Significance
Ribosomopathies are paradoxical: They first appear as diseases caused by too few cells but later present as diseases caused by too many. Here, we show that the presence of too few cells is caused by a quality control system that eliminates mutant ribosomes before they are allowed to translate mRNAs. Genetic suppression of this quality control system increases the amount of ribosomes available to cells. However, this 60S subunit deficiency results in release of defective ribosomes into the translationally active pool. A genetic model is presented describing how these types of defects may result in cancer, resolving the ribosomopathy paradox.
Keywords: frameshifting, L10/uL16
Abstract
Ribosomopathies are a class of diseases caused by mutations that affect the biosynthesis and/or functionality of the ribosome. Although they initially present as hypoproliferative disorders, such as anemia, patients have elevated risk of hyperproliferative disease (cancer) by midlife. Here, this paradox is explored using the rpL10-R98S (uL16-R98S) mutant yeast model of the most commonly identified ribosomal mutation in acute lymphoblastic T-cell leukemia. This mutation causes a late-stage 60S subunit maturation failure that targets mutant ribosomes for degradation. The resulting deficit in ribosomes causes the hypoproliferative phenotype. This 60S subunit shortage, in turn, exerts pressure on cells to select for suppressors of the ribosome biogenesis defect, allowing them to reestablish normal levels of ribosome production and cell proliferation. However, suppression at this step releases structurally and functionally defective ribosomes into the translationally active pool, and the translational fidelity defects of these mutants culminate in destabilization of selected mRNAs and shortened telomeres. We suggest that in exchange for resolving their short-term ribosome deficits through compensatory trans-acting suppressors, cells are penalized in the long term by changes in gene expression that ultimately undermine cellular homeostasis.
Ribosomopathies are a family of congenital diseases that are linked to genetic defects in ribosomal proteins or ribosome biogenesis factors. They are characterized by pleiotropic abnormalities that include birth defects, heart and lung diseases, connective tissue disorders, anemia, ataxia, and mental retardation (reviewed in ref. 1). Although each ribosomopathy presents a unique pathological spectrum, the inherited forms are characterized by bone marrow failure and anemia early in life, followed by elevated cancer risk by middle age. For example, although childhood anemia is one of the cardinal symptoms of the genetically inherited disease Diamond–Blackfan anemia, these patients have a fivefold higher lifetime risk of cancer than the general population and a 30- to 40-fold higher risk of developing acute myeloid leukemia, osteosarcoma, or colon cancer (reviewed in refs. 2, 3). Similarly, patients with X-linked dyskeratosis are predisposed to myeloid leukemia and a variety of solid tumors (4), whereas patients with 5q− syndrome are at higher risk of developing acute myeloid leukemia (reviewed in ref. 5). In the genetically tractable zebrafish model, heterozygous loss-of-function mutations in several ribosomal proteins cause development of peripheral nerve sheet tumors (6). Somatically acquired mutations in ribosomal proteins are also implicated in cancer: ∼10% of children with T-cell acute lymphoblastic leukemia (T-ALL) were found to harbor somatic mutations in the ribosomal protein of the large subunit (LSU) 10, 5, and 22 (RPL10, RPL5, and RPL22) (7). [Note that the proteins encoded by these genes are also named uL16, uL18, and eL22, respectively, under the newly proposed uniform ribosomal protein nomenclature (8).] A separate study identified heterozygous deletions in the region of chromosome 1p that contains RPL22 (eL22) in an additional 10% of patients with T-ALL (9). The model of ribosomal proteins as targets for somatic mutations in cancer is further supported by the finding that two ribosomal protein genes (RPL5/uL18 and RPL22/eL22) are included in the list of 127 genes identified as significantly mutated in cancer in the context of the first Cancer Genome Atlas pan-cancer analysis in 12 tumor types (10).
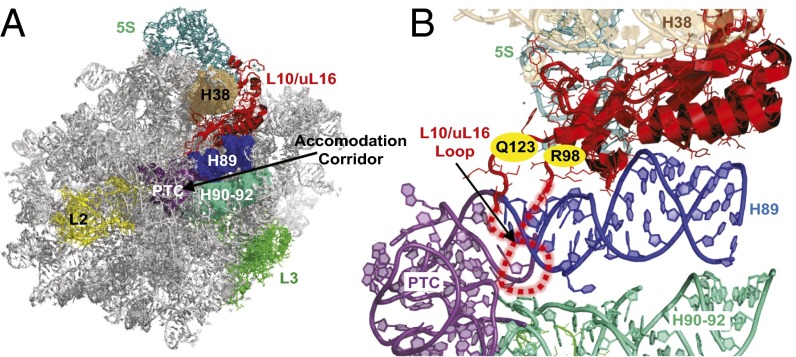
Ribosomopathies present an intriguing paradox: Although patients initially present with hypoproliferative disorders (e.g., anemias, bone marrow failure), those who survive to middle age often develop hyperproliferative diseases (i.e., cancers). The link between ribosome defects and hypoproliferative disease phenotypes has been extensively studied: The current working hypothesis is that impaired ribosome biogenesis activates a “ribosomal stress” cascade, activating the cellular TP53 pathway and resulting in cell cycle arrest and cell death (11). However, activation of TP53 does not explain why ribosomal defects are associated with hyperproliferative diseases, particularly cancer. Mutations in the ribosomal protein gene RPL10/uL16 were recently identified in patients with T-ALL (7). The T-ALL–associated RPL10/uL16 mutations occurred almost exclusively in residue arginine 98 (R98), with the exception of one patient harboring the Q123P mutation, which lies adjacent to R98 within the rpL10/uL16 3D structure (Fig. 1). Both residues are at the base of an essential flexible loop in rpL10 that closely approaches the peptidyltransferase center in the catalytic core in the ribosome (12). In addition to its role in catalysis (13, 14), rpL10/uL16 plays an important role in the late stages of 60S subunit biogenesis. After initial production of the separate ribosomal subunits in the nucleus, immature and functionally inactive pre-60S subunits are exported to the cytoplasm, where they undergo additional maturation events (15), including incorporation of rpL10/uL16, before they can associate with mature 40S subunits and engage in protein synthesis (16). Among the critical set of final 60S maturation steps is the release of the antiassociation factor Tif6, followed by release of Nmd3, the primary export adaptor for the pre-60S subunit in yeast and in humans (17, 18). Tif6 release requires the tRNA structural mimic Sdo1p (19) and the GTPase Efl1, a paralog of eukaryotic elongation factor 2 (eEF2) (20). We have suggested that structural rearrangements of the internal loop of rpL10/uL16 coordinate this final maturation process, resulting in a test drive of the pre-60S subunit to ensure that only properly functioning subunits are allowed to enter the pool of translationally active ribosomes (13, 21). Defective ribosomes carrying mutations in rpL10/uL16 specifically fail in this test drive, leading to their degradation through a molecular pathway that is yet to be characterized. Beyond 60S maturation, rpL10/uL16 plays an important role in coordinating intersubunit rotation and controlling allosteric rearrangements within the ribosome, helping to ensure the directionality and fidelity of protein synthesis (13).
Fig. 1.
Localization of rpL10 and the loop in the LSU. (A) rpL10/uL16 in the context of the crown view of the LSU. (B) Close-up view of rpL10/uL16 and the local environment. The flexible loop structure is indicated by dashed red lines, and the positions of R98 and Q123 are indicated. rpL10/uL16 is situated between helices 38 and 89, and it is located in close proximity to several functional centers of the LSU, including the peptidyltransferase center (PTC), aa-tRNA accommodation corridor, and elongation factor binding site. Images were generated using PyMOL.
rpL10/uL16 is highly conserved among eukaryotes: The yeast and human proteins are interchangeable, and residue 98 is invariantly an arginine (22). Human RPL10/uL16 is located on the X chromosome, and is therefore expressed as a single-copy gene in males. Thus, the haploid yeast model is an excellent mimic of the situation in the cells of a patient with T-ALL. Yeast cells expressing rpl10-R98S, rpl10-R98C, and rpl10-H123P (corresponding to Q123 in human rpL10/uL16) as the sole forms of rpL10/uL16 displayed proliferative defects. Further, polysome profiling revealed increased ratios of free 60S and 40S subunits vs. monosomes, markedly reduced polysomes, and the presence of halfmers in these mutants, suggesting defects in both ribosome biogenesis and subunit joining (7). Tif6 and Nmd3 both accumulated in the cytoplasm in the mutant cells, indicating a defect in their release from the cytoplasmic 60S (7). Thus, all of the rpl10/uL16 mutations appeared to affect 60S biogenesis at the Efl1-dependent quality control step. Consistent with the yeast-based observations, mouse lymphoid cells expressing rpl10-R98S displayed slower proliferation rates than cells expressing WT RPL10/uL16 and conferred defective polysome profiles (7).
The studies presented in the current report use the yeast rpl10-R98S mutant to elucidate the structural, biochemical, and trans-lational fidelity defects that may lead to carcinogenesis. This mutant perturbs the structural equilibrium of ribosomes toward the “rotated state.” At the biochemical level, this underlying structural defect alters the affinity of mutant ribosomes for a specific set of trans-acting ligands. In turn, the biochemical defects affect translational fidelity, promoting elevated rates of −1 programmed ribosomal frameshifting (−1 PRF) and impaired recognition of termination codons. Globally increased rates of −1 PRF result in a decreased abundance of cellular mRNAs that harbor operational −1 PRF signals (23, 24). These −1 PRF signal-containing mRNAs include EST1, EST2, STN1, and CDC13, which play central roles in yeast telomere maintenance (23). In rpl10-R98S cells, the steady-state abundances of these mRNAs are decreased, resulting in telomere shortening. A spontaneously acquired trans-acting mutant suppresses the ribosome biogenesis defects of the rpl10-R98S mutant, thereby reestablishing high levels of ribosome production and cell proliferation. Importantly, however, suppression of the biogenesis and growth impairment defects fails to suppress the profound structural, biochemical, and translational fidelity defects of rpL10-R98S ribosomes. These findings suggest that suppression of the growth defect results from bypassing the test drive. Although the suppressor mutation enables cells to grow at normal rates, genetic suppression comes at the cost of releasing functionally defective ribosomes into the translationally active pool. We propose two different but not mutually exclusive models for how somatically acquired rpL10/uL16 mutations may promote cancer: (i) Mutant ribosomes may drive altered gene expression programs, promoting T-ALL, or (ii) the suppressor mutations may themselves be the drivers of T-ALL.
Results
The rpl10-R98S Proliferation and Biogenesis Defects Are Suppressed by the NMD3-Y379D Mutant.
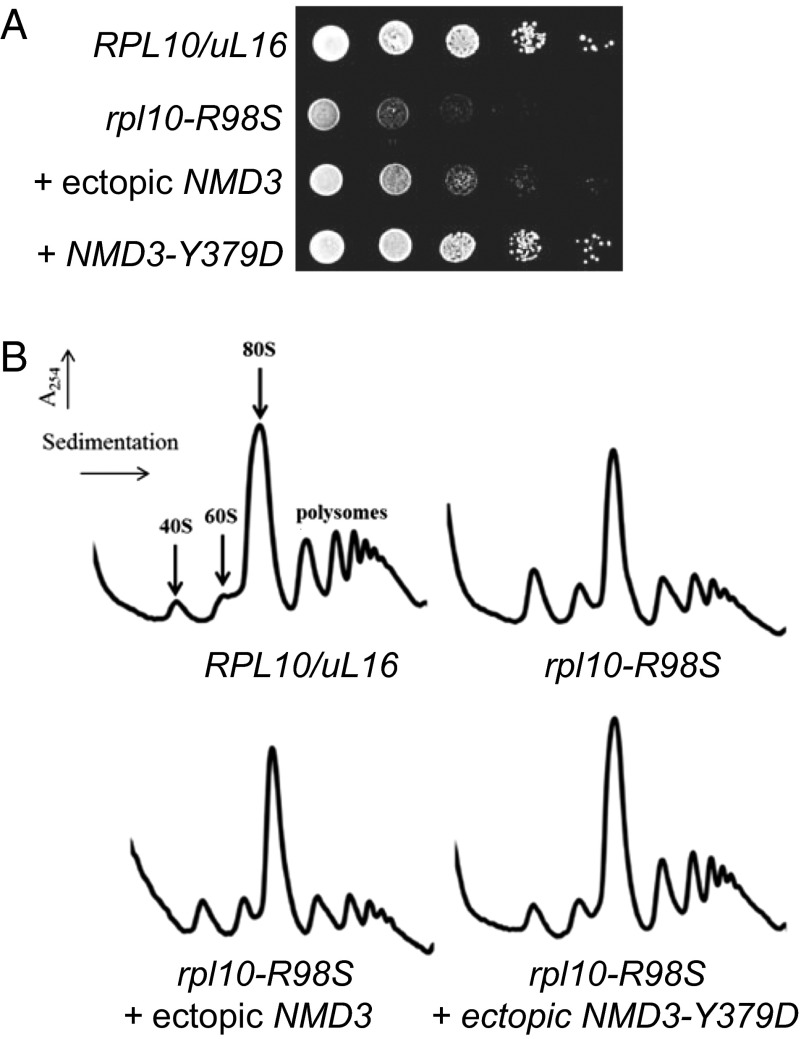
We previously showed that the rpl10-R98S mutation severely impairs growth in yeast (7). However, we have noted that yeast cells carrying the rpl10-R98S mutation readily acquire suppressing mutations that compensate for the growth defect. Sequence analyses identified a mutation in the export adapter NMD3, NMD3-Y379D, which suppresses both the growth (Fig. 2A) and the ribosome biogenesis defects (Fig. 2B) of rpl10-R98S cells. NMD3-Y379D was identified as a dominant extragenic suppressor of the rpl10-R98S defect. Additionally, aneuploidy for chromosome VIII-bearing NMD3 was identified as able to dominantly suppress the rpl10-R98S defect, as is increased gene dosage of NMD3 (Fig. 2).
Fig. 2.
(A) Overexpression of NMD3 or coexpression of NMD3-Y379D suppresses the rpl10-R98S growth defect. A 10-fold dilution spot assay of isogenic strains demonstrates that the rpl10-R98S growth defect is suppressed by ectopic expression of NMD3 and by the NMD3-Y379D mutation. (B) Overexpression of NMD3 or coexpression of NMD3-Y379D suppresses the rpl10-R98S biogenesis defect. Sucrose density gradient analysis. Indicated strains were grown in glucose to repress genomic RPL10/uL16 for 6 h before cells were harvested. Extracts were prepared, and nine A260 units were sedimented through 7–47% sucrose gradients.
The rpl10/uL16-R98S Mutation Alters Ribosome Rotational Status, and This Defect Is Not Suppressed by Coexpression of NMD3-Y379D.
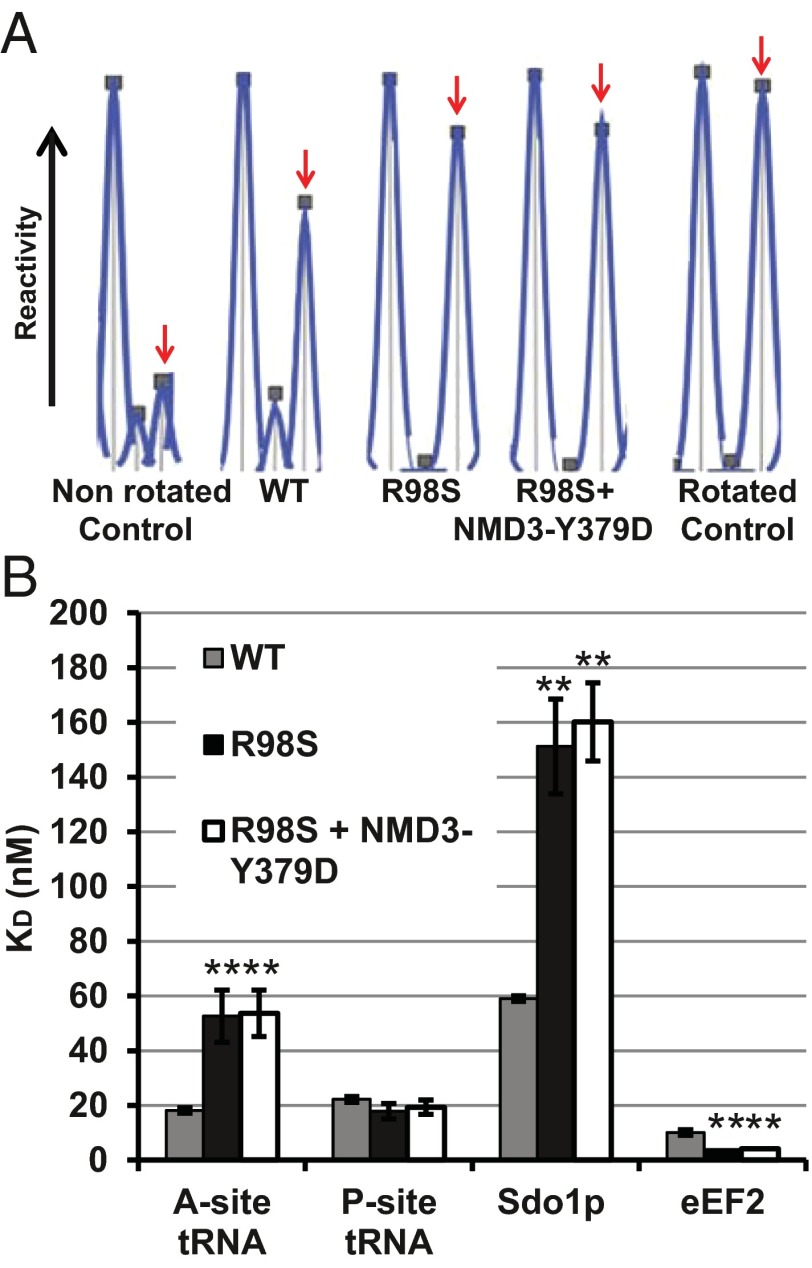
As revealed by extensive rRNA chemical modification analyses, the internal loop of rpL10 is a central driver of ribosomal rotation (13). In the current study, analysis of the landmark B7a intersubunit bridge was used to monitor the rotational status of the mutant ribosomes. This universally conserved bridge is formed by the interaction between A2207 (Escherichia coli A1847) in the LSU rRNA and G913 (E. coli A702) in the small ribosomal subunit (SSU) rRNA in the nonrotated state. This interaction is disrupted upon rotation, rendering both bases susceptible to chemical modification. WT salt-washed, empty ribosomes are distributed in an ∼50:50 equilibrium between these two rotational states (25). To monitor the effects of the R98S mutation on ribosome rotational status, 1-methyl-7-nitroisatoic anhydride (1M7) was used to probe A2207 of the LSU side of the B7a intersubunit bridge in purified empty isogenic WT and mutant ribosomes, as well as in fully rotated and nonrotated WT ribosome controls. The extent to which these bases were modified was quantitatively assessed and normalized using high-throughput selective 2′-hydroxyl acylation analyzed by primer extension (hSHAPE) (26, 27). The increased reactivity of A2207 to 1M7 (area under the peaks) reveals that rpL10-R98S ribosomes shift the structural equilibrium toward the rotated state (Fig. 3A). Importantly, this rotational disequilibrium is not suppressed by coexpression of NMD3-Y379.
Fig. 3.
(A) Coexpression of NMD3-Y379D does not suppress the rpl10-R98S rotational defect. Reactivity peaks obtained by hSHAPE after probing of the landmark base A2207 (arrows) at the LSU side of the B7a intersubunit bridge with 1M7. The control ribosomes were assembled as previously reported (13, 54). The nonrotated control is empty WT ribosomes containing Ac-aa-tRNA in the P-site. The rotated control is empty WT ribosomes containing deacylated tRNAPhe + eEF2-5'-guanyl-imidodiphosphate (GDPNP). (B) Coexpression of NMD3-Y379D does not suppress the rpl10-R98S biochemical defects. Steady-state binding of indicated ligands to ribosomes isolated from cells expressing WT, rpl10-R98S, and rpl10-R98S + NMD3-Y379D. Dissociation constants were obtained from binding assays of elongation ternary complex to the A-site and Ac-aa-tRNA to the P-site as monitored by filter binding. Sdo1p binding was monitored by measuring levels of [32P]-labeled Sdo1 associated with ribosomes, and eEF2 binding was monitored by the extent of [14C]-ADP ribosylation of unbound protein. Bars indicate SEM (n = 4). *P < 0.05; **P < 0.01.
The rpl10-R98S Mutation Alters Ribosomal Affinity for Specific Ligands, and This Defect Is Not Suppressed by Coexpression of NMD3-Y379D.
Recruitment of trans-acting factors to the ribosome is influenced by its rotational status, and other rpl10/uL16 mutants that perturb ribosome rotation alter the steady-state binding affinities for these factors (13). Similarly, rpL10/uL16-R98S ribosomes promoted approximately threefold lower affinity for elongation ternary complex ([14C]-Phe-tRNAPhe•eEF1A•GTP) and approximately threefold increased affinity for eEF2, both of which are ligands that specifically interact with the ribosomal A-site (Fig. 3B and Fig. S1). Comparable to other rpL10/uL16 loop mutants, rpL10/uL16-R98S ribosomes did not affect binding of the model P-site ligand, acetylated-[14C]Phe-tRNAPhe (Ac-aa-tRNA), to the ribosomal P-site. However, akin to other rpl10/uL16 mutants, R98S mutant ribosomes had an ∼2.5-fold lower affinity for Sdo1p, a ligand also previously shown to bind at the ribosomal P-site (13). Notably, none of these biochemical defects was suppressed by expression of NMD3-Y379D (Fig. 3B). This finding indicates that although NMD3-Y379D can suppress the ribosome biogenesis defect, it does not correct the altered ribosomal ligand affinity conferred by the rpl10-R98S mutation. Thus, the effects of the R98S mutant on ribosome biogenesis can be separated from the effects on ribosome biochemistry.
Translational Fidelity Defects of the rpl10-R98S Mutant Are Not Suppressed by NMD3-Y379D.
In general, ribosomes must maintain a translational reading frame to translate the genetic code faithfully. However, there are a growing number of examples of cis-acting mRNA sequences that direct elongating ribosomes to shift the reading frame: This is called PRF (28). Different classes of elements can direct ribosomes to slip by one base in either the 5′ (−) or 3′ (+) direction (−1 PRF and +1 PRF, respectively). Numerous studies have revealed mechanistic differences between the two: Most −1 PRF events require slippage of both the A- and P-site tRNA, whereas only the P-site tRNA is involved in +1 PRF, directed by the yeast Ty1 and Ty3 retrotransposable elements (28). Consistent with the biochemical data indicating a large subunit A-site–specific tRNA binding defect, the rpL10-R98S mutation stimulated −1 PRF by ∼2.6-fold, whereas +1 PRF remained unchanged (Fig. S2A). Termination codon recognition, which requires efficient recruitment of the eRF1/eRF3 complex to the ribosomal A-site (29), was also compromised in rpl10-R98S cells (an approximately twofold increase) (Fig. S2B). Discrimination between sense and missense tRNAs during elongation, a process that occurs in the A-site of the small subunit, was only slightly affected in rpl10-R98S mutant cells. Consistent with the structural and biochemical findings, coexpression of NMD3-Y379D did not suppress either the −1 PRF or termination codon misreading defects (Fig. S2). To determine whether or not these genetic and biochemical trends were unique to the R98S mutant, we assayed the effects of a second trans-acting suppressor of a different rpl10 mutant that exhibits similar phenotypic, biochemical, and functional defects. Similar to the results described above, the TIF6-V192F mutant suppressed the growth and biogenesis defects of rpl10-S104D mutant cells (13) but did not suppress the ternary complex binding or −1 PRF defects (Fig. S3).
Translational Fidelity Defects of the rpl10-R98S Mutant Affect Yeast Telomere Maintenance.
Computational analyses suggest that ∼10% of eukaryotic cellular mRNAs harbor operational −1 PRF signals (30). Over 95% of these frameshift events are predicted to function as mRNA destabilizing elements through the nonsense-mediated mRNA decay pathway by directing translating ribosomes to premature termination codons (31). More recently, −1 PRF was implicated in telomere maintenance in yeast (23). The effects of the rpl10-R98S mutant on −1 PRF directed by sequences located in the yeast EST1, EST2, STN1, and CDC13 mRNAs were quantitatively monitored in live cells using dual-luciferase reporter–based assays. In rpl10-R98S cells, rates of −1 PRF promoted by all of these cis-acting elements were uniformly enhanced approximately twofold to threefold and, consistent with the biochemical findings, NMD3-Y379D did not suppress these −1 PRF defects (Fig. 4A). Similar to results observed with other yeast mutants that promote globally increased rates of −1 PRF (23), the steady-state abundances of endogenous EST1, EST2, STN1, and CDC13 mRNAs were significantly reduced in rpl10-R98S cells as monitored by quantitative RT-PCR (Fig. 4B). Consistent with −1 PRF data, the NMD3-Y379D mutant did not suppress the effects of the rpl10-R98S mutant on the abundance of these mRNAs. A quantitative PCR analysis revealed that the yeast telomeric DNA repeat sequence was ∼25% less abundant in rpl10-R98S cells, suggestive of a telomere maintenance defect (Fig. 4C). Coexpression of NMD3-Y379D did not correct this telomere defect, consistent with the inability of this mutation to suppress the biochemical and translational fidelity defects of the rpl10-R98S mutation (Fig. 4C).
Fig. 4.
Coexpression of NMD3-Y379D does not suppress the rpl10-R98S telomere maintenance defects. (A) The −1 PRF directed by sequences in the following yeast genes: EST1 (signal beginning at nucleotide 1,272), EST2 (signal beginning at nucleotide 1,251), STN1 (signal beginning at nucleotide 1,203), and CDC13 (signal beginning at nucleotide 1,272). (B) Expression of endogenous EST1, EST2, STN1, and CDC13 mRNAs was monitored by quantitative RT-PCR. Bars indicate SEM (n = 4). (C) Coexpression of NMD3-Y379D does not suppress the rpl10-R98S telomere length defects. The abundance of telomere repeat sequences was quantified by PCR, with the single-copy reference gene SGS1 as the loading control. Bars indicate SEM (n = 9). *P < 0.05; **P < 0.01.
Discussion
Although defects in the translational machinery have previously been linked to cancer, the concept that somatic cells can acquire a defect in the ribosome machinery is relatively novel and there is currently little knowledge regarding the molecular mechanisms by which such a defect may promote (a specific type of) cancer. In particular, how a mutation like rpl10-R98S, which has a negative impact on cell growth, can promote a hyperproliferative disease presents a paradox.
Because rpL10/uL16 is an essential ribosomal protein, we reasoned that a first step in identifying the role of the rpl10-R98S mutation in leukemia development should require an in-depth characterization of the effects of the mutant protein on the ribosome. Whereas we previously showed that fewer mature ribosomes are present in rpl10-R98S cells (due to the ribosome biogenesis defect) (7), the current study reveals that this mutation also has a severe impact on ribosomes at both the structural and functional levels by causing malfunction of the rpL10 internal loop. Although previously described mutations that shift the rotational equilibrium were located at the tip of the loop (13), the T-ALL mutations localize at its base, possibly changing the general flexibility and dynamic nature of the loop, and thus its ability to influence ribosomal rotational status. We document in this work that the rpl10/uL16-R98S mutation impairs aa-tRNA binding to the A-site and Sdo1p at the P-site, that it increases affinity for eEF2, and that it decreases the fidelity of the A-site–associated aspects of protein translation.
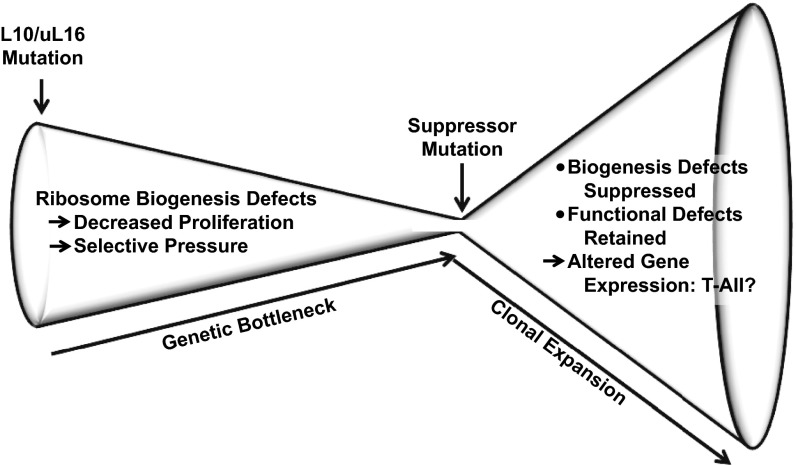
Previously, we showed that NMD3 overexpression partially rescued the proliferation defect observed in rpl10-R98S cells (7). Here, we demonstrate that the spontaneous suppressor NMD3-Y379D also compensates for the proliferation and ribosome biogenesis defects of rpl10-R98S. Importantly, the coexpression of NMD-Y379D only suppressed the ribosome biogenesis and proliferation defects of rpl10-R98S cells but did not alter the intrinsic structural and biochemical defects of rpL10/uL16-R98S mutant ribosomes. Similarly, a different suppressor, TIF6-V192F, is able to suppress the ribosome biogenesis defect conferred by the rpl10-S104D mutant but did not affect any of the downstream translation-associated functions. These findings suggest a “selective pressure”-based model of the connection between ribosomopathies and carcinogenesis (Fig. 5). Given the importance of rpL10 in cytoplasmic maturation of the LSU, mutations in this protein lead to accumulation of defective pre-60S subunits in the cytoplasm. Many of these subunits fail the Efl1- and Sdo1-dependent test drive (21) and are likely removed by the nonfunctional ribosome decay (NRD) apparatus. The resulting dearth of 60S subunits explains the hypoproliferation phenotype caused by expression of rpL10-R98S. We suggest that this 60S subunit deficit also exerts pressure on cell populations to select for mutations that suppress the biogenesis defects or otherwise bypass NRD. Such suppressors could subvert the quality control mechanisms and serve to increase the abundance of 60S subunits, reversing the proliferation defect. However, such suppression comes at the risk of allowing cells to use defective ribosomes. The use of such defective ribosomes in translation ultimately drives altered gene expression patterns, as evidenced, in part, by decreased abundances of −1 PRF signal-containing mRNAs. In yeast, cellular −1 PRF signal containing mRNAs include four that encode proteins implicated in telomere maintenance. Although we have used telomere maintenance to illustrate how defects in ribosome function can lead to altered gene expression, we anticipate that other pathways are also affected in yeast. Studies in bacteria have identified an unexpectedly large number and diversity of recoding events (32–34), suggesting that such recoding is pervasive in biology. Although we have demonstrated that ∼10% of mammalian mRNAs harbor strong candidate −1 PRF signals, our studies also show that this class of cis-acting mRNA elements evolves quickly and does not appear to be well conserved (30). Thus, identifying the critical pathways that are misregulated in T-ALL is a remaining challenge. In sum, the analysis presented here illuminates a pathway from genotype to phenotype, and we suggest that suppression of the ribosome biogenesis defects represents a critical event in the progression of these cells to T-ALL. While providing a temporary fix by bypassing the primary quality checkpoint in ribosome assembly, suppressing mutations that reduce translational fidelity are ultimately themselves the drivers of disease because they directly promote altered cellular gene expression programs by enhancing utilization of endogenous “error-prone” sites (i.e., those that promote frameshifting and termination codon read-through). As such, endogenous error-prone sites may be of general importance for other “later life” phenotypes involving impaired cellular homeostasis.
Fig. 5.
Model of T-ALL progression. A mutation in RPL10/uL16 results in the inability of the pre-60S subunits to pass the quality control checkpoint, leading to decreased ribosome assembly and proliferation. A ribosome biogenesis suppressor can arise due to selective pressure allowing the bypass the quality test drive, thereby boosting production of defective ribosomes. Continued defective translation ultimately leads to an altered gene expression profile and the onset of T-ALL.
The ribosome biogenesis pathways are highly conserved from yeast to humans, begging the question of what kinds of ribosome biogenesis defects may render cells more prone to this transition from hypo- to hyperproliferative growth states. It is reasonable to suppose that the list of ribosomal protein mutants that could promote such a transition would be limited to those mutants involved in late stages of subunit maturation. This is because proteins involved in early stages of LSU biogenesis are more likely to be required for the overall structural integrity of the subunit and mutations in such proteins would not be tolerated. Although analysis of ribosomal protein genes linked to known ribosomopathies that affect LSU biogenesis is consistent with this idea, this trend does not apply to those involved in SSU biogenesis (Fig. S4A). Inspection of yeast atomic resolution structures reveals that the ribosomopathy linked ribosomal proteins are all globular proteins located on the cytoplasmic/solvent-accessible surfaces of the subunits (35) (Fig. S4 B and C). These types of proteins are not involved in stabilizing interdomain rRNA interactions and are not located in regions of the ribosome directly associated with the central functions of mRNA decoding and peptidyl transfer. Thus, this structural analysis suggests that only those mutants that allow a ribosomal subunit to become mature enough to engage the translational apparatus yet still allow sufficient perturbations of gene expression fall into the category of ribosomopathogenic proteins. The two trans-acting factors linked to ribosomopathies, SBDS (Shwachman–Bodian–Diamond syndrome protein) and DKC1 (dyskerin), are also consistent with this theory: SBDS is involved in the late, cytoplasmic test drive of pre-60S subunits (36), and rRNA modifications, although occurring early during rRNA transcription, merely “fine-tune” ribosome structure rather than play central roles in domain folding (37–39). Given the large numbers of small nucleolar ribonuclear particles (snoRNPs) (and the snoRNAs and proteins that constitute them) involved in rRNA modification, plus the scores of other protein factors involved in ribosome biogenesis, it is conceivable that this model of ribosome-based transition from hypoproliferative to hyperproliferative growth may be more widespread than previously appreciated.
The next phase of research will be to determine the nature of compensatory mutations that may be occurring in human RPL10/uL16-R98S T-ALL cells. Studies in our laboratories using cells derived from patients with RPL10/uL16-R98S T-ALL have not uncovered point mutations or copy number changes affecting late 60S biogenesis factors, such as NMD3, EIF6, SBDS, EFTUD1, or LSG1. This is not an exhaustive list of ribosome biogenesis factors: Recent results indicate that mammalian cells may have additional proteins involved in ribosome biogenesis (40). Mutations in other biological pathways downstream of the primary defect could also potentially prevent defective ribosomes from freely entering the translationally active pool. For example, the still poorly understood NRD pathway lies genetically downstream of the pre-60S test drive (41). Mutants in this process would be expected to enable defective ribosomes similarly to enter the translational pool. Moreover, suppressing mutations by themselves may promote cellular transformation by yet other mechanisms.
In this work, we focused on the role of rpL10/uL16 on ribosomal function. Mammalian ribosomal proteins have also been shown to have functions independent of protein translation, however [reviewed in (42, 43)]. In particular, RPL10/uL16 has been linked to regulation of the oncogenic transcription factor JUN (44) and the V-Yes-1 Yamaguchi sarcoma viral oncogene homolog 1 (45). It is possible that alteration of one of these or of additional unknown “extraribosomal” functions of RPL10/uL16 may also contribute to the pathological role of RPL10/uL16-R98S. Although such alternative explanations remain important subjects for investigation, the findings presented in the current work support that RPL10/uL16-R98S promotes defective translational fidelity. The role of these defects in oncogenesis should now be explored.
Materials and Methods
Media, Strains, Plasmids, and Genetic Manipulation.
E. coli DH5 alpha was used to amplify plasmid DNA. Transformation of E. coli and yeast and preparation of yeast growth media (YPAD = 1% Bacto yeast extract, 2% Bacto peptone, 2% glucose, 0.004% adenine sulfate, and synthetic dropout medium) were as reported earlier (46, 47). Restriction enzymes were obtained from Promega and Roche Applied Science. DNA sequencing was performed by Genewiz.
The haploid Saccharomyces cerevisiae strain AJY1437 (MATα rpl10::KanMX lysΔ0 met15Δ0 leu2Δ0 ura3Δ0 his3Δ0) containing WT RPL10/uL16 on a centromeric URA3 vector (pAJ392) has previously been described (21). In AJY3222, the WT vector was replaced by WT RPL10/uL16 on a centromeric LEU2 vector (pAJ2522) through standard 5-fluoroorotic acid shuffling techniques (48). AJY3209 harbors pAJ2609, a centromeric LEU2 vector expressing the rpl10-S104D allele. Similarly, AJY2784 contains pAJ2726, which expresses the rpl10-R98S mutant from a centromeric LEU2 vector. Identification of suppressing mutations has previously been described (21). In suppression studies, AJY3209 was transformed with pAJ2240, which expresses TIF6-V192F from a centromeric URA3 vector; AJY2784 was transformed with pAJ2805, which expresses NMD3-Y379D from a centromeric URA3 vector. Dilution spot assays were performed by growing yeast to midlog growth phase in liquid culture and spotting them in 10-fold serial dilutions from 105 to 101 cfu per spot on appropriate media, followed by a 2-d incubation at 30 °C or as appropriate.
Translational Fidelity and Polysome Analyses.
Standard dual-luciferase reporter plasmids were used to monitor −1 PRF, +1 PRF, suppression of UAA, and suppression of an AGC near-cognate serine codon in place of the cognate AGA codon in the firefly luciferase catalytic site, as previously described (49–51). The reporters were expressed from high-copy URA3/LEU2-based plasmids (pJD375, pJD376, pJD376, pJD431, pJD642, and pJD643), and sample readings were collected using a GloMax Multi-Microplate luminometer (Promega). Sucrose density gradient analysis was carried out as described (21).
Ribosome Biochemistry and Structural Probing.
Purification of active 80S ribosomes using cysteine-charged sulfolink columns was performed as described (13). eEF1A preparation, purification of aminoacyl-tRNA synthetases, charging of tRNAPhe (Sigma) with [14C]-phenylalanine (PerkinElmer), and purification of aminoacyl-tRNA and acetylated aminoacyl-tRNA were carried out as described (52, 53). Filter binding assays to determine steady-state dissociation rates (Kds) of [14C]-Phe-tRNAPhe•eEF1A•GTP to the ribosomal A-site and Ac-[14C]-Phe-tRNAPhe to the ribosomal P-site were performed as described (13). The Kds obtained from binding assays of eEF2, as monitored by extent of [14C]-ADP ribosylation of unbound protein, and [32P]-Sdo1p, as monitored by levels of radiolabel incorporation in ribosomes, were as described (13). Kd values were calculated using GraphPad Prism software fitted to a single binding site with ligand depletion models. rRNA structural probing experiments of the landmark B7a intersubunit bridge with 1M7 were performed as recently described using hSHAPE and primer 2632 (13). To prepare “nonrotated” control ribosomes, empty WT 80S ribosomes were incubated with N-acetyl-phenylalanyl tRNAPhe. To prepare “rotated” control ribosomes, empty WT 80S ribosomes were first incubated with deacylated tRNAPhe, followed by eEF2 and GDPNP as described (13, 54).
mRNA Abundance and Telomere Length Analyses.
Quantitative PCR experiments to assay mRNA abundance were carried out as described (23). Similar methods were used to quantify telomere length in yeast cells. Genomic DNA was isolated from midlogarithmic cell cultures using the “smash and grab” yeast DNA preparation method as described (55). Each DNA sample was diluted serially. Quantitative polymerase chain reactions to determine yeast telomere content (T) relative to the single-copy reference gene SGS1 (S) were performed using the Bio-Rad iTaqTM Universal SYBR Green system utilizing primer pairs complementary to telomere repeats (listed in Table S1), and cycle threshold values were determined. The T/S ratios were calculated from three experimental replicates at each of three DNA concentrations (100, 200, and 400 ng). The Student t test for two-tailed P value calculations was used throughout.
Supplementary Material
Acknowledgments
We thank Ashton Belew, Sharmishtha Musalgaonkar, Alicia Bowen, Suna P. Gülay, and Ryan Kobylarz for technical and intellectual assistance. This work was supported by Public Health Service Grants 2 R01 GM058859-11 and 3 R01 GM053655-15S1 (to J.D.D.) and by Public Health Service Grants 2 RO1 GM53655 and 3 R01 GM053655-15S1 (to A.W.J.). S.O.S. was partially supported by National Institutes of Health Training Grant T32GM080201.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. A.H. is a guest editor invited by the Editorial Board.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1400247111/-/DCSupplemental.
References
- 1.Narla A, Ebert BL. Ribosomopathies: Human disorders of ribosome dysfunction. Blood. 2010;115(16):3196–3205. doi: 10.1182/blood-2009-10-178129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chirnomas SD, Kupfer GM. The inherited bone marrow failure syndromes. Pediatr Clin North Am. 2013;60(6):1291–1310. doi: 10.1016/j.pcl.2013.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Vlachos A, Rosenberg PS, Atsidaftos E, Alter BP, Lipton JM. Incidence of neoplasia in Diamond Blackfan anemia: A report from the Diamond Blackfan Anemia Registry. Blood. 2012;119(16):3815–3819. doi: 10.1182/blood-2011-08-375972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Alter BP, et al. Malignancies and survival patterns in the National Cancer Institute inherited bone marrow failure syndromes cohort study. Br J Haematol. 2010;150(2):179–188. doi: 10.1111/j.1365-2141.2010.08212.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Post SM, Quintás-Cardama A. Closing in on the pathogenesis of the 5q- syndrome. Expert Rev Anticancer Ther. 2010;10(5):655–658. doi: 10.1586/era.10.44. [DOI] [PubMed] [Google Scholar]
- 6.Amsterdam A, et al. Many ribosomal protein genes are cancer genes in zebrafish. PLoS Biol. 2004;2(5):E139. doi: 10.1371/journal.pbio.0020139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.De Keersmaecker K, et al. Exome sequencing identifies mutation in CNOT3 and ribosomal genes RPL5 and RPL10 in T-cell acute lymphoblastic leukemia. Nat Genet. 2013;45(2):186–190. doi: 10.1038/ng.2508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Ban N, et al. (2014) A new system for naming ribosomal proteins. Curr Opin Struct Biol, 10.1016/j.sbi.2014.01.002. [DOI] [PMC free article] [PubMed]
- 9.Rao S, et al. Inactivation of ribosomal protein L22 promotes transformation by induction of the stemness factor, Lin28B. Blood. 2012;120(18):3764–3773. doi: 10.1182/blood-2012-03-415349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kandoth C, et al. Mutational landscape and significance across 12 major cancer types. Nature. 2013;502(7471):333–339. doi: 10.1038/nature12634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Raiser DM, Narla A, Ebert BL. The emerging importance of ribosomal dysfunction in the pathogenesis of hematologic disorders. Leuk Lymphoma. 2014;55(3):491–500. doi: 10.3109/10428194.2013.812786. [DOI] [PubMed] [Google Scholar]
- 12.Ben-Shem A, et al. The structure of the eukaryotic ribosome at 3.0 Å resolution. Science. 2011;334(6062):1524–1529. doi: 10.1126/science.1212642. [DOI] [PubMed] [Google Scholar]
- 13.Sulima SO, et al. Eukaryotic rpL10 drives ribosomal rotation. Nucleic Acids Res. 2014;42(3):2049–2063. doi: 10.1093/nar/gkt1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tate WP, Brown CM. Translational termination: “Stop” for protein synthesis or “pause” for regulation of gene expression. Biochemistry. 1992;31(9):2443–2450. doi: 10.1021/bi00124a001. [DOI] [PubMed] [Google Scholar]
- 15.Panse VG, Johnson AW. Maturation of eukaryotic ribosomes: Acquisition of functionality. Trends Biochem Sci. 2010;35(5):260–266. doi: 10.1016/j.tibs.2010.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.West M, Hedges JB, Chen A, Johnson AW. Defining the order in which Nmd3p and Rpl10p load onto nascent 60S ribosomal subunits. Mol Cell Biol. 2005;25(9):3802–3813. doi: 10.1128/MCB.25.9.3802-3813.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ho JH, Kallstrom G, Johnson AW. Nascent 60S ribosomal subunits enter the free pool bound by Nmd3p. RNA. 2000;6(11):1625–1634. doi: 10.1017/s1355838200001291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Trotta CR, Lund E, Kahan L, Johnson AW, Dahlberg JE. Coordinated nuclear export of 60S ribosomal subunits and NMD3 in vertebrates. EMBO J. 2003;22(11):2841–2851. doi: 10.1093/emboj/cdg249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ng CL, et al. Conformational flexibility and molecular interactions of an archaeal homologue of the Shwachman-Bodian-Diamond syndrome protein. BMC Struct Biol. 2009;9:32. doi: 10.1186/1472-6807-9-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bécam AM, Nasr F, Racki WJ, Zagulski M, Herbert CJ. Ria1p (Ynl163c), a protein similar to elongation factors 2, is involved in the biogenesis of the 60S subunit of the ribosome in Saccharomyces cerevisiae. Mol Genet Genomics. 2001;266(3):454–462. doi: 10.1007/s004380100548. [DOI] [PubMed] [Google Scholar]
- 21.Bussiere C, Hashem Y, Arora S, Frank J, Johnson AW. Integrity of the P-site is probed during maturation of the 60S ribosomal subunit. J Cell Biol. 2012;197(6):747–759. doi: 10.1083/jcb.201112131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Klauck SM, et al. Mutations in the ribosomal protein gene RPL10 suggest a novel modulating disease mechanism for autism. Mol Psychiatry. 2006;11(12):1073–1084. doi: 10.1038/sj.mp.4001883. [DOI] [PubMed] [Google Scholar]
- 23.Advani VM, Belew AT, Dinman JD. Yeast telomere maintenance is globally controlled by programmed ribosomal frameshifting and the nonsense-mediated mRNA decay pathway. Translation (Austin) 2013;1(1):e24418. doi: 10.4161/trla.24418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Belew AT, Advani VM, Dinman JD. Endogenous ribosomal frameshift signals operate as mRNA destabilizing elements through at least two molecular pathways in yeast. Nucleic Acids Res. 2011;39(7):2799–2808. doi: 10.1093/nar/gkq1220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fischer N, Konevega AL, Wintermeyer W, Rodnina MV, Stark H. Ribosome dynamics and tRNA movement by time-resolved electron cryomicroscopy. Nature. 2010;466(7304):329–333. doi: 10.1038/nature09206. [DOI] [PubMed] [Google Scholar]
- 26.Wilkinson KA, et al. High-throughput SHAPE analysis reveals structures in HIV-1 genomic RNA strongly conserved across distinct biological states. PLoS Biol. 2008;6(4):e96. doi: 10.1371/journal.pbio.0060096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Leshin JA, Heselpoth R, Belew AT, Dinman J. High throughput structural analysis of yeast ribosomes using hSHAPE. RNA Biol. 2011;8(3):478–487. doi: 10.4161/rna.8.3.14453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dinman JD. Mechanisms and implications of programmed translational frameshifting. Wiley Interdiscip Rev RNA. 2012;3(5):661–673. doi: 10.1002/wrna.1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhouravleva G, et al. Termination of translation in eukaryotes is governed by two interacting polypeptide chain release factors, eRF1 and eRF3. EMBO J. 1995;14(16):4065–4072. doi: 10.1002/j.1460-2075.1995.tb00078.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Belew AT, Hepler NL, Jacobs JL, Dinman JD. PRFdb: A database of computationally predicted eukaryotic programmed -1 ribosomal frameshift signals. BMC Genomics. 2008;9:339. doi: 10.1186/1471-2164-9-339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jacobs JL, Belew AT, Rakauskaite R, Dinman JD. Identification of functional, endogenous programmed -1 ribosomal frameshift signals in the genome of Saccharomyces cerevisiae. Nucleic Acids Res. 2007;35(1):165–174. doi: 10.1093/nar/gkl1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gurvich OL, et al. Sequences that direct significant levels of frameshifting are frequent in coding regions of Escherichia coli. EMBO J. 2003;22(21):5941–5950. doi: 10.1093/emboj/cdg561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sharma V, et al. A pilot study of bacterial genes with disrupted ORFs reveals a surprising profusion of protein sequence recoding mediated by ribosomal frameshifting and transcriptional realignment. Mol Biol Evol. 2011;28(11):3195–3211. doi: 10.1093/molbev/msr155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Antonov I, Coakley A, Atkins JF, Baranov PV, Borodovsky M. Identification of the nature of reading frame transitions observed in prokaryotic genomes. Nucleic Acids Res. 2013;41(13):6514–6530. doi: 10.1093/nar/gkt274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jenner L, et al. Crystal structure of the 80S yeast ribosome. Curr Opin Struct Biol. 2012;22(6):759–767. doi: 10.1016/j.sbi.2012.07.013. [DOI] [PubMed] [Google Scholar]
- 36.Lo KY, et al. Defining the pathway of cytoplasmic maturation of the 60S ribosomal subunit. Mol Cell. 2010;39(2):196–208. doi: 10.1016/j.molcel.2010.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Decatur WA, Fournier MJ. rRNA modifications and ribosome function. Trends Biochem Sci. 2002;27(7):344–351. doi: 10.1016/s0968-0004(02)02109-6. [DOI] [PubMed] [Google Scholar]
- 38.Baxter-Roshek JL, Petrov AN, Dinman JD. Optimization of ribosome structure and function by rRNA base modification. PLoS ONE. 2007;2(1):e174. doi: 10.1371/journal.pone.0000174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jack K, et al. rRNA pseudouridylation defects affect ribosomal ligand binding and translational fidelity from yeast to human cells. Mol Cell. 2011;44(4):660–666. doi: 10.1016/j.molcel.2011.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tafforeau L, et al. The complexity of human ribosome biogenesis revealed by systematic nucleolar screening of Pre-rRNA processing factors. Mol Cell. 2013;51(4):539–551. doi: 10.1016/j.molcel.2013.08.011. [DOI] [PubMed] [Google Scholar]
- 41.Cole SE, LaRiviere FJ, Merrikh CN, Moore MJ. A convergence of rRNA and mRNA quality control pathways revealed by mechanistic analysis of nonfunctional rRNA decay. Mol Cell. 2009;34(4):440–450. doi: 10.1016/j.molcel.2009.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dinman JD. The eukaryotic ribosome: Current status and challenges. J Biol Chem. 2009;284(18):11761–11765. doi: 10.1074/jbc.R800074200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Warner JR, McIntosh KB. How common are extraribosomal functions of ribosomal proteins? Mol Cell. 2009;34(1):3–11. doi: 10.1016/j.molcel.2009.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Inada H, Mukai J, Matsushima S, Tanaka T. QM is a novel zinc-binding transcription regulatory protein: Its binding to c-Jun is regulated by zinc ions and phosphorylation by protein kinase C. Biochem Biophys Res Commun. 1997;230(2):331–334. doi: 10.1006/bbrc.1996.5955. [DOI] [PubMed] [Google Scholar]
- 45.Oh HS, Kwon H, Sun SK, Yang C-H. QM, a putative tumor suppressor, regulates proto-oncogene c-yes. J Biol Chem. 2002;277(39):36489–36498. doi: 10.1074/jbc.M201859200. [DOI] [PubMed] [Google Scholar]
- 46.Ito H, Fukuda Y, Murata K, Kimura A. Transformation of intact yeast cells treated with alkali cations. J Bacteriol. 1983;153(1):163–168. doi: 10.1128/jb.153.1.163-168.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Inoue H, Nojima H, Okayama H. High efficiency transformation of Escherichia coli with plasmids. Gene. 1990;96(1):23–28. doi: 10.1016/0378-1119(90)90336-p. [DOI] [PubMed] [Google Scholar]
- 48.Boeke JD, LaCroute F, Fink GR. A positive selection for mutants lacking orotidine-5′-phosphate decarboxylase activity in yeast: 5-Fluoro-orotic acid resistance. Mol Gen Genet. 1984;197(2):345–346. doi: 10.1007/BF00330984. [DOI] [PubMed] [Google Scholar]
- 49.Harger JW, Dinman JD. An in vivo dual-luciferase assay system for studying translational recoding in the yeast Saccharomyces cerevisiae. RNA. 2003;9(8):1019–1024. doi: 10.1261/rna.5930803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Plant EP, et al. Differentiating between near- and non-cognate codons in Saccharomyces cerevisiae. PLoS ONE. 2007;2(6):e517. doi: 10.1371/journal.pone.0000517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Grentzmann G, Ingram JA, Kelly PJ, Gesteland RF, Atkins JF. A dual-luciferase reporter system for studying recoding signals. RNA. 1998;4(4):479–486. [PMC free article] [PubMed] [Google Scholar]
- 52.Meskauskas A, Petrov AN, Dinman JD. Identification of functionally important amino acids of ribosomal protein L3 by saturation mutagenesis. Mol Cell Biol. 2005;25(24):10863–10874. doi: 10.1128/MCB.25.24.10863-10874.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Meskauskas A, Dinman JD. A molecular clamp ensures allosteric coordination of peptidyltransfer and ligand binding to the ribosomal A-site. Nucleic Acids Res. 2010;38(21):7800–7813. doi: 10.1093/nar/gkq641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Stern S, Moazed D, Noller HF. Structural analysis of RNA using chemical and enzymatic probing monitored by primer extension. Methods Enzymol. 1988;164:481–489. doi: 10.1016/s0076-6879(88)64064-x. [DOI] [PubMed] [Google Scholar]
- 55.Rose MD, Winston F, Hieter P. Methods in Yeast Genetics. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1990. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.