Google Scholar]
- Gilles-Gonzalez M. A., Gonzalez G., Perutz M. F., Kiger L., Marden M. C., Poyart C. Heme-based sensors, exemplified by the kinase FixL, are a new class of heme protein with distinctive ligand binding and autoxidation. Biochemistry. 1994 Jul 5;33(26):8067–8073. doi: 10.1021/bi00192a011. [DOI] [PubMed] [Google Scholar]
- Gilles-Gonzalez M. A., Gonzalez G. Regulation of the kinase activity of heme protein FixL from the two-component system FixL/FixJ of Rhizobium meliloti. J Biol Chem. 1993 Aug 5;268(22):16293–16297. [PubMed] [Google Scholar]
- Green J., Guest J. R. Activation of FNR-dependent transcription by iron: an in vitro switch for FNR. FEMS Microbiol Lett. 1993 Oct 15;113(2):219–222. doi: 10.1111/j.1574-6968.1993.tb06517.x. [DOI] [PubMed] [Google Scholar]
- Hidalgo E., Demple B. An iron-sulfur center essential for transcriptional activation by the redox-sensing SoxR protein. EMBO J. 1994 Jan 1;13(1):138–146. doi: 10.1002/j.1460-2075.1994.tb06243.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoover T. R., Santero E., Porter S., Kustu S. The integration host factor stimulates interaction of RNA polymerase with NIFA, the transcriptional activator for nitrogen fixation operons. Cell. 1990 Oct 5;63(1):11–22. doi: 10.1016/0092-8674(90)90284-l. [DOI] [PubMed] [Google Scholar]
- Khoroshilova N., Beinert H., Kiley P. J. Association of a polynuclear iron-sulfur center with a mutant FNR protein enhances DNA binding. Proc Natl Acad Sci U S A. 1995 Mar 28;92(7):2499–2503. doi: 10.1073/pnas.92.7.2499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kullik I., Toledano M. B., Tartaglia L. A., Storz G. Mutational analysis of the redox-sensitive transcriptional regulator OxyR: regions important for oxidation and transcriptional activation. J Bacteriol. 1995 Mar;177(5):1275–1284. doi: 10.1128/jb.177.5.1275-1284.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H. S., Narberhaus F., Kustu S. In vitro activity of NifL, a signal transduction protein for biological nitrogen fixation. J Bacteriol. 1993 Dec;175(23):7683–7688. doi: 10.1128/jb.175.23.7683-7688.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morett E., Segovia L. The sigma 54 bacterial enhancer-binding protein family: mechanism of action and phylogenetic relationship of their functional domains. J Bacteriol. 1993 Oct;175(19):6067–6074. doi: 10.1128/jb.175.19.6067-6074.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moshiri F., Kim J. W., Fu C., Maier R. J. The FeSII protein of Azotobacter vinelandii is not essential for aerobic nitrogen fixation, but confers significant protection to oxygen-mediated inactivation of nitrogenase in vitro and in vivo. Mol Microbiol. 1994 Oct;14(1):101–114. doi: 10.1111/j.1365-2958.1994.tb01270.x. [DOI] [PubMed] [Google Scholar]
- Santero E., Hoover T. R., North A. K., Berger D. K., Porter S. C., Kustu S. Role of integration host factor in stimulating transcription from the sigma 54-dependent nifH promoter. J Mol Biol. 1992 Oct 5;227(3):602–620. doi: 10.1016/0022-2836(92)90211-2. [DOI] [PubMed] [Google Scholar]
- Thorneley R. N., Ashby G. A. Oxidation of nitrogenase iron protein by dioxygen without inactivation could contribute to high respiration rates of Azotobacter species and facilitate nitrogen fixation in other aerobic environments. Biochem J. 1989 Jul 1;261(1):181–187. doi: 10.1042/bj2610181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodley P., Drummond M. Redundancy of the conserved His residue in Azotobacter vinelandii NifL, a histidine autokinase homologue which regulates transcription of nitrogen fixation genes. Mol Microbiol. 1994 Aug;13(4):619–626. doi: 10.1111/j.1365-2958.1994.tb00456.x. [DOI] [PubMed] [Google Scholar]