Abstract
Homeobox genes comprise a super-family of evolutionarily conserved genes that play essential roles in controlling body plan specification and cell fate determination. Substantial evidence indicates that leukemogenesis is driven by abnormal expression of homeobox genes that control hematopoiesis. In solid tumors, aberrant expression of homeobox genes has been increasingly found to modulate diverse processes such as cell proliferation, cell death, metastasis, angiogenesis and DNA repair. This review discusses how homeobox genes are deregulated in solid tumors and the functional significance of this deregulation in the hallmarks of cancer.
Keywords: homeobox genes, transcription factors, solid tumors, tumorigenesis, hallmarks
1. Introduction
Homeobox genes were first discovered in Drosophila by the ability of their mutations to cause formation of body parts in inappropriate contexts (Gehring & Hiromi, 1986; McGinnis & Krumlauf, 1992). In humans, mutations in homeobox genes cause a wide range of complex developmental abnormalities including limb malformations and sensory defects (Mortlock & Innis, 1997; Ruf et al., 2004). Most of our current understanding of the functions of homeobox genes in mammalian embryonic development has come from studies of knock-out mice. Different sets of homeobox genes control skeletal patterning, limb and craniofacial morphogenesis, and development of virtually all organ systems (McGinnis & Krumlauf, 1992; Panganiban & Rubenstein, 2002; Christensen et al., 2008). In adults, homeobox genes also regulate tissue regeneration, and play critical roles in controlling self-renewal and differentiation of hematopoietic progenitors (Argiropoulos & Humphries, 2007).
1.1. Genomic organization of mammalian homeobox genes
Mammalian homeobox genes are categorized into several families that are named after their homologs in the fly. Examples include DLX (distal-less), PAX (paired), MSX (muscle segment), CDX (caudal), EN (engrailed) and OTX (orthodenticle). The evolution of homeobox gene families and the phylogenetic relationships between family members have been the focus of extensive study (Sumiyama et al., 2003; Garcia-Fernàndez, 2005; Lemons & McGinnis, 2006). Most homeobox genes are dispersed throughout the genome, whereas the members of the mammalian HOX and DLX gene families are organized in clusters. The HOX family comprises 39 genes that are tandemly arranged in four clusters (HOXA, HOXB, HOXC, HOXD) located on different chromosomes (McGinnis & Krumlauf, 1992; Pearson et al., 2005). The six members of the DLX family are arranged in bigene clusters that are located upstream of the HOX clusters (Sumiyama et al., 2003). The temporal and spatial expression of HOX genes is tightly coupled to their clustered physical organization. In general, HOX genes that are located at the 3′ end of the clusters are expressed early in development and in anterior body regions, whereas the genes at the 5′ end of clusters are expressed later and in more posterior body regions (McGinnis & Krumlauf, 1992; Pearson et al., 2005).
1.2. Structural features of homeoproteins
Proteins that are encoded by homeobox genes are often called ‘homeoproteins’ and are widely regarded to function as transcription factors (Gehring et al., 1994; Biggin & McGinnis, 1997). A few homeoproteins also exhibit non-transcriptional activities such as controlling mRNA export, translation and protein stability (Dubnau & Struhl, 1996; Topisirovic et al., 2005; Aoki et al., 2011). A hallmark of homeoproteins is their DNA-binding domain, termed the homeodomain, which binds DNA elements containing a TAAT motif (Gehring et al., 1994). The helix-turn-helix structure of the homeodomain is highly conserved across homeoproteins. Binding affinity and specificity of homeoproteins for different target gene promoters is mediated by diversity in their amino acid residues and by interactions with other transcription factors (Gehring et al., 1994; Biggin & McGinnis, 1997; Chariot et al., 1999). Transcriptional target selectivity of homeoproteins is also mediated by additional conserved motifs that are unique to different families. For example, HOX proteins contain a motif that mediates interactions with PBX co-factors (Chang et al., 1995). PAX proteins contain an additional, signature DNA-binding domain called the paired domain (Robson et al., 2006), whereas ZEB family members contain zinc finger domains in addition to the homeodomain (Vandewalle et al., 2009).
2. Mechanisms that cause homeobox gene deregulation in tumors
Overexpression or down-regulated expression of many homeobox genes has been observed in a wide variety of malignancies (Abate-Shen, 2002; Samuel & Naora, 2005; Shah & Sukumar, 2010). Translocation-induced fusions of HOX genes in hematologic malignancies are well-documented (Samuel & Naora, 2005; Argiropoulos & Humphries, 2007). One example is the t(7;11)(p15;p15) translocation in acute myeloid leukemia that results in the fusion of the HOXA9 protein to the amino-terminus of the nuclear pore complex protein NUP98 (Borrow et al., 1996; Nakamura et al., 1996). In contrast to hematologic malignancies, the mechanisms that cause homeobox gene deregulation in solid tumors are less understood. Translocations that result in fusion of PAX3 and PAX7 to the FKHR transcription factor have been identified in alveolar rhabdomyosarcoma (Galili et al., 1993; Davis et al., 1994). Translocation-induced fusion of PAX8 to the peroxisome proliferator-activated receptor γ1 (PPARγ1) occurs in thyroid follicular tumors (Kroll et al., 2000). However, as discussed below, deregulation of other homeobox genes in solid tumors has been attributable to other types of genomic aberrations and to epigenetic mechanisms.
2.1. Loss of heterozygosity and gene amplification
Several homeobox genes with tumor-suppressive properties localize to ‘hotspots’ that undergo loss of heterozygosity (LOH) in cancers [Table I]. One example is NKX3.1 that maps to 8p21. This region is deleted in ~80% of prostate cancers (He et al., 1997) and also frequently undergoes allelic loss in prostatic intraepithelial neoplasia (PIN) (Emmert-Buck et al., 1995). Inactivation of Nkx3.1 in mice induces PIN-like lesions and cooperates with Pten loss to induce carcinoma (Kim et al., 2002a; Kim et al, 2002b). To date, only a few overexpressed homeobox genes have been found to localize to regions that are amplified in tumors. The HOXB gene cluster and the DLX4 gene map to 17q21.3-q22, a region that is amplified in ~10% of breast cancers (Hyman et al., 2002). However, HOXB7 and DLX4 are over-expressed in >50% of breast cancers (Man et al., 2005; Wu et al., 2006), indicating that gene amplification is not the sole mechanism underlying the overexpression of these genes.
Table I.
Examples of chromosomal aberrations in homeobox gene-containing loci in solid tumors
| Homeobox gene | Chromosomal region | Lesion/Tumor type | References |
|---|---|---|---|
| LOH | |||
| NKX3.1 | 8p21 | PIN, Prostate cancers | Emmert-Buck et al. (1995); He et al. (1997) |
| BARX2 | 11q24-q25 | Ovarian cancers | Gabra et al. (1996) |
| CUTL1 | 7q22 | Uterine leiomyomas | Zeng et al. (1997) |
| LAGY (HOPX) | 4q11-13.1 | Lung cancers | Chen et al. (2003) |
| Gene Amplification | |||
| HOXB7 | 17q21.3-q22 | Breast, ovarian cancers | Wu et al. (2006); Naora et al. (2001) |
| DLX4 (BP1) | 17q21.3-q22 | Breast, ovarian cancers | Man et al. (2005), Hara et al. (2007) |
2.2. DNA methylation and chromatin modification
Aberrant methylation of CpG islands frequently occurs in tumors, and is the most commonly identified mechanism that silences expression of homeobox genes in solid tumors [Table II]. Many CpG islands within HOX gene clusters are methylated in lung cancers (Rauch et al., 2007). In a study by Tommasi and colleagues (2009), one third of the CpG islands that were identified in genome-wide analysis of DNA methylation to be hypermethylated in early-stage breast cancers were associated with homeobox genes. These authors also identified that ~50% of the hypermethylated genes overlapped with known Polycomb targets (Tommasi et al., 2009). Polycomb group proteins form multi-protein complexes that dynamically alter chromatin structure by modifying specific residues in histone tails and recruit DNA methyltransferases that methylate DNA (Mills, 2010). Polycomb-mediated repression is a principal mechanism by which HOX gene expression is tightly regulated during development (Soshnikova & Duboule, 2009). EZH2, a component of the Polycomb Repressive Complex 2 (PRC2), is overexpressed in breast cancers and several other types of solid tumors (Mills, 2010). Trithorax group proteins counteract Polycomb-mediated silencing and their levels are also altered in various cancers (Mills, 2010). Aberrant expression of Polycomb and Trithorax group proteins might therefore be an important mechanism by which multiple HOX genes are deregulated in tumors.
Table II.
Examples of tumor-suppressive homeobox genes that are methylated in solid tumors
| Homeobox gene | Tumor type | References |
|---|---|---|
| HOXA5 | breast | Raman et al. (2000) |
| HOXA9 | breast | Reynolds et al. (2006) |
| HOXA10 | endometrial | Yoshida et al. (2006) |
| HOXD10 | gastric | Wang et al. (2012b) |
| CDX2 | colorectal | Kawai et al. (2005) |
2.3. Non-coding RNAs
Repression of homeobox gene expression by non-coding RNAs has been the subject of extensive focus in the developmental biology field and has attracted increasing attention in the context of cancer. Long noncoding RNAs and microRNAs are present in the intergenic regions of HOX clusters and control HOX gene expression through both cis- and trans- acting mechanisms (Lemons & McGinnis, 2006; Yekta et al., 2008). One striking example is the long non-coding RNA HOTAIR that is located in the HOXC locus. HOTAIR interacts with and targets PRC2 to the HOXD locus (Rinn et al., 2007). HOTAIR expression in primary breast tumors has been found to be a strong predictor of metastasis and promotes metastasis by inducing genome-wide re-targeting of PRC2 to an occupancy pattern resembling that of embryonic fibroblasts (Gupta et al., 2010). Several microRNAs have been identified that either promote or suppress tumor progression through targeting specific homeobox genes. miR-10b is highly expressed in metastatic breast cancer cells and promotes metastasis by inhibiting translation of HOXD10 mRNA, resulting in increased expression of the pro-metastatic gene RHOC (Ma et al., 2007). Conversely, miR-185 is down-regulated in breast cancer cells, and inhibits tumor cell growth by repressing SIX1, a homeobox gene that induces cyclin A1 expression (Imam et al., 2010). miR-31 has been found to be down-regulated in cancer-associated fibroblasts (CAFs) and inhibits the ability of CAFs to stimulate tumor cell invasiveness by targeting the homeobox gene SATB2 (Aprelikova et al., 2010).
3. Functional significance of homeobox genes in the established hallmarks of cancer
Studies to date have revealed that homeobox genes have either tumor-suppressive or tumor-promoting properties depending on the context of their expression (Abate-Shen, 2002; Samuel & Naora, 2005; Shah & Sukumar, 2010). The expression patterns and functional properties of homeobox genes in solid tumors fall into two broad categories. In the first category are homeobox genes whose expression is normally maintained in differentiated adult tissues, but is down-regulated in tumors [Figure 1]. These homeobox genes often exhibit tumor-suppressive properties. In the second category are homeobox genes that are expressed in tumors derived from tissues in which these genes are normally expressed during embryonic development (i.e. ‘reactivated’ or overexpressed as compared to the normal adult tissue type) [Figure 1]. Less commonly, homeobox genes can be expressed in tumors derived from a lineage in which these genes are not normally expressed during development. Homeobox genes that fall in this second category often have tumor-promoting properties. Despite numerous reports of their aberrant expression, the mechanisms of many homeobox genes in tumors are poorly understood. Whereas homeoproteins have highly selective functions in vivo, they exhibit promiscuous DNA-binding in vitro (Biggin & McGinnis, 1997) and consequently only a few bona fide transcriptional targets have been identified. However, recent studies have revealed a variety of mechanisms by which homeobox genes control key processes that constitute the core hallmarks of cancer.
Figure 1. Trends in relationships between expression patterns and functional significance of homeobox genes in tumors.
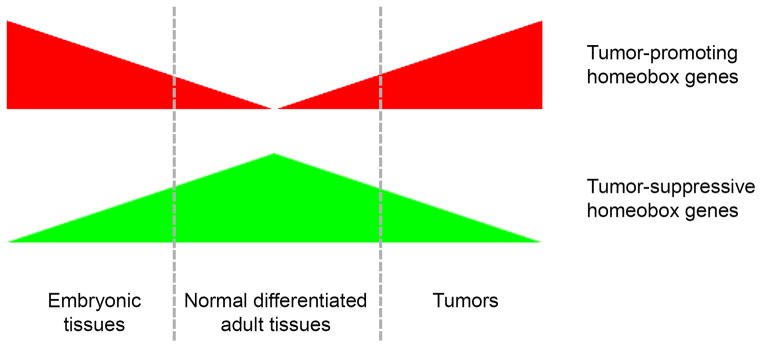
Homeobox genes that are expressed in embryonic tissues and are ‘reactivated’ in tumors (red) tend to have tumor-promoting properties. Homeobox genes whose expression is normally maintained in differentiated adult tissues but is down-regulated in tumors (green) often exhibit tumor-suppressive properties.
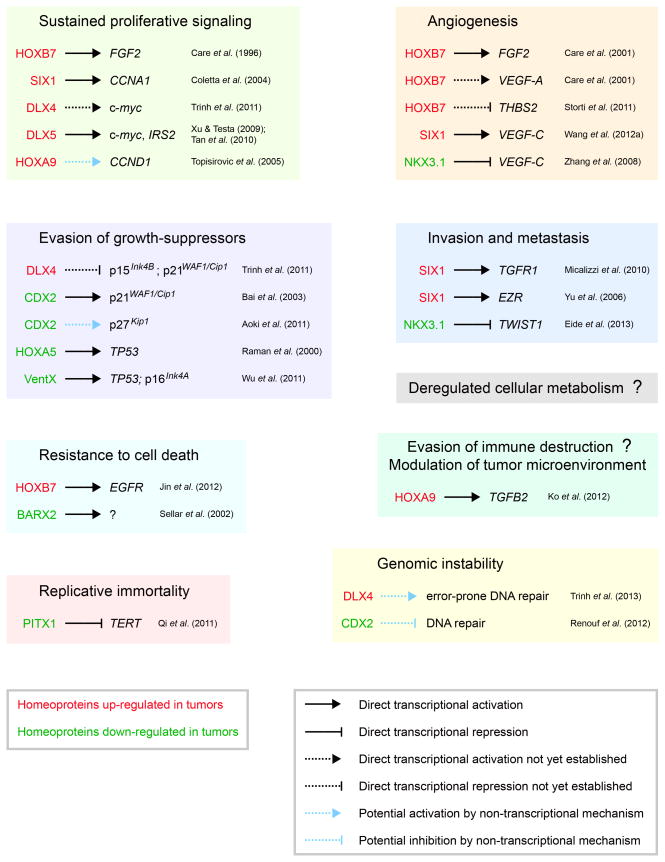
3.1. Sustained proliferative signaling
The ability to sustain chronic proliferation is a well-established hallmark of cancer cells (Hanahan & Weinberg, 2000). One important mechanism by which proliferation is sustained is through autocrine stimulation by growth factors such as fibroblast growth factor-2 (FGF-2). HOXB7 induces transcription of the FGF-2 gene and promotes growth of melanomas, breast and ovarian cancers (Caré et al., 1996; Caré et al., 1998; Naora et al., 2001). Several homeoproteins stimulate tumor cell growth by activating transcription of genes that promote cell cycle progression [Figure 2]. For example, DLX5 stimulates proliferation of lung cancer cells by directly activating c-myc transcription (Xu & Testa, 2009). DLX5 has also been found to stimulate ovarian cancer cell growth by activating transcription of the gene encoding insulin receptor substrate 2 (IRS-2), an oncogenic signaling adaptor protein, and thereby enhancing AKT signaling (Tan et al., 2010). Interestingly, HOXA9 has been reported to induce cyclin D1 expression in leukemic cells by binding the translation initiation factor eIF4E and stimulating eIF4E-dependent export of cyclin D1 mRNA (Topisirovic et al., 2005).
Figure 2. Mechanisms of aberrant homeobox gene expression in the hallmarks of cancer and enabling characteristics.
Examples of homeoproteins that are up- or down-regulated in tumors, their functional significance, transcriptional targets and effector genes.
3.2. Evasion of growth-suppressors
In addition to sustaining growth-promoting signals, cancer cells circumvent signals that inhibit growth (Hanahan & Weinberg, 2000). Several homeoproteins transcriptionally activate genes that induce cell cycle arrest and their expression is decreased in tumors [Figure 2]. CDX2 inhibits proliferation of intestinal epithelial cells and its expression is down-regulated in colorectal tumors (Ee et al., 1995). CDX2 induces expression of the cyclin-dependent kinase (CDK) inhibitor p21WAF1/Cip1 through transcriptional activation (Bai et al., 2003), but stabilizes protein levels of the CDK inhibitor p27Kip1 through a non-transcriptional mechanism (Aoki et al., 2011). Transforming growth factor-β (TGF-β) induces G1 arrest in normal cells by activating a transcriptional program that represses c-Myc and induces p21WAF1/Cip1 and p15Ink4B (Siegel & Massagué, 2003). Many tumors are resistant to the anti-proliferative effect of TGFβ and this resistance has been attributed to TGF-β receptor or Smad4 mutations in several but not all types of tumors (Siegel & Massagué, 2003). One mechanism by which tumors that lack mutations in the TGF-β signaling pathway might become resistant to the anti-proliferative effect of TGF-β is through overexpression of the homeoprotein DLX4. DLX4 blocks the anti-proliferative effect of TGF-β in part by binding to Smad4 and preventing Smad4 from forming transcriptional complexes with Smad2 and Smad3 (Trinh et al., 2011). In addition, DLX4 impairs the DNA-binding ability of Sp1 and induces expression of c-Myc which in turn represses p15Ink4B and p21WAF1/Cip1 transcription (Trinh et al., 2011). Intriguingly, DLX2, another DLX family member, counteracts the growth-inhibitory effect of TGF-β via different mechanisms. DLX2 acts as a transcriptional repressor of TGF-β type II receptor expression and promotes cell survival by inducing expression of betacellulin, a member of the epidermal growth factor family (Yilmaz et al., 2011).
3.3. Resistance to cell death
Cancer cells have evolved adaptive strategies to circumvent cell death programs that are triggered by physiological stresses (Hanahan & Weinberg, 2000). One selective advantage is the ability to survive as floating cells in body fluids. Ovarian cancer cells are often present in ascites as floating aggregates and this aggregation is thought to promote tumor cell survival (Lengyel, 2010). HOXA10 has been found to promote assembly of ovarian cancer cells into aggregates and to enable these cells to evade anoikis (Ko et al., 2010). Another selective advantage for tumor cells is the ability to evade cell death induced by chemotherapeutic agents. Down-regulation of the homeobox gene BARX2 has been implicated in platinum-resistance of ovarian cancer cells (Sellar et al., 2002). HOXB7 has been reported to render breast cancer cells resistant to tamoxifen by transcriptional activation of epidermal growth factor receptor expression (Jin et al., 2012). Trinh et al (2013) recently identified that DLX4 induces expression of topoisomerase IIα (TOP2α), but decreases the sensitivity of tumor cells to TOP2α-targeting drugs such as etoposide and doxorubicin by stimulating DNA repair and thereby reducing the level of drug-induced DNA damage. These two activities of DLX4 provided a possible explanation as to why some TOP2α-overexpressing tumors are not highly sensitive to drugs that target this enzyme. Together, these studies indicate that loss of expression or overexpression of specific homeobox genes in tumors might contribute to acquired chemoresistance, and that expression levels of these genes might potentially serve as predictors of responsiveness to therapy.
3.4. Replicative immortality
Whereas normal cells undergo only a limited number of cycles of cell division, cancer cells are capable of limitless replicative potential (Hanahan & Weinberg, 2000). Substantial evidence indicates the central role of telomeres in the capability for unlimited proliferation and that levels of telomerase, the specialized DNA polymerase which adds telomere repeat segments to the ends of telomeric DNA, are increased in tumor cells (Hanahan & Weinberg, 2011). The ability of homeobox genes to confer limitless replicative potential in solid tumors is not known. One possible negative regulator is PITX1. PITX1 has been described as having tumor-suppressive properties and represses transcription of the gene encoding telomerase reverse transcriptase that controls telomerase activity (Qi et al., 2011).
3.5. Angiogenesis
Angiogenesis is a well-established hallmark of cancer (Hanahan & Weinberg, 2000). Several homeoproteins have been identified to promote tumor growth and progression by inducing expression of pro-angiogenic growth factors [Figure 2]. DLX4 has been found to induce expression of vascular endothelial growth factor A (VEGF-A) and FGF-2 and to increase tumor microvessel density in mouse xenograft models of ovarian cancer (Hara et al., 2007). HOXB7 also induces VEGF-A and FGF-2 expression and stimulates angiogenesis in breast cancer and multiple myeloma (Caré et al., 2001; Storti et al., 2011). In addition, HOXB7 inhibits expression of the anti-angiogenic protein thrombospondin-2 (Storti et al., 2011). Overexpression of SIX1 in breast cancer has been found to promote lymphangiogenesis by inducing expression of VEGF-C (Wang et al., 2012a). Conversely, the ability of NKX3.1 to inhibit VEGF-C expression has been thought to be a mechanism by which loss of NKX3.1 in prostate cancer leads to lymphangiogenesis (Zhang et al., 2008).
3.6. Invasion and metastasis
An extensively studied hallmark of cancer is the ability of tumor cells to invade adjacent tissues and colonize distant sites (Hanahan & Weinberg, 2000). A fundamental and initial step of this capability in carcinomas is the loss of epithelial features and acquisition of mesenchymal features. It is well-established that TGF-β promotes invasion and metastasis by inducing epithelial-to-mesenchymal transition (EMT) (Thiery et al., 2009). Several homeoproteins that are overexpressed in tumors, such as HOXB7 and SIX1, induce EMT and promote invasiveness (Wu et al., 2006; Micalizzi et al., 2010). Conversely, HOXA10 induces mesenchymal-to-epithelial transition and inhibits invasiveness, and its expression is silenced in high-grade endometrial carcinomas (Yoshida et al., 2006). EMT is orchestrated by a repertoire of transcription factors that are induced by TGF-β (Thiery et al., 2009). These include the Snail, Twist and ZEB families of transcription factors. Several homeoproteins mediate their pro- or anti- metastatic effects through direct transcriptional regulation of genes that control the EMT program [Figure 2]. In prostate cancer cells, NKX3.1 represses TWIST1 transcription (Eide et al., 2013). SIX1 induces EMT in breast cancer cells by activating transcription of the gene encoding the TGF-β type I receptor (Micalizzi et al., 2010). SIX1 also promotes metastasis of rhabdomyosarcoma by inducing expression of the cytoskeletal protein ezrin (Yu et al., 2006). The EMT program has been coupled to the acquisition of cancer stem cell (CSC) features (Mani et al., 2008). It is possible that homeobox genes that regulate EMT might also control CSC features. Other homeobox genes might control the self-renewal capability of CSCs. Nanog is a homeobox gene that promotes pluripotency of mouse embryonic stem cells (Mitsui et al., 2003) and has been also found to control ‘stemness’ of glioma stem cells (Zbinden et al., 2010).
4. Functional significance of homeobox genes in the emerging hallmarks and enabling characteristics of cancer
In recent years, two additional traits have emerged as important hallmarks of cancer (Hanahan & Weinberg, 2011). One involves the capability to deregulate cellular metabolism in order to support tumor growth. The second emerging hallmark is the capability of cancer cells to evade immune destruction. In addition, two characteristics have been described that facilitate the acquisition of both the established and emerging hallmarks of cancer (Hanahan & Weinberg, 2011). These enabling characteristics are the development of genomic instability and tumor-promoting inflammation. Although the mechanisms of many homeobox genes have not yet been precisely defined, recent studies have provided insight into how homeobox genes might control these emerging hallmarks of cancer and enabling characteristics.
4.1. Deregulated cellular metabolism
The control of the metabolic switch in cancer cells to glycolysis has been the focus of extensive investigation (Hanahan & Weinberg, 2011). To date, there is no conclusive evidence that deregulation of homeobox genes drives tumor growth by reprogramming energy metabolism. However, several studies have demonstrated the significance of specific homeobox genes in controlling glucose levels and responses to metabolic stress. Deficiency of Sax2, a homeobox gene that is predominantly expressed in the brainstem, has been found to decrease fat and glycogen storage and blood glucose levels (Simon et al., 2007). Expression of DLX2 in tumor cells is induced by glucose deprivation and mediates metabolic stress-induced cell death (Lee et al., 2011). One important mechanism by which metabolism in cancer cells is reprogrammed is through increased expression of glucose transporters that increase glucose uptake (Hanahan & Weinberg, 2011). The gene encoding glucose transporter type 2 is a direct transcriptional target of PDX1, a homeoprotein that controls embryonic development of the pancreas and differentiation of insulin-producing islet β cells (Waeber et al., 1996).
4.2. Evasion of immune destruction and modulation of the tumor microenvironment
Tumor growth is controlled by dynamic interplay between tumor cells and a variety of cell types in the stroma such as fibroblasts, endothelial cells and immune cells (Tlsty & Coussens, 2006). In a recent study, Ko et al (2012) demonstrated that expression of HOXA9 in ovarian cancer cells induces normal peritoneal fibroblasts and mesenchymal stem cells (MSCs) to acquire features of CAFs that promoted growth of tumor and endothelial cells. This tumor-promoting effect of HOXA9 was attributed in substantial part to its transcriptional activation of the gene encoding TGF-β2. HOXA9-induced, tumor-derived TGF-β2 acted in a paracrine manner on peritoneal fibroblasts and MSCs and induced these cells to express mitogenic and pro-inflammatory growth factors (Ko et al., 2012). TGF-β ligands also inhibit proliferation and function of lymphocytes, and down-regulate major histocompatibility antigens on tumor cells (Li et al., 2006). An implication of the study of Ko et al (2012) is that aberrant expression of a homeoprotein can, through regulating expression of tumor-derived factors, promote an inflammatory microenvironment that is permissive for tumor growth and also potentially enables tumors to escape immune destruction.
4.3. DNA repair and genomic instability
Genomic instability endows cancer cells with genetic alterations that drive tumor progression and can stem from defects in components of the genome maintenance machinery that detect and repair DNA damage (Hanahan & Weinberg, 2011). One mechanism by which aberrantly expressed homeobox genes can potentially lead to genomic instability is through cell cycle checkpoint deregulation. SIX1 overexpression has been reported to lead to genomic instability in breast cancer cells by attenuating the DNA damage–induced G2 cell cycle checkpoint via its induction of cyclin A1 expression (Coletta et al., 2008). Repair of DNA double-strand breaks (DSBs) by non-homologous end-joining (NHEJ) is error-prone and misrepair of DSBs can lead to genomic instability (Kasparek & Humphrey, 2011). The canonical NHEJ pathway is initiated by the binding of Ku heterodimers to DNA ends which then recruit other factors to form a complex that enables ligation of DNA ends with little or no homology (Kasparek & Humphrey, 2011). Recent studies have revealed intriguing non-transcriptional functions of several homeoproteins in repairing DSBs. CDX2, HOXB7 and DLX4 have been reported to interact with Ku proteins but have strikingly different effects. CDX2, which is often down-regulated in colon cancer cells, has been found to inhibit end-joining activity (Renouf et al., 2012). In contrast, HOXB7 and DLX4, which are overexpressed in breast and ovarian cancers, stimulate end-joining activity (Rubin et al., 2007; Trinh et al., 2013). Furthermore, Trinh et al (2013) found that DLX4 increases both the frequency and magnitude of erroneous end-joining. Because NHEJ depends on DNA ends being held in close alignment in order for end-processing and ligation to occur, it is possible that the interaction of DLX4 with Ku proteins increases erroneous repair by altering the alignment of DNA ends.
5. Clinical implications and future directions
In summary, increasing evidence indicates the functional significance of overexpression or loss of expression of distinct sets of homeobox genes in the hallmarks and enabling characteristics of cancer [Figure 2]. However, it is not clear whether and how a given homeobox gene controls an acquired capability or enabling characteristic in a tissue-specific manner or in cells of different lineages. For example, HOXB7 has tumor-promoting properties in several different types of tumors (Caré et al., 1996; Caré et al., 1998; Naora et al., 2001; Storti et al., 2011). On the other hand, HOXA9 has tumor-promoting properties in ovarian cancer (Ko et al., 2012), but has tumor-suppressive properties in breast cancer (Reynolds et al., 2006). The mechanisms that cause aberrant expression of homeobox genes in solid tumors also require further investigation. Despite their functional significance, homeoproteins do not represent ideal therapeutic targets that can be readily inhibited with high specificity as these proteins are transcription factors that share tracts of homology with other family members. On the other hand, further study of the mechanisms of homeoproteins, their transcriptional targets and other downstream effectors could provide important insights into focal points for therapeutic intervention. In addition, recent studies have demonstrated the importance of several homeobox genes in chemoresistance and raise the possibility that expression levels of these genes could serve as predictors of responsiveness to therapy. Because homeobox genes play essential functions in lineage-specification, their expression patterns in tumors could also provide valuable information for differential diagnosis when used in the appropriate clinical settings.
Acknowledgments
Research in Dr. Naora’s laboratory is supported by grants from the Cancer & Prevention Research Institute of Texas (RP120390) and NIH/NCI (CA141078).
References
- Abate-Shen C. Deregulated homeobox gene expression in cancer: cause or consequence? Nat Rev Cancer. 2002;2(10):777–785. doi: 10.1038/nrc907. [DOI] [PubMed] [Google Scholar]
- Aoki K, Kakizaki F, Sakashita H, Manabe T, Aoki M, Taketo MM. Suppression of colonic polyposis by homeoprotein CDX2 through its nontranscriptional function that stabilizes p27Kip1. Cancer Res. 2011;71(2):593–602. doi: 10.1158/0008-5472.CAN-10-2842. [DOI] [PubMed] [Google Scholar]
- Aprelikova O, Yu X, Palla J, Wei BR, John S, Yi M, Stephens R, Simpson RM, Risinger JI, Jazaeri A, Niederhuber J. The role of miR-31 and its target gene SATB2 in cancer-associated fibroblasts. Cell Cycle. 2010;9(21):4387–4398. doi: 10.4161/cc.9.21.13674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Argiropoulos B, Humphries RK. Hox genes in hematopoiesis and leukemogenesis. Oncogene. 2007;26(47):6766–6776. doi: 10.1038/sj.onc.1210760. [DOI] [PubMed] [Google Scholar]
- Bai YQ, Miyake S, Iwai T, Yuasa Y. CDX2, a homeobox transcription factor, upregulates transcription of the p21/WAF1/CIP1 gene. Oncogene. 2003;22(39):7942–7949. doi: 10.1038/sj.onc.1206634. [DOI] [PubMed] [Google Scholar]
- Biggin MD, McGinnis W. Regulation of segmentation and segmental identity by Drosophila homeoproteins: the role of DNA binding in functional activity and specificity. Development. 1997;124(22):4425–4433. doi: 10.1242/dev.124.22.4425. [DOI] [PubMed] [Google Scholar]
- Borrow J, Shearman AM, Stanton VP, Becher R, Collins T, Williams AJ, Dubé I, Katz F, Kwong YL, Morris C, Ohyashiki K, Toyama K, Rowley J, Housman DE. The t(7;11)(p15;p15) translocation in acute myeloid leukaemia fuses the genes for nucleoporin NUP98 and class I homeoprotein HOXA9. Nat Genet. 1996;12(2):159–167. doi: 10.1038/ng0296-159. [DOI] [PubMed] [Google Scholar]
- Caré A, Silvani A, Meccia E, Mattia G, Stoppacciaro A, Parmiani G, Peschle C, Colombo MP. HOXB7 constitutively activates basic fibroblast growth factor in melanomas. Mol Cell Biol. 1996;16(9):4842–4851. doi: 10.1128/mcb.16.9.4842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caré A, Silvani A, Meccia E, Mattia G, Peschle C, Colombo MP. Transduction of the SkBr3 breast carcinoma cell line with the HOXB7 gene induces bFGF expression, increases cell proliferation and reduces growth factor dependence. Oncogene. 1998;16(25):3285–3289. doi: 10.1038/sj.onc.1201875. [DOI] [PubMed] [Google Scholar]
- Caré A, Felicetti F, Meccia E, Bottero L, Parenza M, Stoppacciaro A, Peschle C, Colombo MP. HOXB7: a key factor for tumor-associated angiogenic switch. Cancer Res. 2001;61(17):6532–6539. [PubMed] [Google Scholar]
- Chang CP, Shen WF, Rozenfeld S, Lawrence HJ, Largman C, Cleary ML. Pbx proteins display hexapeptide-dependent cooperative DNA binding with a subset of Hox proteins. Genes Dev. 1995;9(6):663–674. doi: 10.1101/gad.9.6.663. [DOI] [PubMed] [Google Scholar]
- Chariot A, Gielen J, Merville MP, Bours V. The homeodomain-containing proteins: an update on their interacting partners. Biochem Pharmacol. 1999;58(12):1851–1857. doi: 10.1016/s0006-2952(99)00234-8. [DOI] [PubMed] [Google Scholar]
- Chen Y, Petersen S, Pacyna-Gengelbach M, Pietas A, Petersen I. Identification of a novel homeobox-containing gene, LAGY, which is downregulated in lung cancer. Oncology. 2003;64(4):450–458. doi: 10.1159/000070306. [DOI] [PubMed] [Google Scholar]
- Christensen KL, Patrick AN, McCoy EL, Ford HL. The Six family of homeobox genes in development and cancer. Adv Cancer Res. 2008;101:93–126. doi: 10.1016/S0065-230X(08)00405-3. [DOI] [PubMed] [Google Scholar]
- Coletta RD, Christensen K, Reichenberger KJ, Lamb J, Micomonaco D, Huang L, Wolf DM, Müller-Tidow C, Golub TR, Kawakami K, Ford HL. The Six1 homeoprotein stimulates tumorigenesis by reactivation of cyclin A1. Proc Natl Acad Sci USA. 2004;101(17):6478–6483. doi: 10.1073/pnas.0401139101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coletta RD, Christensen KL, Micalizzi DS, Jedlicka P, Varella-Garcia M, Ford HL. Six1 overexpression in mammary cells induces genomic instability and is sufficient for malignant transformation. Cancer Res. 2008;68(7):2204–2213. doi: 10.1158/0008-5472.CAN-07-3141. [DOI] [PubMed] [Google Scholar]
- Davis RJ, D’Cruz CM, Lovell MA, Biegel JA, Barr FG. Fusion of PAX7 to FKHR by the variant t(1;13)(p36;q14) translocation in alveolar rhabdomyosarcoma. Cancer Res. 1994;54(11):2869–2872. [PubMed] [Google Scholar]
- Dubnau J, Struhl G. RNA recognition and translational regulation by a homeodomain protein. Nature. 1996;379(6567):694–699. doi: 10.1038/379694a0. [DOI] [PubMed] [Google Scholar]
- Ee HC, Erler T, Bhathal PS, Young GP, James RJ. Cdx-2 homeodomain protein expression in human and rat colorectal adenoma and carcinoma. Am J Pathol. 1995;147(3):586–592. [PMC free article] [PubMed] [Google Scholar]
- Eide T, Ramberg H, Glackin C, Tindall D, Taskén KA. TWIST1, a novel androgen-regulated gene, is a target for NKX3-1 in prostate cancer cells. Cancer Cell Int. 2013;13(1):4. doi: 10.1186/1475-2867-13-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emmert-Buck MR, Vocke CD, Pozzatti RO, Duray PH, Jennings SB, Florence CD, Zhuang Z, Bostwick DG, Liotta LA, Linehan WM. Allelic loss on chromosome 8p12-21 in microdissected prostatic intraepithelial neoplasia. Cancer Res. 1995;55(14):2959–2962. [PubMed] [Google Scholar]
- Gabra H, Watson JE, Taylor KJ, Mackay J, Leonard RC, Steel CM, Porteous DJ, Smyth JF. Definition and refinement of a region of loss of heterozygosity at 11q23.3-q24.3 in epithelial ovarian cancer associated with poor prognosis. Cancer Res. 1996;56(5):950–954. [PubMed] [Google Scholar]
- Galili N, Davis RJ, Fredericks WJ, Mukhopadhyay S, Rauscher FJ, Emanuel BS, Rovera G, Barr FG. Fusion of a fork head domain gene to PAX3 in the solid tumour alveolar rhabdomyosarcoma. Nat Genet. 1993;5(3):230–235. doi: 10.1038/ng1193-230. [DOI] [PubMed] [Google Scholar]
- Garia-Fernàndez J. The genesis and evolution of homeobox gene clusters. Nat Rev Genet. 2005;6(12):881–892. doi: 10.1038/nrg1723. [DOI] [PubMed] [Google Scholar]
- Gehring WJ, Hiromi Y. Homeotic genes and the homeobox. Annu Rev Genet. 1986;20:147–173. doi: 10.1146/annurev.ge.20.120186.001051. [DOI] [PubMed] [Google Scholar]
- Gehring WJ, Qian YQ, Billeter M, Furukubo-Tokunaga K, Schier AF, Resendez-Perez D, Affolter M, Otting G, Wüthrich K. Homeodomain-DNA recognition. Cell. 1994;78(2):211–223. doi: 10.1016/0092-8674(94)90292-5. [DOI] [PubMed] [Google Scholar]
- Gupta RA, Shah N, Wang KC, Kim J, Horlings HM, Wong DJ, Tsai MC, Hung T, Argani P, Rinn JL, Wang Y, Brzoska P, Kong B, Li R, West RB, van de Vijver MJ, Sukumar S, Chang HY. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature. 2010;464(7291):1071–1076. doi: 10.1038/nature08975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000;100(1):57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- Hanahan D, Weinberg RA. Hallmarks of cancer: The next generation. Cell. 2011;144(5):646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- Hara F, Samuel S, Liu J, Rosen D, Langley RR, Naora H. A homeobox gene related to Drosophila distal-less promotes ovarian tumorigenicity by inducing expression of vascular endothelial growth factor and fibroblast growth factor-2. Am J Pathol. 2007;170(5):1594–1606. doi: 10.2353/ajpath.2007.061025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He WW, Sciavolino PJ, Wing J, Augustus M, Hudson P, Meissner PS, Curtis RT, Shell BK, Bostwick DG, Tindall DJ, Gelmann EP, Abate-Shen C, Carter KC. A novel human prostate-specific, androgen-regulated homeobox gene (NKX3.1) that maps to 8p21, a region frequently deleted in prostate cancer. Genomics. 1997;43(1):69–77. doi: 10.1006/geno.1997.4715. [DOI] [PubMed] [Google Scholar]
- Hyman E, Kauraniemi P, Hautaniemi S, Wolf M, Mousses S, Rozenblum E, Ringnér M, Sauter G, Monni O, Elkahloun A, Kallioniemi OP, Kallioniemi A. Impact of DNA amplification on gene expression patterns in breast cancer. Cancer Res. 2002;62(21):6240–6245. [PubMed] [Google Scholar]
- Imam JS, Buddavarapu K, Lee-Chang JS, Ganapathy S, Camosy C, Chen Y, Rao MK. MicroRNA-185 suppresses tumor growth and progression by targeting the Six1 oncogene in human cancers. Oncogene. 2010;29(35):4971–4979. doi: 10.1038/onc.2010.233. [DOI] [PubMed] [Google Scholar]
- Jin K, Kong X, Shah T, Penet MF, Wildes F, Sgroi DC, Ma XJ, Huang Y, Kallioniemi A, Landberg G, Bieche I, Wu X, Lobie PE, Davidson NE, Bhujwalla ZM, Zhu T, Sukumar S. The HOXB7 protein renders breast cancer cells resistant to tamoxifen through activation of the EGFR pathway. Proc Natl Acad Sci USA. 2012;109(8):2736–2741. doi: 10.1073/pnas.1018859108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasparek TR, Humphrey TC. DNA double-strand break repair pathways, chromosomal rearrangements and cancer. Semin Cell Dev Biol. 2011;22(8):886–897. doi: 10.1016/j.semcdb.2011.10.007. [DOI] [PubMed] [Google Scholar]
- Kawai H, Tomii K, Toyooka S, Yano M, Murakami M, Tsukuda K, Shimizu N. Promoter methylation downregulates CDX2 expression in colorectal carcinomas. Oncol Rep. 2005;13(3):547–551. [PubMed] [Google Scholar]
- Kim MJ, Bhatia-Gaur R, Banach-Petrosky WA, Desai N, Wang Y, Hayward SW, Cunha GR, Cardiff RD, Shen MM, Abate-Shen C. Nkx3.1 mutant mice recapitulate early stages of prostate carcinogenesis. Cancer Res. 2002a;62(11):2999–3004. [PubMed] [Google Scholar]
- Kim MJ, Cardiff RD, Desai N, Banach-Petrosky WA, Parsons R, Shen MM, Abate-Shen C. Cooperativity of Nkx3.1 and Pten loss of function in a mouse model of prostate carcinogenesis. Proc Natl Acad Sci USA. 2002b;99(5):2884–2889. doi: 10.1073/pnas.042688999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ko SY, Barengo N, Ladanyi A, Lee JS, Marini F, Lengyel E, Naora H. HOXA9 promotes ovarian cancer growth by stimulating cancer-associated fibroblasts. J Clin Invest. 2012;122(10):3603–3617. doi: 10.1172/JCI62229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ko SY, Lengyel E, Naora H. The Müllerian HOXA10 gene promotes growth of ovarian surface epithelial cells by stimulating epithelial-stromal interactions. Mol Cell Endocrinol. 2010;317(1–2):112–119. doi: 10.1016/j.mce.2009.12.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kroll TJ, Sarraf P, Pecciarini L, Chen CJ, Mueller E, Spiegelman BM, Fletcher JA. PAX8-PPARgamma1 fusion oncogene in human thyroid carcinoma. Science. 2000;289 (5483):1357–1360. doi: 10.1126/science.289.5483.1357. [DOI] [PubMed] [Google Scholar]
- Lee SY, Jeon HM, Kim CH, Ju MK, Bae HS, Park HG, Lim SC, Han SI, Kang HS. Homeobox gene Dlx-2 is implicated in metabolic stress-induced necrosis. Mol Cancer. 2011;10:113. doi: 10.1186/1476-4598-10-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemons D, McGinnis W. Genomic evolution of Hox gene clusters. Science. 2006;313(5795):1918–1922. doi: 10.1126/science.1132040. [DOI] [PubMed] [Google Scholar]
- Lengyel E. Ovarian cancer development and metastasis. Am J Pathol. 2010;177(3):1053–1064. doi: 10.2353/ajpath.2010.100105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li MO, Wan YY, Sanjabi S, Robertson AK, Flavell RA. Transforming growth factor-β regulation of immune responses. Annu Rev Immunol. 2006;24:99–146. doi: 10.1146/annurev.immunol.24.021605.090737. [DOI] [PubMed] [Google Scholar]
- Ma L, Teruya-Feldstein J, Weinberg RA. Tumour invasion and metastasis initiated by microRNA-10b in breast cancer. Nature. 2007;449(7163):682–688. doi: 10.1038/nature06174. [DOI] [PubMed] [Google Scholar]
- Man YG, Fu SW, Schwartz A, Pinzone JJ, Simmens SJ, Berg PE. Expression of BP1, a novel homeobox gene, correlates with breast cancer progression and invasion. Breast Cancer Res Treat. 2005;90(3):241–247. doi: 10.1007/s10549-004-4492-9. [DOI] [PubMed] [Google Scholar]
- Mani SA, Guo W, Liao MJ, Eaton EN, Ayyanan A, Zhou AY, Brooks M, Reinhard F, Zhang CC, Shipitsin M, Campbell LL, Polyak K, Brisken C, Yang J, Weinberg RA. The epithelial-mesenchymal transition generates cells with properties of stem cells. Cell. 2008;133(4):704–715. doi: 10.1016/j.cell.2008.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGinnis W, Krumlauf R. Homeobox genes and axial patterning. Cell. 1992;68(2):283–302. doi: 10.1016/0092-8674(92)90471-n. [DOI] [PubMed] [Google Scholar]
- Micalizzi DS, Wang CA, Farabaugh SM, Schiemann WP, Ford HL. Homeoprotein Six1 increases TGF-beta type I receptor and converts TGF-beta signaling from suppressive to supportive for tumor growth. Cancer Res. 2010;70(24):10371–10380. doi: 10.1158/0008-5472.CAN-10-1354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mills AA. Throwing the cancer switch: reciprocal roles of polycomb and trithorax proteins. Nat Rev Cancer. 2010;10(10):669–682. doi: 10.1038/nrc2931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitsui K, Tokuzawa Y, Itoh H, Segawa K, Murakami M, Takahashi K, Maruyama M, Maeda M, Yamanaka S. The homeoprotein Nanog is required for maintenance of pluripotency in mouse epiblast and ES cells. Cell. 2003;113(5):631–642. doi: 10.1016/s0092-8674(03)00393-3. [DOI] [PubMed] [Google Scholar]
- Mortlock DP, Innis JW. Mutation of HOXA13 in hand-foot-genital syndrome. Nat Genet. 1997;15(2):179–180. doi: 10.1038/ng0297-179. [DOI] [PubMed] [Google Scholar]
- Nakamura T, Largaespada DA, Lee MP, Johnson LA, Ohyashiki K, Toyama K, Chen SJ, Willman CL, Chen IM, Feinberg AP, Jenkins NA, Copeland NG, Shaughnessy JD. Fusion of the nucleoporin gene NUP98 to HOXA9 by the chromosome translocation t(7;11)(p15;p15) in human myeloid leukaemia. Nat Genet. 1996;12(2):154–158. doi: 10.1038/ng0296-154. [DOI] [PubMed] [Google Scholar]
- Naora H, Yang YQ, Montz FJ, Seidman JD, Kurman RJ, Roden RB. A serologically identified tumor antigen encoded by a homeobox gene promotes growth of ovarian epithelial cells. Proc Natl Acad Sci USA. 2001;98(7):4060–4065. doi: 10.1073/pnas.071594398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panganiban G, Rubenstein JL. Developmental functions of the Distal-less/Dlx homeobox genes. Development. 2002;129(19):4371–4386. doi: 10.1242/dev.129.19.4371. [DOI] [PubMed] [Google Scholar]
- Pearson JC, Lemons D, McGinnis W. Modulating Hox gene functions during animal body patterning. Nat Rev Genet. 2005;6(12):893–904. doi: 10.1038/nrg1726. [DOI] [PubMed] [Google Scholar]
- Qi DL, Ohhira T, Fujisaki C, Inoue T, Ohta T, Osaki M, Ohshiro E, Seko T, Aoki S, Oshimura M, Kugoh H. Identification of PITX1 as a TERT suppressor gene located on human chromosome 5. Mol Cell Biol. 2011;31(8):1624–1636. doi: 10.1128/MCB.00470-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raman V, Martensen SA, Reisman D, Evron E, Odenwald WF, Jaffee E, Marks J, Sukumar S. Compromised HOXA5 function can limit p53 expression in human breast tumours. Nature. 2000;405(6789):974–978. doi: 10.1038/35016125. [DOI] [PubMed] [Google Scholar]
- Rauch T, Wang Z, Zhang X, Zhong X, Wu X, Lau SK, Kernstine KH, Riggs AD, Pfeifer GP. Homeobox gene methylation in lung cancer studied by genome-wide analysis with a microarray-based methylated CpG island recovery assay. Proc Natl Acad Sci USA. 2007;104(13):5527–5532. doi: 10.1073/pnas.0701059104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renouf B, Soret C, Saandi T, Delalande F, Martin E, Vanier M, Duluc I, Gross I, Freund JN, Domon-Dell C. Cdx2 homeoprotein inhibits non-homologous end joining in colon cancer but not in leukemia cells. Nucleic Acids Res. 2012;40(8):3456–3469. doi: 10.1093/nar/gkr1242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynolds PA, Sigaroudinia M, Zardo G, Wilson MB, Benton GM, Miller CJ, Hong C, Fridlyand J, Costello JF, Tlsty TD. Tumor suppressor p16INK4A regulates polycomb-mediated DNA hypermethylation in human mammary epithelial cells. J Biol Chem. 2006;281(34):24790–24802. doi: 10.1074/jbc.M604175200. [DOI] [PubMed] [Google Scholar]
- Rinn JL, Kertesz M, Wang JK, Squazzo SL, Xu X, Brugmann SA, Goodnough LH, Helms JA, Farnham PJ, Segal E, Chang HY. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell. 2007;129(7):1311–1323. doi: 10.1016/j.cell.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robson EJ, He SJ, Eccles MR. A panorama of PAX genes in cancer and development. Nat Rev Cancer. 2006;6(1):52–62. doi: 10.1038/nrc1778. [DOI] [PubMed] [Google Scholar]
- Rubin E, Wu X, Zhu T, Cheung JC, Chen H, Lorincz A, Pandita RK, Sharma GG, Ha HC, Gasson J, Hanakahi LA, Pandita TK, Sukumar S. A role for the HOXB7 homeodomain protein in DNA repair. Cancer Res. 2007;67(4):1527–1535. doi: 10.1158/0008-5472.CAN-06-4283. [DOI] [PubMed] [Google Scholar]
- Ruf RG, Xu PX, Silvius D, Otto EA, Beekmann F, Muerb UT, Kumar S, Neuhaus TJ, Kemper MJ, Raymond RM, Brophy PD, Berkman J, Gattas M, Hyland V, Ruf EM, Schwartz C, Chang EH, Smith RJ, Stratakis CA, Weil D, Petit C, Hildebrandt F. SIX1 mutations cause branchio-oto-renal syndrome by disruption of EYA1-SIX1-DNA complexes. Proc Natl Acad Sci USA. 2004;101(21):8090–8095. doi: 10.1073/pnas.0308475101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samuel S, Naora H. Homeobox gene expression in cancer: insights from developmental regulation and deregulation. Eur J Cancer. 2005;41(16):2428–2437. doi: 10.1016/j.ejca.2005.08.014. [DOI] [PubMed] [Google Scholar]
- Sellar GC, Watt KP, Li L, Nelkin BD, Rabiasz GJ, Porteous DJ, Smyth JF, Gabra H. The homeobox gene BARX2 can modulate cisplatin sensitivity in human epithelial ovarian cancer. Int J Oncol. 2002;21(5):929–933. [PubMed] [Google Scholar]
- Shah N, Sukumar S. The Hox genes and their roles in oncogenesis. Nat Rev Cancer. 2010;10(5):361–371. doi: 10.1038/nrc2826. [DOI] [PubMed] [Google Scholar]
- Siegel PM, Massagué J. Cytostatic and apoptotic actions of TGF-beta in homeostasis and cancer. Nat Rev Cancer. 2003;3(11):807–821. doi: 10.1038/nrc1208. [DOI] [PubMed] [Google Scholar]
- Simon R, Lufkin T, Bergemann AD. Homeobox gene Sax2 deficiency causes an imbalance in energy homeostasis. Dev Dyn. 2007;236(10):2792–2799. doi: 10.1002/dvdy.21320. [DOI] [PubMed] [Google Scholar]
- Soshnikova N, Duboule D. Epigenetic regulation of vertebrate Hox genes: a dynamic equilibrium. Epigenetics. 2009;4(8):537–540. doi: 10.4161/epi.4.8.10132. [DOI] [PubMed] [Google Scholar]
- Storti P, Donofrio G, Colla S, Airoldi I, Bolzoni M, Agnelli L, Abeltino M, Todoerti K, Lazzaretti M, Mancini C, Ribatti D, Bonomini S, Franceschi V, Pistoia V, Lisignoli G, Pedrazzini A, Cavicchi O, Neri A, Rizzoli V, Giuliani N. HOXB7 expression by myeloma cells regulates their pro-angiogenic properties in multiple myeloma patients. Leukemia. 2011;25(3):527–537. doi: 10.1038/leu.2010.270. [DOI] [PubMed] [Google Scholar]
- Sumiyama K, Irvine SQ, Ruddle FH. The role of gene duplication in the evolution and function of the vertebrate Dlx/distal-less bigene clusters. J Struct Funct Genomics. 2003;3(1–4):151–159. [PubMed] [Google Scholar]
- Tan Y, Cheung M, Pei J, Menges CW, Godwin AK, Testa JR. Upregulation of DLX5 promotes ovarian cancer cell proliferation by enhancing IRS-2-AKT signaling. Cancer Res. 2010;70(22):9197–9206. doi: 10.1158/0008-5472.CAN-10-1568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiery JP, Acloque H, Huang RY, Nieto MA. Epithelial-mesenchymal transitions in development and disease. Cell. 2009;139(5):871–890. doi: 10.1016/j.cell.2009.11.007. [DOI] [PubMed] [Google Scholar]
- Tlsty TD, Coussens LM. Tumor stroma and regulation of cancer development. Annu Rev Pathol. 2006;1:119–150. doi: 10.1146/annurev.pathol.1.110304.100224. [DOI] [PubMed] [Google Scholar]
- Tommasi S, Karm DL, Wu X, Yen Y, Pfeifer GP. Methylation of homeobox genes is a frequent and early epigenetic event in breast cancer. Breast Cancer Res. 2009;11(1):R14. doi: 10.1186/bcr2233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Topisirovic I, Kentsis A, Perez JM, Guzman ML, Jordan CT, Borden KL. Eukaryotic translation initiation factor 4E activity is modulated by HOXA9 at multiple levels. Mol Cell Biol. 2005;25(3):1100–1112. doi: 10.1128/MCB.25.3.1100-1112.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trinh BQ, Barengo N, Naora H. Homeodomain protein DLX4 counteracts key transcriptional control mechanisms of the TGF-β cytostatic program and blocks the anti-proliferative effect of TGF-β. Oncogene. 2011;30(24):2718–2729. doi: 10.1038/onc.2011.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trinh BQ, Ko SY, Barengo N, Lin SY, Naora H. Dual functions of the homeoprotein DLX4 in modulating responsiveness of tumor cells to topoisomerases II-targeting drugs. Cancer Res. 2013;73(2):1000–1010. doi: 10.1158/0008-5472.CAN-12-3538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandewalle C, Van Roy F, Berx G. The role of the ZEB family of transcription factors in development and disease. Cell Mol Life Sci. 2009;66(5):773–787. doi: 10.1007/s00018-008-8465-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waeber G, Thompson N, Nicod P, Bonny C. Transcriptional activation of the GLUT2 gene by the IPF-1/STF-1/IDX-1 homeobox factor. Mol Endocrinol. 1996;10(11):1327–1334. doi: 10.1210/mend.10.11.8923459. [DOI] [PubMed] [Google Scholar]
- Wang CA, Jedlicka P, Patrick AN, Micalizzi DS, Lemmer KC, Deitsch E, Casás-Selves M, Harrell JC, Ford HL. SIX1 induces lymphangiogenesis and metastasis via upregulation of VEGF-C in mouse models of breast cancer. J Clin Invest. 2012a;122(5):1895–1906. doi: 10.1172/JCI59858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Chen S, Xue M, Zhong J, Wang X, Gan L, Lam EK, Liu X, Zhang J, Zhou T, Yu J, Jin H, Si J. Homeobox D10 gene, a candidate tumor suppressor, is downregulated through promoter hypermethylation and associated with gastric carcinogenesis. Mol Med. 2012b;18:389–400. doi: 10.2119/molmed.2011.00172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X, Chen H, Parker B, Rubin E, Zhu T, Lee JS, Argani P, Sukumar S. HOXB7, a homeodomain protein, is overexpressed in breast cancer and confers epithelial-mesenchymal transition. Cancer Res. 2006;66(19):9527–9534. doi: 10.1158/0008-5472.CAN-05-4470. [DOI] [PubMed] [Google Scholar]
- Wu X, Gao H, Ke W, Hager M, Xiao S, Freeman MR, Zhu Z. VentX trans-activates p53 and p16ink4a to regulate cellular senescence. J Biol Chem. 2011;286(14):12693–12701. doi: 10.1074/jbc.M110.206078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J, Testa JR. DLX5 (distal-less homeobox 5) promotes tumor cell proliferation by transcriptionally regulating MYC. J Biol Chem. 2009;284(31):20593–20601. doi: 10.1074/jbc.M109.021477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yekta S, Tabin CJ, Bartel DP. MicroRNAs in the Hox network: an apparent link to posterior prevalence. Nat Rev Genet. 2008;9(10):789–796. doi: 10.1038/nrg2400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yilmaz M, Maass D, Tiwari N, Waldmeier L, Schmidt P, Lehembre F, Christofori G. Transcription factor Dlx2 protects from TGFβ-induced cell-cycle arrest and apoptosis. EMBO J. 2011;30(21):4489–4499. doi: 10.1038/emboj.2011.319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida H, Broaddus R, Cheng W, Xie S, Naora H. Deregulation of the HOXA10 homeobox gene in endometrial carcinoma: Role in epithelial-mesenchymal transition. Cancer Res. 2006;66(2):889–897. doi: 10.1158/0008-5472.CAN-05-2828. [DOI] [PubMed] [Google Scholar]
- Yu Y, Davicioni E, Triche TJ, Merlino G. The homeoprotein six1 transcriptionally activates multiple protumorigenic genes but requires ezrin to promote metastasis. Cancer Res. 2006;66(4):1982–1989. doi: 10.1158/0008-5472.CAN-05-2360. [DOI] [PubMed] [Google Scholar]
- Zbinden M, Duquet A, Lorente-Trigos A, Ngwabyt SN, Borges I, Ruiz i Altaba A. NANOG regulates glioma stem cells and is essential in vivo acting in a cross-functional network with GLI1 and p53. EMBO J. 2010;29(15):2659–2674. doi: 10.1038/emboj.2010.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng WR, Scherer SW, Koutsilieris M, Huizenga JJ, Filteau F, Tsui LC, Nepveu A. Loss of heterozygosity and reduced expression of the CUTL1 gene in uterine leiomyomas. Oncogene. 1997;14(19):2355–2365. doi: 10.1038/sj.onc.1201076. [DOI] [PubMed] [Google Scholar]
- Zhang H, Muders MH, Li J, Rinaldo F, Tindall DJ, Datta K. Loss of NKX3.1 favors vascular endothelial growth factor-C expression in prostate cancer. Cancer Res. 2008;68(21):8770–8778. doi: 10.1158/0008-5472.CAN-08-1912. [DOI] [PMC free article] [PubMed] [Google Scholar]