Abstract
Background:
Various markers are used to identify the unique sub-population of breast cancer cells with stem cell properties. Whether these markers are expressed in all breast cancers, identify the same population of cells, or equate to therapeutic response is controversial.
Methods:
We investigated the expression of multiple cancer stem cell markers in human breast cancer samples and cell lines in vitro and in vivo, comparing across and within samples and relating expression with growth and therapeutic response to doxorubicin, docetaxol and radiotherapy.
Results:
CD24, CD44, ALDH and SOX2 expression, the ability to form mammospheres and side-population cells are variably present in human cancers and cell lines. Each marker identifies a unique rather than common population of cancer cells. In vivo, cells expressing these markers are not specifically localized to the presumptive stem cell niche at the tumour/stroma interface. Repeated therapy does not consistently enrich cells expressing these markers, although ER-negative cells accumulate.
Conclusions:
Commonly employed methods identify different cancer cell sub-populations with no consistent therapeutic implications, rather than a single population of cells. The relationships of breast cancer stem cells to clinical parameters will require identification of specific markers or panels for the individual cancer.
Keywords: breast cancer stem cells, mammospheres, side-population, therapy resistance
Current opinion proposes that tumours contain a sub-population of cells with stem cell properties that are responsible for maintaining tumour growth. These ‘cancer stem cells' (CSCs) or ‘cancer-initiating cells' are considered to have unlimited proliferative potential and to divide slowly and asymmetrically, whereas the resulting progenitor cells divide rapidly, progressively lose proliferative potential and undergo limited differentiation to form the heterogeneous populations of cells that are seen in tumours. Although the hypothesis remains controversial (Lawson et al, 2009; Rosen and Jordan, 2009), putative CSCs have been identified in human cancers, including breast cancer, on the basis of the ability of specific cell populations to initiate tumour formation when transplanted into xenograft models. The clinically important implication of the CSC hypothesis is that CSCs are critical therapeutic targets, because they are uniquely able to reform the tumour. Moreover, CSCs have been reported to exhibit enhanced resistance to therapy through a variety of mechanisms (Bao et al, 2006; Phillips et al, 2006; Eyler and Rich, 2008; Lawson et al, 2009). Therefore, identification of CSCs is important for understanding tumour biology and should have clinical relevance.
Breast CSCs were first identified as a CD24−/low/CD44+population with enhanced ability to initiate tumour growth when xenografted into immunocompromised mice (Al-Hajj et al, 2003). Subsequent studies identified several other markers, such as aldehyde dehydrogenase 1 (ALDH1), CD133, Sox2, CK5, alpha-6 integrin/CD49f, beta-1 integrin/CD29 or lack of estrogen receptor (ER); cells exhibiting these phenotypes are also found in breast cancer cell lines and may mark cells with enhanced tumour initiation ability (Ginestier et al, 2007; Fillmore and Kuperwasser, 2008; Wright et al, 2008; Charafe-Jauffret et al, 2009; Lawson et al, 2009; Kabos et al, 2011; Leis et al, 2012). Other markers for breast CSCs include the identification of a side-population of cells that exclude uptake of dyes due to the presence of membrane transporters, or the ability to grow as anchorage-independent spheres (Christgen et al, 2007; Engelmann et al, 2008; Grimshaw et al, 2008; Kok et al, 2009; Harrison et al, 2010).
Although the existence of a specific sub-population of cells with stem cell or cancer-initiating properties and therapeutic resistance has gained considerable support, conflicting data also exist indicating that not all cancers contain a specific marker, or that an individual marker necessarily associates with therapy resistance or patient outcome (Kok et al, 2009; Stingl, 2009; Pajic et al, 2010; Ahmed et al, 2012; Tan et al, 2013). Given the plethora of approaches used to identify CSCs in different studies, the inter-relationships of the identified cell populations with each other is also unclear and might account for some of these discrepancies. Therefore, to gain further insight into the nature of breast CSCs and the inter-relationships of CSC markers with therapy response, the aim of this study was to employ multiple methods to identify breast CSCs in human samples and cell lines in vitro and in vivo, and compare the expression of these markers with each other and in relation to therapy.
Materials and methods
Human breast cancer samples
Metastatic deposits of 33 cancers with axillary lymph node metastasis greater than 2 mm in size (at least pN1) were retrieved from the histopathological records of consecutive ductal breast carcinoma cases diagnosed in Brno, Czech Republic. All samples had been collected at surgery, fixed in formalin and processed to paraffin wax for diagnostic histopathology. The status of ER alpha, progesterone receptor and HER2 was determined by immunohistochemistry (Table 1). HER2 gene amplification status was confirmed by fluorescence in situ hybridization according to the manufacturer's instructions (Abbott PathVysion HER2 DNA Probe Kit, Maidenhead, UK). Permission for the use of human tissues was granted following local ethical committee review and all patients gave consent for the use of their tissue for research.
Table 1. Antibodies used for immunohistochemistry.
| Antigen | Supplier, clone/Cat # | Dilution IHC |
|---|---|---|
| ALDH |
Becton–Dickinson, #611165 |
1/2000 |
| CD44 |
Novocastra, NVL-CD44 |
1/4000 |
| Sox2 |
Cell Signalling Technology #3579 |
1/200 |
| EMA |
Dako, E29 |
1/5000 |
| SMA |
Dako, 1A4 |
1/1500 |
| ERα |
LabVision, SP2 |
1/500 |
| PR | LabVision SP2 | 1/2000 |
Abbreviations: ALDH=aldehyde dehydrogenase; CD=cluster of differentiation; EMA=epithelial membrane antigen; ER=estrogen receptor; IHC=immunohistochemistry; PR=progesterone receptor; SMA=smooth muscle actin.
Cell culture and murine xenografts
The luminal-type human breast cancer cell lines MCF7, T47D and ZR75-1 (steroid receptor positive); SKBR3 (HER2+); and triple negative cell lines MDAMB231 (mesenchymal) and MDAMB468 (epithelial, EGFR+) were obtained from ATCC, ECACC or DSMZ and grown in DMEM, RPMI (for T47D) or McCoys 5A (for SKBR3) with 10% fetal bovine serum (all from Invitrogen, Life Technologies Ltd., Paisley, UK) at 37 °C with 5% CO2. Cells were routinely passaged at least every three days and all assays were performed when cells were sub-confluent. For mammosphere culture, cells were plated in dishes previously coated overnight with 1% poly(2-hydroxy-ethyl-methacrylate) in 90% ethanol (Sigma, Poole, UK). Mammosphere growth medium contained DMEM/F12 without serum but with B27 (Invitrogen) and SingleQuots (Lonza Biologics, Slough, UK) as growth factor supplements (Harrison et al, 2010). In some experiments, cells or mammospheres were collected by centrifugation, fixed overnight in formalin, re-suspended in agarose and processed into paraffin wax using standard histological procedures. Colony-forming efficiencies were measured by plating cells at low density depending on the cell line and counting the number of colonies after growth for 12–15 days.
For xenografting, MCF7 or MDAMB468 cells were re-suspended in a 50 : 50 mixture of culture medium and matrigel (Becton–Dickinson) and injected subcutaneously into female immunocompromised mice supplemented with slow release 17β-estradiol pellets (for MCF7), as previously described (Appleyard et al, 2012). Mice were housed under aseptic conditions in individually ventilated cages. All animal procedures were carried out under project licence 60/3405 after local ethical review and according to the guidelines of the UKCCR.
Chemotherapy and radiotherapy in vitro and in vivo
Cultured cells were treated three times with doxorubicin, docetaxel (Sigma-Aldrich, Gillingham, UK) or ionizing radiation. For chemotherapy, cells were exposed to the drug for 2 h, washed and incubated in a fresh medium for 48 h, followed by two further rounds of treatment. Cells were collected 48 h after the third treatment. For ionizing radiation, cells were treated with a standard clinical radiotherapy dose of 2 Gy using a CIS Bio International 637 caesium irradiator (0.4 Gy min−1). Radiation was repeated daily for a total of three treatments and cells were collected 48 h after the third exposure. Control cells were maintained under the same conditions but without irradiation or exposure to chemotherapeutic agents. In addition, established MCF7 xenografts were treated with doxorubicin at the maximum tolerated dose once a week for three weeks. Residual tumours were excised and fixed in 10% neutral buffered formalin before processing to paraffin wax.
Immunohistochemistry
Cells grown on glass slides were fixed in −20 °C acetone/methanol (1 : 1) for 10 min at room temperature, air-dried and stored at −80 °C. Sections of formalin-fixed paraffin-embedded human breast cancer sample, cultured cell pellets, spheroids or tumour xenografts were de-waxed and antigen retrieval performed by boiling for 15 min in citric acid buffer (10 mM, pH 6.0) in a microwave oven. Primary antibodies (Table 1) were applied overnight at 4 °C and were detected with biotinylated secondary antibody and avidin/biotinylated peroxidase complex (Vector Laboratories Ltd., Peterborough, UK) with DAB (Sigma) as chromogen. Nuclei were counterstained with haematoxylin. For dual peroxidase staining, mouse and rabbit primary antibodies and detection reagents were applied sequentially. The first antigen was detected with DAB containing nickel sulphate to produce a blue/grey reaction product and the second antigen was detected with DAB (brown). These sections were mounted without counterstaining.
Flow cytometry and FACS
Cells (106 in 1% bovine serum albumin in PBS) were stained with FITC-conjugated mouse anti-human CD44 and R-Phycoerythrin-conjugated mouse anti-human CD24 (BD Bioscience, Oxford, UK) at 1/100 dilution at 4 °C for 30 min. Aldehyde dehydrogenase activity was measured using the ALDEFLUOR assay (STEMCELL Technologies, Grenoble, France). Cells were incubated in ALDEFLUOR reagent with or without DEAB (ALDH inhibitor) at 37 °C for 40 min, centrifuged and re-suspended in assay buffer. In some experiments, PE-conjugated mouse anti-human CD24 (BD Bioscience, Oxford, UK) and Alexa Fluor 647-CD44 (AbD Serotec, Kidlington, UK) were added. For assessment of side-population, cells were stained with Hoechst 33342 (5 μg ml−1) at 37 °C for 90 min with or without reserpine (10 μM, as a negative control; Sigma-Aldrich, Dorset, UK), washed and re-suspended in PBS. CellQuest Pro (BD) and Summit v.4 (Dako, Glostrup, Denmark) software were applied for data acquisition and analysis, respectively, using measurements from 10 000 cells in each experiment. Cells were sorted using a FACSVantage (BD Bioscience) either directly into tissue culture plates or into centrifuge tubes for further culture.
Statistical tests
For immunostaining, data were collected by counting the percentages of positive cells in at least 10 high-power fields. Student's t-test was applied for cell culture assays, whereas Mann–Whitney test was used for xenograft analyses due to uneven distribution of positive cells in tumour material. All experimental data were obtained from three experimental repeats and P-values <0.05 were considered significant. Bar graphs show mean values with 95% confidence intervals.
Results
Variable expression and lack of correlation of CSC markers in human breast cancers
To investigate the relationships of various markers of the breast cancer stem cell population, we examined axillary node metastatic deposits of primary cancer samples. Immunohistochemistry showed highly variable levels of expression of CD44, Sox2 and ALDH1 in different tumours; 27% of tumours did not express CD44 in any tumour cells and the remaining tumours showed up to 80% CD44+ cells; Sox2 was present in 73% of tumours; and ALDH1+ tumour cells in only 43% of the tumours studied (note that ALDH1 was commonly seen in stromal cells, as previously reported (Resetkova et al, 2010) and CD44 was expressed by a subset of lymphocytes (see Supplementary Figure 1), but only tumour cells were included in this study). We also observed a highly heterogeneous distribution of staining in different areas of the section, where some areas contained ∼100% of positive cells whereas other areas of the same section showed no positive cells for a given marker (see Supplementary Figure 1 for an example of SOX2 staining localized to a specific area of tumour cells).
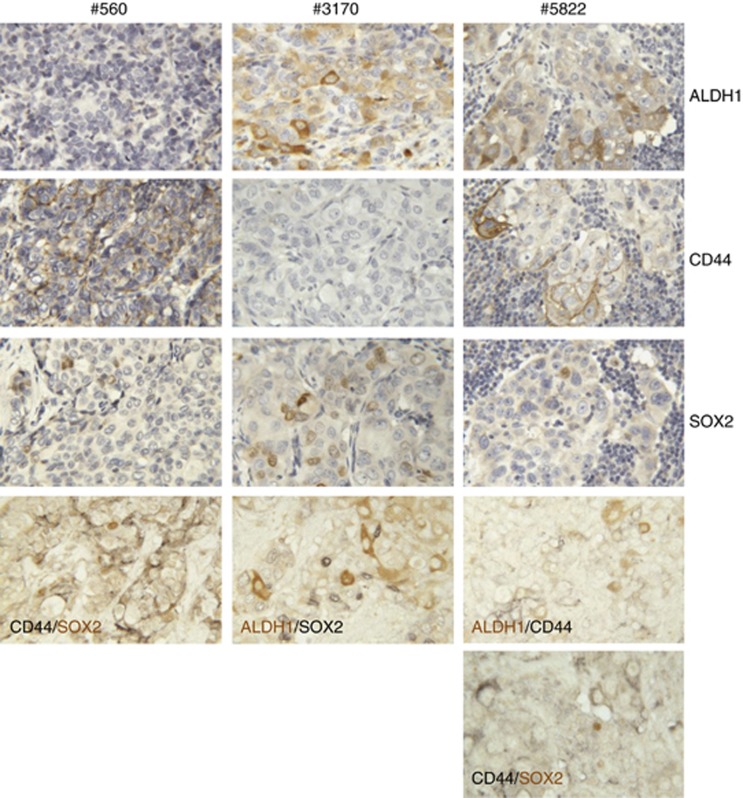
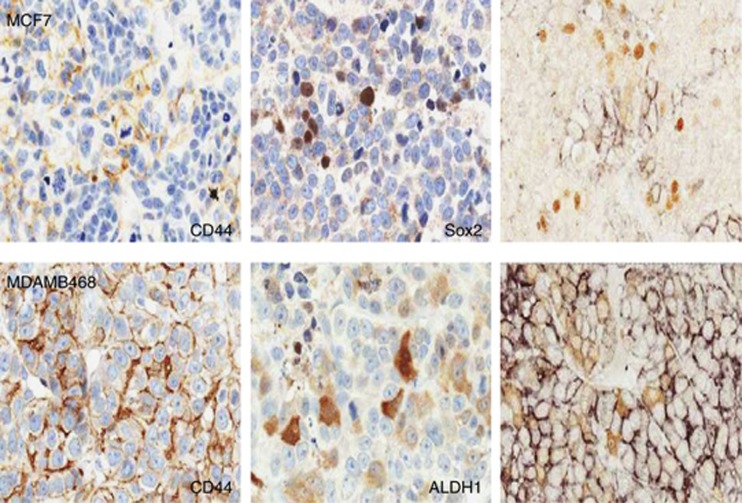
To investigate whether each of these markers was associated with the same sub-population of cells, we employed double-labelling immunohistochemistry on samples. Figure 1 shows three different tumours, two of which contained cells positive for two of the three markers and one that contained cells positive for all three markers. Double labelling of the first two tumours showed that individual cells expressed either one marker or the other, but not both markers. In the third tumour, no co-localization of CD44 was seen with either of the two other markers, ALDH1 or SOX2 (Figure 1). Thus, each marker identifies a unique sub-population of cancer cells, rather than identifying the same population of cells.
Figure 1.
Marker expression and co-localization in human breast cancers. Immunocytochemistry of three different human breast cancers (#560, #3170 and #5822). Single antigen detection of ALDH1, CD44 and SOX2 are shown for each cancer (positive reaction is brown, nuclei are counterstained blue). Also shown are double labelling for the indicated antigens in each cancer using a black chromogen to identify one antigen and brown for the second antigen, as indicated on the individual photomicrographs (no nuclear counterstain).
Expression of CSC markers and CSC phenotypes in cell lines
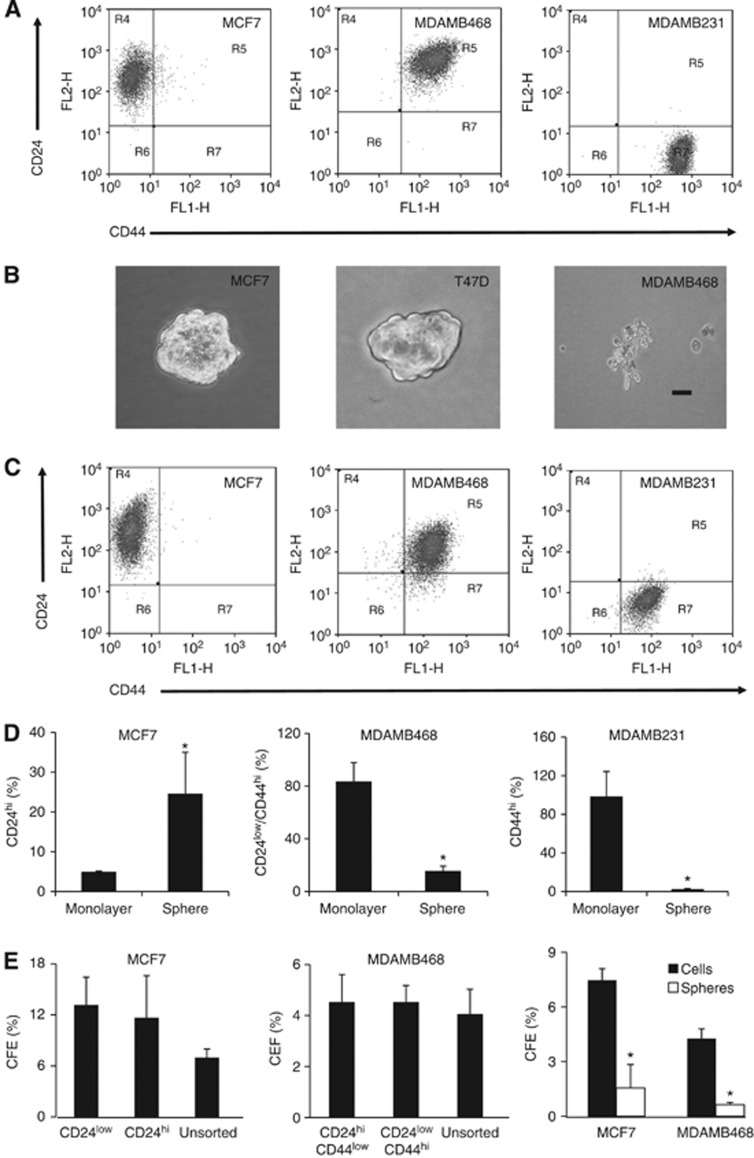
Given that the data from human cancers indicated that the three CSC markers studies identified a unique rather than common cell population, we investigated the relationships of marker expression with other CSC phenotypes and with therapeutic response in cell line models. FACS analysis of breast cancer cell lines showed highly variable expression of CD44 and CD24 in different cell lines (Figure 2a). MCF7 cells grown as monolayers were strongly positive for CD24 with undetectable CD44, MDAMB468 cells showed high level expression of both markers and MDAMB231 cells expressed high levels of CD44 but not CD24. When grown under mammosphere culture conditions, only MCF7 and T47D of the cell lines tested formed three-dimensional-structured spheroids, compared with the loosely packed and irregular-shaped clusters formed by MDAMB468 and MDAMB231 cells in suspension (Figure 2b), as previously reported (Rappa et al, 2008). To cross-correlate CD24/CD44 phenotypes in monolayer and suspension culture conditions, we compared the representative population associated with each cell line (i.e. changes in CD24 expression in MCF7, a change in the CD24low/CD44high population in MDAMB468 and the CD44high population in MDAMB231). MCF7 spheres showed a five-fold increase in CD24high cells compared with cells grown as monolayer (Figure 2c and d; P=0.0268, n=3) but no change in the CD24low population. In MDAMB468, we demonstrated a four-fold reduction of CD24low/CD44high cells in suspension cultures compared with monolayers (Figure 2c and d; P=0.015; n=3) and CD44 was decreased (P=0.0103, n=3) in MDAMB231 cells growing in suspension culture conditions compared with monolayers (Figure 2c and d). There were no statistically significant differences in colony-forming efficiencies of cells sorted according to their CD24/CD44-expression profiles. Cells growing in suspension had reduced colony-forming efficiencies compared with cells maintained as monoloayers (Figure 2e).
Figure 2.
CD24/CD44 expression in monolayer and mammosphere growth conditions. (A) FACS profiles of the indicated cell lines grown as monolayers and stained for CD24 and CD44. (B) Photomicrographs of cells maintained under mammosphere growth conditions (bar=100μm). (C) FACS profiles of cells maintained under mammosphere growth conditions. (D) The percentages of cells with a CD24hi phenotype in MCF7 cells grown as monolayers or mammospheres; of MDAMB468 monolayer or suspension cells with a CD24low/CD44hi phenotype; of the CD44high population in MDAMB231 cells grown as monolayer or spheres. (E) Colony-forming efficiency (CFE) of FACS-sorted MCF7 or MDAMB468 cells on the basis of CD24/CD44 expression, or of monolayers compared with cells growing under mammosphere culture conditions before plating. Data are mean values with 95% confidence intervals; *P<0.05, n=3.
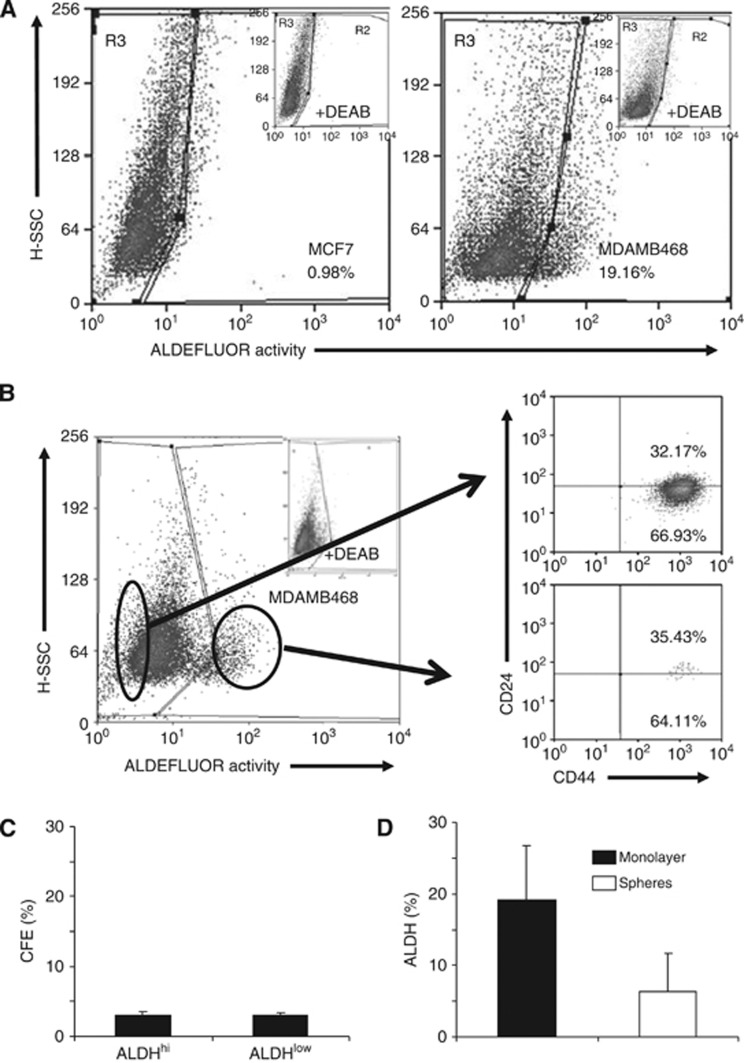
ALDH activity was highly variable in the cells tested, being weaker in MCF7 cells compared with MDAMB468 (Figure 3a), whereas all SKBR3 cells growing as a monolayer showed substantial ALDH activity (data not shown). ALDEFLUOR-positive and negative populations of MDAMB468 cells did not show differential expression of either CD44 or CD24 (Figure 3b) and did not have different cloning efficiencies (Figure 3c). The percentage of cells with high ALDEFLUOR activity was three-fold lower in MDAMB468 cells under suspension culture conditions than in monolayers (Figure 3d), although this was not statistically significant.
Figure 3.
ALDH activity and correlation with CD24/CD44 or colony-forming efficiency. (A) FACS profiles of ALDH activity in MCF7 and MDAMB468 cells grown as monolayers. The insets show fluorescence values in the presence of the inhibitor DEAB, used to set the R3 gate. (B) MDAMB468 cells were FACS-sorted into populations with the highest or lowest ALDH activity and stained for CD24 and CD44. (C) Colony-forming efficiencies of the FACS-sorted cells. (D) The percentages of MDAMB468 cells showing ALDH activity after growth as monolayers or under mammosphere growth conditions.
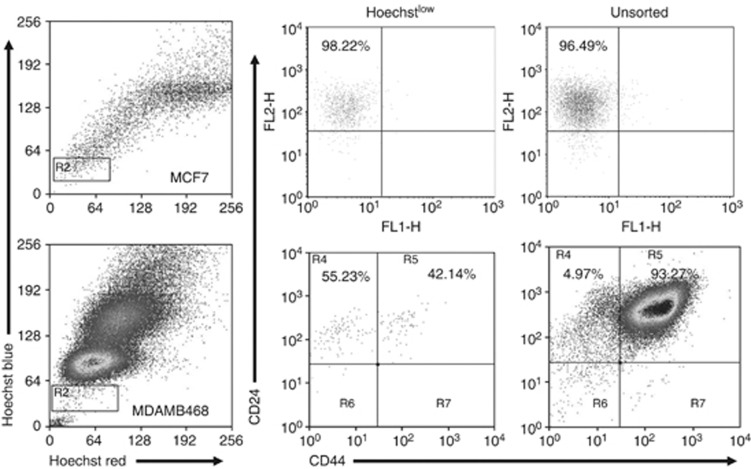
A typical side-population is identified as a tail-shaped tip spreading out sideways from the main population, exhibiting low fluorescent signal but relatively high blue/red signal ratio after staining with Hoechst dye. None of the cell lines exhibited this distinctive pattern (Figure 4), in agreement with published data where a true SP was identified in only four of sixteen tested lines, although a Hoechstlow population could be seen more often (Christgen et al, 2007). In the absence of a true SP, we sorted the Hoechstlow populations as used previously (Engelmann et al, 2008) and measured the expression of CD44 and CD24. In MCF7, there were no significant changes in expression of CD24 or CD44 in cells with the lowest Hoechst fluorescence compared with unsorted cells (Figure 4). In MDAMB468 cells, the Hoechstlow population contained a higher proportion (10-fold increase) of cells with low CD44 levels than the total population (Figure 4).
Figure 4.
Hoechstlow populations do not correlate with CD24/CD44 expression. Top panels represent data from MCF7 cells and the lower panels are from MDAMB468 cells. The left column shows FACS profiles of Hoechst staining. Cells were sorted on the basis of low fluorescence (gate R2) or were not sorted and analysed for CD24 and CD44 expression (middle and right panels).
Expression of markers in xenografted cells
A number of studies have indicated that CSCs, like normal stem cells, occupy a specific niche within the tumour microenvironment, next to the stromal interface. To investigate whether cells expressing putative CSC markers showed such location preference and to investigate co-expression of markers, immunohistochemistry was used to study the expression of CD44, ALDH1 and Sox2 in xenografts. MCF7 xenografts did not express ALDH1, but expressed CD44 in 2.17±1.3% and Sox2 in 3.63±3.4% of cells. Double labelling showed that Sox2 and CD44 were expressed in different cells, rather than the same cell sub-population (Figure 5 top panel). Sox2 was not expressed in MDAMB468 xenografts, CD44 in 95.88+1.4% cells and ALDH in 8.47±7.6% of cells. Because virtually all MDAMB468 tumour cells were CD44+, ALDH1 was expressed in a sub-population of these cells (Figure 5 bottom panel). Cells expressing these markers were randomly distributed in tumour xenografts and were not localized specifically at the tumour/stroma interface (Figure 5).
Figure 5.
Marker expression and co-localization in tumor xenografts. Immunocytochemistry for CD44 and Sox2 in MCF7 xenografts and for CD44 and ALDH1 in MDAMB468 xenografts as indicated. The image at the right-hand side of the two upper panels shows double staining of the two markers with CD44 in black and the other marker in brown. Single-label staining is counterstained with haematoxylin; no counterstain has been applied to double-labelled sections.
Effects of therapy on marker expression
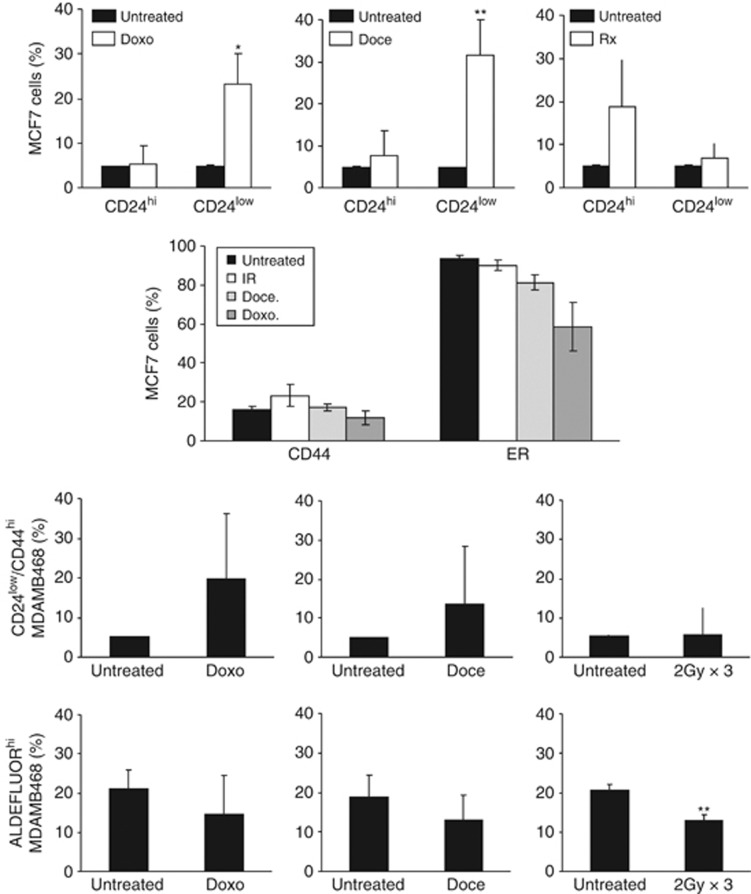
Monolayer cells were exposed to three cycles of radiotherapy or chemotherapy. MCF7 cells that survived treatment with chemotherapy showed an increase in the percentage of cells with low level CD24 expression (P<0.05 for doxorubicin and P<0.01 for docetaxel), whereas there was a non-statistically significant increase in the CD24hi population following radiotherapy. The percentage of cells expressing CD44 was unchanged by any of the three treatments, but both chemotherapeutic agents increased the percentage of ER-negative cells (P<0.001 for each, Figure 6 upper panels). In MDAMB468, there were non-statistically significant increases in the percentages of cells with the CD24low/CD44high phenotype after chemotherapy but not radiotherapy. The percentages of MDAMB468 cells with ALDEFLUOR activity were not increased after repeated chemotherapy exposures, and were decreased following ionizing radiation (P<0.01) (Figure 6 lower panels).
Figure 6.
The effects of repeated therapy on CSC markers. MCF7 or MDAMB468 cells were treated three times with Doxorubicin (Doxo), Docetaxel (Doce) or with ionizing radiation (Rx). In the top panel; the percentages of MCF7 cells showing a CD24hi or CD24low phenotype are shown with or without treatment (the expression levels are defined by gating on the 5% of untreated cells with the highest or lowest expression). In addition, the percentages of cells stained with CD44 or ER are shown underneath. For MDAMB468 cells, data are shown for the percentages of cells with a CD24low/CD44hi phenotype (third panel) or that have ALDH activity (bottom panel). Data are mean values with 95% confidence intervals, n=3 for each. *P<0.05; **P<0.01.
Discussion
This examination of putative CSC markers in breast cancer demonstrated that individual markers are not always expressed in an individual cancer, nor co-expressed in the same cells. We were also unable to identify specific sub-populations in many cell lines and found that relative expression levels of different markers correlate neither with each other, nor with therapy resistance. These data indicate that commonly used markers of the CSC sub-population of breast cancer are not expressed in all tumours. Although many studies have reported expression of individual markers or marker combinations in association with various clinical and biological characteristics, our data are supported by previous observations that not all tumours or cell lines contain a CD24−/low/CD44+ population (Sheridan et al, 2006; Grimshaw et al, 2008; Wright et al, 2008; Hwang-Verslues et al, 2009; Park et al, 2010) or contain cells that can form mammospheres (Farnie et al, 2007; Grimshaw et al, 2008). Similarly, only a minority of primary tumours and two-thirds of breast cancer cell lines have any ALDH1+ cells, and a distinct SP is seen in only 25% of breast cancer cell lines (Christgen et al, 2007; Tan et al, 2013). Further evidence for divergent marker expression has come from experimental studies, where individual murine breast cancers that arise in a single model are marked by either CD133 expression or the CD24low/CD44+ phenotype, but not both (Wright et al, 2008), whereas CSCs in different models either express ITGB3 or do not express this specific marker (Vaillant et al, 2008). However, because these data are derived from either single markers or single-model systems, the relationships between marker expression in each situation has remained unclear. In the present study, we comprehensively examined a number of different markers in a range of primary material and cell lines and the data indicate that none of the markers employed can be considered as a universal marker applicable to the identification of a CSC population in breast cancer cell lines or metastatic breast cancer.
Secondly, by employing multiple methods in vitro, in vivo and in human samples, we have shown that an individual cancer commonly contains distinct cell populations expressing different CSC markers. These data indicate that each marker identifies a different cell sub-population, making the precise biology of each population uncertain. Similar observations have been made in more limited studies comparing expression of markers in specific circumstances, such as a lack of correlation between CD24/CD44 populations and mammosphere forming ability (Grimshaw et al, 2008), the dye-excluding population and expression of either CD24 or CD44 (Zhou et al, 2007), and between CD44/CD24 and ALDH1 (Charafe-Jauffret et al, 2009; Stingl, 2009). Consideration of these findings makes it unclear which of these populations, if any, are authentic CSCs. In this regard, we were also able to investigate the position of putative CSCs in vivo, on the basis that, similar to normal stem cells, CSCs localize to the tumour/stroma interface that forms the stem cell niche (Calabrese et al, 2007; Prince and Ailles, 2008; Korkaya et al, 2011; Liu et al, 2011). However, we found that CD44, Sox2 or ALDH1 cells are not localized specifically to the stromal interface in either breast cancer xenografts or human breast cancers.
Finally, a variable effect of therapy was demonstrated on putative CSC populations in vitro. Although many studies have indicated that CSCs are therapy-resistant, it has also been shown that ER+ tumours with mammosphere gene expression profiles have a better prognosis (Kok et al, 2009), whereas CD24 expression is a marker of poor prognosis (Kristiansen et al, 2003; Ahmed et al, 2012). In different studies, expression of ALDH1 is not a predictor of outcome (Tan et al, 2013), is not increased following treatment (Resetkova et al, 2010), or ALDH1+ cells are enriched following treatment but CD24/CD44 populations are not altered (Tanei et al, 2009). Similarly, although the CD24/CD49f population of murine breast cancer has CSC properties, these cells are not therapy-resistant (Pajic et al, 2010) and isolated CSCs from cell lines commonly do not show enhanced radioresistance (Al-Assar et al, 2009; Kim et al, 2012). It is likely that at least some of these discrepancies relate to the use of individual therapeutic regimes associated with a single marker in different studies. In our work, we applied repeated treatment to selectively remove the bulk of tumour cells that are relatively therapy-sensitive and thereby enrich for a therapy-resistant sub-population (Phillips et al, 2006; Kabos et al, 2011), using three different treatments on two different cell lines with different marker expression. We found that the two cell lines tested show distinctive changes in CSC marker expression according to the precise therapy employed. The only consistent finding across therapeutic agents was an expansion of ER-negative cells after exposure of the luminal cancer cell line, MCF7, to all three therapies. These data support the observations that lack of ER in individual cells in ER+ breast cancer is a marker of a therapy-resistant population of CSCs (Kabos et al, 2011) and selection of cells that will be unresponsive to endocrine therapy could account for the delayed recurrence of initially ER+ breast cancer as ER− disease observed in clinical practice (Thompson et al, 2010; Moussa et al, 2012).
In summary, these findings indicate that there are no universal markers that identify a common breast CSC population or a therapy-resistant population in all breast cancers, or even in a given subtype of breast cancer. Taken together with the observations that CSC populations are highly dynamic and influenced by their surrounding microenvironment (He et al, 2011; Korkaya et al, 2011; Scheel et al, 2011; Chaffer et al, 2013) the heterogeneity of breast CSCs likely reflects the diversity of onocogenic events, cell of origin and tumour microenvironmental factors operating in an individual cancer. However, such diversity and plasticity of marker expression makes it unlikely that there will be a simple relationship between the identified CSC population and the clinical course of breast cancer, accounting for the discrepant results often obtained using marker approaches. It may therefore be more relevant to focus on fundamental stem cell properties such as asymmetric division or tumour initiation rather than expression of individual markers as assessment methodology. In addition, the evidence for plasticity of stem cell populations indicates that the measurement of any CSC population, however defined, at a single point in time is unlikely to have predictive therapeutic value.
Acknowledgments
We are grateful to R Clarke, Manchester, for advice on mammosphere culture conditions and to the Tayside Tissue Bank for processing samples. This work was supported by a University of Dundee PhD studentship (YL, AMT and PJC). We also acknowledge the financial support of Breast Cancer Research Scotland (AMT, MVA and KM). RN is supported by European Regional Development Fund and the State Budget of the Czech Republic (RECAMO, CZ.1.05/2.1.00/03.0101).
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on British Journal of Cancer website (http://www.nature.com/bjc)
This work is published under the standard license to publish agreement. After 12 months the work will become freely available and the license terms will switch to a Creative Commons Attribution-NonCommercial-Share Alike 3.0 Unported License.
Supplementary Material
References
- Ahmed MA, Aleskandarany MA, Rakha EA, Moustafa RZ, Benhasouna A, Nolan C, Green AR, Ilyas M, Ellis IO. A CD44(−)/CD24(+) phenotype is a poor prognostic marker in early invasive breast cancer. Breast Cancer Res Treat. 2012;133:979–995. doi: 10.1007/s10549-011-1865-8. [DOI] [PubMed] [Google Scholar]
- Al-Assar O, Muschel RJ, Mantoni TS, McKenna WG, Brunner TB. Radiation response of cancer stem-like cells from established human cell lines after sorting for surface markers. Int J Radiat Oncol Biol Phys. 2009;75:1216–1225. doi: 10.1016/j.ijrobp.2009.07.001. [DOI] [PubMed] [Google Scholar]
- Al-Hajj M, Wicha MS, Benito-Hernandez A, Morrison SJ, Clarke MF. Prospective identification of tumorigenic breast cancer cells. Proc Natl Acad Sci USA. 2003;100:3983–3988. doi: 10.1073/pnas.0530291100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Appleyard MV, Murray KE, Coates PJ, Wullschleger S, Bray SE, Kernohan NM, Fleming S, Alessi DR, Thompson AM. Phenformin as prophylaxis and therapy in breast cancer xenografts. Br J Cancer. 2012;106:1117–1122. doi: 10.1038/bjc.2012.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao S, Wu Q, McLendon RE, Hao Y, Shi Q, Hjelmeland AB, Dewhirst MW, Bigner DD, Rich JN. Glioma stem cells promote radioresistance by preferential activation of the DNA damage response. Nature. 2006;444:756–760. doi: 10.1038/nature05236. [DOI] [PubMed] [Google Scholar]
- Calabrese C, Poppleton H, Kocak M, Hogg TL, Fuller C, Hamner B, Oh EY, Gaber MW, Finklestein D, Allen M, Frank A, Bayazitov IT, Zakharenko SS, Gajjar A, Davidoff A, Gilbertson RJ. A perivascular niche for brain tumour stem cells. Cancer Cell. 2007;11:69–82. doi: 10.1016/j.ccr.2006.11.020. [DOI] [PubMed] [Google Scholar]
- Chaffer CL, Marjanovic ND, Lee T, Bell G, Kleer CG, Reinhardt F, D'Alessio AC, Young RA, Weinberg RA. Poised chromatin at the ZEB1 promoter enables breast cancer cell plasticity and enhances tumorigenicity. Cell. 2013;154:61–74. doi: 10.1016/j.cell.2013.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charafe-Jauffret E, Ginestier C, Iovino F, Wicinski J, Cervera N, Finetti P, Hur MH, Diebel ME, Monville F, Dutcher J, Brown M, Viens P, Xerri L, Bertucci F, Stassi G, Dontu G, Birnbaum D, Wicha MS. Breast cancer cell lines contain functional cancer stem cells with metastatic capacity and a distinct molecular signature. Cancer Res. 2009;69:1302–1313. doi: 10.1158/0008-5472.CAN-08-2741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christgen M, Ballmaier M, Bruchhardt H, von Wasielewski R, Kreipe H, Lehmann U. Identification of a distinct side population of cancer cells in the Cal-51 human breast carcinoma cell line. Mol Cell Biochem. 2007;306:201–212. doi: 10.1007/s11010-007-9570-y. [DOI] [PubMed] [Google Scholar]
- Engelmann K, Shen H, Finn OJ. MCF7 side population cells with characteristics of cancer stem/progenitor cells express the tumor antigen MUC1. Cancer Res. 2008;68:2419–2426. doi: 10.1158/0008-5472.CAN-07-2249. [DOI] [PubMed] [Google Scholar]
- Eyler CE, Rich JN. Survival of the fittest: cancer stem cells in therapeutic resistance and angiogenesis. J Clin Oncol. 2008;26:2839–2845. doi: 10.1200/JCO.2007.15.1829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farnie G, Clarke RB, Spence K, Pinnock N, Brennan K, Anderson NG, Bundred NJ. Novel cell culture technique for primary ductal carcinoma in situ: role of Notch and epidermal growth factor receptor signaling pathways. J Natl Cancer Inst. 2007;99:616–627. doi: 10.1093/jnci/djk133. [DOI] [PubMed] [Google Scholar]
- Fillmore CM, Kuperwasser C. Human breast cancer cell lines contain stem-like cells that self-renew, give rise to phenotypically diverse progeny and survive chemotherapy. Breast Cancer Res. 2008;10:R25. doi: 10.1186/bcr1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ginestier C, Hur MH, Charafe-Jauffret E, Monville F, Dutcher J, Brown M, Jacquemier J, Viens P, Kleer CG, Liu S, Schott A, Hayes D, Birnbaum D, Wicha MS, Dontu G. ALDH1 is a marker of normal and malignant human mammary stem cells and a predictor of poor clinical outcome. Cell Stem Cell. 2007;1:555–567. doi: 10.1016/j.stem.2007.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimshaw MJ, Cooper L, Papazisis K, Coleman JA, Bohnenkamp HR, Chiapero-Stanke L, Taylor-Papadimitriou J, Burchell JM. Mammosphere culture of metastatic breast cancer cells enriches for tumorigenic breast cancer cells. Breast Cancer Res. 2008;10:R52. doi: 10.1186/bcr2106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison H, Farnie G, Howell SJ, Rock RE, Stylianou S, Brennan KR, Bundred NJ, Clarke RB. Regulation of breast cancer stem cell activity by signaling through the Notch4 receptor. Cancer Res. 2010;70:709–718. doi: 10.1158/0008-5472.CAN-09-1681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He K, Xu T, Goldkorn A. Cancer cells cyclically lose and regain drug-resistant highly tumorigenic features characteristic of a cancer stem-like phenotype. Mol Cancer Ther. 2011;10:938–948. doi: 10.1158/1535-7163.MCT-10-1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang-Verslues WW, Kuo WH, Chang PH, Pan CC, Wang HH, Tsai ST, Jeng YM, Shew JY, Kung JT, Chen CH, Lee EY, Chang KJ, Lee WH. Multiple lineages of human breast cancer stem/progenitor cells identified by profiling with stem cell markers. PLoS One. 2009;4:e8377. doi: 10.1371/journal.pone.0008377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabos P, Haughian JM, Wang X, Dye WW, Finlayson C, Elias A, Horwitz KB, Sartorius CA. Cytokeratin 5 positive cells represent a steroid receptor negative and therapy resistant subpopulation in luminal breast cancers. Breast Cancer Res Treat. 2011;128:45–55. doi: 10.1007/s10549-010-1078-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SY, Rhee JG, Song X, Prochownik EV, Spitz DR, Lee YJ. Breast cancer stem cell-like cells are more sensitive to ionizing radiation than non-stem cells: role of ATM. PLoS One. 2012;7:e50423. doi: 10.1371/journal.pone.0050423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kok M, Koornstra RH, Margarido TC, Fles R, Armstrong NJ, Linn SC, Van't Veer LJ, Weigelt B. Mammosphere-derived gene set predicts outcome in patients with ER-positive breast cancer. J Pathol. 2009;218:316–326. doi: 10.1002/path.2544. [DOI] [PubMed] [Google Scholar]
- Korkaya H, Liu S, Wicha MS. Breast cancer stem cells, cytokine networks, and the tumor microenvironment. J Clin Invest. 2011;121:3804–3809. doi: 10.1172/JCI57099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kristiansen G, Winzer KJ, Mayordomo E, Bellach J, Schluns K, Denkert C, Dahl E, Pilarsky C, Altevogt P, Guski H, Dietel M. CD24 expression is a new prognostic marker in breast cancer. Clin Cancer Res. 2003;9:4906–4913. [PubMed] [Google Scholar]
- Lawson JC, Blatch GL, Edkins AL. Cancer stem cells in breast cancer and metastasis. Breast Cancer Res Treat. 2009;118:241–254. doi: 10.1007/s10549-009-0524-9. [DOI] [PubMed] [Google Scholar]
- Leis O, Eguiara A, Lopez-Arribillaga E, Alberdi MJ, Hernandez-Garcia S, Elorriaga K, Pandiella A, Rezola R, Martin AG. Sox2 expression in breast tumours and activation in breast cancer stem cells. Oncogene. 2012;31:1354–1365. doi: 10.1038/onc.2011.338. [DOI] [PubMed] [Google Scholar]
- Liu S, Ginestier C, Ou SJ, Clouthier SG, Patel SH, Monville F, Korkaya H, Heath A, Dutcher J, Kleer CG, Jung Y, Dontu G, Taichman R, Wicha MS. Breast cancer stem cells are regulated by mesenchymal stem cells through cytokine networks. Cancer Res. 2011;71:614–624. doi: 10.1158/0008-5472.CAN-10-0538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moussa O, Purdie C, Vinnicombe S, Thompson AM. Biomarker discordance: prospective and retrospective evidence that biopsy of recurrent disease is of clinical utility. Cancer Biomark. 2012;12:231–239. doi: 10.3233/CBM-130314. [DOI] [PubMed] [Google Scholar]
- Pajic M, Kersbergen A, van Diepen F, Pfauth A, Jonkers J, Borst P, Rottenberg S. Tumor-initiating cells are not enriched in cisplatin-surviving BRCA1;p53-deficient mammary tumor cells in vivo. Cell Cycle. 2010;9:3780–3791. doi: 10.4161/cc.9.18.13002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park SY, Lee HE, Li H, Shipitsin M, Gelman R, Polyak K. Heterogeneity for stem cell-related markers according to tumor subtype and histologic stage in breast cancer. Clin Cancer Res. 2010;16:876–887. doi: 10.1158/1078-0432.CCR-09-1532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips TM, McBride WH, Pajonk F. The response of CD24(-/low)/CD44+ breast cancer-initiating cells to radiation. J Natl Cancer Inst. 2006;98:1777–1785. doi: 10.1093/jnci/djj495. [DOI] [PubMed] [Google Scholar]
- Prince ME, Ailles LE. Cancer stem cells in head and neck squamous cell cancer. J Clin Oncol. 2008;26:2871–2875. doi: 10.1200/JCO.2007.15.1613. [DOI] [PubMed] [Google Scholar]
- Rappa G, Mercapide J, Anzanello F, Prasmickaite L, Xi Y, Ju J, Fodstad O, Lorico A. Growth of cancer cell lines under stem cell-like conditions has the potential to unveil therapeutic targets. Exp Cell Res. 2008;314:2110–2122. doi: 10.1016/j.yexcr.2008.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Resetkova E, Reis-Filho JS, Jain RK, Mehta R, Thorat MA, Nakshatri H, Badve S. Prognostic impact of ALDH1 in breast cancer: a story of stem cells and tumor microenvironment. Breast Cancer Res Treat. 2010;123:97–108. doi: 10.1007/s10549-009-0619-3. [DOI] [PubMed] [Google Scholar]
- Rosen JM, Jordan CT. The increasing complexity of the cancer stem cell paradigm. Science. 2009;324:1670–1673. doi: 10.1126/science.1171837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheel C, Eaton EN, Li SH, Chaffer CL, Reinhardt F, Kah KJ, Bell G, Guo W, Rubin J, Richardson AL, Weinberg RA. Paracrine and autocrine signals induce and maintain mesenchymal and stem cell states in the breast. Cell. 2011;145:926–940. doi: 10.1016/j.cell.2011.04.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheridan C, Kishimoto H, Fuchs RK, Mehrotra S, Bhat-Nakshatri P, Turner CH, Goulet R, Jr, Badve S, Nakshatri H. CD44+/CD24- breast cancer cells exhibit enhanced invasive properties: an early step necessary for metastasis. Breast Cancer Res. 2006;8:R59. doi: 10.1186/bcr1610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stingl J. Detection and analysis of mammary gland stem cells. J Pathol. 2009;217:229–241. doi: 10.1002/path.2457. [DOI] [PubMed] [Google Scholar]
- Tan EY, Thike AA, Tan PH. ALDH1 expression is enriched in breast cancers arising in young women but does not predict outcome. Br J Cancer. 2013;109:109–113. doi: 10.1038/bjc.2013.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanei T, Morimoto K, Shimazu K, Kim SJ, Tanji Y, Taguchi T, Tamaki Y, Noguchi S. Association of breast cancer stem cells identified by aldehyde dehydrogenase 1 expression with resistance to sequential Paclitaxel and epirubicin-based chemotherapy for breast cancers. Clin Cancer Res. 2009;15:4234–4241. doi: 10.1158/1078-0432.CCR-08-1479. [DOI] [PubMed] [Google Scholar]
- Thompson AM, Jordan LB, Quinlan P, Anderson E, Skene A, Dewar JA, Purdie CA. Prospective comparison of switches in biomarker status between primary and recurrent breast cancer: the Breast Recurrence In Tissues Study (BRITS) Breast Cancer Res. 2010;12:R92. doi: 10.1186/bcr2771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaillant F, Asselin-Labat ML, Shackleton M, Forrest NC, Lindeman GJ, Visvader JE. The mammary progenitor marker CD61/beta3 integrin identifies cancer stem cells in mouse models of mammary tumorigenesis. Cancer Res. 2008;68:7711–7717. doi: 10.1158/0008-5472.CAN-08-1949. [DOI] [PubMed] [Google Scholar]
- Wright MH, Calcagno AM, Salcido CD, Carlson MD, Ambudkar SV, Varticovski L. Brca1 breast tumors contain distinct CD44+/CD24- and CD133+ cells with cancer stem cell characteristics. Breast Cancer Res. 2008;10:R10. doi: 10.1186/bcr1855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J, Wulfkuhle J, Zhang H, Gu P, Yang Y, Deng J, Margolick JB, Liotta LA, Petricoin E, 3rd, Zhang Y. Activation of the PTEN/mTOR/STAT3 pathway in breast cancer stem-like cells is required for viability and maintenance. Proc Natl Acad Sci USA. 2007;104:16158–16163. doi: 10.1073/pnas.0702596104. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.