Abstract
Gene silencing approaches have the potential to become a powerful curative tool for a variety of monogenic diseases caused by gain-of-function mutations. Classical osteogenesis imperfecta (OI), a dominantly inherited bone dysplasia, is characterized in its more severe forms by synthesis of structurally abnormal type I collagen, which exerts a negative effect on extracellular matrix. Specific suppression of the mutant (Mut) allele would convert severe OI forms to the mild type caused by a quantitative defect in normal collagen. Here, we describe the in vitro and ex vivo investigation of a small interfering RNA (siRNA) approach to allele-specific gene silencing using Mut Col1a1 from the Brtl mouse, a well-characterized model for classical human OI. A human embryonic kidney cell line, which expresses the firefly luciferase gene, combined with either wild-type or Mut Brtl Col1a1 exon 23 sequences, was used for the first screening. The siRNAs selected based on their specificity and the corresponding short hairpin RNAs (shRNAs) subcloned in a lentiviral vector were evaluated ex vivo in Brtl fibroblasts for their effect on collagen transcripts and protein. A preferential reduction of the Mut allele of up to 52% was associated with about 40% decrease of the Mut protein, with no alteration of cell proliferation. Interestingly, a downregulation of HSP47, a specific collagen chaperone known to be upregulated in some OI cases, was detected. Our data support further testing of shRNAs and their delivery by lentivirus as a strategy to specifically suppress the Mut allele in mesenchymal stem cells of OI patients for autologous transplantation.
Keywords: allele-specific Col1a1 silencing, siRNA, shRNA, osteogenesis imperfecta, lentiviral vector
Introduction
For an actual cure of genetic diseases, gene therapeutic approaches that provide normal DNA/RNA or correct the DNA mutation represent the treatment of choice. For recessive diseases, even partial replacement of normal transcripts eliminated by null mutations could restore physiological function and relieve the disease phenotype. Dominant disorders, however, are far more complicated because the product of the mutant (Mut) allele negatively affects normal protein function; thus, simply adding extra copies of the normal allele into cells is not efficient. In principle, these diseases require complete allele-specific suppression of the abnormal allele, without affecting expression of its normal counterpart. This is further complicated by the fact that the causative mutation for most single gene diseases varies in sequence and location within the causative gene.
Allele-specific gene silencing by RNA interference (RNAi) is a cutting-edge tool suited to address these problems.1 Small interfering RNA (siRNAs) are double-stranded RNA molecules first identified as a sequence-specific mRNA-interfering species in C. elegans2 and plants,3 and then recognized as a powerful genetic knock-down system in mammalian cells. However, only a few studies have reported successful discrimination between single-nucleotide variants within mRNAs by siRNAs in monogenic dominant disorders, including osteogenesis imperfecta (OI).4, 5, 6, 7, 8 Among these investigations, two demonstrated allele preferential suppression by siRNA in cells expressing both wild type (WT) and Mut alleles.6, 8 No algorithm guarantees siRNAs efficiency, requiring the screening of multiple candidates. A set of siRNA molecules can be rapidly evaluated against a specific aberrant transcript in vitro, and then the promising structures are validated in cell culture assays. Following siRNA sequence selection, a genomic cassette is cloned into appropriate vectors to produce short hairpin RNAs (shRNA), to be processed into siRNA by the cellular machinery.9, 10 Lentivirus-derived vectors enable the high level of stable shRNA essential for gene silencing as a therapeutic strategy for human genetic diseases.
Classical OI is a bone dysplasia caused by mutations in the COL1A1 and COL1A2 genes, coding respectively for the α1 and α2 chains of type I collagen, the major protein of bone extracellular matrix (ECM).11 Over 85% of molecular defects causing classical OI are single-nucleotide changes causing substitution of one of the glycine residue that occur every third residue in the helical Gly-X-Y collagen sequence and are necessary for proper collagen folding.12 No curative treatment is available for OI; physiotherapy for rehabilitation and surgery for bone fixation are the standards of care.13 The administration of bisphosphonates is widely used to increase bone mass by reducing bone turnover, although treatment duration and side effects are still under evaluation.14 More recently, novel pharmacological approaches, such as treatment with anti-RANKL or sclerostin antibodies have come under evaluation, either in preclinical or clinical studies.15, 16
Structural collagen defects are generally associated with moderate-to-severe, even lethal, clinical outcomes (OI type IV, III, II, respectively, based on the Sillence classification17) whereas quantitative defects, because of the presence of a null COL1A1 allele, are responsible for the mildest form of the disease (OI type I). Thus, for clinically significant OI caused by structural defects in collagen, suppression of the Mut allele could in principle convert severe types III and IV to the minimally symptomatic OI type I, a situation particularly favorable to therapeutic strategies based on gene silencing. The promising preliminary data on stem cell transplantation in both murine models18, 19, 20 and human OI21, 22 contributes to an appealing possibility for correcting the patient's own stem cells in vitro, and transplanting them back to the patient, after checking chromosomal rearrangements23 or off-target effects24 in vitro.
In this report, we have utilized the Brtl OI mouse (Col1a1tm1.1Jcm, MGI: 2158863), a well-characterized murine model for the moderately severe dominant form of human OI, which carries a typical glycine substitution in half of its α1(I) collagen chains, to evaluate allele-specific silencing as well as Mut collagen protein suppression. We present in vitro screening of various siRNA against the Brtl Mut Col1a1 allele, as well as ex vivo testing of siRNA and shRNA efficiency and specificity at both transcript and protein levels in cultured fibroblasts.
Materials and methods
Cell cultures
Primary dermal fibroblasts were isolated from newborn Brtl mouse skin, as previously described, and used at low passages (from P3 to P7).25 The HEK 293 FT cells derive from a human kidney cell line modified for optimal production of lentiviral particles (Invitrogen, Cergy-Pontoise, France). Cells were cultured at 37 °C with 5% CO2 in Dulbecco's modified Eagle's medium (DMEM, Lonza, Cologne, Germany) containing 2 mM glutamine and supplemented with 10% fetal bovine serum (FBS, Hyclone Perbio, Bezons, France).
RNAi molecules
Small interfering RNA
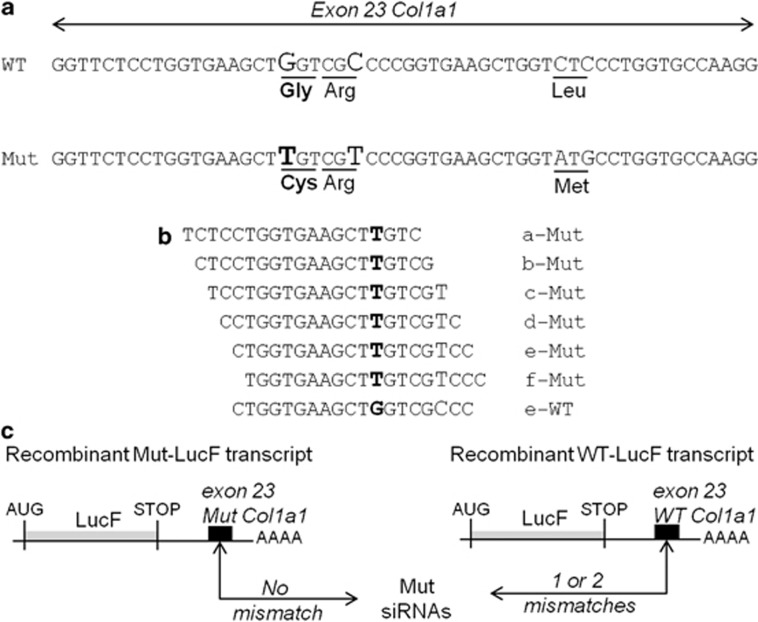
All siRNAs were purchased with 3′ overhanging dTdT from Eurogentec (Angers, France). SiRNAs named a- to f-Mut were designed to target the Mut Col1a1 exon 23 of the Brtl mouse (Figures 1a and b). SiRNA target sequences differed by at least four nucleotides from any known murine gene as shown by BLAST database searches. Three siRNA controls were used: (1) a negative control, named LacZ-siRNA targeting the bacterial galactosidase gene (sense strand 5′-GUGACCAGCGAAUACCUGU-3′), but showing no significant homology to any mouse mRNA sequence according to BLAST database searches, (2) a positive control, named LucF-siRNA (sense strand 5′-CUUACGCUGAGUACUUCGA-3′), designed against the firefly luciferase and (3) a second positive control, indicated as e-WT siRNA, which targeted WT murine Col1a1 exon 23 (Figure 1b).
Figure 1.
Strategy for siRNA validation against the Mut Brtl Col1a1 transcript. (a) Alignment of the sequences of the WT and Mut murine Col1a1 (NM_007742.3) exon 23. Nucleotide variations between WT and Mut sequences are shown in larger characters and the one causing the glycine 349 substitution to cysteine is in bold (b). Sense strands of 19 nucleotides are indicated for the six customer-designed Mut siRNAs to target the sequence around the causative mutation and the WT siRNA. (c) Representation of the recombinant transcripts coding the firefly luciferase and containing either WT or Mut Col1a1 exon 23 sequences (black boxes) in its 3' untranslated region, named recombinant WT-LucF or Mut-LucF transcript, respectively. The number of mismatches between the Mut siRNAs and Col1a1 sequences is indicated.
Short hairpin RNAs
For LacZ- and Mut-shRNAs synthesis, sequences of corresponding siRNAs were used to design 2 oligonucleotides (Eurogentec) with an overlapping sequence as described by Chang et al.26
Generation of Col1a1 WT- or Mut-LucF-HEK cells for siRNA screening
Oligonucleotide duplexes (sequences in Figure 1a, Sigma, Saint-Quentin Fallavier, France) corresponding to 54 base pairs of the WT or the Mut Brtl Col1a1 exon 23 sequence were cloned at the MluI site in the 3' untranslated region of the firefly luciferase gene (LucF) in the pLNT-LucF plasmid.27 Using the ViraPower Lentiviral Expression System (Invitrogen, Saint Aubin, France), the resulting plasmids were used for lentiviral particles production. To measure lentiviral titration and transduction efficiency, EGFP encoding particles were coproduced utilizing pFG12.28 In all, 2–10 EGFP-viral units per cell were added to 2 × 103 HEK 293 FT cells. Transduction efficiency was assessed by EGFP expression analysis by flow cytometry (FC500, Beckman Coulter, Villepinte, France) and luciferase activity measurement, as previously described.27
siRNA transient transfection
For siRNA transfection, 104 HEK-LucF-Col1a1 cells were seeded in 96-well plates. After 24 h, 33 nM of siRNA mixed with 0.75 μl of lipofectamine 2000 reagent (Invitrogen) was added to the cells in triplicate assays. Concerning the primary Brtl fibroblasts, 104 cells were seeded in 24-well plates and transfected 24 h later in triplicate using 1 or 10 nM siRNA mixed with 2 μl of interferin (PolyPLus transfection, Saint Quentin Yvelines, France). Luciferase activity or Col1a1 relative expression were measured 48 h after transfection.
Generation of fibroblasts stably expressing shRNAs
Oligonucleotides corresponding to LacZ- and Mut-shRNAs were annealed and cloned into the pSUPER vector, 3' to the H1 promoter sequence.26 The H1 promoter and the DNA cassette-coding shRNAs were then subcloned in pFG12, following a protocol previously described.29 The resulting constructs were used to produce lentiviral particles. In all, 2–20 EGFP-viral units per cell were added to 3 × 104 Brtl primary fibroblasts. After 7 days in culture, 106 fibroblasts were analyzed by flow cytometry for EGFP expression, and an enriched EGFP+ cell population was generated by cell sorting with 96–98% of purity (FacsAria flow cytometer using Diva software, BD Biosciences, Le Pont de Claix, France).
Proliferation assay
Untransduced, LacZ-shRNA and F-Mut-shRNA-transduced Brtl primary fibroblasts plated at 1 × 104 cells per well in 96-well plates were used for the proliferation assay. Proliferation was measured by pulse labeling in the last 4 h of culture with the CellTiter 96 AQueous One Solution Assay (Promega, Milan, Italy), according to the manufacturer's protocol.
RNA isolation and real-time allele-specific PCR
Total RNA was extracted from modified Brtl fibroblasts (after siRNA transient transfection or shRNA transduction) and unmodified fibroblasts using the Nucleospin kit (Macherey-Nagel, Hoerdt, France). First-strand complementary DNA was synthesized from 100 ng of total RNA in a final volume of 40 μl by the cDNA Archive kit (Applied Biosystem, Monza, Italy). The expression of the Col1a1 gene was evaluated by real-time PCR (qPCR) using the Assay-on-Demand primers–probe set Mm00801666-g1 (Applied Biosystems) in 25 μl final volume, using Mx3000P instrumentation (Stratagene, Cornaredo, Milan, Italy). The expression level of the target gene was normalized to the housekeeping gene Gapdh (Mm99999915_g1, Applied Biosystems) and the relative expression was calculated using the ΔΔCt method. The evaluation of the specific expression of each Col1a1 allele was then performed by means of allele-specific real-time PCR as described in Forlino et al.30
Collagen analysis
F-Mut shRNA transduced and parental Brtl fibroblasts were plated (2.5 × 105/3 cm Petri dishes) and allowed to adhere for 24 h. Collagen synthesis was stimulated by incubating the cells for 24 h before protein recovery with 100 μg/ml sodium ascorbate in DMEM containing 1% FBS. Type I collagen was extracted and purified from media and cell layer as described previously.31 The purified collagen was dissolved in 0.5 M sodium carbonate, pH 8.3, labeled using DyLight550 NHS ester (Thermo Scientific, Euroclone, Milan, Italy) for 20 min at room temperature and separated by 6% SDS-PAGE. The gels were scanned for DyLight fluorescence using Versa Doc 3000 (Bio-Rad, Milan, Italy) and analyzed by Quanty One software (Bio-Rad).
Western blotting
Total proteins were extracted from untransduced, LacZ-shRNA and F-Mut-shRNA-transduced Brtl primary fibroblasts plated in T25 flasks at 5 × 105 cell density and collected at confluency using RIPA buffer (Sigma Aldrich, Milan, Italy) with protease inhibitors (130 mM benzamidine, 2 mM NEM, 5 mM EDTA, 1 mM PMSF). In all, 60 μg of total proteins were separated by SDS-PAGE in denaturing conditions using 10% gels and electro-transferred to a PVDF membrane (Amersham, GE Healthcare, Chalfont St Giles, UK) for 2 h at 100 V. Incubation with primary antibodies against HSP47 and α-tubulin (Santa Cruz Biotechnology, Santa Cruz, CA, USA) and signal detection were performed as previously described.32
Statistical analysis
Statistical comparisons were performed by t-test analysis. A P-value <0.05 was considered statistically significant (two-sided). The analyses were performed using SigmaPlot Statistics 11.0 (SxST, Milan, Italy).
Results
In vitro efficiency and specificity screening of siRNA sequences targeting the Mut Col1a1 exon 23
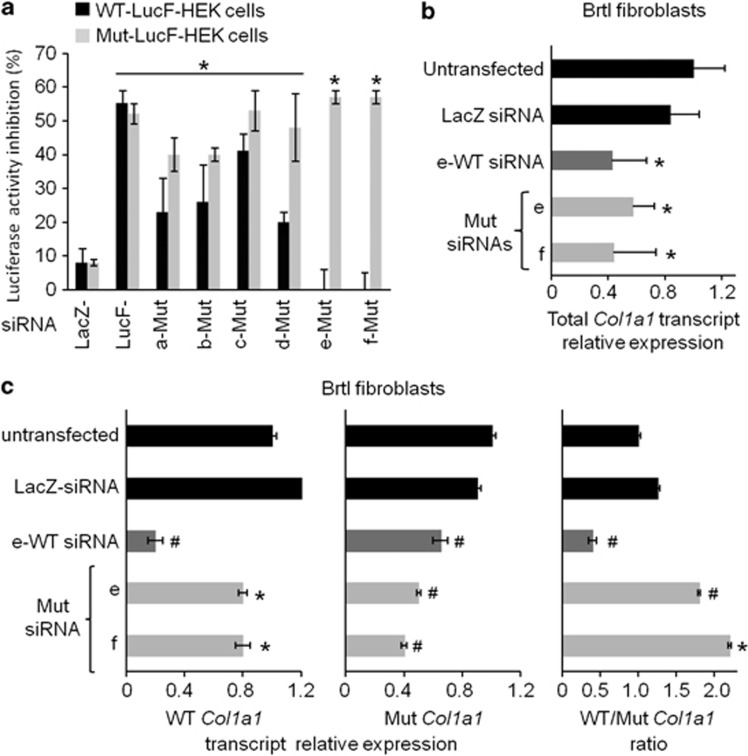
Either WT or Mut Brtl Col1a1 exon 23 sequences were cloned into the 3' untranslated region of the firefly luciferase gene (LucF) in pLNT-LucF (Figures 1a and c).27 Using these plasmids, lentiviral particles were produced to stably transduce HEK 293 cells. We selected two modified HEK 293 cell lines that express similar levels of the recombinant LucF transcript associated with either WT or Mut exon 23 (data not shown). These cell lines were named WT-LucF-HEK and Mut-LucF-HEK, respectively. Six customer-specified siRNAs (a- to f-Mut siRNAs) were designed to target 19 nucleotides of the Mut Col1a1 exon 23 with perfect sequence complementarity, but varied positioning encompassing the causative mutation. Two siRNAs (a-Mut and b-Mut) contain only one mismatch with the WT sequence, corresponding to the causative mutation (c.1546G>T). However, four others (c- to f-Mut) have two mismatches, as they include a second single base change originally introduced in the Brtl construct to create a GUC ribozyme cleavage site (c.1551C>T; Figures 1b and c).25, 33 These siRNAs were transfected into both WT-LucF-HEK and Mut-LucF-HEK cell lines. Silencing of the LucF transcripts was assessed by luciferase activity, as compared with a negative LacZ-siRNA control with no complementary target in mammalian cells, and to a positive LucF-siRNA control, which targeted the LucF open reading frame sequence (Figure 2a). Under transfection conditions, which yielded a 50% inhibition of the luciferase activity from LucF-siRNA, the six Mut siRNAs yielded a similar inhibition (40–53%) of the recombinant LucF transcript containing the Mut Col1a1 exon 23. From a corresponding set of transfections into WT-LucF-HEK cells, e-Mut and f-Mut siRNAs did not produce significant suppression of luciferase activity, indicating that the activation of the RNAi pathway by these molecules was highly specific for the Mut sequence. In contrast, a-, b-, c- and d-Mut siRNAs induced a stronger inhibition of the recombinant WT-LucF transcript (20–41%) than of LacZ-siRNA (8%). These RNAi molecules would generate poor specificity for the Mut sequence.
Figure 2.
In vitro screening of designed siRNA sequences against the Mut Col1a1 exon 23. (a) Inhibition of luciferase activity in HEK 293 cells, which were modified to express either recombinant WT-LucF or Mut-LucF transcript (named WT-LucF-HEK or Mut-LucF-HEK, respectively). The results were obtained 24 h after transfection of LacZ-siRNA, LucF-siRNA or siRNAs designed against the Mut Col1a1 exon 23 (Mut siRNAs a to f). They are expressed as percentage inhibition and compared with baseline luciferase activity of untransfected LucF-HEK cells. Mut siRNA e and f showed the strongest and specific Luciferase inhibition against the Mut allele. Results are expressed as mean±SD of three independent experiments (*P<0.05 as compared with LacZ-siRNA-transfected cells). (b) The relative expression of the total Col1a1 transcript and (c) WT and Mut Col1a1 transcripts were assessed in Brtl fibroblasts transfected with 10 nM LacZ-siRNA, e-WT siRNA or siRNA e-Mut, f-Mut against the Mut Col1a1 and in untransfected cells by real-time PCR. Ratio of WT/Mut Col1a1 transcripts is also shown. Although Mut siRNA e and f significantly inhibited both WT and Mut allele, the WT/Brtl allele ratio was only significantly increased in cells transfected with e-Mut and f-Mut siRNA and not in untransfected cells. The results are expressed as mean±SD of a triplicate experiments (*P<0.05; #P<10−4, referred to untransfected cells).
The e- and f-Mut siRNAs increased the WT/Mut Col1a1 expression ratio in Brtl fibroblasts
The siRNA screening in the HEK 293 cell models did not challenge e-Mut and f-Mut siRNA to discriminate their target sequences within the full Col1a1 transcript and within a context of competition between WT and Mut transcripts. Accordingly, it was now necessary that the selective siRNA molecules be tested for efficiency and specificity in the more physiological environment of fibroblasts from the knock-in Brtl mice. When Brtl fibroblasts were transfected with 10 nM e- or f-Mut siRNA or e-WT siRNA, the overall Col1a1 expression in total RNA was reduced by 43%, 57% and 58%, respectively, compared with untransfected cells (Figure 2b). In comparison, transfection with LacZ-siRNA yielded 17% nonspecific suppression of total Col1a1 (Figure 2b). The specificity of these siRNAs was then evaluated by allele-specific qPCR. The e- and f-Mut siRNAs produced 57% and 65% reduction of the Mut Col1a1 transcript versus 24% and 28% reduction of WT transcripts, respectively, when compared with untransfected cells (Figure 2c). This relative specificity significantly increased the WT/Mut ratio to 1.8 and 2.1 for e-Mut and f-Mut siRNA, respectively, in comparison with the 1.2 transcript ratio obtained after LacZ-siRNA transfection and the 1.0 transcript ratio of untransfected Brtl fibroblasts. Similar results were also observed using only 1 nM siRNAs. Total Col1a1 expression was inhibited by 66% and 46%, respectively, following e-Mut and f-Mut siRNA transfection, with relative Mut allele-specific reduction increasing the normal/Mut Col1a1 ratio to 2.4-fold for the e-Mut and 1.4-fold for the f-Mut siRNA (LacZ-siRNA: 0.8, data not shown).
The E- and F-Mut shRNAs reduced Col1a1 expression, but only F-Mut increased WT/Mut Col1a1 expression ratio in Brtl fibroblasts
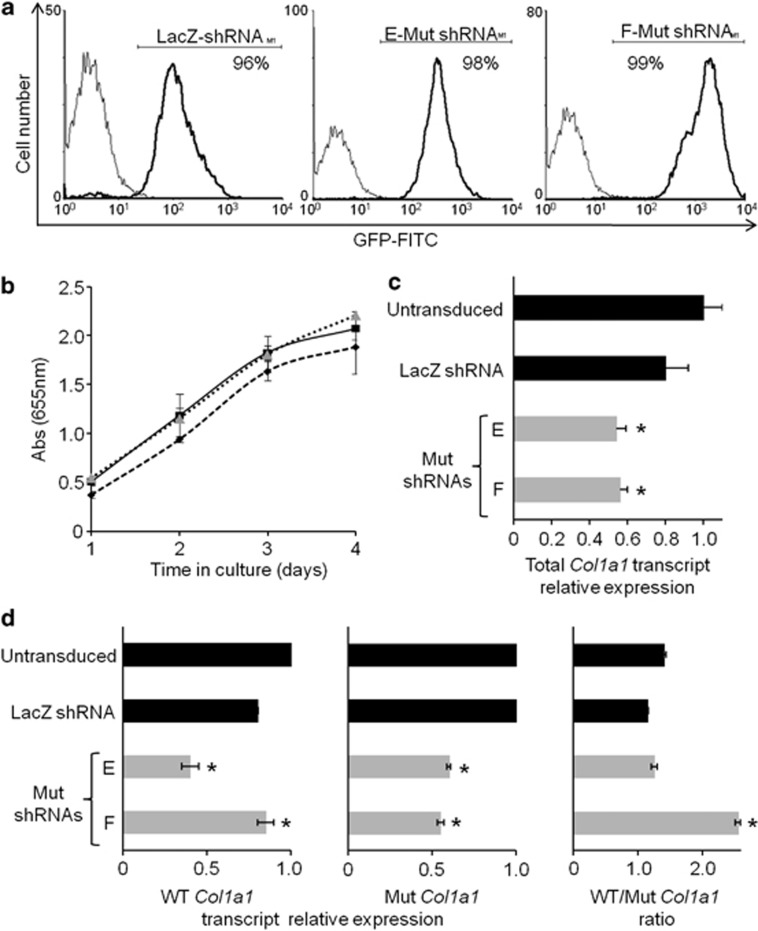
For therapeutic purposes, a substantial inhibition of the Mut transcript, combined with a moderate inhibition of normal allele expression, might be sufficient to correct the ECM of OI patients.34 To this end, we designed cDNA cassettes for E-Mut and F-Mut shRNA synthesis corresponding to e-Mut and f-Mut siRNAs sequences. Lentiviral particles were produced using the pFG12 plasmids containing EGFP and either LacZ-, E-Mut or F-Mut shRNA sequences, and used to stably modify Brtl fibroblasts. The lentivirus transductions of E-Mut or F-Mut shRNA produced efficient modification of cells as shown by flow cytometry analysis (Figure 3a). No changes in cell morphology or density were observed by bright field microscopy after transduction. Furthermore, shRNAs did not affect the proliferation of transduced cells (Figure 3b).
Figure 3.
Stable downregulation of Mut Col1a1 transcript in Brtl fibroblasts following transduction with LacZ-, E-Mut or F-Mut shRNA-expressing lentivirus. (a) Flow cytometry analysis of EGFP demonstrated a similar transduction efficiency for all shRNAs used. Percentages of EGFP positive cells are indicated. (b) Cell viability evaluation by MTT assay. Untransduced (diamond) and shLacZ (square) or F-Mut shRNA (triangle) transduced Brtl fibroblasts were tested at different time points. Each point was measured in triplicate and each assay was repeated in three independent experiments. No difference in proliferation was detected among the untransduced and transduced cells. (c) The relative expression of the total Col1a1 transcript and (d) WT and Mut Col1a1 transcripts were assessed in untransduced and transduced Brtl fibroblasts. Ratio of WT/Mut Col1a1 transcripts is shown. Although shRNA E-Mut and F-Mut significantly inhibited both WT and Mut allele, the WT/Brtl allele ratio was significantly increased only in cells transduced with F-Mut shRNA. The results are shown as mean±SD of a triplicate experiment (*P<0.05, referred to untransduced cells).
The suppression of total Col1a1 transcripts because of stable expression of the shRNAs was investigated. Both E-Mut and F-Mut shRNAs showed comparable decrease of Col1a1 expression compared with untransduced cells (46% and 44% decrease, respectively), which is approximately double the 20% reduction in Col1a1 expression obtained when LacZ-shRNA was used (Figure 3c). Interestingly, and in contrast to the in vitro screening results, only F-Mut shRNA was found to have allele-specific suppression by qPCR, with 52% suppression of the Mut allele and only 14% silencing of the normal Col1a1 transcript. In stably transduced fibroblasts, E-Mut shRNA induced a 51% and 45% inhibition of normal and Mut alleles, respectively (Figure 3d). As a consequence, the normal/Mut Col1a1 transcript ratio is similar for E-Mut or LacZ-shRNA expression, as well as unmodified fibroblasts, whereas Brtl fibroblasts expressing F-Mut shRNAs showed a significant increase of the normal/Mut Col1a1 transcript ratio to 2.5.
The F-Mut shRNA improved the COL1 protein synthesis in Brtl fibroblasts
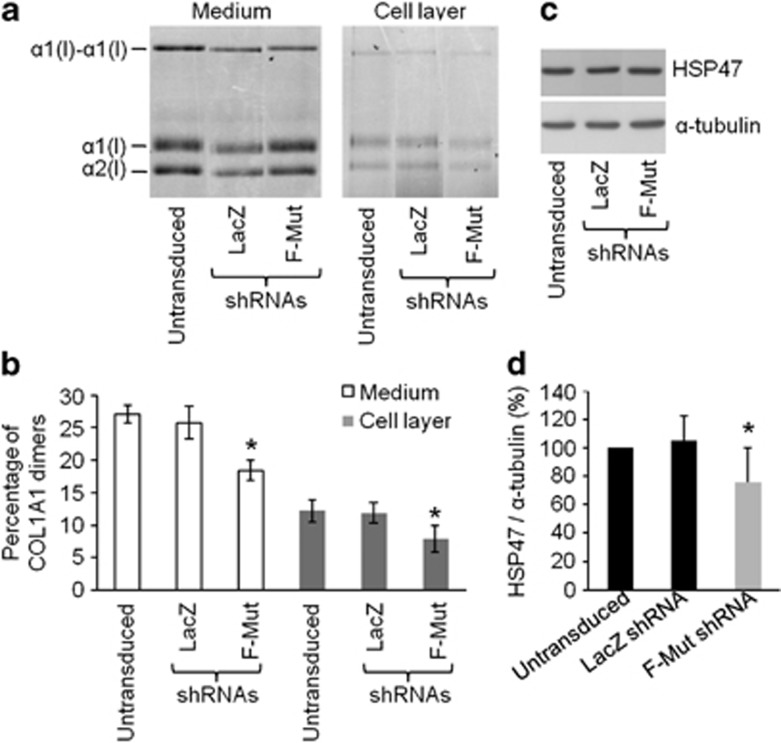
Collagen protein from Brtl fibroblasts transduced with LacZ-shRNA or F-Mut shRNA cassettes was compared with collagen from untransduced cells (Figure 4a). Collagen from media and cell layer was fluorescently labeled and electrophoresed under non-reducing conditions by SDS-PAGE. Measurement of the percentage of α1(I)2 Mut dimers was used to evaluate the proportion of Mut collagen. We found that Mut dimers were reduced from 27.1±1.4% of total secreted α1(I) in untransduced or 25.8±2.5% in LacZ-shRNA-transduced cells, to 18.4±1.6% of total α1(I) in cells with F-Mut shRNA, nearly attaining the expected 15% of total based on F-mut transcript levels (Figure 4b). The cell layer dimer band was significantly reduced (Figure 4b). We also evaluated the ratio of α1(I)/α2(I) bands, which was indeed increased in F-Mut shRNA-transduced cells approaching the expected value of 2 (Supplementary Figure 1). These data demonstrated that the reduction in transcript was reflected at the protein level.
Figure 4.
Evaluation at protein level of collagen chains and HSP47 expression in F-Mut shRNA-expressing Brtl fibroblasts. (a) Representative SDS-PAGE of type I collagen from medium and cell lysates obtained from untransduced and LacZ and F-Mut shRNA-transduced Brtl fibroblasts. (b) Densitometric analysis of the Mut α1(I)–α1(I) dimer percentage as represented in panel a. The Mut collagen was significantly reduced only in Brtl fibroblasts transduced with F-Mut shRNA both in medium and cell layer fractions. The reported values are expressed as mean±SD of three independent experiments. (c) Representative western blot of HSP47 performed on cell lysates obtained from untransduced and LacZ and F-Mut shRNA-transduced Brtl fibroblasts. α tubulin is used as loading control. (d) Densitometry of HSP47 with respect to α tubulin as detected by western blot. HSP47 expression was reduced significantly only in Brtl fibroblasts transduced with F-Mut shRNA. The reported values are expressed as mean±SD of three independent experiments. *P<0.05 referred to F-Mut shRNA-transduced cells versus untransduced fibroblasts and LacZ shRNA-transduced cells.
We also investigated the expression level of HSP47, a specific procollagen molecular chaperone, whose expression was shown to be increased in Brtl cells.32 Interestingly, HSP47 evaluated by western blot demonstrated significant reduction (about 25%) in F-Mut-transduced Brtl cells compared with untransduced or LacZ-transduced Brtl fibroblasts (Figures 4c and d).
Discussion
The highly repetitious primary sequence of type I collagen, along with over a thousand distinct single base substitutions identified in OI patients, presents a significant challenge to an allele-specific targeting approach to treat this disease.12 Nevertheless, previous silencing attempts involving single-nucleotide polymorphisms have been reported against both the 3' untranslated region of COL1A1 and the N-telopeptide coding COL1A2 sequences with encouraging preferential, if not absolute, allele specificity.5, 6
Although antisense oligos achieved a decrease of the Mut α2(I) chain both at transcript and protein levels, this strategy failed to demonstrate specificity because of unacceptable decreases of normal mRNA.35 Specificity was improved using hammerhead ribozymes directed against helical region of both α1(I) and α2(I) chains, in cell-free assays as well as in patient fibroblasts.36 In this study, we have targeted the Mut exonic sequence and the Mut primary fibroblasts from the OI Brtl murine model with si/shRNAs.
The efficiency and specificity of siRNAs are strongly dependent on the position of the mutation along the silencing sequence.37 In general, mismatches in the region 3' to the seed sequence of the siRNA promoted robust single-nucleotide discrimination.38 Nevertheless Ohnishi et al.39 showed that optimal positioning of a single-nucleotide variation depends on the target sequence itself. In this study, we tested a series of six 19 nt siRNAs, named a to f, covering various positions surrounding the causative sequence, as well as an additional silent mutation present in the Mut Brtl allele. All siRNAs designed against the Mut sequence reduced the luciferase expression by targeting the exon 23 reporter alleles, but only two of them, siRNAs e and f showed both high efficiency and high specificity against the Mut construct. SiRNAs a and b, which contained only the G>T causative mutation located in the seed region were potent but poorly specific. Similarly, siRNA c, which contained the causative mutation in the seed region and the silent base change at its 5' end, was not specific. From previous studies, a 5'-end mismatch within the antisense strand could have increased the potency of siRNA;40, 41 however, we observed that siRNA c, which presented such mismatches, was not more efficient than other tested siRNAs. We then compared the efficiency and specificity of siRNA d and e, in which both nucleotide changes were in the seed region, with siRNA f, in which the silent nucleotide change was in the seed region and the causative mutation was 3' of this sequence. Interestingly, siRNAs c and d had two mismatches but poor specificity, supporting the idea that the position of a second mismatch is more important for allele specificity than its simple occurrence. We are aware that the presence of two closely positioned mutations is an unusual situation in OI, but these results further support the idea that introduction of a second mismatch in a siRNA molecule could increase its specificity without affecting efficiency, as already demonstrated in OI using mismatches in the hammerhead ribozymes binding arm as well as in other diseases.33
The two most promising siRNAs determined by the in vitro luciferase assay, namely e and f, were also efficient against the full-length collagen gene in Brtl primary fibroblasts. Both silencing molecules were able to specifically downregulate over 50% of the Mut allele, although they caused a 24–28% inhibition of the WT allele. Overall, this yielded an approximately twofold increase in the WT versus Mut allele ratio.
To evaluate the practical long-term suitability of a silencing molecule as a potential OI therapy, the next required step was to stably express siRNAs in cells. Injections of siRNAs would not be a suitable therapy because their effect is short-lived (2–7 days after injection) and bone tissue delivery is itself a challenge. To achieve stable expression in cells, we used a lentivirus vector system to integrate the siRNA sequence as an shRNA-expressing cassette into the murine genome. Only one of the selected RNAi molecules functioned properly in stably transduced cells, specifically suppressing 52% of the Mut allele with only 14% reduction of WT allele expression. The glycine to cysteine causative substitution (p.Gly420Cys) allowed us to accurately determine for the first time suppression of the Mut protein, using quantitation of the Mut dimeric α1–α1 collagen. A significant reduction of Mut collagen was detected. Our results resemble the data reported by Dawson et al. using a ribozyme-based antisense approach against a Mut Col1a2 allele in patient fibroblasts.33 We speculate that a reduction of Mut protein in Brtl cells could produce a decrease of Mut molecules in the collagen fibrils. Of note, we recently demonstrated that a 2% increase of normal collagen in Brtl bone ECM following in utero transplantation ameliorated the Brtl bone tissue outcome.32
Many inherited skeletal dysplasias are associated with mutations in ECM components and their molecular basis has conventionally been attributed to the presence of an abnormal ECM and impairment of its structural integrity. It is now clear that many mutant proteins are retained intracellularly and accumulate in the ER, causing ER stress and triggering endoplasmic reticulum stress signaling, that, in turn, contributes to the skeletal outcomes.42 We recently described ER stress in Brtl fibroblast and calvarial bone, as well as in MSCs differentiating toward the osteoblastic lineage.30, 31, 32 In the latter case, we showed an increase in HSP47, a specific molecular chaperone known to bind folded type I procollagen molecules in the ER, similar to previous reports in some OI cases.43 Thus, we evaluated the expression of HSP47 following silencing and found a significant reduction in F-Mut shRNA-transduced Brtl cells, compared with control fibroblasts, as might be expected based on about 40% total collagen suppression. This result allows us to speculate that the suppression of the Mut collagen may also ameliorate the ER stress caused by aberrant collagen retention.
Using the endogenous processing machinery, our optimized F-shRNA construct allowed for high potency and sustainable effects. Over 85% of OI patients carry a point mutation either in COL1A1 or COL1A2 and could in principle be treated by this silencing approach. Our data represent a proof of principle, but the issue of si/shRNA delivery in humans remains to be addressed. The choice of lentivirus as delivery system for patient therapy is still a subject of debate. Encouraging results were obtained in a clinical trial using lentiviral vectors, with integration in specific genomic regions not associated with oncogenic selection.44, 45 The availability of Brtl mice will allow us to test such an approach for siRNA delivery by transducing Brtl mesenchymal stem cells with the lentiviral vector expressing the effective F-shRNA and transplanting them back to Brtl, as previously described.20 Other novel tools may allow us to more safely manipulate mammalian cells than the use of lentivirus vectors. The identification in both human and mouse cells of specific and ‘safe' loci for exogenous DNA integration, the AAVS1 and Rosa26, respectively, is particularly appealing.46
We demonstrated for the first time that an RNAi approach can be systematically designed and successfully applied in vitro to develop efficient and allele-specific suppression of even a highly repetitive gene, such as type I collagen, thus opening the potential use of such a gene therapy approach for OI treatment.
Acknowledgments
This study was supported by the Région des Pays de la Loire (JG/ND/RECH N 660, to JR), the Agence Nationale de la Recherche 2007 Project N R07196NS, PRIN20094C2H2M to AR, Cariplo 2011-0270 to AF and AFM TELETHON N.16017 to AF and VT.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on European Journal of Human Genetics website (http://www.nature.com/ejhg)
Supplementary Material
References
- Miller VM, Xia H, Marrs GL, et al. Allele-specific silencing of dominant disease genes. Proc Natl Acad Sci USA. 2003;100:7195–7200. doi: 10.1073/pnas.1231012100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391:806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- Hamilton AJ, Baulcombe DC. A species of small antisense RNA in posttranscriptional gene silencing in plants. Science. 1999;286:950–952. doi: 10.1126/science.286.5441.950. [DOI] [PubMed] [Google Scholar]
- Ding H, Schwarz DS, Keene A, et al. Selective silencing by RNAi of a dominant allele that causes amyotrophic lateral sclerosis. Aging Cell. 2003;2:209–217. doi: 10.1046/j.1474-9728.2003.00054.x. [DOI] [PubMed] [Google Scholar]
- Millington-Ward S, McMahon HP, Allen D, et al. RNAi of COL1A1 in mesenchymal progenitor cells. Eur J Hum Genet. 2004;12:864–866. doi: 10.1038/sj.ejhg.5201230. [DOI] [PubMed] [Google Scholar]
- Lindahl K, Rubin CJ, Kindmark A, Ljunggren O. Allele dependent silencing of COL1A2 using small interfering RNAs. Int J Med Sci. 2008;5:361–365. doi: 10.7150/ijms.5.361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klootwijk RD, Savelkoul PJ, Ciccone C, et al. Allele-specific silencing of the dominant disease allele in sialuria by RNA interference. Faseb J. 2008;22:3846–3852. doi: 10.1096/fj.08-110890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loy RE, Lueck JD, Mostajo-Radji MA, Carrell EM, Dirksen RT. Allele-specific gene silencing in two mouse models of autosomal dominant skeletal myopathy. PLoS One. 2012;7:e49757. doi: 10.1371/journal.pone.0049757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brummelkamp TR, Bernards R, Agami R. A system for stable expression of short interfering RNAs in mammalian cells. Science. 2002;296:550–553. doi: 10.1126/science.1068999. [DOI] [PubMed] [Google Scholar]
- Paddison PJ, Caudy AA, Bernstein E, Hannon GJ, Conklin DS. Short hairpin RNAs (shRNAs) induce sequence-specific silencing in mammalian cells. Genes Dev. 2002;16:948–958. doi: 10.1101/gad.981002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forlino A, Cabral WA, Barnes AM, Marini JC. New perspectives on osteogenesis imperfecta. Nat Rev Endocrinol. 2011;7:540–557. doi: 10.1038/nrendo.2011.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marini JC, Forlino A, Cabral WA, et al. Consortium for osteogenesis imperfecta mutations in the helical domain of type I collagen: regions rich in lethal mutations align with collagen binding sites for integrins and proteoglycans. Hum Mutat. 2007;28:209–221. doi: 10.1002/humu.20429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monti E, Mottes M, Fraschini P, et al. Current and emerging treatments for the management of osteogenesis imperfecta. Ther Clin Risk Manag. 2010;6:367–381. doi: 10.2147/tcrm.s5932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung MS, Glorieux FH. Osteogenesis imperfecta: update on presentation and management. Rev Endocr Metab Disord. 2008;9:153–160. doi: 10.1007/s11154-008-9074-4. [DOI] [PubMed] [Google Scholar]
- Sinder BP, Eddy MM, Ominsky MS, Caird MS, Marini JC, Kozloff KM. Sclerostin antibody improves skeletal parameters in a Brtl/+ mouse model of osteogenesis imperfecta. J Bone Miner Res. 2013;28:73–80. doi: 10.1002/jbmr.1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semler O, Netzer C, Hoyer-Kuhn H, Becker J, Eysel P, Schoenau E. First use of the RANKL antibody denosumab in osteogenesis imperfecta type VI. J Musculoskelet Neuronal Interact. 2012;12:183–188. [PubMed] [Google Scholar]
- Sillence DO, Senn A, Danks DM. Genetic heterogeneity in osteogenesis imperfecta. J Med Genet. 1979;16:101–116. doi: 10.1136/jmg.16.2.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F, Wang X, Niyibizi C. Distribution of single-cell expanded marrow derived progenitors in a developing mouse model of osteogenesis imperfecta following systemic transplantation. Stem Cells. 2007;25:3183–3193. doi: 10.1634/stemcells.2007-0466. [DOI] [PubMed] [Google Scholar]
- Guillot PV, Abass O, Bassett JH, et al. Intrauterine transplantation of human fetal mesenchymal stem cells from first-trimester blood repairs bone and reduces fractures in osteogenesis imperfecta mice. Blood. 2008;111:1717–1725. doi: 10.1182/blood-2007-08-105809. [DOI] [PubMed] [Google Scholar]
- Panaroni C, Gioia R, Lupi A, et al. In utero transplantation of adult bone marrow decreases perinatal lethality and rescues the bone phenotype in the knockin murine model for classical, dominant osteogenesis imperfecta. Blood. 2009;114:459–468. doi: 10.1182/blood-2008-12-195859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horwitz EM, Prockop DJ, Gordon PL, et al. Clinical responses to bone marrow transplantation in children with severe osteogenesis imperfecta. Blood. 2001;97:1227–1231. doi: 10.1182/blood.v97.5.1227. [DOI] [PubMed] [Google Scholar]
- Le Blanc K, Gotherstrom C, Ringden O, et al. Fetal mesenchymal stem-cell engraftment in bone after in utero transplantation in a patient with severe osteogenesis imperfecta. Transplantation. 2005;79:1607–1614. doi: 10.1097/01.tp.0000159029.48678.93. [DOI] [PubMed] [Google Scholar]
- Prockop DJ, Brenner M, Fibbe WE, et al. Defining the risks of mesenchymal stromal cell therapy. Cytotherapy. 2010;12:576–578. doi: 10.3109/14653249.2010.507330. [DOI] [PubMed] [Google Scholar]
- Jackson AL, Bartz SR, Schelter J, et al. Expression profiling reveals off-target gene regulation by RNAi. Nat Biotechnol. 2003;21:635–637. doi: 10.1038/nbt831. [DOI] [PubMed] [Google Scholar]
- Forlino A, Porter FD, Lee EJ, Westphal H, Marini JC. Use of the Cre/lox recombination system to develop a non-lethal knock-in murine model for osteogenesis imperfecta with an alpha1(I) G349C substitution. Variability in phenotype in BrtlIV mice. J Biol Chem. 1999;274:37923–37931. doi: 10.1074/jbc.274.53.37923. [DOI] [PubMed] [Google Scholar]
- Chang JT. An economic and efficient method of RNAi vector constructions. Anal Biochem. 2004;334:199–200. doi: 10.1016/j.ab.2004.07.018. [DOI] [PubMed] [Google Scholar]
- Rousseau J, Escriou V, Perrot P, et al. Advantages of bioluminescence imaging to follow siRNA or chemotherapeutic treatments in osteosarcoma preclinical models. Cancer Gene Ther. 2010;17:387–397. doi: 10.1038/cgt.2009.89. [DOI] [PubMed] [Google Scholar]
- Qin XF, An DS, Chen IS, Baltimore D. Inhibiting HIV-1 infection in human T cells by lentiviral-mediated delivery of small interfering RNA against CCR5. Proc Natl Acad Sci USA. 2003;100:183–188. doi: 10.1073/pnas.232688199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chipoy C, Brounais B, Trichet V, et al. Sensitization of osteosarcoma cells to apoptosis by oncostatin M depends on STAT5 and p53. Oncogene. 2007;26:6653–6664. doi: 10.1038/sj.onc.1210492. [DOI] [PubMed] [Google Scholar]
- Forlino A, Tani C, Rossi A, et al. Differential expression of both extracellular and intracellular proteins is involved in the lethal or nonlethal phenotypic variation of BrtlIV, a murine model for osteogenesis imperfecta. Proteomics. 2007;7:1877–1891. doi: 10.1002/pmic.200600919. [DOI] [PubMed] [Google Scholar]
- Forlino A, Kuznetsova NV, Marini JC, Leikin S. Selective retention and degradation of molecules with a single mutant alpha1(I) chain in the Brtl IV mouse model of OI. Matrix Biol. 2007;26:604–614. doi: 10.1016/j.matbio.2007.06.005. [DOI] [PubMed] [Google Scholar]
- Gioia R, Panaroni C, Besio R, et al. Impaired osteoblastogenesis in a murine model of dominant osteogenesis imperfecta: a new target for osteogenesis imperfecta pharmacological therapy. Stem Cells. 2012;30:1465–1476. doi: 10.1002/stem.1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grassi G, Forlino A, Marini JC. Cleavage of collagen RNA transcripts by hammerhead ribozymes in vitro is mutation-specific and shows competitive binding effects. Nucleic Acids Res. 1997;25:3451–3458. doi: 10.1093/nar/25.17.3451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabral WA, Marini JC. High proportion of mutant osteoblasts is compatible with normal skeletal function in mosaic carriers of osteogenesis imperfecta. Am J Hum Genet. 2004;74:752–760. doi: 10.1086/383252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, Marini JC. Antisense oligodeoxynucleotides selectively suppress expression of the mutant alpha 2(I) collagen allele in type IV osteogenesis imperfecta fibroblasts. A molecular approach to therapeutics of dominant negative disorders. J Clin Invest. 1996;97:448–454. doi: 10.1172/JCI118434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawson PA, Marini JC. Hammerhead ribozymes selectively suppress mutant type I collagen mRNA in osteogenesis imperfecta fibroblasts. Nucleic Acids Res. 2000;28:4013–4020. doi: 10.1093/nar/28.20.4013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rettig GR, Behlke MA. Progress toward in vivo use of siRNAs-II. Mol Ther. 2012;20:483–512. doi: 10.1038/mt.2011.263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz DS, Ding H, Kennington L, et al. Designing siRNA that distinguish between genes that differ by a single nucleotide. PLoS Genet. 2006;2:e140. doi: 10.1371/journal.pgen.0020140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohnishi Y, Tamura Y, Yoshida M, Tokunaga K, Hohjoh H. Enhancement of allele discrimination by introduction of nucleotide mismatches into siRNA in allele-specific gene silencing by RNAi. PLoS One. 2008;3:e2248. doi: 10.1371/journal.pone.0002248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz DS, Hutvagner G, Du T, Xu Z, Aronin N, Zamore PD. Asymmetry in the assembly of the RNAi enzyme complex. Cell. 2003;115:199–208. doi: 10.1016/s0092-8674(03)00759-1. [DOI] [PubMed] [Google Scholar]
- Bumcrot D, Manoharan M, Koteliansky V, Sah DW. RNAi therapeutics: a potential new class of pharmaceutical drugs. Nat Chem Biol. 2006;2:711–719. doi: 10.1038/nchembio839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsang KY, Chan D, Bateman JF, Cheah KS. In vivo cellular adaptation to ER stress: survival strategies with double-edged consequences. J Cell Sci. 2010;123:2145–2154. doi: 10.1242/jcs.068833. [DOI] [PubMed] [Google Scholar]
- Kojima T, Miyaishi O, Saga S, Ishiguro N, Tsutsui Y, Iwata H. The retention of abnormal type I procollagen and correlated expression of HSP 47 in fibroblasts from a patient with lethal osteogenesis imperfecta. J Pathol. 1998;184:212–218. doi: 10.1002/(SICI)1096-9896(199802)184:2<212::AID-PATH996>3.0.CO;2-Z. [DOI] [PubMed] [Google Scholar]
- Cartier N, Hacein-Bey-Abina S, Bartholomae CC, et al. Hematopoietic stem cell gene therapy with a lentiviral vector in X-linked adrenoleukodystrophy. Science. 2009;326:818–823. doi: 10.1126/science.1171242. [DOI] [PubMed] [Google Scholar]
- Biffi A, Bartolomae CC, Cesana D, et al. Lentiviral vector common integration sites in preclinical models and a clinical trial reflect a benign integration bias and not oncogenic selection. Blood. 2011;117:5332–5339. doi: 10.1182/blood-2010-09-306761. [DOI] [PubMed] [Google Scholar]
- Perez-Pinera P, Ousterout DG, Brown MT, Gersbach CA. Gene targeting to the ROSA26 locus directed by engineered zinc finger nucleases. Nucleic Acids Res. 2012;40:3741–3752. doi: 10.1093/nar/gkr1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.