Significance
The aggregation of mutant proteins is pathologically implicated in a large number of neuropathies, including Huntington disease and ALS. Although the appearance of protein aggregates is known to sequester other proteins, how this results in the gain-of-function toxicity in these diseases is unclear. Here, we show that the aggregation of disease-associated proteins causes the reversible collapse of clathrin-mediated endocytosis (CME) and inhibits the internalization of membrane receptors that affect neuronal function. CME inhibition occurs through aggregate-mediated sequestration of the molecular chaperone heat shock cognate protein 70, which is essential for CME. We propose that a toxic “tug-of-war” occurs between aggregates and endogenous client proteins for available chaperones, leading to the collapse of multiple cellular processes in neurodegeneration and other protein conformation diseases.
Keywords: proteostasis, chaperone-dependent processes, protein misfolding, HSP70
Abstract
Protein conformational diseases exhibit complex pathologies linked to numerous molecular defects. Aggregation of a disease-associated protein causes the misfolding and aggregation of other proteins, but how this interferes with diverse cellular pathways is unclear. Here, we show that aggregation of neurodegenerative disease-related proteins (polyglutamine, huntingtin, ataxin-1, and superoxide dismutase-1) inhibits clathrin-mediated endocytosis (CME) in mammalian cells by aggregate-driven sequestration of the major molecular chaperone heat shock cognate protein 70 (HSC70), which is required to drive multiple steps of CME. CME suppression was also phenocopied by HSC70 RNAi depletion and could be restored by conditionally increasing HSC70 abundance. Aggregation caused dysregulated AMPA receptor internalization and also inhibited CME in primary neurons expressing mutant huntingtin, showing direct relevance of our findings to the pathology in neurodegenerative diseases. We propose that aggregate-associated chaperone competition leads to both gain-of-function and loss-of-function phenotypes as chaperones become functionally depleted from multiple clients, leading to the decline of multiple cellular processes. The inherent properties of chaperones place them at risk, contributing to the complex pathologies of protein conformational diseases.
Many neurodegenerative diseases are characterized by protein misfolding and aggregation (1–5). Although the underlying disease origins may be genetically inherited or manifest sporadically, as exemplified by Huntington disease and ALS, respectively, the pathologies of these maladies all share the common molecular occurrence of protein aggregation (6). A network of protein folding and clearance mechanisms (the proteostasis network) is proposed to maintain a healthy proteome for normal cellular function (7, 8). Central to the proteostasis network are molecular chaperones and cochaperones, a diverse group of proteins that modulate the synthesis, folding, transport, and degradation of proteins (7). The conformations of aggregation-prone proteins are subject to multiple layers of regulation by the proteostasis network; however, as evidenced by the widespread pathologies of protein conformational diseases, the aggregation propensity of proteins associated with these diseases ultimately overwhelms the proteostasis machineries, thus initiating a cascade of cellular dysfunction (9–11).
It is increasingly common for diseases of protein aggregation to be described as the result of gain-of-function toxicity. This toxicity is largely attributed to the dominant appearance of diverse aggregate species and the subsequent aberrant association of various proteostasis network components and other metastable proteins with these aggregates. This position is supported by experiments using immunohistochemical, biochemical, and MS methods on diseased patient tissues, as well as on numerous cellular and animal model systems (12–16). Some of these molecular interactions, such as those between aggregates and proteasomal subunits, appear irreversible, suggesting a permanent sequestration of these proteins. The association of molecular chaperones with aggregates, on the other hand, appears transient (17, 18), indicating that chaperones may be functionally recognizing aggregates as substrates for potential disaggregation and refolding (19).
Beyond refolding of toxic misfolded proteins, chaperones are also essential for the folding of endogenous metastable client proteins, as well as in the assembly and disassembly of functional protein complexes. Thus, chaperones regulate a wide range of essential cellular processes, including gene expression, vesicular trafficking, and signal transduction (20–25). This dual role of chaperones suggests that a “competition” may arise between aggregates and endogenous protein clients for finite chaperone resources in situations where aggregates have accumulated. It has been proposed that such an imbalance may trigger the onset of many neurodegenerative diseases (10, 26), and recent studies report that polyglutamine (polyQ)-based aggregates can sequester and inhibit the function of a low-abundance cochaperone, Sis1p/DNAJB1, in protein degradation (27).
Here, we show that diverse disease-associated aggregates sequester the highly abundant major chaperone heat shock cognate protein 70 (HSC70) to the point of functional collapse of an essential cellular process, clathrin-mediated endocytosis (CME). Importantly, aggregate-driven CME inhibition is reversible and can be rescued by nominally increasing HSC70 levels. Aggregate-driven chaperone depletion may help explain the phenotypic complexities displayed in protein conformational diseases.
Results
We tested the chaperone competition hypothesis by monitoring the effect of protein aggregation on CME, where the chaperone HSC70 is required for multiple steps of the pathway (28). Beyond maintaining the general solubility of the CME machinery, HSC70 is specifically required for both the disassembly of the clathrin coat from endocytosed vesicles (20, 29–31) and in the recycling of coat components back to the plasma membrane. The relative rates of CME can be quantified in individual mammalian cells by measuring the levels of internalized fluorescently labeled transferrin (32, 33). It is well established that internalization of transferrin through binding to the transferrin receptor relies solely on the CME pathway.
Different disease-associated misfolded proteins were analyzed for their effects on CME. Previous studies have shown that aggregation of expanded polyQ negatively affects endocytosis in yeast and in human HEK 293 cells (34). We verified and extended upon these observations by analyzing expanded polyQ, whose length expansion induces aggregation in multiple unrelated disease proteins (17), as well as polyQ-expanded huntingtin (Htt) exon 1 fragment and ataxin-1 (Atx1), which are specifically associated with Huntington disease and spinocerebellar ataxia type I, respectively (12, 14). In addition, mutant superoxide dismutase-1 (SOD1) A4V, associated with familial ALS, was tested (35). The aggregation characteristics of these proteins have been studied extensively and range from highly ordered amyloid fibrils, as seen with pure polyQ, to structurally disordered aggregates, as exemplified by mutant SOD1 (18, 36, 37). These proteins have also been shown to localize and aggregate in varying degrees to the cytoplasmic or nuclear compartment.
When PC-3 cells were transiently transfected to express these aggregation-prone proteins as GFP fusions, a heterogeneous population of cells resulted, where the protein appeared diffuse or formed bright punctae. Fluorescence recovery after photobleaching (FRAP) experiments were performed to establish the aggregation state of each WT and mutant protein. As reported previously (17, 18), when the aggregation-prone Q82 or mutant SOD1 A4V exhibited a visually diffuse appearance, its relative diffusion kinetics remained unchanged from that of soluble Q19 or WT SOD1, respectively (Fig. S1). Both Q82 and SOD1 A4V were highly immobile, however, when found within bright punctae, indicating aggregation. Although small, misfolded oligomeric intermediates have been proposed to represent the toxic species, these intermediates could not be distinguished from the soluble population; nevertheless, in our subsequent experiments, proteins with diffuse distribution were defined as soluble, whereas those coalesced into foci were defined as insoluble and aggregated.
Protein Aggregation Broadly Inhibits CME.
The internalization of transferrin was unaffected in human PC-3 cells transiently expressing WT Q19 or in cells expressing soluble Q82 (Fig. 1 A and B). However, CME was substantially reduced in cells containing Q82 aggregates (Fig. 1B, arrows), with quantification of internalized transferrin fluorescence showing a 63 ± 11% reduction in aggregate-containing cells compared with cells expressing soluble Q19 or Q82 (Fig. 1C). CME inhibition was also observed in cells containing aggregated forms of polyQ-expanded Htt exon 1 (Htt Q53); these cells exhibited 50 ± 15% reduced levels of internalized transferrin compared with cells with soluble Htt Q23 or Htt Q53 protein (Fig. S2 A–C). Down-regulation of CME was not limited to polyQ-induced aggregation; cells with aggregated mutant SOD1 A4V also exhibited a 50 ± 10% decrease in levels of internalized transferrin compared with cells expressing the soluble WT or mutant protein (Fig. 1 E–G). CME was not strongly inhibited in cells where Q82, SOD1 A4V, or Htt Q53 maintained solubility but was expressed at similar levels to where the proteins had aggregated (Fig. 1 D and H and Fig. S2D). These results indicate that CME inhibition is not due to general overexpression of the heterologous GFP-fused proteins but, rather, is specifically associated with the presence of diverse protein aggregates.
Fig. 1.
CME is reduced in cells with protein aggregates. Internalization of Alexa555-conjugated transferrin was assayed by fluorescence confocal microscopy in cells transiently (A–H) or stably (I–K) expressing various aggregation-prone proteins fused to GFP. (A and B) Internalized Alexa555-transferrin (Tf, Right) in human PC-3 cells transiently expressing soluble or aggregation-prone Q19-EGFP or Q82-EGFP, respectively (Left). Images shown are maximal projections of a z-series of fluorescence confocal slices through the entire cell volume. Arrows indicate cells containing aggregates. Note the aggregate-containing cells in B have reduced fluorescence levels of transferrin compared with untransfected or soluble Q82-expressing cells. (Scale bar, 10 μm.) (C) Quantification of the effect of relative Q82 expression levels, as measured by sum fluorescence intensities of EGFP divided by the cell area, on transferrin internalization. Quantification for each cell was measured by taking the sum fluorescence intensity of the z-series through the total cell volume, divided by the area of the cell. Shown are the mean values, with error bars representing SDs. P values were determined using a Student t test. (D) Scatterplot depicts quantification of internalized Alexa555-transferrin in untransfected and Q19-EGFP–, or Q82-EGFP–expressing cells. (E and F) As in A and B, but in PC-3 cells expressing WT (wt) or aggregation-prone mutant (A4V) SOD1-AcGFP, respectively. Arrows indicate cells containing aggregates. The cell labeled with an asterisk contains minute aggregates and is further magnified in Fig. S1E. (G) Quantification, as in C, of internalized transferrin in SOD1-expressing cells. (H) Effect of relative expression levels on transferrin uptake, as in D, in SOD1(A4V)-expressing cells. (I and J) As in A and B, but in Neuro2a cells induced to express stably a single integrated copy of EGFP-fused Q2- or Q82-expanded Atx-1. (J) Two separate image samples are shown for GFP (1, 2) and transferrin (1′, 2′) fluorescence. Arrows indicate cells containing aggregates. (K) Quantification, as in C, of internalized transferrin in Neuro2a cells induced to express Atx-1 Q2 or Q82. Dox, doxycycline.
Aggregate subcellular localization has also been proposed to have varying consequences on disease pathology (5), and the aggregation-prone proteins tested above localize and aggregate differentially to the cytoplasm or the nucleus. PolyQ and Htt exon 1 fragments distribute and aggregate in both compartments, whereas mutant SOD1 aggregates preferentially in the cytoplasm. We next tested whether nuclear aggregation alone could also inhibit CME. Additionally, given that the initial experiments were performed in human prostate cancer cells using transient expression, we sought to establish whether aggregate-driven CME occurs in different cell types and upon expression at lower levels.
To extend upon these initial observations, we established a neuronal cell model in which a polyQ disease protein with a nuclear localization signal can be conditionally expressed. Stable Neuro2a cell lines were generated in which a single gene copy of EGFP-fused Atx1 Q2 or Q82 is chromosomally integrated and can be induced to express with doxycycline. The expression and subsequent aggregation frequency of Atx1 Q82 were significantly lower than in cells transiently expressing Q82, Htt Q53, or SOD1 A4V; moreover, Atx1 Q82 localized to and aggregated within the nucleus. Nevertheless, transferrin internalization was reduced in aggregate-containing cells compared with those expressing Atx1 Q2 or soluble Atx1 Q82 (Fig. 1 J vs. I), where quantification showed a significant decrease of 58 ± 15% in internalized transferrin levels (Fig. 1K).
Collectively, these results demonstrate the widespread inhibitory nature of protein aggregation on CME. This suppression occurs over a wide range of aggregating protein expression levels, independent of cytoplasmic or nuclear aggregate localization, and in cells of neuronal and nonneuronal origins.
CME Inhibition Is Associated with Aggregate Sequestration of HSC70.
To determine the molecular basis for aggregate-driven CME inhibition, we first examined whether the localization of transferrin receptor at the plasma membrane or the receptor’s ability to bind transferrin was altered. Cells were incubated with Alexa Fluor-conjugated transferrin at 4 °C, washed, and then immediately fixed, because transferrin binds with cell surface-exposed receptors at high affinity but fails to become endocytosed under these conditions (32). Cells containing Q82, SOD1 A4V, or Htt Q53 aggregates showed no appreciable difference of substrate-bound receptor at the plasma membrane compared with cells containing soluble protein (Fig. S3 A–C). These results support the idea that CME inhibition is not caused by defective synthesis, trafficking, or substrate-binding ability of the transferrin receptor itself.
Because aggregates can physically sequester a variety of metastable endogenous proteins, the CME inhibition observed could be through aberrant interaction of other proteins of the endocytosis machinery. These factors include clathrin heavy chains (CHCs) and the adaptor-related protein complex 2 (AP-2), which constitute the coat (38–40), as well as HSC70 and its associated J-domain cochaperone auxilin (29, 41). These components have been identified to associate with aggregation-prone disease proteins by biochemistry or MS (16, 42); thus, we tested whether aggregates were sequestering these components in intact cells.
PC-3 cells transiently expressing Q82 and assayed for transferrin internalization were immunostained for endogenous components that are essential for CME. Neither mu2, an AP-2 subunit, nor CHC was recruited to Q82 aggregates in the majority of cells, where CME was concomitantly affected (Fig. S4 A and B, arrows). On rare occasions, mu2 and CHC colocalized with aggregation; however, this finding was observed only in cells expressing the highest levels of Q82 (Fig. S5 A and B, Right).
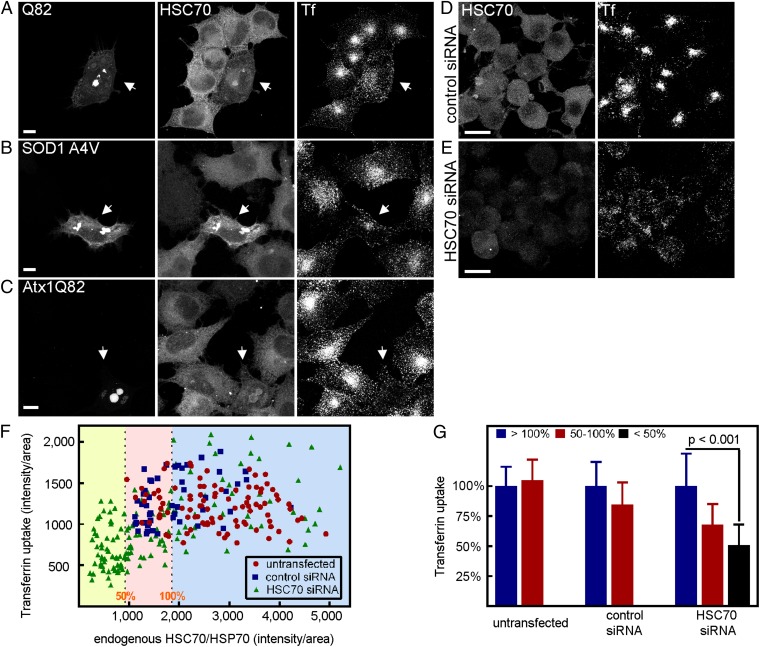
Next, the subcellular localization of endogenous auxilin and HSC70 was probed in aggregate-containing cells. The distribution of cyclin G-activated kinase (GAK), the ubiquitously expressed form of auxilin (43, 44), was unaffected in Q82 aggregate-containing cells (Fig. S4C, arrows). However, HSC70 redistributed to aggregates of Q82, SOD1 A4V, or Atx1 Q82 (Fig. 2 A–C, Center). All cells in which HSC70 was redistributed to aggregates also exhibited decreased levels of internalized transferrin (Fig. 2 A–C, Right). The titration of HSC70 to aggregates also did not depend on aggregate localization, because HSC70 redistributed to aggregates in the cytoplasm or the nucleus (Fig. 2 B vs. C). Often, a decrease in the diffuse cytoplasmic pool of this chaperone was observed when the aggregates were prominent. However, this type of colocalization analysis likely underestimates the amount of HSC70 that is functionally occupied, because HSC70 will also bind misfolded monomeric or small oligomeric proteins dispersed throughout the nucleus or cytoplasm that cannot be resolved by fluorescence microscopy. Nevertheless, HSC70-aggregate colocalization reliably predicted CME inhibition. These results suggest that the aggregates titrate HSC70 away from the CME pathway underlying aggregate-driven CME inhibition. Moreover, HSC70, relative to other endocytosis factors, appears to be preferentially affected in aggregate-driven inhibition of CME.
Fig. 2.
Depletion of HSC70 by protein aggregate relocalization or by RNAi inhibits CME. (A) PC-3 cells transiently expressing Q82-EGFP (Left) were assayed for internalized Alexa555-transferrin (Right) and endogenous HSC70 localization (Center). HSC70 was detected using specific antibodies by indirect immunofluorescence. Arrows point to an aggregate-containing cells. Note that the diffuse pool of HSC70 appears reduced in this cell. (Scale bar, 10 μm.) (B) As in A, but in PC-3 cells expressing mutant SOD1 (A4V)-AcGFP. Arrows point to an aggregate-containing cells. (C) As in A, but in PC-3 cells expressing EGFP-Atx-1 Q82. Arrows point to an aggregate-containing cells. Neuro2a cells transfected with control (D) or specific siRNAs targeting HSC70 (E) were assayed for Alexa555-transferrin internalization (Right) and immunostained for endogenous HSC70 (Left). Note the decreased levels of HSC70 and corresponding low transferrin levels in E. (F) Quantification of internalized transferrin levels in Neuro2a cells that were untransfected or transfected with control or HSC70 siRNA oligonucleotides. Shown are the relative HSC70 expression level values of individual cells plotted against their corresponding internalized transferrin fluorescence values as calculated in G. The plot background has been divided into three sections based on relative HSC70 levels normalized to the average of the control siRNA cells (yellow, <50%; pink, 50–100%; and blue, >100%) to show the number of cells that fall into each category. The three binned categories are those depicted in G. (G) Quantification of internalized transferrin levels in Neuro2a cells that were untransfected or treated with control or HSC70 siRNA oligonucleotides. Cells were grouped into three categories based on relative HSC70 expression levels and normalized to that of control siRNA transfected cells. Internalized transferrin fluorescence was analyzed as in Fig. 1. Shown are mean values of transferrin levels with error bars representing SDs. P values were determined using a Student t test.
RNAi Depletion of HSC70 Phenocopies Aggregate-Driven CME Inhibition.
If relocalization of HSC70 to aggregates alone is the basis for diminished CME, then the direct depletion of HSC70 expression should have the same effect. We therefore depleted endogenous HSC70 in Neuro2a cells using specific siRNA oligonucleotides, assayed for transferrin internalization, and probed for relative HSC70 levels in individual cells using specific antibodies and indirect immunofluorescence. siRNA transfection leads to variable levels of HSC70 depletion within the cell population (Fig. 2F), affording the opportunity to establish whether there was a critical level of HSC70 necessary for CME inhibition. Cells were sorted into three groups according to their HSC70 expression levels relative to that of control siRNA-treated cells, and the amounts of internalized transferrin were quantified (Fig. 2 F and G). Although internalized transferrin levels remained unchanged in cells with near-normal amounts of HSC70, cells with over a 50% reduction in HSC70 expression had significant CME inhibition (compare Fig. 2 E and D), with a 51 ± 17% decrease in internalized transferrin fluorescence compared with nondepleted cells. Cells with intermediate-lower levels of HSC70 (50–100%) also showed reduced, but more variable, effects on CME. These results were further supported with depletion experiments using two other siRNA oligonucleotides targeting different regions of HSC70 (Fig. S6). Taken together, these experiments show that CME is sensitive to fluctuations in HSC70 levels, and that functional depletion of HSC70 by its relocalization to aggregates is likely the basis for CME inhibition.
Increasing HSC70 Levels Restores CME in Aggregate-Containing Cells.
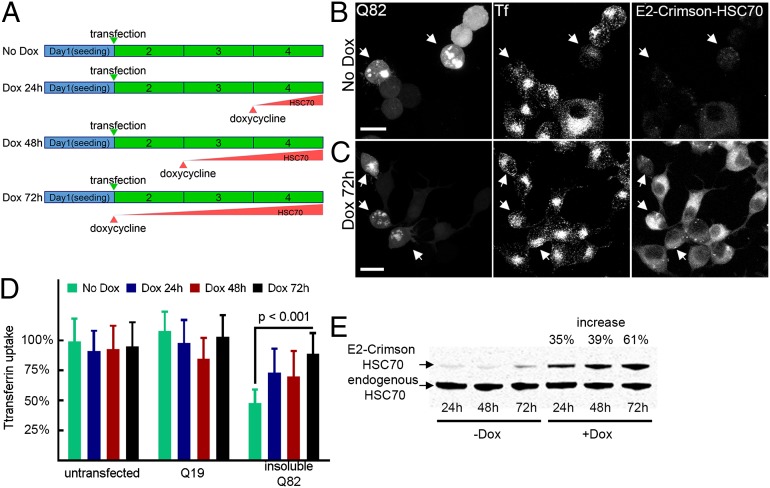
The transient nature of HSC70–aggregate interactions (17) led to the intriguing possibility that aggregate-driven CME disruption may be reversed by increasing the levels of HSC70. To address this possibility, a conditional expression system was used, where HSC70 levels could be elevated in a controlled manner. A stable Neuro2a cell line was generated containing a single chromosomally integrated copy of E2-Crimson fused HSC70, whose expression is under the control of a doxycycline-inducible promoter. This expression system allowed the homogeneous expression of E2-Crimson HSC70 in all cells after induction (Fig. S7).
Upon transient expression of Q82 in noninduced cells, we observed a marked aggregation-dependent decrease in transferrin internalization as before (Fig. 3B; quantification in Fig. 3D). Cells were next treated with doxycycline over a period of 24–72 h to allow for different levels of E2-Crimson HSC70 to accumulate (Fig. 3A; compare Fig. 3 C vs. B, Right), where the increase in total HSC70 levels ranged from 35 to 61%, respectively (Fig. 3E). Although Q82 aggregates persisted at these levels of HSC70, there was a 70–90% improvement in CME when comparing internalized transferrin levels with noninduced Q82 aggregate-containing cells (compare Fig. 3 C vs. B; quantification in Fig. 3D). Moreover, the cells with the earliest and highest expression of E2-Crimson HSC70 exhibited the most effective restoration of CME.
Fig. 3.
Increasing HSC70 in a concentration-dependent manner rescues CME defects in protein aggregate-containing cells. (A) Schematic of Dox-induced E2-Crimson HSC70 expression relative to Q82 transfection. (B and C) Neuro2a cells containing a Dox-inducible, single integrated copy of E2-Crimson HSC70 were transiently transfected to express Q82-EGFP. Note that the cells containing Q82 aggregates (Left) without induction of E2-Crimson HSC70 (B, Right) show decreased levels of Alexa555-transferrin (Center), compared with cells where E2-Crimson HSC70 has been induced (C, Right). Arrows indicate cells with aggregates. (Scale bar, 10 μm.) (D) Quantification of internalized transferrin fluorescence in Neuro2a cells expressing induced E2-Crimson HSC70 for the indicated time periods. (E) Relative protein levels of induced E2-Crimson fused and endogenous HSC70 at different time points were detected by immunoblotting with specific antibodies recognizing both HSC70 and HSP70. The combined levels of endogenous and E2-Crimson HSC70 were quantified for each induced time point compared with control nontreated cells cultured for the same time period.
These experiments show that CME can be restored by conditional expression of HSC70 to near-WT levels in cells expressing expanded polyQ, revealing that aggregate-driven inhibition of CME is reversible. Notably, the recovery of CME also occurs without the visible suppression of aggregates, thus separating aggregation from cellular toxicity.
Direct Role for Aggregate-Driven CME Inhibition in Neuropathologies.
To investigate whether aggregate-driven CME inhibition could contribute directly to neurodegenerative disease pathologies, we monitored the trafficking of a neuron-specific cargo, AMPA receptor, which mediates synaptic transmission and has essential roles in synaptic plasticity (45–47). AMPA receptors at the cell surface interact directly with AP-2 and can undergo CME constitutively or upon ligand activation (48–50). To test for effects of aggregation on ligand-activated CME of the AMPA receptor, a subunit of the AMPA receptor, HA-tagged GluR2, was transiently coexpressed with Q19 or Q82 in Neuro2a cells. GluR2 was surface-labeled with anti-HA antibodies before endocytosis was stimulated by the addition of insulin (49, 50). These cells were simultaneously tested for transferrin internalization before fixation and processing for immunofluorescence. In cells coexpressing Q82, the appearance of aggregation reduced the internalization of both HA-tagged GluR2 and transferrin compared with that in Q19-expressing cells (compare Fig. 4 A vs. B, Lower and C).
Fig. 4.
Simultaneous inhibition of transferrin and AMPA receptor endocytosis in aggregate-containing cells. (A) Neuro2a cells were cotransfected with plasmids encoding both HA-GluR2 and Q19-EGFP for 48 h. (Left) Cells expressing Q19-EGFP were labeled with anti-HA antibody for AMPA receptor at the cell surface. Cells were then treated without (Upper) or with (Lower) 3 μM insulin for 15 min, and followed by transferrin uptake for another 15 min (Right). (Center) Localization of anti-HA antibody was probed through immunofluorescence. Arrowheads point to Q19-expressing cells. (B, Left) As in A, but transiently expressing Q82-EGFP. The arrows point to an aggregate-containing cell. Note that whereas cells without protein aggregates showed a normal rate of transferrin uptake and GluR2 internalization (arrowheads), the cells with aggregates showed defects in both processes (arrows). (Scale bar, 10 μm.) (C) Enlarged images show cells labeled with asterisks in A and B.
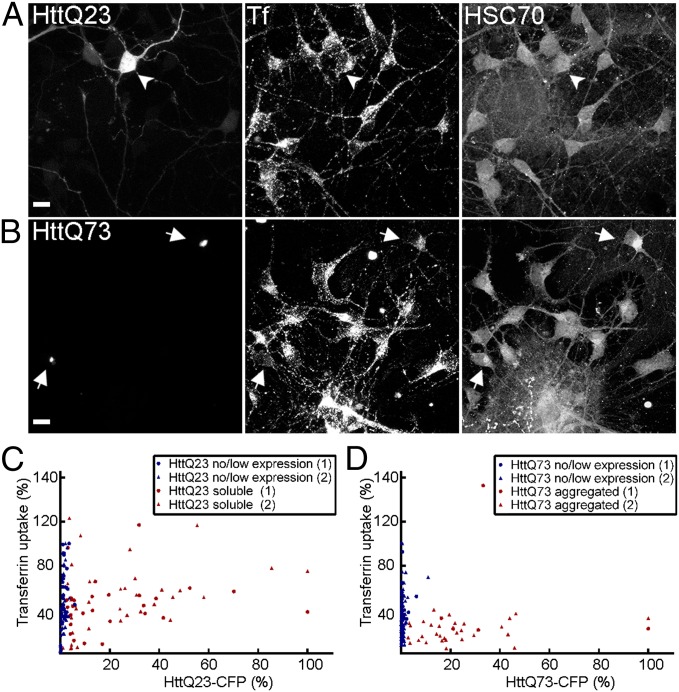
We then investigated whether aggregate-driven CME inhibition could be recapitulated in a Huntington disease model based on primary neurons (51). In this model, mouse primary neurons are isolated from the cortex and striatum, two key brain regions implicated in the initiation and progression of Huntington disease, and cocultured on support layers of glial cells to mimic the intercellular interactions and tissue environment found in vivo. These corticostriatal cocultures showed larger variance in transferrin endocytosis among control nontransfected neurons compared with cell lines (Figs. 5 C and D), perhaps reflecting intrinsic variability in postmitotic primary neurons. In cocultured neurons expressing soluble Htt exon 1 Q23-CFP, the range of transferrin uptake compared with that in control nonexpressing neurons was not noticeably affected (Fig. 5A; quantification in Fig. 5C). In contrast, there was a marked reduction in CME in neurons containing mutant Htt exon 1 Q73-CFP aggregates compared with nonexpressing cells (Fig. 5B, Center; quantification in Fig. 5D); this CME reduction coincided with the recruitment of endogenous HSC70 to aggregation (Fig. 5B, Right, arrows).
Fig. 5.
HSC70 recruitment to HttQ73 aggregates in primary CNS neurons is accompanied by inhibition of CME. Primary corticostriatal neuronal cocultures were nucleofected to express either Htt exon 1 Q23-CFP (A) or Htt exon 1 Q73-CFP (B), assayed for Alexa555-transferrin internalization, and immunostained for endogenous HSC70 localization. Images shown are maximal projections of a z-series of fluorescence confocal slices through the entire cell volume. In B, note that HSC70 colocalizes with Htt Q73 aggregates (arrows) and that neurons expressing these aggregates show reduced levels of transferrin uptake. Arrowheads and arrows point to neurons expressing HttQ23 or aggregated HttQ73 in A and B, respectively. (Scale bar, 10 μm.) (C) Scatterplot of fluorescence quantification of internalized transferrin as related to Htt exon 1 Q23-CFP levels, as in Fig. 1D, in individual primary neurons. Neurons were grouped into nonexpressing vs. expressing categories. Both transferrin and CFP fluorescence were normalized to the highest values of the no expression control and CFP-expressing cells, respectively, and data from two imaging trials (labeled as 1 and 2) of the same sample were plotted. (D) As in C, except for neurons expressing Htt exon 1 Q73-CFP. Neurons were grouped into nonexpressing/low soluble expression vs. aggregated categories.
The dysregulation of AMPA receptor in aggregate-containing neuronal tissue culture cells and the inhibitory effects of Htt aggregation on CME in primary corticostriatal cocultures indicate that aggregate-driven CME has direct implications for cytotoxicity in neurodegenerative disease.
Discussion
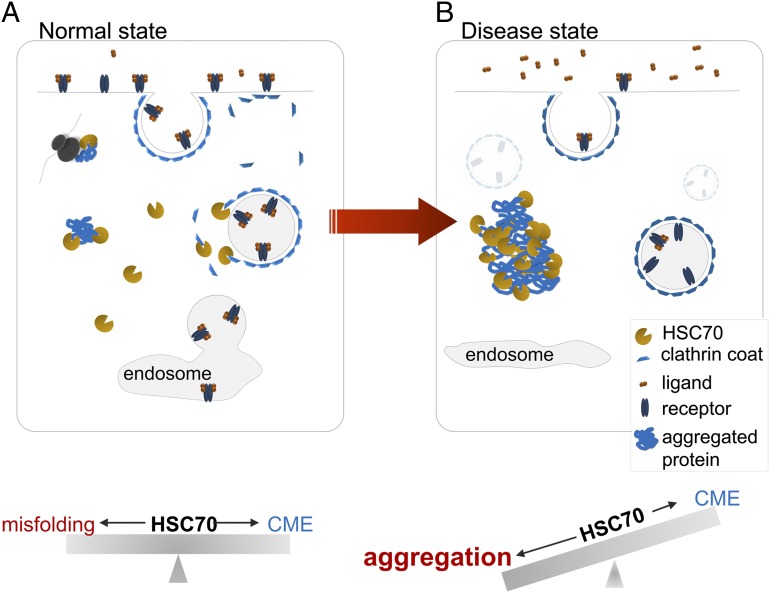
We have demonstrated that aggregation of neurodegenerative disease-associated proteins shifts the intracellular distribution of molecular chaperones, leading to the functional inhibition of an essential cellular function as exemplified by aggregate-induced sequestration of HSC70 from CME (Fig. 6). Whereas our studies reveal the primary effects of aggregation on CME are through the functional loss of HSC70, our results do not preclude the possibility that aggregation could eventually affect other aspects of endocytosis during aggregation progression, for example, by direct sequestration of other CME components or vesicle recycling to the cell surface as reported by others (34, 52). Given the role of HSC70 in maintaining the solubility of metastable coat components, as well in membrane receptor recycling (53–55), sequestration of this chaperone may help explain why these steps are also affected. Future studies using more sensitive methods will be illuminating to understand the exact kinetics and step(s) of the endocytic process that are inhibited, and whether different types of misfolding or aggregating species have specific effects on CME. Nevertheless, the ability to assay transferrin internalization directly provides a highly sensitive, straightforward, and physiologically relevant measure to quantify the downstream toxicity of misfolding and aggregation in various cell-based and tissue models of proteinopathy.
Fig. 6.
Model depicts chaperone competition between protein aggregates and the CME pathway. (A) Under normal conditions, HSC70 is at sufficiently high levels to mediate CME, as well as basal protein client folding. (B) Under disease conditions, where protein aggregates have accumulated and cytosolic HSC70 levels have been titrated, CME is inhibited, but this inhibition can be restored by increasing chaperone levels.
Our studies incorporated the use of cell culture models overexpressing various diseased proteins under the control of exogenous promoters. Our rationale for using this approach was to gain a simplified, “accelerated” view of the pathological consequence of protein aggregation in disease. Aggregation kinetics of many misfolded proteins occur more quickly at higher concentrations (56), and the use of intact brain tissue would be immensely challenging, given that the localization of aggregates could be distant from the synapses of the same neuron, making it difficult to deconvolve chaperone competition effects within a single neuron. Consequently, our experiments do not reflect the endogenous expression levels or temporal onset of protein misfolding and aggregation in these age-related diseases, where the accumulation and consequences of protein aggregation manifest after many years. The aberrant engagement of HSC70 by the misfolded proteins in our model systems may therefore be exaggerated. The threshold and onset of CME decline by chaperone competition are likely different across the multitude of distinct cell types within the diseased brain, given the complex interconnected network of neural, glial, and vascular tissues within the brain (more discussion below). Nonetheless, our experiments offer an important proof-of-principle demonstration of the downstream consequences of chaperone competition by disease-associated protein aggregates. Indeed, HSP70 members have been shown to coaggregate with inclusion bodies in Huntington disease and ALS brain samples (14, 15), and our results show that CME is sensitive to relatively modest changes in HSC70 levels. Thus, we propose that chaperone abundance is likely a rate-limiting factor in protein conformational diseases.
The specific inhibition of CME by aggregate-mediated chaperone competition has significant implications for neurodegenerative disease pathologies, given the central role of CME-mediated trafficking of vesicles and receptors in neurons (57). Synaptic neuronal transmission depends on CME to recycle synaptic vesicle components at the presynaptic membrane; at the postsynapse, CME regulates neuronal signaling outcomes by down-regulation of activated receptors, as well as by activation of compartmentalized signaling along the endosomal pathway (57, 58). The downstream effects of CME inhibition in neuronal cells are demonstrated here by decreased internalization of AMPA receptors, which are required for long-term, potentiation-based memory (49, 59). Inhibition of AMPA receptor endocytosis could cause detrimental changes in synapses of many neuronal proteinopathies. Aberrant activity of other receptors is also implicated in disease, as exemplified by misregulated glutamate receptor activity in Huntington disease (60).
More broadly, we propose that chaperone competition by protein aggregation will lead to the inhibition of other chaperone-dependent molecular processes and the failure of interconnected networks contributing to cellular dysfunction and disease. Our present work has focused on the function of HSC70 in CME, but this chaperone also has major roles in a large number of other essential processes, including protein synthesis and polypeptide targeting to organelles, such as the mitochondria, lysosomes, and proteasomes (21, 23, 25). Therefore, down-regulation of HSC70 likely affects these cellular processes and the activities of innumerable client proteins. Likewise, other chaperones, cochaperones, and proteostasis components—including HSP90, DNAJ, HSP110, BAG, CHIP, as well as the chaperonins, p97, and subunits of the proteasome—have been reported to associate with aggregates (16, 42), thus implicating numerous other downstream pathways affected by aggregate-driven chaperone competition.
We anticipate that the functional depletion of chaperones by protein aggregates will have selective but differential effects on other chaperone-dependent molecular and cellular processes, and will likely vary spatiotemporally. Each chaperone-dependent pathway is anticipated to have different sensitivities to the level of chaperone depletion, depending on the cellular abundance of specific client proteins, their requirements for chaperone activity, and their relative role in the respective pathway. The exact threshold for functional chaperone depletion will likely vary across cell and tissue types, because chaperone and client levels will differ within and among cells and tissues. This variability is supported by the heterogeneous effects of aggregation observed in our corticostriatal coculture experiments, and we anticipate this effect to be even more complex in the diseased brain, which consists of a complex interconnected network of distinct neural, glial, and vascular tissues. In addition, the consequences of chaperone competition will likely differ during the course of development, during adulthood, and throughout aging, where the relative concentrations and requirements for specific chaperones may change.
We propose that many aspects behind the heterogeneity of molecular pathologies in proteinopathies may arise from the complex downstream consequences of chaperone competition. Gain-of-function toxicity and loss-of-function phenotypes are ultimately inseparable outcomes of protein aggregation, given the inherent nature of chaperones. Aggregate-driven chaperone competition may help explain the phenotypic complexity of protein misfolding diseases. The beneficial consequence of increasing chaperone levels in cells containing misfolded protein, as exemplified by HSC70 and CME, offers a potential therapeutic approach to restore specific disease-affected molecular processes.
Materials and Methods
DNA Plasmids and Antibodies.
Plasmids encoding Q19-EGFP, Q82-EGFP, Htt (exon 1) Q23-EGFP, and Htt (exon 1) Q53-EGFP were described previously (17). Plasmids encoding SOD1 WT-AcGFP and SOD1 (A4V)-AcGFP were obtained from Addgene (ID26406 and ID26408). For Flp-In TREX plasmids, ORFs of human Atx1 Q2 or Q82 were first cloned into the pEGFP-C2 vector using the EcoRI and HindIII sites. Regions encoding EGFP-Atx1 Q2 or Q82 were subcloned into pcDNA5-FRT-TO vector (Life Technologies) between the AflII and NotI sites. The ORF of human HSC70 was PCR-amplified with the forward primer, GCGCTCGAGCTATGTCCAAGGGACCTGCAG, and the reverse primer, GCGGGATCCTTAATCAACCTCTTCAATGGTGGG, and the PCR fragment was cloned into pE2-Crimson-C1 (Clontech) using the XhoI and BamHI restriction sites. The regions encoding E2-Crimson HSC70 were then subcloned into pcDNA5-FRT-TO vector between the AflII and NotI sites. The HA-GluR2 ORF was obtained by digesting pGW1-HA-GluR2 plasmid with HindIII and SalI, where the resulting fragment was blunt-ended by Klenow fragment and ligated into the EcoRV site of the pcDNA5-FRT-TO vector (50). Htt exon 1 constructs gWiz-HttN90 Q73-CFP and gWiz-HttN90 Q23-CFP have been described previously (51, 61).
The specificity of the monoclonal 3a3 antibody that recognizes HSC70, as well as stress-inducible HSP70 used for indirect immunofluorescence and immunoblotting experiments, has been described previously (62). Commercial antibodies recognizing the following were used: anti-HA (monoclonal 12CA5, from hybridoma culture supernatant), anti-CHC (monoclonal TD1; Santa Cruz Biotechnology), anti–AP-2 component mu2 adaptin (monoclonal AC1M11; Abcam), anti-GAK/auxilin 2 (monoclonal G10; Santa Cruz Biotechnology), and various Alexa Fluor-conjugated secondary antibodies (Life Technologies). The specificities of 12CA5, TD1, and AC1M11 monoclonal antibodies have been previously described (63–65). Anti-GAK monoclonal antibody specifically recognizes amino acids 1–360 of rat GAK, which is conserved in the human protein.
Cell Culture and Transient Transfection.
Cells were maintained at 37 °C and 5% (vol/vol) CO2. For transfection, human PC3 prostate adenocarcinoma cells cultured in DMEM/F12 medium containing 10% (vol/vol) FBS were seeded at 3 × 105 in 12-well plates overnight. The cells were transfected with 500 ng of DNA plasmid for 6 h using XtremeGENE transfection reagent (Roche) and then reseeded and cultured on glass coverslips for 72 h before analysis.
Doxycycline-Inducible Flp-In T-REX Neuro2a Cell Lines.
Neuro2a-FRT-TREX cells (a gift from G. Matsumoto, Brain Science Institute, RIKEN, Saitama, Japan) were cotransfected with pcDNA5/FRT/TO- EGFP-Atx-1 Q2, EGFP-Atx-1 Q82, or E2 Crimson-HSC70 and Flp recombinase-encoding plasmid pOG44. Stable clones were selected using hygromycin B (100 μg/mL). Cells were maintained in complete DMEM/10% FBS supplemented with 50 μg/mL hygromycin B and 20 μg/mL blasticidin.
For induced expression of EGFP-Atx-1 Q2 or Q82, cells were seeded onto polylysine-coated coverslips in 12-well plates. Doxycycline was added at a final concentration of 1 μg/mL for 72 h to induce protein expression, and it was refreshed after 48 h. For experiments where E2-Crimson HSC70 and Q19-EGFP or Q82-EGFP were coexpressed, cells were seeded at a density of ∼5 × 105 into 12-well plates overnight. Q19-EGFP or Q82-EGFP plasmid (500 ng) was transfected into cells using XtremeGENE transfection reagent and then reseeded 6 h later on polylysine-coated coverslips in 12-well plates for a total incubation time of 72 h. Doxycycline (final concentration of 1 μg/mL) was added at the indicated time points after Q19-EGFP or Q82-EGFP transfection.
siRNA-Mediated HSC70 Depletion.
Mouse HSC70 was depleted using the following predesigned Silencer Select siRNA oligonucleotides (Life Technologies): HSC70 siRNA-1 (catalog no. S125649), HSC70 siRNA-2 (catalog no. S125651), and HSC70 siRNA-3 (catalog no. S67830), as well as the Silencer Select Negative Control 1 siRNA (catalog no. 4390843). Neuro2a cells were transfected with 10 nM total siRNA using Lipofectamine 2000 (Life Technologies) and subcultured onto polylysine-coated coverslips 24 h later in 12-well plates. Transferrin internalization was assayed an additional 24 h afterward.
Transferrin Internalization Assay.
Transferrin internalization was performed as described previously (66). Briefly, cells starved for 30 min in prewarmed, serum-free DMEM were incubated for 10 min with Alexa555-conjugated transferrin (10 μg/mL; Life Technologies) in prewarmed DMEM and 0.1% BSA for 10 min. The cells were then transferred on ice, and extracellular transferrin was removed by washing with ice-cold PBS, followed by a 5-min incubation with cold glycine buffer [0.1 M glycine (pH 2.5), 150 mM NaCl]. Cells were then fixed for immunofluorescence. Distribution of transferrin receptor at the cell surface was examined by incubating cells on ice with transferrin for 30 min, after which the cells were rinsed with ice-cold PBS and then immediately fixed with paraformaldehyde for microscopy.
Indirect Immunofluorescence.
Paraformaldehyde-fixed cells were permeabilized in PBS containing 0.1% Triton X-100 and 1% (wt/vol) BSA (PBS-BSA) for 10 min. Cells were sequentially incubated with primary antibodies and Alexa Fluor-conjugated secondary antibodies in PBS-BSA for 1 h at room temperature, with three PBS washes after each antibody treatment. Coverslips were mounted using Fluoromount-G (Southern Biotech).
Microscopy and Image Analysis.
Images were acquired using an inverted Leica SP5 II laser scanning confocal microscope equipped with a 63×/1.4 N.A. or 100×/1.44 N.A. oil immersion lens and HyD detectors. Sixteen-bit z-series of confocal sections (step size = 0.42 μm for 63× objective and 0.29 μm for 100× objective) were acquired in the photon-counting mode using Leica Applied Suite Advanced Fluorescence (LAS AF) software, and these acquisition parameters were kept identical across all samples. For image presentation, eight-bit maximal projections of the z-series were created using ImageJ (National Institutes of Health), and brightness was adjusted across the entire image using Volocity software (PerkinElmer). Images shown are representative of three independent experiments.
For quantification of Alexa555-transferrin internalization or endogenous HSC70/HSP70 expression levels in tissue culture cells, raw 16-bit z-series were first converted to sum slice projections, which represent the total sum value of the integrated fluorescence intensities of the entire z-series, using ImageJ. Individual cells were next outlined as regions of interest (ROIs), and the total integrated fluorescence intensities (Icell) were measured, using Volocity software. The mean intensity of an ROI outside of the cell was also measured to serve as background intensity (Iback), and the relative fluorescence intensity (Irel) was calculated as
where A represents the pixel area of the ROI.
Resultant data were normalized against the average values of control samples, compiled using Microsoft Excel and displayed using GraphPad Prism. For transferrin internalization assays, it was empirically determined that the cells with the 15% highest and lowest fluorescence intensities reliably represented mechanically broken or dead cells, respectively, so these cells were excluded from the analyses. For each condition, at least 500 cells in total were analyzed. Three independent experiments were performed, and representative data from one experiment are shown.
HA-GluR2 Internalization Assay.
Neuro2a cells were cotransfected with plasmids encoding either Q19 or Q82 and HA-GluR2 for 72 h. Plasma membrane-localized HA-GluR2 was labeled by incubating live cells with anti-HA antibody in DMEM at 37 °C for 10 min. Cells were washed in DMEM and then incubated with 3 μM insulin at 37 °C for 15 min. The cells were then assayed for transferrin internalization, fixed, permeabilized with TX-100 (Sigma), and immunostained with secondary Alexa Fluor-labeled mouse antibodies to label the internalized cell surface AMPA receptor (49).
Primary Mouse Neuronal Culture, Nucleofection, and Transferrin Internalization.
Striatal and cortical neurons were harvested from embryonic day (E) 18 C57BL/6J mice as previously described (51). Animals were euthanized in accordance with NIH guidelines and under Duke University Medical Center Institutional Animal Care and Use Committee approval and oversight. Cortical and striatal tissues were dissected from E18 mouse brains and enzymatically dissociated with papain/DNase 1 (Worthington Biochemical Corp.). A total of 5 × 106 cells of each neuronal type were electroporated separately with either the Htt exon 1 gWiz-HttN90 Q23-CFP or gWiz-HttN90 Q73-CFP construct using the Mouse Neuron Nucleofector Kit and Amaxa electroporation device (both from Lonza). Cortical and striatal neurons were then mixed and plated at equal densities onto poly-d-lysine–coated coverslips previously seeded with glial feeder layers. Cultures were grown in Neurobasal medium supplemented with B27 (Life Technologies), 2 mM glutamine (Glutamax; Life Technologies), 10 mM KCl, and 5 μg/mL gentamicin.
For transferrin internalization, 3-d in vitro cultures were starved for 1 h in Neurobasal medium without B27 supplement before being incubated with Alexa555-conjugated transferrin (10 μg/mL) for 15 min. The cultures were washed twice in ice-cold PBS, followed by a 60-s incubation in cold acid buffer (500 mM sodium chloride, 200 mM acetic acid), and then fixed with 4% (wt/vol) paraformaldehyde/4% (wt/vol) sucrose for 10 min. Neuronal cultures were processed for indirect immunofluorescence as described above, except cells were preincubated in blocking solution [5% (vol/vol) goat serum, 2% (wt/vol) BSA, 0.1% Triton X-100 in PBS] for 1 h prior, and were incubated with primary anti-HSC/HSP70 antibodies (monoclonal 3a3) overnight.
For imaging, 12-bit z-series through the entire cell volume were acquired using a Zeiss LSM 510 laser confocal microscope equipped with a 63×/1.4 N.A. oil immersion lens and LSM software (Zeiss). Images were quantified for transferrin internalization and Htt-CFP fluorescence as described above, except that ROIs of individual neurons were confined to the cell body and excluded most neuronal processes due to excessive overlap with neighboring cells.
FRAP.
FRAP experiments were conducted using an inverted Leica SP5 II laser scanning confocal microscope with a 63×/1.4 N.A. oil immersion lens, a photomultiplier tube detector, and an argon 488-nm laser line. PC-3 cells were transfected as described above, and cells were maintained at 37 °C and 5% CO2 using a Tokai imaging chamber (Tokai Hit) for all experiments. LAS AF software was used for image acquisition and raw data analysis. For acquisition, 12-bit, 512 × 512-resolution images were obtained, and a small rectangular ROI was photobleached using 100% laser power for three iterations. All magnification, photobleaching, and acquisition parameters were kept identical across all samples except for the gain, which was adjusted when appropriate so that the cells’ fluorescence would fall within the detector’s dynamic range.
For FRAP quantification, the mean fluorescence intensities of three ROIs were measured: the photobleached region (Ip), a region outside of the cell to check for overall background fluorescence (Ib), and a separate unbleached cell to correct for overall fluorescence fluctuations within the acquisition period (Ic). Microsoft Excel was used to calculate the normalized fluorescence intensity (I) for each individual FRAP experiment using the equation
where t represents time.
For data presentation, the mean averages and SEs of each sample were plotted using GraphPad Prism.
Protein Expression and Immunoblotting.
Neuro2a E2-Crimson HSC70 cells were seeded in 10-cm dishes, and protein expression was induced with doxycycline for the indicated periods of time. Cells were lysed and clarified by centrifugation as described before (67). Ten micrograms of total protein was separated by SDS/PAGE using a 4–12% (wt/vol) gradient gel (BioRad) and transferred to PVDF membranes. Membranes were sequentially probed with primary anti-HSC70/HSP70 antibodies (3a3) and secondary Alexa Fluor-labeled antibodies, and were visualized within the dynamic range using a Typhoon 9400 imaging system (GE). Quantification of protein bands was performed on raw 12-bit depth displays using ImageJ.
Supplementary Material
Acknowledgments
We thank G. Matsumoto for providing Neuro2a-FRT-TREX cells; Y. Wang for the HA-GluR2 construct; the Northwestern Biological Imaging Facility for the use of microscopes and software; L. Kaltenbach and M. Van Kanegan for help with corticostriatal cocultures; anonymous reviewers for helpful comments; and J. Kirstein-Miles, J. Labbadia, and C. Nussbaum-Krammer for critical reading of the manuscript. Y.S. is a Howard Hughes Medical Institute postdoctoral fellow of the Damon Runyon Cancer Foundation (DRG 2086-11). This work was supported by the Cure Huntington's Disease Initiative Foundation, Inc., National Institutes of Health (NIH) Grant NS080514 (to D.C.L.), the Chicago Biomedical Consortium, the Ellison Medical Foundation, and NIH Grants GM038109, GM081192, AG026647, and NS047331 (to R.I.M.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1321811111/-/DCSupplemental.
References
- 1.Chartier-Harlin MC, et al. Early-onset Alzheimer’s disease caused by mutations at codon 717 of the beta-amyloid precursor protein gene. Nature. 1991;353(6347):844–846. doi: 10.1038/353844a0. [DOI] [PubMed] [Google Scholar]
- 2.Bruijn LI, et al. ALS-linked SOD1 mutant G85R mediates damage to astrocytes and promotes rapidly progressive disease with SOD1-containing inclusions. Neuron. 1997;18(2):327–338. doi: 10.1016/s0896-6273(00)80272-x. [DOI] [PubMed] [Google Scholar]
- 3.Davies SW, et al. Formation of neuronal intranuclear inclusions underlies the neurological dysfunction in mice transgenic for the HD mutation. Cell. 1997;90(3):537–548. doi: 10.1016/s0092-8674(00)80513-9. [DOI] [PubMed] [Google Scholar]
- 4.DiFiglia M, et al. Aggregation of huntingtin in neuronal intranuclear inclusions and dystrophic neurites in brain. Science. 1997;277(5334):1990–1993. doi: 10.1126/science.277.5334.1990. [DOI] [PubMed] [Google Scholar]
- 5.Skinner PJ, et al. Ataxin-1 with an expanded glutamine tract alters nuclear matrix-associated structures. Nature. 1997;389(6654):971–974. doi: 10.1038/40153. [DOI] [PubMed] [Google Scholar]
- 6.Soto C. Unfolding the role of protein misfolding in neurodegenerative diseases. Nat Rev Neurosci. 2003;4(1):49–60. doi: 10.1038/nrn1007. [DOI] [PubMed] [Google Scholar]
- 7.Hartl FU, Bracher A, Hayer-Hartl M. Molecular chaperones in protein folding and proteostasis. Nature. 2011;475(7356):324–332. doi: 10.1038/nature10317. [DOI] [PubMed] [Google Scholar]
- 8.Balch WE, Morimoto RI, Dillin A, Kelly JW. Adapting proteostasis for disease intervention. Science. 2008;319(5865):916–919. doi: 10.1126/science.1141448. [DOI] [PubMed] [Google Scholar]
- 9.Hay DG, et al. Progressive decrease in chaperone protein levels in a mouse model of Huntington’s disease and induction of stress proteins as a therapeutic approach. Hum Mol Genet. 2004;13(13):1389–1405. doi: 10.1093/hmg/ddh144. [DOI] [PubMed] [Google Scholar]
- 10.Gidalevitz T, Ben-Zvi A, Ho KH, Brignull HR, Morimoto RI. Progressive disruption of cellular protein folding in models of polyglutamine diseases. Science. 2006;311(5766):1471–1474. doi: 10.1126/science.1124514. [DOI] [PubMed] [Google Scholar]
- 11.Ben-Zvi A, Miller EA, Morimoto RI. Collapse of proteostasis represents an early molecular event in Caenorhabditis elegans aging. Proc Natl Acad Sci USA. 2009;106(35):14914–14919. doi: 10.1073/pnas.0902882106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cummings CJ, et al. Chaperone suppression of aggregation and altered subcellular proteasome localization imply protein misfolding in SCA1. Nat Genet. 1998;19(2):148–154. doi: 10.1038/502. [DOI] [PubMed] [Google Scholar]
- 13.Chai Y, Koppenhafer SL, Bonini NM, Paulson HL. Analysis of the role of heat shock protein (Hsp) molecular chaperones in polyglutamine disease. J Neurosci. 1999;19(23):10338–10347. doi: 10.1523/JNEUROSCI.19-23-10338.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jana NR, Tanaka M, Wang Gh, Nukina N. Polyglutamine length-dependent interaction of Hsp40 and Hsp70 family chaperones with truncated N-terminal huntingtin: their role in suppression of aggregation and cellular toxicity. Hum Mol Genet. 2000;9(13):2009–2018. doi: 10.1093/hmg/9.13.2009. [DOI] [PubMed] [Google Scholar]
- 15.Watanabe M, et al. Histological evidence of protein aggregation in mutant SOD1 transgenic mice and in amyotrophic lateral sclerosis neural tissues. Neurobiol Dis. 2001;8(6):933–941. doi: 10.1006/nbdi.2001.0443. [DOI] [PubMed] [Google Scholar]
- 16.Olzscha H, et al. Amyloid-like aggregates sequester numerous metastable proteins with essential cellular functions. Cell. 2011;144(1):67–78. doi: 10.1016/j.cell.2010.11.050. [DOI] [PubMed] [Google Scholar]
- 17.Kim S, Nollen EA, Kitagawa K, Bindokas VP, Morimoto RI. Polyglutamine protein aggregates are dynamic. Nat Cell Biol. 2002;4(10):826–831. doi: 10.1038/ncb863. [DOI] [PubMed] [Google Scholar]
- 18.Matsumoto G, Stojanovic A, Holmberg CI, Kim S, Morimoto RI. Structural properties and neuronal toxicity of amyotrophic lateral sclerosis-associated Cu/Zn superoxide dismutase 1 aggregates. J Cell Biol. 2005;171(1):75–85. doi: 10.1083/jcb.200504050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rampelt H, et al. Metazoan Hsp70 machines use Hsp110 to power protein disaggregation. EMBO J. 2012;31(21):4221–4235. doi: 10.1038/emboj.2012.264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schlossman DM, Schmid SL, Braell WA, Rothman JE. An enzyme that removes clathrin coats: Purification of an uncoating ATPase. J Cell Biol. 1984;99(2):723–733. doi: 10.1083/jcb.99.2.723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chiang HL, Terlecky SR, Plant CP, Dice JF. A role for a 70-kilodalton heat shock protein in lysosomal degradation of intracellular proteins. Science. 1989;246(4928):382–385. doi: 10.1126/science.2799391. [DOI] [PubMed] [Google Scholar]
- 22.Bronk P, et al. Drosophila Hsc70-4 is critical for neurotransmitter exocytosis in vivo. Neuron. 2001;30(2):475–488. doi: 10.1016/s0896-6273(01)00292-6. [DOI] [PubMed] [Google Scholar]
- 23.Meacham GC, Patterson C, Zhang W, Younger JM, Cyr DM. The Hsc70 co-chaperone CHIP targets immature CFTR for proteasomal degradation. Nat Cell Biol. 2001;3(1):100–105. doi: 10.1038/35050509. [DOI] [PubMed] [Google Scholar]
- 24.Freeman BC, Yamamoto KR. Disassembly of transcriptional regulatory complexes by molecular chaperones. Science. 2002;296(5576):2232–2235. doi: 10.1126/science.1073051. [DOI] [PubMed] [Google Scholar]
- 25.Young JC, Hoogenraad NJ, Hartl FU. Molecular chaperones Hsp90 and Hsp70 deliver preproteins to the mitochondrial import receptor Tom70. Cell. 2003;112(1):41–50. doi: 10.1016/s0092-8674(02)01250-3. [DOI] [PubMed] [Google Scholar]
- 26.Sakahira H, Breuer P, Hayer-Hartl MK, Hartl FU. Molecular chaperones as modulators of polyglutamine protein aggregation and toxicity. Proc Natl Acad Sci USA. 2002;99(Suppl 4):16412–16418. doi: 10.1073/pnas.182426899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Park SH, et al. PolyQ proteins interfere with nuclear degradation of cytosolic proteins by sequestering the Sis1p chaperone. Cell. 2013;154(1):134–145. doi: 10.1016/j.cell.2013.06.003. [DOI] [PubMed] [Google Scholar]
- 28.Eisenberg E, Greene LE. Multiple roles of auxilin and hsc70 in clathrin-mediated endocytosis. Traffic. 2007;8(6):640–646. doi: 10.1111/j.1600-0854.2007.00568.x. [DOI] [PubMed] [Google Scholar]
- 29.Ahle S, Ungewickell E. Auxilin, a newly identified clathrin-associated protein in coated vesicles from bovine brain. J Cell Biol. 1990;111(1):19–29. doi: 10.1083/jcb.111.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fotin A, et al. Structure of an auxilin-bound clathrin coat and its implications for the mechanism of uncoating. Nature. 2004;432(7017):649–653. doi: 10.1038/nature03078. [DOI] [PubMed] [Google Scholar]
- 31.Braell WA, Schlossman DM, Schmid SL, Rothman JE. Dissociation of clathrin coats coupled to the hydrolysis of ATP: Role of an uncoating ATPase. J Cell Biol. 1984;99(2):734–741. doi: 10.1083/jcb.99.2.734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Harding C, Heuser J, Stahl P. Receptor-mediated endocytosis of transferrin and recycling of the transferrin receptor in rat reticulocytes. J Cell Biol. 1983;97(2):329–339. doi: 10.1083/jcb.97.2.329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Motley A, Bright NA, Seaman MN, Robinson MS. Clathrin-mediated endocytosis in AP-2-depleted cells. J Cell Biol. 2003;162(5):909–918. doi: 10.1083/jcb.200305145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Meriin AB, et al. Aggregation of expanded polyglutamine domain in yeast leads to defects in endocytosis. Mol Cell Biol. 2003;23(21):7554–7565. doi: 10.1128/MCB.23.21.7554-7565.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rosen DR, et al. Mutations in Cu/Zn superoxide dismutase gene are associated with familial amyotrophic lateral sclerosis. Nature. 1993;362(6415):59–62. doi: 10.1038/362059a0. [DOI] [PubMed] [Google Scholar]
- 36.Scherzinger E, et al. Self-assembly of polyglutamine-containing huntingtin fragments into amyloid-like fibrils: Implications for Huntington’s disease pathology. Proc Natl Acad Sci USA. 1999;96(8):4604–4609. doi: 10.1073/pnas.96.8.4604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.DiDonato M, et al. ALS mutants of human superoxide dismutase form fibrous aggregates via framework destabilization. J Mol Biol. 2003;332(3):601–615. doi: 10.1016/s0022-2836(03)00889-1. [DOI] [PubMed] [Google Scholar]
- 38.Kirchhausen T. Adaptors for clathrin-mediated traffic. Annu Rev Cell Dev Biol. 1999;15:705–732. doi: 10.1146/annurev.cellbio.15.1.705. [DOI] [PubMed] [Google Scholar]
- 39.Kirchhausen T. Clathrin. Annu Rev Biochem. 2000;69:699–727. doi: 10.1146/annurev.biochem.69.1.699. [DOI] [PubMed] [Google Scholar]
- 40.Owen DJ, Collins BM, Evans PR. Adaptors for clathrin coats: Structure and function. Annu Rev Cell Dev Biol. 2004;20:153–191. doi: 10.1146/annurev.cellbio.20.010403.104543. [DOI] [PubMed] [Google Scholar]
- 41.Ungewickell E, et al. Role of auxilin in uncoating clathrin-coated vesicles. Nature. 1995;378(6557):632–635. doi: 10.1038/378632a0. [DOI] [PubMed] [Google Scholar]
- 42.Kirstein-Miles J, Scior A, Deuerling E, Morimoto RI. The nascent polypeptide-associated complex is a key regulator of proteostasis. EMBO J. 2013;32(10):1451–1468. doi: 10.1038/emboj.2013.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Greener T, Zhao X, Nojima H, Eisenberg E, Greene LE. Role of cyclin G-associated kinase in uncoating clathrin-coated vesicles from non-neuronal cells. J Biol Chem. 2000;275(2):1365–1370. doi: 10.1074/jbc.275.2.1365. [DOI] [PubMed] [Google Scholar]
- 44.Umeda A, Meyerholz A, Ungewickell E. Identification of the universal cofactor (auxilin 2) in clathrin coat dissociation. Eur J Cell Biol. 2000;79(5):336–342. doi: 10.1078/S0171-9335(04)70037-0. [DOI] [PubMed] [Google Scholar]
- 45.Anggono V, Huganir RL. Regulation of AMPA receptor trafficking and synaptic plasticity. Curr Opin Neurobiol. 2012;22(3):461–469. doi: 10.1016/j.conb.2011.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shepherd JD, Huganir RL. The cell biology of synaptic plasticity: AMPA receptor trafficking. Annu Rev Cell Dev Biol. 2007;23:613–643. doi: 10.1146/annurev.cellbio.23.090506.123516. [DOI] [PubMed] [Google Scholar]
- 47.Carroll RC, Beattie EC, von Zastrow M, Malenka RC. Role of AMPA receptor endocytosis in synaptic plasticity. Nat Rev Neurosci. 2001;2(5):315–324. doi: 10.1038/35072500. [DOI] [PubMed] [Google Scholar]
- 48.Kastning K, et al. Molecular determinants for the interaction between AMPA receptors and the clathrin adaptor complex AP-2. Proc Natl Acad Sci USA. 2007;104(8):2991–2996. doi: 10.1073/pnas.0611170104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lee SH, Liu L, Wang YT, Sheng M. Clathrin adaptor AP2 and NSF interact with overlapping sites of GluR2 and play distinct roles in AMPA receptor trafficking and hippocampal LTD. Neuron. 2002;36(4):661–674. doi: 10.1016/s0896-6273(02)01024-3. [DOI] [PubMed] [Google Scholar]
- 50.Man HY, et al. Regulation of AMPA receptor-mediated synaptic transmission by clathrin-dependent receptor internalization. Neuron. 2000;25(3):649–662. doi: 10.1016/s0896-6273(00)81067-3. [DOI] [PubMed] [Google Scholar]
- 51.Kaltenbach LS, et al. Composite primary neuronal high-content screening assay for Huntington’s disease incorporating non-cell-autonomous interactions. J Biomol Screen. 2010;15(7):806–819. doi: 10.1177/1087057110373392. [DOI] [PubMed] [Google Scholar]
- 52.Richards P, et al. Dendritic spine loss and neurodegeneration is rescued by Rab11 in models of Huntington’s disease. Cell Death Differ. 2011;18(2):191–200. doi: 10.1038/cdd.2010.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Newmyer SL, Schmid SL. Dominant-interfering Hsc70 mutants disrupt multiple stages of the clathrin-coated vesicle cycle in vivo. J Cell Biol. 2001;152(3):607–620. doi: 10.1083/jcb.152.3.607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Jiang RF, Greener T, Barouch W, Greene L, Eisenberg E. Interaction of auxilin with the molecular chaperone, Hsc70. J Biol Chem. 1997;272(10):6141–6145. doi: 10.1074/jbc.272.10.6141. [DOI] [PubMed] [Google Scholar]
- 55.Lee DW, Zhao X, Zhang F, Eisenberg E, Greene LE. Depletion of GAK/auxilin 2 inhibits receptor-mediated endocytosis and recruitment of both clathrin and clathrin adaptors. J Cell Sci. 2005;118(Pt 18):4311–4321. doi: 10.1242/jcs.02548. [DOI] [PubMed] [Google Scholar]
- 56.Vendruscolo M, Knowles TP, Dobson CM. Protein solubility and protein homeostasis: A generic view of protein misfolding disorders. Cold Spring Harb Perspect Biol. 2011;3(12) doi: 10.1101/cshperspect.a010454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.McMahon HT, Boucrot E. Molecular mechanism and physiological functions of clathrin-mediated endocytosis. Nat Rev Mol Cell Biol. 2011;12(8):517–533. doi: 10.1038/nrm3151. [DOI] [PubMed] [Google Scholar]
- 58.von Zastrow M, Sorkin A. Signaling on the endocytic pathway. Curr Opin Cell Biol. 2007;19(4):436–445. doi: 10.1016/j.ceb.2007.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ahmadian G, et al. Tyrosine phosphorylation of GluR2 is required for insulin-stimulated AMPA receptor endocytosis and LTD. EMBO J. 2004;23(5):1040–1050. doi: 10.1038/sj.emboj.7600126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Labbadia J, Morimoto RI. Huntington’s disease: Underlying molecular mechanisms and emerging concepts. Trends Biochem Sci. 2013;38(8):378–385. doi: 10.1016/j.tibs.2013.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Reinhart PH, et al. Identification of anti-inflammatory targets for Huntington’s disease using a brain slice-based screening assay. Neurobiol Dis. 2011;43(1):248–256. doi: 10.1016/j.nbd.2011.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Carr VM, Murphy SP, Morimoto RI, Farbman AI. Small subclass of rat olfactory neurons with specific bulbar projections is reactive with monoclonal antibodies to the HSP70 heat shock protein. J Comp Neurol. 1994;348(1):150–160. doi: 10.1002/cne.903480109. [DOI] [PubMed] [Google Scholar]
- 63.Wilson IA, et al. The structure of an antigenic determinant in a protein. Cell. 1984;37(3):767–778. doi: 10.1016/0092-8674(84)90412-4. [DOI] [PubMed] [Google Scholar]
- 64.Näthke IS, et al. Folding and trimerization of clathrin subunits at the triskelion hub. Cell. 1992;68(5):899–910. doi: 10.1016/0092-8674(92)90033-9. [DOI] [PubMed] [Google Scholar]
- 65.Robinson MS. 100-kD coated vesicle proteins: Molecular heterogeneity and intracellular distribution studied with monoclonal antibodies. J Cell Biol. 1987;104(4):887–895. doi: 10.1083/jcb.104.4.887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kirchhausen T, Macia E, Pelish HE. Use of dynasore, the small molecule inhibitor of dynamin, in the regulation of endocytosis. Methods Enzymol. 2008;438:77–93. doi: 10.1016/S0076-6879(07)38006-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Calamini B, et al. Small-molecule proteostasis regulators for protein conformational diseases. Nat Chem Biol. 2012;8(2):185–196. doi: 10.1038/nchembio.763. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.