SUMMARY
Background
INF2 is a formin protein with the unique ability to accelerate both actin polymerization and depolymerization, the latter requiring filament severing. Mutations in INF2 lead to the kidney disease focal segmental glomerulosclerosis (FSGS) and the neurological disorder Charcot-Marie Tooth Disease (CMTD).
Results
Here, we compare the severing mechanism of INF2 with that of the well-studied severing protein, cofilin. INF2, like cofilin, binds stoichiometrically to filament sides and severs in a manner that requires phosphate release from the filament. In contrast to cofilin, however, INF2 binds ADP and ADP-Pi filaments equally well. Furthermore, two-color TIRF microscopy reveals that a low number of INF2 molecules, as few as a single INF2 dimer, are capable of severing, while measurable cofilin-mediated severing requires more extensive binding. Hence, INF2 is a more potent severing protein than cofilin. While a construct containing the FH1 and FH2 domains alone has some severing activity, addition of the C-terminal region increases severing potency by 40-fold, and we show that the WH2-resembling DAD motif is responsible for this increase. Helical 3D reconstruction from electron micrographs at 20 Å resolution provides a structure of filament-bound INF2, showing that the FH2 domain encircles the filament.
Conclusions
We propose a severing model in which FH2 binding and phosphate release causes local filament deformation, allowing the DAD to bind adjacent actin protomers, further disrupting filament structure.
INTRODUCTION
The actin cytoskeleton plays roles in many cellular processes, including migration, cytokinesis, phagocytosis, and organelle dynamics [1]. The diversity of actin-based processes arises from modulation of filament assembly and organization by specific actin-binding proteins, including formins. Formins use their dimeric Formin Homology 2 (FH2) domains to nucleate new filaments, and remain at the fast-growing barbed ends of these filaments as they elongate [2, 3]. The FH1 domain accelerates elongation through its interaction with the actin monomer binding protein, profilin[4]. The diversity of formins, including 15 mammalian proteins, provides the potential for extensive variation in assembly of actin-based structures [1, 5].
Several formins also interact with actin through C-terminal sequences, which provide additional abilities to influence actin dynamics [6, 7]. The vertebrate formin INF2 is particularly noteworthy, in that its long C-terminal region (300 amino acids) allows acceleration of both actin polymerization and depolymerization[8]. INF2’s C-terminus contains a Diaphanous Auto-regulatory Domain (DAD) close to the FH2, which binds actin monomers similar to WASp Homology 2 (WH2) motifs[8].
In cells, INF2 exists as two isoforms varying at their extreme C-termini. The INF2-nonCAAX variant is cytosolic, and plays a role in maintaining Golgi integrity [9] and in directed vesicular transport [10, 11]. The INF2-CAAX variant is tightly bound to ER, and acts in mitochondrial fission by assembling highly transient actin filaments at the ER/mitochondrial interface [12, 13]. These functions are medically important, since INF2 mutations lead to the kidney disease, focal segmental glomerulosclerosis (FSGS) and the neuropathy, Charcot-Marie Tooth Disease (CMTD) [14, 15].
To understand INF2’s physiological roles, it is critical to clarify its effects on actin biochemically. Prior work suggested that INF2-mediated filament depolymerization requires a severing step, which is dependent on ATP hydrolysis and phosphate release from actin protomers in the filament[8]. The need for phosphate release is not unique to INF2. Cofilin, a well-characterized severing protein, binds and severs filaments mainly after phosphate release [16– 18]. Under both cellular and experimental conditions, ATP hydrolysis (0.3 sec−1) and phosphate release (0.002 sec−1) lag significantly behind polymerization (10 µM−1sec−1), resulting in preferential cofilin binding to aged filament segments [19–22]. Upon binding, cofilin changes filament twist, creating strain at the boundary between cofilin-decorated and undecorated regions [23–25]. Cofilin binding also accelerates phosphate release from adjacent actin subunits, rendering binding cooperative [20, 21, 23, 26, 27].
We show that INF2’s severing mechanism is distinct from that of cofilin. INF2 binds stoichiometrically to filament sides in a phosphate-independent manner. This binding step consists of opening the FH2 dimer and then encirclement of the filament. INF2 binds at discrete sites throughout the filament length and, in a step dependent on phosphate release, severs at sites of binding. We propose a model for INF2-mediated severing, involving filament disruption both by FH2 binding and DAD insertion.
RESULTS
INF2 severs along the length of the filament
Previous data suggested that INF2 severs filaments, but did not show direct evidence for severing nor reveal the position of binding or severing events [8]. We used a two-color TIRF-microscopy assay containing fluorescently-labeled actin and GFP-INF2-FFC (containing the FH1, FH2, and C-terminal regions, Figure 1) to visualize INF2 severing events in real-time. Due to INF2’s potent actin nucleation activity, we first polymerized actin alone for a fixed time before washing out the remaining monomers and adding GFP-INF2-FFC. This procedure allows the production of longer filaments, more amenable to severing analysis than the short filaments generated by INF2-mediated nucleation.
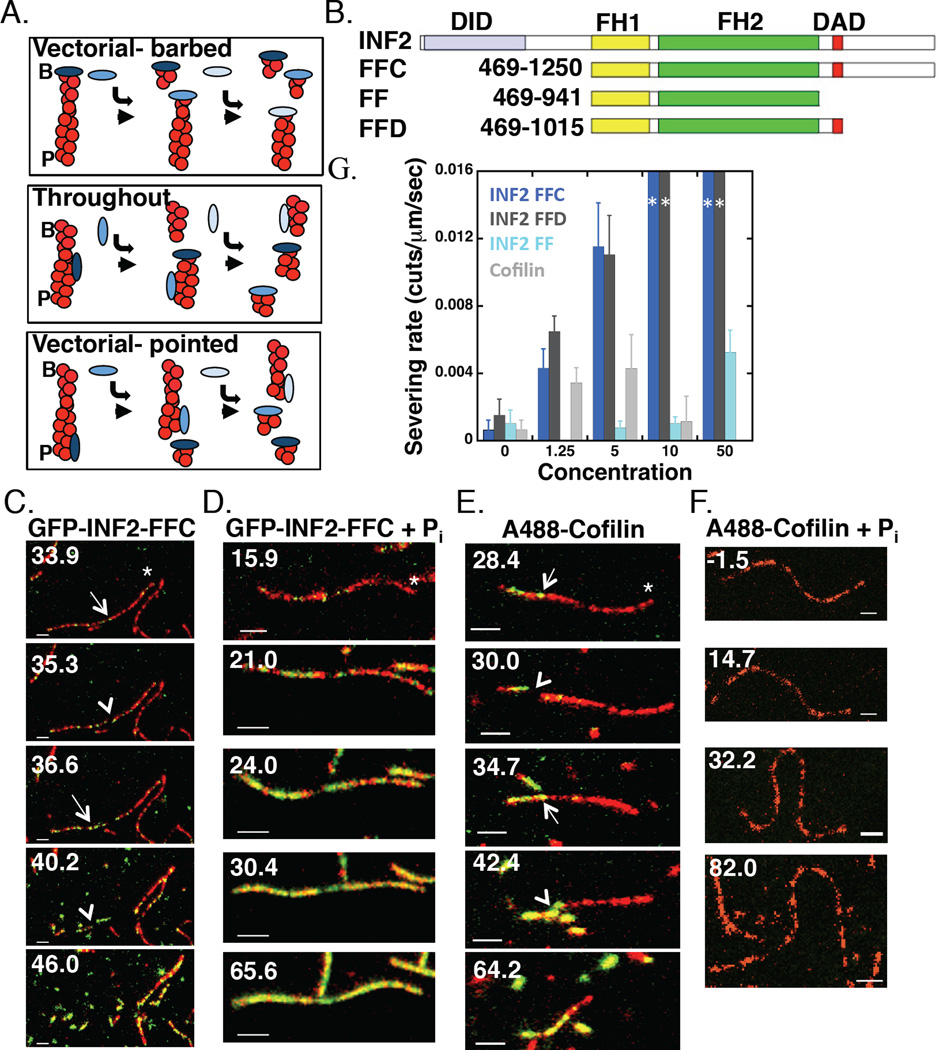
Figure 1. INF2 severs filaments throughout their length.
(A)Three models for INF2 severing. I) Vectorial-barbed: INF2 binds barbed end and removes a short filament fragment from that end. II) Throughout: INF2 binds and severs at random positions along filament side, creating two longer fragments. INF2 subsequently binds the newly created barbed end. III) Vectorial-pointed: INF2 binds and severs at ADP-actin protomers near pointed end. B, barbed. P, pointed.
(B) Bar diagram showing human INF2 constructs (lengths in amino acids). DID-Diaphanous Inhibitory Domain, FH1- Formin Homology 1, FH2- Formin Homology 2, DAD- Diaphanous Autoregulatory Domain.
(C) Two-color simultaneous TIRF microscopy montage of GFP-INF2-FFC (10nM) binding actin filament sides (1µM, 20% TAMRA initially) and severing on DDS/F127 treated slides. Time indicates seconds after INF2 addition (and wash-out of actin monomers). Asterisk indicates barbed end, arrow indicates GFP punctum correlated with severing, arrowhead indicates severing event. Scale bar 2 µm. Corresponds to Movie 1.
(D)As in (C) but with GFP-INF2-FFC (100nM) + 10mM phosphate. Corresponds to Movie 2.
(E) As in (C) but with A488-Cofilin (5nM). Corresponds to Movie 3.
(F) As in (C) but with A488-cofilin (100nM) + 10mM phosphate. Corresponds to Movie 4.
(G)Concentration dependence of severing rate for INF2-FFC, INF2-FFD, INF2-FF, and cofilin on DDS/F127 treated slides. N values: 18, 50, 48 severing events (7, 6, 6 filaments) for 0, 1.25, and 5nM INF2-FFC (dark blue); 15, 29, 40 events (6, 5, 7 filaments) for 0, 1.25, and 5nM INF2-FFD (dark grey); 9, 14, 15, 23 events (5, 6, 5, 5 filaments) for 0, 5, 10, and 50nM INF2-FF (light blue); 18, 42, 21, 4 events (7, 7, 5, 7 filaments) for 0, 1.25, 5, and 10nM cofilin (light grey). For 10 and 50 nM INF2-FFC and INF2-FFD, 10 and 6 filaments were examined respectively, but severing events were too rapid to quantify (depicted by dark blue or dark grey bar to the maximum rate). The highest concentrations tested (1 µM INF2-FFC and 500nM INF2-FFD) produce similar results.
We anticipated three possible results regarding severing position (Figure 1A): I) severing occurs vectorially from the barbed end, due to INF2’s barbed end binding; II) severing occurs throughout the filament length, at some distance from either filament end; or III) severing occurs predominantly toward the pointed end, due to the need for phosphate release [8]. Our results support model II, with severing occurring at many positions along the filament (Figure 1C and Movie 1). This tendency to sever throughout the filament is independent of the TIRF coverslip preparation technique, and occurs with unlabeled INF2-FFC as well as with GFP-INF2-FFC (Figure S1).
We compared severing by INF2 to that by cofilin, whose severing mechanism is better understood. Cofilin-mediated severing occurs predominately at boundaries between cofilin-bound and unbound regions (Figure 1E and Movie 3) [21]. Accordingly, the concentration dependence of cofilin severing is bi-modal, with reduced severing at low and high cofilin concentrations [28–30]. Cofilin displays similar severing tendency in our system (Figure 1G, S1). In contrast, INF2-mediated severing frequency increases continuously with INF2 concentration (Figure 1G) until all filaments are reduced to unresolvable lengths. Deletion mutagenesis of the C-terminus shows that inclusion of DAD alone is sufficient for robust severing activity, with INF2 FFC and FFD constructs displaying similar severing rates (Figure 1G, S1). Removal of the entire C-terminus causes a dramatic decrease in severing activity for the resulting INF2-FF construct, with no measurable severing at lower concentrations and a severing rate at 50 nM approaching that of 1.25 nM FFC (Figure 1G, S1). Additionally, INF2-FFC severing activity is significantly more potent than cofilin’s at most concentrations tested (Figure 1G).
INF2 binds throughout the filament but severs preferentially toward the pointed end
The TIRF assays also show that GFP-INF2-FFC binds filament sides, similar to cofilin. Previously, we showed that the presence of phosphate inhibits INF2-mediated severing [8], suggesting that ATP hydrolysis and phosphate release by actin protomers in the filament is necessary for severing. Phosphate inhibits cofilin-mediated severing by decreasing cofilin’s side-binding affinity significantly (Figure 1F and Movie 4) [16]. In contrast, GFP-INF2-FFC still binds filaments in the presence of phosphate (Figure 1D and Movie 2). To confirm and extend this result, we used actin filament co-sedimentation assays. INF2-FFC binds filaments with comparable affinity in the presence and absence of phosphate (Figure S1). In addition, INF2 binding saturates at a 1:1 ratio (INF2 monomer:actin monomer) in both cases (Figure S1). INF2-FF binds with 5-fold lower affinity, but still reaches saturation (not shown). We conclude that, in clear contrast to cofilin, INF2 binds filament sides in a manner independent of phosphate release, suggesting that phosphate release allows severing downstream of INF2 binding.
We used two-color TIRF microscopy to examine the pattern of GFP-INF2-FFC side binding. Initially, GFP-INF2-FFC binds at discrete sites without preference for a particular filament end in the presence or absence of phosphate (Figure 1C–D, S2). Most puncta are stably bound, with 77% and 97% of puncta remaining throughout the 3-minute acquisition time (Figure 2A). In the presence of phosphate, progressive addition of puncta results in full filament saturation with GFP-INF2-FFC at a uniform intensity (Figure 1D). In the absence of phosphate, saturating GFP-INF2-FFC occurs in some segments, but is largely counteracted by severing (Figure 1C).
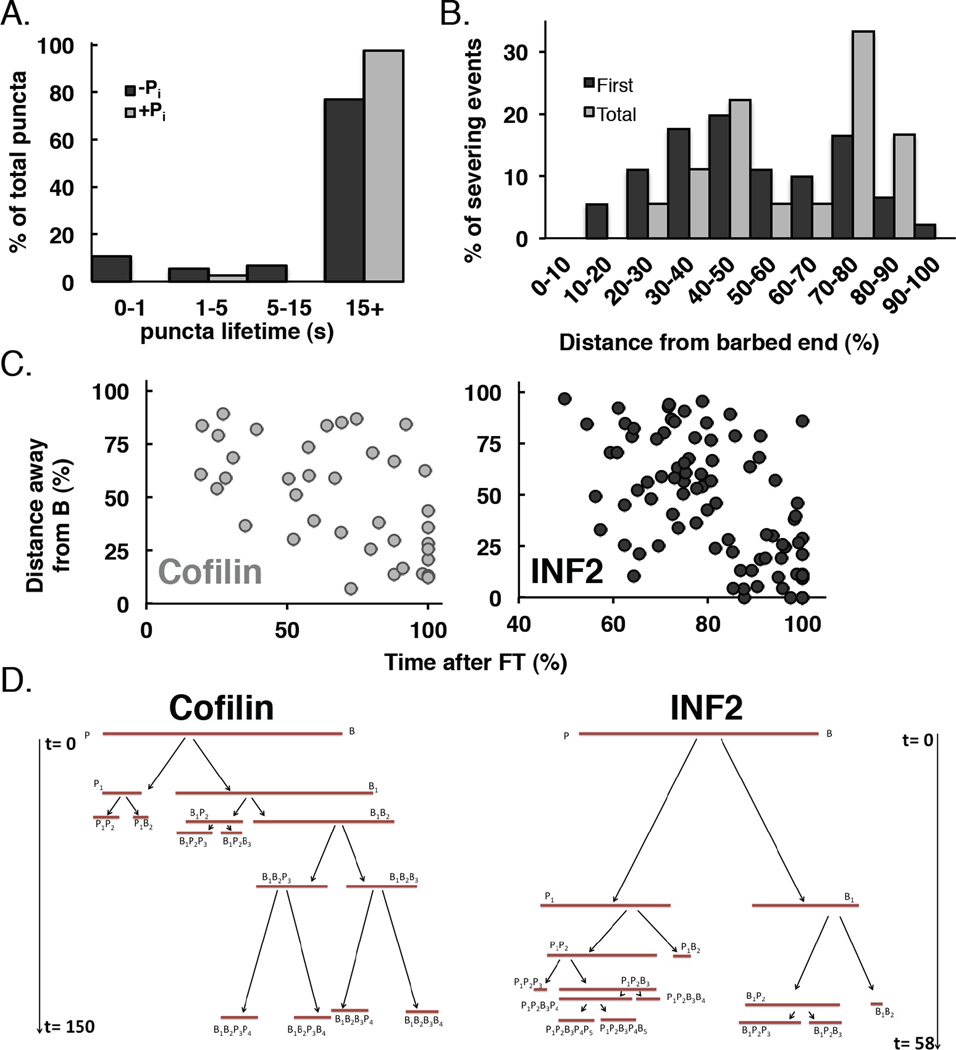
Figure 2. INF2 binds throughout the filament but severs towards pointed end.
(A)Lifetimes of filament-bound puncta in (A). >15s represents puncta that did not dissociate during the observation period (180 sec). n=74 and 39 puncta (13 and 6 filaments) in the absence and presence of phosphate respectively.
(B)Histogram of severing positions for GFP-INF2-FFC (5–20nM) on DDS/F127 treated slides. 18 filaments (91 severing events) quantified (average length 12.3µm, range 5.83–24.22µm at time = 0). Barbed end fragments were measured as a percentage of total filament length.
(C) Correlation of time of severing with respect to distance from original barbed end (as % of original length). n=80 and 38 severing events (11 and 7 filaments) for GFP-INF2-FFC and A488-cofilin, respectively. Filaments were12.5 and 7.86µm mean length (range 6.49–21.59 and 4.55–11.05µm) prior to INF2-FFC or cofilin addition respectively. Severing events were observed for 57.5–112.3 and 68.6–170.8 s after addition of INF2 or cofilin respectively. Note x-axis scale is different between INF2 and cofilin.
(D)Tree map diagraming severing events on individual filaments with time (sec) on the y-axis. t=0 denotes time of cofilin or INF2-FFC addition. B and P denote barbed and pointed ends. Severed filament fragments are named based on severing history with subscripts denoting lineage (B1 being the first barbed end fragment, B1B2 being the second barbed end fragment originating from the first barbed end fragment, and so forth). Original filament lengths: 11.1 (cofilin) and 13.6 µm (INF2). Time and sizes drawn to scale. Additional examples in Figure S2.
The distribution of GFP-INF2-FFC puncta throughout the filament supports our observation that INF2-mediated severing is not vectorial. Quantification of severing events with respect to original or newly created barbed ends further shows that severing occurs throughout the filament (Figure 2B, S2). However, our results reported previously and in this publication, showing that severing depends upon phosphate release, suggest that severing should occur preferentially toward the pointed end. We analyzed severing positional kinetics in more detail to determine the distance of a severing event from the original filament barbed end as a function of time after INF2 addition to the assay. This analysis shows that both INF2 and cofilin display preferences for severing nearer the pointed end at early time points, with events closer to the barbed end increasing at later time points (Figure 2C). Tracking the progressive severing of individual filaments reveals a similar pattern, but also shows that INF2-mediated severing is less vectorial than cofilin, with some initial severing events occurring more towards the barbed end (Figure 2D, S2). Severing often occurs more rapidly than would be predicted from ATP hydrolysis and phosphate release rate constants (Figure S2), which might imply that INF2 binding could enhance phosphate release locally.
INF2 binds filament sides as single dimers and severs at binding sites
Having established that GFP-INF2-FFC binds filament sides as discrete puncta, we next examined the relationship between INF2 binding and filament severing in more detail by asking two questions: how many INF2 molecules are required for severing, and how rapidly does severing occur after INF2 binding? To calibrate GFP-INF2-FFC fluorescence, we first monitored GFP intensity on the barbed ends of elongating filaments, assuming dimeric GFP-INF2-FFC binding the barbed end, since INF2 is dimeric in solution [8], like other formins [31]. Barbed end GFP intensity varies ~2-fold over time (Figure S3), possibly due to varying distance from the coverslip or GFP flickering. Using this calibration, the majority of diffraction-limited side-bound puncta appear to represent GFP-INF2-FFC dimers in the absence or presence of phosphate (Figure 3A).
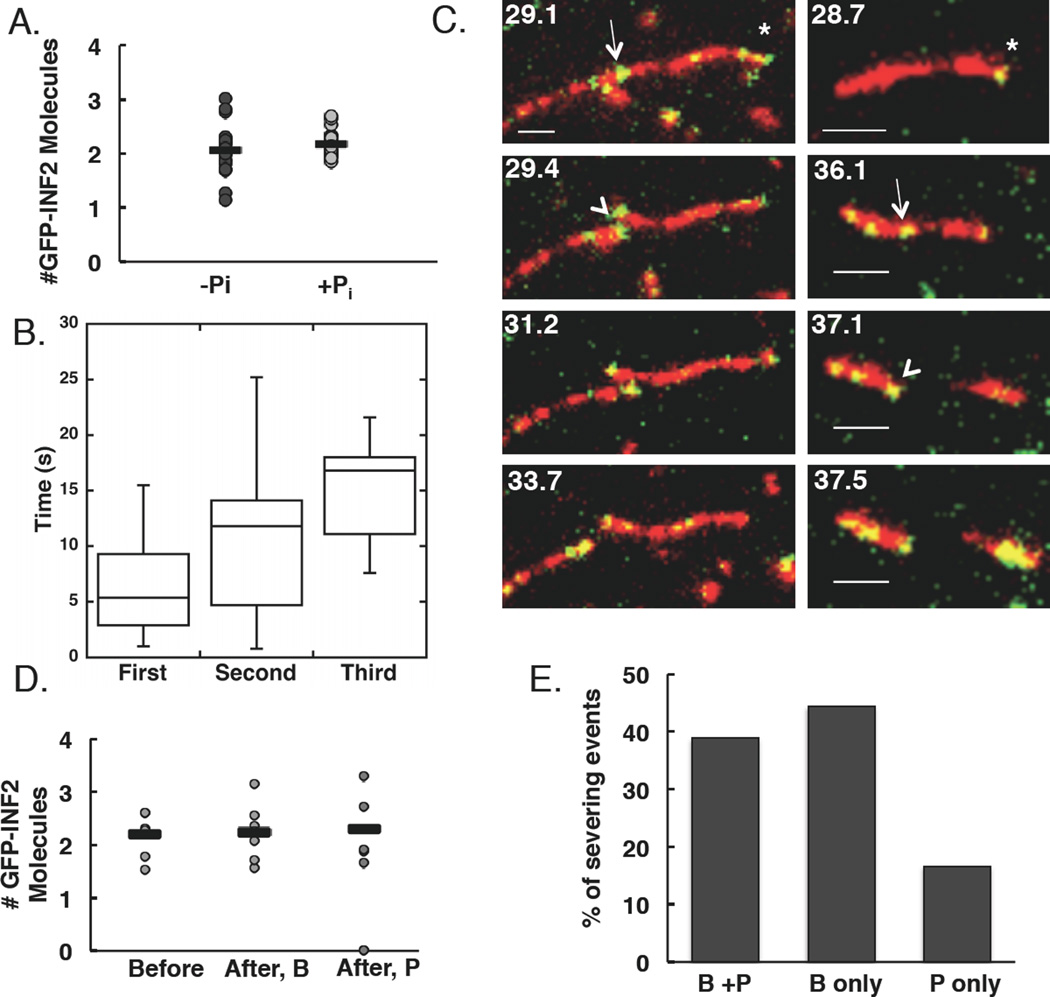
Figure 3. INF2 binds filament sides as single dimers and severs at binding sites.
(A)Number of GFP-INF2-FFC molecules localized at diffraction-limited puncta (n=28 and 25 puncta (8 and 6 filaments) in the absence and presence of phosphate respectively). Averages (bars) are 2.06 and 2.17 molecules in the absence and presence of phosphate respectively.
(B) Box and whisker quantification of severing time after punctum binding for first, second, and third severing events. Line indicates mean, box represents values 25–75% of totals, whiskers indicate full range of values. n= 14, 9, and 5 events.
(C) Two examples of severing at sites where an apparent single GFP-INF2-FFC dimer (green) is bound to TAMRA-actin (red). Asterisk indicates barbed end, arrow indicates punctum correlated with severing, arrowhead indicates severing event. Left panels: GFP-INF2 puncta present at both new barbed and pointed ends after severing. In right panels: GFP-INF2 punctum only at new barbed end after severing. Scale bar 2 µm. Corresponds to Movies 5 and 6.
(D)Distribution of GFP-INF2-FFC localization after severing for events occurring at sites where two GFP-INF2-FFC were located prior to severing. After, B = new barbed end. After, P = new pointed end. Bars represent averages from 7 events. Note: two events result in punctum localization at only new barbed end.
(E) Localization of puncta after a severing event with respect to newly created barbed or pointed ends. All severing events included, regardless of the number of GFP-INF2-FFC molecules present before the event. n=18 filaments.
We next investigated the kinetic relationship between GFP-INF2-FFC binding and severing, quantifying initial severing events in greater detail than subsequent events because individual puncta were more clearly discernable at early time points. Severing typically occurs at sites where an INF2 punctum is present (18 out of 19 events), but there is a significant delay (6.5±4.5 sec) between binding and severing (Figure 3B). Often, severing does not occur at the first binding site, and on average 7±3 puncta bind to the filament prior to the first severing event. For subsequent severing events, GFP-INF2-FFC is always present at the severing site, either as a punctum or as a uniform coating on that region of the filament (n=14 severing events, 11 filaments), with INF2 arriving on average 10.7±7.6 and 15±5 seconds prior to severing for second and third events, respectively (Figure 3B).
The number of GFP-INF2-FFC molecules present at the severing site is variable, but appears to be as low as two (or one INF2 dimer, Figure 3D), although variability in the calibration standard (Figure S3) introduces some uncertainty here. After severing, INF2 remains at the new barbed end (Figure 3C right, 3E and Movie 5). However, in several cases INF2 is also present at the new pointed end immediately after severing (Figure 3C left, 3D–E and Movie 6), suggesting that a second INF2 dimer may have added during severing, although we were unable to observe this directly. When all severing events are analyzed, INF2 remains at the new barbed end 83% of the time, with 47% also having INF2 at the new pointed end (Figure 3E). In a minority of cases, INF2 is only at the new pointed end (Figure 3E).
INF2’s FH2 domain encircles the filament
Our results show that INF2 binds filament sides and dissociates with an extremely slow off-rate. How does this binding occur? FH2 domains are dimeric and adopt a “donut”-like conformation, with barbed end-interacting residues on the inner face of the donut [31, 32]. We expected that INF2’s FH2 domain might bind filament sides through two possible mechanisms: I) by encircling the filament and binding through residues on the inner face of the donut, similar to those used at the barbed end; or II) by binding the filament side through residues on the outer face of the donut (Figure 4A).
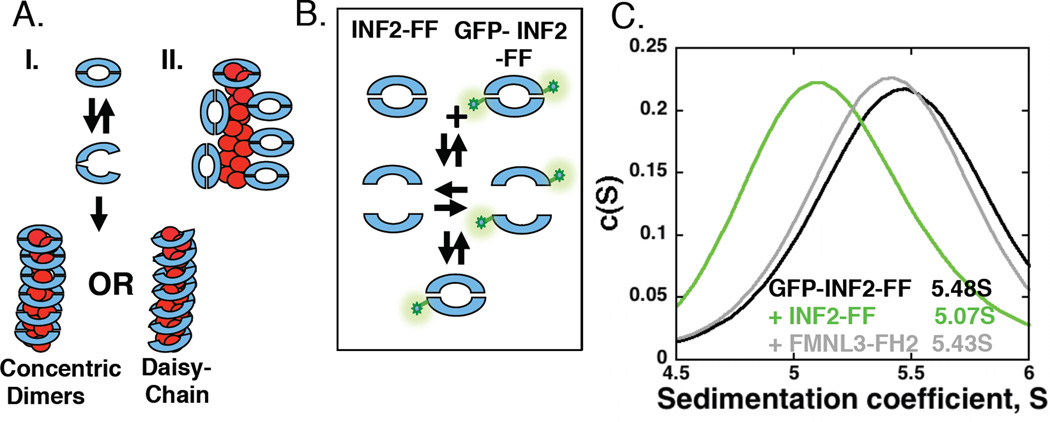
Figure 4. INF2 is a dissociating dimer that binds stoichiometrically to filament sides.
(A)Two models for INF2 binding filament sides. I. FH2 dimer partially dissociates in order to encircle the filament. Encirclement could occur either as concentric dimers or in a daisy-chain, where the lasso-region of one FH2 domain interacts with the post region of an adjacent FH2. II. FH2 dimer binds using interactions between the exterior of the dimer and the filament side. Two possible side-binding orientations shown, as well as a barbed end-bound dimer.
(B) Schematic representation of heterogenous dimer formation in velocity analytical ultracentrifucation experiments. If dimer dissociation is appreciable, a heterogeneous dimer should form upon mixing GFP-labeled and unlabeled FH2 domains, resulting in a shift in S value for the GFP signal.
(C)Velocity analytical ultracentrifugation profiles of GFP-INF2-FF (2µM) alone or mixed with unlabeled INF2-FF (20µM) or FMNL3-FH2 (20µM).
If model I is correct and INF2 encircles the filament, the INF2 FH2 dimer might be able to dissociate because it is unlikely that the dimer could “slide” along the filament over micron distances once bound to the barbed end. To test INF2 FH2 dimer dissociation, we conducted sedimentation velocity experiments using purified GFP-INF2-FF and unlabeled INF2-FF, and detecting fluorescence at 490nm to follow the GFP (Figure 4B). GFP-INF2-FF alone has a sedimentation coefficient of approximately 5.4S (Figure 4C, Table 1). In the presence of excess unlabeled INF2-FF, the sedimentation coefficient of GFP-INF2-FF shifts appreciably, to a value of 5 (Figure 4C). This result suggests that the INF2 dimer can dissociate, then re-associate to form the heterogeneous dimer. In contrast, FMNL3 FH2 does not cause an appreciable shift in the sedimentation coefficient of GFP-INF2-FF, suggesting that these dimers cannot efficiently re-associate with each other. These results are similar to those obtained using GFP-INF2-FFC (Table 1), and suggest that the INF2 FH2 domain dimer is capable of dissociating, then re-associating around the filament.
Table 1. Velocity analytical ultra analysis of INF2 FH2-dimer stability Sedimentation Coefficients (S).
Results from multiple experiments tracking either GFP-INF2-FF or GFP-INF2-FFC sedimentation velocity (at 490 nm absorbance) alone or in the presence of unlabeled INF2-FF, FMNL3-FH2 domain, or mDia1-FH2 domain. A plot of run 2 is shown in Figure 4B.
| Run | GFP-INF2 FF | GFP-INF2 FFC | +INF2 FF | +FMNL3 FH2 | +mDia1 FH2 |
|---|---|---|---|---|---|
| 1 | 5.33 | - | 5.03 | 5.28 | - |
| 2 | 5.48 | - | 5.07 | 5.34 | - |
| 3 | 5.38 | - | 4.92 | - | 5.28 |
| 4 | 5.73 | 5.23 | 5.53 | - | |
| 5 | 5.68 | 5.13 | - | - | |
| 6 | 5.63 | 5.13 | - | 5.50 |
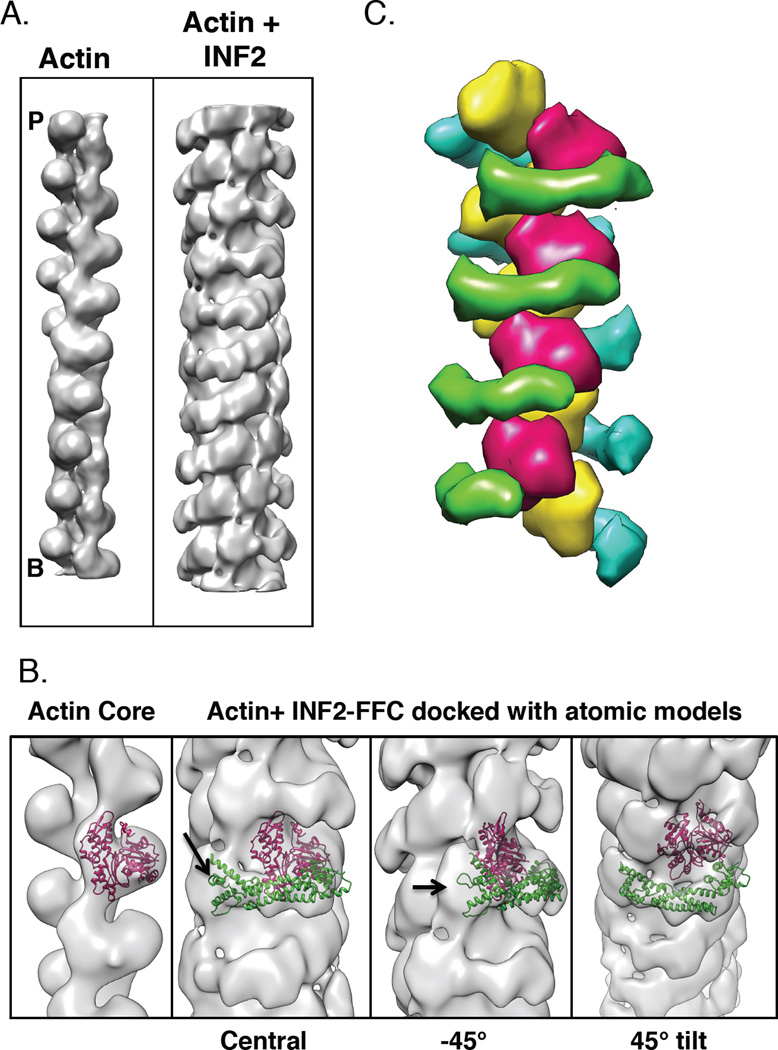
To test directly the binding conformation of INF2 on actin filaments, we imaged complexes of INF2-FFC and actin filaments by negative staining electron microscopy in the presence of beryllium fluoride (a stably-bound phosphate analogue [33]) and phalloidin. Helical 3D reconstruction of this complex results in a structure at 20 Å resolution. The final reconstruction reveals an inner core of higher density surrounded by an outer region of lower density (Figure 5A). The lower density of the outer region may result from the blurring effect of the variable twist of actin and/or partial occupancy of formin.
Figure 5. Examination of INF2-FFC binding actin filaments by electron microscopy.
A) 3D reconstructions from negative staining EM images of bare (left) and INF2-FFC bound (right) actin filaments. Barbed (B) and pointed (P) ends indicated.
B) Zoomed images of full actin/INF2-FFC reconstruction (inner and outer densities) with actin monomer (PDB: 3MFP, pink ribbon) on inner density alone (left panel) or on INF2-bound filaments with FH2 crystal backbone ribbon (PDB 4EAH, green ribbon). Note density unaccounted for by either actin or FH2 (arrow). Three angles shown.
C) Schematic model of FH2 (green, blue) binding to filament (red, yellow), based on structural data in agreement with color scheme of [34].
The inner core can be fit with a model of filamentous actin structure (PDB 3MFP) on a per protomer basis (Figure 5B). The helical twist per protomer of this reconstructed filament is 166.65°, similar to that of actin alone. However, the helical rise per protomer identified by the program is 28.0 Å, which is 1.5% greater than the 27.6 Å rise of actin alone. Apart from the increased helical rise, the conformation of the protomers does not appear to be altered by INF2-FFC binding at this resolution.
The outer region of lower density is attributed to INF2. This density pattern describes a spiral around the core, at an orientation approximately perpendicular to the filament axis. Part of the density for actin subdomain 1 contributes to the continuity of the spiral. The outer density is at approximately the same position as in a recent crystal structure of an FH2 domain bound to two actin monomers (PDB 4EAH from [31]). When the FH2 portion of this crystal structure is docked into this outer density, it occupies most of the density that does not correspond to actin (Figure 5B). This conformation encircles the filament. Other conformations in which the FH2 binds without encirclement fit poorly into the outer density. There is a region of unaccounted density close to the C-terminus of the fitted FH2 crystal structure. This density may include a portion of the C-terminus, which extends 300 residues beyond the FH2, or the FH1 domain.
In principle, this binding configuration would allow for many of the FH2-actin binding interactions observed in the previous crystal structure (PDB 4EAH), including the interaction between INF2’s knob helix D and the cleft between actin subdomains 1 and 3. Figure 5C shows a model of the binding pattern based on these structural data. At this resolution, we cannot differentiate between INF2 dimers binding as stacked concentric circles or in a daisy-chain pattern, with oligomerization by lasso/post interactions between adjacent FH2s (Figure 4A).
DISCUSSION
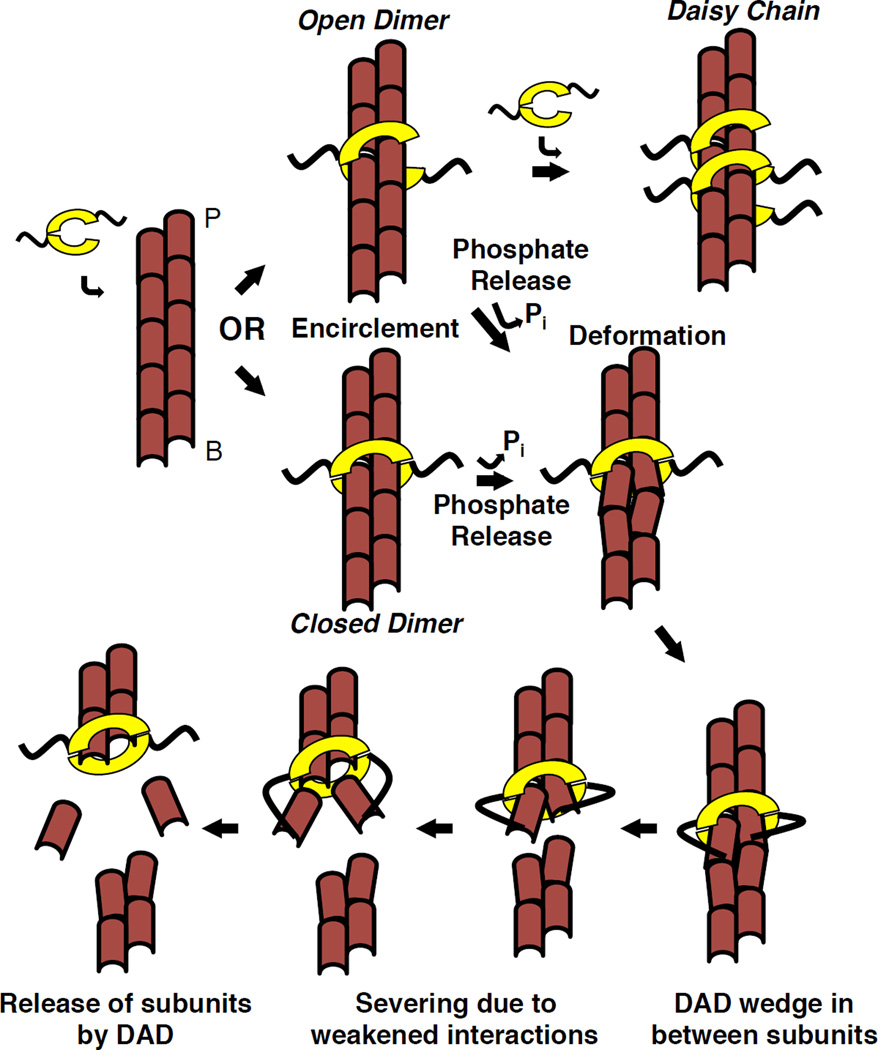
In this work, we elucidate basic features of INF2’s actin severing mechanism, and propose a working model for severing (Figure 6). INF2 binds filament sides in a manner independent of phosphate release. Side binding occurs through filament encirclement by INF2’s FH2 domain, requiring release of at least one FH2-FH2 interaction in the FH2 dimer, which might re-associate or associate with an adjacent FH2. FH2 binding causes a small change in filament rise, but a bigger change might occur upon phosphate release, when the bound FH2 alters filament structure to cause a “weak point” in the filament, making it susceptible to severing. The altered filament structure also enhances the ability of INF2’s DAD to bind actin protomers adjacent to the bound FH2. DAD binding further weakens the filament, thus enhancing severing rate. One INF2-FFC dimer is sufficient to carry out this severing reaction.
Figure 6. Model of INF2 severing mechanism.
INF2 dimer partially dissociates, then encircles the filament, either as a closed ring or an open dimer that can form a daisy-chain. Subsequent steps shown in the closed ring conformation, but could also occur as the open dimer. INF2 binding causes a change in helical rise. Upon phosphate release, INF2 causes further local filament deformation, partially exposing the DAD binding site between subdomains 1 and 3 of adjacent actin protomers. DAD binding destabilizes the filament by wedging between protomers. These destabilizations lead to severing at this weak point. Upon severing, INF2 remains bound to the new barbed end, and DAD releases bound actin monomers removed from filament. Our data suggest that DAD binding to the barbed end side of the FH2 is favored.
In our model, as in cofilin-based severing models, the purpose of INF2 is to weaken actin protomer interactions. The thermal motion of the filament then causes breakage at this site [34]. INF2 weakens protomer interactions in two ways: by FH2-induced conformational change in filament structure and by DAD binding disrupting protomer-protomer interactions. We will discuss each of the steps below.
Actin filament binding by INF2’s FH2 domain
Using electron microscopy, we find that INF2’s FH2 domain encircles the actin filament, and through TIRFM, we show that bound dimers have low off-rates. Using analytical ultracentrifugation, we show that INF2’s FH2 is capable of dissociation and re-association. Our results do not give an indication of dissociation dynamics, but the fact that we observe both transient and stable GFP-INF2-FFC puncta on filament sides suggests that some binding events are non-productive. The stability of most GFP-INF2-FFC puncta suggests an extensive interaction, such as encirclement. The resolution of the actin/INF2 EM structure presented here is insufficient to determine whether INF2-FFC forms an oligomeric daisy-chain similar to Bni1p, or binds as independent concentric dimers. Since as few as single GFP-INF2-FFC dimers are capable of severing, oligomerization does not appear to be essential for severing.
Some FH2 domains have been shown to possess the ability to dissociate, including FMNL1, FMNL3, and mDia2, while the FH2 domains of other formins (mDia1, Bni1p) appear to undergo little dissociation [35, 36]. However, even the apparently stable Bni1p FH2 domain has the ability to encircle actin, as evidenced by the crystal structure of Bni1p-FH2 in an oligomeric chain helically encircling two non-helical strands of actin monomers [32, 35]. These results suggest that even FH2s that exist as stable dimers in solution might dissociate under some conditions. It is possible that actin filaments might accelerate FH2 dissociation rate to enhance encirclement.
Changes to filament structure upon INF2-FH2 side binding
We postulate that FH2 binding alters filament structure, resulting in two outcomes: 1) weakening of filament structure, leading to the low but measurable severing by the FF construct; and 2) exposure of the DAD binding site on adjacent actin protomers, allowing even greater filament instability. Our EM results show that INF2-FFC induces a small change in filament structure, manifested in a 1.5% change in helical rise. A larger change in filament structure could occur upon phosphate release from the filament, since this causes major changes in filament properties for actin alone [37]. Our EM structure is of INF2-FFC bound to filaments containing the phosphate analogue BeF, chosen because of the higher affinity of FFC for filaments versus FF, and the need to prevent severing. Future studies of filament-bound INF2-FF in the presence and absence of phosphate analogue, and higher resolution cryoEM reconstruction of the INF2/actin complex, will clarify whether phosphate release causes larger changes in INF2-bound filament structure. For cofilin, filament twist change results from alteration in subdomain 2 and other structural elements of individual actin protomers within cofilin-decorated filaments [23, 25].
DAD insertion accelerates severing
INF2-FFC is at least 40-fold more efficient at severing than INF2-FF, based on our kinetic assays by TIRF microscopy. This increased efficiency is due to the DAD sequence, which binds actin monomers similarly to a WH2 motif. Monomer binding by DAD occurs predominately at the groove between subdomains 1 and 3 at the barbed end of actin, a region that is partially occluded in the filament [8, 38, 39]. In our severing model, the conformational change induced by FH2 binding serves to increase exposure of the DAD binding site on adjacent actin protomers. DAD binding further disrupts inter-protomer contacts and compromises filament stability at this site, leading to severing. Our two-color TIRF studies, showing a higher instance of GFP-INF2-FFC on the new barbed end, suggest that the DAD preferentially binds protomers to the barbed end of the FH2. These experiments also show a variable time between INF2 binding and severing. This lag could suggest that the actual severing event is due to thermal motion of the filament, thus is probabilistic in nature.
A low number of INF2 dimers can induce severing
We frequently observe as few as two GFP molecules in the immediate vicinity of a severing event within 0.5 sec of the event, suggesting that a low number of INF2-FFC dimers are sufficient for severing. Due to the noise in our fluorescence calibration (~2-fold), we cannot claim that single FFC dimers are capable of severing. Another source of possible uncertainty in our barbed end calibration standard is that we have no direct evidence that INF2 binds barbed ends as a dimer, but a large amount of evidence for other formins suggest that this is the case. The dependence of severing on INF2-FFC concentration does not reveal any strong negative cooperativity; with rapid severing occurring at the highest INF2 concentrations we are able to test (up to 1 µM). In this feature, INF2 is different from cofilin, for which saturated binding inhibits severing by creating uniform twist [23, 24]. For INF2, small local filament deformations might promote filament instability and severing.
INF2 also presents both similarities and differences to another well-characterized severing protein, gelsolin. Similar to INF2, gelsolin is a barbed end binding protein that can bind filament sides [40]. Also similar to INF2, gelsolin wraps around the filament, making contacts through several motifs including a WH2-like domain. It is unclear whether one gelsolin molecule is capable of severing. However, gelsolin’s severing mechanism might involve long-range changes in filament twist and/or other changes in filament structure [41–43].
INF2 severing in the cellular context
A functional consequence of severing is to produce new filament ends for at least two purposes: 1) priming new filament growth through elongation or Arp2/3 complex mediated branched nucleation, or 2) creating new filament ends for depolymerization. Both cofilin and gelsolin have been proposed to serve both functions depending on the cellular context [40, 44, 45].
Which function does INF2-mediated severing serve? For the INF2-CAAX variant, severing could be a mechanism for rapidly amplifying filaments number after initial nucleation at the ER/mitochondrial interface [13]. Since constitutively active INF2 causes excessive actin polymerization at mitochondrial fission sites [13], INF2 certainly is a contributor to the assembly of these filaments. Alternately, severing might participate in rapid depolymerization of these filaments at the completion of mitochondrial fission, since these filaments appear to be highly transient. In any case, our results set fundamental parameters for understanding INF2 function in cells by showing that single INF2 dimers are capable of severing and that INF2 binds actin filaments by encirclement.
EXPERIMENTAL PROCEDURES
Detailed plasmid construction, protein purification, buffer composition, and analysis methods can be found in the Supplemental Experimental Procedures. Briefly, formins were expressed in Rosetta2 E. coli (Stratagene Inc) as GST fusion proteins, following procedures used previously [8].
TIRF Microscopy
Rabbit skeletal muscle actin (1 µM, 20% TAMRA labeled) diluted in TIRF buffer (1xKMEI, 100mM DTT, 0.2mM ATP, 15mM Glucose, 0.5% Methyl Cellulose, 0.01mg/mL catalase (Sigma C3515), 0.05mg/mL glucose oxidase (Sigma G6125), 0.1% BSA) was polymerized for 7 minutes in flow chambers, at which point indicated concentrations of formin or cofilin were added. The filaments were visualized on a Nikon Eclipse Ti-E inverted microscope, and simultaneous dual-color images were acquired every 100ms with TIRF objective (60× 1.49 N.A.) and two iXON Ultra 897 cameras. Detailed methods can be found in the Supplemental Experimental Procedures.
Velocity Analytical Ultra
Analytical ultracentrifugation was conducted using a Beckman Proteomelab XL-A and an AN-60 rotor. For sedimentation velocity analytical ultracentrifugation, GFP-INF2-FF or FFC (2µM) was mixed with unlabeled INF2-FF or either FMNL3-F or mDIA-F as negative controls (20µM unlabeled protein) in K100MEIDT and centrifuged at 50,000rpm with monitoring at 490nm. Data were analyzed by Sedfit. Sedimentation coefficient reported is that of the major peak (at least 80% of the total analyzed mass) of GFP sedimentation.
Negative Staining Electron Microscopy
To make negatively stained grids, actin (bare or INF2-FFC-decorated) samples are applied to EM grids with continuous carbon, and then stained by 2.5% uranyl acetate solution. Prepared grids are imaged in an FEI Tecnai F20 microscope operated at 200keV acceleration, and images are taken with a Tietz F415 CCD camera (TVIPS) at 50,000 nominal magnification. Filaments from the CCD frames are manually selected with EMAN (ref: http://www.ncbi.nlm.nih.gov/pubmed/10600563) helixboxer module. Helical 3D reconstructions are conducted with IHRSR (doi: 10.1016/j.jsb.2006.05.015) with EMAN as the reconstruction engine. Detailed methods can be found in the Supplemental Experimental Procedures.
Supplementary Material
Highlights.
-
-
INF2’s actin filament severing activity is compared with that of cofilin
-
-
INF2’s FH2 domain dimer dissociates and re-associates around the actin filament
-
-
As few as one INF2 dimer can sever actin filaments
-
-
A 20 Å resolution structure of filament-bound INF2 FH2 domain is provided
ACKNOWLEDGEMENTS
We thank Gary Brouhard and Susanne Bechstedt for vital help in TIRF coverslip preparation and two-color TIRF techniques, Enrique de la Cruz, Hyeran Kang, Dave Kovar, Colleen Skau, and Dave Warshaw for helpful discussions, Shimeng Feng for excellent pilot experiments, and Clement Crien for being around us all. Supported by NIH grants GM069818 and DK88826 to HNH, GM077190 to ER, GM071940 to ZHZ. The TIRF microscopy would not have been possible without an administrative supplement for GM069818 provided by the American Recovery and Reinvestment Act (ARRA). We acknowledge the use of the EICN core facility supported by CNSI at UCLA.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
The authors do not have any financial conflict of interest associated with this work.
REFERENCES
- 1.Chhabra ES, Higgs HN. The many faces of actin: matching assembly factors with cellular structures. Nat Cell Biol. 2007;9:1110–1121. doi: 10.1038/ncb1007-1110. [DOI] [PubMed] [Google Scholar]
- 2.Moseley JB, Sagot I, Manning AL, Xu Y, Eck MJ, Pellman D, Goode BL. A conserved mechanism for Bni1- and mDia1-induced actin assembly and dual regulation of Bni1 by Bud6 and profilin. Mol Biol Cell. 2004;15:896–907. doi: 10.1091/mbc.E03-08-0621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chesarone MA, Goode BL. Actin nucleation and elongation factors: mechanisms and interplay. Curr Opin Cell Biol. 2009;21:28–37. doi: 10.1016/j.ceb.2008.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Paul AS, Pollard TD. Review of the mechanism of processive actin filament elongation by formins. Cell Motil Cytoskeleton. 2009;66:606–617. doi: 10.1002/cm.20379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Higgs HN, Peterson KJ. Phylogenetic analysis of the formin homology 2 domain. Mol Biol Cell. 2005;16:1–13. doi: 10.1091/mbc.E04-07-0565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Heimsath EG, Jr, Higgs HN. The C terminus of formin FMNL3 accelerates actin polymerization and contains a WH2 domain-like sequence that binds both monomers and filament barbed ends. J Biol Chem. 2012;287:3087–3098. doi: 10.1074/jbc.M111.312207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gould CJ, Maiti S, Michelot A, Graziano BR, Blanchoin L, Goode BL. The formin DAD domain plays dual roles in autoinhibition and actin nucleation. Curr Biol. 2011;21:384–390. doi: 10.1016/j.cub.2011.01.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chhabra ES, Higgs HN. INF2 Is a WASP homology 2 motif-containing formin that severs actin filaments and accelerates both polymerization and depolymerization. J Biol Chem. 2006;281:26754–26767. doi: 10.1074/jbc.M604666200. [DOI] [PubMed] [Google Scholar]
- 9.Ramabhadran V, Korobova F, Rahme GJ, Higgs HN. Splice variant-specific cellular function of the formin INF2 in maintenance of Golgi architecture. Mol Biol Cell. 2011;22:4822–4833. doi: 10.1091/mbc.E11-05-0457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Andres-Delgado L, Anton OM, Madrid R, Byrne JA, Alonso MA. Formin INF2 regulates MAL-mediated transport of Lck to the plasma membrane of human T lymphocytes. Blood. 2010;116:5919–5929. doi: 10.1182/blood-2010-08-300665. [DOI] [PubMed] [Google Scholar]
- 11.Madrid R, Aranda JF, Rodriguez-Fraticelli AE, Ventimiglia L, Andres-Delgado L, Shehata M, Fanayan S, Shahheydari H, Gomez S, Jimenez A, et al. The formin INF2 regulates basolateral-to-apical transcytosis and lumen formation in association with Cdc42 and MAL2. Dev Cell. 2010;18:814–827. doi: 10.1016/j.devcel.2010.04.001. [DOI] [PubMed] [Google Scholar]
- 12.Chhabra ES, Ramabhadran V, Gerber SA, Higgs HN. INF2 is an endoplasmic reticulum-associated formin protein. J Cell Sci. 2009;122:1430–1440. doi: 10.1242/jcs.040691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Korobova F, Ramabhadran V, Higgs HN. An actin-dependent step in mitochondrial fission mediated by the ER-associated formin INF2. Science. 2013;339:464–467. doi: 10.1126/science.1228360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brown EJ, Schlondorff JS, Becker DJ, Tsukaguchi H, Tonna SJ, Uscinski AL, Higgs HN, Henderson JM, Pollak MR. Mutations in the formin gene INF2 cause focal segmental glomerulosclerosis. Nat Genet. 2010;42:72–76. doi: 10.1038/ng.505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Boyer O, Nevo F, Plaisier E, Funalot B, Gribouval O, Benoit G, Cong EH, Arrondel C, Tete MJ, Montjean R, et al. INF2 mutations in Charcot-Marie-Tooth disease with glomerulopathy. N Engl J Med. 2011;365:2377–2388. doi: 10.1056/NEJMoa1109122. [DOI] [PubMed] [Google Scholar]
- 16.Maciver SK, Zot HG, Pollard TD. Characterization of actin filament severing by actophorin from Acanthamoeba castellanii. J Cell Biol. 1991;115:1611–1620. doi: 10.1083/jcb.115.6.1611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hayden SM, Miller PS, Brauweiler A, Bamburg JR. Analysis of the interactions of actin depolymerizing factor with G- and F-actin. Biochemistry. 1993;32:9994–10004. doi: 10.1021/bi00089a015. [DOI] [PubMed] [Google Scholar]
- 18.De La Cruz EM. How cofilin severs an actin filament. Biophys Rev. 2009;1:51–59. doi: 10.1007/s12551-009-0008-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Blanchoin L, Pollard TD. Hydrolysis of ATP by polymerized actin depends on the bound divalent cation but not profilin. Biochemistry. 2002;41:597–602. doi: 10.1021/bi011214b. [DOI] [PubMed] [Google Scholar]
- 20.Blanchoin L, Pollard TD. Mechanism of interaction of Acanthamoeba actophorin (ADF/Cofilin) with actin filaments. J Biol Chem. 1999;274:15538–15546. doi: 10.1074/jbc.274.22.15538. [DOI] [PubMed] [Google Scholar]
- 21.Suarez C, Roland J, Boujemaa-Paterski R, Kang H, McCullough BR, Reymann AC, Guerin C, Martiel JL, De la Cruz EM, Blanchoin L. Cofilin tunes the nucleotide state of actin filaments and severs at bare and decorated segment boundaries. Curr Biol. 2011;21:862–868. doi: 10.1016/j.cub.2011.03.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Carlier MF, Pantaloni D, Evans JA, Lambooy PK, Korn ED, Webb MR. The hydrolysis of ATP that accompanies actin polymerization is essentially irreversible. FEBS Lett. 1988;235:211–214. doi: 10.1016/0014-5793(88)81264-x. [DOI] [PubMed] [Google Scholar]
- 23.McGough A, Pope B, Chiu W, Weeds A. Cofilin changes the twist of F-actin: implications for actin filament dynamics and cellular function. J Cell Biol. 1997;138:771–781. doi: 10.1083/jcb.138.4.771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McCullough BR, Grintsevich EE, Chen CK, Kang H, Hutchison AL, Henn A, Cao W, Suarez C, Martiel JL, Blanchoin L, et al. Cofilin-linked changes in actin filament flexibility promote severing. Biophys J. 2011;101:151–159. doi: 10.1016/j.bpj.2011.05.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Galkin VE, Orlova A, Kudryashov DS, Solodukhin A, Reisler E, Schroder GF, Egelman EH. Remodeling of actin filaments by ADF/cofilin proteins. P Natl Acad Sci USA. 2011;108:20568–20572. doi: 10.1073/pnas.1110109108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ressad F, Didry D, Xia GX, Hong Y, Chua NH, Pantaloni D, Carlier MF. Kinetic analysis of the interaction of actin-depolymerizing factor (ADF)/cofilin with G- and F-actins. Comparison of plant and human ADFs and effect of phosphorylation. J Biol Chem. 1998;273:20894–20902. doi: 10.1074/jbc.273.33.20894. [DOI] [PubMed] [Google Scholar]
- 27.Bobkov AA, Muhlrad A, Pavlov DA, Kokabi K, Yilmaz A, Reisler E. Cooperative effects of cofilin (ADF) on actin structure suggest allosteric mechanism of cofilin function. J Mol Biol. 2006;356:325–334. doi: 10.1016/j.jmb.2005.11.072. [DOI] [PubMed] [Google Scholar]
- 28.Pavlov D, Muhlrad A, Cooper J, Wear M, Reisler E. Actin filament severing by cofilin. J Mol Biol. 2007;365:1350–1358. doi: 10.1016/j.jmb.2006.10.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Andrianantoandro E, Pollard TD. Mechanism of actin filament turnover by severing and nucleation at different concentrations of ADF/cofilin. Mol Cell. 2006;24:13–23. doi: 10.1016/j.molcel.2006.08.006. [DOI] [PubMed] [Google Scholar]
- 30.De La Cruz EM. Cofilin binding to muscle and non-muscle actin filaments: isoform-dependent cooperative interactions. J Mol Biol. 2005;346:557–564. doi: 10.1016/j.jmb.2004.11.065. [DOI] [PubMed] [Google Scholar]
- 31.Thompson ME, Heimsath EG, Gauvin TJ, Higgs HN, Kull FJ. FMNL3 FH2-actin structure gives insight into formin-mediated actin nucleation and elongation. Nat Struct Mol Biol. 2013;20:111–118. doi: 10.1038/nsmb.2462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Otomo T, Tomchick DR, Otomo C, Panchal SC, Machius M, Rosen MK. Structural basis of actin filament nucleation and processive capping by a formin homology 2 domain. Nature. 2005;433:488–494. doi: 10.1038/nature03251. [DOI] [PubMed] [Google Scholar]
- 33.Combeau C, Carlier MF. Probing the Mechanism of Atp Hydrolysis on F-Actin Using Vanadate and the Structural Analogs of Phosphate Bef3- and Alf4- J Biol Chem. 1988;263:17429–17436. [PubMed] [Google Scholar]
- 34.Elam WA, Kang H, De La Cruz EM. Biophysics of actin filament severing by cofilin. FEBS Lett. 2013;587:1215–1219. doi: 10.1016/j.febslet.2013.01.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Xu YW, Moseley JB, Sagot I, Poy F, Pellman D, Goode BL, Eck MJ. Crystal structures of a formin homology-2 domain reveal a tethered dimer architecture. Cell. 2004;116:711–723. doi: 10.1016/s0092-8674(04)00210-7. [DOI] [PubMed] [Google Scholar]
- 36.Harris ES, Rouiller I, Hanein D, Higgs HN. Mechanistic differences in actin bundling activity of two mammalian formins, FRL1 and mDia2. J Biol Chem. 2006;281:14383–14392. doi: 10.1074/jbc.M510923200. [DOI] [PubMed] [Google Scholar]
- 37.Kudryashov DS, Reisler E. ATP and ADP actin states. Biopolymers. 2013;99:245–256. doi: 10.1002/bip.22155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chereau D, Kerff F, Graceffa P, Dominguez R. Structural basis of the actin-WH2 domain interaction. Biophys J. 2005;88:10A–11A. [Google Scholar]
- 39.Chereau D, Kerff F, Graceffa P, Grabarek Z, Langsetmo K, Dominguez R. Actin-bound structures of Wiskott-Aldrich syndrome protein (WASP)-homology domain 2 and the implications for filament assembly. P Natl Acad Sci USA. 2005;102:16644–16649. doi: 10.1073/pnas.0507021102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nag S, Larsson M, Robinson RC, Burtnick LD. Gelsolin: The tail of a molecular gymnast. Cytoskeleton (Hoboken) 2013;70:360–384. doi: 10.1002/cm.21117. [DOI] [PubMed] [Google Scholar]
- 41.McGough A, Chiu W, Way M. Determination of the gelsolin binding site on F-actin: Implications for severing and capping. Biophys J. 1998;74:764–772. doi: 10.1016/S0006-3495(98)74001-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Prochniewicz E, Zhang QN, Janmey PA, Thomas DD. Cooperativity in F-actin: Binding of gelsolin at the barbed end affects structure and dynamics of the whole filament. J Mol Biol. 1996;260:756–766. doi: 10.1006/jmbi.1996.0435. [DOI] [PubMed] [Google Scholar]
- 43.Orlova A, Prochniewicz E, Egelman EH. Structural dynamics of F-actin: II. Cooperativity in structural transitions. J Mol Biol. 1995;245:598–607. doi: 10.1006/jmbi.1994.0049. [DOI] [PubMed] [Google Scholar]
- 44.Chan AY, Bailly M, Zebda N, Segall JE, Condeelis JS. Role of cofilin in epidermal growth factor-stimulated actin polymerization and lamellipod protrusion. J Cell Biol. 2000;148:531–542. doi: 10.1083/jcb.148.3.531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bravo-Cordero JJ, Magalhaes MA, Eddy RJ, Hodgson L, Condeelis J. Functions of cofilin in cell locomotion and invasion. Nat Rev Mol Cell Bio. 2013;14:405–415. doi: 10.1038/nrm3609. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.