Abstract
Curcumin has become a compound of interest for its antioxidant and anti-neoplastic properties. This study sought to determine the effect of curcumin administration on cell proliferation and apoptosis in hepatoma cells. SMMC-7721 hepatoma cells were treated with 10, 30, or 90 μM curcumin solution, with DMEM alone (negative control), or with 20 mg/L fluorouracil (positive control). MTT colorimetry detected significant differences in the rates of cell proliferation inhibition following curcumin treatment, with increasing inhibition accompanying increasing doses of curcumin (P < 0.05), compared to the negative control. Similarly, flow cytometry revealed significant differences in the numbers of apoptotic cells following curcumin treatment: increasing doses of curcumin produced increases in the numbers of apoptotic cells (P < 0.05). To determine whether curcumin exerts these effects by altering the Notch signaling pathway, a phenomenon reported for other cancers, relative expression of Notch1 mRNA and protein were determined in curcumin-treated cells. Both mRNA and protein expression of Notch1 decreased with increasing curcumin dose (P < 0.05). Thus, curcumin appears to inhibit proliferation and induce apoptosis in hepatoma cells by altering the Notch signaling pathway.
Keywords: Curcumin, hepatoma cell, Notch signal pathway, proliferation, apoptosis
Introduction
Primary hepatic carcinoma, or liver cancer, is a common cause of cancer death, having both high morbidity [1]. Indeed, primary hepatic cancer typically results in death within 6 to 20 months of diagnosis [2]. This cancer is currently treated using chemotherapy, radiotherapy, and surgery; these treatments, however, have serious side effects and high risks, and the overall therapeutic effect remains limited despite encouraging progress [3,4]. Therefore, continued exploration of alternative treatment options is critical to improving the outcomes of liver cancer.
Curcumin is a phenolic compound derived from root stocks of the rhizoma Curcuma longa, Curcuma aromatica, Curcuma zedoary, and Acorus calamus. A yellow pigment used in cooking, curcumin has been used pharmacologically in traditional Chinese medicine for centuries [5]. Among its various pharmacologic actions, curcumin has anti-neoplastic capabilities: it can inhibit growth, induce apoptosis [6], and reduce invasion and metastasis [7,8] of tumor cells. In addition, curcumin has few side effects and high safety. Thus, this natural compound has become an international research focus for potential cancer therapies [9].
Curcumin appears to act as an anti-neoplastic agent through directly and indirectly targeting cell cycle regulators [10,11]. By binding to transcription factors, cytokines, and growth factors, curcumin and its analogues can mediate cell growth, proliferation, and migration. Further, curcumin can influence signal transduction through the pathways regulating these cell functions by interacting with diverse molecules like TNF-α, COX-2, NF-κB, and several MMPs (matrix metalloproteinases) [12,13].
One signal transduction pathway that appears to be affected by curcumin activity is the Notch signaling pathway, which is highly conserved among invertebrates and vertebrates [14]. Notch signaling influences multiple processes of cell development, including cell differentiation, proliferation, and apoptosis. Additionally, dysregulation of Notch signaling is associated with oncogenesis [15-17]. Interestingly, accumulating evidence demonstrates some effects of curcumin on Notch signaling in the context of cancers, indicating the potential of using curcumin to target Notch signaling as an alternative cancer therapy [18-20].
Here, curcumin was administered in hepatic carcinoma cells to determine the cellular processes affected by the compound and whether it dysregulated Notch signaling.
Materials and methods
Cell culture
Hepatoma SMMC-7721 cells (Wuhan Boster Biotechnology) were cultured in high-glucose DMEM medium (HyClone, USA) containing 10% newborn calf serum (Wuhan Sanli Biotechnology), 100 U/mL penicillin, and 100 U/mL streptomycin at 37°C and 5% CO2. Medium was changed every 24 hours, and cells were passaged at about 90% confluency. Cells were rinsed with PBS (pH 7.2-7.4) and detached using 0.25% trypsin (HyClone) for 2-3 min. Complete medium was used to wash cells. Cells at logarithmic growth phase were used for experiments.
MTT colorimetry
Cells (3 × 105 cells/mL) were transferred to a 96-well plate, with 200 μL suspension per well, and cultured in an incubator (MCO-15AC, SANYO, Japan) for 24 hours at 37°C and 5% CO2. Wells were prepared for addition of curcumin by removing the culture medium. Curcumin powder (formula C12H36O6, relative molecular mass 368.38, purity > 99%, China National Pharmaceutical Group, Shanghai Sinopharm Chemical Reagent Co.) was dissolved in dimethyl sulfoxide (DMSO), to prepare 10 mM stock solution. Curcumin solution was sterilized by membrane filtration (0.22 μm) sterilization, aliquoted, and stored at -20°C. For use, stock solution was thawed and diluted with high-glucose DMEM to final concentrations of 10 μM, 30 μM, and 90 μM; the final concentration of DMSO in the curcumin solution was < 1%.
Curcumin was added, 200 μL per well, to different wells according to concentration: curcumin low-dose group, 10 μM; mid-dose group, 30 μM; and high-dose group, 90 μM. Other wells received the following treatments: negative control group, high-glucose DMEM medium; positive control group, 20 mg/L fluorouracil injection (FU, Shanghai Xudong Haipu Pharmaceutical). Each of the 5 groups comprised 6 sample wells. The medium was changed every 24 hours, followed by drug supplement. After 48 hours in culture, 20 μL of 5 mg/mL fresh thiazolyl blue solution (MTT, Sigma, USA) were added to each well. Following 4 hours in culture, medium was removed and 150 μL DMSO (Sigma, USA) were added to each well. Plates were horizontally vibrated for 10 min before optical absorbance (A) at 490 nm was measured with a microplate reader (Model 680, BIO-RAD, USA). The average of the absorbance values for all six wells was used to calculate the rate of inhibition of proliferation: inhibition rate = (1-absorbance of experimental group / absorbance of control group) × 100%.
Flow cytometry
Cells were prepared in 96-well culture plates as for the MTT assay. Curcumin solution, positive control, and negative control were administered as for the MTT assay. After culturing for 48 hours, cells were detached using trypsin, collected, and suspended at a density of 2.5 × 105 cells/mL. From that, 2 mL cell suspension were centrifuged at 1000 rpm for 5 minutes. Cell pellets were washed twice with cold PBS, filtered through 300 mesh nylon net, and re-suspended in 200 μL binding buffer. For flow cytometry detection, 10 μL Annexin-V were applied to wells for 15 min away from light, then 5 μL propidium iodide were applied for 15 min away from light. Finally, 200 μL binding buffer were added to each well, and cells were detected immediately with a flow cytometer (FACS calibur, BD Biosciences, USA). Results were analyzed with CellQuest (BD Biosciences) software.
Western blotting
SMMC-7721 cells in logarithmic growth phase were used to prepare a suspension of 3 × 105 cells/mL, for culture on a 6-well plate (2 mL suspension in each well) at 37°C and 5% CO2. After 24 hours in culture, medium was discarded and curcumin solution and control solutions (2 mL per well) were added as for previous experiments. After culturing for 48 hours, cells were detached with trypsin and collected. Cells were treated with RIPA lysis solution (Shanghai Beyotime Biotech) to extract total protein, and the BCA method was used to determine protein concentration. Samples were separated with standard polyacrylamide gel electrophoresis, transferred to a PVDF membrane, and immunoblotted with primary antibodies (dilution: 1:1000) and secondary antibodies. Proteins were detected with ECL luminescence. β-actin (Sigma) was used as the internal reference. Experiment was repeated 3 times.
RT-PCR
Total RNA was extracted from SMMC-7721 cells with Trizol (Invitrogen, USA) according to kit instructions. RNA was reverse-transcribed, then cDNA was used as a template for PCR. Primer sequences for Notch1 amplification were as follows: forward, 5’-CAGCGAATCCGAGGACTATG-3’; and reverse, 5’-CAGGCGTGTT GTTCTCACAG-3’. Thermal cycling comprised denaturation at 94°C for 30 seconds, annealing at 55°C for 1 min, and extension at 72°C for 1 min, for 30 cycles. Expected PCR product size for Notch1 was 428 bp. Gapdh was amplified as an internal control using the following primer sequences: forward, 5’-GGGTGATGCTGGTGCTGAGTATGT-3’; and reverse, 5’-AAGAATGGGTGTTG CTGTTGAAGTC-3’. PCR cycling conditions were denaturation at 94°C for 30 seconds, annealing at 55°C for 30 seconds, and extension at 72°C for 1 min, for 25 cycles. PCR product was expected to be 700 bp. PCR products were separated by agarose gel electrophoresis. Gel imaging system was used for observation and photographing. Each test was repeated 3 times.
Statistical analysis
SPSS17.0 statistical software was used for statistical analysis. Measurements are expressed as mean ± standard deviation (x̅ ± s). Single factor analysis of variance (one-way ANOVA) and pair-wise comparisons (SNK-q test method) of inter-group mean values were performed on the test results. The above analyses were performed using two-sided tests, at α of 0.05. P < 0.05 was considered statistically significant.
Results
Cell growth inhibition by curcumin is dose-dependent
Rates of inhibition of cell proliferation, detected by MTT colorimetry, were altered by curcumin administration in a dose-dependent fashion (Table 1). Optical absorbance values in the low-dose, mid-dose, and high-dose curcumin groups and positive control group were all significantly different from that of the negative control group (P < 0.05). Further, as the curcumin dose increased, the absorbance values decreased, while the inhibition rate showed a rising trend, in a dose-dependent manner.
Table 1.
Growth inhibition rates in curcumin-treated hepatoma cells
| Treatment Group | n | Absorbance at 490 nm (A) | Cell growth inhibition rate (%) |
|---|---|---|---|
| Negative control | 6 | 1.535 ± 0.170 | - |
| Low-dose curcumin (10 μM) | 6 | 1.214 ± 0.107* | 79.6 ± 8.2 |
| Mid-dose curcumin (30 μM) | 6 | 1.093 ± 0.071* | 72.1 ± 10.3 |
| High-dose curcumin (90 μM) | 6 | 0.537 ± 0.059* | 35.3 ± 4.8 |
| Positive control | 6 | 0.647 ± 0.044* | 42.8 ± 7.2 |
| F | 100.138 | 45.547 | |
| P | 0.001 | 0.001 |
P < 0.05, compared to the negative control group.
Apoptosis increases in curcumin-treated samples
Flow cytometry with annexin- and PI-labeled hepatoma cells revealed dose-dependent differences in the numbers of apoptotic cells in curcumin-treated hepatoma cultures (Table 2). Total numbers of apoptotic cells were significantly increased in each of the curcumin-treated groups and the positive control group compared with the negative control group (P < 0.05). Further, the early apoptotic cell count increased with the increasing doses of curcumin, while the late apoptotic cell count gradually decreased (P < 0.05).
Table 2.
Apoptosis rates in curcumin-treated hepatoma cells (%)
| Group | n | Early apoptosis | Late apoptosis | Total number of apoptotic cells |
|---|---|---|---|---|
| Negative control | 3 | 4.426 ± 0.391 | 3.644 ± 0.358 | 8.070 ± 0.744 |
| Low-dose curcumin (10 μM) | 3 | 5.856 ± 0.125* | 6.082 ± 0.911* | 11.938 ± 1.036* |
| Mid-dose curcumin (30 μM) | 3 | 24.757 ± 3.365* | 5.919 ± 0.304* | 30.677 ± 3.547* |
| High-dose curcumin (90 μM) | 3 | 56.780 ± 6.500* | 1.648 ± 0.394* | 58.428 ± 6.363* |
| Positive control | 3 | 61.951 ± 3.515* | 0.646 ± 0.236* | 62.596 ± 3.515* |
| F | 170.193 | 71.592 | 144.507 | |
| P | 0.001 | 0.001 | 0.001 |
P<0.05, compared to the negative control group.
Relative Notch1 expression decreases in curcumin-treated cells
To determine the effect of curcumin treatment on Notch1 signaling, mRNA (Figure 1, left) and protein expression (Figure 1, right) of Notch1 were examined in treated hepatoma cells. Compared with the negative control group, both mRNA and protein expression of Notch1 were significantly lower in curcumin-treated and positive control groups (P < 0.05).
Figure 1.
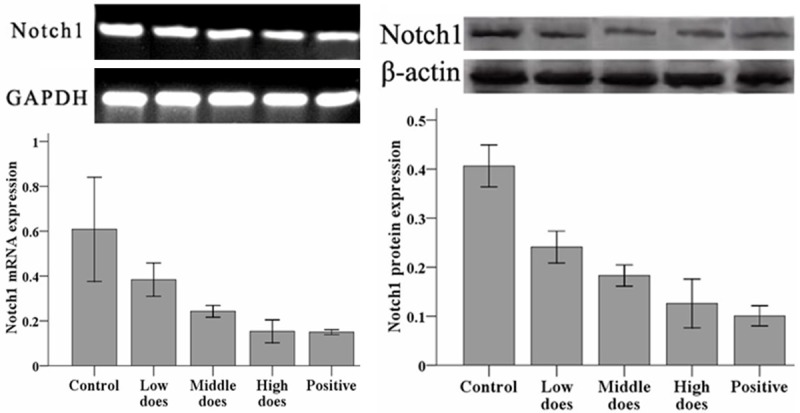
Expression of Notch1 mRNA (left) and protein (right) in curcumin-treated hepatoma cells. Relative expression of Notch1 mRNA was quantified by RT-PCR and compared with Gapdh. Relative expression of Notch1 protein was quantified by Western blotting and compared with β-actin.
Discussion
Liver cancer involves a large number of genetic and physiological changes, which can affect the survival and proliferation of liver cancer cells. The most common change occurs in the Notch signaling pathway [21]. Notch signaling plays critical roles in embryonic development, organ formation, and tissue balance [14]. Importantly, Notch1 expression is up-regulated in gastric cancer, esophageal cancer, and other tumors [16,17,22,23], suggesting its potential involvement in tumorigenesis, particularly within the digestive tract.
The main active ingredient of curcumin is turmeric, an inexpensive and abundant compound often used in food coloring. Studies have shown that curcumin can inhibit the growth and transformation of cells, has a variety of potential molecular targets, and may act through different signaling pathways [5]. Further, it has been demonstrated to reduce tumor capabilities to grow and metastasize [11-13]. Additionally, curcumin appears to interact with Notch signaling [18-20,24-26]. Here, curcumin administration in hepatoma cells in vitro resulted in increased inhibition of proliferation, increased apoptosis, and decreased Notch1 expression. Indeed, each of these features correlated with the dose of curcumin, with more dramatic effects at higher doses. Therefore, curcumin might inhibit proliferation and induce apoptosis of liver cancer cells through directly or indirectly inhibiting Notch1 expression. Further studies are needed to determine the mechanism(s) by which curcumin regulates Notch1 expression and how these changes lead to increased apoptosis and inhibition of cell proliferation.
Disclosure of conflict of interest
None.
References
- 1.Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61:69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- 2.Bosch FX, Ribes J, Díaz M, Cleries R. Primary liver cancer: worldwide incidence and trends. Gastroenterology. 2004;127:S5–S16. doi: 10.1053/j.gastro.2004.09.011. [DOI] [PubMed] [Google Scholar]
- 3.Oishi N, Wang XW. Novel therapeutic strategies for targeting liver cancer stem cells. Int J Biol Sci. 2011;7:517–535. doi: 10.7150/ijbs.7.517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Woo HY, Heo J. Sorafenib in liver cancer. Expert Opin Pharmacother. 2012;13:1059–1067. doi: 10.1517/14656566.2012.679930. [DOI] [PubMed] [Google Scholar]
- 5.Asher GN, Spelman K. Clinical utility of curcumin extract. Altern Ther Health Med. 2013;19:20–22. [PubMed] [Google Scholar]
- 6.Choudhuri T, Pal S, Agwarwal ML, Das T, Sa G. Curcumin induces apoptosis in human breast cancer cells through p53-dependent Bax induction. FEBS Lett. 2002;512:334–340. doi: 10.1016/s0014-5793(02)02292-5. [DOI] [PubMed] [Google Scholar]
- 7.Menon LG, Kuttan R, Kuttan G. Anti-metastatic activity of curcumin and catechin. Cancer Lett. 1999;141:159–165. doi: 10.1016/s0304-3835(99)00098-1. [DOI] [PubMed] [Google Scholar]
- 8.Chen HW, Yu SL, Chen JJ, Li HN, Lin YC, Yao PL, Chou HY, Chien CT, Chen WJ, Lee YT, Yang PC. Anti-invasive gene expression profile of curcumin in lung adenocarcinoma based on a high throughput microarray analysis. Mol Pharmacol. 2004;65:99–110. doi: 10.1124/mol.65.1.99. [DOI] [PubMed] [Google Scholar]
- 9.Gupta SC, Kismali G, Aggarwal BB. Curcumin, a component of turmeric: from farm to pharmacy. Biofactors. 2013;39:2–13. doi: 10.1002/biof.1079. [DOI] [PubMed] [Google Scholar]
- 10.Shehzad A, Lee J, Lee YS. Curcumin in various cancers. Biofactors. 2013;39:56–68. doi: 10.1002/biof.1068. [DOI] [PubMed] [Google Scholar]
- 11.Kunnumakkara AB, Anand P, Aggarwal BB. Curcumin inhibits proliferation, invasion, angiogenesis and metastasis of different cancers through interaction with multiple cell signaling proteins. Cancer Lett. 2008;269:199–225. doi: 10.1016/j.canlet.2008.03.009. [DOI] [PubMed] [Google Scholar]
- 12.Gupta SC, Prasad S, Kim JH, Patchva S, Webb LJ, Priyadarsini IK, Aggarwal BB. Multitargeting by curcumin as revealed by molecular interaction studies. Nat Prod Rep. 2011;28:1937–1955. doi: 10.1039/c1np00051a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shehzad A, Lee YS. Molecular mechanisms of curcumin action: signal transduction. Biofactors. 2013;39:27–36. doi: 10.1002/biof.1065. [DOI] [PubMed] [Google Scholar]
- 14.Bray S. Notch signalling: a simple pathway becomes complex. Nat Rev Mol Cell Biol. 2006;7:678–689. doi: 10.1038/nrm2009. [DOI] [PubMed] [Google Scholar]
- 15.Dang TP. Notch, apoptosis and cancer. Adv Exp Med Biol. 2012;727:199–209. doi: 10.1007/978-1-4614-0899-4_15. [DOI] [PubMed] [Google Scholar]
- 16.Geissler K, Zach O. Pathways involved in Drosophila and human cancer development: the Notch, Hedgehog, Wingless, Runt, and Trithorax pathway. Ann Hematol. 2012;91:645–669. doi: 10.1007/s00277-012-1435-0. [DOI] [PubMed] [Google Scholar]
- 17.Hu YY, Zheng MH, Zhang R, Liang YM, Han H. Notch signaling pathway and cancer metastasis. Adv Exp Med Biol. 2012;727:186–198. doi: 10.1007/978-1-4614-0899-4_14. [DOI] [PubMed] [Google Scholar]
- 18.Wang Z, Zhang Y, Banerjee S, Li Y, Sarkar FH. Notch-1 down-regulation by curcumin is associated with the inhibition of cell growth and the induction of apoptosis in pancreatic cancer cells. Cancer. 2006;106:2503–2513. doi: 10.1002/cncr.21904. [DOI] [PubMed] [Google Scholar]
- 19.Kunnumakkara AB, Anand P, Aggarwal BB. Curcumin inhibits proliferation, invasion, angiogenesis and metastasis of different cancers through interaction with multiple cell signaling proteins. Cancer Lett. 2008;269:199–225. doi: 10.1016/j.canlet.2008.03.009. [DOI] [PubMed] [Google Scholar]
- 20.Ning LI, Wentworth L, Chen H, Weber SM. Down-regulation of Notch1 signaling inhibits tumor growth in human hepatocellular carcinoma. Am J Transl Res. 2009;1:358–366. [PMC free article] [PubMed] [Google Scholar]
- 21.Wang ZW, Li Y, Sarkar FH. Notch signaling proteins: legitimate targets for cancer therapy. Curr Protein Pept Sci. 2010;11:398–408. doi: 10.2174/138920310791824039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Han JX, Hendzel MJ, Allalunis-Turner J. Notch signaling as a therapeutic target for breast cancer treatment? Breast Cancer Res. 2011;13:210. doi: 10.1186/bcr2875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li DW, Wu Q, Peng ZH, Yang ZR, Wang Y. Expression and significance of Notch1 and PTEN in gastric cancer. Ai Zheng. 2007;26:1183–1187. [PubMed] [Google Scholar]
- 24.Liu JN, Zhang ZM, Chen C. Expression of ERCC1, P27, Cyclin E in gastric cancer and its clinical significance. Cancer Res Prev Treat. 2010;37:540–543. [Google Scholar]
- 25.Milacic V, Banerjee V, Landis-Piwowar KR, Sarkar FH, Majumdar AP, Dou QP. Curcumin inhibits the proteasome activity in human colon cancer cells in vitro and in vivo. Cancer Res. 2008;68:7283–7292. doi: 10.1158/0008-5472.CAN-07-6246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Subramaniam D, Ponnurangam S, Ramamoorthy P, Standing D, Battafarano RJ, Anant S, Sharma P. Curcumin induces cell death in esophageal cancer cells through modulating notch signaling. PLoS One. 2012;7:e30590. doi: 10.1371/journal.pone.0030590. [DOI] [PMC free article] [PubMed] [Google Scholar]


