Abstract
Background
Promising developments in porcine islet xenotransplantation could resolve the donor pancreas shortage for type 1 diabetic patients. Using GTKO donor pigs with multiple transgenes should extend xenoislet survival via reducing complement activation, thrombus formation, and the requirement for exogenous immune suppression. Studying the xenoantibody response to GTKO/hCD55/hCD59/hHT islets in the pig-to-baboon model, and comparing it with previously analyzed responses, would allow the development of inhibitory reagents capable of targeting conserved idiotypic regions.
Methods
We generated IgM heavy and light chain gene libraries from ten untreated baboons and three baboons at 28 days following transplantation of GTKO/hCD55/hCD59/hHT pig neonatal islet cell clusters with immunosuppression. Flow cytometry was used to confirm the induction of a xenoantibody response. IgM germline gene usage was compared pre and post transplant. Homology modeling was used to compare the structure of xenoantibodies elicited after transplantation of GTKO/hCD55/hCD59/hHT pig islets with those induced by GTKO and wild type pig endothelial cells without further genetic modification.
Results
IgM xenoantibodies that bind to GTKO pig cells and wild type pig cells were induced after transplantation. These anti-nonGal antibodies were encoded by the IGHV3-66*02 (Δ28%) and IGKV1-12*02 (Δ25%) alleles, for the immunoglobulin heavy and light chains, respectively. IGHV3-66 is 86.7% similar to IGHV3-21 which was elicited by rhesus monkeys in response to GTKO endothelial cells. Heavy chain genes most similar to IGHV3-66 were found to utilize the IGHJ4 gene in 85% of V-D regions analyzed. However, unlike the wild type response, a consensus complementary determining region 3 was not identified.
Conclusions
Additional genetic modifications in transgenic GTKO pigs do not substantially modify the structure of the restricted group of anti-nonGal xenoantibodies that mediate induced xenoantibody responses with or without immunosuppression. The use of this information to develop new therapeutic agents to target this restricted response will likely be beneficial for long term islet cell survival and for developing targeted immunosuppressive regimens with less toxicity.
Keywords: Immunogenetics, Antibody, Pancreatic islets, Genetically modified animal, Sus scrofa domestica, Baboon
Introduction
Allotransplantation has provided an effective treatment for patients with type 1 diabetes (T1D) and its debilitating chronic complications (1, 2). Due to the current shortage of donor pancreatic islets, however, transplantation of porcine islets is being considered as a potential alternative (3-6). Survival for over one year with diabetes reversal has now been reported in diabetic NHPs using porcine islets in combination with chronic immunosuppression (3, 4, 6).
Immune-mediated rejection remains a challenge to the survival of genetically modified porcine xenografts (5-9). In pre-clinical studies, various immunosuppressive regimens have been developed to facilitate porcine to primate islet cell transplants, but the most successful regimens represent an immunosuppressive burden that is greater than that currently used in human allotransplantation (7, 10-12). The use of α 1,3 galactosyltransferase gene knockout (GTKO) neonatal porcine islets reduces the immune response and improves the rate of return to normoglycemia (5). Genetic modifications such as the expression of human complement regulatory proteins, hCD55 and hCD59 in transgenic pig donors reduces the rate of complement activation (6, 13-15) and the introduction of additional transgenes may make it feasible to further prolong graft survival. Nevertheless, genetic modification alone is not likely to be sufficient to mitigate rejection given the profound immune barrier existing between the human and pig species. Combination therapies, including those that are directed at xenoantibodies, will need to be developed to improve xenograft survival beyond what is currently achievable using existing strategies.
Our laboratory has defined a selected, restricted usage of immunoglobulin heavy chain variable (IGHV) and immunoglobulin kappa light chain variable (IGKV) genes that encode xenoantibodies in multiple settings (16-19). Although we have previously identified the IGHV utilized in response to GTKO endothelial cells in non-immunosuppressed rhesus monkeys, this report identifies both the immunoglobulin (Ig) heavy and light chain genes that encode the xenoantibodies induced in baboons responding to transplantation with GTKO/hCD55/hCD59/hHT transgenic porcine neonatal islet cell clusters (NICC). Amino acid residues which contribute the binding site of anti-nonGal xenoantibodies are likely to be conserved between systems. Thus, the goal of our study is to identify the structure of xenoantibodies that initiate antibody-mediated injury after genetically modified porcine islet cell xenotransplantation, compare these with previously identified antibody sequences, and ultimately to use this information to rationally design selective immunosuppressive interventions directed at mitigating humoral rejection.
Materials and Methods
Preparation of cDNA libraries and analysis of immunoglobulin gene usage
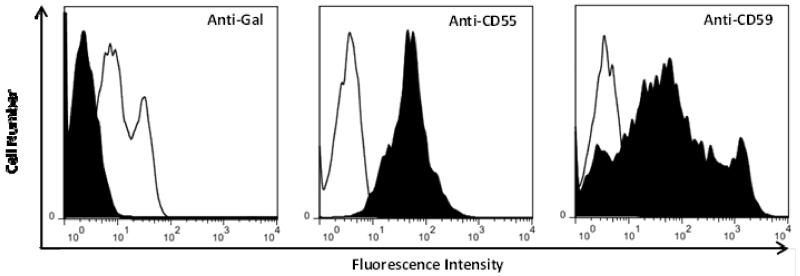
GTKO donor pigs (Sus scrofa) transgenic for hCD55, hCD59, and hHT were generated by Cowan et al (8, 13, 14) and porcine neonatal islet cell clusters (NICC) from these animals were produced and transplanted (10,000 IEQ/Kg) into baboons (Papio hamadryas) at the Westmead laboratories. Prior to NICC isolation and transplantation, expression of Gal, CD55 and CD59 on peripheral blood leukocytes from GTKO/hCD55/hCD59/hHTF was analysed by flow cytometry (Figure 1) as previously described (13) with the exception that Gal expression was detected using a fluorescein isothiocynate conjugated anti-Gal monoclonal antibody (GT6-27-23) which was prepared from a hybridoma kindly provided by Dr. Guerard Byrne, Nextran, Princeton NJ, USA. Baboons were treated with a typical allotransplant immunosuppressive protocol; including a combination of induction with ATG and ongoing treatment with mycophenolate mofetil and tacrolimus. However, because antibodies developed after transplantation, we sought the opportunity to analyze this response. Serum and peripheral blood cell samples from a total of 10 baboons were used to prepare cDNA libraries for the analysis of the immune repertoire in control and transplanted animals. cDNA libraries representing genes encoding xenoantibodies before and after transplantation were constructed as previously described (16). Nested PCR reactions were performed to amplify the product of somatic recombination of either the heavy chain V, D, and J antibody gene progenitors or the light chain V and J antibody gene progenitors (17-20). Ig heavy chain libraries were prepared by amplifying cDNA using a μ-chain-specific primer (Cμ; 5′-GGG AAA AGG GTT GGG GCG GAT GCA-3′) and a framework region (FWR) specific primer for the IgVH 3 family (VH3; 5′-GAG GTG CAG CTG GTG GAG TCT GG-3′). The first PCR reaction for the IgM libraries was then subjected to one additional nested reaction. VH primer (5′-TCT GGG GGA GGC TTG GTC-3′) and Cμ primer pairs were used in the nested PCR. The primers used to amplify immunoglobulin kappa light chain genes were kappa 5 (5′-CAG ATG GCG GGA AGA TGA AG-3′) and CR50 (5′-AGC TCC TGG GGC T(GC) CT(AG)(AC) TGC-3′), which is aligned to the FWR1 of the light chain. The first nested PCR was performed using the CR50 and kappa 4 (5′-ACA GAT GGT GCA GCC ACA G-3′) primers. The second nested PCR reaction was amplified using kappa 4 paired with the primer (5′-TCT GCA TCT GTA GGA GAC-3′).
Figure 1. Flow cytometric analysis of transgene expression of peripheral blood from a GTKO/hCD55/hCD59/hHTF pig (black), compared with a wild type non transgenic pig (white).
GTKO/hCD55/hCD59/hHTF pigs do not express αGal and strongly express human CD55 and human CD59.
The PCR was performed in a TECHNE TC-4000 PCR System Thermocycler for one cycle of 94°C 5min followed by 35 cycles of 94°C for 30 s, 53°C for 30 S, 72°C for 60 s and one cycle of 72°C for 7 min. The PCR products were verified by size on a 2% agarose gel, cloned into the TA 2.1 vector (Invitrogen, Carlsbad, CA), transformed into MAX Efficiency DH5α™ T1R competent cells (Invitrogen), DNA was isolated and the clones were sequenced by the City of Hope DNA sequencing core facility. A minimum of 40 heavy and 40 light chain sequences were obtained per transplanted animal. Sequencing results were analyzed using the NCBI website (http://www.ncbi.nlm.nih.gov/igblast/) to identify the closest human germline progenitor genes. Pre and post transplant Ig gene usage was compared in each animal and in a series of control animals, allowing us to define the normal repertoire of Ig gene usage in baboons and to identify changes in the relative frequency of usage of specific IGHV and IGKV genes.
Flow cytometry
Xenoantibody levels in the sera of recipient baboons were determined at 28 days after transplantation of genetically modified porcine NICC. Heat-inactivated baboon serum samples were diluted 1/10 and were incubated at room temperature with endothelial cells from both GTKO and wild type pigs. Cells were washed twice with cold FACS buffer and incubated with FITC conjugated goat (Fab’) anti-human IgM (Southern Biotech, Birmingham, AL) or FITC conjugated goat anti-human IgG (γ-chain specific) (Sigma, St. Louis, MO). The cells were washed, resuspended in cold FACS buffer and analyzed on a FACSCalibur Flow Cytometer (Becton Dickinson, San Diego, CA) using FlowJo software (Tree stars, Ashland, Oregon).
Homology modeling
Homology modeling, along with further in silico refinement, was used to compare post transplant anti-nonGal xenoantibodies induced by rhesus monkeys in response to GTKO pig endothelial cells without additional genetic modifications (19) and GTKO/hCD55/hCD59/hHT porcine islets. Antibody models were prepared using the Discovery Studio 3.5 software suite (Accelrys, San Diego CA) using representative sequences derived from post transplant IgM xenoantibodies. Each antibody FWR was modeled based on homology using Modeller and crystal structures deposited in the Protein Data Bank (RCSB.org). Antibody complementarity determining regions (CDRs) were modeled separately using the three crystal structures with the highest degree of sequence homology available for each CDR. Ab initio structural refinement as well as molecular dynamic simulations were used to optimize prediction of the heavy chain CDR3 which had the lowest percent homology in each case. For visual comparison, heavy chain models were aligned by the α-carbons and colored by amino acid.
Results
Immunoglobulin heavy and light chain gene usage in untreated baboons
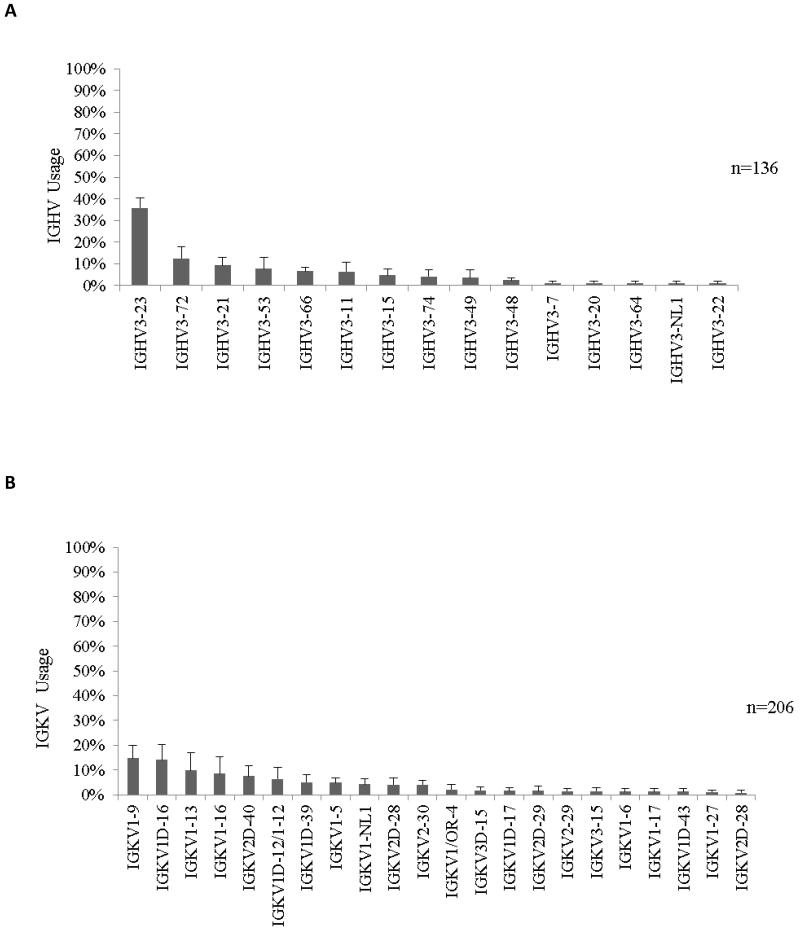
The distribution of heavy and kappa light chain Ig germline gene usage in ten untreated baboons including the three recipient animals was analyzed to identify normal variability within the baboon colony (Figure 2). The Ig heavy chain gene that was used most frequently in untreated baboons most closely resembled human IGHV3-23. The light chain genes used most frequently were most similar to human IGKV1D-16 and IGKV1-9 germline progenitors. There is little information currently available reporting the sequence of baboon germline genes, however, in our experience, the sequences encoding immunoglobulin genes in baboons are very similar to human immunoglobulin gene sequences.
Figure 2. The frequency of IGVH3 and IGKV gene usage in untreated baboons.
(A) The IGHV3-23 germline gene is most frequently utilized in the normal baboon repertoire. The distribution of IGHV3 germline gene usage was determined by sequencing IgVH3 family genes in ten untreated baboons including 3 which later received transplants. (B) IgKV germline gene usage in untreated baboons was determined by sequencing immunoglobulin light chain gene libraries in ten baboons. The IGKV1D-16 and IGKV1-9 germline genes were most frequently used in these animals. Data is represented as average usage per animal ± the standard error of the mean. N= the number of colonies sequenced.
Restricted xenoantibody response to porcine islet transplantation
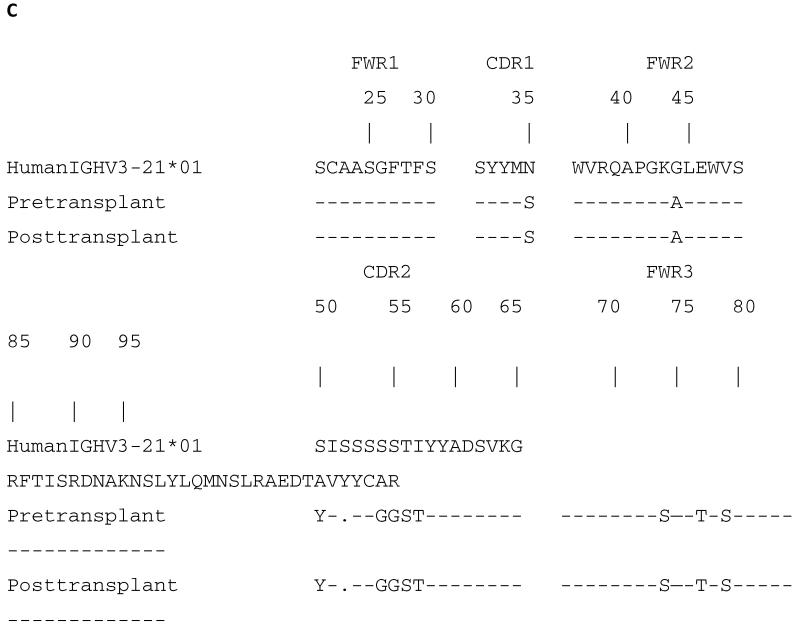
At 28 days after transgenic porcine NICC transplantation, we observed a greater than 2-fold increase in the usage of the IGKV1-12*02 and IGHV3-66*02 light and heavy chain genes relative to pre transplant levels (Figure 3A). Both pre and post transplant levels were determined independently in each baboon. The results were consistent in all animals. Figure 3 shows representative cDNA sequences from libraries that were prepared pre and post transplantation. The germline gene used most frequently to encode xenoantibodies in this model was IGHV3-66, a gene that is closely related but not identical to IGHV3-21, the germline progenitor that encodes xenoantibodies elicited by non-immunosuppressed rhesus monkeys after immunization with GTKO pig endothelial cells without additional genetic modifications. The similarity in the Ig gene sequence when comparing xenoantibodies to GTKO islet cells and IGHV3-21 is shown in Figure 3 B, C. These genes are 86.7% similar at the amino acid level. The xenoantibodies induced in response to transplantation of genetically modified porcine islets are also 92.0% similar to the human IGHV3-11*01 allele encoding human xenoantibodies to wild type pig solid organs or islet cells (17). The light chain gene encoding xenoantibodies to either wild type or genetically modified GTKO cells is identical (21). Representative amino acid sequences from pre transplant and post transplant samples are shown aligned to human IGKV1-12*02 (IGKV1D-12*02) (Figure 4). As demonstrated by flow cytometry in Figure 5, induced xenoantibodies present in the serum at 28 days after transplantation with genetically modified GTKO porcine islets also bind to wild type pig cells. The fact that the response induced by NICC transplantation can be measured using endothelial cells indicates there is likely a prominent common nonGal antigen expressed on both of these cell types. Furthermore, the high degree of sequence similarity in the induced xenoantibodies that bind Gal and nonGal targets supports the concept of an induced, structurally related xenoantibody response initiated after pig cell xenotransplantation.
Figure 3. The anti-nonGal xenoantibody response to genetically modified porcine islets is restricted.
(A) The usage of the immunoglobulin heavy chain gene IGHV3-66*02 was elevated from 7%±0% in pretransplant baboon serum to 35%±6% post-islet cell transplantation. (B) The nucleotide sequence of four representative IGVH genes encoding IgM anti-nonGal xenoantibodies is shown. The IGVH genes encoding anti-nonGal porcine islet xenoantibodies is shown compared to the sequence of the IGHV3-21*01 germline gene which encodes anti-nonGal xenoantibodies to GTKO pig endothelial cells without multiple genetic modifications. (C) The amino acid sequence of IGVH genes encoding anti-nonGal xenoantibodies to genetically modified pig islets is shown. The regions of the CDR1 and CDR2 which are conserved may be relevant for contact with nonGal xenoantigen. (−) Identical residues when compared with the closest human germline gene. FWR (framework region) CDR (complementary determining region).
Figure 4. The immunoglobulin light chain germline genes encoding anti-nonGal xenoantibodies and anti-gal xenoantibodies is identical.
(A) The usage of the IGKV1-12*02 germline gene was 11%±6% in pre-transplant serum and 36%±5% in post-transplant serum. (B) The nucleotide sequence of four representative IGKV genes encoding IgM xenoantibodies following porcine islet transplantation is shown compared to the closest human progenitor, the IGKV1-12*02 germline gene. (C) The amino acid sequence of representative pre and post-transplant samples is shown aligned with the amino acid sequence of the human IGKV1-12*02 gene. The residues which contribute to the gal carbohydrate binding sites of anti-gal xenoantibody are reported in ref (20). Many of these site are conserved on both the heavy and light chain suggesting anti-nonGal and anti-gal xenoantibodies may have similar antibody-antigen interactions. (−)Identical residues when compared with the closest human germline gene. FWR (framework region) CDR (complementary determining region).
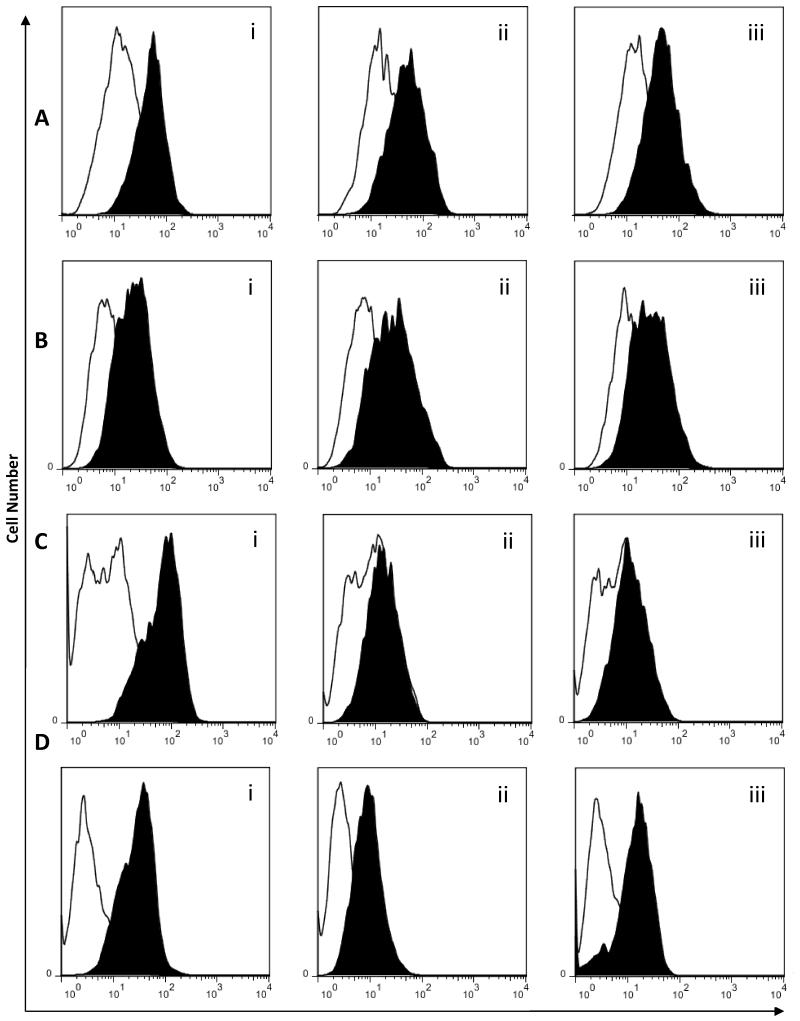
Figure 5. Xenoantibody responses pre (white) and 28 days post (black) transplantation in the sera of three baboons (i, ii, iii) which received genetically modified GTKO porcine islets.
Induced IgM (A&B) and IgG (C&D) xenoantibodies bind to (A&C) wild type pig (1.4-8.5 fold increase in Mean Fluorescence Intensity or MFI)and (B&D) GTKO pig endothelial cells (1.7-5.5 fold increase in MFI).
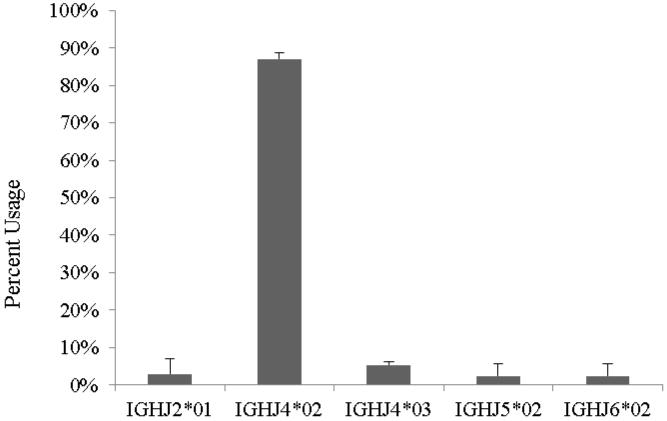
J segment usage
A restricted usage of heavy chain joining (IGHJ) genes was noted when comparing post transplant cDNA libraries with those prepared from pre transplant animals. A diverse selection of immunoglobulin kappa joining (IGKJ) genes were represented pre transplant. The IGHJ4*02 allele accounted for 85% of J segment usage in 40 sequenced cDNA samples per animal examined with a functional IGHV gene (Figure 6). Heavy chain genes most similar to the human IGHV3-66*2 germline gene were found to utilize the IGHJ4*02 allele in 85% of V-D regions analyzed. However, unlike the heavy chain utilized in response to wild type pig solid organs or islet cells (17), our analysis of the encoded antibody sequences indicates that there is no consensus CDR3 amino acid sequence utilized in the anti-nonGal xenoantibody response.
Figure 6. Immunoglobulin J gene usage following islet cell cluster transplantation in baboons.
The IGHJ gene usage was restricted to IGHJ4*02 in all baboons after porcine islet cell transplantation (87%±2%).
Comparison of heavy chain structure in GTKO xenotransplantation
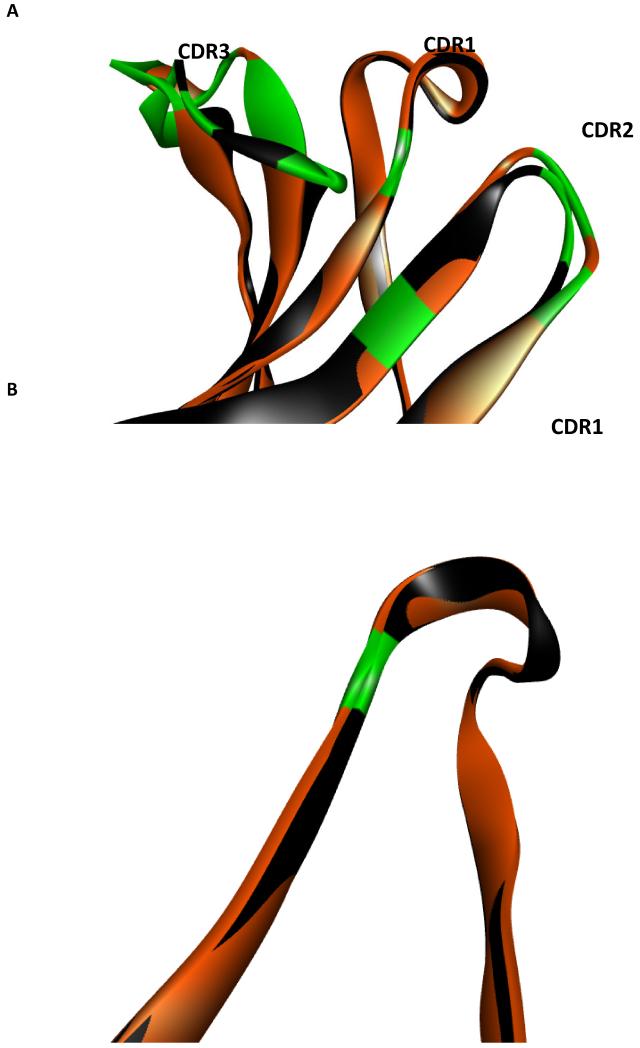
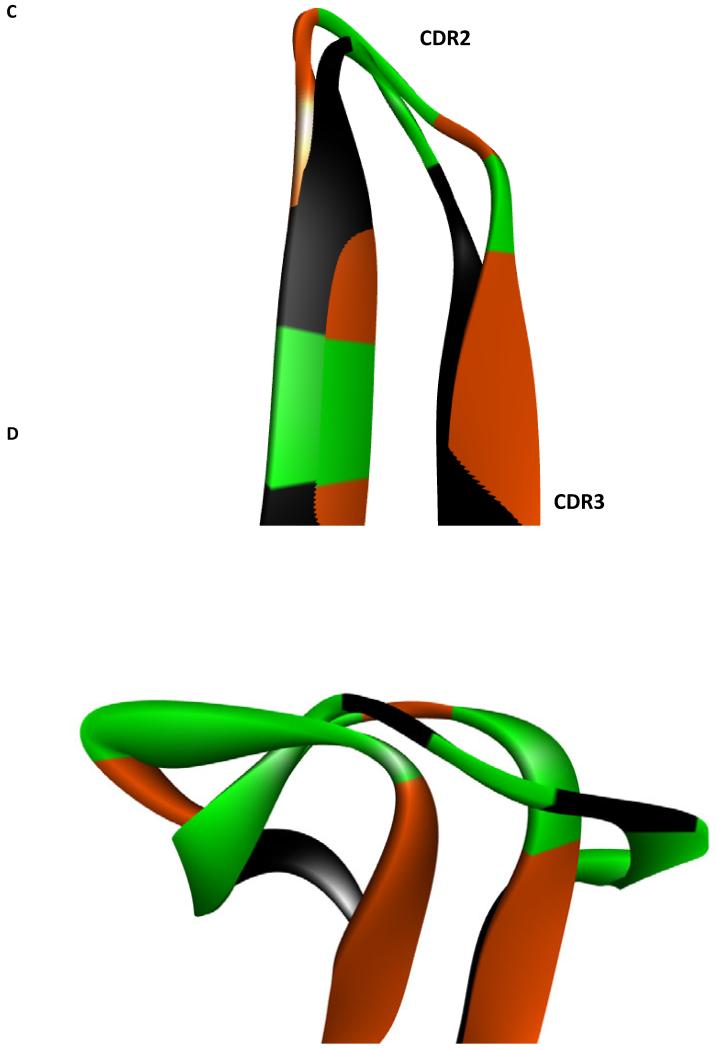
There is predicted to be a high degree of structural homology between the heavy chains utilized in the xenoantibody responses of rhesus monkeys responding against GTKO endothelial cells (19) and baboons responding against GTKO/hCD55/hCD59/hHT transgenic porcine islets (Figure 7). Within the heavy chain variable region, the CDR1 segments are nearly identical (Figure 7b) while only moderate differences exist when comparing the CDR2 segments (Figure 7c). No structural homology is evident when comparing the CDR3 segments (Figure 7d). Thus, amino acid residues which are conserved are likely to make contact with a common antigen.
Figure 7. Structural alignment of modeled anti-non gal heavy chain antibodies.
Structural alignment of modeled xenoantibody heavy chains associated with the immune response induced by GTKO endothelial cells (orange) and GTKO islet cells (black). Residues which are not conserved are highlighted in green on both heavy chains. The high degree of structural similarity suggests conserved amino acids likely make contact with xenoantigen. (A) Complementary determining regions (CDRs) are depicted together. For emphasis, the (B) CDR1, (C) CDR2, and (D) CDR3 are depicted individually. The CDR1 and CDR2 display a high degree of structural similarity. In contrast, the CDR3 does not display this structural similarity. GTKO islet associated CDR3; GTKO endothelial cell associated CDR3. Alignment is by the α-carbon.
Discussion
Our study has demonstrated that the IGVH genes encoding the restricted xenoantibody response to GTKO/hCD55/hCD59/hHT transgenic islets in the pig to baboon model are closely related, but not identical, to IGVH genes encoding the induced xenoantibody response to GTKO endothelial cells without further transgenic modification. A restricted usage of IGHJ4*02 was noted in both models however, the amino acid sequence of the CDR3 segment was not conserved as shown by both sequence analysis and modeling. The light chain genes encoding xenoantibodies, in contrast, are identical when comparing induced xenoantibodies responding to GTKO pig cells or wild type pig cells.
The development of GTKO pigs has played an important role in reducing the xenoantibody response after xenograft transplantation (5, 22, 23). Xenotransplantation using GTKO donors abrogates hyperacute rejection, significantly extending heart survival (22), and resulting in the rapid return of euglycemia (5) after pig heart and islet transplantation in baboons, respectively. The more recent generation of GTKO pigs with additional genetic modifications improves xenograft survival in cardiac models (15) and reduces early islet loss as well (6).
A significant xenoantibody response is induced at 28 days after transplantation of genetically modified GTKO porcine NICC while under immunosuppression. This finding is consistent with recent data from the Kirk laboratory using Gal-deficient pig neonatal islets (5). Transplantation of Gal-deficient islets improves the rate of achieving insulin independence in diabetic NHP, but rejection is not prevented. Elimination of anti-nonGal xenoantibodies may be required to prolong graft survival after porcine islet cell cluster xenotransplantation.
We report here that specific heavy (IGHV3-66) and light chain (IGKV1-12) gene usage was elevated 28% in baboons after transplantation with porcine islet cell clusters. These antibodies pre-exist in the immune repertoire of baboons, albeit at relatively low levels. Monoclonal antibodies encoded by the IGHV3-66*01 can react to a highly conserved influenza A virus epitope (24), and interestingly, both the IGHV3-21 and IGHV3-66 heavy chain IGHV genes in germline formation can contribute to the production of antibodies which inhibit clotting factor VIII in patients with hemophilia A (25). It is currently unknown whether the immune response contributes to coagulative dysfunction in xenotransplantation and further investigation is warranted.
Moderate differences that were identified when comparing the IGHV genes encoding antibodies elicited by GTKO/hCD55/hCD59/hHT transgenic islet cells compared to GTKO pig endothelial cells could be due to a difference in the epitopes expressed. It is also possible that the additional genetic modifications may have mildly influenced the surface antigenicity of the transplanted pig cells. Alternatively, subtle differences between rhesus monkey and baboon IGHV genes encoding immune responses could account for these minor differences in IGHV gene usage. Structural analysis indicates that within the heavy chain, the CDR1 sequences of Ig genes encoding anti-nonGal and anti-Gal xenoantibodies are highly conserved (19, 20, 22, 26). In contrast, the heavy chain CDR2 segment of anti-nonGal xenoantibodies demonstrates mild variability while the CDR3 segment demonstrates no similarity. Those regions which are structurally preserved are more likely to make contact with the relevant nonGal xenoantigen(s). Designing reagents which target these regions may be a promising strategy for interfering with antibody-antigen interactions.
In conclusion, this is the first study to identify the immunoglobulin genes encoding the xenoantibody response following transplantation of GTKO/hCD55/hCD59/hHT transgenic pig islets to baboons. Grafts stemming from different cell types, tissues or organs induce a restricted anti-nonGal xenoantibody response encoded by a small group of structurally restricted immunoglobulin gene progenitors. Potent immunosuppressive regimens may be substantially reduced in pig islet xenotransplantation by blocking these initial IgM responses in a targeted fashion using anti-idiotypic antibodies or small molecular inhibitors selective for these antibodies.
Acknowledgements
We thank Tanya Haddidin, Kathleen Kiernan and Ivan Harnden for technical assistance. This project was funded by NIH grant 7R01AI052079-06 (MKJ).
Abbreviations
- AVR
acute vascular rejection
- α-Gal
Galalpha1-3Galbeta1-4GlcNAc-R
- CDR
complementary determining region
- DAF
decay accelerating factor
- DXR
delayed xenograft rejection
- FWR
framework region
- GTKO
α 1,3 Galactosyltransferase gene knockout
- HAR
hyperacute rejection
- hHT
human α 1,2 fucosyl-transferase
- HRF
homologous restriction factor
- Ig
immunoglobulin
- IGHJ
immunoglobulin heavy chain joining region
- IGHV
immunoglobulin heavy chain variable region
- IGKV
immunoglobulin kappa chain variable region
- NICC
neonatal islet cell clusters
- NHP
nonhuman primate
- T1D
Type 1 Diabetic
- MFI
mean fluorescence intensity
References
- 1.JOHNSON PR, JONES KE. Pancreatic islet transplantation. Semin Pediatr Surg. 2012;21:272–280. doi: 10.1053/j.sempedsurg.2012.05.012. [DOI] [PubMed] [Google Scholar]
- 2.RYAN EA, PATY BW, SENIOR PA, et al. Five-year follow-up after clinical islet transplantation. Diabetes. 2005;54:2060–2069. doi: 10.2337/diabetes.54.7.2060. [DOI] [PubMed] [Google Scholar]
- 3.VAN DER WINDT DJ, BOTTINO R, CASU A, et al. Long-term controlled normoglycemia in diabetic non-human primates after transplantation with hCD46 transgenic porcine islets. Am J Transplant. 2009;9:2716–2726. doi: 10.1111/j.1600-6143.2009.02850.x. [DOI] [PubMed] [Google Scholar]
- 4.HECHT G, EVENTOV-FRIEDMAN S, ROSEN C, et al. Embryonic pig pancreatic tissue for the treatment of diabetes in a nonhuman primate model. Proc Natl Acad Sci USA. 2009;106:8659–8664. doi: 10.1073/pnas.0812253106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.THOMPSON P, BADELL IR, LOWE M, et al. Islet Xenotransplantation Using Gal-Deficient Neonatal Donors Improves Engraftment and Function. Am J Transplant. 2011;11:2593–2602. doi: 10.1111/j.1600-6143.2011.03720.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.AYARES D, PHELPS C, VAUGHT T, et al. Multi-transgenic pigs for xenoislet transplantation. Xenotransplantation. 2013;20:46–46. [Google Scholar]
- 7.SATYANANDA V, HARA H, EZZELARAB MB, et al. New Concepts of Immune Modulation in Xenotransplantation. Transplantation. 2013 doi: 10.1097/TP.0b013e31829bbcb2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.LE BAS-BERNARDET S, TILLOU X, POIRIER N, et al. Xenotransplantation of Galactosyl-Transferase Knockout, CD55, CD59, CD39, and Fucosyl-Transferase Transgenic Pig Kidneys Into Baboons. Transplant P. 2011;43:3426–3430. doi: 10.1016/j.transproceed.2011.09.024. [DOI] [PubMed] [Google Scholar]
- 9.HISASHI Y, YAMADA K, KUWAKI K, et al. Rejection of cardiac xenografts transplanted from alpha1,3-galactosyltransferase gene-knockout (GalT-KO) pigs to baboons. Am J Transplant. 2008;8:2516–2526. doi: 10.1111/j.1600-6143.2008.02444.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.MOHIUDDIN MM, CORCORAN PC, SINGH AK, et al. B-cell depletion extends the survival of GTKO.hCD46Tg pig heart xenografts in baboons for up to 8 months. Am J Transplant. 2012;12:763–771. doi: 10.1111/j.1600-6143.2011.03846.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.MOHIUDDIN MM, SINGH AK, CORCORAN PC, et al. Role of anti-CD40 antibody-mediated costimulation blockade on non-Gal antibody production and heterotopic cardiac xenograft survival in a GTKO.hCD46Tg pig-to-baboon model. Xenotransplantation. 2013 doi: 10.1111/xen.12066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.EZZELARAB MB, EKSER B, ECHEVERRI G, et al. Costimulation blockade in pig artery patch xenotransplantation - a simple model to monitor the adaptive immune response in nonhuman primates. Xenotransplantation. 2012;19:221–232. doi: 10.1111/j.1399-3089.2012.00711.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.COWAN PJ, AMINIAN A, BARLOW H, et al. Renal Xenografts From Triple-Transgenic Pigs Are Not Hyperacutely Rejected But Cause Coagulopathy in Non-Immunosuppressed Baboons. Transplantation. 2000;69:2504–2515. doi: 10.1097/00007890-200006270-00008. [DOI] [PubMed] [Google Scholar]
- 14.WESTALL GP, LEVVEY BJ, SALVARIS E, et al. Sustained function of genetically modified porcine lungs in an ex vivo model of pulmonary xenotransplantation. J Heart Lung Transpl. 2013;32:1123–1130. doi: 10.1016/j.healun.2013.07.001. [DOI] [PubMed] [Google Scholar]
- 15.MCGREGOR CG, RICCI D, MIYAGI N, et al. Human CD55 expression blocks hyperacute rejection and restricts complement activation in Gal knockout cardiac xenografts. Transplantation. 2012;93:686–692. doi: 10.1097/TP.0b013e3182472850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.KEARNS-JONKER M, SWENSSON J, GHIUZELI C, et al. The Human Antibody Response to Porcine Xenoantigens Is Encoded by IGHV3-11 and IGHV3-74 IgVH Germline Progenitors. J Immunol. 1999;163:4399–4412. [PubMed] [Google Scholar]
- 17.KLEIHAUER A, GREGORY CR, BORIE DC, et al. Identification of the VH genes encoding xenoantibodies in non-immunosuppressed rhesus monkeys. Immunology. 2005;116:89–102. doi: 10.1111/j.1365-2567.2005.02204.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.ZAHORSKY-REEVES J, GREGORY C, CRAMER D, et al. Similarities in the immunoglobulin response and VH gene usage in rhesus monkeys and humans exposed to porcine hepatocytes. BMC Immunology. 2006;7:3. doi: 10.1186/1471-2172-7-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.KIERNAN K, HARNDEN I, GUNTHART M, et al. The anti-non-gal xenoantibody response to xenoantigens on gal knockout pig cells is encoded by a restricted number of germline progenitors. Am J Transplant. 2008;8:1829–1839. doi: 10.1111/j.1600-6143.2008.02337.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.KEARNS-JONKER M, BARTENEVA N, et al. Use of molecular modeling and site-directed mutagenesis to define the structural basis for the immune response to carbohydrate xenoantigens. BMC Immunology. 2007;8:3. doi: 10.1186/1471-2172-8-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.FISCHER-LOUGHEED JY, TARANTAL AF, SHULKIN I, et al. Gene therapy to inhibit xenoantibody production using lentiviral vectors in non-human primates. Gene Ther. 2007;14:49–57. doi: 10.1038/sj.gt.3302818. [DOI] [PubMed] [Google Scholar]
- 22.KUWAKI K, TSENG YL, DOR F, et al. Heart transplantation in baboons using 1,3-galactosyl transferase gene-knockout pigs as donors: initial experience. Nat Med. 2005;11:29–31. doi: 10.1038/nm1171. [DOI] [PubMed] [Google Scholar]
- 23.PHELPS CJ, KOIKE C, VAUGHT TD, et al. Production of alpha 1,3-galactosyltransferase-deficient pigs. Science. 2003;299:411–414. doi: 10.1126/science.1078942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.GRANDEA AG, OLSEN OA, COX TC, et al. Human antibodies reveal a protective epitope that is highly conserved among human and nonhuman influenza A viruses. Proc Natl Acad Sci. 2010;107:12658–12663. doi: 10.1073/pnas.0911806107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.GHARAGOZLOU S, KARDAR GA, RABBANI H, SHOKRI F. Molecular analysis of the heavy chain variable region genes of human hybridoma clones specific for coagulation factor VIII. Thromb Haemost. 2005;94:1131–1137. doi: 10.1160/TH05-06-0445. [DOI] [PubMed] [Google Scholar]
- 26.AGOSTINO M, SANDRIN MS, THOMPSON PE, YURIEV E, RAMSLAND PA. In silico analysis of antibody-carbohydrate interactions and its application to xenoreactive antibodies. Mol Immunol. 2009;47:233–246. doi: 10.1016/j.molimm.2009.09.031. [DOI] [PubMed] [Google Scholar]