Abstract
Background:
Oesophageal squamous cell carcinoma (ESCC) is the predominant subtype of oesophageal carcinoma in China, with the overall 5-year survival rate of <10%. The current tumour-node-metastasis (TNM) staging system has become so complex that it is not easy to use in the life expectancy assessment. We aim to combine clinical variables and biomarkers to develop and validate a relative simple and reliable model, named the FENSAM, for ESCC prognosis.
Methods:
To build the FENSAM, we analysed 22 potential prognostic factors from 461 patients, including 9 biomarkers (Ezrin, Fascin, desmocollin 2 (DSC2), pFascin, activating transcription factor 3 (ATF3), connective-tissue growth factor (CTGF), neutrophil gelatinase-associated lipocalin (NGAL), NGAL receptor (NGALR), and cysteine-rich angiogenic protein 61 (CYR61)) and other 13 clinical variables. We selected significant factors associated with survival of ESCC patients, and used them to build our FENSAM model. We then obtained the hazard risk score of the model to classify ESCC patients. In addition, we validated the model in an independent cohort of 290 patients from the same hospital. The predictive performance of the model was assessed by the Area under the Receiver Operating Characteristic Curve (AUC) and Kaplan–Meier survival analysis.
Results:
We found six markers significantly associated with survival of ESCC patients (Ezrin, Fascin, ATF3, surgery extent, N-stage, and M-stage). They were combined to create a novel four-stage FENSAM model for patients' classification. FENSAM possessed a high classification precision similar to the TNM staging system, but with a much simpler model. The efficiency of FENSAM was evaluated by different quantiles of AUC and the results of survival analysis. The validation result demonstrated the potential of the FENSAM model to improve classification accuracy for ESCC patients
Conclusions:
FENSAM provides an alternative classifier for ESCC patients with a high classification precision using a simple model.
Keywords: FENSAM, TNM stage system, biomarker, ESCC, LASSO
Oesophageal carcinoma (EC) is one of the most malignant gastrointestinal cancers worldwide (Sterne et al, 2009b; Peters et al, 2010a). China is one of the countries with high EC risk. Oesophageal carcinoma has a unique geographic distribution in China, including Chaoshan in Guangdong province, the southern region of Fujian province, Linzhou in Henan province, Cixian in Hebei province, and Yangcheng in Shanxi province (Su et al, 2001). The annual average crude incidence of EC in Chaoshan was >100/100 000 people from 1995 to 2004 (Umar and Fleischer, 2008). Oesophageal squamous cell carcinoma (ESCC) is the predominant subtype of EC in China (Pisani et al, 1999; Lehrbach et al, 2003), with an overall 5-year survival rate of <10% (Takeno et al, 2007).
One of the most commonly used staging systems for ESCC is the tumour-node-metastasis (TNM) system, which was proposed by the American Joint Committee in Cancer (AJCC) in 1988, then revised in 2009 (the 7th edition) (ohlschlager et al, 2010). The TNM staging system does not reflect the whole cancer progression status because the clinical and pathologic findings are gathered for staging at the time of surgery (Kunisaki et al, 2005; Goldstraw et al, 2007), and the system has become complex (Varotti et al, 2005).
As with the development of cancer biology, clinical oncology has greatly changed. Cancer biology will hasten the development of new prognostic and predictive models that may be more relevant in some cases (Zisman et al, 2001).
Recent advances in molecular biology have demonstrated the benefit of molecular profiling in clarifying subtypes of cancer pertinent to cancer progression (Wang et al, 1999; Takeno et al, 2007). Peters et al (2010b) proposed five gene signatures closely associated with an overall survival rate in oesophageal adenocarcinoma, with good external validation. Chen et al (2007) considered four genes as the prognostic genetic signature in non-small-cell lung cancer. However, these authors did not use critical clinical and pathological information in their studies. With univariate survival analysis, survival with ESCC was suggested to be associated with the proteins Ezrin, Fascin, pFascin, connective-tissue growth factor (CTGF), cysteine-rich angiogenic protein 61 (CYR61), neutrophil gelatinase-associated lipocalin (NGAL), NGAL receptor (NGALR), desmocollin 2 (DSC2), and activating transcription factor 3 (ATF3) (Wang et al, 1999; Zhang et al, 2006; Chen et al, 2007; Gao et al, 2009; Xie et al, 2009, 2010, 2011; Fang et al, 2010; Zhao et al, 2010; Du et al, 2011).
To identify important factors, we performed a retrospective review of 461 patients with ESCC to evaluate independent prognostic factors. A novel model was built by using pathology information with other important clinical indicators and it validated the simple-to-use FENSAM model in a contemporary independent cohort.
Patients and methods
Patients
We included two independent cohorts of patients with ESCC in this study. A total of 596 patients were enrolled to construct the classifier. We excluded 135 patients who had >8 missing biomarkers each, resulting in a final learning cohort of 461 patients. All patients had undergone surgical resection at the Central Hospital of Shantou City, Guangdong province, China, from 1987 to 1997. The validation cohort included 290 patients from the same hospital during 2007–2010. The clinicopathological characteristics of patients are in Table 3 in the Supplementary Appendix. The clinicopathological characteristics were comparable in the two cohorts (Table 3 in the Supplementary Appendix).
Computerised Tomography (CT) was used to confirm ESCC in patients. Patients were excluded if they declined to undergo surgical resection or sign the informed consent. We excluded patients who died from diseases other than ESCC. In addition, surgery was not possible for patients who were confirmed by CT as at TNM stage IV from 2007 to 2010, so these patients were not included in the test cohort.
Because of inconvenient communication at Shantou in 1990s, follow-up data were mainly collected from medical records and mail after surgical resection. Nonetheless, all patients in the two cohorts were followed up for 3–10 years or until death.
Tissue microarray and immunohistochemistry
The study was approved by the ethics committee of the Central Hospital of Shantou City. Pathology was used to help clinicians with proper assessment. Tissue microarray (TMA) and immunohistochemistry were performed as described (Xie et al, 2009; Du et al, 2011). Briefly, we obtained two or more tissue cores from each specimen that was 1.8 mm in diameter and 1.0–3.0 mm in length depending on the tissue depth in the donor block. Each core was set into a new paraffin block. The antibodies used for incubation are in Table 4 in the Supplementary Appendix. The Polymer Detection Systems for Immuno-Histological Staining Kit and the Liquid Substrate kit (ZSGB-BIO, Beijing, China) were used for immunohistochemistry. Each section was independently assessed by two pathologists who were blinded to patient data.
The immunohistochemical staining results were assigned a maximum score considering both the intensity of staining and the proportion of tumour cells showing unequivocal positive reactions (see Figures 1–3 in the Supplementary Appendix). Positive reactions were defined as brown signals in cell cytoplasm, nucleus, and membrane. The intensity of staining was 0, no staining; 1, weak staining; 2, moderate staining; and 3, strong staining. Tumour cell area was defined as positive staining, for scores of 0, <5% of tumour cells; 1, 5–25% of tumour cells; 2, 26–50% of tumour cells; 3, 51–75% of tumour cells; and 4, 75–100% of tumour cells. A final score was calculated by multiplying positive reactions and tumour cell area, for a total score ranging from 0 to 12.
Statistical analysis
Multivariate analysis of survival and model selection
The association between survival and each of 22 potential prognostic factors was evaluated by the univariate Cox proportional hazards regression analysis. The hazard ratio determined which markers were associated with survival among patients with high vs low frequency of ESCC (see Table 1 in the Supplementary Appendix). Combined the results with previous studies, 12 associated prognostic significant factors were applied as initiate covariates for multivariate Cox proportional hazards model (tumour size, surgery extent, differentiation, T-stage, N-stage, M-stage, Ezrin, CYR61, DSC2, Fascin, pFascin, and ATF3). To select significant prognostic factors associated with a survival rate, we used a standard variable-selection procedure, LASSO (Datta et al, 2007), in a multivariate Cox model and Bayesian Information Criteria to select a proper tuning parameter in LASSO (O'Hara and Sillanpaa, 2009; Alhamzawi and Yu, 2012). Six variables were found to be independent and significant predictors of risk of death. We used a linear combination of the protein-expression values weighted by the regression coefficient to determine a hazard risk score (RS) for each patient (for details, see Methods section in the Supplementary Appendix). We applied K-means clustering method according to the RSs for the six selected variables to set the cutoff values (Ferreira et al, 2007). These cutoff values were used to classify patients into four ordered subgroups.
To compare with the traditional TNM stage system, we repeated the following procedure 200 times. At each time, 75% of the data (n=346) was chosen as training data to build the FENSAM, with the remaining data (n=115) as the validation data. The 200 P-values for the log-rank test were recorded, with box plots used to depict the difference between these 2 stages. To further evaluate the FENSAM and the TNM stage system, we used Receiver Operating Characteristic (ROC) curve and Kaplan–Meier survival analyses of the four subgroups along with the log-rank statistics for the 200 P-values. For more details, see Methods section in the Supplementary Appendix.
Validation
According to these results, we validated the model with an independent cohort of 290 patients by ROC curve analysis and the log-rank test.
Results
Combining the selected variables to build a model
Previous univariate survival analyses suggested that 12 markers could be a potential prognostic factor in ESCC (Zhang et al, 2006; Gao et al, 2009; Xie et al, 2009, 2010, 2011; Fang et al, 2010; Zhao et al, 2010; Du et al, 2011) (see Tables 1 and 2 in the Supplementary Appendix).
Table 1 lists the estimated coefficients for the characteristics in the Cox proportional-hazards model. We selected six variables, and all except ATF3 were positively associated with a survival rate. N-stage had the largest absolute effect size, 0.763, so the hazard ratio for N-stage was 2.145 (=exp(0.763)); in other words, patients with N-stage=1 may die at about twice the rate per unit time as patients with N-stage=0. In addition, ATF3 was negatively associated with survival.
Table 1. Estimated coefficients for variables used in the FENSAM (Fascin Ezrin N M Surgery extent ATF3) modela.
| Variables | Surgery extent | N-stage | M-stage | Ezrin | Fascin | ATF3 |
|---|---|---|---|---|---|---|
| 0.357 | 0.763 | 0.112 | 0.056 | 0.026 | −0.063 |
Abbreviation: ATF3=activating transcription factor 3.
Least absolute shrinkage and selection operator (LASSO) was applied to select significant variables, with tuning parameters determined by Bayesian information criteria. The variables with coefficients >0 were included.
The RSs for the selected characteristics were calculated from the estimated coefficients in Table 1. The RS-based FENSAM system is presented in Table 2. The optimal cutoffs calculated by the clustering method were 0.359, 0.797, and 1.304, respectively. Among the 461 patients, 75 were classified as having ESCC as stage I, 132 stage II, 119 stage III, and 138 stage IV.
Table 2. Thresholds of the FENSAM (Fascin Ezrin N M Surgery extent ATF3) stages for ESCCa.
| FENSAM stage | Risk score range | No. of patients |
|---|---|---|
| I |
<0.359 |
75 |
| II |
0.359–0.797 |
132 |
| III |
0.797–1.304 |
119 |
| IV | >1.304 | 138 |
Abbreviation: ESCC=oesophageal squamous cell carcinoma.
The k-means clustering technique was applied to calculate the optimal cutoff points.
The median area under the ROC curve (AUC) values for the FENSAM and TNM systems are in Figure 1A. The AUC was larger for the FENSAM than the TNM system (0.6829 vs 0.6196). Comparison of other quantiles is depicted in Figure 1B. The median AUC values of the FENSAM were larger than the TNM stage.
Figure 1.
Receiver operating characteristic (ROC) curve analysis of the FENSAM and the TNM stage with the test data. (A) ROC curve for the FENSAM and the TNM stage. (B) Box plots for median AUC for the log-rank test of the FENSAM and TNM stage systems.
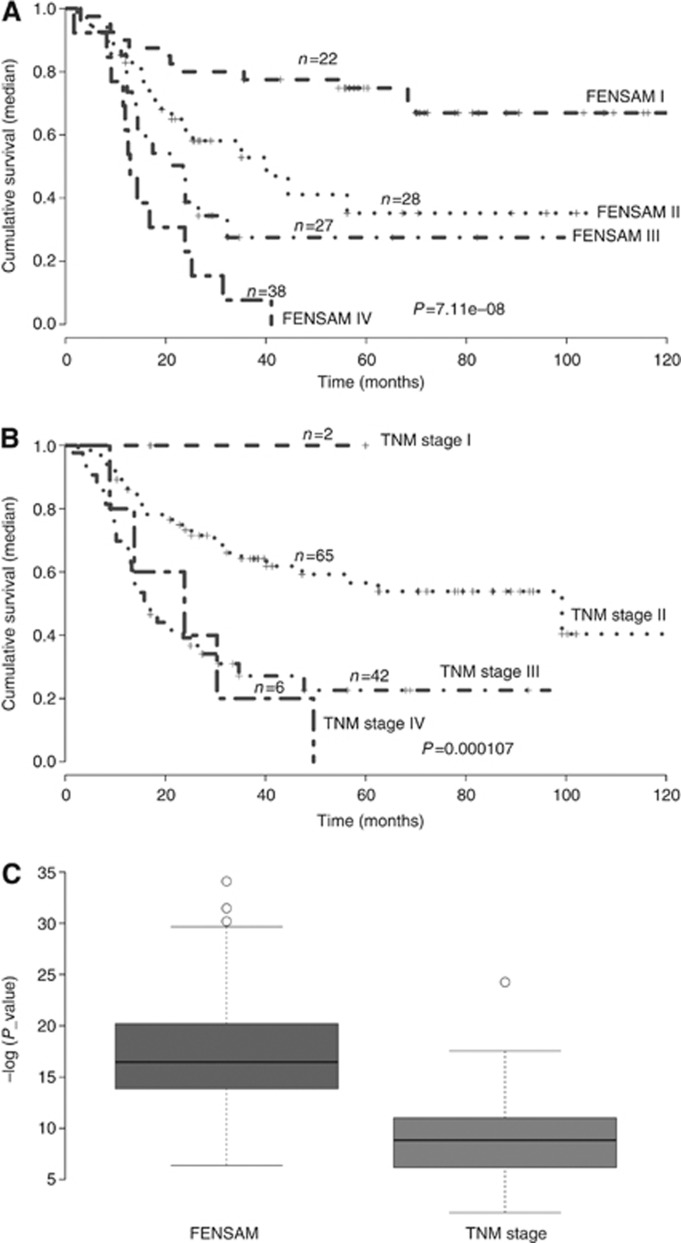
Survival was lower by the FENSAM than by the TNM system (P=7.11e−8 and P=1.07e−3) (Figure 2A and B). For survival time<30 months, survival was indistinguishable between TNM stage III and IV (Figure 2B) but was distinct between FENSAM III and IV (Figure 2A). Furthermore, the 5-year survival rate for TNM stages I–IV was 0.9%, 53.91%, 35.7%, and 1.0%, respectively, and for FENSAM stages I–IV was 20.9%, 37.4%, 19.1%, and 22.6%, respectively. In addition, the negative log-transformed P-values were larger with the FENSAM than the TNM system (Figure 2C). Detailed comparisons based on Kaplan–Meier survival analysis are described (see Figures 4–7 in the Supplementary Appendix).
Figure 2.
Comparison of the FENSAM and the TNM stage systems with the test data by Kaplan–Meier analysis. (A) FENSAM model. (B) TNM stage model. (C) Box plots for log-transformed P-values (−log 10 (P-value)) from the log-rank test with the 200 test data sets for the FENSAM and TNM models.
Validation
In the validation data set, no patient was at TNM stage IV, whereas 14 patients were classified as FENSAM stage IV, and all survived<3 years (Figure 3A and B). Therefore, the FENSAM stage coincided with the practical experience of clinicians and could increase the accuracy of ESCC prognosis. In addition, AUC scores were greater for FENSAM than for TNM stages (0.678 vs 0.626) (Figure 3C).
Figure 3.
The predictive ability of the FENSAM as compared with the TNM stage model by Kaplan–Meier analysis in the validation cohort (A and B) and ROC curves (C).
Discussion
Oesophageal squamous cell carcinoma is the predominant subtype of EC in China, with an overall 5-year survival rate of <10%. The staging system is based on the anatomic extent of the cancer. We aim to combine clinical variables and biomarkers to develop and validate a single, reliable model, named FENSAM, for ESCC prognosis. To build the FENSAM, we analysed 22 potential prognostic factors from 461 patients. We used multivariate analysis and variable selection to select significant factors associated with survival of ESCC patients. These selected factors were used to build our FENSAM model. We then obtained the hazard RS of the model to classify ESCC patients. In addition, we validated the model in an independent cohort of 290 patients from the same hospital. The predictive performance of the model was assessed by AUC and Kaplan–Meier survival analysis.
In short, FENSAM performed better than the TNM staging system based on our validation data. Although both the learning and validation data were collected from the same hospital and with some missing data, the FENSAM's superior performance compared with TNM is noteworthy and further validation using multi-centre data will enhance the generalisation of our result.
Missing data present a ubiquitous problem in everyday clinical practice, and incomplete data are removed in the data analysis. We should pay more attention to the issue of missing data, which can introduce bias and lead to inaccurate conclusions. Although a variety of ad-hoc approaches are commonly used to handle missing data, some of these approaches are usually statistically invalid and also lead to serious bias (Sterne et al, 2009a). Rubin's multiple imputation (MI) method (Sterne et al, 2009a) (see Methods section in the Supplementary Appendix), a general approach, can improve the validity of medical research. This study featured a lot of missing data, especially for the nine biochemical indicators in the test data set. In particular, data for Fascin, pFascin, and ATF3 featured more than 300 missing values, each with a missing rate of >50% (Table 3). Since we focussed on how the biomarkers influenced ESCC prognosis in this study, we removed data for patients with missing data for more than eight missing biomarkers. As a result, we included only 461 patients in our prediction model. Admittedly, all these missing data undermine the validity of our results. To reduce bias and increase efficiency as much as possible, the EMB algorithm combining the classic EM algorithm and a bootstrap approach was applied to impute data.
Table 3. The number of missing values for oesophageal squamous cell carcinoma (ESCC) cohort at the central hospital of Shantou from 1987 to 1999.
| Time | Status | Age | Blood groups | Therapy method | Tumour size | Differentiation | Surgery extent | Smoking | Alcohol | Gender | Tumour site | T-stage | N-stage | M-stage | Ezrin | CTGF | CYR61 | NGAL | NGALR | DSC2 | Fascin | pFascin | ATF3 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 0 | 0 | 7 | 5 | 78 | 78 | 6 | 15 | 20 | 0 | 11 | 28 | 2 | 8 | 291 | 234 | 234 | 273 | 255 | 292 | 342 | 342 | 300 |
Abbreviations: ATF3=activating transcription factor 3; CTGF=connective-tissue growth factor; CYR61=cysteine-rich angiogenic protein 61; DSC2=desmocollin 2; NGAL=neutrophil gelatinase-associated lipocalin; NGALR=NGAL receptor.
Previously, six biomarkers (Ezrin, CYR61, DSC2, Fascin, pFascin, and ATF3) and six other clinicopathological characteristics (tumour size, surgery extent, differentiation, T-stage, N-stage, and M-stage) were found associated with the overall survival of ESCC patients by univariate COX proportional hazards model and in the previous studies (Wang et al, 1999; Zhang et al, 2006; Chen et al, 2007; Gao et al, 2009; Xie et al, 2009, 2010, 2011; Fang et al, 2010; Zhao et al, 2010; Du et al, 2011). However, our multivariate analysis revealed only Ezrin, Fascin, ATF3, N-stage and M-stage and surgery extent associated with survival. Both Ezrin and Fascin are actin-binding proteins that are overexpressed in ESCC and have key roles in the progression of ESCC (Xie et al, 2005, 2009). ATF3 is a transcription factor involved in cell-cycle progression, invasiveness, and metastasis of cancer cells. We recently revealed that ATF3 was involved in Ezrin or Fascin-mediated proliferation and invasiveness of ESCC cells (Xie et al, 2009, 2010). Therefore, molecules involved in a certain biological process or signal cascade may help in developing a cancer staging system. As well, in our model, coefficients were higher for clinicopathological characteristics than for biomarkers, so regional lymph-node involvement and distant metastasis were still predominant prognostic factors in ESCC.
Previous studies considered TNM clinical stage as the best prognostic indicator and T-, N-, and M-stage factors were associated with ESCC prognosis on univariate analysis in most previous studies (Goldstraw et al, 2007). However, our study showed that only N- and M-stages were independent prognostic factors on multivariate analysis. This result was consistent with several previous studies (Ikeda et al, 1999; Kuo et al, 2003).
By an extensive literature review, we selected the six biomarkers in the present study—three biomarkers (Ezrin, Fascin, and ATF3) and three clinical indicators (surgery extent, N- and M-stages)—as prognostic factors in ESCC. In addition, with the development of molecular techniques, more new biomarkers might emerge, which might be used as novel clinical indicators and provide higher specificity for the diagnosis and prognosis of ESCC.
The prognostic significance of ESCC classification is important. We can use prognosis to evaluate clinical outcome, guide treatment decisions, and account for the results of clinical trials (Zisman et al, 2001). The classification method we used could better separate patients into prognostic groups and improve the accuracy of the survival prediction on the basis of easily accessible clinical indicators and biomarkers. The six variables are readily available, are statistically significant prognostic factors, and were easily validated by confirmatory studies. FENSAM is based on pathology response rate, clinical indicators, and pathologic markers (Mittendorf et al, 2011; Yi et al, 2011) and is reliable and easy to use. On the basis of a simple model, FENSAM provides an alternative classifier for ESCC patients with a high precision. The method could be used with other malignant tumours.
The FENSAM is available at our website: http://escc.bio.med.stu.edu.cn/member/login.php. Our data sets are available upon request.
Acknowledgments
We thank Mrs Zepeng Du and Lingling Sun at the Institute of Oncologic Pathology, Shantou University Medical College for the assistance in obtaining data. We also thank Mr Shaoming Ou at the Network and Information Center, Shantou University Medical College. This work was supported by grants from the Major State Basic Research Development Program (20110171110037), the Doctoral Fund of Ministry of education of China (RFDP 20110171110037), NSFC for Excellent Young Scholar (11322108), NCET (12-0559), the overseas and Hong Kong, Macau Young Scholars Collaborative Research Fund from NSFC (11328103), the Natural Science Foundation of China (11271383), the National High Technology Research and Development Program of China (2012AA02A503 and 2012AA02A209), the National Basic Research Program (2012CB526608), the Natural Science Foundation of China-GuangDong Joint Fund (U0932001), the National Science Foundation of China (81071737) and the US National Institute on Drug Abuse (R01DA016750).
Author contributions
HT and XW conceived the concept for this study; HT, CW, and XG discussed and performed the analyses; HT and HZ wrote the manuscript; EL and LX developed the study design; ZW carried out surgery conduct and data collection; JS and QZ carried out review; JX and BC were responsible for immunohistochemistry, pathological data analysis, and interpretation; XX produced the data from tissue microarrays and immunohistochemistry; JW was responsible for follow-up trace; XW and HZ supervised the data analysis. All authors had input into the data interpretation and preparation of the final report for publication.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on British Journal of Cancer website (http://www.nature.com/bjc)
This work is published under the standard license to publish agreement. After 12 months the work will become freely available and the license terms will switch to a Creative Commons Attribution-NonCommercial-Share Alike 3.0 Unported License.
Supplementary Material
References
- Alhamzawi R, Yu KM. Variable selection in quantile regression via Gibbs sampling. J Appl Stat. 2012;39 (4:799–813. [Google Scholar]
- Chen HY, Yu SL, Chen CH, Chang GC, Chen CY, Yuan A, Cheng CL, Wang CH, Terng HJ, Kao SF, Chan WK, Li HN, Liu CC, Singh S, Chen WJ, Chen JJ, Yang PC. A five-gene signature and clinical outcome in non-small-cell lung cancer. N Engl J Med. 2007;356 (1:11–20. doi: 10.1056/NEJMoa060096. [DOI] [PubMed] [Google Scholar]
- Datta S, Le-Rademacher J, Datta S. Predicting patient survival from microarray data by accelerated failure time modeling using partial least squares and LASSO. Biometrics. 2007;63 (1:259–271. doi: 10.1111/j.1541-0420.2006.00660.x. [DOI] [PubMed] [Google Scholar]
- Du ZP, Lv Z, Wu BL, Wu ZY, Shen JH, Wu JY, Xu XE, Huang Q, Shen J, Chen HB, Li EM, Xu LY. Neutrophil gelatinase-associated lipocalin and its receptor: independent prognostic factors of oesophageal squamous cell carcinoma. J Clin Pathol. 2011;64 (1:69–74. doi: 10.1136/jcp.2010.083907. [DOI] [PubMed] [Google Scholar]
- Fang WK, Gu W, Li EM, Wu ZY, Shen ZY, Shen JH, Wu JY, Pan F, Lv Z, Xu XE, Huang Q, Xu LY. Reduced membranous and ectopic cytoplasmic expression of DSC2 in esophageal squamous cell carcinoma: an independent prognostic factor. Hum Pathol. 2010;41 (10:1456–1465. doi: 10.1016/j.humpath.2010.04.003. [DOI] [PubMed] [Google Scholar]
- Ferreira I, Boreham CA, Twisk JWR, Gallagher AM, Young IS, Murray LJ, Stehouwer CDA. Clustering of metabolic syndrome risk factors and arterial stiffness in young adults: the Northern Ireland Young Hearts Project. J Hypertens. 2007;25 (5:1009–1020. doi: 10.1097/HJH.0b013e3280a94e76. [DOI] [PubMed] [Google Scholar]
- Gao S-Y, Li E-M, Cui L, Lu X-F, Meng L-Y, Yuan H-M, Xie J-J, Du Z-P, Pang J-X, Xu L-Y. Sp1 and AP-1 regulate expression of the human gene VIL2 in esophageal carcinoma cells. J Biol Chem. 2009;284 (12:7995–8004. doi: 10.1074/jbc.M809734200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstraw P, Crowley J, Chansky K, Giroux DJ, Groome PA, Rami-Porta R, Postmus PE, Rusch V, Sobin L. The IASLC Lung Cancer Staging Project: proposals for the revision of the TNM stage groupings in the forthcoming (seventh) edition of the TNM Classification of malignant tumours. J Thorac Oncol. 2007;2 (8:706–714. doi: 10.1097/JTO.0b013e31812f3c1a. [DOI] [PubMed] [Google Scholar]
- Ikeda G, Isaji S, Chandra B, Watanabe M, Kawarada Y. Prognostic significance of biologic factors in squamous cell carcinoma of the esophagus. Cancer. 1999;86 (8:1396–1405. doi: 10.1002/(sici)1097-0142(19991015)86:8<1396::aid-cncr3>3.0.co;2-h. [DOI] [PubMed] [Google Scholar]
- Kunisaki C, Akiyama H, Nomura M, Matsuda G, Otsuka Y, Ono HA, Shimada H. Developing an appropriate staging system for esophageal carcinoma. J Am Coll Surg. 2005;201 (6:884–890. doi: 10.1016/j.jamcollsurg.2005.07.002. [DOI] [PubMed] [Google Scholar]
- Kuo K-T, Chow K-C, Wu Y-C, Lin C-S, Wang H-W, Li W-Y, Wang L-S. Clinicopathologic significance of cyclooxygenase-2 overexpression in esophageal squamous cell carcinoma. Ann Thorac Surg. 2003;76 (3:909–914. doi: 10.1016/s0003-4975(03)00717-3. [DOI] [PubMed] [Google Scholar]
- Lehrbach DM, Nita ME, Cecconello I. Molecular aspects of esophageal squamous cell carcinoma carcinogenesis. Arq Gastroenterol. 2003;40 (4:256–261. doi: 10.1590/s0004-28032003000400011. [DOI] [PubMed] [Google Scholar]
- Mittendorf EA, Jeruss JS, Tucker SL, Kolli A, Newman LA, Gonzalez-Angulo AM, Buchholz TA, Sahin AA, Cormier JN, Buzdar AU, Hortobagyi GN, Hunt KK. Validation of a novel staging system for disease-specific survival in patients with breast cancer treated with neoadjuvant chemotherapy. J Clin Oncol. 2011;29 (15:1956–1962. doi: 10.1200/JCO.2010.31.8469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Hara RB, Sillanpaa MJ. A review of Bayesian variable selection methods: what, how and which. Bayesian Anal. 2009;4 (1:85–117. [Google Scholar]
- ohlschlager J, Wittekind C, Theegarten D. [New TNM classification of malignant lung tumours] Pathologe. 2010;31 (5:355–360. doi: 10.1007/s00292-010-1303-4. [DOI] [PubMed] [Google Scholar]
- Peters CJ, Rees JR, Hardwick RH, Hardwick JS, Vowler SL, Ong CA, Zhang C, Save V, O'Donovan M, Rassl D, Alderson D, Caldas C, Fitzgerald RC.2010A 4-gene signature predicts survival of patients with resected adenocarcinoma of the esophagus, junction, and gastric cardia Gastroenterology 139(61995–2004.e15. [DOI] [PubMed] [Google Scholar]
- Peters CJ, Rees JRE, Hardwick RH, Hardwick JS, Vowler SL, Ong CAJ, Zhang CS, Save V, O'Donovan M, Rassl D, Alderson D, Caldas C, Fitzgerald RC, Stra OCCM A 4-gene signature predicts survival of patients with resected adenocarcinoma of the esophagus, junction, and gastric cardia. Gastroenterology. 2010;139 (6:1995–2004. doi: 10.1053/j.gastro.2010.05.080. [DOI] [PubMed] [Google Scholar]
- Pisani P, Parkin DM, Bray F, Ferlay J. Estimates of the worldwide mortality from 25 cancers in 1990. Int J Cancer. 1999;83 (1:18–29. doi: 10.1002/(sici)1097-0215(19990924)83:1<18::aid-ijc5>3.0.co;2-m. [DOI] [PubMed] [Google Scholar]
- Sterne JA, White IR, Carlin JB, Spratt M, Royston P, Kenward MG, Wood AM, Carpenter JR. Multiple imputation for missing data in epidemiological and clinical research: potential and pitfalls. BMJ. 2009;338:b2393. doi: 10.1136/bmj.b2393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sterne JAC, White IR, Carlin JB, Spratt M, Royston P, Kenward MG, Wood AM, Carpenter JR. Multiple imputation for missing data in epidemiological and clinical research: potential and pitfalls. Br Med J. 2009;339:b2393. doi: 10.1136/bmj.b2393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su M, Lu SM, Tian DP, Zhao H, Li XY, Li DR, Zheng ZC. Relationship between ABO blood groups and carcinoma of esophagus and cardia in Chaoshan inhabitants of China. World J Gastroenterol. 2001;7 (5:657–661. doi: 10.3748/wjg.v7.i5.657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeno S, Noguchi T, Takahashi Y, Fumoto S, Shibata T, Kawahara K. Assessment of clinical outcome in patients with esophageal squamous cell carcinoma using TNM classification score and molecular biological classification. Ann Surg Oncol. 2007;14 (4:1431–1438. doi: 10.1245/s10434-006-9286-3. [DOI] [PubMed] [Google Scholar]
- Umar SB, Fleischer DE. Esophageal cancer: epidemiology, pathogenesis and prevention. Nat Clin Pract Gastr. 2008;5 (9:517–526. doi: 10.1038/ncpgasthep1223. [DOI] [PubMed] [Google Scholar]
- Varotti G, Ramacciato G, Ercolani G, Grazi GL, Vetrone G, Cescon M, Del Gaudio M, Ravaioli M, Ziparo V, Lauro A, Pinna A. Comparison between the fifth and sixth editions of the AJCC/UICC TNM staging systems for hepatocellular carcinoma: multicentric study on 393 cirrhotic resected patients. Eur J Surg Oncol. 2005;31 (7:760–767. doi: 10.1016/j.ejso.2005.04.008. [DOI] [PubMed] [Google Scholar]
- Wang LS, Chow KC, Chi KH, Liu CC, Li WY, Chiu JH, Huang MH. Prognosis of esophageal squamous cell carcinoma: analysis of clinicopathological and biological factors. Am J Gastroenterol. 1999;94 (7:1933–1940. doi: 10.1111/j.1572-0241.1999.01233.x. [DOI] [PubMed] [Google Scholar]
- Xie JJ, Xu LY, Wu JY, Shen ZY, Zhao Q, Du ZP, Lv Z, Gu W, Pan F, Xu XE, Xie D, Li EM. Involvement of CYR61 and CTGF in the fascin-mediated proliferation and invasiveness of esophageal squamous cell carcinomas cells. Am J Pathol. 2010;176 (2:939–951. doi: 10.2353/ajpath.2010.090118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie JJ, Xu LY, Wu ZY, Li LY, Xu XE, Wu JY, Huang Q, Li EM. Expression of cysteine-rich 61 is correlated with poor prognosis in patients with esophageal squamous cell carcinoma. Eur J Surg Oncol. 2011;37 (8:669–674. doi: 10.1016/j.ejso.2011.05.009. [DOI] [PubMed] [Google Scholar]
- Xie JJ, Xu LY, Xie YM, Zhang HH, Cai WJ, Zhou F, Shen ZY, Li EM. Roles of ezrin in the growth and invasiveness of esophageal squamous carcinoma cells. Int J Cancer. 2009;124 (11:2549–2558. doi: 10.1002/ijc.24216. [DOI] [PubMed] [Google Scholar]
- Xie JJ, Xu LY, Zhang HH, Cai WJ, Mai RQ, Xie YM, Yang ZM, Niu YD, Shen ZY, Li EM. Role of fascin in the proliferation and invasiveness of esophageal carcinoma cells. Biochem Biophys Res Commun. 2005;337 (1:355–362. doi: 10.1016/j.bbrc.2005.09.055. [DOI] [PubMed] [Google Scholar]
- Yi M, Mittendorf EA, Cormier JN, Buchholz TA, Bilimoria K, Sahin AA, Hortobagyi GN, Gonzalez-Angulo AM, Luo S, Buzdar AU, Crow JR, Kuerer HM, Hunt KK. Novel staging system for predicting disease-specific survival in patients with breast cancer treated with surgery as the first intervention: time to modify the current American Joint Committee on Cancer staging system. J Clin Oncol. 2011;29 (35:4654–4661. doi: 10.1200/JCO.2011.38.3174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Xu L, Xiao D, Xie J, Zeng H, Cai W, Niu Y, Yang Z, Shen Z, Li E. Fascin is a potential biomarker for early-stage oesophageal squamous cell carcinoma. J Clin Pathol. 2006;59 (9:958–964. doi: 10.1136/jcp.2005.032730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Q, Shen J-H, Shen Z-Y, Wu Z-Y, Xu X-E, Xie J-J, Wu J-Y, Huang Q, Lu X-F, Li E-M, Xu L-Y. Phosphorylation of fascin decreases the risk of poor survival in patients with esophageal squamous cell carcinoma. J Histochem Cytochem. 2010;58 (11:979–988. doi: 10.1369/jhc.2010.955765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zisman A, Pantuck AJ, Dorey F, Said JW, Shvarts O, Quintana D, Gitlitz BJ, deKernion JB, Figlin RA, Belldegrun AS. Improved prognostication of renal cell carcinoma using an integrated staging system. J Clin Oncol. 2001;19 (6:1649–1657. doi: 10.1200/JCO.2001.19.6.1649. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.