Significance
Our in vitro analyses reveal that dystrophin, the protein absent in Duchenne muscular dystrophy patients, binds microtubules with high affinity and pauses microtubule polymerization, whereas utrophin, the autosomal homologue of dystrophin thought to mirror many known functions of dystrophin, has no activity in either assay. We also report that transgenic utrophin overexpression does not correct subsarcolemmal microtubule lattice disorganization, physical inactivity after mild exercise, or loss of torque production after in vivo eccentric contraction in dystrophin-deficient skeletal muscle. Finally, our data demonstrate that microtubule lattice disorganization contributes to the greater eccentric contraction-induced injury experienced by dystrophin-deficient skeletal muscle, demonstrating a phenotype of dystrophin deficiency that utrophin-based therapy may not be able to correct.
Abstract
Dystrophin and utrophin are highly similar proteins that both link cortical actin filaments with a complex of sarcolemmal glycoproteins, yet localize to different subcellular domains within normal muscle cells. In mdx mice and Duchenne muscular dystrophy patients, dystrophin is lacking and utrophin is consequently up-regulated and redistributed to locations normally occupied by dystrophin. Transgenic overexpression of utrophin has been shown to significantly improve aspects of the disease phenotype in the mdx mouse; therefore, utrophin up-regulation is under intense investigation as a potential therapy for Duchenne muscular dystrophy. Here we biochemically compared the previously documented microtubule binding activity of dystrophin with utrophin and analyzed several transgenic mouse models to identify phenotypes of the mdx mouse that remain despite transgenic utrophin overexpression. Our in vitro analyses revealed that dystrophin binds microtubules with high affinity and pauses microtubule polymerization, whereas utrophin has no activity in either assay. We also found that transgenic utrophin overexpression does not correct subsarcolemmal microtubule lattice disorganization, loss of torque production after in vivo eccentric contractions, or physical inactivity after mild exercise. Finally, our data suggest that exercise-induced inactivity correlates with loss of sarcolemmal neuronal NOS localization in mdx muscle, whereas loss of in vivo torque production after eccentric contraction-induced injury is associated with microtubule lattice disorganization.
Duchenne muscular dystrophy (DMD) is a lethal X-linked disease found in approximately 1 of every 4,000 live male births and is caused by loss-of-function mutations in the DMD gene encoding dystrophin (1, 2). The dystrophin protein consists of an amino-terminal tandem calponin-homology actin binding domain, a large rod domain composed of spectrin-like repeats and flexible hinge regions, within which lie a second actin binding domain and the neuronal nitric oxide synthase (nNOS) binding domain, and cysteine-rich and carboxyl-terminal domains (3–9). Dystrophin is enriched at subsarcolemmal protein assemblies known as costameres, where it couples actin and intermediate filaments to a membrane-associated glycoprotein complex (10–12). Through its many protein interactions, dystrophin is hypothesized to protect against contraction-induced muscle damage via radial force transmission and stabilization of the sarcolemma (13–15). Additionally, through both direct and indirect interactions via proteins such as ankyrin-B and obscurin, dystrophin has been found to bind microtubules to form a rectilinear lattice beneath the sarcolemma (16–20).
The commonly used mdx mouse model of DMD encodes a nonsense mutation in exon 23 of the DMD transcript ablating dystrophin protein expression (21, 22). Similar to DMD patients, the mdx mouse displays widespread muscle weakness, undergoes repetitive rounds of muscle degeneration and regeneration, and is highly susceptible to contraction-induced injury, losing almost all force-generating capacity after as few as five eccentric contractions (22–25). However, although the mdx mouse recapitulates some phenotypes associated with DMD, it has a normal lifespan and presents with an overall milder phenotype than do DMD patients (26). This relatively mild dystrophic phenotype is partially attributed to up-regulation of the protein utrophin. Much like dystrophin, utrophin contains an amino-terminal tandem calponin-homology domain that binds actin, a large central rod domain, and cysteine-rich and carboxyl-terminal domains (27–29). Because of its structural and functional similarity to dystrophin, utrophin is under active investigation as a potential therapy for DMD (30, 31). In fact, increased utrophin levels have been shown to correlate with improved prognosis in a small cohort of DMD patients (32). Furthermore, transgenic overexpression of utrophin in the Fiona-mdx mouse rescues many of the dystrophic phenotypes of the mdx mouse (33).
Although the mdx mouse expresses increased utrophin, these levels fail to restore nNOS to the sarcolemma or rescue the disorganized microtubule lattice that results owing to loss of dystrophin (16, 19, 34, 35), indicating that utrophin cannot fully compensate for the lack of dystrophin. There are two main possibilities why endogenously up-regulated utrophin fails to retain microtubule lattice organization. The first possibility is that the microtubule binding function of dystrophin is not conserved in utrophin. A precedent for this hypothesis has already been shown in regard to nNOS. In normal muscle, dystrophin localizes nNOS to the sarcolemma, where it functions to regulate vasodilation during active muscle contraction (36–40). However, in dystrophic muscle, the loss of nNOS localization to the sarcolemma correlates with several pathophysiologic phenotypes, including a dramatic reduction in physical activity after mild exercise (37, 38). The transgenic utrophin overexpression mouse model does not restore the localization of nNOS to the sarcolemma, making this the only thus-far documented mdx phenotype not rescued in the Fiona-mdx mouse (9, 34). The second possibility is that the amount of endogenous utrophin up-regulation in the mdx mouse is simply insufficient to restore microtubule lattice organization. In support of this hypothesis, endogenous utrophin up-regulation in the mdx mouse is insufficient to mechanically anchor costameric actin filaments to the sarcolemma, whereas transgenic utrophin overexpression in the Fiona-mdx mouse restores coupling (29). Thus, the lack of microtubule lattice organization in mdx muscle may be due to an absence of microtubule binding activity in utrophin or to insufficient utrophin expression.
Here we show that microtubule lattice disorganization persists in the presence of transgenically overexpressed utrophin owing to the absence of intrinsic microtubule binding activity in utrophin. Additionally, we show that the microtubule binding domain of dystrophin resides within spectrin-like repeats 20–23. Moreover, we show that the loss of nNOS localization contributes to loss of physical activity after mild exercise independent of microtubule organization, whereas microtubule disorganization contributes to torque loss after eccentric contraction-induced injury in vivo. Our data further define the distinct pathophysiological consequences of nNOS mislocalization and microtubule disorganization associated with dystrophinopathy that are not rescued by the surrogate utrophin.
Results
Utrophin Lacks Microtubule Binding Activity in Vivo and in Vitro.
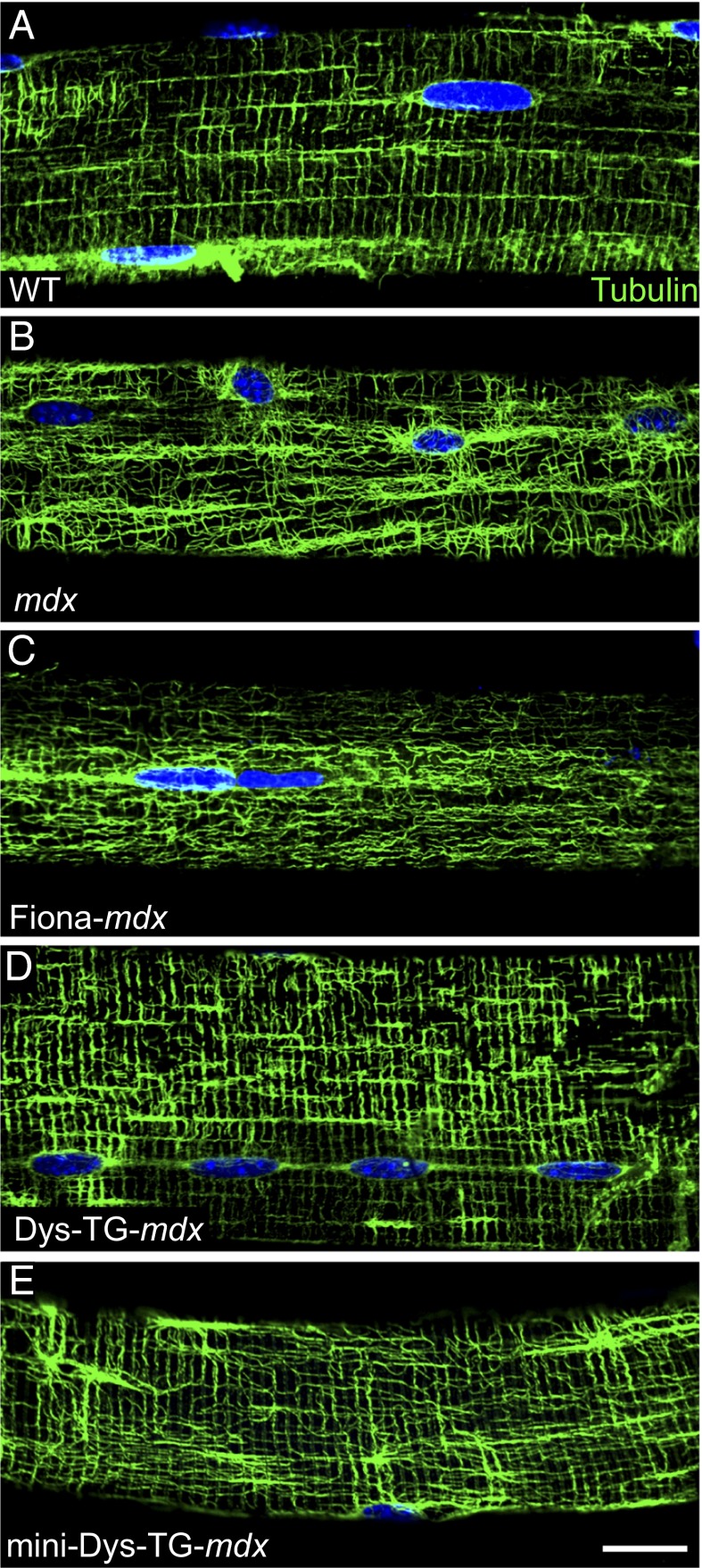
To determine whether overexpression of utrophin could rescue microtubule disorganization in dystrophin-deficient mouse extensor digitorum longus (EDL) muscle fibers, we compared mdx mice transgenically overexpressing utrophin (Fiona-mdx), nearly full-length dystrophin (Dys-TG-mdx), or a miniaturized dystrophin (mini-Dys-TG-mdx) lacking spectrin-like repeats 4 through 19 (33, 41, 42). Western blotting of quadriceps muscle lysates verified that all three transgenic proteins are overexpressed above WT levels (Fig. S1). Unlike WT mice, which show rectilinear microtubule lattice organization (Fig. 1A), the subsarcolemmal microtubule lattice in the Fiona-mdx mouse remained highly disorganized, with morphology similar to that observed in mdx muscle (Fig. 1 B and C). Conversely, the Dys-TG-mdx and mini-Dys-TG-mdx mice showed rectilinear microtubule lattice organization comparable to that of WT mice (Fig. 1 D and E). Additionally, in agreement with previously published data (43), we find that mdx mice have a significantly denser subsarcolemmal microtubule network than all other mouse lines (Fig. S2A; density = 61.5% ± 3.4%). Conversely, WT and Dys-TG-mdx mice display the least dense networks (27.8% ± 1.8% and 31.8% ± 1.5%, respectively), whereas the Fiona-mdx and mini-Dys-TG-mdx mice display intermediate network densities (48.4% ± 1.7% and 45%.8 ± 2.0%, respectively). WT and Dys-TG-mdx were significantly different from Fiona-mdx and mini-Dys-TG-mdx mice, and all lines were significantly different from mdx mice (P < 0.01 for all statistically significant comparisons). Moreover, we quantitatively analyzed microtubule lattice images using a recently developed program (44) for analyzing microtubule directionality (TeDT). In agreement with previously published data (19, 20, 35, 44, 45), the directionality histograms (Fig. S2B) resulting from TeDT analysis revealed a significant decrease in the presence of transversely oriented microtubules (centered around 90°) in mdx and Fiona-mdx mice that were present in WT, Dys-TG-mdx, and mini-Dys-TG-mdx mice (P < 0.05). There was no significant difference between any of the mouse lines with respect to the longitudinally oriented microtubules (0° and 180°). These data indicate that transgenic utrophin overexpression is insufficient to restore normal microtubule lattice organization in mdx skeletal muscle and confirm that the absence of dystrophin mainly results in loss of the transverse microtubule element of the subsarcolemmal microtubule lattice.
Fig. 1.
Microtubule organization in 6-mo-old mouse EDL muscle fibers. (A) When dystrophin is present (WT), microtubules are organized into a rectilinear lattice beneath the sarcolemma. (B) In the absence of dystrophin (mdx), the microtubule lattice becomes disordered. (C) Transgenic overexpression of utrophin in the absence of dystrophin (Fiona-mdx) does not rescue the disorganized microtubule lattice. (D) Transgenic overexpression of nearly full-length dystrophin (Dys-TG-mdx) or (E) mini-dystrophin (mini-Dys-TG-mdx) in the absence of endogenous dystrophin rescues the microtubule lattice. Images are representative of those obtained for n ≥ 10 fibers from each of n ≥ 3 mice per genotype. (Scale bar, 20 µm.)
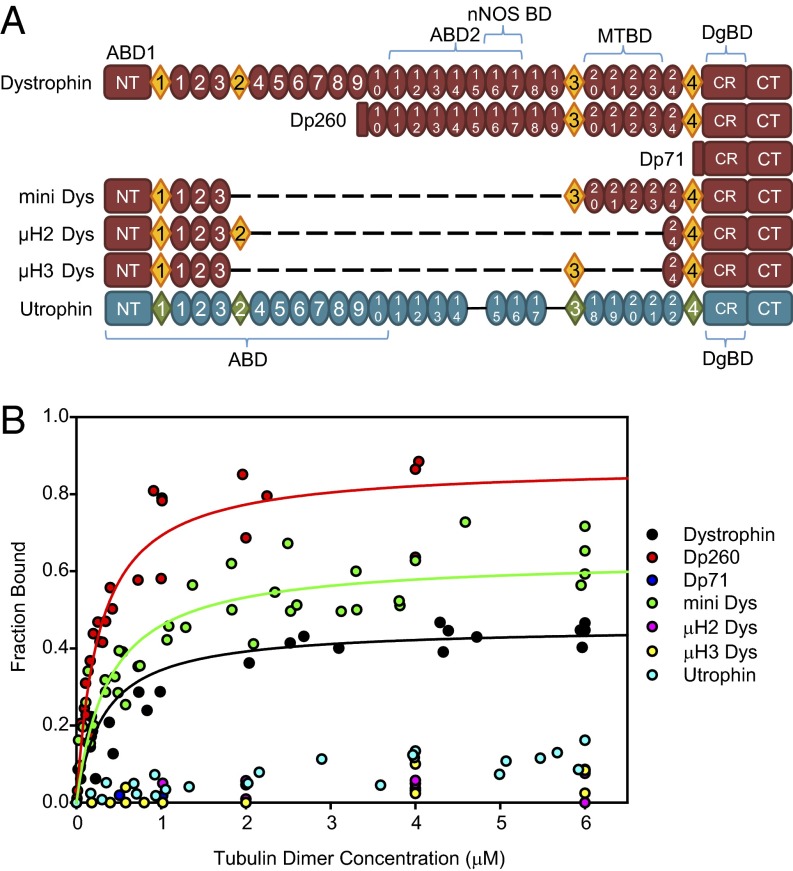
To further understand why utrophin overexpression failed to restore microtubule lattice organization in mdx muscle, we measured the microtubule binding activities for an array of naturally occurring dystrophin isoforms, therapeutically relevant internally truncated dystrophin constructs, and utrophin using an established high-speed microtubule cosedimentation assay (Fig. 2A). Only full-length dystrophin (KD = 0.33 µM), Dp260 (KD = 0.26 µM), and mini Dys (KD = 0.38 µM) showed specific microtubule binding, whereas all other constructs, including utrophin, exhibited no measureable specific binding (Fig. 2B). These data indicate that utrophin overexpression fails to restore microtubule lattice organization in mdx muscle (Fig. 1C) because it lacks the microtubule binding activity intrinsic to dystrophin. Additionally, on the basis of the minimal region common to the dystrophin constructs that bind microtubules, we hypothesize that spectrin-like repeats 20–23 encode the microtubule binding domain of dystrophin.
Fig. 2.
Domain structure and microtubule binding properties of dystrophin and utrophin. (A) Schematic representation of dystrophin and utrophin protein constructs. Ovals represent spectrin-like repeats, diamonds represent hinge regions. Homologous repeats are aligned. Dystrophin repeats 15 and 19 do not have homologous repeats in utrophin. ABD, actin binding domain; nNOS BD, nNOS binding domain; MTBD, microtubule binding domain; DgBD, β-dystroglycan binding domain; NT, N terminus; CT, C terminus; CR, cysteine rich domain. (B) Microtubule binding curves of constructs assayed. Dystrophin binds microtubules with high affinity (KD = 0.33 µM), as do Dp260 (KD = 0.26 µM) and mini-dystrophin (KD = 0.38 µM), whereas all other dystrophin constructs and utrophin display no specific microtubule binding.
Dystrophin Acts as a Molecular Guidepost to Organize the Microtubule Lattice.
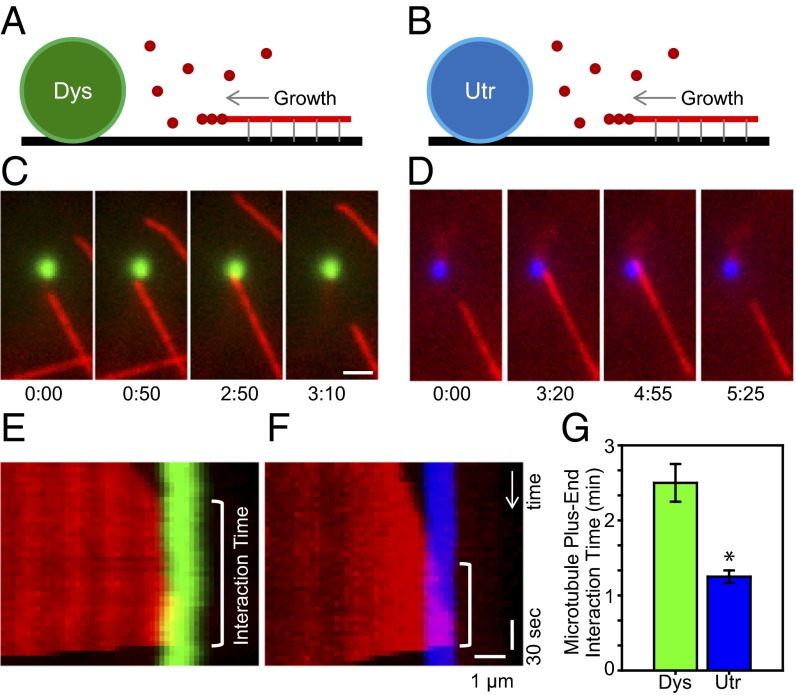
To investigate the regulation of microtubule localization/organization, we coupled either dystrophin or utrophin to fluorescent microparticles and imaged the dystrophin/microtubule or utrophin/microtubule interaction as free fluorescent tubulin was allowed to polymerize off immobilized seeds toward these protein-coated microparticles (46). Kymographs of the image data revealed that microtubules significantly paused when in contact with a dystrophin-coated microparticle (Fig. 3 A, C, E, and G), whereas microtubule pausing was not observed when microtubules encountered a utrophin-coated microparticle (Fig. 3 B, D, F, and G). These in vitro data support recent in vivo studies suggesting that dystrophin may act as a molecular guidepost to define domains along which microtubules preferentially grow (45). Our results indicate that utrophin does not recapitulate this function of dystrophin.
Fig. 3.
Effect of dystrophin and utrophin on microtubule localization. (A and B) Schematic representation of assay performed. Dystrophin (Dys, green) or utrophin (Utr, blue). (C) Time-lapse images of a direct dystrophin/microtubule interaction. (Scale bar, 2 µm.) (D) Time-lapse images of a direct utrophin/microtubule interaction. (E) Kymograph of the data presented in C. (F) Kymograph of the data presented in D. (G) The dystrophin/microtubule (n = 6) interaction time is significantly greater than the utrophin/microtubule (n = 8) interaction time. Data are presented as means ± SE. *P < 0.001.
To better understand the cellular role of dystrophin binding to microtubules, we performed experiments to address the possibility that dystrophin may regulate tubulin dynamics and/or microtubule bundling. The rate of tubulin polymerization was not measurably different in the absence or presence of dystrophin (2.42 mOD/min and 2.46 mOD/min for tubulin and tubulin plus dystrophin, respectively), or utrophin (2.38 mOD/min), suggesting that neither dystrophin nor utrophin have any effect on tubulin polymerization (Fig. S3A). Over the course of 30 min, the rate of tubulin depolymerization (−8.75%/min) was no different regardless of whether dystrophin or utrophin were present (−8.01%/min and −6.92%/min, respectively; Fig. S3B), suggesting that neither dystrophin nor utrophin has any effect on microtubule depolymerization. Last, we incubated both red and green fluorescently labeled microtubules in the absence or presence of dystrophin or utrophin. Microtubule bundling induced by tau was detected as yellow fluorescence, indicating the close apposition of separately labeled red and green microtubules. Although neither dystrophin nor utrophin induced detectable microtubule bundling activity in vitro (Fig. S3C), it remains possible that dystrophin can bundle microtubules in vivo, although we (Fig. 1) and others (43) have measured greater microtubule density in dystrophin-deficient mdx muscle.
Microtubule Disorganization and nNOS Mislocalization Correlate with Distinct Pathophysiological Phenotypes.
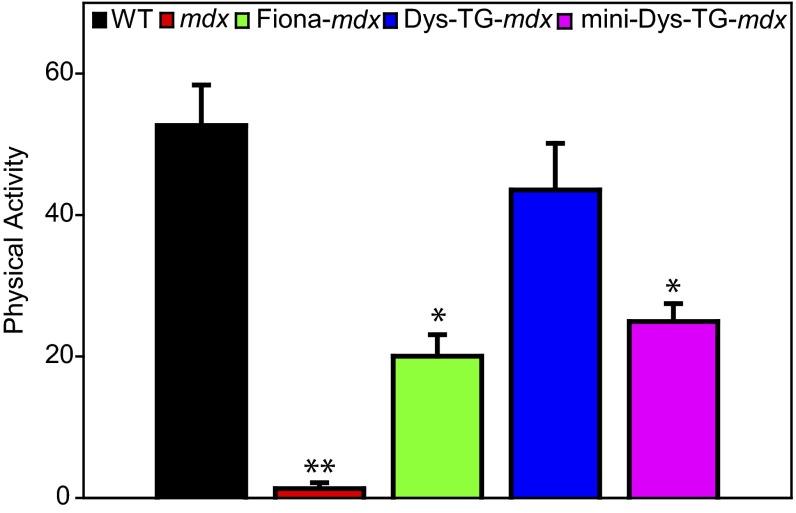
The mdx mouse shows greatly reduced physical activity after mild exercise that has been attributed to a lack of contraction-induced signaling from sarcolemmal-localized nNOS (37–40, 47). Although transgenic overexpression of utrophin in the Fiona-mdx mouse clearly fails to restore sarcolemmal nNOS localization (34), physical activity levels after mild exercise have not yet been measured. Similar to previously reported data (47), we found that mdx mice were significantly less active compared with WT mice after mild exercise (Fig. 4; P < 0.001). The Fiona-mdx mice showed intermediate postexercise physical activity that was significantly greater than in mdx mice (P = 0.029) but also significantly less than that of WT (P = 0.004) and Dys-TG-mdx mice (P = 0.036). Because Fiona-mdx mice were significantly more active than mdx mice after mild exercise, our data support the idea that the underlying mechanism of postexercise inactivity in mdx muscle is more complex and attributable to more than solely the loss of sarcolemmal nNOS localization (37). Interestingly, the mini-Dys-TG-mdx mouse, which shows rescued microtubule morphology (Fig. 1E) but lacks the nNOS binding domain (34), exhibited postexercise activity that mirrored the activity of the Fiona-mdx mouse (P = 0.685), suggesting that microtubule lattice disorganization does not contribute to exercise-induced inactivity.
Fig. 4.
Physical activity of mice after mild treadmill exercise. WT mice exhibited ∼50% of their initial cage activity after mild exercise, whereas postexercise activity in mdx mice dropped more than 95% of their initial activity. Transgenic overexpression of dystrophin in mdx mice (Dys-TG-mdx) restored postexercise activity levels similar to that in WT, whereas transgenic overexpression of utrophin (Fiona-mdx) or mini-dystrophin (mini-Dys-TG-mdx) yielded intermediate (∼25%) restoration of activity. *Statistically different from WT, Dys-TG-mdx, and mdx; **statistically different from all other lines.
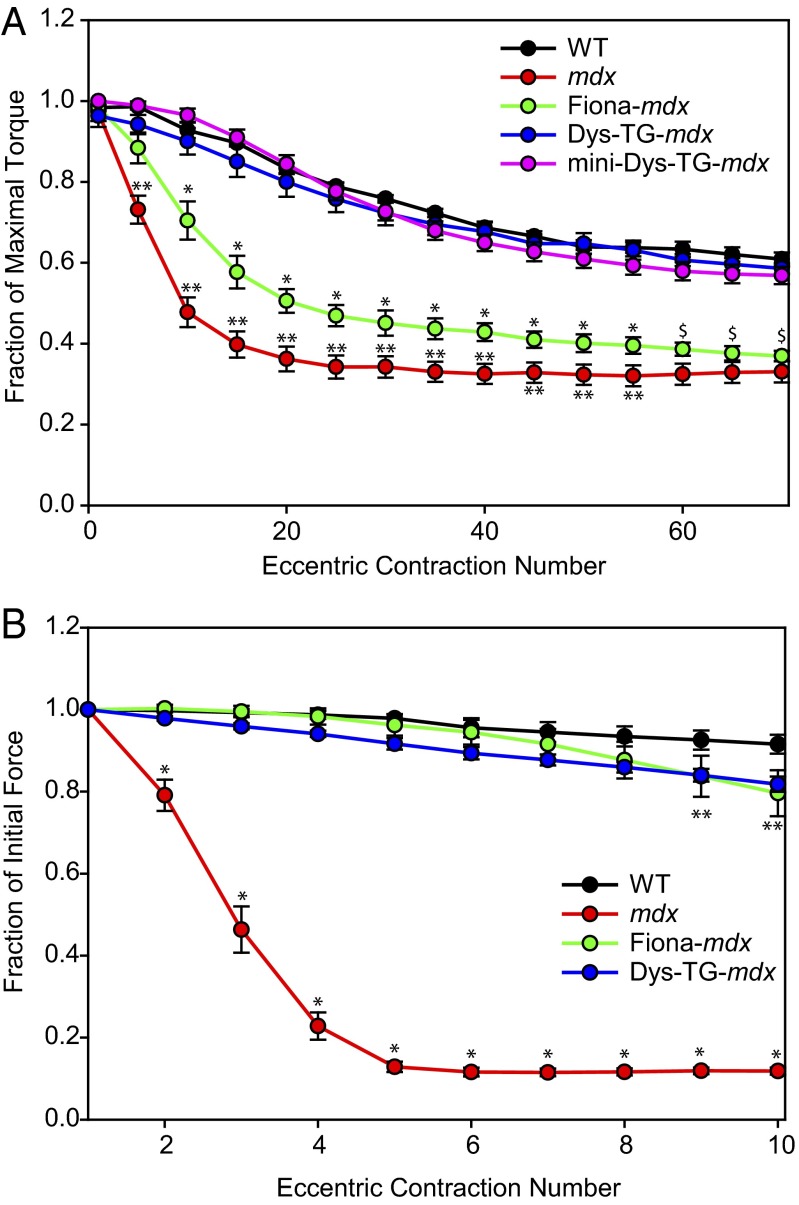
Moving from the whole mouse to intact muscle groups, we measured in vivo torque production by the anterior crural muscles (EDL, extensor hallicus longus, and tibialis anterior) in each mouse line in response to repeated rounds of eccentric contraction-induced injury. This in vivo assay is designed to reveal more subtle differences between the WT, mdx, and transgenic mdx lines in resistance to more stringent mechanical stresses (48, 49). Overall, the effect of genotype on torque loss was dependent on contraction number (Fig. 5A; significant interaction between genotype and contraction, P < 0.001). Compared with WT, mdx mice exhibited a drastic loss in torque production, whereas Dys-TG-mdx and mini-Dys-TG-mdx mice both showed losses of torque production equivalent to that in WT mice. Interestingly, Fiona-mdx mice showed an intermediate loss of torque production that was significantly different from both mdx and the other mouse lines evaluated over the first 60 eccentric contractions. Over the last 10 eccentric contractions, Fiona-mdx mice began to become indistinguishable from mdx mice (P = 0.057, 0.145, and 0.233 for contractions 60, 65, and 70, respectively), possibly indicating transient protection from contraction-induced injury in the Fiona-mdx mice that wanes to mdx levels as contractions continue. These data suggest that microtubule lattice disorganization contributes to contraction-induced injury because Fiona-mdx mice, which lack both nNOS and have disordered microtubules, were sensitive to injury, whereas mini-Dys-TG-mdx mice, which also lack nNOS but have rescued microtubule organization, were resistant to injury. This also suggests that nNOS localization to the sarcolemma or lack thereof has no bearing on susceptibility to contraction-induced injury. However, Fiona-mdx mice were significantly more protected from injury than mdx mice, indicating that the underlying mechanism of contraction-induced injury in mdx muscle is more complex and cannot solely be attributed to microtubule lattice disorganization.
Fig. 5.
In vivo and ex vivo contraction-induced injury loss by anterior crural muscles. (A) Compared with WT mice, mdx mice showed drastic loss of torque during a series of eccentric contractions. Dys-TG-mdx and mini-Dys-TG-mdx mice showed much less loss of torque, analogous to that of WT mice. Conversely, the Fiona-mdx mice showed intermediate loss of torque, comparable to the intermediate loss of physical activity seen in Fig. 4. *Statistically different from WT, Dys-TG-mdx, mini-Dys-TG-mdx, and mdx; **statistically different from WT, Dys-TG-mdx, Fiona-mdx, and mini-Dys-TG-mdx; $statistically different from WT, Dys-TG-mdx, and mini-Dys-TG-mdx but not mdx. (B) Significant loss of force by mdx muscles at contractions 2 through 10 compared with WT (P < 0.001). EDL muscles from Fiona-mdx and Dys-TG-mdx mice were protected from force loss and were not different from WT at contractions 2 through 8 (P > 0.05). At contractions 9 and 10, Fiona-mdx (P = 0.031 and P = 0.002 for contractions 9 and 10, respectively) and Dys-TG-mdx (P = 0.014 and P = 0.005 for contractions 9 and 10, respectively) began to show differences from WT but were not different from each other (P = 0.945 and P = 0.532 for contractions 9 and 10, respectively).
Last, we measured ex vivo contractile function in isolated EDL muscles from the various mouse lines. The contraction-induced injury protocol exposed muscles to repeated eccentric contractions (25). Overall, the effect of genotype on force loss was dependent on contraction number (Fig. 5B and Table S1; significant interaction between genotype and contraction, P < 0.001). The mdx EDL showed a statistically significant loss of force production after a single eccentric contraction, which plateaued to 15% of WT force production by the fifth eccentric contraction. Consistent with previous studies (33, 38, 41), isolated Fiona-mdx and Dys-TG-mdx EDL muscles revealed no statistically significant force loss compared with WT EDL muscles, even after 10 consecutive eccentric contractions.
Discussion
Multiple groups have shown that proper microtubule function is necessary for myotube formation (50–52) and that the microtubule (+) end binding protein EB1 is necessary for this process (53). Additionally, it is known that vesicular trafficking and organelle localization require proper microtubule function in muscle cells (20, 54). Despite these insights into microtubule function, little is known about the relevance of microtubule tethering and lattice organization in skeletal muscle. Studies in other cell types have shown that the CLASP family of proteins is responsible for tethering microtubule (+) ends to the cell cortex, thereby organizing them spatially and inhibiting their depolymerization (55–57). These studies provide a basis from which to speculate that dystrophin may play a similar tethering role in skeletal muscle. In the present study, we find that dystrophin does not affect tubulin polymerization, microtubule depolymerization, or microtubule bundling, but does indeed tether growing microtubules in an in vitro assay. Our data provide a molecular basis for a recent in vivo microtubule dynamics study that hypothesized a molecular guidepost function for dystrophin in organizing the subsarcolemmal microtubule lattice in adult skeletal muscle fibers (45).
In the absence of dystrophin, the subsarcolemmal microtubule lattice becomes disorganized (19, 20, 35, 44), and this disorganization contributes to the dystrophic phenotype of the mdx mouse (43, 58). Like dystrophin, utrophin couples membrane-bound dystrophin-associated proteins with costameric actin filaments, suggesting that up-regulation of utrophin could compensate for the loss of dystrophin (28–31). However, here we show that utrophin exhibits no microtubule binding activity in vitro and that transgenic overexpression of utrophin fails to rescue microtubule lattice disorganization in mdx muscle. BLASTp analysis of the microtubule binding repeats 20–23 in dystrophin reports only 42% identity to the homologous utrophin repeats. These data may have implications for therapies that aim to up-regulate utrophin in DMD patients in an attempt to compensate for the lack of dystrophin because these therapies will not rescue microtubule lattice organization.
Our previous data led us to hypothesize that microtubule binding activity is localized to spectrin-like repeat 24 through the first third of the WW domain (19). However, the cosedimentation assay used previously (19) was performed on skeletal muscle lysates from mice expressing endogenous dystrophin or transgenic dystrophin constructs, which cannot distinguish between direct and indirect interactions, and dystrophin also interacts with microtubules via ankyrin-B (16, 59). Here, using purified recombinant dystrophin proteins and tubulin in a high-speed cosedimentation assay, we identified spectrin-like repeats 20–23 in the direct binding interaction of dystrophin with microtubules.
Microtubule lattice derangement in the mdx mouse has recently been linked to an increase in reactive oxygen species (ROS) and aberrant calcium regulation, providing evidence for a microtubule-associated pathophysiology in DMD (43, 58). Here, we report that microtubule disorganization significantly contributes by additional mechanisms to the pathophysiology of the mdx mouse. Independent of sarcolemmal nNOS localization, we conclude that the in vivo torque loss of mdx mice in response to contraction-induced injury is associated with microtubule lattice disorganization. Because excess ROS signaling and increased intracellular calcium are thought to contribute to contraction-induced injury, our results support a link between these and microtubule lattice disorganization. Additionally, previous studies have shown that mitochondria move to sites of damage in healthy muscle, presumably along microtubules (60). Therefore, microtubule lattice disorganization may affect how mitochondria respond to injury, thus leading to increased ROS signaling and impaired calcium handling. Moreover, subsarcolemmal mitochondria are less dense in mdx muscle, supporting the idea that they are not functioning properly (61). Thus, it is possible that microtubule disorganization may impair mitochondrial localization and function, limiting the buffering capacity of the cell and rendering it more susceptible to contraction-induced damage.
In addition to the differences in microtubule binding of dystrophin and utrophin, it has been previously established that dystrophin localizes nNOS to the sarcolemma, whereas utrophin does not. Mice lacking sarcolemmal nNOS exhibit dramatically reduced cage activity after mild exercise (47). Although sarcolemmal nNOS localization is not rescued by the endogenous utrophin overexpression in mdx muscle or by transgenic utrophin overexpression in Fiona-mdx muscle (34), we show that postexercise activity in mdx mice is partially rescued by transgenic utrophin overexpression. Whereas mdx mice lose almost all physical activity after mild exercise, Fiona-mdx mice retain approximately half of WT physical activity levels after mild exercise. To us, this indicates that although nNOS almost certainly contributes to restoration of physical activity after exercise, it cannot be the sole contributing factor. We therefore posit that the decreased rounds of muscle degeneration and regeneration, the increased sarcolemmal stability, and the lack of many other dystrophic phenotypes in the Fiona-mdx mouse at least partially contribute to this increased physical activity after mild exercise.
In summary, we show that utrophin lacks the intrinsic microtubule binding activity of dystrophin repeats 20–23, illustrating another key functional difference between dystrophin and utrophin, which should be taken into consideration in the development of utrophin-based therapies for treating DMD. Additionally, we provide data in support of a molecular guidepost function for dystrophin, which would serve to anchor microtubules to the costamere as a means to guide microtubule lattice organization in skeletal muscle.
Materials and Methods
All animals were housed and treated in accordance with the standards set by the University of Minnesota Institutional Animal Care and Use Committee. Fiona-mdx mice were kind gifts from Kay E. Davies (University of Oxford, Oxford, UK). Dys-TG-mdx and mini-Dys-TG-mdx mice were kind gifts from Jeffrey S. Chamberlain (University of Washington, Seattle, WA). Detailed descriptions of reagents, EDL fiber imaging, mouse physiological assays, protein expression and purification, and microtubule assays are provided in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank Drs. Wenhua Liu and Evelyn Ralston from the Light Image Section of National Institute of Arthritis and Musculoskeletal and Skin Diseases (NIAMS)/National Institutes of Health (NIH) for providing the directionality analysis program (TeDT). This study was supported by NIAMS Grant RO1 AR042423 (to J.M.E.). J.J.B. was supported by the NIH Training Program in Muscle Research (AR007612) and a University of Minnesota Doctoral Dissertation Fellowship. Muscle functional assessments were supported by NIH Grant P30 AR057220.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1323842111/-/DCSupplemental.
References
- 1.Mendell JR, et al. Evidence-based path to newborn screening for Duchenne muscular dystrophy. Ann Neurol. 2012;71(3):304–313. doi: 10.1002/ana.23528. [DOI] [PubMed] [Google Scholar]
- 2.Hoffman EP, Brown RH, Jr, Kunkel LM. Dystrophin: The protein product of the Duchenne muscular dystrophy locus. Cell. 1987;51(6):919–928. doi: 10.1016/0092-8674(87)90579-4. [DOI] [PubMed] [Google Scholar]
- 3.Koenig M, Monaco AP, Kunkel LM. The complete sequence of dystrophin predicts a rod-shaped cytoskeletal protein. Cell. 1988;53(2):219–228. doi: 10.1016/0092-8674(88)90383-2. [DOI] [PubMed] [Google Scholar]
- 4.Koenig M, Kunkel LM. Detailed analysis of the repeat domain of dystrophin reveals four potential hinge segments that may confer flexibility. J Biol Chem. 1990;265(8):4560–4566. [PubMed] [Google Scholar]
- 5.Way M, Pope B, Cross RA, Kendrick-Jones J, Weeds AG. Expression of the N-terminal domain of dystrophin in E. coli and demonstration of binding to F-actin. FEBS Lett. 1992;301(3):243–245. doi: 10.1016/0014-5793(92)80249-g. [DOI] [PubMed] [Google Scholar]
- 6.Bork P, Sudol M. The WW domain: A signalling site in dystrophin? Trends Biochem Sci. 1994;19(12):531–533. doi: 10.1016/0968-0004(94)90053-1. [DOI] [PubMed] [Google Scholar]
- 7.Ponting CP, Blake DJ, Davies KE, Kendrick-Jones J, Winder SJ. ZZ and TAZ: New putative zinc fingers in dystrophin and other proteins. Trends Biochem Sci. 1996;21(1):11–13. [PubMed] [Google Scholar]
- 8.Rybakova IN, Ervasti JM. Dystrophin-glycoprotein complex is monomeric and stabilizes actin filaments in vitro through a lateral association. J Biol Chem. 1997;272(45):28771–28778. doi: 10.1074/jbc.272.45.28771. [DOI] [PubMed] [Google Scholar]
- 9.Lai Y, et al. Dystrophins carrying spectrin-like repeats 16 and 17 anchor nNOS to the sarcolemma and enhance exercise performance in a mouse model of muscular dystrophy. J Clin Invest. 2009;119(3):624–635. doi: 10.1172/JCI36612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rybakova IN, Amann KJ, Ervasti JM. A new model for the interaction of dystrophin with F-actin. J Cell Biol. 1996;135(3):661–672. doi: 10.1083/jcb.135.3.661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rybakova IN, Patel JR, Ervasti JM. The dystrophin complex forms a mechanically strong link between the sarcolemma and costameric actin. J Cell Biol. 2000;150(5):1209–1214. doi: 10.1083/jcb.150.5.1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ervasti JM. Costameres: The Achilles’ heel of Herculean muscle. J Biol Chem. 2003;278(16):13591–13594. doi: 10.1074/jbc.R200021200. [DOI] [PubMed] [Google Scholar]
- 13.Pardo JV, Siliciano JD, Craig SW. A vinculin-containing cortical lattice in skeletal muscle: Transverse lattice elements (“costameres”) mark sites of attachment between myofibrils and sarcolemma. Proc Natl Acad Sci USA. 1983;80(4):1008–1012. doi: 10.1073/pnas.80.4.1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Street SF. Lateral transmission of tension in frog myofibers: A myofibrillar network and transverse cytoskeletal connections are possible transmitters. J Cell Physiol. 1983;114(3):346–364. doi: 10.1002/jcp.1041140314. [DOI] [PubMed] [Google Scholar]
- 15.Danowski BA, Imanaka-Yoshida K, Sanger JM, Sanger JW. Costameres are sites of force transmission to the substratum in adult rat cardiomyocytes. J Cell Biol. 1992;118(6):1411–1420. doi: 10.1083/jcb.118.6.1411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ayalon G, Davis JQ, Scotland PB, Bennett V. An ankyrin-based mechanism for functional organization of dystrophin and dystroglycan. Cell. 2008;135(7):1189–1200. doi: 10.1016/j.cell.2008.10.018. [DOI] [PubMed] [Google Scholar]
- 17.Ayalon G, et al. Ankyrin-B interactions with spectrin and dynactin-4 are required for dystrophin-based protection of skeletal muscle from exercise injury. J Biol Chem. 2011;286(9):7370–7378. doi: 10.1074/jbc.M110.187831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Randazzo D, et al. Obscurin is required for ankyrinB-dependent dystrophin localization and sarcolemma integrity. J Cell Biol. 2013;200(4):523–536. doi: 10.1083/jcb.201205118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Prins KW, et al. Dystrophin is a microtubule-associated protein. J Cell Biol. 2009;186(3):363–369. doi: 10.1083/jcb.200905048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ralston E, Lu Z, Ploug T. The organization of the Golgi complex and microtubules in skeletal muscle is fiber type-dependent. J Neurosci. 1999;19(24):10694–10705. doi: 10.1523/JNEUROSCI.19-24-10694.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sicinski P, et al. The molecular basis of muscular dystrophy in the mdx mouse: a point mutation. Science. 1989;244(4912):1578–1580. doi: 10.1126/science.2662404. [DOI] [PubMed] [Google Scholar]
- 22.Bulfield G, Siller WG, Wight PAL, Moore KJ. X chromosome-linked muscular dystrophy (mdx) in the mouse. Proc Natl Acad Sci USA. 1984;81(4):1189–1192. doi: 10.1073/pnas.81.4.1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Carlson CG, Makiejus RV. A noninvasive procedure to detect muscle weakness in the mdx mouse. Muscle Nerve. 1990;13(6):480–484. doi: 10.1002/mus.880130603. [DOI] [PubMed] [Google Scholar]
- 24.Moens P, Baatsen PH, Maréchal G. Increased susceptibility of EDL muscles from mdx mice to damage induced by contractions with stretch. J Muscle Res Cell Motil. 1993;14(4):446–451. doi: 10.1007/BF00121296. [DOI] [PubMed] [Google Scholar]
- 25.Petrof BJ, Shrager JB, Stedman HH, Kelly AM, Sweeney HL. Dystrophin protects the sarcolemma from stresses developed during muscle contraction. Proc Natl Acad Sci USA. 1993;90(8):3710–3714. doi: 10.1073/pnas.90.8.3710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Partridge TA. The mdx mouse model as a surrogate for Duchenne muscular dystrophy. FEBS J. 2013;280(17):4177–4186. doi: 10.1111/febs.12267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Blake DJ, Weir A, Newey SE, Davies KE. Function and genetics of dystrophin and dystrophin-related proteins in muscle. Physiol Rev. 2002;82(2):291–329. doi: 10.1152/physrev.00028.2001. [DOI] [PubMed] [Google Scholar]
- 28.Matsumura K, Ervasti JM, Ohlendieck K, Kahl SD, Campbell KP. Association of dystrophin-related protein with dystrophin-associated proteins in mdx mouse muscle. Nature. 1992;360(6404):588–591. doi: 10.1038/360588a0. [DOI] [PubMed] [Google Scholar]
- 29.Rybakova IN, Patel JR, Davies KE, Yurchenco PD, Ervasti JM. Utrophin binds laterally along actin filaments and can couple costameric actin with sarcolemma when overexpressed in dystrophin-deficient muscle. Mol Biol Cell. 2002;13(5):1512–1521. doi: 10.1091/mbc.01-09-0446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hirst RC, McCullagh KJA, Davies KE. Utrophin upregulation in Duchenne muscular dystrophy. Acta Myol. 2005;24(3):209–216. [PubMed] [Google Scholar]
- 31.Fairclough RJ, Wood MJ, Davies KE. Therapy for Duchenne muscular dystrophy: Renewed optimism from genetic approaches. Nat Rev Genet. 2013;14(6):373–378. doi: 10.1038/nrg3460. [DOI] [PubMed] [Google Scholar]
- 32.Kleopa KA, Drousiotou A, Mavrikiou E, Ormiston A, Kyriakides T. Naturally occurring utrophin correlates with disease severity in Duchenne muscular dystrophy. Hum Mol Genet. 2006;15(10):1623–1628. doi: 10.1093/hmg/ddl083. [DOI] [PubMed] [Google Scholar]
- 33.Tinsley J, et al. Expression of full-length utrophin prevents muscular dystrophy in mdx mice. Nat Med. 1998;4(12):1441–1444. doi: 10.1038/4033. [DOI] [PubMed] [Google Scholar]
- 34.Li D, et al. Sarcolemmal nNOS anchoring reveals a qualitative difference between dystrophin and utrophin. J Cell Sci. 2010;123(Pt 12):2008–2013. doi: 10.1242/jcs.064808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Percival JM, et al. rAAV6-microdystrophin rescues aberrant Golgi complex organization in mdx skeletal muscles. Traffic. 2007;8(10):1424–1439. doi: 10.1111/j.1600-0854.2007.00622.x. [DOI] [PubMed] [Google Scholar]
- 36.Chang WJ, et al. Neuronal nitric oxide synthase and dystrophin-deficient muscular dystrophy. Proc Natl Acad Sci USA. 1996;93(17):9142–9147. doi: 10.1073/pnas.93.17.9142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Percival JM, Anderson KNE, Huang P, Adams ME, Froehner SC. Golgi and sarcolemmal neuronal NOS differentially regulate contraction-induced fatigue and vasoconstriction in exercising mouse skeletal muscle. J Clin Invest. 2010;120(3):816–826. doi: 10.1172/JCI40736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Percival JM, Anderson KNE, Gregorevic P, Chamberlain JS, Froehner SC. Functional deficits in nNOSmu-deficient skeletal muscle: myopathy in nNOS knockout mice. PLoS ONE. 2008;3(10):e3387. doi: 10.1371/journal.pone.0003387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Thomas GD, Shaul PW, Yuhanna IS, Froehner SC, Adams ME. Vasomodulation by skeletal muscle-derived nitric oxide requires alpha-syntrophin-mediated sarcolemmal localization of neuronal nitric oxide synthase. Circ Res. 2003;92(5):554–560. doi: 10.1161/01.RES.0000061570.83105.52. [DOI] [PubMed] [Google Scholar]
- 40.Thomas GD, et al. Impaired metabolic modulation of alpha-adrenergic vasoconstriction in dystrophin-deficient skeletal muscle. Proc Natl Acad Sci USA. 1998;95(25):15090–15095. doi: 10.1073/pnas.95.25.15090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Crawford GE, et al. Assembly of the dystrophin-associated protein complex does not require the dystrophin COOH-terminal domain. J Cell Biol. 2000;150(6):1399–1410. doi: 10.1083/jcb.150.6.1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li S, et al. A highly functional mini-dystrophin/GFP fusion gene for cell and gene therapy studies of Duchenne muscular dystrophy. Hum Mol Genet. 2006;15(10):1610–1622. doi: 10.1093/hmg/ddl082. [DOI] [PubMed] [Google Scholar]
- 43.Khairallah RJ, et al. Microtubules underlie dysfunction in Duchenne muscular dystrophy. Sci Signal. 2012;5(236):ra56. doi: 10.1126/scisignal.2002829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Liu W, Ralston E. A new directionality tool for assessing microtubule pattern alterations. Cytoskeleton (Hoboken) 2014 doi: 10.1002/cm.21166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Oddoux S, et al. Microtubules that form the stationary lattice of muscle fibers are dynamic and nucleated at Golgi elements. J Cell Biol. 2013;203(2):205–213. doi: 10.1083/jcb.201304063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hendricks AG, et al. Dynein tethers and stabilizes dynamic microtubule plus ends. Curr Biol. 2012;22(7):632–637. doi: 10.1016/j.cub.2012.02.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kobayashi YM, et al. Sarcolemma-localized nNOS is required to maintain activity after mild exercise. Nature. 2008;456(7221):511–515. doi: 10.1038/nature07414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Baltgalvis KA, Call JA, Nikas JB, Lowe DA. Effects of prednisolone on skeletal muscle contractility in mdx mice. Muscle Nerve. 2009;40(3):443–454. doi: 10.1002/mus.21327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ingalls CP, Warren GL, Lowe DA, Boorstein DB, Armstrong RB. Differential effects of anesthetics on in vivo skeletal muscle contractile function in the mouse. J Appl Physiol (1985) 1996;80(1):332–340. doi: 10.1152/jappl.1996.80.1.332. [DOI] [PubMed] [Google Scholar]
- 50.Saitoh O, Arai T, Obinata T. Distribution of microtubules and other cytoskeletal filaments during myotube elongation as revealed by fluorescence microscopy. Cell Tissue Res. 1988;252(2):263–273. doi: 10.1007/BF00214368. [DOI] [PubMed] [Google Scholar]
- 51.Chang W, et al. Alteration of the C-terminal amino acid of tubulin specifically inhibits myogenic differentiation. J Biol Chem. 2002;277(34):30690–30698. doi: 10.1074/jbc.M204930200. [DOI] [PubMed] [Google Scholar]
- 52.Perez OD, Chang YT, Rosania G, Sutherlin D, Schultz PG. Inhibition and reversal of myogenic differentiation by purine-based microtubule assembly inhibitors. Chem Biol. 2002;9(4):475–483. doi: 10.1016/s1074-5521(02)00131-x. [DOI] [PubMed] [Google Scholar]
- 53.Zhang T, et al. Microtubule plus-end binding protein EB1 is necessary for muscle cell differentiation, elongation and fusion. J Cell Sci. 2009;122(Pt 9):1401–1409. doi: 10.1242/jcs.039255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ralston E, Ploug T, Kalhovde J, Lomo T. Golgi complex, endoplasmic reticulum exit sites, and microtubules in skeletal muscle fibers are organized by patterned activity. J Neurosci. 2001;21(3):875–883. doi: 10.1523/JNEUROSCI.21-03-00875.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Akhmanova A, et al. Clasps are CLIP-115 and -170 associating proteins involved in the regional regulation of microtubule dynamics in motile fibroblasts. Cell. 2001;104(6):923–935. doi: 10.1016/s0092-8674(01)00288-4. [DOI] [PubMed] [Google Scholar]
- 56.Mimori-Kiyosue Y, et al. CLASP1 and CLASP2 bind to EB1 and regulate microtubule plus-end dynamics at the cell cortex. J Cell Biol. 2005;168(1):141–153. doi: 10.1083/jcb.200405094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lansbergen G, et al. CLASPs attach microtubule plus ends to the cell cortex through a complex with LL5β. Dev Cell. 2006;11(1):21–32. doi: 10.1016/j.devcel.2006.05.012. [DOI] [PubMed] [Google Scholar]
- 58.Prosser BL, Ward CW, Lederer WJ. X-ROS signaling: Rapid mechano-chemo transduction in heart. Science. 2011;333(6048):1440–1445. doi: 10.1126/science.1202768. [DOI] [PubMed] [Google Scholar]
- 59.Davis JQ, Bennett V. Brain ankyrin. A membrane-associated protein with binding sites for spectrin, tubulin, and the cytoplasmic domain of the erythrocyte anion channel. J Biol Chem. 1984;259(21):13550–13559. [PubMed] [Google Scholar]
- 60.Sharma N, et al. Use of quantitative membrane proteomics identifies a novel role of mitochondria in healing injured muscles. J Biol Chem. 2012;287(36):30455–30467. doi: 10.1074/jbc.M112.354415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Percival JM, Siegel MP, Knowels G, Marcinek DJ. Defects in mitochondrial localization and ATP synthesis in the mdx mouse model of Duchenne muscular dystrophy are not alleviated by PDE5 inhibition. Hum Mol Genet. 2013;22(1):153–167. doi: 10.1093/hmg/dds415. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.