Abstract
There is now good evidence that non-coding sequence variants are involved in the heritability of many common complex traits. The current ‘gold standard' approach for assessing functionality is the in vitro reporter gene assay to assess allelic differences in transcriptional activity, usually followed by electrophoretic mobility shift assays to assess allelic differences in transcription factor binding. Although widely used, these assays have inherent limitations, including the lack of endogenous chromatin context. Here we present a more contemporary approach to assessing functionality of non-coding sequence variation within the Vanin-1 (VNN1) promoter. By combining ‘gold standard' assays with in vivo assessments of chromatin accessibility, we greatly increase our confidence in the statistically assigned functional relevance. The standard assays revealed the −137 single nucleotide variant to be functional but the −587 variant to have no functional relevance. However, our in vivo tests show an allelic difference in chromatin accessibility surrounding the −587 variant supporting strong functional potential at both sites. Our approach advances the identification of functional variants by providing strong in vivo biological evidence for function.
Keywords: pantetheinase, allele-specific chromatin arrangement, cardiovascular disease, functional variant
INTRODUCTION
There is growing evidence that non-coding genetic variation has an important role in the determination of individual susceptibility to complex disease.1, 2 Single nucleotide variants (SNVs) account for approximately 90% of all known sequence variation, most of which is located in non-coding regions of the genome.3 It is therefore likely that a significant number of SNVs that influence phenotype, that is, functional variants, actually lie outside amino-acid coding regions of genes and affect the regulation of gene expression as opposed to protein sequence and function.4, 5 Current estimates from the 1000 genomes data indicate that there are as many non-coding functional variants as coding ones.3 One of the greatest challenges currently facing genetics is determining which non-coding genetic variants have a functional effect on gene expression rather than being neutral.
The primary purpose of the study is to identify the functional variants that have been identified previously using an integrative genetics approach. We have previously used this approach to identify novel cis-acting genes that correlate with HDL cholesterol levels, a predictive determinant for cardiovascular disease.6 Leukocyte-based RNA samples from 1240 individuals in the San Antonio Family Heart Study (SAFHS)7 were used to obtain genome-wide transcriptional profiles. Analysis identified 67 cis-regulated transcripts whose expression levels significantly correlated with HDL-C levels. One gene, Vanin-1 (VNN1 (MIM No. 603570)), showed strong support for both cis-regulation and positive correlation with HDL-C levels. VNN1 is a pantetheine hydrolase (pantetheinase)8 that catalyses the hydrolysis of pantetheine to pantothenic acid (Vitamin B5) and cysteamine, a potent antioxidant.9, 10 Administration of pantethine, the dimer of pantetheine, to hyperlipidemic subjects results in the lowering of serum TG, low-density lipoproteins and ApoB, whilst increasing HDL-C and ApoA levels,11, 12 supporting our data implicating Vanin-1 in lipid biosynthesis (for a review, see Kaskow et al13).
The objective of the current study was the identification and molecular characterization of the functional cis-acting sequence variation in the promoter of VNN1. The ‘gold standard' approach for determining whether cis-regulatory elements such as SNVs regulate gene expression consists of the reporter gene assay14, 15, 16 and the electrophoretic mobility shift assay (EMSA).17, 18 In the reporter gene assay, allelic promoter regions are cloned upstream of a reporter gene such as luciferase, transfected into relevant cell lines and assayed for reporter gene activity as a measure of transcriptional activity. Differences in activity between the allelic constructs are considered to reflect functional differences. The EMSA is used to detect allelic differences in transcription factor binding when comparing nucleotide sequence probes carrying specific SNVs (for a review of these approaches, see Knight19) Although these in vitro approaches have traditionally been considered adequate to ascertain the functionality of sequence variation, there are many variables that can confound the results and render the ascertainment of function inconclusive, as we have shown previously.20 This led us to develop strategies that assess function in an in vivo context.21 Here we use traditional approaches and also further develop in vivo approaches22 to validate statistically prioritized variants within the VNN1 promoter. In addition, we illustrate the potential of a novel, robust method for assessing variant's functionality on a genome-wide level23 that we have developed based on next-generation sequencing technology. The combination of the ‘gold standard' approaches and our more rigorous in vivo tests allows for a more confident assessment of whether a variant does indeed regulate gene expression and yields insights into the allele-specific mechanism of action.
METHODS
Nucleotide numbering
Numbering of nucleotides reflects transcription initiation site numbering as defined before.6 The transcription initiation site is defined as the nucleotide 13 basepairs upstream of the A of the ATG translational initiation codon of VNN1 (GenBank accession number NG_012147.1). In reference to the hg19 genome, the translation initiation codon is located at position chr6:133035174 with −137 (rs4897612:G>T; −150) at chr6:133035325 and −587 (rs2050153:G>A; −600) at chr6:133035775 according to 1-based numbering. The alternative translation initiation numbering is provided in brackets at the first mention in the text. All SNVs can be found in the dbSNP database.24
Statistical genetics
The description of the genotyping and statistical genetic methods used to prioritize the VNN1 promoter variants have been previously described by Goring et al.6 The population stratification, measured genotype, quantitative trait disequilibrium test (QTDT) and quantitative trait linkage disequilibrium (QTLD) results for the −137 SNP (rs4897612:G>T) were computed on the inverse-normalized VNN1 mRNA phenotype using the qtld command in the SOLAR computer package.25 This procedure performs a test for population stratification as well as the commonly used association tests, QTDT26 and the measured genotype test.27 The QTDT procedure is not limited to the scoring of allele transmission from parents to offspring but assesses the entire pedigree structure of the SAFHS cohort, thus increasing the power to detect an association.28 The measured genotype test uses a standard threshold model assuming an underlying normal distribution of liability. The measured genotype graph for the −137 SNP was generated using geom_boxplot in ggplot2.29
Cell culture
The Jurkat E6-1 cell line (ATCC No. TIB-152) was cultured in RPMI-1640 with L-glutamine (Life Technologies Co., Grand Island, NY, USA) supplemented with 100 g/ml of penicillin/streptomycin (Life Technologies Co.) and 10% heat inactivated FBS (Life Technologies Co.). Cells were maintained at 37 °C with 5% CO2 according to cell-specific guidelines. Epstein Bar Virus -transformed lymphoblastoid cell lines were created from the SAFHS cohort. Cell lines A04210, A01223, A01518, A02238, A00263, A01721 and A01897 were maintained as described above.
Reporter gene assays
To create reporter gene constructs, approximately 2 kilobasepairs (kb) of the VNN1 proximal promoter from −2021 to +14 was cloned into the pGL4.10 vector between the Acc65I and HindIII restriction cut sites immediately upstream of the firefly luciferase reporter gene. A construct representing the −587G/−137G haplotype was initially generated and subsequently each variant was mutated to the other allele using the Quikchange II Site-Directed mutagenesis kit (Agilent Technologies Pty Ltd, Melbourne, VIC, Australia) according to the manufacturer's instructions. Genotypes were confirmed by sequencing.
Mycoplasma-free Jurkat E6-1 cells were transiently transfected using the Amaxa Nucleofector II system (Lonza Inc., Walkersville, MD, USA) according to the manufacturer's instructions. In brief, 2 × 106 cells were harvested on day 7 of growth and resuspended in solution V. One microgram of DNA and 100 ng pRL-TK control vector was transfected per reaction using the setting X-001. Twenty-four hours post transfection, the cells were harvested, and luciferase activity was measured using the Dual luciferase Reporter Assay System (Promega Co., Sydney, NSW, Australia). Statistical significance was determined by the Student's unpaired t-test. P<0.05 was considered significant.
EMSA
Complementary 5′ biotinylated oligonucleotides representing both allelic forms of the promoter variants were obtained commercially (Operon Biotechnologies, Inc., Huntsville, AL, USA). The sequences of the EMSA oligonucleotides VNN137T, VNN137G, VNN587G and VNN587A: 5′ are shown in Table 1. Nuclear extract preparation, binding reactions and gel analysis and detection were as described previously.6, 30 For EMSA supershift reactions, 500 ng human Sp1 supershift-grade antibody (Santa Cruz Biotechnology Inc., Santa Cruz, CA, USA) was incubated with the nuclear extract for 30 min on ice, then incubated with the labeled oligonucleotide for 30 min on ice before analysis.
Table 1. Sequences of EMSA and PCR oligonucleotides used in this study.
| Name | Oligonucleotide sequence (5′–3′) |
|---|---|
| VNN137T | ATCTTCACAAATTACTGCAATAAGGTAGGGTCTTTTGTTATGTAACAATG |
| VNN137G | ATCTTCACAAATTACTGCAATAAGGGAGGGTCTTTTGTTATGTAACAATG |
| VNN587G | GAGAGGCGGAGGTTGCAGTGAGCCGAGATTTCGCCACTGCACTCTAGCCT |
| VNN587A | GAGAGGCGGAGGTTGCAGTGAGCCAAGATTTCGCCACTGCACTCTAGCCT |
| −137 Chart F | CCTAGCGAACTAATGTACATAGAGTTCTTGAG |
| −137 Chart R | AACGCTATGATTTCATAGCATTGTTACATAAC |
| −587 Chart F | GCTACTTGGGAGGCTGAGG |
| −587 Chart R | CTTGTCACCCAGGCTAGAGTGC |
| VNNB F | CAGTGAGCCTGTACCCCTGG |
| VNNB R | TATTCTCCCCATGTGTGTGCC |
Chromatin arrangement assay
Chromatin arrangement assays were performed as previously described22, 31, 32 with the following modifications. Nuclei were prepared and digested with nuclease using approximately 1 × 107 genotyped cells from the SAFHS cohort. DNA were purified using the QIAamp DNA blood mini kit (Qiagen Pty Ltd, Doncaster, VIC, Australia). Quantitative real-time PCR was performed using 100 ng DNA, 0.5 μM of primers (−137 Chart F and −137 Chart R or −587 Chart F and −587 Chart R—see Table 1) and SensiMix SYBR No Rox (Bioline (Aust) Pty Ltd, Alexandria, NSW, Australia) in a final reaction volume of 20 μl. Thermal cycling conditions for the CFX96 Real-Time Detection System (Bio-Rad Laboratories Inc., Hercules, CA, USA) were as follows: 95 °C for 10 min; followed by 40 cycles of 95 °C for 15 s, 58 °C for 15 s and 72 °C for 15 s. Percentage of chromatin accessibility was calculated as a ratio of relative abundance of digested to undigested amplicon, a measurement calculated using the CFX Manager software (Bio-Rad Laboratories). Statistics were performed using GraphPad Prism v4 (GraphPad Software, San Diego, CA, USA). Statistical significance was determined by the Student's unpaired t-test. P<0.05 was considered significant.
Bisulfite sequencing
Genomic DNA was isolated from cultured human leukocyte cell lines and purified using DNA Blood mini columns (Qiagen). Purified DNA was treated with bisulfite to convert unmethylated cytosine bases into uracils and subsequently purified using EpiTect spin columns (Qiagen). The converted DNA was enriched for the VNN1 gene promoter by PCR using primers VNNB F and VNNB R (Table 1) flanking the predicted CpG island (between −758 to −496 upstream of the VNN1 transcriptional start site). The amplification products were cloned into the pCR2.1-TOPO vector (Life Technologies Co.) for sequencing (Australian Genome Research Facility, Perth, WA, Australia). Sequences were analyzed using the online analysis program Bisulfite Sequencing DNA Methylation Analysis (BISMA) to identify the methylation state of predicted CG dinucleotides in CpG islands.33
Chromatin arrangement–sequencing (CHA-seq)
Nuclei were isolated from 1 × 107 cells of an immortalized leukocyte cell line (ID: A01721) heterozygous at −137 and −587 of VNN1 identified by our previous sequence analysis of the VNN1 promoter.6 Isolated nuclei were digested with micrococcal nuclease (New England Biolabs Inc., Ipswich, MA, USA) for 20 min at 37 °C, and DNA purified using DNA Blood mini columns (Qiagen). DNA fragment sizes, including mononucleosomal and dinucleosomal fragments as well as intervening sizes (140–280 basepairs), were isolated following agarose gel electrophoresis to maximize the capture of non-nucleosomal nuclease sensitivity. Purified fragments were prepared for next-generation sequencing using the NEBNext DNA sample prep kit (New England Biolabs) according to the manufacturer's protocol. The prepared library was enriched for VNN1 sequences using the SureSelect DNA capture array (Agilent Technologies Inc., Santa Clara CA, USA), then sequenced on an Illumina Genome Analyser II (Illumina Inc., San Diego, CA, USA). The sequence data were analyzed using the Integrative Genomics Viewer (IGV) 2.0 software.34
RESULTS
Identification of putative functional variants within the VNN1 promoter
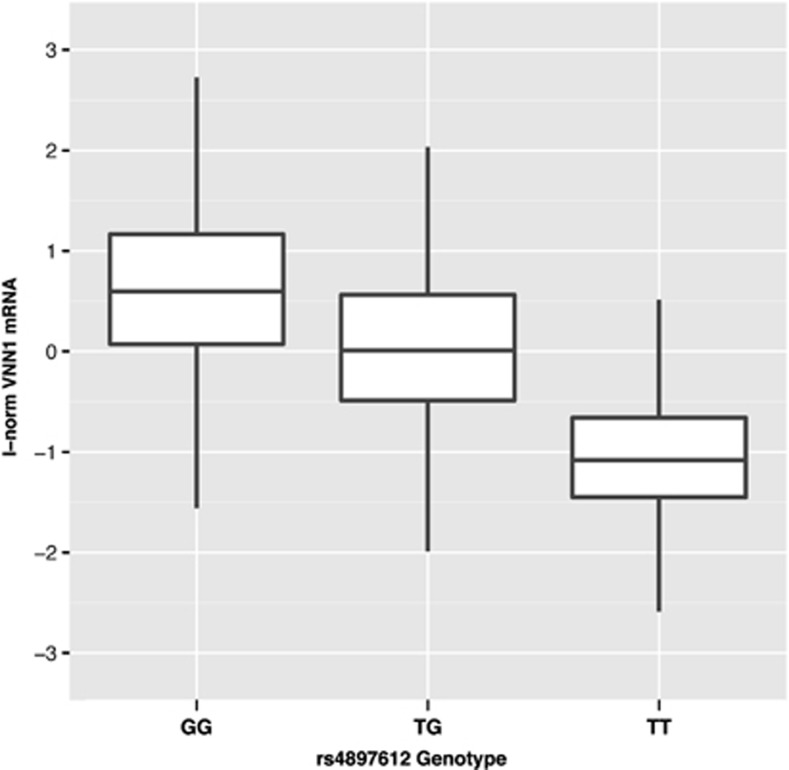
To identify cis-acting functional elements within the VNN1 promoter, we previously re-sequenced 2 kb of proximal promoter upstream from the VNN1 transcriptional start site and identified 22 promoter variants in the SAFHS founder population.6 The 6 best candidate variants were typed in all 1240 SAFHS samples to assess association with VNN1 transcript level in the larger population. Analysis revealed five variants strongly associated with VNN1 transcript abundance; the −137 (rs4897612:G>T) and −587 (rs2050153:G>A) variants showing the strongest relationship. In the current study, the population stratification, measured genotype, QTDT and QTLD results for the −137 variant were computed on the inverse-normalized VNN1 mRNA phenotype. The measured genotype results for the −137 variant, demonstrating an additive genetic effect, are shown in Figure 1. The strong linkage disequilibrium (LD) between the −137 and −587 SNPs (ρ2=0.99) impeded further statistical prioritization, as the two variants were providing essentially identical genetic information. Functional validation was necessary to resolve whether one or both of the variants was responsible for influencing the expression of the mRNA.
Figure 1.
Box and whisker plot for the measured genotype analysis of the −137 SNP on VNN1 mRNA abundance in leukocytes from the SAFHS participants. The absence of the G allele strongly impairs expression (P=5.7 × 10−83). The mRNA phenotype has been inverse-normalized for this analysis, resulting in a population mean of zero, with units approximating the SDs.
‘Gold standard' functional tests of the −137 and −587 VNN1 promoter SNPs
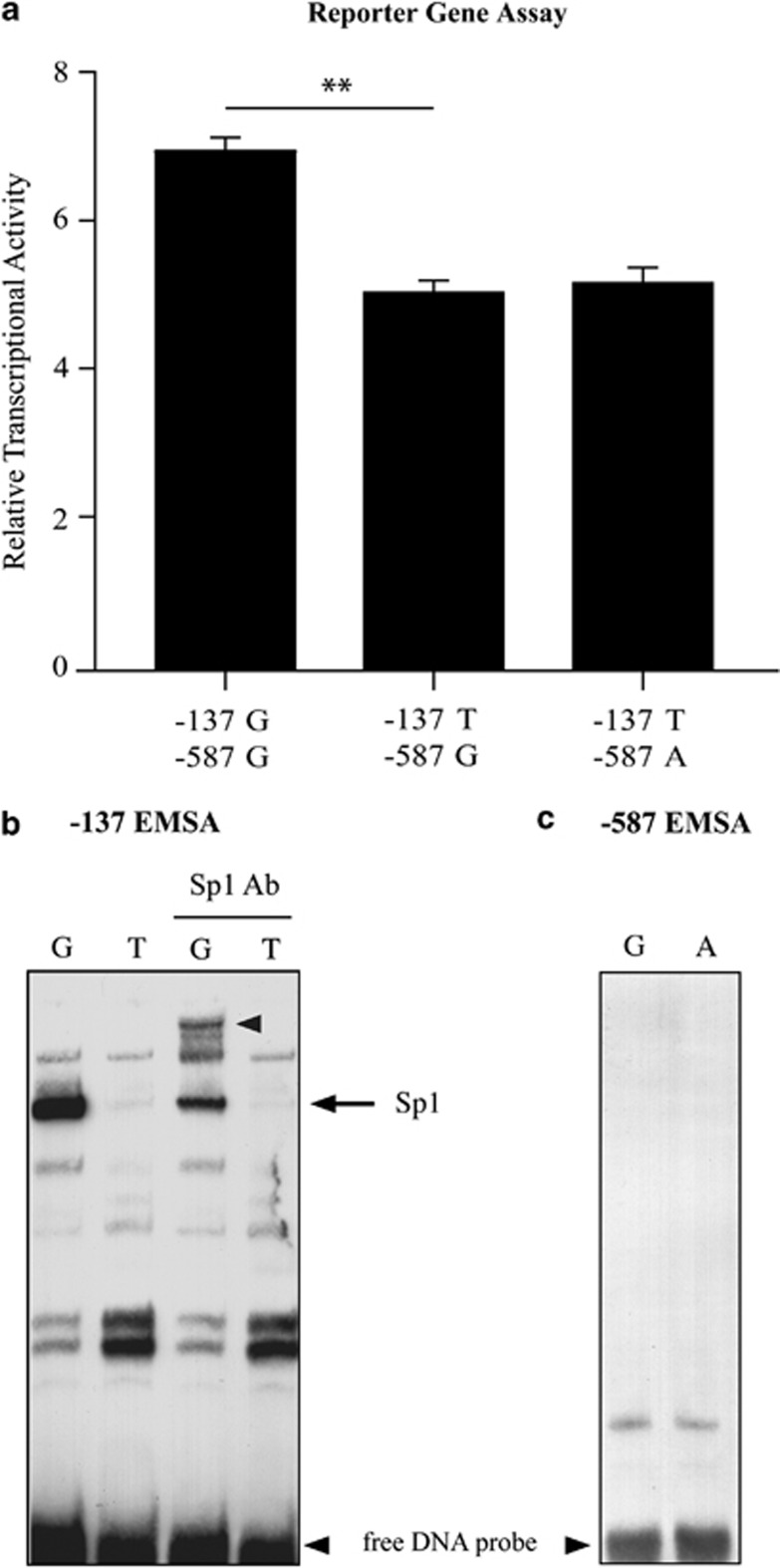
In a previous study, we reported the identification of several SNVs within the proximal promoter of the VNN1 gene that showed significant association. The variant most highly correlated with VNN1 transcript levels in leukocytes (−137) was also shown by EMSA to exhibit differential nuclear factor binding supporting a potential regulatory role.6 To more fully assess the functional attributes of the −137 SNP and the next most significantly correlated variant at position −587, we have now used the ‘gold standard' approaches for characterizing variant functionality. Data from the interrogation of the ENCODE databases using RegulomeDB35 did not provide any convincing evidence that the SNPs were located in a transcriptionally active region, although there is a paucity of ENCODE data for lymphocytes, the cell type with which the original study was done. Also, no ChIP-seq data were available for transcription factor Sp1, which we have tentatively identified as being relevant previously,6 and no evidence was found for the binding of miRNA or other non-coding RNAs. For the gold standard tests, a 2-kb region of the VNN1 promoter was inserted upstream of a firefly luciferase reporter gene. A construct representing the major −137T haplotype (−137T/−587G) was generated initially and subsequently mutated to the other allele to allow the independent assessment of variants at each site. Reporter gene analysis indicated that the −137G allele showed significantly higher expression (1.36-fold, P=0.005) than the T allele (Figure 2a). The functional attributes of the −137 alleles were also tested in the context of both −587 alleles. The results indicated that the −587 SNP alleles did not influence transcriptional activity differences due to the −137 alleles in this assay.
Figure 2.
Gold standard assays assessing −137 and −587 SNP functionality. (a) Relative transcriptional activity of VNN1 promoter variant haplotypes transfected in Jurkat E6-1 cells is shown as a measure of firefly luciferase activity normalized to renilla activity. (n=3, bars represent mean±SEM, **P<0.005) (b) EMSA of the −137 variant and surrounding sequences. Approximately 20 fmol radiolabeled probe was incubated with 3 μg Jurkat nuclear extract, and analyzed by gel electrophoresis. Unbound labeled DNA probe is indicated. Binding of a specific complex to the G allele is shown and on addition of Sp1 antibody results in the upward shift of this complex as shown by the arrows. (c) EMSA of the −587 SNP shows no allelic difference in transcription factor binding.
To determine whether the allelic difference in transcriptional activity related to differences in transcription factor binding, EMSA was performed. The results confirmed our previous results that showed allele-specific difference in transcription factor binding at the −137 SNP. As previous bioinformatic analysis indicated the presence of a putative Sp1 transcription factor binding site in the −137G region,6 supershift EMSA was done using an anti-Sp1 antibody. The major complex interacting with the −137G sequence was supershifted with this antibody, indicating that the major −137G complex contained the activating transcription factor Sp1 (Figure 2b). This is consistent with the reporter gene analysis where the −137G allelic reporter showed an increase in transcriptional activity compared with the −137T allele. In contrast, no binding was seen for either the −587 G or A allele (Figure 2c). Taken together, these ‘gold standard' tests would indicate that the −137 SNP is functional, whereas the −587 variant is not.
Allele-specific chromatin accessibility analysis of the −137 and −587 SNPs
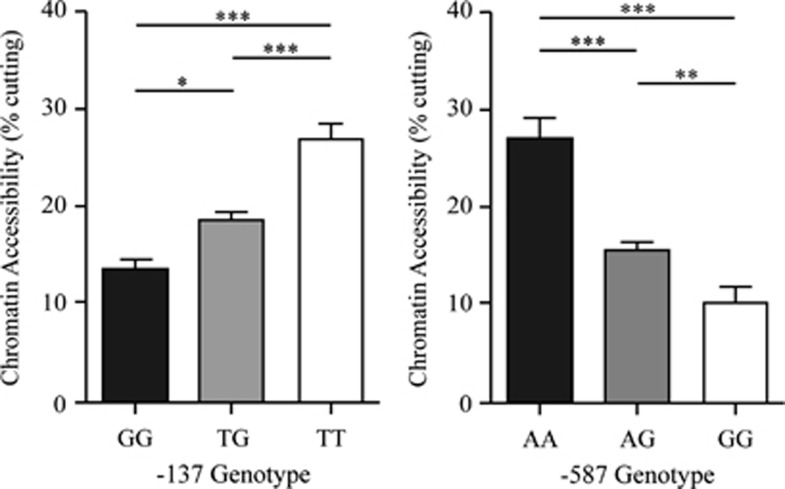
One limitation of the ‘gold standard' approaches is that they fail to assess variant function in their in vivo chromosomal context. As chromatin context is dependent on DNA sequence per se as well as methylation status, variants can potentially alter the conformation of the surrounding chromatin and result in distinct transcriptional potential and hence alter the regulation of a gene. Given that the −587 G variant allele occurs as part of a CpG dinucleotide and is a potential site for methylation, the development of a chromatin accessibility assay was particularly important in assigning function. To determine the endogenous chromatin conformation around both the −137 and −587 SNPs, genotyped lymphoblastoid cell lines from the SAFHS population were used to perform chromatin arrangement by real-time PCR (CHART-PCR) assays.22, 32 The results showed that −587 A/A homozygotes displayed a greater chromatin accessibility (27%) around the −587 site than −587 G/G homozygotes (10%, Figure 3). As expected, the −587 heterozygotes showed an intermediate level of accessibility (16%). Similar results were obtained for the −137 SNP, where the T allele (27%) showed a greater accessibility than the G allele (13%) with intermediate levels for the heterozyote −137 G/T lines (19%). Overall, the results indicate that both the −587 and the −137 SNPs differentially influence chromatin arrangement and potentially the expression of VNN1.
Figure 3.
Chromatin accessibility differences at the −137 and −587 SNPs. MNase-treated cell lines of the genotypes shown were used to calculate chromatin accessibility expressed as a ratio of undigested/MNase digested (percentage of cutting). The data were obtained from five different cell lines for each genotype assayed in triplicate. Significance was determined by the Student's t-test. Error bars represent mean±SEM, *P<0.05, **P<0.005, ***P<0.001.
Allele-specific methylation status of the −587 SNP region
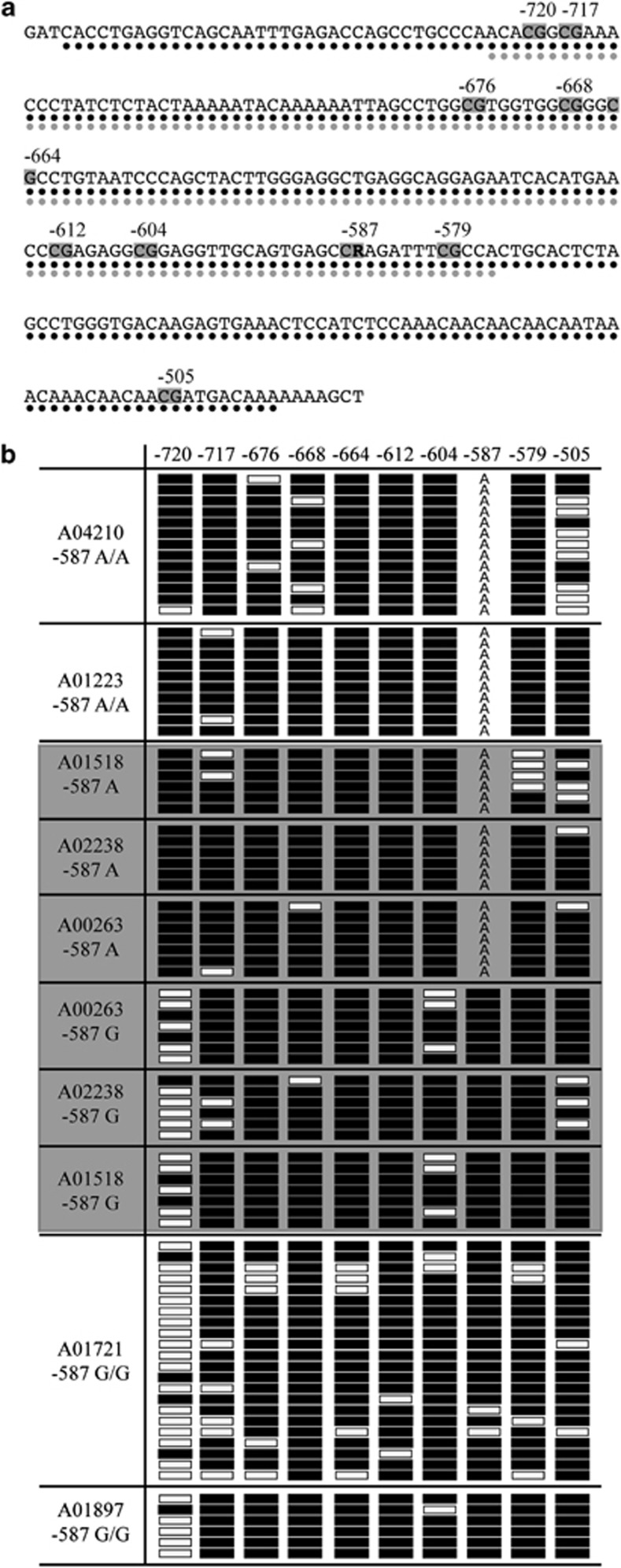
Bioinformatic analysis36 of the VNN1 promoter region surrounding the −587 and −137 SNPs indicated the presence of a CpG island encompassing the −587 site, the predicted extent of which was different for each allele. For the −587 G allele (which is in LD with the −137 T allele), the region was 264 bp in length, and for the A allele, a predicted 204 bp region (Figure 4a). Although no data were available for lymphocytes, ENCODE data also supported the presence of a CpG island encompassing the −587 variant in brain tissue.35 As the chromatin accessibility assay results indicated that the −587 G variant was in a region of condensed chromatin, it was of interest to determine whether the −587 G nucleotide methylation status was directly responsible for the difference predicted. We therefore performed bisulfite sequence analysis on genotyped lymphoblastoid cell lines from the SAHFS population to assess the DNA methylation status of the VNN1 promoter CpG island in the region in which the −587 SNP was embedded (Figure 4b). The −587 G site in both homozygotes and heterozygotes shows near complete cytosine methylation (on the opposite strand). In fact, all CpG dinucleotides displayed heavy methylation, except at the −720 (−733) CpG site. For this site, the methylation pattern correlated with −587 genotype. On haplotypes carrying the −587 G allele, the −720 CpG site was generally unmethylated. Conversely, when the −587 A allele is present, the −720 CpG site was almost exclusively cytosine methylated. This pattern was present even in the heterozygous lines, indicating that the allele-specific pattern is not cell line specific but is rather determined by genotype. The functional consequence of this is yet to be determined but is likely responsible for the −587 allelic differences apparent in chromatin arrangement.
Figure 4.
Methylation status of the −587 SNP. (a) Bioinformatic predictions of CpG island length. Black circles represent a predicted CpG island length of 264 bp when the −587 G allele is present. Grey circles represent a CpG island length of 204 bp when the −587 A allele is present. Grey boxes indicated methylated CG dinucleotides. R=G or A. (b) Bisulfite sequence analysis on genotyped lymphoblastoid lines from the SAFHS population. Line designations and −587 genotype are shown on the left. Black boxes indicate methylation. White boxes indicate no methylation. ‘A' represents the alternate allele at −587, which is not a substrate for CpG methylation. The grey shading indicates heterozygote cell lines. Near complete methylation is seen at the −587 G allele. The methylation status at −587 appears to effect methylation at −720, a non-variant site.
Assessment of chromatin arrangement at the −587 SNP using a novel genome-wide assay
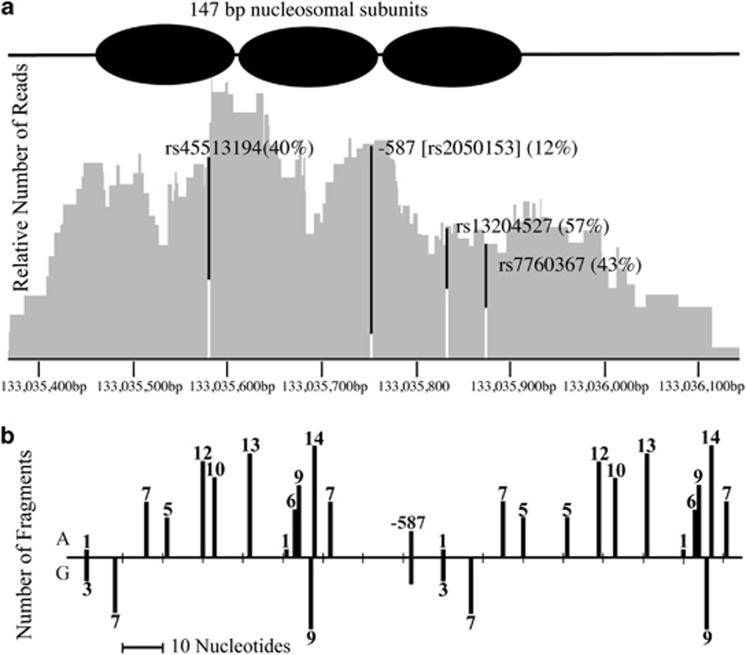
As our results suggested that the effects of allelic variation may be manifest at regions remote from the actual site of sequence difference, we have translated the CHART-PCR assay into a genome-wide format (CHA-seq) in order to assess allele-specific differences in chromatin arrangement across an extended genomic region. A micrococcal nuclease-digested genomic library was prepared from a −587 G/A heterozygous cell line from the SAFHS cohort. The library was subjected to next-generation sequencing to map the nuclease digestion sites. The results showed that the region surrounding the −587 SNP displayed a periodicity in the number of nuclease cut sites consistent with the positioning of the nucleosomes in the region (Figure 5a). Comparison of the number of sequenced library fragments ending at a particular nucleotide (and hence mapping the sites of digestion by the nuclease) for each −587 variant indicated an allele-specific bias in the digestion pattern (Figures 5a and b). Only 12% of the total number of fragments carried −587 G, with the majority carrying the A allele (88%). This is consistent with the region of the haplome carrying the −587G allele being subject to greater chromatin condensation than that carrying the A allele.
Figure 5.
Assessment of the chromatin arrangement across the VNN1 promoter using CHA-seq. (a) Integrated Genomics Viewer output of the region surrounding the −587 SNP (rs2050153) of the VNN1 promoter. Shown is the total coverage across the region and the percentage of reads carrying each allele for the variants located in the region. The −587A allele is shown in black and the −587G allele in white. Three putatively non-functional variants (rs45513194:C>G; −430, rs13204527:G>A; −681, and rs7760367:A>G; −723) are shown and act as controls. Also shown are the putative positions of the nucleosomes based on sequence coverage. (b) Sites of digestion by Micrococcal nuclease are plotted across the −587 VNN1 promoter region. The number of sequenced fragments ending at each nucleotide are indicated for each −587 allele (A or G).
DISCUSSION
The ‘gold standard' functional tests have been useful over the past two decades as a test for determining the functionality of genetic variants. However, both the reporter gene and EMSA assays are unreliable in a number of respects. We have developed strategies that initially involves a statistical prioritization of sequence variants based on comprehensive genotyping and correlation with transcript expression levels,6 and in the current study, application of the ‘gold standard' functional tests followed by novel in vivo tests that quantitate differences in chromatin arrangement as a measure of functional difference. Our strategy serves to greatly increase confidence in assigning function to a particular variant.
Our results indicated that the two SNPs within the VNN1 promoter, previously predicted to be functional following extensive statistical interrogation of transcript levels from the SAFHS,6 were functional. The ‘gold standard' assays established that the −137 SNP was functional and showed a higher level of transcriptional activity for the −137G allele that is consistent with the ability of this allele to preferentially bind the activating transcription factor Sp1 in EMSA. In contrast, the −587 SNP alleles displayed no differences in either transcriptional activity or transcription factor binding.
However, application of our in vivo tests show a significant allelic difference in chromatin accessibility surrounding the −587 SNP and consequently indicates strong functional potential at this variant also. As one of the alleles at −587 is contained within a CG dinucleotide and hence is a potential site of cytosine methylation, bioinformatic analysis of the region surrounding the −587 SNP revealed allele-specific differences in the extent of the CpG island predicted to be present in the VNN1 promoter. Assessment of the methylation status of the region surrounding the −587 SNP revealed an allele-specific methylation pattern at a single CG dinucleotide approximately 120 bp upstream of the −587 SNP, establishing that allele-specific chromatin differences can be seen at sites remote from the actual genetic variation. Similar results have been observed previously,37 where two PBL and one CD34-positive cell samples showed a similar pattern to our lymphoblastoid cell lines, of differential CpG methylation at the −720 site. However, in contrast to our results, the results from the previous study suggest that the genotype at −587 does not correlate with −720 methylation status. Given the almost complete LD between −587 and −137 in our SAFHS population, it is possible that the effects seen at the −720 site are due to allele-specific events at the −137 site. In partial support of this, the reporter gene analysis shows that the allele-specific effects seen for the −137 SNP are independent of −587 genotype.
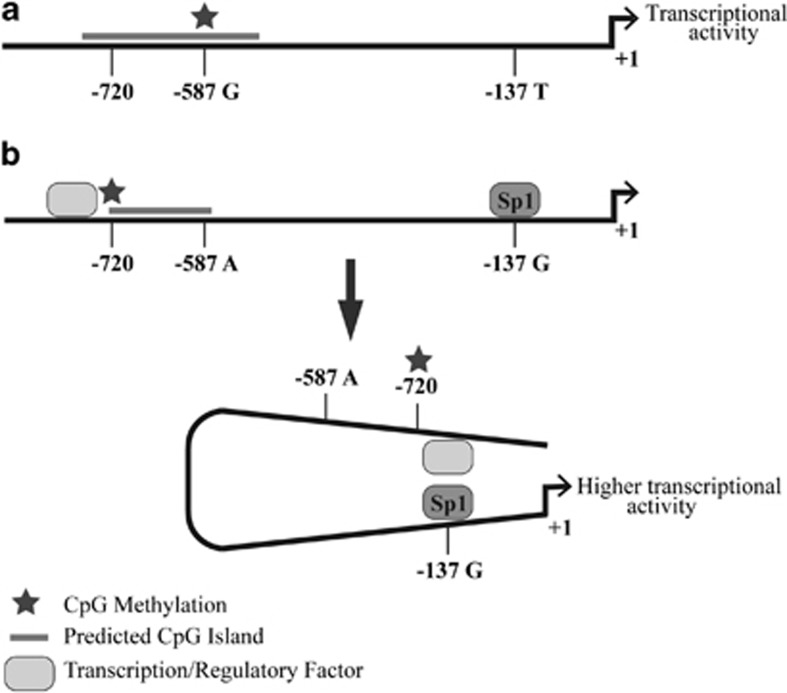
A model to provide a possible explanation for the allele-specific methylation events is that −137G allele-specific Sp1 transcription factor binding leads to allele-specific chromatinization of a restricted promoter region upstream of the VNN1 gene and consequent increased gene expression (Figure 6). We propose that regulatory factor/s are able bind to a region upstream of the −720 site in the −137G allele due to this restricted chromatinization (Figure 6b), whereas in the −137T allele the extended CpG island blocks binding (Figure 6a). The upstream factor may serve to increase transcription via interaction with the downstream Sp1 binding at −137 (Figure 6b) or otherwise by remodeling the chromatin at the CpG island leading to derepressed Sp1-mediated activation. It is likely that the genotype at the −137 site dictates both the occupancy of the Sp1 site and also the chromatin conformation of the VNN1 upstream promoter region and serves to regulate genotype-specific VNN1 expression.
Figure 6.
Model of the interplay between variants at the −137 and −587 sites and the upstream CpG island of the VNN1 promoter. (a) The −137T/−587G haplotype induces methylation of a region that extends past the −720 site blocking binding of regulatory factors to sites upstream. (b) In the −137G/−587A haplotype, the −720 site is methylated but the −587 site is not, leading to a restricted CpG island methylation which allows binding of regulatory factors upstream of −720. Also, the activating transcription factor Sp1 is able to bind at the −137G site. This has the effect of enhancing transcription by direct or indirect interaction between the upstream regulatory factor and the Sp1 transcription factor.
In any case, the results obtained from this study provide a platform to investigate the allele-specific regulation of the VNN1 gene and the influence of the genetic variation on CVD-related phenotypes. We will use the functional cis-regulatory SNP/s identified here to define, by association analyses in our lymphocyte transcriptome data set, those genes that are obligately downstream of VNN1. Identification of −137 as a true functional SNP also allows a simplified re-evaluation of genetic associations with multiple CVD risk-related phenotypes (including triglycerides, HDL-C, carotid wall thickness).
As we have shown previously,38 the functional effects of SNP variation are often only manifest under specific tissue-specific or stimulus-specific conditions. Although our current studies were carried out in genotyped lymphocytes or lymphocyte cell lines, a cell type that is easily obtained, many functional studies will be limited due to the context in which a functional effect will be seen. One of the potential limitations of our in vivo assays, which require the isolation of nuclei from genotyped cells, is the availability of the appropriate cell or tissue type, but results from our own expression QTL studies6 and those of others39 indicate that significant functional information can be obtained for genes that are not normally expressed in the cell type used.
Our approach to the identification of non-coding functional variation, involving statistical prioritization of variants that are associated with transcript levels of the locus, followed by the application of a comprehensive suite of standard as well as new in vivo tests for function, will greatly improve the positive identification of non-coding functional variants by reducing the levels of false-positive ascertainment in addition to providing more compelling evidence with respect to the mechanism of action.
Acknowledgments
This study was supported by the National Institutes of Health Grant HL93537 (to EKM and LJA).
The authors declare no conflict of interest.
References
- Chorley BN, Wang X, Campbell MR, Pittman GS, Noureddine MA, Bell DA. Discovery and verification of functional single nucleotide polymorphisms in regulatory genomic regions: Current and developing technologies. Mutat Res. 2008;659:147–157. doi: 10.1016/j.mrrev.2008.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knight JC. Allele-specific gene expression uncovered. Trends Genet. 2004;20:113–116. doi: 10.1016/j.tig.2004.01.001. [DOI] [PubMed] [Google Scholar]
- The_1000_Genomes_Project_Consortium A map of human genome variation from population-scale sequencing. Nature. 2010;467:1061–1073. doi: 10.1038/nature09534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buckland PR. The importance and identification of regulatory polymorphisms and their mechanisms of action. Biochim Biophys Acta. 2006;1762:17–28. doi: 10.1016/j.bbadis.2005.10.004. [DOI] [PubMed] [Google Scholar]
- Cooper GM, Shendure J. Needles in stacks of needles: finding disease-causal variants in a wealth of genomic data. Nat Rev Genet. 2011;12:628–640. doi: 10.1038/nrg3046. [DOI] [PubMed] [Google Scholar]
- Goring HHH, Curran JE, Johnson MP, et al. Discovery of expression QTLs using large-scale transcriptional profiling in human lymphocytes. Nat Genet. 2007;39:1208–1216. doi: 10.1038/ng2119. [DOI] [PubMed] [Google Scholar]
- Mitchell BD, Kammerer CM, Blangero J, et al. Genetic and environmental contributions to cardiovascular risk factors in Mexican Americans—The San Antonio Family Heart Study. Circulation. 1996;94:2159–2170. doi: 10.1161/01.cir.94.9.2159. [DOI] [PubMed] [Google Scholar]
- Dupre S, Graziani MT, Rosei MA, Fabi A, Delgross. E. Enzymatic breakdown of pantethine to pantothenic acid and cystamine. Eur J Biochem. 1970;16:571–578. doi: 10.1111/j.1432-1033.1970.tb01119.x. [DOI] [PubMed] [Google Scholar]
- Aurrand-Lions M, Galland F, Bazin H, Zakharyev VM, Imhof BA, Naquet P. Vanin-1, a novel GPI-linked perivascular molecule involved in thymus homing. Immunity. 1996;5:391–405. doi: 10.1016/s1074-7613(00)80496-3. [DOI] [PubMed] [Google Scholar]
- Pitari G, Malergue F, Martin F, et al. Pantetheinase activity of membrane-bound Vanin-1: lack of free cysteamine in tissues of Vanin-1 deficient mice. FEBS Lett. 2000;483:149–154. doi: 10.1016/s0014-5793(00)02110-4. [DOI] [PubMed] [Google Scholar]
- Bocos C, Herrera E. Pantethine stimulates lipolysis in adipose tissue and inhibits cholesterol and fatty acid synthesis in liver and intestinal mucosa in the normolipidemic rat. Environ Toxicol Pharmacol. 1998;6:59–66. doi: 10.1016/s1382-6689(98)00020-9. [DOI] [PubMed] [Google Scholar]
- Wittwer CT, Gahl WA, Butler JD, Zatz M, Thoene JG. Metabolism of pantethine in cystinosis. J Clin Invest. 1985;76:1665–1672. doi: 10.1172/JCI112152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaskow BJ, Proffitt JM, Blangero J, Moses EK, Abraham LJ. Diverse biological activities of the vascular non-inflammatory molecules—the Vanin pantetheinases. Biochem Biophys Res Commun. 2012;417:653–658. doi: 10.1016/j.bbrc.2011.11.099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorman CM, Moffat LF, Howard BH. Recombinant genomes which express chloramphenicol acetyltransferase in mammalian cells. Mol Cell Biol. 1982;2:1044–1051. doi: 10.1128/mcb.2.9.1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gould SJ, Subramani S. Firefly luciferase as a tool in molecular and cell biology. Anal Biochem. 1988;175:5–13. doi: 10.1016/0003-2697(88)90353-3. [DOI] [PubMed] [Google Scholar]
- Mercola M, Goverman J, Mirell C, Calame K. Immunoglobulin heavy-chain enhancer requires one or more tissue-specific factors. Science. 1985;227:266–270. doi: 10.1126/science.3917575. [DOI] [PubMed] [Google Scholar]
- Fried M, Crothers DM. Equilibria and kinetics of lac repressor-operator interactions by polyacrylamide gel electrophoresis. Nucleic Acids Res. 1981;9:6505–6525. doi: 10.1093/nar/9.23.6505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garner MM, Revzin A. A gel electrophoresis method for quantifying the binding of proteins to specific DNA regions: application to components of the Escherichia coli lactose operon regulatory system. Nucleic Acids Res. 1981;9:3047–3060. doi: 10.1093/nar/9.13.3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knight JC. Functional implications of genetic variation in non-coding DNA for disease susceptibility and gene regulation. Clin Sci (Lond) 2003;104:493–501. doi: 10.1042/CS20020304. [DOI] [PubMed] [Google Scholar]
- Karimi M, Goldie LC, Cruickshank MN, Moses EK, Abraham LJ. A critical assessment of the factors affecting reporter gene assays for promoter SNP function: a reassessment of −308 TNF polymorphism function using a novel integrated reporter system. Eur J Hum Genet. 2009;17:1454–1462. doi: 10.1038/ejhg.2009.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karimi M, Goldie LC, Ulgiati D, Abraham LJ. Integration site-specific transcriptional reporter gene analysis using Flp recombinase targeted cell lines. Biotechniques. 2007;42:217–224. doi: 10.2144/000112317. [DOI] [PubMed] [Google Scholar]
- Cruickshank MN, Karimi M, Mason RL, et al. Transcriptional effects of a lupus-associated polymorphism in the 5' untranslated region (UTR) of human complement receptor 2 (CR2/CD21) Mol Immunol. 2012;52:165–173. doi: 10.1016/j.molimm.2012.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birney E, Lieb JD, Furey TS, Crawford GE, Iyer VR. Allele-specific and heritable chromatin signatures in humans. Hum Mol Genet. 2010;19:R204–R209. doi: 10.1093/hmg/ddq404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherry ST, Ward MH, Kholodov M, et al. dbSNP: the NCBI database of genetic variation. Nucleic Acids Res. 2001;29:308–311. doi: 10.1093/nar/29.1.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almasy L, Blangero J. Multipoint quantitative-trait linkage analysis in general pedigrees. Am J Hum Genet. 1998;62:1198–1211. doi: 10.1086/301844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abecasis GR, Cookson WO, Cardon LR. Pedigree tests of transmission disequilibrium. Eur J Hum Genet. 2000;8:545–551. doi: 10.1038/sj.ejhg.5200494. [DOI] [PubMed] [Google Scholar]
- Boerwinkle E, Chakraborty R, Sing CF. The use of measured genotype information in the analysis of quantitative phenotypes in man. I. Models and analytical methods. Ann Hum Genet. 1986;50:181–194. doi: 10.1111/j.1469-1809.1986.tb01037.x. [DOI] [PubMed] [Google Scholar]
- Havill LM, Dyer TD, Richardson DK, Mahaney MC, Blangero J. The quantitative trait linkage disequilibrium test: a more powerful alternative to the quantitative transmission disequilibrium test for use in the absence of population stratification. BMC Genet. 2005;6 (Suppl 1:S91. doi: 10.1186/1471-2156-6-S1-S91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickham H. ggplot: Elegant Graphics for Data Analysis. Springer Inc: New York; 2009. [Google Scholar]
- Franchina M, Woo AJ, Dods J, et al. The CD30 gene promoter microsatellite binds transcription factor Yin Yang 1 (YY1) and shows genetic instability in anaplastic large cell lymphoma. J Pathol. 2008;214:65–74. doi: 10.1002/path.2258. [DOI] [PubMed] [Google Scholar]
- Cruickshank M, Fenwick E, Abraham LJ, Ulgiati D. Quantitative differences in chromatin accessibility across regulatory regions can be directly compared in distinct cell-types. Biochem Biophys Res Commun. 2008;367:349–355. doi: 10.1016/j.bbrc.2007.12.121. [DOI] [PubMed] [Google Scholar]
- Rao S, Procko E, Shannon MF. Chromatin remodeling, measured by a novel real-time polymerase chain reaction assay, across the proximal promoter region of the IL-2 gene. J Immunol. 2001;167:4494–4503. doi: 10.4049/jimmunol.167.8.4494. [DOI] [PubMed] [Google Scholar]
- Rohde C, Zhang Y, Reinhardt R, Jeltsch A. BISMA-fast and accurate bisulfite sequencing data analysis of individual clones from unique and repetitive sequences. BMC Bioinformatics. 2010;11:230. doi: 10.1186/1471-2105-11-230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson JT, Thorvaldsdottir H, Winckler W, et al. Integrative genomics viewer. Nat Biotechnol. 2011;29:24–26. doi: 10.1038/nbt.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyle AP, Hong EL, Hariharan M, et al. Annotation of functional variation in personal genomes using RegulomeDB. Genome Res. 2012;22:1790–1797. doi: 10.1101/gr.137323.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takai D, Jones PA. Comprehensive analysis of CpG islands in human chromosomes 21 and 22. Proc Natl Acad Sci USA. 2002;99:3740–3745. doi: 10.1073/pnas.052410099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerkel K, Spadola A, Yuan E, et al. Genomic surveys by methylation-sensitive SNP analysis identify sequence-dependent allele-specific DNA methylation. Nat Genet. 2008;40:904–908. doi: 10.1038/ng.174. [DOI] [PubMed] [Google Scholar]
- Kroeger KM, Steer JH, Joyce DA, Abraham LJ. Effects of stimulus and cell type on the expression of the −308 tumour necrosis factor promoter polymorphism. Cytokine. 2000;12:110–119. doi: 10.1006/cyto.1999.0529. [DOI] [PubMed] [Google Scholar]
- Morley M, Molony CM, Weber TM, et al. Genetic analysis of genome-wide variation in human gene expression. Nature. 2004;430:743–747. doi: 10.1038/nature02797. [DOI] [PMC free article] [PubMed] [Google Scholar]