Significance
The plant hormone abscisic acid (ABA) acts both as a developmental signal and as an integrator of environmental cues such as drought and cold. ABA perception recruits an ABA-binding regulatory component [regulatory component of ABA receptor (RCAR)/PYR1/PYL] and an associated protein phosphatase 2C (PP2C). Phytohormone binding to the RCAR inactivates the phosphatase activity of the coreceptor, permitting phosphorelay of the ABA signal via downstream protein kinases. The amino acid residues of the ABA-binding pocket are conserved among RCAR members; however, RCAR7 has several changes in these residues, and its function as an ABA receptor has remained elusive. Here we show a bona fide ABA receptor function for RCAR7 and identify structural constraints that contribute to specific pairing of RCAR7 with PP2Cs.
Abstract
The plant hormone abscisic acid (ABA) acts both as a developmental signal and as an integrator of environmental cues such as drought and cold. ABA perception recruits an ABA-binding regulatory component [regulatory component of ABA receptor (RCAR)/PYR1/PYL] and an associated protein phosphatase 2C (PP2C). Phytohormone binding inactivates the phosphatase activity of the coreceptor, permitting phosphorelay of the ABA signal via downstream protein kinases. RCARs and PP2C coreceptors are represented by small protein families comprising 14 and 9 members in Arabidopsis, respectively. The specificity of the RCAR–PP2C interaction and the constraints contributing to specific combinations are poorly understood. In this contribution, we analyzed RCAR7/PYL13, which is characterized by three variant amino acid residues in the conserved ABA-binding pocket. RCAR7 regulated the phosphatase activity of the PP2Cs ABI1, ABI2, and PP2CA in vitro at nanomolar ABA levels; however, it was unable to regulate the structurally related hypersensitive to ABA 1 (HAB1). Site-directed mutagenesis of HAB1 established ABA-dependent regulation by RCAR7. Conversion of the noncanonical amino acid residues of RCAR7 into the consensus ABA-binding pocket did not perceptibly change receptor function. Ectopic expression of RCAR7 in Arabidopsis resulted in ABA hypersensitivity affecting gene regulation, seed germination, and stomatal closure. The RCAR7 loss-of-function mutant revealed no changes in ABA responses, similar to the RCAR9 knockout line, whereas the combined deficiency of RCAR7 and RCAR9 resulted in ABA-insensitive seed germination. The study shows a role of RCAR7 in early plant development, proves its ABA receptor function, and identifies structural constraints of RCAR7–PP2C interaction.
The phytohormone abscisic acid (ABA) is a major regulator of plant development and environmental responses. ABA accumulates in response to water deficit and triggers rapid physiological responses for homeostasis of plants’ water status. The immediate response includes stomatal closure to limit transpiration and initiation of adaptive gene expression (1, 2), including protection of the photosynthetic machinery (3). ABA perception occurs by soluble ABA receptors acting in concert with protein phosphatases 2C (PP2Cs) of clade A (1, 4–6). Biochemical, genetic, and structural analysis revealed a holo-receptor complex in which the PP2C coreceptor is required for both high-affinity binding of ABA and negative control of downstream ABA signaling. The ABA-binding proteins, referred to as regulatory component of ABA receptor (RCAR) or PYR1/PYL proteins, constitute a protein family of 14 members in Arabidopsis subdivided into three groups according to their sequence homology (7, 8). ABA binding stabilizes the RCAR–PP2C complex and releases PP2C downstream targets from the latent inhibition by the protein phosphatase (9, 10); however, a few RCAR members are capable of regulating PP2Cs in the absence of ABA (11). ABA holo-receptors strongly discriminate against both nonphysiological ABA stereoisomers (8, 12) and ABA biosynthetic precursors (13), but some receptor complexes tolerate binding of structurally unrelated ABA agonists (7, 14–16).
Ligand binding in the hydrophobic cavity of RCARs triggers a conformational change in two adjacent loops, referred to as “gate” and “latch” (17–19). Conserved amino acid residues in the RCAR molecule and a conserved tryptophan residue found in clade A PP2Cs coordinate the phytohormone by direct ionic interaction and/or water-mediated hydrogen bonds. ABA coreceptors present in the cytosol and nucleus (8, 20, 21) target protein kinases, transcription factors, and ion channels for ABA-dependent regulation (22–26). The PP2Cs negatively regulate SNF1-related kinases such as OST1/SnrK2.6/SnrK2E, SnrK2.2/SnrK2D, and SnRK2.3/SnrK2I (27, 28). Downstream phosphorylation targets are nuclear-localized transcription factors such as the ABA response element-binding factors and membrane-localized anion channels (28–32). ABA coreceptors can form stable protein complexes with RCARs and SnRK2, which remain associated with PP2C by virtue of ionic and hydrophobic interactions reminiscent of signalosome complexes (9–11, 33, 34).
The Arabidopsis genome encodes nine PP2Cs of clade A (35). Interaction studies with RCARs indicate a large number of possible receptor–coreceptor combinations, although the specificity of holo-receptor formation has not been systematically explored (7, 21, 34, 36, 37). Site-directed mutagenesis of RCARs and PP2Cs identified critical amino acid residues required for interaction (7, 38–40). Biochemical analyses revealed distinct ABA sensitivities controlled both by PP2C as well as the RCAR component (8, 11, 12, 21). Interesting in this context is the RCAR7 family member, which appears to differ from the majority of other RCARs in that it fails to bind to the hypersensitive to ABA 1 (HAB1) PP2C in a heterologous system (36). Of the 14 RCARs, RCAR7 is the only one that has a variant ABA-binding pocket, with three nonconsensus amino acids, and an ABA receptor function of this member has remained elusive. Recently, an ABA-independent inhibition of PP2Cs by RCAR7 has been postulated (41, 42).
In this study, we show a bona fide ABA receptor function of RCAR7. RCAR7 regulates a number of coreceptors such as PP2CA and ABA insensitive 2 (ABI2) by inhibiting phosphatase activity in an ABA-dependent manner, although not the structurally related HAB1. Structural constraints that contribute to the specific discrimination of HAB1 have been elucidated.
Results
Conserved Amino Acid Residues at the ABA Binding Site and Alterations in RCAR7.
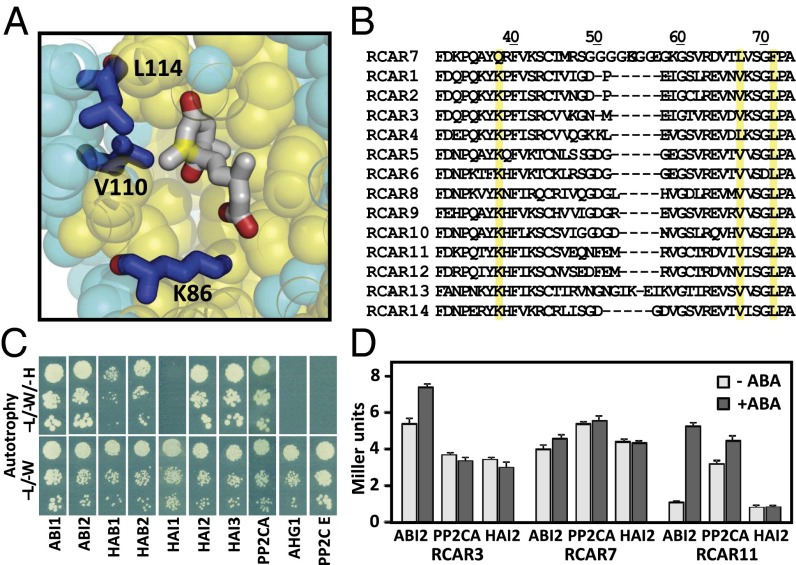
Analyses of RCAR–ABA crystal structures (17–19) identified a number of highly conserved amino acid residues that form the inner RCAR cavity for binding ABA (Fig. 1A). Alignment of the primary RCAR structures reveals that RCAR7 is exceptional among the ABA receptors because, only in this member, three amino acid residues of the ABA-binding pocket differ from the consensus. Two of these changes are specific for RCAR7, namely the glutamine residue at position 38 and the phenylalanine at position 71, replacing the prototypical lysine and leucine residues, respectively (Fig. 1B). The third variation is also found in RCAR4, and is a replacement of a conserved valine residue by leucine at position 67 of RCAR7. The amino acid substitutions are expected to cause only minor physicochemical changes. Phenylalanine is somewhat more bulky than leucine and glutamine is less basic than lysine, whereas leucine and valine are quite similar. Given the function and strict conservation of these amino acids in the other RCARs, these differences in RCAR7 still might interfere with ABA binding and, hence, with phosphatase regulation and receptor function.
Fig. 1.
ABA-binding pocket of RCAR7 and interaction with PP2Cs. (A) ABA bound to RCAR12. Amino acid residues of RCAR12 lining the ABA-binding pocket are shown as yellow spheres, whereas those farther away from the ligand (gray molecule) are shown in cyan. Three RCAR-conserved amino acid residues of the ABA-binding pocket and divergent in RCAR7 are highlighted as blue residues. The view shows a cut through the RCAR12–ABI1 crystal structure (19) using PyMOL (www.pymol.org) such that K86, V110, and L114 are visible. The methyl group of the C2′ from the ABA carbon ring (marked in yellow) provides a hydrophobic interaction with RCAR residues. (B) Alignment of the primary structure of the RCAR family. A partial sequence is shown and the positions of Q38, L67, and F71 in RCAR7 are emphasized in yellow, which are divergent from the consensus. (C) Y2H analysis of RCAR7 interaction with all nine clade A PP2Cs. The histidine-autotrophic growth (-H) shown on leucine- (-L) and tryptophane- (-W) deficient media indicates RCAR7–PP2C interaction. PP2CE is a clade E PP2C that served as a negative control. (D) Quantitative β-glucosidase assays of yeast lines expressing different RCAR–PP2C Y2H combinations grown in absence and presence of 30 μM ABA. The activity is given in Miller units (mean ± SD).
RCAR7 Binding to Specific PP2Cs in Yeast.
To examine a putative receptor function of RCAR7, we performed a first indicative experiment by testing whether RCAR7 is able to interact with known ABA coreceptor PP2Cs. We analyzed all nine clade A PP2Cs for binding to RCAR7 using the yeast two-hybrid (Y2H) system, including a clade E PP2C as a negative control (Fig. 1C). The analysis revealed clear interaction of RCAR7 with six ABA coreceptors. In the presence of RCAR7 and coexpressed ABI1, ABI2, HAB2, highly ABA-induced PP2C gene 2 (HAI2), HAI3, or PP2CA, yeast colonies were capable of histidine-autotrophic growth but not in the absence of RCAR7 expression (Fig. S1), indicating a robust interaction of the Arabidopsis proteins in yeast. The other members of the PP2C subfamily, HAB1, HAI1, and AHG1, were not able to support histidine-autotrophic growth, similar to the negative control (Fig. 1C). Interestingly, HAB1, which is closely related to the other ABI1-subfamily PP2Cs (35), does not clearly interact with RCAR7 in this system. ABI2, PP2CA, and HAI2 interacted with RCAR7 irrespective of the presence of ABA, as indicated by the LacZ reporter activity, similar to RCAR3 and different from RCAR11 (Fig. 1D).
Control of ABA Signaling by RCAR7 Expression.
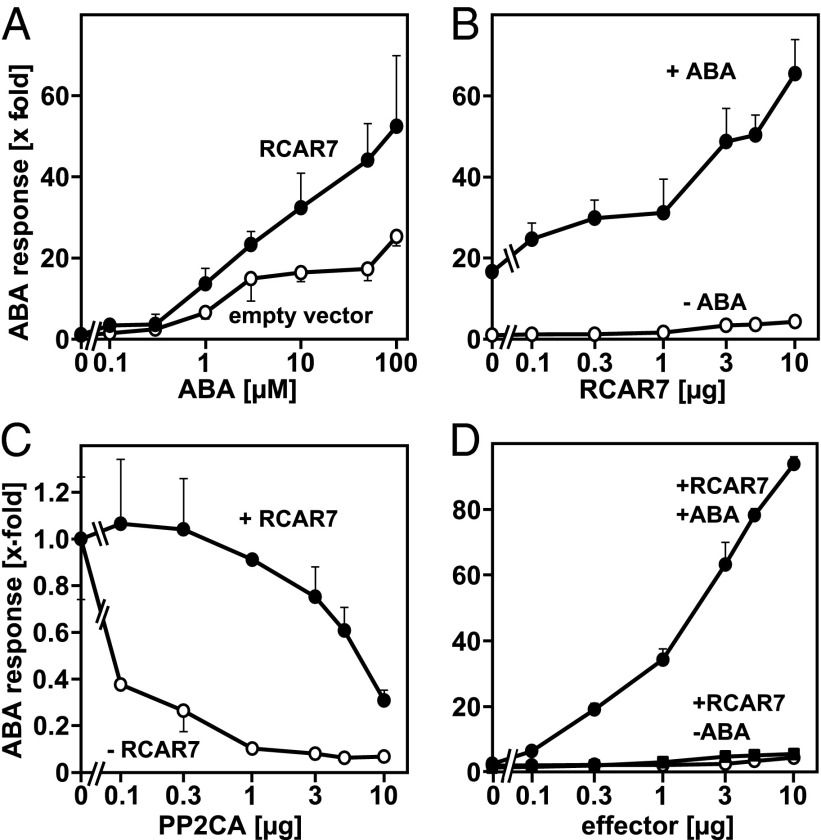
The results indicate an interaction of RCAR7 with most clade A PP2Cs and are compatible with a role of RCAR7 in ABA signaling. To examine the predicted function of RCAR7 in Arabidopsis, ABA-dependent signaling was analyzed in vivo using mesophyll protoplasts. Ectopic expression of RCAR7 consistently stimulated the ABA response approximately twofold in the presence of 1 μM or higher levels of exogenous ABA compared with the empty effector cassette (Fig. 2A). The presence of 5 μM ABA induced ABA-responsive reporter expression by a factor of 17 in the absence of ectopically expressed RCAR7 (Fig. 2B), whereas coexpression of various levels of RCAR7 effector further increased the ABA response, and an induction up to 65-fold was observed relative to the non–ABA-treated control. The RCAR7-mediated increase of ABA signaling was still moderate and did not exceed a factor of 4, similar to the fourfold stimulatory effect of RCAR7 in the absence of exogenous ABA.
Fig. 2.
Regulation of ABA signaling by RCAR7 in Arabidopsis protoplasts. (A) RCAR7- and ABA-dependent activation of ABA signaling in Arabidopsis cells. Protoplasts were transfected with the RCAR7 expression cassette or the empty vector, and induction of ABA-responsive luciferase expression was expressed relative to the control (without ABA and RCAR7). (B) Regulation of ABA response by different effector levels of RCAR7 in the absence or presence of 5 µM ABA (note the ABA-evoked induction in the absence of RCAR7 expression). (C) Inhibition of ABA-induced reporter expression by ectopic PP2CA expression without and with RCAR7 transfection (5 µg effector DNA) in the presence of 10 µM ABA. The reporter expression transfected without PP2CA was 2.8 kRLU (relative light unit)/RFU (relative fluorescence unit) for cells without RCAR7 and 8.0 kRLU/RFU for cells with RCAR7 expression, and both values were set to 1. (D) Activation of PP2CA-inhibited ABA response is dependent on ABA and RCAR7. The ABA response was inhibited by PP2CA expression (1 µg DNA). The inhibition was counteracted by RCAR7 in the presence and absence of 5 µM exogenous ABA. The data are expressed as fold induction compared with the response without RCAR7 and ABA (set to 1). The ABA response with 5 µM exogenous ABA and varying levels of the empty vector cassette is shown as an additional control (open circles). Activities were measured as RLUs using the ABA-responsive pRD29B::luciferase reporter and normalized to RFUs of a constitutively expressed control gene. Each data point represents the mean value of at least three independent transfections (mean ± SD).
The moderate inductive action of RCAR7 throughout the entire ABA range indicated that either RCAR7 is an ineffective ABA response regulator compared with RCAR1 and RCAR3 (8, 12, 27) or PP2Cs targeted by RCAR7 were limiting in the mesophyll cells. To rule out the latter possibility, we expressed PP2CA in Arabidopsis protoplasts as a robust interactor of RCAR7 (Fig. 1D). The clade A PP2Cs are characterized as negative regulators of the ABA response. Consistently, ABA-mediated reporter activation (10 μM ABA) was gradually attenuated as the amount of PP2CA effector was increased (Fig. 2C). Coexpression of RCAR7 efficiently rescued the ABA response. For example, transfection of protoplasts with 0.3 μg DNA of PP2CA effector plasmid caused a decline of the ABA response by ∼70%, whereas concomitant expression of RCAR7 (5 μg DNA) fully restored it. Subsequently, Arabidopsis cells were cotransfected with increasing levels of RCAR7 and with a constant level of PP2CA effector (1 μg; Fig. 2D), causing a 90% attenuation of the ABA response without RCAR7, as shown in Fig. 2C. In clear difference from the moderate induction of the ABA response by RCAR7 observed in the first analysis (Fig. 2A), RCAR7 now stimulated ABA signaling by up to 90-fold in the presence of exogenous ABA, whereas in the absence of ABA or RCAR7 a maximal enhancement of the hormone response by a factor of 4 occurred. The results reveal an RCAR7- and ABA-dependent activation of ABA signaling by targeting PP2CA.
RCAR7-Dependent PP2C Regulation.
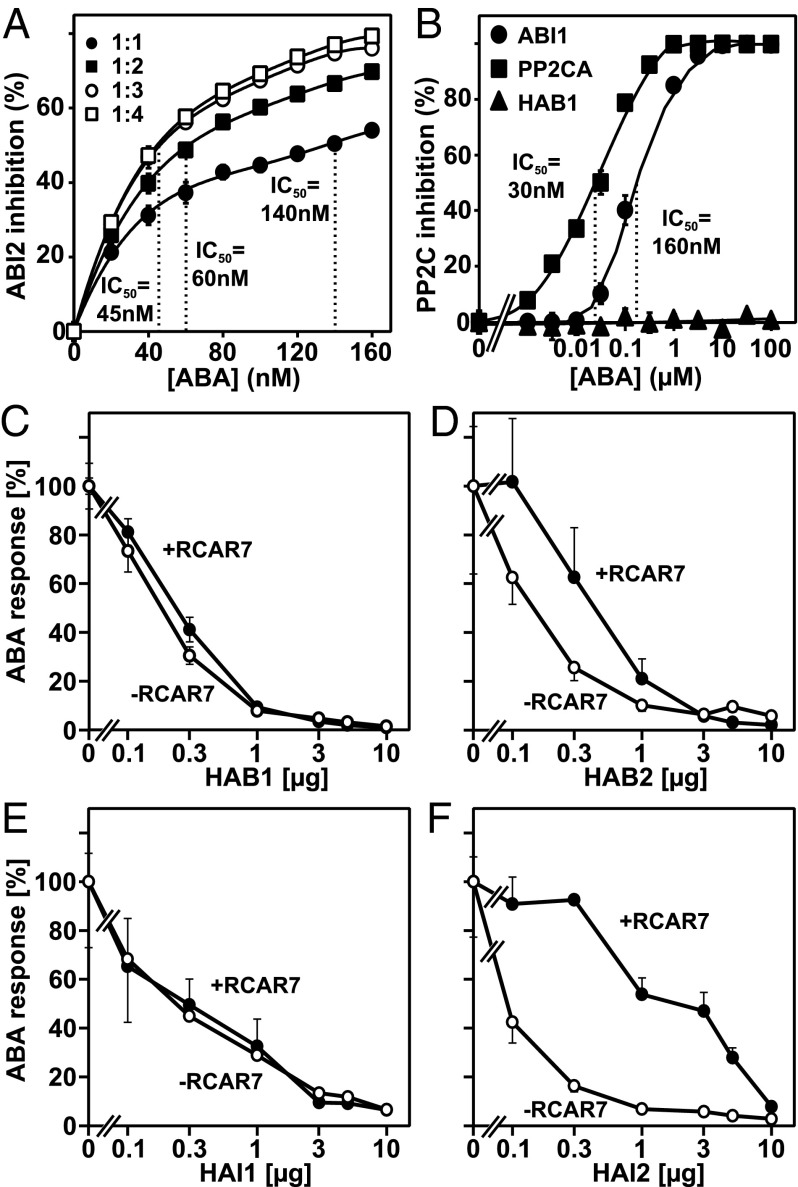
The in vivo analysis showed a positive regulatory function of RCAR7 in ABA-responsive gene expression at micromolar ABA levels comparable to other characterized RCAR members (8, 12, 27). Hence, the RCAR7 changes in amino acid residues conserved among the RCAR family might not impair ABA binding. To address this question, purified RCAR7 protein was examined for ABA-mediated inhibition of PP2C activity. Inhibition of phosphatase activity by RCARs requires the formation of a ternary complex consisting of the heteromeric protein and the phytohormone. This complex formation is a first-order reaction at constant ABA levels and, consequently, the concentration of the proteinaceous reactants and the stoichiometry of RCAR and PP2C affect the IC50 values for ABA (12, 13). We used the PP2C ABI2 at a 50 nM concentration and RCAR7 in an equimolar or up to fourfold higher concentration. In all cases, ABI2 was efficiently inhibited by nanomolar levels of ABA and the IC50 values ranged from 45 to 140 nM with 60 ± 9 nM ABA at a twofold molar excess of RCAR7 (Fig. 3A).
Fig. 3.
ABA receptor function. (A) RCAR7-mediated inhibition of ABI2 phosphatase activity by ABA. ABI2 protein (50 nM) was incubated in the presence of equimolar and two-, three-, and fourfold excess of RCAR7 in the presence of nanomolar ABA concentrations. The IC50-ABA for each ratio is depicted. (B) Specific regulation of PP2Cs by RCAR7. The experiment was performed as mentioned in A at a twofold molar excess of RCAR7 over ABI1, PP2CA, and HAB1. HAB1 phosphatase activity is not regulated by RCAR7, even at 0.1 mM ABA (mean ± SD). (C–F) Analysis of RCAR7-mediated rescue of ABA signaling attenuated by PP2C expression. The analysis was conducted as mentioned in Fig. 2C, except that the PP2CA effector was replaced by HAB1 (C), HAB2 (D), HAI1 (E), and HAI2 (F).
The ABA sensitivity of the heteromeric complex is known to differ between a specific RCAR member and the interacting PP2C (12, 21). We compared three other clade A PP2Cs with ABI2 in their RCAR7- and ABA-mediated inhibition. ABI1 had an IC50 value of 160 ± 19 nM ABA, clearly reduced compared with ABI2 at a 1:2 ratio of PP2C to ABA receptor, whereas the RCAR7–PP2CA complex revealed a higher ABA sensitivity and the IC50 value was shifted to 30 ± 5 nM ABA. PP2CA inhibition was strictly dependent on both RCAR7 and ABA (Fig. S2A). HAB1 was not inhibited in the presence of RCAR7 even at 100 µM ABA (Fig. 3B); however, inhibition of this PP2C occurred efficiently by RCAR3, 8, and 11 (Fig. S2B). The noninteracting property of HAB1 with RCAR7 was also observed in yeast interaction studies (Fig. 1) (36) and corroborated in ABA signaling analyses (Fig. 3C). Arabidopsis protoplasts transfected with increasing HAB1 effector levels showed a clear inhibition of ABA signaling. Cotransfection of RCAR7 did not rescue ABA signaling (Fig. 3C), in contrast to the analysis with PP2CA (Fig. 2C) and HAB2 (Fig. 3D). An observation comparable to the HAB1 experiment was made for HAI1 (Fig. 3E), whereas the HAI2-imposed inhibition of the ABA response was counteracted by RCAR7 (Fig. 3F). The RCAR7-dependent ABA signaling analysis reflects the interaction data obtained in vitro for HAB1, and the study is consistent with the interaction analysis in yeast for HAB1, HAB2, HAI1, and HAI2 (Fig. 1C).
RCAR7 Variants with Consensus Amino Acid Residues.
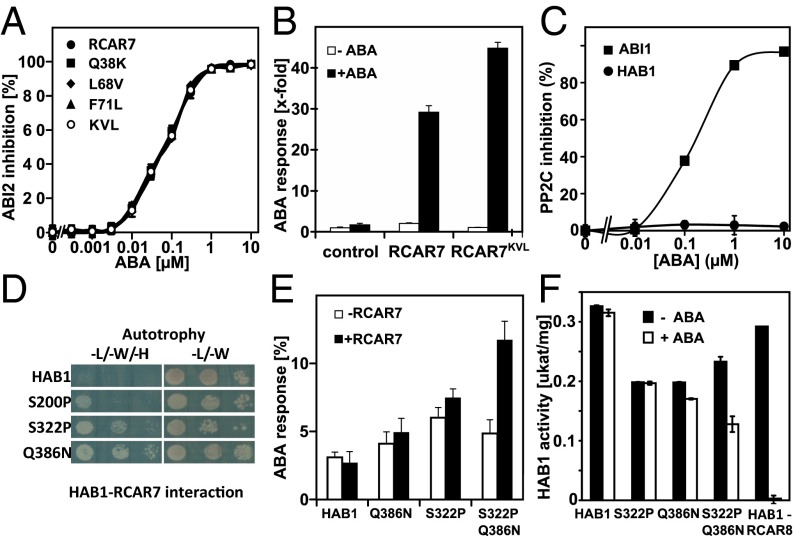
The high affinity of RCAR7 and specific PP2Cs for ABA is illustrated by low IC50 ABA values (Fig. 3 A and B). To address the effect of the noncanonical amino acid residues in RCAR7 on receptor function, the three nonconserved RCAR7 amino acids were singly converted into the consensus residues of the RCAR family. Subsequently, the consequence of the conversion was examined in vitro by analyzing RCAR7-mediated ABI2 regulation. None of the three changes altered the inhibitory action (Fig. 4A), and RCAR7Q38K, RCAR7L67V, and RCAR7F71L were indistinguishable from RCAR7 in the ABA-dependent regulation of phosphatase activity. The triple-variant RCAR7KVL also regulated ABI2 indistinguishable from RCAR7, indicating that the change of the amino acid residues had no apparent effect on the ABA sensitivity of the ABI2–RCAR7 complex. Assessment of RCAR7KVL capacity to stimulate ABA signaling in the presence of coexpressed PP2CA showed a highly active variant similar in activity to RCAR7 (Fig. 4B). Thus, the restoration of the amino acid sequence discrepancies in RCAR7 to the conserved ABA-binding pocket revealed no major effect on the ABA receptor’s function to regulate ABA responses.
Fig. 4.
RCAR7 and HAB1 variants. (A) Changing the ABA-binding pocket to the consensus residues does not affect ABA-dependent RCAR7 regulation of ABI2. The single-amino acid variants of RCAR7 in which Q38, L68, and F71 were exchanged for K, V, and L, respectively, and the triple-modified form RCAR7KVL were indistinguishable from RCAR7 in their capacity to inhibit PP2C (50 nM) in the presence of ABA. (B) Activation of ABA signaling by the triple-altered RCAR7KVL compared with RCAR7. The analysis was performed in protoplasts expressing PP2CA (1 µg DNA) and the RCAR7 effector with or without exogenous ABA (5 µM). The experiments without RCAR expression served as controls, and the ABA response in the absence of ABA was set to 1 (n = 3). (C) The RCAR7KVL variant is not able to regulate HAB1 but rather ABI1 protein phosphatase in the presence of ABA. The in vitro analysis was performed as described in Fig. 3A. (D) Yeast two-hybrid analysis of the interaction of RCAR7 variants with HAB1. The single-amino acid variants of HAB1 in which S200, S322, and Q386 were exchanged for P, P, and N, respectively, revealed enhanced growth on selective media (-H) supporting an RCAR7–PP2C interaction. (E) Partial rescue of HAB1-imposed inhibition of ABA signaling by changing two amino acid residues in HAB1S322P,Q386N. The analysis is performed as mentioned in B, but using RCAR7 and various HAB1 versions (HAB1S322P, HAB1Q385N, HAB1S322P,Q386N) as effectors. The analysis was conducted in triplicate (mean ± SD), and four independent analyses showed regulation by HAB1S322P,Q386N in the presence of 0.1 mM ABA. (F) ABA-mediated inhibition of HAB1S322P,Q386N by RCAR7 in the presence of 0.1 mM ABA. The HAB1–RCAR8 complex served as a control for HAB1 regulation.
Conversion of HAB1 into an RCAR7-Regulated Target.
The strong discrimination of HAB1 by RCAR7 was surprising, because HAB1 is highly similar in primary structure to other RCAR7-regulated PP2Cs such as HAB2, ABI1, and ABI2. RCAR7KVL is able to regulate ABA-dependent PP2CA but not HAB1 (Fig. 4C), indicating that the three nonconsensus amino acids of RCAR7 are not key for the observed PP2C selectivity.
To identify structural components critical for RCAR7–PP2C interaction, crystal structures of trimeric RCAR–PP2C–ABA receptor complexes (11, 17–19) were scrutinized for RCAR-contacting amino acid residues of PP2Cs. Three variations in the primary structure of HAB1 were scored, which might contribute to the discrimination in RCAR7 interaction, including a conserved asparagine residue of clade A PP2C that interacts with the closed latch structure of RCARs. This asparagine is found in ABI1 and ABI2 but is changed into glutamine in HAB1 (Q386) and HAB2. Whereas glutamine and asparagine are chemically quite similar, the more bulky glutamine residue might generate interaction constraints that are compensated in HAB2 but not in HAB1. In addition, S200 and S322 of HAB1 replace PP2C-conserved proline residues. Hence, the three changes were introduced into HAB1 and the variants were analyzed for recovery of RCAR7 protein interaction by the Y2H system (Fig. 4D). The S322P and Q386N modifications of HAB1 significantly improved RCAR7 interaction, as indicated by histidine-autotrophic yeast growth. Subsequently, HAB1S322P and HAB1Q386N were analyzed for rescue of ABA signaling by RCAR7 in Arabidopsis (Fig. 4E). The HAB1 versions showed little to no RCAR7-dependent rescue of ABA signaling (P = 0.2), but combining both changes in HAB1S322P,Q386N allowed a moderate RCAR7-mediated rescue (P < 0.001). To corroborate the findings, the HAB1 point-mutated forms were analyzed for ABA-mediated RCAR7 regulation in vitro. None of the versions showed ABA regulation by RCAR7 in the nanomolar ABA range (Fig. 4F), as observed for other PP2Cs (Fig. S2). In the presence of high ABA levels (100 μM), RCAR7 regulated HAB1Q386N marginally and HAB1S322P,Q386N half-maximally. The in vivo and in vitro analyses indicate a partial recovery of RCAR7 regulation by combined changes of two amino acid residues in HAB1.
Function of RCAR7 in Seed Germination.
The biochemical and molecular biological characterization supports a bona fide receptor function for RCAR7. ABA controls a number of physiological processes such as seed germination, root growth, and stomatal closure in which RCAR7 might be involved. To study the physiological processes regulated by RCAR7, we analyzed overexpressors and knockout lines.
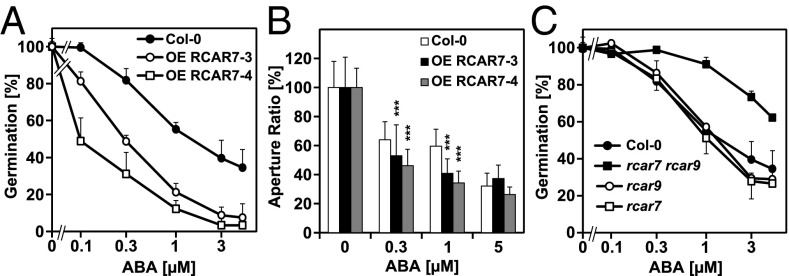
Several independent Arabidopsis lines were generated that ectopically expressed RCAR7. All of these exhibited ABA-hypersensitive germination compared with the parental line. The analysis of two representative lines is depicted in Fig. 5A, which revealed a shift in the IC50 values from 1.5 μM ABA in the parental line to 0.1 and 0.3 μM ABA in the two RCAR7 lines. Transcript analysis revealed a low expression of RCAR7 in wild-type seeds and seedlings, whereas ectopic expression resulted in more than 10- and 100-fold higher levels, respectively (Fig. S3A). ABA hypersensitivity was pronounced for the germination response and was also detectable in stomatal control (P < 0.001; Fig. 5B). Root growth regulation was not altered (Fig. S3B). Comparable analyses of a line with homozygous T-DNA insertion in the exon of the RCAR7 gene showed no significant change in ABA insensitivity during germination (Fig. 5C) and root growth regulation (Fig. S3C) as well as stomatal closing (Fig. S3D). RCAR single knockouts scarcely show significant phenotypic differences due to genetic redundancy, and only multiple RCAR-deficient lines gave rise to robust ABA-insensitive phenotypes (7). We therefore generated a double mutant between RCAR7 and an insertion allele of the closely related RCAR9. The homozygous double mutant rcar7rcar9 was ABA-insensitive with respect to seed germination (Fig. 5C). The IC50 values of the parental lines including the Columbia (Col-0) wild type were ∼1.5 μM ABA, whereas the IC50 value was shifted to above 5 μM ABA in the double mutant. Root growth (Fig. S3C) and stomatal regulation in response to ABA (Fig. S3D) were not changed in the double mutant. The data indicate that RCAR7 in combination with RCAR9 contributes to ABA-mediated germination inhibition in Arabidopsis.
Fig. 5.
Control of ABA responses in Arabidopsis plants. (A) Ectopic expression of RCAR7 (OE RCAR7) resulted in ABA-hypersensitive inhibition of seed germination. The germination rate of two representative lines is presented in comparison with the parental line (Col-0). (B) Stomatal response of the lines shown in A exposed to ABA (n > 20 per data point, mean ± SD). ***P < 0.001 indicates a significant difference from the Col-0 control (two-way ANOVA with posthoc Tukey test). (C) Seed germination of the rcar7 and rcar9 mutants in comparison with Col-0. The double mutant rcar7rcar9 exhibited ABA-insensitive germination (n > 90 per data point, mean ± SD).
Discussion
ABA perception and signaling profoundly impact on the plant phosphoproteome (43), metabolism, and gene expression (44). The specific roles played by the different RCAR–PP2C receptor combinations within the network are still poorly understood. In this study, we unequivocally corroborated a bona fide ABA receptor function of RCAR7 based on high ABA sensitivity of RCAR7–PP2C complexes, its ABA-dependent activation of ABA-responsive gene expression, and RCAR7-mediated changes in ABA responses of plants. In two recent studies, RCAR7 has been claimed to selectively inhibit PP2CA independent of ABA and not to bind ABA because of changes in conserved amino acid residues involved in ABA binding (41, 42). We found no evidence for this claim. In our hands, RCAR7 inhibited PP2CA, ABI2, and ABI1 only in the presence of both RCAR7 and ABA in the receptor assay (Fig. 3 and Fig. S2). Q38 of RCAR7 has been postulated to disrupt coordination of the carboxyl group of ABA mediated by a conserved lysine residue in other RCARs, and the bulky phenylalanine residue of RCAR7 at position 71 might generate a “steric clash” with the methyl group of ABA (41). RCAR7Q38L, RCAR7L67V, RCAR7F71L, and the triple-modified RCAR7KVL were indistinguishable in the ABA-sensitive inhibition of ABI2 phosphatase activity, with half-maximal inactivation at 60 nM ABA (Fig. 4A). The results indicate that the amido group of Q38 can substitute for carboxyl binding via the amino group of lysine, as found for a folate carrier (45), and that F71 does not compromise receptor function. In line with our observation, exchange of valine for a leucine residue in RCAR11 corresponding to L67 in RCAR7 did not inhibit PP2C interaction (40).
The functionality of RCAR7 and RCAR7KVL as positive regulators of ABA response was corroborated by the ABA- and RCAR7-dependent rescue of PP2C-inhibited ABA signaling. The analysis uncovered different efficiencies of RCAR7 to antagonize clade A PP2C action. A more than 80% inhibition of ABA signaling imposed by PP2CA and HAI2 expression was suppressed by concomitant expression of RCAR7, whereas the ABA receptor was ineffective in counteracting HAB1 and HAI1. The failure of RCAR7 to counteract HAB1 and HAI1 in vivo might be accounted for by the incapability of binding to both PP2Cs, as indicated by yeast interaction analysis. The incapacity of RCAR7 to interact with HAB1 was also observed by Bhaskara et al. (36), and is reflected in the complete insensitivity of HAB1 for ABA regulation by RCAR7. Whereas RCAR7 inhibited PP2CA, ABI2, and ABI1 at nanomolar ABA levels, this ABA receptor was unable to detectably regulate HAB1 even at 0.1 mM ABA concentration. RCAR1, 3, 8, and 11 efficiently inhibited HAB1 phosphatase activity in an ABA-sensitive manner. HAB1 is highly similar in structure to HAB2, ABI1, and ABI2, which have been shown in this study to interact with RCAR7 either in yeast, plant cells, or in vitro. The interaction of the receptor with the coreceptor has features of a pseudosubstrate binding to PP2C, and the contact sites are conserved (9, 35). Several amino acid residues of HAB1 located at the structural interface are different in ABI1 and ABI2, and introducing such ABI1/ABI2-conserved residues into HAB1 (HAB1S322P and HAB1Q386N) allowed RCAR7 binding, as indicated by yeast interaction analysis. These single-amino acid changes were not sufficient to cause significant regulation of HAB1 activity in vivo and in vitro; however, combining both alterations allowed ABA-sensitive regulation by RCAR7. The amino acids are located at the outer rim of the PP2C–RCAR interaction surface (Fig. S4A). The Q386 residue of HAB1 is positioned next to the conserved tryptophan residue W385 (Fig. S4 B–D) that interlocks with the ABA receptor and is involved in ABA coordination (17–19, 39). The change of this glutamine Q386 to the less bulky asparagine residue seems to contribute to a better interaction of RCAR7 and HAB1. However, the low ABA sensitivity of the RCAR7–HAB1S322P,Q386N complex implies additional structural constraints for efficient HAB1 regulation. The crystal structure of RCAR7–PP2CA revealed an additional interaction domain provided by a zinc-finger domain present in PP2CA and HAI-member PP2Cs (41), which might provide an explanation for the high affinity of PP2CA in vitro and the high efficiency of RCAR7 to antagonize PP2CA and HAI2 action on ABA signaling. Several studies identified amino acid residues of RCARs (7, 14, 40, 46) that alter interaction with PP2Cs, but none have reported so far on successful alteration of PP2C to establish RCAR binding. For clade A PP2Cs, a number of mutations are known to reduce or obliterate the interaction with RCARs, which results in ABA-insensitive coreceptors such as the classical abi1-1 (ABI1G180D) and abi2-1 (ABI2G168D) proteins or mutations in the ABA-sensing tryptophan residue (7, 8, 39).
PP2CA is a negative regulator of ABA-sensitive seed germination and stomatal closure (29, 37, 47, 48). Deficiency of RCAR7 would restrain the inhibitory action on PP2CA by this ABA receptor. The ABA-insensitive germination of the rcar7rcar9 double mutant indicates the redundant function of both RCARs in this developmental process. Interestingly, the rcar7 line was unaffected in the stomatal response to ABA, whereas ectopic expression of RCAR7 enhanced ABA sensitivity of stomata and seed germination, as also observed by an independent study (42).
Understanding ABA signaling is key for a rational approach to improving crop performance under a limiting water supply. The complex interaction network of the ABA receptors with their cognate coreceptors provides a major challenge in elucidating the molecular details of ABA action. This study provides insights into structural constraints critical for ABA receptor–coreceptor specificity, which will aid in future attempts to explore and design the regulatory capacity of distinct receptor complexes.
Methods
Plant Material and Chemicals.
Chemicals were obtained from Sigma-Aldrich and J.T. Baker, and ABA was obtained from Lomon Bio Technology. The Arabidopsis lines were provided by the Nottingham Arabidopsis Stock Center, including the knockout lines for RCAR7 (SALK 071488) and RCAR9 (SM 3.17321) in the Col-0 background. The double mutant rcar7rcar9 was generated by crossing the parental lines and selecting for homozygous knockout lines by insertion-specific PCR analysis (primers: RCAR7: 5′-ggcctcttcgctattacgc-3′, 5′-tcggtgacggtgataattca-3′, and 5′-tggttcacgtagtgggccatcg-3′; RCAR9: 5′-CTCTCTTTCCTAATCCACATACTTGC-3′, 5′-GCTACGAGAGCTGTACAAGAAGAAATGG-3′, and 5′-CTTATTTCAGTAAGAGTGTGGGGTTTTGG-3′). RCAR7 overexpressors were established by transferring an expression cassette for RCAR7 under control of the 35S promoter into Arabidopsis and selecting for homozygous lines as carried out for RCAR1 (8). RCAR7 transcript abundance was determined by RT-PCR analysis (Bio-Rad; CFX96) using ubiquitin 10 as reference and the primers 5′-TGGCGGTAACCATAGGCTTGC-3′ and 5′-TAGTTCCTTCCGGCACATCC-3′ for RCAR7 and 5′-GGCCTTGTATAATCCCTGATGAATAAG-3′ and 5′-AAAGAGATAACAGGAACGGAAACATAGT-3′ for UBI10. RNA extraction and subsequent cDNA biosynthesis from seeds imbibed in water and kept at 4 °C for 1 d and from 4-d-old seedlings were performed as described (3). Col-0 plants were grown in a perlite-soil mixture at 23 °C under long-day conditions with 16 h of light (250 µE⋅m−2⋅s−1) (20).
Physiological Analysis.
Stomatal apertures were analyzed as described (8) with minor modifications. Briefly, epidermal peels were generated, immobilized on translucent adhesive tape, and immediately mounted on a microscope slide for photography with a digital camera (Olympus Soft Imaging System Solution F-View II) using an Olympus BX61 microscope. Stomatal apertures were measured using digital images and imaging software (ImageJ 1.45s; http://rsb.info.nih.gov/ij). Germination and root growth assays were performed in the presence of various ABA levels, as reported (13, 49).
Receptor Assays.
Expression, purification, and analysis of RCARs and PP2Cs were performed as described (13). RCAR and PP2Cs were expressed in Escherichia coli strain M15 and purified to homogeneity. HAB1 versions were generated by site-directed mutagenesis according to the manufacturer’s instruction (QuikChange; Stratagene). All constructs used were verified for correctness by DNA sequencing. RCAR7 expression yielded 0.05 mg protein/L of E. coli culture, whereas the yields for PP2CA and HAB1 were 2.5 and 0.5 mg, respectively. The PP2Cs had a specific protein phosphatase activity of 0.45 μkat/mg PP2CA, 0.32 μkat/mg HAB1, 0.21 μkat/mg ABI1, and 0.27 μkat/mg ABI2. The receptor assay was performed with 50 nM PP2C and a twofold molar excess of RCAR unless otherwise stated. Values are means ± SDs of four replicates. Yeast two-hybrid tests were carried out for analysis of receptor–coreceptor interaction as described (3). HAB1 (AT1G72770), HAB2 (AT1G17550), PP2CA (AT3G11410), HAI1 (AT5G59220), HAI2 (AT1G07430), HAI3 (AT2G29380), AHG3 (AT3G11410), and PP2CE (AT5G27930) were amplified from a cDNA library as mentioned (8), cloned into the pGAD vector (Clontech), and expressed as activation domain fusion proteins. [Data accessible via The Arabidopsis Information Resource (www.arabidopsis.org).] All constructs were verified by DNA sequence analysis.
ABA Signaling in Protoplasts.
Preparation and analysis of Arabidopsis protoplasts were performed as described (20). Arabidopsis protoplasts of the ABA-deficient aba2-1 mutant (105 protoplasts in 0.1 mL) were transfected with 5 µg DNA of reporter construct (pRD29B::LUC), 3 µg of p35S::GUS plasmid as a control for internal normalization of expression, and various levels of effector expression cassettes as indicated. The effector cassettes drive expression of RCAR and PP2C cDNA under control of the 35S promoter (20), and all constructs were verified by DNA sequence analysis. The total amount of transfected DNA per assay remained constant by supplementation with DNA of the empty effector cassette. The protoplast suspensions were incubated in the presence or absence of ABA immediately after transfection. Assays were done in three replicates per data point.
Statistical Analysis.
Statistical analyses were performed using R 2.15.1 software (http://www.r-project.org). All experiments were repeated at least three times with similar results.
Supplementary Material
Acknowledgments
We are grateful to Johanna Berger, Christoph Heidersberger, and Christian Kornbauer for technical assistance. We thank the Deutsche Forschungsgemeinschaft (EG938, SFB924) and ForPlanta for financial support.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1322085111/-/DCSupplemental.
References
- 1.Cutler SR, Rodriguez PL, Finkelstein RR, Abrams SR. Abscisic acid: Emergence of a core signaling network. Annu Rev Plant Biol. 2010;61:651–679. doi: 10.1146/annurev-arplant-042809-112122. [DOI] [PubMed] [Google Scholar]
- 2.Kim TH, Böhmer M, Hu H, Nishimura N, Schroeder JI. Guard cell signal transduction network: Advances in understanding abscisic acid, CO2, and Ca2+ signaling. Annu Rev Plant Biol. 2010;61:561–591. doi: 10.1146/annurev-arplant-042809-112226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yang Y, et al. Fibrillin expression is regulated by abscisic acid response regulators and is involved in abscisic acid-mediated photoprotection. Proc Natl Acad Sci USA. 2006;103(15):6061–6066. doi: 10.1073/pnas.0501720103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Raghavendra AS, Gonugunta VK, Christmann A, Grill E. ABA perception and signalling. Trends Plant Sci. 2010;15(7):395–401. doi: 10.1016/j.tplants.2010.04.006. [DOI] [PubMed] [Google Scholar]
- 5.Hauser F, Waadt R, Schroeder JI. Evolution of abscisic acid synthesis and signaling mechanisms. Curr Biol. 2011;21(9):R346–R355. doi: 10.1016/j.cub.2011.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Miyakawa T, Fujita Y, Yamaguchi-Shinozaki K, Tanokura M. Structure and function of abscisic acid receptors. Trends Plant Sci. 2013;18(5):259–266. doi: 10.1016/j.tplants.2012.11.002. [DOI] [PubMed] [Google Scholar]
- 7.Park SY, et al. Abscisic acid inhibits type 2C protein phosphatases via the PYR/PYL family of START proteins. Science. 2009;324(5930):1068–1071. doi: 10.1126/science.1173041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ma Y, et al. Regulators of PP2C phosphatase activity function as abscisic acid sensors. Science. 2009;324(5930):1064–1068. doi: 10.1126/science.1172408. [DOI] [PubMed] [Google Scholar]
- 9.Soon FF, et al. Molecular mimicry regulates ABA signaling by SnRK2 kinases and PP2C phosphatases. Science. 2012;335(6064):85–88. doi: 10.1126/science.1215106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xie T, et al. Molecular mechanism for inhibition of a critical component in the Arabidopsis thaliana abscisic acid signal transduction pathways, SnRK2.6, by protein phosphatase ABI1. J Biol Chem. 2012;287(1):794–802. doi: 10.1074/jbc.M111.313106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hao Q, et al. The molecular basis of ABA-independent inhibition of PP2Cs by a subclass of PYL proteins. Mol Cell. 2011;42(5):662–672. doi: 10.1016/j.molcel.2011.05.011. [DOI] [PubMed] [Google Scholar]
- 12.Szostkiewicz I, et al. Closely related receptor complexes differ in their ABA selectivity and sensitivity. Plant J. 2010;61(1):25–35. doi: 10.1111/j.1365-313X.2009.04025.x. [DOI] [PubMed] [Google Scholar]
- 13.Kepka M, et al. Action of natural abscisic acid precursors and catabolites on abscisic acid receptor complexes. Plant Physiol. 2011;157(4):2108–2119. doi: 10.1104/pp.111.182584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Peterson FC, et al. Structural basis for selective activation of ABA receptors. Nat Struct Mol Biol. 2010;17(9):1109–1113. doi: 10.1038/nsmb.1898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Okamoto M, et al. Activation of dimeric ABA receptors elicits guard cell closure, ABA-regulated gene expression, and drought tolerance. Proc Natl Acad Sci USA. 2013;110(29):12132–12137. doi: 10.1073/pnas.1305919110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cao M, et al. An ABA-mimicking ligand that reduces water loss and promotes drought resistance in plants. Cell Res. 2013;23(8):1043–1054. doi: 10.1038/cr.2013.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Melcher K, et al. A gate-latch-lock mechanism for hormone signalling by abscisic acid receptors. Nature. 2009;462(7273):602–608. doi: 10.1038/nature08613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yin P, et al. Structural insights into the mechanism of abscisic acid signaling by PYL proteins. Nat Struct Mol Biol. 2009;16(12):1230–1236. doi: 10.1038/nsmb.1730. [DOI] [PubMed] [Google Scholar]
- 19.Miyazono K, et al. Structural basis of abscisic acid signalling. Nature. 2009;462(7273):609–614. doi: 10.1038/nature08583. [DOI] [PubMed] [Google Scholar]
- 20.Moes D, Himmelbach A, Korte A, Haberer G, Grill E. Nuclear localization of the mutant protein phosphatase abi1 is required for insensitivity towards ABA responses in Arabidopsis. Plant J. 2008;54(5):806–819. doi: 10.1111/j.1365-313X.2008.03454.x. [DOI] [PubMed] [Google Scholar]
- 21.Santiago J, et al. Modulation of drought resistance by the abscisic acid receptor PYL5 through inhibition of clade A PP2Cs. Plant J. 2009;60(4):575–588. doi: 10.1111/j.1365-313X.2009.03981.x. [DOI] [PubMed] [Google Scholar]
- 22.Vlad F, et al. Protein phosphatases 2C regulate the activation of the Snf1-related kinase OST1 by abscisic acid in Arabidopsis. Plant Cell. 2009;21(10):3170–3184. doi: 10.1105/tpc.109.069179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Umezawa T, et al. Type 2C protein phosphatases directly regulate abscisic acid-activated protein kinases in Arabidopsis. Proc Natl Acad Sci USA. 2009;106(41):17588–17593. doi: 10.1073/pnas.0907095106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Brandt B, et al. Reconstitution of abscisic acid activation of SLAC1 anion channel by CPK6 and OST1 kinases and branched ABI1 PP2C phosphatase action. Proc Natl Acad Sci USA. 2012;109(26):10593–10598. doi: 10.1073/pnas.1116590109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lynch T, Erickson BJ, Finkelstein RR. Direct interactions of ABA-insensitive(ABI)-clade protein phosphatase(PP)2Cs with calcium-dependent protein kinases and ABA response element-binding bZIPs may contribute to turning off ABA response. Plant Mol Biol. 2012;80(6):647–658. doi: 10.1007/s11103-012-9973-3. [DOI] [PubMed] [Google Scholar]
- 26.Rodrigues A, et al. ABI1 and PP2CA phosphatases are negative regulators of Snf1-related protein kinase1 signaling in Arabidopsis. Plant Cell. 2013;25(10):3871–3884. doi: 10.1105/tpc.113.114066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fujii H, et al. In vitro reconstitution of an abscisic acid signalling pathway. Nature. 2009;462(7273):660–664. doi: 10.1038/nature08599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fujita Y, et al. Three SnRK2 protein kinases are the main positive regulators of abscisic acid signaling in response to water stress in Arabidopsis. Plant Cell Physiol. 2009;50(12):2123–2132. doi: 10.1093/pcp/pcp147. [DOI] [PubMed] [Google Scholar]
- 29.Lee SC, Lan W, Buchanan BB, Luan S. A protein kinase-phosphatase pair interacts with an ion channel to regulate ABA signaling in plant guard cells. Proc Natl Acad Sci USA. 2009;106(50):21419–21424. doi: 10.1073/pnas.0910601106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Geiger D, et al. Activity of guard cell anion channel SLAC1 is controlled by drought-stress signaling kinase-phosphatase pair. Proc Natl Acad Sci USA. 2009;106(50):21425–21430. doi: 10.1073/pnas.0912021106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sirichandra C, et al. The Arabidopsis ABA-activated kinase OST1 phosphorylates the bZIP transcription factor ABF3 and creates a 14-3-3 binding site involved in its turnover. PLoS ONE. 2010;5(11):e13935. doi: 10.1371/journal.pone.0013935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Geiger D, et al. Guard cell anion channel SLAC1 is regulated by CDPK protein kinases with distinct Ca2+ affinities. Proc Natl Acad Sci USA. 2010;107(17):8023–8028. doi: 10.1073/pnas.0912030107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nishimura N, et al. PYR/PYL/RCAR family members are major in-vivo ABI1 protein phosphatase 2C-interacting proteins in Arabidopsis. Plant J. 2010;61(2):290–299. doi: 10.1111/j.1365-313X.2009.04054.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Antoni R, et al. PYRABACTIN RESISTANCE1-LIKE8 plays an important role for the regulation of abscisic acid signaling in root. Plant Physiol. 2013;161(2):931–941. doi: 10.1104/pp.112.208678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fuchs S, Grill E, Meskiene I, Schweighofer A. Type 2C protein phosphatases in plants. FEBS J. 2013;280(2):681–693. doi: 10.1111/j.1742-4658.2012.08670.x. [DOI] [PubMed] [Google Scholar]
- 36.Bhaskara GB, Nguyen TT, Verslues PE. Unique drought resistance functions of the highly ABA-induced clade A protein phosphatase 2Cs. Plant Physiol. 2012;160(1):379–395. doi: 10.1104/pp.112.202408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Antoni R, et al. Selective inhibition of clade A phosphatases type 2C by PYR/PYL/RCAR abscisic acid receptors. Plant Physiol. 2012;158(2):970–980. doi: 10.1104/pp.111.188623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Melcher K, et al. Identification and mechanism of ABA receptor antagonism. Nat Struct Mol Biol. 2010;17(9):1102–1108. doi: 10.1038/nsmb.1887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dupeux F, et al. Modulation of abscisic acid signaling in vivo by an engineered receptor-insensitive protein phosphatase type 2C allele. Plant Physiol. 2011;156(1):106–116. doi: 10.1104/pp.110.170894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mosquna A, et al. Potent and selective activation of abscisic acid receptors in vivo by mutational stabilization of their agonist-bound conformation. Proc Natl Acad Sci USA. 2011;108(51):20838–20843. doi: 10.1073/pnas.1112838108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li W, et al. Molecular basis for the selective and ABA-independent inhibition of PP2CA by PYL13. Cell Res. 2013;23(12):1369–1379. doi: 10.1038/cr.2013.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhao Y, et al. The unique mode of action of a divergent member of the ABA-receptor protein family in ABA and stress signaling. Cell Res. 2013;23(12):1380–1395. doi: 10.1038/cr.2013.149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kline KG, Barrett-Wilt GA, Sussman MR. In planta changes in protein phosphorylation induced by the plant hormone abscisic acid. Proc Natl Acad Sci USA. 2010;107(36):15986–15991. doi: 10.1073/pnas.1007879107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Böhmer M, Schroeder JI. Quantitative transcriptomic analysis of abscisic acid-induced and reactive oxygen species-dependent expression changes and proteomic profiling in Arabidopsis suspension cells. Plant J. 2011;67(1):105–118. doi: 10.1111/j.1365-313X.2011.04579.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Deng Y, et al. Role of lysine 411 in substrate carboxyl group binding to the human reduced folate carrier, as determined by site-directed mutagenesis and affinity inhibition. Mol Pharmacol. 2008;73(4):1274–1281. doi: 10.1124/mol.107.043190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pizzio GA, et al. The PYL4 A194T mutant uncovers a key role of PYR1-LIKE4/PROTEIN PHOSPHATASE 2CA interaction for abscisic acid signaling and plant drought resistance. Plant Physiol. 2013;163(1):441–455. doi: 10.1104/pp.113.224162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kuhn JM, Boisson-Dernier A, Dizon MB, Maktabi MH, Schroeder JI. The protein phosphatase AtPP2CA negatively regulates abscisic acid signal transduction in Arabidopsis, and effects of abh1 on AtPP2CA mRNA. Plant Physiol. 2006;140(1):127–139. doi: 10.1104/pp.105.070318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yoshida T, et al. ABA-hypersensitive germination3 encodes a protein phosphatase 2C (AtPP2CA) that strongly regulates abscisic acid signaling during germination among Arabidopsis protein phosphatase 2Cs. Plant Physiol. 2006;140(1):115–126. doi: 10.1104/pp.105.070128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Christmann A, Weiler EW, Steudle E, Grill E. A hydraulic signal in root-to-shoot signalling of water shortage. Plant J. 2007;52(1):167–174. doi: 10.1111/j.1365-313X.2007.03234.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.