Significance
Upon virus infection, host cells detect viral nucleic acids and induce antiviral innate immune responses by producing cytokines. The retinoic acid inducible gene-1 (RIG-I)-like receptors are cytoplasmic sensors for RNA viruses that mediate antiviral innate immunity. In this study, we identify K homology (KH) domain- and really interesting new gene (RING)-finger domain-containing MEX3C as a molecule that regulates RIG-I function. MEX3C preferentially colocalizes with viral RNA and RIG-I in cytoplasmic granules in infected cells and mediates K63-linked ubiquitination of RIG-I, which is important for activation of downstream signaling. We show that MEX3C is required for cytokine production after infection with RNA viruses that are detected by RIG-I by generating MEXC-deficient mice. Our study demonstrates a critical role of MEX3C in induction of RIGI-mediated antiviral innate immune responses.
Keywords: signal transduction, cytoplasmic puncta
Abstract
The RIG-I–like receptors, retinoic acid inducible gene-1 (RIG-I), melanoma differentiation-associated protein 5, and laboratory of genetics and physiology-2, are cytoplasmic sensors for RNA viruses that mediate the antiviral innate immune responses. We demonstrate that really interesting new gene-finger domain- and K homology domain-containing MEX3C regulates RIG-I function. MEX3C colocalizes with RIG-I in the stress granules of virally infected cells, and its overexpression causes the lysine-63–linked ubiquitination of RIG-I and activates IFN-β promoter. Embryonic fibroblast cells, macrophages, and conventional dendritic cells derived from Mex3c-deficient mice showed defective production of type I IFN after infection with RNA viruses that are recognized by RIG-I. These results demonstrate that MEX3C is an E3 ubiquitin ligase that modifies RIG-I in stress granules and plays a critical role in eliciting antiviral immune responses.
The RIG-I–like receptors (RLRs), which include retinoic acid inducible gene-1 (RIG-I), melanoma differentiation-associated protein 5 (MDA5), and laboratory of genetics and physiology-2 (LGP2), detect a signature of viral RNAs and induce antiviral innate immune responses. RIG-I and MDA5 contain tandem caspase-recruitment domains (CARDs) at their N termini, which mediate downstream signaling; a central DExD/H helicase domain with an ATP-binding motif; and a C-terminal region known as the “RNA-binding domain” (1, 2). They sense RNAs derived from different RNA viruses. RIG-I is essential for the recognition of vesicular stomatitis virus (VSV), Newcastle disease virus (NDV), and Japanese encephalitis virus (JEV), whereas MDA5 is essential for the recognition of encephalomyocarditis virus (EMCV) and Mengo virus (1, 3). RLRs also differentially recognize synthesized RNA molecules. Whereas MDA5 senses long poly(I:C), RIG-I senses digested, relatively short (approximately 300 bp) poly(I:C) and RNAs synthesized by T7 polymerase, which carr a 5′ppp RNA (4, 5).
RIG-I and MDA5 trigger intracellular signaling pathways via an adaptor, IPS-1 (also known as MAVS/CARDIF/VISA) (6). IPS-1 localizes to the outer mitochondrial membrane and forms large aggregates that activate downstream signaling pathways (7). Downstream from IPS-1, transcription factors IRF3 and NF-κB are activated to induce type I IFN and inflammatory cytokines for host defense (1, 8). It was shown that tripartite motif 25 (TRIM25) induced the lysine 63 (K63)-linked ubiquitination of the CARD of RIG-I at lysine 172 or the generation of unanchored K63-linked polyubiquitin chains (9). Riplet (also known as REUL or RNF135) has been shown to ubiquitinate the C terminus of RIG-I to facilitate RIG-I–mediated viral recognition (10, 11).
Recent studies demonstrated that RLRs accumulate in cytoplasmic granules containing stress granule (SG) markers after viral infection (12, 13). These granules, called “antiviral stress granules” (avSGs), are distinguishable from canonical SGs and contain viral RNA, viral nucleocapsid protein, and RNA-dependent protein kinase (PKR). AvSGs are reported to be a platform for the detection of viruses (12, 13). However, the relationship between RLR-mediated antiviral responses and the formation of avSGs is poorly understood.
Here we identified the RNA-binding and RING-containing protein MEX3C, that localizes to SGs and triggers RIG-I-dependent antiviral responses by ubiquitination of RIG-I.
Results
Identification of Mex3c.
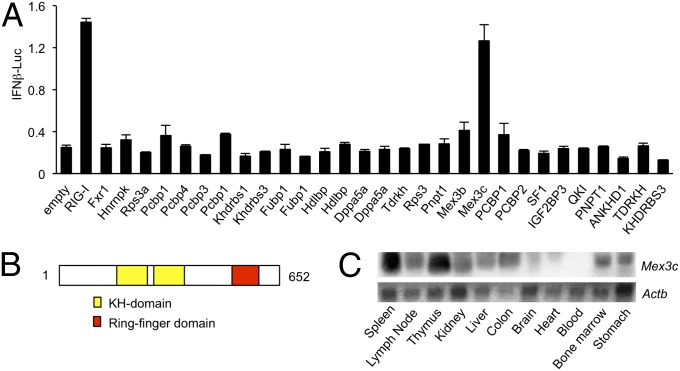
To search for molecules that regulate the induction of type I IFN, we overexpressed a series of proteins carrying the K homology (KH) domain, a well-known RNA-binding domain, in HEK293 cells together with a reporter plasmid driven by the promoter of the IFN-β gene and measured the reporter gene expression with a luciferase assay. Of the proteins tested, MEX3C was found to increase the IFN-β (IFNB) promoter activity (Fig. 1A). MEX3C contains two KH domains in its middle portion and one RING-finger domain in its C-terminal region (Fig. 1B). This protein forms a family with the homologous proteins MEX3A, -B, and -D (14). Mex3c is expressed in many tissues, with particularly strong expression in the spleen and thymus (Fig. 1C).
Fig. 1.
Identification of Mex3c. (A) HEK293 cells were transfected with the indicated plasmids containing the KH domain, together with an IFNB-promoter-driven reporter plasmid, and the luciferase activity was determined. Data are from three independent experiments. The results shown are means ± SD (n = 3). (B) Schematic representation of mouse MEX3C protein. (C) Total RNA from the indicated mouse tissues was extracted and subjected to Northern blotting to examine the expression of Mex3c and β-actin (Actb).
MEX3C Mediates the Ubiquitination and Activation of RIG-I.
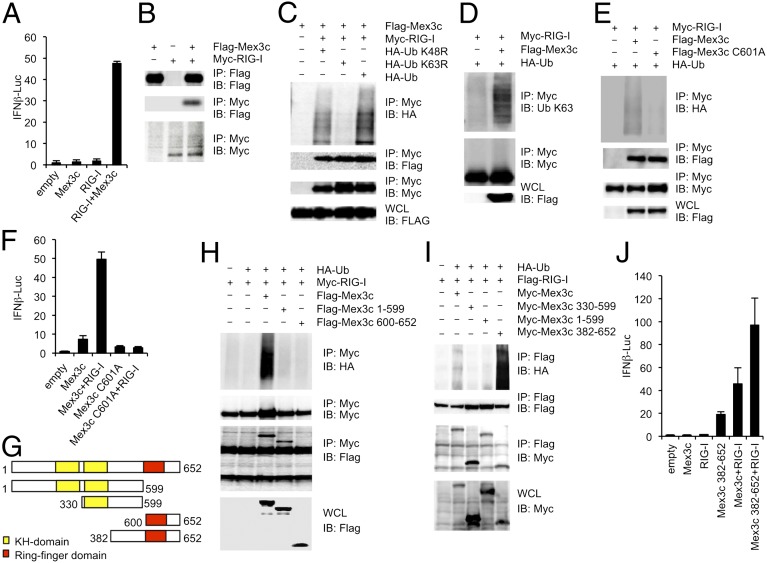
To determine the roles of MEX3C in the antiviral innate immune responses, we overexpressed MEX3C and RIG-I together with an IFNB promoter-driven reporter plasmid in HEK293 cells and measured the reporter gene expression. Whereas the overexpression of either MEX3C or RIG-I alone did not cause the expression of the reporter gene, the overexpression of both proteins together markedly enhanced the reporter gene expression (Fig. 2A). Next, we overexpressed Myc–RIG-I and Flag–MEX3C in HEK293 cells and examined their interaction with a coimmunoprecipitation assay (Fig. 2B). Flag–MEX3C was coprecipitated with Myc–RIG-I. Notably, an anti-Myc antibody detected smeared bands of Myc–RIG-I protein in cells coexpressing MEX3C. To examine whether the smeared bands were attributable to ubiquitination, we overexpressed Flag–MEX3C and Myc–RIG-I together with hemagglutinin–ubiquitin (HA–Ub) or HA–Ub with a point mutation at Lys-48 or -63 that converted it to Arg (K48R or K63R, respectively). An anti-HA antibody detected smeared bands in the cells expressing HA–Ub or HA–Ub K48R, whereas these bands were not observed in the cells expressing HA–Ub K63R (Fig. 2C). Furthermore, an immunoblot analysis with an anti–Ub K63 antibody detected smeared bands in anti-Myc immunoprecipitates prepared from cells overexpressing Myc–RIG-I and Flag–MEX3C (Fig. 2D). When a point mutation at the cysteine residue at position 601 in the RING-finger domain of MEX3C was substituted with alanine (C601A), it abrogated RIG-I ubiquitination (Fig. 2E). Notably, the overexpression of Flag–MEX3C C601A did not enhance the RIG-I–mediated IFNB promoter activity (Fig. 2F). These findings suggest that E3 ubiquitin ligase activity is required for the MEX3C-mediated enhancement of RIG-I function. In contrast, MEX3C failed to induce the ubiquitination of MDA5 (Fig. S1).
Fig. 2.
MEX3C-mediated RIG-I ubiquitination. (A, F, and J) HEK293 cells were transfected with the indicated plasmids together with an IFNB-promoter-driven reporter plasmid, and the luciferase activity was determined. Data are from three independent experiments. The results shown are means ± SD (n = 3). (B–E, H, and I) HEK293 cells were transfected with the indicated plasmids. Cell lysates were immunoprecipitated (IP) with anti-Flag or anti-Myc antibody and immunoblotted (IB) with the indicated antibodies. (G) Schematic representation of full-length MEX3C and the MEX3C deletion mutants (encoding amino acids 1–599, 330–599, 600–652, and 382–652).
We next generated a series of deletion mutants of MEX3C to determine the region responsible for its interaction with RIG-I (Fig. 2G). MEX3C 1–599, which contains the KH domains, was coprecipitated with RIG-I, whereas MEX3C 600–652, which encodes the RING-finger domain, failed to associate with RIG-I (Fig. 2H). RIG-I ubiquitination was not induced by either MEX3C 1–599 or 600–652 (Fig. 2H), suggesting that although MEX3C 1–599 binds to RIG-I, it is insufficient to cause RIG-I ubiquitination. Next, we explored the function of the region between the KH domains and the RING-finger domain of MEX3C. Full-length of MEX3C and MEX3C 330–599, 1–599, and 382–652 all associated with RIG-I (Fig. 2I), suggesting that MEX3C 382–599 is responsible for binding to RIG-I. Notably, the overexpression of MEX3C 382–652, which does not contain the KH domains but does contain the RIG-I–binding region and the RING-finger domain, induced RIG-I ubiquitination more strongly than full-length MEX3C. Furthermore, the overexpression of MEX3C 382–652 induced IFNB promoter activation more efficiently than full-length MEX3C (Fig. 2J). These findings suggest that the KH domains negatively regulate the MEX3C-mediated ubiquitination of RIG-I.
MEX3C Delivers the K63-Linked Polyubiquitin Moiety to RIG-I CARDs.
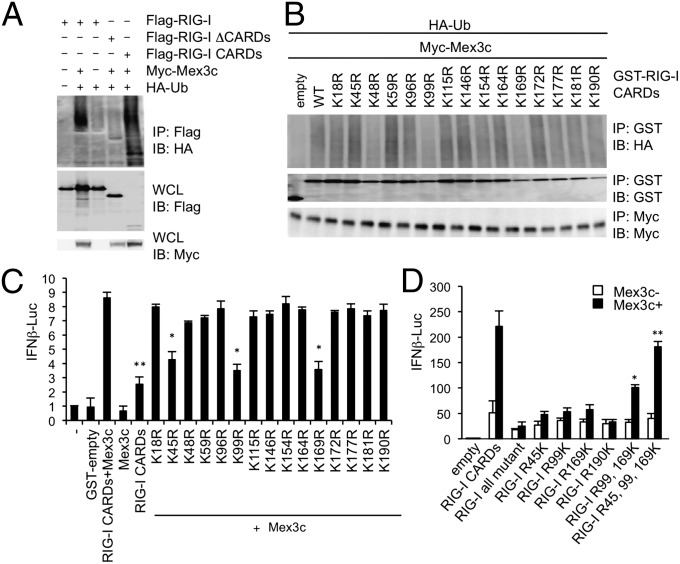
We investigated the target residues of RIG-I that are ubiquitinated by MEX3C. MEX3C overexpression caused the ubiquitination of RIG-I CARDs but not ΔCARDs (Fig. 3A). We then mutated individual lysine residues in the CARDs to arginine and tested RIG-I ubiquitination (Fig. 3B). Ubiquitination of a RIG-I by MEX3C was reduced by a point mutation at K48, K99, or K169 (Fig. 3B). The MEX3C-mediated enhancement of the IFNB promoter activity decreased when K99 or K169 in RIG-I was mutated but not when K48 was mutated (Fig. 3C). This finding, together with those shown in Fig. 3B, suggests that the ubiquitination of RIG-I at K99 and K169 by MEX3C is important for the activation of the IFNB promoter. However, the reason for reduced activity of the IFNB promoter due to mutation of K45 remains unclear.
Fig. 3.
Identification of the lysine residues in RIG-I required for signaling. (A and B) Cell lysates prepared from HEK293 cells transfected with the indicated plasmids were immunoprecipitated and blotted with the indicated antibodies. (C and D) HEK293 cells were transfected with the indicated plasmids together with an IFNB-promoter-driven reporter plasmid, and the luciferase activity was measured. Data are from three independent experiments. The results shown are means ± SD (n = 3). *P < 0.05 and **P < 0.005 compared with lane 3 in C (RIG-I CARDs+MEX3C) or lane 6 in D (RIG-I all mutant with black bar).
Next, we generated mutant RIG-I CARDs in which all of the lysine residues were substituted with arginine (RIG-I all-mutant) and found that the overexpression of this mutant failed to enhance the IFNB promoter activity, even in the presence of MEX3C, whereas the IFNB promoter activation mediated by the overexpression of RIG-I CARDs was enhanced by the coexpression of MEX3C (Fig. 3D). We then resubstituted the arginine residues in RIG-I all-mutant with lysines. The overexpression of either RIG-I R45K, R99K, R169K, or R190K failed to restore the MEX3C-mediated IFNB promoter activation. However, the overexpression of the RIG-I R99,169K mutant, in which the arginine residues at 99 and 169 were restored to lysine, partially restored the MEX3C-mediated enhancement of the IFNB promoter activation. Furthermore, mutation of the arginines at 45, 99, and 169 to lysine (R45,99,169K) restored the IFNB promoter activity to a level similar to that induced by the RIG-I CARDs (Fig. 3D).
MEX3C Colocalizes with RIG-I in avSGs.
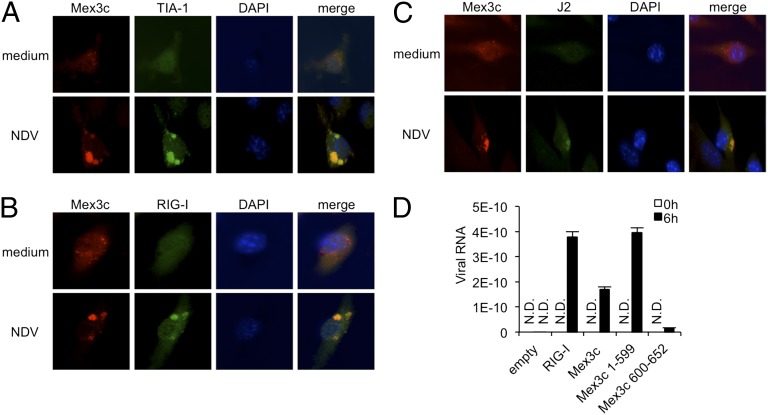
We determined cellular localization of MEX3C. In unstimulated murine bone marrow-derived macrophages (BMDMs), MEX3C localized to cytoplasmic puncta (Fig. 4A). After NDV infection, MEX3C accumulated in enlarged cytoplasmic structures, where it colocalized with TIA-1, a marker of SGs (Fig. 4A). These findings suggest that MEX3C translocates from cytoplasmic puncta to TIA-1–positive SGs after viral infection. Next we investigated the localization of endogenous MEX3C and RIG-I. In unstimulated BMDMs, RIG-I was diffusely present in the cytoplasm, whereas MEX3C was localized in dot-like structures in the cytoplasm (Fig. 4B). After NDV infection, both RIG-I and MEX3C accumulated in cytoplasmic puncta (Fig. 4B). These findings suggest that MEX3C is incorporated into RIG-I–positive avSGs. To investigate the cellular localization of viral RNA, we stained NDV-infected BMDMs with the anti-dsRNA antibody J2 and found that MEX3C colocalized with J2-positive puncta (Fig. 4C). Furthermore, in HeLa cells transiently transfected with MEX3C-expressing constructs and 5′-ppp RNA, MEX3C and 5′-ppp RNA were present in the same puncta (Fig. S2). These findings suggest that MEX3C, RIG-I, and viral RNA accumulate in SGs in virally infected cells.
Fig. 4.
Cellular localization of MEX3C and RIG-I. (A–C) Murine BMDMs were left untreated (medium) or infected with NDV. The cells prepared after 6-h infection were immunostained with the indicated antibodies. Image shown is the representative of three independent experiments. (D) HEK293 cells were transfected with constructs encoding Flag–RIG-I, Flag–MEX3C WT, or Flag-MEX3C KH domains or RING domain. After 30 h the cells were infected with NDV. The cell lysates prepared after 6-h infection were immunoprecipitated with anti-Flag antibody, and the viral RNAs in the anti-Flag immunoprecipitates were evaluated by qPCR. Data are from three independent experiments. The results shown are means ± SD (n = 3).
The observations that MEX3C contains KH domains and colocalizes with viral RNA suggest that MEX3C interacts with viral RNA. To test this proposition, we transiently overexpressed Flag–RIG-I, Flag–MEX3C, the Flag–MEX3C KH domains, or the Flag–MEX3C RING domain in HEK293 cells. After NDV infection, the presence of viral RNAs in the anti-Flag immunoprecipitates was verified by quantitative PCR (qPCR). Viral RNAs were detected in the immunoprecipitates prepared from cells transfected with constructs expressing RIG-I and MEX3C or the MEX3C KH domains but not in those prepared from cells expressing the MEX3C RING domain (Fig. 4E), suggesting that MEX3C directly or indirectly interacts with viral RNA via its KH domains.
MEX3C Is Critical for Innate Immune Responses Against RNA Virus Infections.
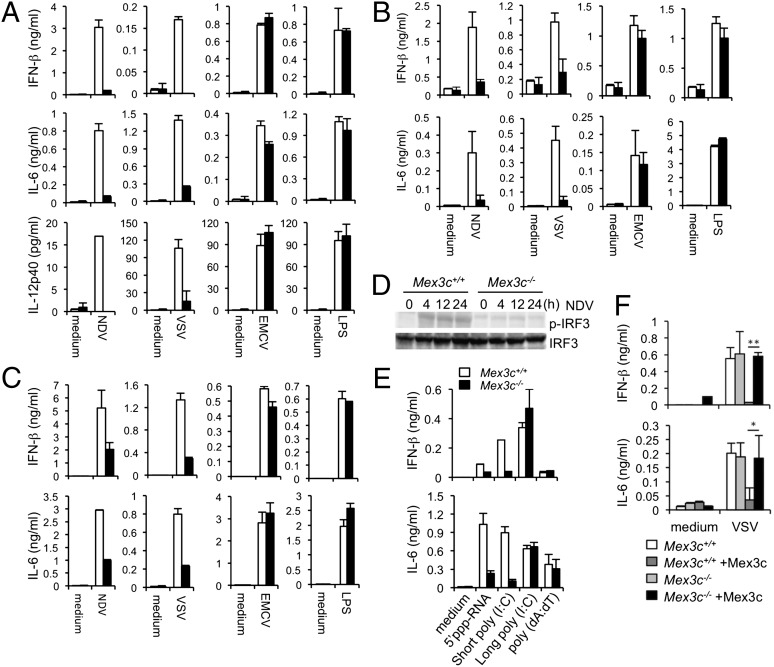
To examine the physiological contribution of MEX3C to antiviral immune responses, we generated Mex3c-deficient mice (Fig. S3). The Mex3c−/− mice were born in Mendelian ratios, although consistent with a previous report approximately one-fifth of the mice were runts, which was attributed to growth disturbance (15). The populations of peritoneal macrophages (PECs) and dendritic cells (DCs) were unimpaired in Mex3c-deficient mice (Fig. S4). We examined the production of cytokines in response to RNA virus infection. The production of IFN-β, IL-6, and IL-12p40 after infection with NDV or VSV was abrogated by Mex3c-deficient PECs (Fig. 5A). In contrast, the production of these cytokines after infection with EMCV, which is recognized by MDA5, or stimulation with LPS, a ligand for TLR4, was not impaired in the Mex3c-deficient PECs. Defective IFN-β and IL-6 production after NDV or VSV infection was also observed in Mex3c-deficient bone marrow-derived conventional DCs (cDCs) (Fig. 5B) or embryonic fibroblast (MEF) cells (Fig. 5C). Furthermore, the phosphorylation of IRF3 induced by NDV infection was impaired in Mex3c-deficient PECs (Fig. 5D). The up-regulation of IL-6 and IFN-β mRNAs by NDV infection was also impaired in the Mex3c-deficient PECs, whereas their up-regulation by LPS stimulation was similar in the WT and Mex3c-deficient cells (Fig. S5A). The production of IL-6 and IFN-β against shortened poly(I:C) and 5′-ppp RNA generated by T7 polymerase, agonists of RIG-I, but not against long poly(I:C), an agonist of MDA5, was severely impaired in the Mex3c-deficient macrophages (Fig. 5E). The production of IFN-β and IL-6 in response to the transfection of dsDNA [poly(dA:dT)] was not altered in the absence of Mex3c (Fig. 5E). We transduced WT or Mex3c-deficient MEFs with MEX3C retrovirus construct and found that MEX3C transduction restored their IFN-β and IL-6 production (Fig. 5F). To rule out the possibility that the inability of Mex3c-deficient cells to respond to RIG-I agonists results from the loss of RIG-I expression, we examined their RIG-I expression with an immunoblot, which showed that RIG-I expression was similar in the WT and Mex3c-deficient MEFs (Fig. S5B). We next infected WT and Mex3c-deficient mice with JEV, which is sensed by RIG-I (3). IFN-α production in the serum was reduced in the Mex3c-deficient mice (Fig. S6A). Furthermore, the levels of NDV RNA expression were elevated in the Mex3c-deficient PECs relative to those in the WT cells (Fig. S6B).
Fig. 5.
Impaired cytokine production in Mex3c-deficient mice. (A–C) PECs (A), cDCs (B), or MEF cells (C) from Mex3c+/+ (white bar) and Mex3c−/− (black bar) mice were stimulated with LPS (100 ng/mL) or treated with NDV (multiplicity of infection [moi] 0.5), VSV (moi 3), or EMCV (moi 0.01) for 24 h. The concentrations of IFN-β, IL-6, and IL-12p40 in the culture supernatants were measured with ELISAs. (D) PECs from Mex3c+/+ and Mex3c−/− mice were treated with NDV (moi 0.5) for the indicated periods, and the cell lysates were subjected to immunoblot with antibodies directed against p-IRF3 and IRF3. (E and F) PECs from Mex3c+/+ and Mex3c−/− mice were transfected with 5′pppRNA, short poly(I:C), long poly(I:C), or poly(dA:dT) for 24 h (E) or MEF cells retrovirally transduced with a construct encoding MEX3C were treated with VSV (moi 3) for 24 h (F). The concentrations of IFN-β and IL-6 in the culture supernatants were measured with ELISAs. Data are from three independent experiments (A–C, E, and F). The results shown are means ± SD (n = 3) (A–C, E, and F). *P < 0.05 and **P < 0.005 compared with controls (F).
Plasmacytoid DCs (pDCs) play important roles in the detection of viral infections and in the production of type I IFN. In pDCs, TLR7 is the predominant sensor of RNA viruses. IFN-β production after treatment with NDV or VSV was similar in the WT and Mex3c-deficient pDCs (Fig. S6C), suggesting that MEX3C does not participate in TLR7-mediated IFN-β production in pDCs.
Dispensable Role of MEX3C in SG Formation.
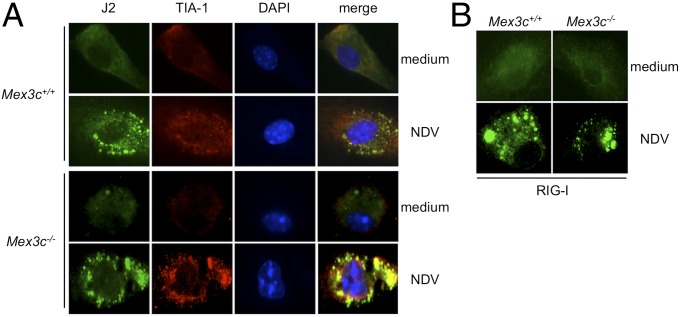
We next investigated whether MEX3C is required for SG formation and RIG-I trafficking. J2-positive and TIA-1–positive puncta was found to colocalize in both WT and Mex3c-deficient cells infected with NDV (Fig. 6A). Moreover, RIG-I translocation to the cytoplasmic puncta in response to NDV infection was also found in MEX3C-deficient cells (Fig. 6B). These findings suggest that MEX3C is not essential for the trafficking of RIG-I or the formation of SGs in virally infected cells.
Fig. 6.
MEX3C is not essential for avSG formation. (A and B) BMDMs from Mex3c+/+ and Mex3c−/− mice were left untreated (medium) or treated with NDV for 24 h and immunostained with the indicated antibodies. Image shown is the representative of three independent experiments.
Discussion
Here we have shown that the antiviral responses induced by RIG-I are impaired by Mex3c deficiency in mice. The MEX3 proteins are evolutionarily conserved and mediate the posttranslational regulation of diverse biological processes, including hypertension (16), development, differentiation (17–19), cancer (20), and the immune responses (21, 22). There are four MEX3 proteins in mammals, and they are thought to play similar roles to that of the Caenorhabditis elegans Mex3 protein (23), which is a translational repressor that binds to the Mex3 recognition element in the 3′ UTRs of target genes (14). It has been reported that Mex3c-deficient mice display growth retardation after birth, which may be caused by the increased translation of insulin-like growth factor 1 mRNA (24). Another study demonstrated that MEX3C regulates the expression of the HLA-A2 gene by binding to its 3′ UTR (19) and degrading its transcripts in a ubiquitin-dependent manner, contributing to NK cell function (21). MEX3C has also been identified as a suppressor of chromosomal instability in cancer (20). Here we demonstrated that MEX3C binds to viral RNA and regulates RIG-I–mediated signaling. Thus, MEX3C may bind to a number of RNAs, including self-RNAs and viral RNAs. Identification of the target RNAs of MEX3C and clarification of the MEX3C-mediated RNA recognition mechanisms must be addressed in the future.
Currently the role of avSGs in the host defense against viral infections is unclear. Recent studies have shown that RLRs and viral RNAs relocate from the cytoplasm to the SGs after viral infection (12). It has been reported that cells deficient in PKRs fail to form SGs after RNA viral infection and exhibit reduced type I IFN production (12). Nonstructural protein 1 of influenza A virus and leader protein of Theiler’s murine encephalomyelitis virus inhibit both avSG formation and type I IFN induction. These findings imply that avSGs act as RNA virus recognition sites and in the initiation of signaling (12). In contrast, another study implied that SG formation is not essential for type I IFN production after viral infection, especially by viruses that are recognized by MDA5 (13). Therefore, the roles of avSGs in virus-induced type I IFN responses are still controversial. Here we demonstrated that avSG formation and RIG-I trapping in the avSGs in response to RNA virus infection was unimpaired in Mex3c-deficient cells. However, MEX3C is required for the activation of RIG-I–dependent signaling. RIG-I directly interacts via its C-terminal region with the viral RNA (24, 25), and MEX3C can also bind to viral RNA. Together, these data prompt us to speculate that viral RNA trapped in avSGs behaves as a hub upon which both MEX3C and RIG-I are assembled and that the consequent proximity of RIG-I and MEX3C allows MEX3C to trigger RIG-I ubiquitination and activation.
Several E3 ubiquitin ligases that target RIG-I for ubiquitination have been identified. Riplet has been shown to mediate the K63-linked polyubiquitination of the C-terminal region of RIG-I (10, 11). The RIG-I C-terminal region masks its CARDs to prevent downstream signaling in unstimulated condition. When RIG-I binds to viral RNA, Riplet triggers RIG-I ubiquitination. This results in the recruitment of the signaling complex involving downstream molecules, including TBK1, IKKi, and NEMO (26). In contrast, TRIM25 has been shown to mediate the ubiquitination of lysine 172 of the RIG-I CARD (9). Another study has shown that TRIM25 does not induce RIG-I ubiquitination but instead mediates the generation of unanchored ubiquitin chains associated with RIG-I in virally infected cells, which are important in both RIG-I oligomerization and its interaction with IPS-1/MAVS (27). We have shown that MEX3C mediates the K63-linked polyubiquitination of the CARDs of RIG-I. A deficiency of TRIM25, Riplet, or MEX3C in mice impaired the RIG-I–mediated antiviral response, indicating that the coordinated activation of these three molecules is required for the RIG-I–mediated antiviral immune responses.
Our results suggest that lysines 48, 99, and 169 of RIG-I are required for Mex3-mediated ubiquitination (Fig. 3B), which differs from TRIM25-mediated ubiquitination, which occurs on lysine 172. K172R did not affect the MEX3C-mediated increase of IFNB promoter activity in our assay (Fig. 3C), suggesting that the functions of MEX3C and TRIM25 in terms of RIG-I activation are not redundant. Our reporter analysis suggested that lysine 48 is not essential for the MEX3C-mediated enhancement of RIG-I–dependent IFNB promoter activation, whereas the lysine at 99 and 169 are required (Fig. 3C). This suggests that the MEX3C-mediated ubiquitination of lysine 99 and 169 of RIG-I is essential for the antiviral innate immune response. However, it remains unclear why the K45R mutant showed reduced IFNB promoter activation, although it underwent relatively normal ubiquitination (Fig. 3B). It is possible that the mutation of lysine 45 influences the conformation of RIG-I, making it unable to interact with IPS-1 or other downstream signaling molecules. Alternatively, other ubiquitin ligases that ubiquitinate RIG-I K45 may exist.
Our analysis of a series of deletion mutants of MEX3C demonstrated that MEX3C 382–652 but not 600–652 coprecipitated with RIG-I, suggesting that MEX3C 382–599, the region between the KH and RING-finger domains, is responsible for its interaction with RIG-I. Notably, RIG-I ubiquitination was increased in cells overexpressing MEX3C 382–652 compared with that in cells overexpressing full-length MEX3C (Fig. 2I). Similarly, MEX3C 382–652 augmented RIG-I–mediated IFNB promoter activity more potently than full-length MEX3C (Fig. 2J). These findings suggest that MEX3C 382–652, which lacks the KH domain, acts as an active form of the protein that constitutively binds to and ubiquitinates RIG-I, irrespective of RNA binding, and that the KH domains prevent the activation of MEX3C unless an RNA virus is present. Once the avSGs are formed, viral RNA binding to the KH domains of MEX3C may induce conformational changes in MEX3C, allowing it to become active.
Viruses have developed strategies to evade the host innate immune system. Therefore, some viruses can be expected to target MEX3C for inactivation, presumably through its degradation or cleavage, or the inhibition of its binding to or ubiquitination of RIG-I. In the future, understanding the evasion mechanisms of these viruses will allow the development of new antiviral drugs. Moreover, the aberrant recognition of self-RNAs by the innate immune system may cause autoimmune diseases, such as systemic lupus erythematosus, by constitutively producing type I IFN, which potentiates adaptive immunity, suggesting a link between the aberrant function of MEX3C and autoimmunity. This must be also investigated in the future.
Materials and Methods
Reagents.
ELISA kits were purchased from R&D Systems. Long poly(I:C) was purchased by GE Healthcare. Poly(dA:dT) was purchased from Sigma. 5′ppp-RNA and short poly(I:C) were synthesized as described previously (4, 28). Anti-Flag M2-peroxidase monoclonal and anti-c-Myc monoclonal antibodies were from Sigma. Monoclonal antibody J2 was from English & Scientific Consulting. Anti-RIG-I and anti-HA-Tag (C29F4) rabbit monoclonal antibodies were purchased from Cell Signaling. Anti-K63-linkage-specific polyubiquitin rabbit monoclonal antibody was purchased from Millipore. Anti-TIA-1 and horseradish peroxidase-conjugated anti-β-actin antibodies were purchased from Santa Cruz Biotechnology. LPS from Salmonella minnesota, Hoechst 33342, Alexa Fluor 488-conjugated phalloidin, Alexa Fluor 568-conjugated goat anti-rabbit IgG antibody (A21246), Alexa Fluor 568-conjugated goat anti-mouse IgG antibody (A21236), Alexa Fluor 488-conjugated goat anti-rabbit IgG antibody (A11070), and Alexa Fluor 488-conjugated goat anti-mouse IgG antibody (A11029) were from Invitrogen. Anti-MEX3C monoclonal antibody was from Eurofins MWG Operon.
Mice, Cells, and Viruses.
Mex3c−/− mice were generated as described in SI Materials and Methods. All animal experiments were performed with the approval of the Animal Research Committee of the Research Institute for Microbial Diseases (Osaka University). PECs were isolated from the peritoneal cavities of mice 3 d after injection with 2 mL of 4.0% (wt/vol) thioglycollate medium (Sigma). Bone marrow-derived DCs and macrophages were prepared from the femurs and tibias of mice. Briefly, the cells were cultured in RPMI medium 1640 supplemented with 10% FCS, 100 μM 2-mercaptoethanol, and 10 ng/mL murine GM-CSF (PeproTech) or 40 ng/mL murine M-CSF (PeproTech), and the cells were collected after 6 d to be used as GM-CSF–induced cDCs and BMDMs, respectively (29). MEF cells were isolated from WT or Mex3c-deficient embryo (day 13.5). NDV, VSV, JEV, and EMCV have been described elsewhere (3).
Plasmids and Primers.
Full-length MEX3C and MEX3C deletion mutant cDNAs encoding amino acids 1–599, 330–599, 600–652, and 382–652 were inserted in the pFlag–CMV2 vector (Sigma) or the pcDNA 3.1(+)–Myc expression vector (Invitrogen). A point mutation (K601R) was introduced into MEX3C using the QuikChange II XL Site-Directed Mutagenesis Kit (Stratagene). Flag-tagged RIG-I, RIG-I ΔCARDs, RIG-I CARDs, and MDA5 CARDs were described previously (5). Plasmids encoding GST-fused RIG-I CARDs with point mutations (WT-RIG-I-CARDs, K99R, K172R, K181R, and K190R) were obtained from Jae U. Jung (University of Southern California, Los Angeles). RIG-I CARDs with point mutations (K18R, K45R, K48R, K59R, K96R, K115R, K146R, K154R, K164R, K169R, and K177R) were prepared using the QuikChange II XL Site-Directed Mutagenesis Kit. DNA fragments encoding RIG-I mutants containing point mutations in both the CARD domains, in which lysine was substituted with arginine, were synthesized by Hokkaido System Science Co., Ltd. The point mutation vectors for all mutant RIG-I proteins (R45K, R99K, R169K, R190K, R99,169K, and R45,99,169K) were prepared using the QuikChange II XL Site-Directed Mutagenesis Kit. The plasmids encoding the KH domain were obtained from Mammalian Gene Collection cDNA (OPEN Biosystems). RNase inhibitor (N261A) was purchased from Promega. Primer sequences used in this study were follows. MEX3C forward: ATGCTGTCCCACGCCTAC; MEX3C reverse: AGTGCTTTAATTTTACAACCCTGG; β-actin forward: TGTCACCAACTGGGACGATA; β-actin reverse: AACACAGCCTGGATGGCTAC; GAPDH forward: TGACGGCCCAAGATGCCCTTCAG. TaqMan probe detecting NDV is described in ref. 30.
Luciferase Assay.
HEK293 cells (5 × 105) were transfected with the pGL3–IFN-β promoter luciferase reporter plasmid (0.1 μg per well) together with plasmids encoding the KH domain, which were obtained from Mammalian Gene Collection cDNA (OPEN Biosystems) (Fig. 1A) or plasmids encoding RIG-I, MEX3C-WT, mutant MEX3C, or empty control plasmid (0.25 μg per well) with Lipofectamine 2000 (Invitrogen). After 24 h the cells were infected with NDV; 24 h later the cells were lysed, and the luciferase activity in the lysates was determined with the Dual-Luciferase Reporter Assay System (Promega). The Renilla luciferase construct pRL-TK (0.05 μg per well) was simultaneously transfected as an internal control.
Immunoprecipitation and Immunoblot Analysis.
HEK293 cells (1 × 106) were transfected with the indicated plasmids (1 μg per well). After 30 h the cells were lysed with lysis buffer [1% (vol/vol) Nonidet P-40, 150 mM NaCl, 20 mM Tris⋅HCl (pH 7.5), and 1 mM EDTA]. The cell lysates were then immunoprecipitated with anti-Flag or anti-Myc antibody. Immunoprecipitates or whole-cell lysates were immunoblotted with the indicated antibodies. For detection of ubiquitination, cell lysates were boiled with 1% SDS for 10 min to remove nonspecific ubiquitination before immunoprecipitation.
Immunostaining.
BMDMs seeded on cover glass were infected with NDV for 24 h. The cells were fixed with paraformaldehyde and permeabilized with 0.5% Triton-X 100 and then subjected to immunocytochemistry. Samples were examined under a fluorescence laser scanning confocal BZ-9000 fluorescence microscope (KEYENCE).
Quantitative Real-Time PCR.
HEK293 cells (1 × 106) were transfected with the indicated plasmids (1 μg per well). After 30 h the cells were infected with NDV [multiplicity of infection (moi) 5] for 6 h. The cell lysates were then immunoprecipitated with anti-Flag antibody in RNase free lysis buffer. After washing, RNA in the immunoprecipitates was extracted using ZR Viral RNA Kit (Zumo Research), and cDNA was synthesized using ReverTra Ace qPCR RT Master Mix (Toyobo). qPCR analysis was performed using 770Sequence Detector (Applied Biosystems).
Retroviral Transduction of MEF Cells.
MEF cells were isolated from WT or Mex3c-deficient embryo (day 13.5). Viruses were produced by Plat-E packaging cells (5.5 × 106) transfected with plasmid (12 μg per dish) encoding MEX3C or empty control plasmid with FuGENE 6 (Roche). After 72 h the cells were maintained in medium with puromycin (2.5 μg/mL; InvivoGen) for 4 d to select the stably transfected cells.
Supplementary Material
Acknowledgments
We thank M. Shiokawa and Y. Fujiwara for technical assistance, the members of the Laboratory of Host Defense for valuable discussion, E. Kamada and M. Kageyama for secretarial assistance, and Dr. Jae U. Jung’s group for providing us with a series of TRIM25 plasmids. This study was supported by a KAKENHI Grant-in-Aid for Research Activity (A) of the Japanese Ministry of Education, Culture, Sports, Science and Technology (to T.K.); the Cabinet Office, Government of Japan, and the Japan Society for the Promotion of Science Funding Program for World-Leading Innovative Research and Development on Science and Technology “FIRST Program” (to S.A.); and National Institutes of Health Grant P01-AI070167 (to S.A.).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1401674111/-/DCSupplemental.
References
- 1.Takeuchi O, Akira S. Pattern recognition receptors and inflammation. Cell. 2010;140(6):805–820. doi: 10.1016/j.cell.2010.01.022. [DOI] [PubMed] [Google Scholar]
- 2.Yoneyama M, Fujita T. RNA recognition and signal transduction by RIG-I-like receptors. Immunol Rev. 2009;227(1):54–65. doi: 10.1111/j.1600-065X.2008.00727.x. [DOI] [PubMed] [Google Scholar]
- 3.Kato H, et al. Differential roles of MDA5 and RIG-I helicases in the recognition of RNA viruses. Nature. 2006;441(7089):101–105. doi: 10.1038/nature04734. [DOI] [PubMed] [Google Scholar]
- 4.Kato H, et al. Length-dependent recognition of double-stranded ribonucleic acids by retinoic acid-inducible gene-I and melanoma differentiation-associated gene 5. J Exp Med. 2008;205(7):1601–1610. doi: 10.1084/jem.20080091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hornung V, et al. 5′-Triphosphate RNA is the ligand for RIG-I. Science. 2006;314(5801):994–997. doi: 10.1126/science.1132505. [DOI] [PubMed] [Google Scholar]
- 6.Loo YM, Gale M., Jr Immune signaling by RIG-I-like receptors. Immunity. 2011;34(5):680–692. doi: 10.1016/j.immuni.2011.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hou F, et al. MAVS forms functional prion-like aggregates to activate and propagate antiviral innate immune response. Cell. 2011;146(3):448–461. doi: 10.1016/j.cell.2011.06.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Iwasaki H, et al. The IκB kinase complex regulates the stability of cytokine-encoding mRNA induced by TLR-IL-1R by controlling degradation of regnase-1. Nat Immunol. 2011;12(12):1167–1175. doi: 10.1038/ni.2137. [DOI] [PubMed] [Google Scholar]
- 9.Gack MU, et al. TRIM25 RING-finger E3 ubiquitin ligase is essential for RIG-I-mediated antiviral activity. Nature. 2007;446(7138):916–920. doi: 10.1038/nature05732. [DOI] [PubMed] [Google Scholar]
- 10.Oshiumi H, Matsumoto M, Hatakeyama S, Seya T. Riplet/RNF135, a RING finger protein, ubiquitinates RIG-I to promote interferon-beta induction during the early phase of viral infection. J Biol Chem. 2009;284(2):807–817. doi: 10.1074/jbc.M804259200. [DOI] [PubMed] [Google Scholar]
- 11.Oshiumi H, et al. The ubiquitin ligase Riplet is essential for RIG-I-dependent innate immune responses to RNA virus infection. Cell Host Microbe. 2010;8(6):496–509. doi: 10.1016/j.chom.2010.11.008. [DOI] [PubMed] [Google Scholar]
- 12.Onomoto K, et al. Critical role of an antiviral stress granule containing RIG-I and PKR in viral detection and innate immunity. PLoS ONE. 2012;7(8):e43031. doi: 10.1371/journal.pone.0043031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Langereis MA, Feng Q, van Kuppeveld FJ. MDA5 localizes to stress granules, but this localization is not required for the induction of type I interferon. J Virol. 2013;87(11):6314–6325. doi: 10.1128/JVI.03213-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Buchet-Poyau K, et al. Identification and characterization of human Mex-3 proteins, a novel family of evolutionarily conserved RNA-binding proteins differentially localized to processing bodies. Nucleic Acids Res. 2007;35(4):1289–1300. doi: 10.1093/nar/gkm016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kawai T, Akira S. Toll-like receptor and RIG-I-like receptor signaling. Ann N Y Acad Sci. 2008;1143:1–20. doi: 10.1196/annals.1443.020. [DOI] [PubMed] [Google Scholar]
- 16.Guzmán B, et al. Implication of chromosome 18 in hypertension by sibling pair and association analyses: putative involvement of the RKHD2 gene. Hypertension. 2006;48(5):883–891. doi: 10.1161/01.HYP.0000244085.52918.a0. [DOI] [PubMed] [Google Scholar]
- 17.Draper BW, Mello CC, Bowerman B, Hardin J, Priess JR. MEX-3 is a KH domain protein that regulates blastomere identity in early C. elegans embryos. Cell. 1996;87(2):205–216. doi: 10.1016/s0092-8674(00)81339-2. [DOI] [PubMed] [Google Scholar]
- 18.Jiao Y, et al. Mex3c mutation reduces adiposity and increases energy expenditure. Mol Cell Biol. 2012;32(21):4350–4362. doi: 10.1128/MCB.00452-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jiao Y, Bishop CE, Lu B. Mex3c regulates insulin-like growth factor 1 (IGF1) expression and promotes postnatal growth. Mol Biol Cell. 2012;23(8):1404–1413. doi: 10.1091/mbc.E11-11-0960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Burrell RA, et al. Replication stress links structural and numerical cancer chromosomal instability. Nature. 2013;494(7438):492–496. doi: 10.1038/nature11935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cano F, Lehner PJ. A novel post-transcriptional role for ubiquitin in the differential regulation of MHC class I allotypes. Mol Immunol. 2013;55(2):135–138. doi: 10.1016/j.molimm.2012.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cano F, et al. The RNA-binding E3 ubiquitin ligase MEX-3C links ubiquitination with MHC-I mRNA degradation. EMBO J. 2012;31(17):3596–3606. doi: 10.1038/emboj.2012.218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pereira B, Le Borgne M, Chartier NT, Billaud M, Almeida R. MEX-3 proteins: Recent insights on novel post-transcriptional regulators. Trends Biochem Sci. 2013;38(10):477–479. doi: 10.1016/j.tibs.2013.08.004. [DOI] [PubMed] [Google Scholar]
- 24.Kowalinski E, et al. Structural basis for the activation of innate immune pattern-recognition receptor RIG-I by viral RNA. Cell. 2011;147(2):423–435. doi: 10.1016/j.cell.2011.09.039. [DOI] [PubMed] [Google Scholar]
- 25.Takahasi K, et al. Nonself RNA-sensing mechanism of RIG-I helicase and activation of antiviral immune responses. Mol Cell. 2008;29(4):428–440. doi: 10.1016/j.molcel.2007.11.028. [DOI] [PubMed] [Google Scholar]
- 26.Oshiumi H, Miyashita M, Matsumoto M, Seya T. A distinct role of Riplet-mediated K63-Linked polyubiquitination of the RIG-I repressor domain in human antiviral innate immune responses. PLoS Pathog. 2013;9(8):e1003533. doi: 10.1371/journal.ppat.1003533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jiang X, et al. Ubiquitin-induced oligomerization of the RNA sensors RIG-I and MDA5 activates antiviral innate immune response. Immunity. 2012;36(6):959–973. doi: 10.1016/j.immuni.2012.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ranjan P, et al. 5’PPP-RNA induced RIG-I activation inhibits drug-resistant avian H5N1 as well as 1918 and 2009 pandemic influenza virus replication. Virol J. 2010;7:102. doi: 10.1186/1743-422X-7-102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hemmi H, Kaisho T, Takeda K, Akira S. The roles of Toll-like receptor 9, MyD88, and DNA-dependent protein kinase catalytic subunit in the effects of two distinct CpG DNAs on dendritic cell subsets. J Immunol. 2003;170(6):3059–3064. doi: 10.4049/jimmunol.170.6.3059. [DOI] [PubMed] [Google Scholar]
- 30.Matsui K, et al. Cutting edge: Role of TANK-binding kinase 1 and inducible IkappaB kinase in IFN responses against viruses in innate immune cells. J Immunol. 2006;177(9):5785–5789. doi: 10.4049/jimmunol.177.9.5785. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.