Abstract
Background:
Aberrant global DNA methylation is shown to increase cancer risk. LINE-1 has been proven a measure of global DNA methylation. The objectives of this study were to assess the association between LINE-1 methylation level and bladder cancer risk and to evaluate effect modification by environmental and genetic factors.
Methods:
Bisulphite-treated leukocyte DNA from 952 cases and 892 hospital controls was used to measure LINE-1 methylation level at four CpG sites by pyrosequencing. Logistic regression model was fitted to estimate odds ratios (ORs) and 95% confidence intervals (95% CIs). Interactions between LINE-1 methylation levels and environmental and genetic factors were assessed.
Results:
The risk of bladder cancer followed a nonlinear association with LINE-1 methylation. Compared with subjects in the middle tertile, the adjusted OR for subjects in the lower and the higher tertiles were 1.26 (95% CI 0.99–1.60, P=0.06) and 1.33 (95% CI 1.05–1.69, P=0.02), respectively. This association significantly increased among individuals homozygous for the major allele of five single-nucleotide polymorphisms located in the phosphatidylethanolamine N-methyltransferase gene (corrected P-interaction<0.05).
Conclusions:
The findings from this large-scale study suggest that both low and high levels of global DNA methylation are associated with the risk of bladder cancer.
Keywords: bladder cancer, LINE-1 repetitive sequences, DNA methylation, one-carbon metabolism, epigenetic–gene interaction
Bladder cancer is one of the most common cancers in the developed countries mainly affecting men (Parkin, 2008). In Spain, the incidence rate of bladder cancer has been increasing in the past three decades and the ratio of men to women developing the disease is 7 : 1 (Izarzugaza et al, 2010). Recurrence is a major problem in the management of bladder cancer necessitating frequent follow-ups, thus making it one of the most expensive cancer types to treat (Lotan et al, 2008). Established risk factors for bladder cancer include tobacco smoking, occupational exposures to aromatic amines, arsenic in drinking water (Murta-Nascimento et al, 2007; Volanis et al, 2010) and genetic variants of NAT2 and GSTM1; the latter two have been shown to significantly modify the risk conferred by tobacco smoking (Garcia-Closas et al, 2005; Rothman et al, 2010). Recently, additional common genetic variants on CBX6-APOBEC3A, CCNE1, MYC, PSCA, SLC14A1, TERT-CLPTM1L, TMEM129-TACC-FGFR3, TP63 and UGT1A cluster have been associated with risk of bladder cancer (Rothman et al, 2010; Garcia-Closas et al, 2011).
Genome-wide altered epigenetic states are common findings in several malignancies, including bladder tumours. These epigenetic states include global DNA hypomethylation and gene promoter methylation, post-translational histone modifications and deregulation of microRNAs (Esteller, 2008; Taby and Issa, 2010). Global DNA hypomethylation in cancer is mostly found in repetitive sequences of the genome such as long interspersed nuclear element 1 (LINE-1) and Alu elements (Esteller, 2008). It has been suggested that methylation represses the transcription of these repetitive regions to maintain genomic stability and prevent mutations, deletions and insertions (Esteller, 2008). In bladder cancer, genomic instability has been observed at different stages of the disease (Florl and Schulz, 2008). There is evidence that LINE-1 hypomethylation could also induce the expression of the MET oncogene in bladder cancer and normal tissues (Wolff et al, 2010). Global DNA methylation measured in peripheral blood cells has been used to investigate its relationship with cancer risk, although results have been inconclusive (Woo and Kim, 2012). Studies investigating the role of genomic DNA methylation (measured by direct quantification of 5-methylcytosine as well as at CpG sites in LINE-1 sequences) and bladder cancer have inconclusively shown an inverse association between genomic DNA methylation and risk of bladder cancer (Moore et al, 2008; Wilhelm et al, 2010; Cash et al, 2012). Whether environmental and genetic factors modify the association between global DNA methylation and bladder cancer risk is not known, although it has been suggested that smoking, B vitamins, arsenic and genes involved in the one-carbon metabolism pathway may have a modifier role (Moore et al, 2007; Volanis et al, 2010). Hence, understanding the role of global DNA methylation in bladder cancer development and its interaction with other known and potential risk factors may provide further insight on the aetiology and susceptibility of this tumour.
The objectives of this study were to assess the risk of bladder cancer associated with LINE-1 methylation levels, as a measure of global DNA methylation in peripheral blood, as well as to explore whether personal, lifestyle, environmental and genetic factors modify this association.
Materials and methods
Study population
The Spanish Bladder Cancer/EPICURO Study is a multi-center, hospital-based case–control study conducted in five regions of Spain (Barcelona, Valles, Elche, Tenerife and Asturias) between 1998 and 2001. The design of the study has been previously described elsewhere in detail (Garcia-Closas et al, 2005; Samanic et al, 2006). Briefly, cases and controls were recruited to the study from 18 hospitals. Cases were newly diagnosed subjects with histologically confirmed urothelial carcinoma of the bladder. Controls were patients admitted to the same hospital as cases for reasons not related to exposures of interest and individually matched for age (±5 years), sex and geographic region to cases. From the initial study population of 1219 cases and 1271 controls, 1107 cases and 1032 controls provided blood sample (before treatment was instituted). Of these, 995 cases and 925 controls, all Caucasians and without missing demographic data, had enough granulocyte DNA for sodium bisulphite treatment and subsequent measurement of DNA methylation. Because in 43 cases and 33 controls quantification of DNA methylation failed, the final study population for the current analysis was 952 cases and 892 controls aged from 20 to 81 years. Written informed consent was obtained from each study subject before interview, and the study was approved by the US National Cancer Institute and local institutional review boards.
Data on demographic and personal characteristics, smoking and other known and potential risk factors for bladder cancer were collected by means of a computer-assisted personal interview. Food frequency questionnaires were used to collect data on fruit, vegetable, B vitamins and protein and alcohol intake over the 5 years before interview (Garcia-Closas et al, 2007b).
Analysis of LINE-1 methylation
Bisulphite modification of genomic DNA extracted from granulocytes using standard techniques was performed using EZ-96 DNA METHYLATION-GOLD KIT (Zymo Research, Irvin, CA, USA) following the recommendations of the manufacturer. PCR amplification of bisulphite-modified DNA was carried out using a set of forward and reverse primers reported previously (Estecio et al, 2007). To quantify the methylation level of each of the four CpG sites immediately after the sequencing primer, sequencing of the PCR product was performed by pyrosequencing using PyroMark Q24 System (QIAGEN, Valencia, CA, USA) as recommended by the manufacturer. Methylation level at each CpG site was extracted using PyroMark Application Software version 2.0.6 (QIAGEN), and the value was expressed as the percentage of methylated cytosines over the sum of methylated and unmethylated cytosines. The average methylation level of the four LINE-1 CpG sites was used in the analysis as a marker of the global DNA methylation level. As a quality control measure, 307 randomly selected samples from cases and controls were run in duplicate and the within-sample coefficient of variation was 4.53%. The mean methylation level of the duplicates was used in the analysis.
Genotyping
Genotyping was carried out using DNA isolated form granulocytes as described previously (Garcia-Closas et al, 2005). All genotype assays used in this analysis were done at the Core Genotyping Facility of the Division of Cancer Epidemiology and Genetics, National Cancer Institute, Bethesda, MD, USA. Methods of the SNP genotyping process were described previously (Garcia-Closas et al, 2005, 2007a; Rothman et al, 2010). SNP genotyping was done using Illumina Infinium Human1M-Duo (480 SNPs), Illumina GoldenGate (36 SNPs) (Illumina, San Diego, CA, USA), and TaqMan (8 SNPs) (Applied Biosystems, Foster City, CA, USA) assays. All genotypes included in the study were in Hardy–Weinberg equilibrium in the control population (P>0.05). A total of 524 SNPs and copy number variation in GSTM1 and GSTT1 were assessed in the present study. Of these SNPs, 515 SNPs were located in 24 candidate genes involved in one-carbon metabolism pathway, including DNA methylation and arsenic metabolism (Supplementary Table S1). These pathways are known to influence DNA methylation by altering the cellular concentration of the methyl group donor S-adenosylmethionine (SAM) (Ulrey et al, 2005; Moore et al, 2007; Lee et al, 2009). Additionally, eight SNPs identified in a recent multi-stage genome-wide association study of bladder cancer and a SNP rs1495741 tagging NAT2 acetylator phenotype were also included in the analysis (Rothman et al, 2010). SNPs and variants were investigated for their modifier effect on the association between LINE-1 methylation and bladder cancer risk.
Quantification of trace elements
Methods of quantification of toenail trace element level included in the current study have been previously described (Amaral et al, 2011). Briefly, after cleaning and washing the toenails to remove external contaminants, trace elements were quantified at the Trace Element Analysis Core (Dartmouth College, NH, USA), using inductively coupled plasma-mass spectrometry (Hopkins et al, 2004). Toenails were digested with Optima nitric acid (Fisher Scientific, St Louis, MO, USA), at 105 °C, then with hydrogen peroxide and further heating the dilution with deionised water. All sample preparation steps were recorded gravimetrically. For quality control, each batch of analyses accommodated six standard reference material samples with known trace element content (GBW 07601, powdered human hair) and six analytic blanks, together with the study samples. Overall, concentrations (μg g−1) of 12 trace elements (aluminum, arsenic, cadmium, chromium, copper, iron, lead, manganese, nickel, selenium, vanadium and zinc) were determined.
Statistical analysis
Chi-squared test was used to evaluate the association between case–control status and categorical variables. Normality of LINE-1 methylation distribution was assessed graphically by plotting histograms (Supplementary Figure S1) and formally tested by using Shapiro–Wilk test for normality. The non-parametric Mann–Whitney U-test was used to compare medians of continuous variables in different strata.
To estimate the risk associated with global DNA methylation and examine nonlinear association, LINE-1 methylation level was modeled using restricted cubic splines in a logistic regression model adjusting for age (continuous), sex, region and smoking status (never, occasional, former and current smoker). Cubic spline models are polynomial functions that provide examination of a nonlinear association between a dependent and an explanatory variable. In the restricted cubic spline regression model, methylation level of 59% was set as a reference, and four knots (boundaries) at the 5, 35, 65 and 95 percentiles of LINE-1 methylation level were used to restrict the function to be linear beyond the boundaries at extreme values of LINE-1 methylation (Royston and Sauerbrei, 2007). Upon the visualisation of a nonlinear ‘U-shape' risk pattern from the cubic spline regression (Supplementary Figure S2), LINE-1 methylation levels were categorised into three categories by using tertile cut points (56.68% and 58.65%) based on its distribution among the control population. For subsequent analyses, this variable was used by assigning the middle tertile as the referent category. Unadjusted and adjusted logistic regression models were fitted to estimate odds ratios (ORs) and the corresponding 95% confidence intervals (95% CIs). In addition to LINE-1 methylation, variables included in the basic multivariable adjusted model were age, sex, region and smoking status. Association between LINE-1 methylation, in tertiles, at each of the four LINE-1 CpG sites and risk of bladder cancer was evaluated. Also, and in order to mimic the distribution of LINE-1 methylation levels in tertiles using a continuous variable, the association between the absolute deviation (continuous) from the median LINE-1 methylation level was modeled by logistic regression unadjusted and adjusted for age, sex, region and smoking status. Moreover, the association between LINE-1 methylation and bladder cancer subphenotypes (low-, high-grade non-muscle-invasive bladder cancer (NMIBC) and muscle-invasive bladder cancer (MIBC)) was modeled using multinomial logistic regression. Likelihood ratio test for heterogeneity was used to test whether the ORs for low-, high-grade NMIBC and MIBC are different from each other.
Interaction between LINE-1 methylation and individual covariates (age (<60, 60–69, and⩾70 years), sex, smoking status (never and ever smoker), dietary factors, trace elements (continuous) and SNPs (four modes of inheritance)) was tested by including a multiplicative interaction term into the logistic regression model. For the interaction between LINE-1 methylation and smoking, additional analyses included adjustment among regular smokers for each of the following, as a continuous variable: pack-years, duration of cigarette smoking, and number of cigarettes smoked per day. Significance of the interaction term was tested by comparing the models with and without the interaction term using the likelihood ratio test. To estimate a corrected multiple testing interaction P for each SNP, a permutation test done by randomly sampling 10 000 times was applied by assigning case/control status in a proportion similar to the current study. All statistical tests were two-sided, and a P<0.05 was considered significant. Statistical analyses were carried out by using STATA/SE, version 10.1 (StataCorp, College Station, TX, USA).
Results
The characteristics of cases and controls are shown in Table 1. Subjects were mostly men and smokers. A large proportion of cases (78%) were diagnosed with a NMIBC. The distribution of global DNA methylation was bimodal. The median (interquartile range) LINE-1 methylation levels among cases and controls were 57.60% (4.06%) and 57.44% (3.36%), respectively (P=0.3). The minimum and maximum values of LINE-1 methylation were 28.81% and 85.44% among cases and 37.91% and 85.68% among controls, respectively.
Table 1. Distribution of characteristics of cases and controls with LINE-1 methylation data in the SBC/EPICURO study.
| Cases, n (%) (n=952) | Controls, n (%) (n=892) | P-value | |
|---|---|---|---|
| Age (years), median (IQR) |
68 (13) |
66 (13) |
<0.001a |
|
Sex | |||
| Men | 826 (86.8) | 792 (88.8) | 0.2b |
| Women |
126 (13.2) |
100 (11.2) |
|
|
Region | |||
| Barcelona | 162 (17.0) | 168 (18.8) | 0.7b |
| Valles | 148 (15.5) | 135 (15.1) | |
| Elche | 72 (7.6) | 73 (8.2) | |
| Tenerife | 173 (18.2) | 145 (16.3) | |
| Asturias |
397 (41.7) |
371 (41.6) |
|
|
Smoking status | |||
| Never smoker | 137 (14.5) | 255 (28.7) | <0.001b |
| Occasional smoker | 34 (3.6) | 66 (7.4) | |
| Former smoker | 376 (39.7) | 329 (37.0) | |
| Current smoker | 399 (42.2) | 239 (26.9) | |
| Missing |
6 |
3 |
|
|
Subphenotypes | |||
|
Non-muscle invasive | |||
| Low-risk non-muscle invasive | 523 (57.3) | – | |
| High-risk non-muscle invasive | 185 (20.3) | – | |
| Muscle invasive | 204 (22.4) | – | |
| Missing |
40 |
– |
|
|
Tumour stage | |||
| Ta | 590 (64.7) | – | |
| Tis | 6 (0.7) | – | |
| T1 | 112 (12.3) | – | |
| T2 | 111 (12.2) | – | |
| T3 | 47 (5.1) | – | |
| T4 | 46 (5.0) | – | |
| Missing |
40 |
– |
|
|
Grade | |||
| PUNLMP | 42 (4.6) | – | |
| G1 | 257 (28.2) | – | |
| G2 | 267 (29.3) | – | |
| G3 | 346 (37.9) | – | |
| Missing | 40 | – | |
Abbreviations: IQR=interquartile range; PUNLMP=papillary urothelial neoplam of low malignant potential; Tis=flat carcinoma in situ (CIS).
Mann–Whitney U-test's P-value.
χ2-test's P-value.
Compared with subjects in the middle tertile of LINE-1 methylation, the adjusted ORs were 1.26 (95% CI 0.99–1.60, P=0.06) and 1.33 (95% CI 1.05–1.69, P=0.02) for the lowest and highest tertiles, respectively. These ORs were very similar to the crude ones (Table 2). The associations between methylation level at each of the four CpG positions of LINE-1 and risk of bladder cancer were similar to the results using the average of LINE-1 methylation level (Supplementary Table S2). The association for the absolute deviation from the median LINE-1 methylation level, modeled as a continuous variable and adjusted for age, sex, region and smoking status, was similarly significant (adjusted OR=1.03, 95% CI 1.01–1.05, P=0.008). When further analysis was carried out separately for low-, high-grade NMIBC and MIBC, ORs for LINE-1 methylation levels were similar to the overall OR, and no heterogeneity was found between the three bladder cancer subphenotypes (P>0.05) (Supplementary Table S3).
Table 2. Association between LINE-1 methylation and bladder cancer risk in the SBC/EPICURO study.
| LINE-1 methylation (tertiles) | Cases, n | Controls, n | Crude OR | 95% CI | P-value |
|---|---|---|---|---|---|
| T1 (<56.68%) |
337 |
298 |
1.29 |
(1.02–1.62) |
0.03 |
| T2 (56.68–<58.65%) |
261 |
297 |
1.00 |
Reference |
|
| T3 (⩾58.65%) |
354 |
297 |
1.36 |
(1.08–1.70) |
0.008 |
|
LINE-1 methylation (tertiles) |
Cases,
n |
Controls,
n |
ORa |
95% CI |
P-value |
| T1 (<56.68%) |
335 |
296 |
1.26 |
(0.99–1.60) |
0.06 |
| T2 (56.68–<58.65%) |
260 |
297 |
1.00 |
Reference |
|
| T3 (⩾58.65%) | 351 | 296 | 1.33 | (1.05–1.69) | 0.02 |
Abbreviations: CI=confidence interval; OR=odds ratio.
Model adjusted for age, sex, region and smoking status.
The effect of LINE-1 on bladder cancer risk was not modified by age, sex, tobacco type, nutrient intakes or trace elements (Supplementary Tables S4 and S5). Although the risk of bladder cancer associated with low and high levels of LINE-1 methylation was significant among never smokers, the interaction between LINE-1 methylation and smoking status was not significant (P=0.2; Supplementary Table S4). This interaction was not significant even when comparing never smokers with regular smokers, after adjusting the models for duration of cigarette smoking, cigarettes smoked per day or pack-years among smokers (data not shown). There was no interaction between variation in GSTM1 or GSTT1 or SNPs previously associated with bladder cancer risk and LINE-1 methylation levels. The same is true for most of the SNPs in genes from the one-carbon metabolism pathway (Supplementary Tables S5 and S6). However, the increased risk of bladder cancer among subjects in the lowest and highest tertiles was significantly modified by five SNPs (rs2124344, rs7215833, rs4646340, rs4646350 and rs4646341) in the phosphatidylethanolamine N-methyltransferase (PEMT) gene. Under the dominant mode of inheritance, the risk of bladder cancer being in the lowest or highest tertile of LINE-1 methylation was significantly increased only among individuals homozygous for the major allele of each of the five SNPs (P-interaction⩽4.9 × 10−5; P-interaction⩽0.03 corrected for multiple testing; Table 3).
Table 3. Interaction between LINE-1 methylation and single-nucleotide polymorphisms in phosphatidylethanolamine N-methyltransferase (PEMT) in the SBC/EPICURO study.
|
rs2124344[CC], PEMT, intron |
rs2124344 [CT-TT], PEMT, intron |
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| LINE-1 methylation (tertiles) | Cases | Controls | OR | 95% CI | P | LINE-1 methylation (tertiles) | Cases | Controls | OR | 95% CI | P | P interactiona | Corrected P interactionb |
| T1 (<56.68%) |
148 |
112 |
2.26 |
1.52–3.36 |
5.2 × 10−5 |
T1 (<56.68%) |
179 |
177 |
0.89 |
0.65–1.21 |
0.4 |
1.5 × 10−5 |
0.01 |
| T2 (56.68–<58.65%) |
76 |
132 |
1 (Ref.) |
|
|
T2 (56.68–<58.65%) |
181 |
160 |
1 (Ref.) |
|
|
|
|
| T3 (⩾58.65%) | 158 | 107 | 2.69 | 1.80–4.00 | 1.2 × 10−6 | T3 (⩾58.65%) | 184 | 183 | 0.86 | 0.63–1.18 | 0.4 | ||
|
rs7215833[CC], PEMT, flanking 5′UTR |
rs7215833[CT-TT], PEMT, flanking 5′UTR |
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| LINE-1 methylation (tertiles) | Cases | Controls | OR | 95% CI | P | LINE-1 methylation (tertiles) | Cases | Controls | OR | 95% CI | P | P interactiona | Corrected P interactionb |
| T1 (<56.68%) |
155 |
119 |
2.13 |
1.44–3.14 |
0.0002 |
T1 (<56.68%) |
172 |
171 |
0.89 |
0.65–1.23 |
0.5 |
3.6 × 10−5 |
0.02 |
| T2 (56.68–<58.65%) |
77 |
131 |
1 (Ref.) |
|
|
T2 (56.68–<58.65%) |
180 |
161 |
1 (Ref.) |
|
|
|
|
| T3 (⩾58.65%) | 155 | 101 | 2.62 | 1.76–3.91 | 2.4 × 10−6 | T3 (⩾58.65%) | 187 | 189 | 0.87 | 0.64–1.19 | 0.4 |
|
rs4646340[AA], PEMT, intronc |
rs4646340 [AG-GG], PEMT, intron |
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| LINE-1 methylation (tertiles) | Cases | Controls | OR | 95% CI | P | LINE-1 methylation (tertiles) | Cases | Controls | OR | 95% CI | P | P interactiona | Corrected P interactionb |
| T1 (<56.68%) |
153 |
116 |
2.13 |
1.43–3.16 |
0.0002 |
T1 (<56.68%) |
174 |
174 |
0.92 |
0.67–1.25 |
0.6 |
3.8 × 10−5 |
0.02 |
| T2 (56.68–<58.65%) |
75 |
126 |
1 (Ref.) |
|
|
T2 (56.68–<58.65%) |
182 |
166 |
1 (Ref.) |
|
|
|
|
| T3 (⩾58.65%) | 156 | 99 | 2.69 | 1.79–4.04 | 1.8 × 10−6 | T3 (⩾58.65%) | 186 | 191 | 0.88 | 0.65–1.19 | 0.4 |
|
rs4646350[CC], PEMT,intron |
rs4646350[CT-TT], PEMT,intron |
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| LINE-1 methylation (tertiles) | Cases | Controls | OR | 95% CI | P | LINE-1 methylation (tertiles) | Cases | Controls | OR | 95% CI | P | P interactiona | Corrected P interactionb |
| T1 (<56.68%) |
179 |
175 |
2.16 |
1.45–3.20 |
0.0001 |
T1 (<56.68%) |
179 |
175 |
0.91 |
0.66–1.24 |
0.5 |
4.9 × 10−5 |
0.03 |
| T2 (56.68–<58.65%) |
181 |
162 |
1 (Ref.) |
|
|
T2 (56.68–<58.65%) |
181 |
162 |
1 (Ref.) |
|
|
|
|
| T3 (⩾58.65%) | 186 | 184 | 2.64 | 1.77–3.94 | 2.1 × 10−6 | T3 (⩾58.65%) | 186 | 184 | 0.88 | 0.65–1.20 | 0.4 |
Abbreviations: CI=confidence interval; OR=odds ratio; UTR=untranslated region.
Minor allele frequency—rs2124344=0.36, rs7215833=0.36, rs4646340=0.37, rs4646350=0.36 and rs46341=0.37.
All models are adjusted for age, sex, region and smoking status (never, occasional, former and current smoker).
Uncorrected likelihood ratio test's P-value.
Likelihood ratio test's P-value corrected for multiple testing using permutation test.
rs4646341 was in perfect linkage disequilibrium with rs4646340 (r2=1.0, D'=1.0) and the corrected P-value for interaction of this single-nucleotide polymorphism was 0.037.
Discussion
The current study shows that both low and high levels of LINE-1 methylation are associated with increased risk of bladder cancer corresponding to a nonlinear – ‘U-shaped' – association between global genomic DNA methylation level and bladder cancer risk. This association was significantly modified by the five SNPs in the PEMT gene. Altogether, this points to the complex aetiological mechanisms of DNA methylation in bladder cancer development.
The finding of increased risk of bladder cancer among individuals with low level of methylation is in agreement with previous reports (Moore et al, 2008; Wilhelm et al, 2010; Cash et al, 2012). Moore et al (2008), by analyzing global 5-methylcytosine content of the genome using HpaII, high-performance capillary electrophoresis and densitometry, showed that low level of global DNA methylation was associated with increased risk of bladder cancer in a subsample of the same study. Additionally, two studies also using sodium bisulphite modification of leukocyte DNA, followed by pyrosequencing to quantify methylation levels of four LINE-1 CpG sites, reported a similar inverse association between LINE-1 methylation level and risk of bladder cancer in independent populations (Wilhelm et al, 2010; Cash et al, 2012). An experimental study demonstrated hypomethylation of LINE-1 sequences in urothelial carcinoma cell lines and tumours compared with normal bladder tissue (Jurgens et al, 1996). Another study also showed that hypomethylation of LINE-1 promoter can induce the expression of the MET oncogene in bladder tumours and normal urothelial tissues (Wolff et al, 2010). Interestingly, the current study also shows that higher methylation level was associated with increased risk of bladder cancer. In support of this finding, a recent case–control study that investigated the association between LINE-1 methylation and renal cell carcinoma showed a direct association between methylation and risk of renal cell carcinoma (Liao et al, 2011), while another study that evaluated LINE-1 methylation status in normal and cancerous cells found out sporadic hypermethylation of LINE-1 loci in cancer cells (Phokaew et al, 2008). Although nonlinear associations between exposures and cancer are still uncommonly reported, yet there are recent examples indicating that this more complex relationship is also possible and a fact. Studies that have shown U-shaped risk patterns similar to that observed in this study relate to studies analyzing the association between pancreatic cancer and folate, a primary methyl group donor for DNA methylation modifications (Chuang et al, 2011), telomere length and pancreatic cancer (Skinner et al, 2012), serum sex hormones and prostate cancer (Salonia et al, 2012) and serum vitamin D and prostate cancer (Tuohimaa et al, 2004). Recent studies have demonstrated that methylation blocks the initiation of transcription of repetitive DNA sequences such as LINE-1 and Alu elements, while at the same time allowing transcriptional elongation of the host gene, suggesting that methylation at repetitive sequences might have dual functions in addition to silencing repetitive sequences depending on the location and context of these regions (Jones, 2012).
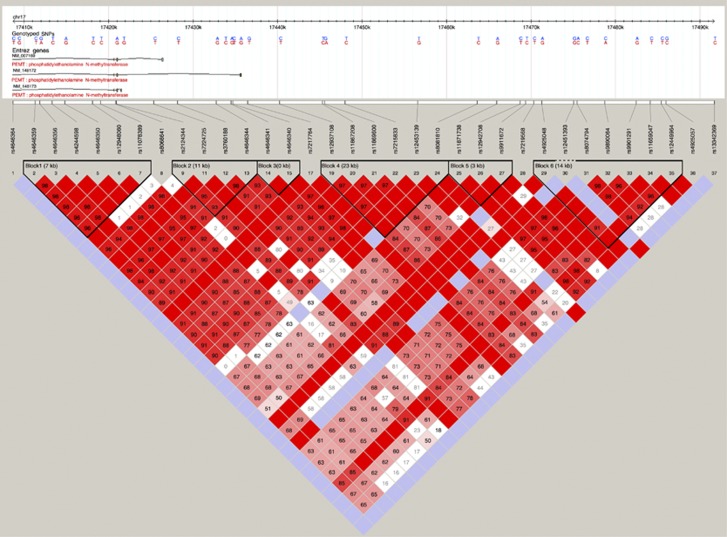
To our knowledge, the present study is the first to show a significant interaction between LINE-1 methylation and SNPs in PEMT gene on the risk of bladder cancer. There is no available data on bladder cancer risk and PEMT polymorphisms. However, a population-based study identified a SNP in a promoter region of PEMT to be associated with increased risk of breast cancer (Xu et al, 2008). PEMT is located within the Smith–Magenis syndrome region on chromosome 17 and encodes for a transmembrane protein involved in important cellular processes, including choline and lipid metabolism, insulin sensitivity and homocysteine levels (Vance et al, 2007). PEMT methylates, using SAM as a substrate, phosphatidylethanolamine to phosphatidylcholine, which accounts for 95% of total choline pool in tissues (Vance et al, 2007; Ueland, 2011). Evidence suggest that phosphatidylcholine metabolism could be involved in malignant transformations (Glunde et al, 2011). Experimental studies have demonstrated altered choline metabolism in tumours, mediated by transcription factors (signaling pathways) associated with oncogenesis such as hypoxia-inducible factors 1 and by the RAS and PI3K/AKT pathways (Glunde et al, 2011). It has been known that these pathways are constitutively activated in the majority of bladder tumours, namely low-grade non-muscle-invasive papillary tumours, which account for 70–80% of bladder cancer cases (Wu, 2005; Lopez-Knowles et al, 2006). On the other hand, upon oxidation choline gives rise to betaine, which promotes remethylation of homocysteine to methionine affecting the concentration of SAM (Ueland, 2011). Therefore, polymorphisms in PEMT could directly affect the one-carbon metabolism pathway and impair DNA methylation. The five SNPs are located in flanking 5′UTR and intron of PEMT (Figure 1). Alternatively, we cannot rule out these associations could be due to chance.
Figure 1.
Linkage disequilibrium plot for single-nucleotide polymorphisms (SNPs) in phosphatidylethanolamine N-methyltransferase (PEMT), including the five SNPs analysed in the current study.
Unlike the previous studies where linear associations were observed between LINE-1 methylation and cancer risk, in this study increased risk of bladder cancer at both extremes of LINE-1 methylation was observed. The risk pattern differences with the other studies could be explained by the possibility of the present study to further characterise the risk pattern due to its larger sample size as well as differences in methods used to quantify DNA methylation. A recent meta-analysis of LINE-1 methylation and bladder cancer found a significant heterogeneity in assays used to quantify methylation (Woo and Kim, 2012). That same analysis also identified significant heterogeneity among studies. It should be noted that LINE-1 methylation levels in the current study population (mean=59.2%) were significantly different from those in the previous two studies on bladder cancer risk and LINE-1 methylation, which reported mean methylation levels of 79.6% (Wilhelm et al, 2010) and 81.9% (Cash et al, 2012). This difference is not surprising and probably results from the fact that distinct CpG sites were analysed. On the other hand, the differences could perhaps be explained by geographic dissimilarities resulting in differential exposure to environmental factors, including diet, or genetic background and stochastic factors present in these populations.
The limitations of this study include the fact that blood samples were collected at the time of interviews, before initiation of treatment. Some individuals were not included in the present analyses owing to the lack of LINE-1 methylation data; however, there was no relevant difference in selected characteristics except age (among controls) and sex (among cases and controls) between those with and without that data (Supplementary Table S7). To minimise the potential bias, all the models were adjusted for age, sex as well as region and smoking status. DNA methylation is tissue specific, and it could be affected by cell type. To evaluate whether there is a difference in methylation level among blood cell types, LINE-1 methylation level was quantified in DNA separately isolated from neutrophils and lymphocytes, the most abundant white blood cell types. There was no difference in LINE-1 methylation level between these cell types, which is in agreement with studies that found significant correlations between LINE-1 methylation assays of different blood cell types among healthy individuals (Wu et al, 2011) as well as bladder cancer patients (van Bemmel et al, 2012). However, the study on bladder cancer patients also showed lack of correlation between global DNA methylation in the blood and bladder tissue (van Bemmel et al, 2012), while another study showed LINE-1 methylation to be consistent from tissue to tissue, indicating that tissue heterogeneity might not be associated with LINE-1 methylation levels (Choi et al, 2009).
The study has also important strengths. First is the sample size: to the best of our knowledge, this is the first large study to investigate the association between LINE-1 methylation and bladder cancer risk (and any other cancer type). Second, the study integrates a wide range of information, including epidemiological, clinical, environmental and genetic data. Third, the method used to quantify LINE-1 methylation has been shown to be a good surrogate marker of global DNA methylation (Yang et al, 2004). Determining DNA methylation levels by pyrosequencing provides reproducible and accurate results (Tost and Gut, 2007; Laird, 2010).
In conclusion, this study showed that both low and high levels of global DNA methylation may be associated with increased risk of bladder cancer and that this risk can be markedly modified by PEMT. Taken together, the results suggest that DNA methylation and the one-carbon metabolism pathway have a relevant role in the development of bladder cancer. Global DNA methylation may have a more complex association with bladder cancer than previously thought. In addition, the findings suggest that DNA methylation measured in peripheral blood cells could be further explored to be used as a susceptibility marker for bladder cancer risk. Nevertheless, further studies in large and independent populations are required.
Acknowledgments
We acknowledge the coordinators, field and administrative workers, technicians and patients of the Spanish Bladder Cancer/EPICURO Study. This work was supported by the Association for International Cancer Research (grant number 09-0780); Fondo de Investigación Sanitaria, Spain (grant numbers 00/0745, PI051436, PI061614, G03/174); Red Temática de Investigación Cooperativa en Cáncer (grant number RD12/0036/0050-RTICC), Spain; European Commission (grant numbers EU-FP7-HEALTH-F2-2008-201663-UROMOL; EU-FP7-HEALTH-F2-2008-201333-DECanBio); Fundación Científica de la AECC, Spain; US National Institutes of Health (grant number USA-NIH-RO1-CA089715); and the Intramural Research Program of the Division of Cancer Epidemiology and Genetics, National Cancer Institute at the National Institutes of Health, USA.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on British Journal of Cancer website (http://www.nature.com/bjc)
This work is published under the standard license to publish agreement. After 12 months the work will become freely available and the license terms will switch to a Creative Commons Attribution-NonCommercial-Share Alike 3.0 Unported License.
Contributor Information
for the Spanish Bladder Cancer/EPICURO Study investigators:
M Kogevinas, N Malats, F X Real, M Sala, G Castaño, M Torà, D Puente, C Villanueva, C Murta-Nascimento, J Fortuny, E López, S Hernández, R Jaramillo, G Vellalta, L Palencia, F Fermández, A Amorós, A Alfaro, G Carretero, J Lloreta, S Serrano, L Ferrer, A Gelabert, J Carles, O Bielsa, K Villadiego, L Cecchini, J M Saladié, L Ibarz, M Céspedes, C Serra, D García, J Pujadas, R Hernando, A Cabezuelo, C Abad, A Prera, J Prat, M Domènech, J Badal, J Malet, R García-Closas, J Rodríguez de Vera, A I Martín, J Taño, F Cáceres, A Carrato, F García-López, M Ull, A Teruel, E Andrada, A Bustos, A Castillejo, J L Soto, A Tardón, J L Guate, J M Lanzas, J Velasco, J M Fernández, J J Rodríguez, A Herrero, R Abascal, C Manzano, T Miralles, M Rivas, M Arguelles, M Díaz, J Sánchez, O González, A Mateos, V Frade, Mieres Asturias, P Muntañola, C Pravia, A M Huescar, F Huergo, and J Mosquera
Supplementary Material
References
- Amaral AF, Porta M, Silverman DT, Milne RL, Kogevinas M, Rothman N, Cantor KP, Jackson BP, Pumarega JA, Lopez T, Carrato A, Guarner L, Real FX, Malats N. Pancreatic cancer risk and levels of trace elements. Gut. 2011;61 (11:1583–1588. doi: 10.1136/gutjnl-2011-301086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cash HL, Tao L, Yuan JM, Marsit CJ, Houseman EA, Xiang YB, Gao YT, Nelson HH, Kelsey KT. LINE-1 hypomethylation is associated with bladder cancer risk among nonsmoking Chinese. Int J Cancer. 2012;130 (5:1151–1159. doi: 10.1002/ijc.26098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi SH, Worswick S, Byun HM, Shear T, Soussa JC, Wolff EM, Douer D, Garcia-Manero G, Liang G, Yang AS. Changes in DNA methylation of tandem DNA repeats are different from interspersed repeats in cancer. Int J Cancer. 2009;125 (3:723–729. doi: 10.1002/ijc.24384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuang SC, Stolzenberg-Solomon R, Ueland PM, Vollset SE, Midttun O, Olsen A, Tjonneland A, Overvad K, Boutron-Ruault MC, Morois S, Clavel-Chapelon F, Teucher B, Kaaks R, Weikert C, Boeing H, Trichopoulou A, Benetou V, Naska A, Jenab M, Slimani N, Romieu I, Michaud DS, Palli D, Sieri S, Panico S, Sacerdote C, Tumino R, Skeie G, Duell EJ, Rodriguez L, Molina-Montes E, Huerta JM, Larranaga N, Gurrea AB, Johansen D, Manjer J, Ye W, Sund M, Peeters PH, Jeurnink S, Wareham N, Khaw KT, Crowe F, Riboli E, Bueno-de-Mesquita B, Vineis P. A U-shaped relationship between plasma folate and pancreatic cancer risk in the European Prospective Investigation into Cancer and Nutrition. Eur J Cancer. 2011;47 (12:1808–1816. doi: 10.1016/j.ejca.2011.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estecio MR, Gharibyan V, Shen L, Ibrahim AE, Doshi K, He R, Jelinek J, Yang AS, Yan PS, Huang TH, Tajara EH, Issa JP. LINE-1 hypomethylation in cancer is highly variable and inversely correlated with microsatellite instability. PLoS One. 2007;2 (5:e399. doi: 10.1371/journal.pone.0000399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esteller M. Epigenetics in cancer. N Engl J Med. 2008;358 (11:1148–1159. doi: 10.1056/NEJMra072067. [DOI] [PubMed] [Google Scholar]
- Florl AR, Schulz WA. Chromosomal instability in bladder cancer. Arch Toxicol. 2008;82 (3:173–182. doi: 10.1007/s00204-008-0280-3. [DOI] [PubMed] [Google Scholar]
- Garcia-Closas M, Malats N, Real FX, Yeager M, Welch R, Silverman D, Kogevinas M, Dosemeci M, Figueroa J, Chatterjee N, Tardon A, Serra C, Carrato A, Garcia-Closas R, Murta-Nascimento C, Rothman N, Chanock SJ. Large-scale evaluation of candidate genes identifies associations between VEGF polymorphisms and bladder cancer risk. PLoS Genet. 2007;3 (2:e29. doi: 10.1371/journal.pgen.0030029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Closas M, Malats N, Silverman D, Dosemeci M, Kogevinas M, Hein DW, Tardon A, Serra C, Carrato A, Garcia-Closas R, Lloreta J, Castano-Vinyals G, Yeager M, Welch R, Chanock S, Chatterjee N, Wacholder S, Samanic C, Tora M, Fernandez F, Real FX, Rothman N. NAT2 slow acetylation, GSTM1 null genotype, and risk of bladder cancer: results from the Spanish Bladder Cancer Study and meta-analyses. Lancet. 2005;366 (9486:649–659. doi: 10.1016/S0140-6736(05)67137-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Closas M, Ye Y, Rothman N, Figueroa JD, Malats N, Dinney CP, Chatterjee N, Prokunina-Olsson L, Wang Z, Lin J, Real FX, Jacobs KB, Baris D, Thun M, De Vivo I, Albanes D, Purdue MP, Kogevinas M, Kamat AM, Lerner SP, Grossman HB, Gu J, Pu X, Hutchinson A, Fu YP, Burdett L, Yeager M, Tang W, Tardon A, Serra C, Carrato A, Garcia-Closas R, Lloreta J, Johnson A, Schwenn M, Karagas MR, Schned A, Andriole G, Jr, Grubb R, 3rd, Black A, Jacobs EJ, Diver WR, Gapstur SM, Weinstein SJ, Virtamo J, Hunter DJ, Caporaso N, Landi MT, Fraumeni JF, Jr, Silverman DT, Chanock SJ, Wu X. A genome-wide association study of bladder cancer identifies a new susceptibility locus within SLC14A1, a urea transporter gene on chromosome 18q12.3. Hum Mol Genet. 2011;20 (21:4282–4289. doi: 10.1093/hmg/ddr342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Closas R, Garcia-Closas M, Kogevinas M, Malats N, Silverman D, Serra C, Tardon A, Carrato A, Castano-Vinyals G, Dosemeci M, Moore L, Rothman N, Sinha R. Food, nutrient and heterocyclic amine intake and the risk of bladder cancer. Eur J Cancer. 2007;43 (11:1731–1740. doi: 10.1016/j.ejca.2007.05.007. [DOI] [PubMed] [Google Scholar]
- Glunde K, Bhujwalla ZM, Ronen SM. Choline metabolism in malignant transformation. Nat Rev Cancer. 2011;11 (12:835–848. doi: 10.1038/nrc3162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopkins WA, Staub BP, Baionno JA, Jackson BP, Roe JH, Ford NB. Trophic and maternal transfer of selenium in brown house snakes (Lamprophis fuliginosus) Ecotoxicol Environ Saf. 2004;58 (3:285–293. doi: 10.1016/S0147-6513(03)00076-9. [DOI] [PubMed] [Google Scholar]
- Izarzugaza MI, Ardanaz E, Chirlaque MD, Font C, Ameijide A, Linares C. Tobacco-related tumours of the lung, bladder and larynx: changes in Spain. Ann Oncol. 2010;21 (Suppl 3:iii52–iii60. doi: 10.1093/annonc/mdq084. [DOI] [PubMed] [Google Scholar]
- Jones PA. Functions of DNA methylation: islands, start sites, gene bodies and beyond. Nat Rev Genet. 2012;13 (7:484–492. doi: 10.1038/nrg3230. [DOI] [PubMed] [Google Scholar]
- Jurgens B, Schmitz-Drager BJ, Schulz WA. Hypomethylation of L1 LINE sequences prevailing in human urothelial carcinoma. Cancer Res. 1996;56 (24:5698–5703. [PubMed] [Google Scholar]
- Laird PW. Principles and challenges of genomewide DNA methylation analysis. Nat Rev Genet. 2010;11 (3:191–203. doi: 10.1038/nrg2732. [DOI] [PubMed] [Google Scholar]
- Lee YL, Xu X, Wallenstein S, Chen J. Gene expression profiles of the one-carbon metabolism pathway. J Genet Genomics. 2009;36 (5:277–282. doi: 10.1016/S1673-8527(08)60115-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao LM, Brennan P, van Bemmel DM, Zaridze D, Matveev V, Janout V, Kollarova H, Bencko V, Navratilova M, Szeszenia-Dabrowska N, Mates D, Rothman N, Boffetta P, Chow WH, Moore LE. LINE-1 methylation levels in leukocyte DNA and risk of renal cell cancer. PLoS One. 2011;6 (11:e27361. doi: 10.1371/journal.pone.0027361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Knowles E, Hernandez S, Malats N, Kogevinas M, Lloreta J, Carrato A, Tardon A, Serra C, Real FX. PIK3CA mutations are an early genetic alteration associated with FGFR3 mutations in superficial papillary bladder tumors. Cancer Res. 2006;66 (15:7401–7404. doi: 10.1158/0008-5472.CAN-06-1182. [DOI] [PubMed] [Google Scholar]
- Lotan Y, Svatek RS, Malats N. Screening for bladder cancer: a perspective. World J Urol. 2008;26 (1:13–18. doi: 10.1007/s00345-007-0223-2. [DOI] [PubMed] [Google Scholar]
- Moore LE, Malats N, Rothman N, Real FX, Kogevinas M, Karami S, Garcia-Closas R, Silverman D, Chanock S, Welch R, Tardon A, Serra C, Carrato A, Dosemeci M, Garcia-Closas M. Polymorphisms in one-carbon metabolism and trans-sulfuration pathway genes and susceptibility to bladder cancer. Int J Cancer. 2007;120 (11:2452–2458. doi: 10.1002/ijc.22565. [DOI] [PubMed] [Google Scholar]
- Moore LE, Pfeiffer RM, Poscablo C, Real FX, Kogevinas M, Silverman D, Garcia-Closas R, Chanock S, Tardon A, Serra C, Carrato A, Dosemeci M, Garcia-Closas M, Esteller M, Fraga M, Rothman N, Malats N. Genomic DNA hypomethylation as a biomarker for bladder cancer susceptibility in the Spanish Bladder Cancer Study: a case-control study. Lancet Oncol. 2008;9 (4:359–366. doi: 10.1016/S1470-2045(08)70038-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murta-Nascimento C, Schmitz-Drager BJ, Zeegers MP, Steineck G, Kogevinas M, Real FX, Malats N. Epidemiology of urinary bladder cancer: from tumor development to patient's death. World J Urol. 2007;25 (3:285–295. doi: 10.1007/s00345-007-0168-5. [DOI] [PubMed] [Google Scholar]
- Parkin DM. The global burden of urinary bladder cancer. Scand J Urol Nephrol Suppl. 2008. pp. 12–20. [DOI] [PubMed]
- Phokaew C, Kowudtitham S, Subbalekha K, Shuangshoti S, Mutirangura A. LINE-1 methylation patterns of different loci in normal and cancerous cells. Nucleic Acids Res. 2008;36 (17:5704–5712. doi: 10.1093/nar/gkn571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothman N, Garcia-Closas M, Chatterjee N, Malats N, Wu X, Figueroa JD, Real FX, Van Den Berg D, Matullo G, Baris D, Thun M, Kiemeney LA, Vineis P, De Vivo I, Albanes D, Purdue MP, Rafnar T, Hildebrandt MA, Kiltie AE, Cussenot O, Golka K, Kumar R, Taylor JA, Mayordomo JI, Jacobs KB, Kogevinas M, Hutchinson A, Wang Z, Fu YP, Prokunina-Olsson L, Burdett L, Yeager M, Wheeler W, Tardon A, Serra C, Carrato A, Garcia-Closas R, Lloreta J, Johnson A, Schwenn M, Karagas MR, Schned A, Andriole G, Jr., Grubb R, 3rd, Black A, Jacobs EJ, Diver WR, Gapstur SM, Weinstein SJ, Virtamo J, Cortessis VK, Gago-Dominguez M, Pike MC, Stern MC, Yuan JM, Hunter DJ, McGrath M, Dinney CP, Czerniak B, Chen M, Yang H, Vermeulen SH, Aben KK, Witjes JA, Makkinje RR, Sulem P, Besenbacher S, Stefansson K, Riboli E, Brennan P, Panico S, Navarro C, Allen NE, Bueno-de-Mesquita HB, Trichopoulos D, Caporaso N, Landi MT, Canzian F, Ljungberg B, Tjonneland A, Clavel-Chapelon F, Bishop DT, Teo MT, Knowles MA, Guarrera S, Polidoro S, Ricceri F, Sacerdote C, Allione A, Cancel-Tassin G, Selinski S, Hengstler JG, Dietrich H, Fletcher T, Rudnai P, Gurzau E, Koppova K, Bolick SC, Godfrey A, Xu Z, Sanz-Velez JI, D García-Prats M, Sanchez M, Valdivia G, Porru S, Benhamou S, Hoover RN, Fraumeni JF, Jr, Silverman DT, Chanock SJ. A multi-stage genome-wide association study of bladder cancer identifies multiple susceptibility loci. Nat Genet. 2010;42 (11:978–984. doi: 10.1038/ng.687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Royston P, Sauerbrei W. Multivariable modeling with cubic regression splines: a principled approach. Stata J. 2007;7 (1:45–70. [Google Scholar]
- Salonia A, Abdollah F, Capitanio U, Suardi N, Briganti A, Gallina A, Colombo R, Ferrari M, Castagna G, Rigatti P, Montorsi F. Serum sex steroids depict a nonlinear u-shaped association with high-risk prostate cancer at radical prostatectomy. Clin Cancer Res. 2012;18 (13:3648–3657. doi: 10.1158/1078-0432.CCR-11-2799. [DOI] [PubMed] [Google Scholar]
- Samanic C, Kogevinas M, Dosemeci M, Malats N, Real FX, Garcia-Closas M, Serra C, Carrato A, Garcia-Closas R, Sala M, Lloreta J, Tardon A, Rothman N, Silverman DT. Smoking and bladder cancer in Spain: effects of tobacco type, timing, environmental tobacco smoke, and gender. Cancer Epidemiol Biomarkers Prev. 2006;15 (7:1348–1354. doi: 10.1158/1055-9965.EPI-06-0021. [DOI] [PubMed] [Google Scholar]
- Skinner HG, Gangnon RE, Litzelman K, Johnson RA, Chari ST, Petersen GM, Boardman LA. Telomere length and pancreatic cancer: a case-control study. Cancer Epidemiol Biomarkers Prev. 2012;21 (11:2095–2100. doi: 10.1158/1055-9965.EPI-12-0671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taby R, Issa JP. Cancer epigenetics. CA Cancer J Clin. 2010;60 (6:376–392. doi: 10.3322/caac.20085. [DOI] [PubMed] [Google Scholar]
- Tost J, Gut IG. DNA methylation analysis by pyrosequencing. Nat Protoc. 2007;2 (9:2265–2275. doi: 10.1038/nprot.2007.314. [DOI] [PubMed] [Google Scholar]
- Tuohimaa P, Tenkanen L, Ahonen M, Lumme S, Jellum E, Hallmans G, Stattin P, Harvei S, Hakulinen T, Luostarinen T, Dillner J, Lehtinen M, Hakama M. Both high and low levels of blood vitamin D are associated with a higher prostate cancer risk: a longitudinal, nested case-control study in the Nordic countries. Int J Cancer. 2004;108 (1:104–108. doi: 10.1002/ijc.11375. [DOI] [PubMed] [Google Scholar]
- Ueland PM. Choline and betaine in health and disease. J Inherit Metab Dis. 2011;34 (1:3–15. doi: 10.1007/s10545-010-9088-4. [DOI] [PubMed] [Google Scholar]
- Ulrey CL, Liu L, Andrews LG, Tollefsbol TO. The impact of metabolism on DNA methylation. Hum Mol Genet. 2005;14 (Spec No 1:R139–R147. doi: 10.1093/hmg/ddi100. [DOI] [PubMed] [Google Scholar]
- van Bemmel D, Lenz P, Liao LM, Baris D, Sternberg LR, Warner AC, Johnson AT, Jones MA, Kida M, Schwenn M, Schned A, Silverman DT, Rothman N, Moore LE. Correlation of LINE-1 methylation levels in patient matched buffy coat, serum, buccal cell and bladder tumor tissue DNA samples. Cancer Epidemiol Biomarkers Prev. 2012;21 (7:1143–1148. doi: 10.1158/1055-9965.EPI-11-1030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vance DE, Li Z, Jacobs RL. Hepatic phosphatidylethanolamine N-methyltransferase, unexpected roles in animal biochemistry and physiology. J Biol Chem. 2007;282 (46:33237–33241. doi: 10.1074/jbc.R700028200. [DOI] [PubMed] [Google Scholar]
- Volanis D, Kadiyska T, Galanis A, Delakas D, Logotheti S, Zoumpourlis V. Environmental factors and genetic susceptibility promote urinary bladder cancer. Toxicol Lett. 2010;193 (2:131–137. doi: 10.1016/j.toxlet.2009.12.018. [DOI] [PubMed] [Google Scholar]
- Wilhelm CS, Kelsey KT, Butler R, Plaza S, Gagne L, Zens MS, Andrew AS, Morris S, Nelson HH, Schned AR, Karagas MR, Marsit CJ. Implications of LINE1 methylation for bladder cancer risk in women. Clin Cancer Res. 2010;16 (5:1682–1689. doi: 10.1158/1078-0432.CCR-09-2983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolff EM, Byun HM, Han HF, Sharma S, Nichols PW, Siegmund KD, Yang AS, Jones PA, Liang G. Hypomethylation of a LINE-1 promoter activates an alternate transcript of the MET oncogene in bladders with cancer. PLoS Genet. 2010;6 (4:e1000917. doi: 10.1371/journal.pgen.1000917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo HD, Kim J. Global DNA hypomethylation in peripheral blood leukocytes as a biomarker for cancer risk: a meta-analysis. PLoS One. 2012;7 (4:e34615. doi: 10.1371/journal.pone.0034615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu HC, Delgado-Cruzata L, Flom JD, Kappil M, Ferris JS, Liao Y, Santella RM, Terry MB. Global methylation profiles in DNA from different blood cell types. Epigenetics. 2011;6 (1:76–85. doi: 10.4161/epi.6.1.13391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu XR. Urothelial tumorigenesis: a tale of divergent pathways. Nat Rev Cancer. 2005;5 (9:713–725. doi: 10.1038/nrc1697. [DOI] [PubMed] [Google Scholar]
- Xu X, Gammon MD, Zeisel SH, Lee YL, Wetmur JG, Teitelbaum SL, Bradshaw PT, Neugut AI, Santella RM, Chen J. Choline metabolism and risk of breast cancer in a population-based study. FASEB J. 2008;22 (6:2045–2052. doi: 10.1096/fj.07-101279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang AS, Estecio MR, Doshi K, Kondo Y, Tajara EH, Issa JP. A simple method for estimating global DNA methylation using bisulfite PCR of repetitive DNA elements. Nucleic Acids Res. 2004;32 (3:e38. doi: 10.1093/nar/gnh032. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.