The maritime palette is blue because light of this hue alone penetrates through the upper ocean. Longer wavelengths of visible light are absorbed by water; red light is found only within the top 5–20 m of coastal ocean, and yellow and green light are lost by ∼50 and ∼70 m, respectively (Fig. 1). Because primary production on land or sea requires sunlight, aquatic photosynthetic organisms are generally found in relatively shallow waters (1). Within this narrow surface layer, prokaryotic and eukaryotic algae with stunning ranges of size and complexity thrive. In PNAS, Rockwell et al. dissect a previously unknown aspect of the manifold ways eukaryotic algae are adapted to their environments (2).
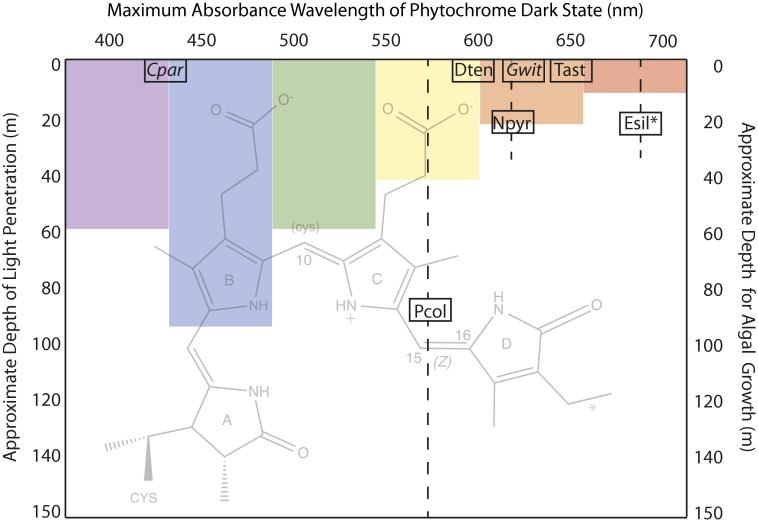
Fig. 1.
Eukaryotic algal phytochromes use phycocyanobilin (PCB) or PΦB and one- or two-cysteine photocycles to sense a wide range of the visible spectrum. The seven phytochromes described by Rockwell et al. in PNAS (2) are plotted according to the maximum absorbance wavelength of their dark state chromophore (x axis) and the approximate midpoint of the depth range (shown as a dashed line) at which they are found in either salt water (plain text) or fresh water (italics) (2). The protein is covalently attached to the A ring of the PCB chromophore (background image, with an asterisk indicating the position of a double bond in PΦB) via a thioether link and, for those examples that are underlined, via a second cysteine to C10 in the dark state only, interrupting the conjugated bond system and blue-shifting the absorbance maximum of the photoproduct. In all cases, photon absorbance causes a photoisomerization of the D ring around the C15=C16 bond. The seven phytochromes tested are encoded in the unicellular prasinophytes Tetraselmis astigmatica (Tast), Dolihomastix tenuilepis (Dten), Nephroselmis pyriformis (Npyr), and Prasinoderma coloniale (Pcol); in the filamentous multicellular brown alga Ectocarpus siliculosus (Esil); and in the fresh water microscopic Glaucophytes Cyanophora paradoxa (Cpar) and Gloeochaete wittrockiana (Gwit). As a schematic guide only, the approximate penetration depths at which 99% of light has been absorbed for various wavelengths in coastal waters is indicated (1).
Marine bacteria carry out photosynthesis using chlorophylls and antennae pigments that are adapted to the available light, whether it be red, green, or blue (3). Chromatic adaptation permits many species of cyanobacteria to adjust the ratio of antennae pigments made to match the ambient light condition (4). Proton and ion pumping rhodopsins in proteobacteria are likewise tuned to the depth of an organism’s habitat, with photocycles driven by either blue or green light (5). Similarly, eukaryotic producers tune their photosynthesis to appropriate wavelengths (6).
In addition to harvesting photons, pigmented proteins can report on the environment by responding to the quality and intensity of light. For example, the visual opsins in fish vary in concordance with habitat depth. In Lake Baikal in Siberia, the deepest waters are home to teleost fish that sense blue (484 nm) light, whereas their shallower-dwelling relatives sense green (516 nm) light most efficiently (7). A similar phenomenon has been observed in marine rock fish (8).
Rhodopsins in animals and microbes are tuned with amino acid substitutions around the 11-cis-retinal chromophore. Human red, green, and blue color vision opsins are encoded in three genes, with the red and green opsin genes in particular derived from a duplication and being closely related in sequence (9). The few tuning residues around the retinal (5) can affect the chromophore conformation, the counterion for the retinal and the chromophore environment (10). Although our understanding of these parameters is incomplete, it has been possible to both engineer blue-shifted rhodopsins based on natural proteins (11) and to design rhodopsin mimics that span absorbance maxima from 425 nm to 624 nm (12).
On land, plants rely on the highly conserved phytochromes to interpret environmental cues from light. These topologically knotted proteins use a linear tetrapyrrole chromophore covalently linked to the protein via a cysteine side chain, and are generally limited to sensing the ratio of red to far-red light (∼650 nm and 700 nm, respectively) (13). Absorbance of a photon leads to repositioning of the terminal pyrrole ring because of a flip about a double bond; the photoproduct itself is relatively stable on the time scale of hours, so phytochromes are photoswitches. Whereas plant phytochromes are exclusively integrators of red and far-red light, their cyanobacterial cousins (chloroplast progenitors) encode not only such classic phytochromes but also multiple photosignaling cyanobacteriochromes (CBCRs), unknotted members of the phytochrome superfamily whose maximum absorbance can vary from the UV to the far red (14). Rather than evolving unique chromophores, the CBCR absorbance maximum and photocycle properties again, as in the rhodopsins, use a single chromophore (Fig. 1) whose absorbance maxima and photocycle features are dictated by the surrounding amino acid pocket.
Eukaryotic algal genome sequences and metagenomic studies have revealed genes for phytochromes in many freshwater and sea-dwelling algae. Moreover, photochemical investigations of two of these revealed classic red/far-red photochemistry (15, 16), and dramatic transcription profile changes were brought on by red light in Saccharina japonica, the important food crop kelp (17). In contrast, genomes of red algae (18) and the single-celled model organism Chlamydomonas reinhardtii (19) uncovered no phytochrome genes. Several postgenome questions remained; in particular, given the staggering diversity of eukaryotic algae, would those with phytochromes have signaling limited to red and far-red light? Or might their phytochromes be active over a wider spectrum and thus have the capability of sensing depth, crowding, cloud cover, or other environmental conditions that impact light quality? In PNAS, Rockwell et al. answer this question with photochemical characterization of seven algal phytochromes, from marine and freshwater single-celled organisms and from a model filamentous brown algae related to important ocean food crops (2). Their data reveal that these phytochromes monitor an astonishing range of the visible spectrum, with dark and photoproduct states that absorb from the deep blue (434 nm) to the near infrared (734 nm) (Fig. 1). Moreover, the absorbance maxima changes from the dark state (Z configuration of bilin 15=16 bond; see Fig. 1) to the photoproduct (E configuration of 15=16 bond) include red shifts of +86 to +258 nm and blue shifts of −133 to −192 nm.
Eukaryotic phytochromes thus share the prokaryotic CBCRs’ large range of absorption maxima. In CBCRs blue-shifted dark states or reverse photocycles are caused by the covalent attachment of a second cysteine to the
Rockwell et al. dissect a previously unknown aspect of the manifold ways eukaryotic algae are adapted to their environments.
methine bridge linking pyrrole rings B and C (Fig. 1). Indeed a two-cysteine–linked ground state is known in at least one prokaryotic phytochrome (14) as well. Rockwell et al. (2) hypothesize that similar two-cysteine photocycles may explain the blue-shifted dark state in Cyanophora paradoxa phytochrome and the reverse far-red/green photocycle in Gloeochaete wittrockiana phytochrome (Fig. 1). That eukaryotic algae as a group have phytochromes whose absorbance maxima in dark and photoproduct states cover a 300-nm–wide range of the visible spectrum is remarkable not only from a basic biology perspective but also because it opens up a new set of possibilities for applications in aquaculture, bioenergy, and water quality control.
To date, there is no experimental link from photochemistry of phytochromes to photobiology in these algal species. The biggest open question is certainly what signaling pathways are directed by these phytochromes. It is tempting to draw a parallel to the rhodopsins and speculate that phytochrome photoproperties are correlated with the available light at an organism’s characteristic depth (Fig. 1). More complete cataloguing of habitat and spectral properties will be needed to determine if depth and phytochrome absorbance maxima are related. If such a correlation does exist it could be confounded by the mixing of microorganisms in the water column. Finally, it will be interesting to find out if many algal species have panvision themselves: that is, multiple phytochrome genes that describe proteins with a combined broad spectral responsiveness, as has already been observed for brown algae (20) and now for the freshwater microalgal Glaucophytes (2). The eukaryotic algae have taught us they can see in color, but the biological roles of their phytochromes remain an important and open question.
Footnotes
The author declares no conflict of interest.
See companion article on page 3871 of issue 10 in volume 111.
References
- 1.Moran MA, Miller WL. Resourceful heterotrophs make the most of light in the coastal ocean. Nat Rev Microbiol. 2007;5(10):792–800. doi: 10.1038/nrmicro1746. [DOI] [PubMed] [Google Scholar]
- 2.Rockwell NC, et al. Eukaryotic algal phytochromes span the visible spectrum. Proc Natl Acad Sci USA. 2014;111(10):3871–3876. doi: 10.1073/pnas.1401871111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Duxbury Z, Schliep M, Ritchie RJ, Larkum AWD, Chen M. Chromatic photoacclimation extends utilisable photosynthetically active radiation in the chlorophyll d-containing cyanobacterium, Acaryochloris marina. Photosynth Res. 2009;101(1):69–75. doi: 10.1007/s11120-009-9466-7. [DOI] [PubMed] [Google Scholar]
- 4.Gutu A, Kehoe DM. Emerging perspectives on the mechanisms, regulation, and distribution of light color acclimation in cyanobacteria. Mol Plant. 2012;5(1):1–13. doi: 10.1093/mp/ssr054. [DOI] [PubMed] [Google Scholar]
- 5.Man D, et al. Diversification and spectral tuning in marine proteorhodopsins. EMBO J. 2003;22(8):1725–1731. doi: 10.1093/emboj/cdg183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Haxo FT, Blinks LR. Photosynthetic action spectra of marine algae. J Gen Physiol. 1950;33(4):389–422. doi: 10.1085/jgp.33.4.389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hunt DM, Fitzgibbon J, Slobodyanyuk SJ, Bowmaker JK. Spectral tuning and molecular evolution of rod visual pigments in the species flock of cottoid fish in Lake Baikal. Vision Res. 1996;36(9):1217–1224. doi: 10.1016/0042-6989(95)00228-6. [DOI] [PubMed] [Google Scholar]
- 8.Sivasundar A, Palumbi SR. Parallel amino acid replacements in the rhodopsins of the rockfishes (Sebastes spp.) associated with shifts in habitat depth. J Evol Biol. 2010;23(6):1159–1169. doi: 10.1111/j.1420-9101.2010.01977.x. [DOI] [PubMed] [Google Scholar]
- 9.Nathans J, Thomas D, Hogness DS. Molecular genetics of human color vision: The genes encoding blue, green, and red pigments. Science. 1986;232(4747):193–202. doi: 10.1126/science.2937147. [DOI] [PubMed] [Google Scholar]
- 10.Sekharan S, Sugihara M, Buss V. Origin of spectral tuning in rhodopsin—It is not the binding pocket. Angew Chem Int Ed Engl. 2007;46(1–2):269–271. doi: 10.1002/anie.200603306. [DOI] [PubMed] [Google Scholar]
- 11.Sudo Y, et al. A blue-shifted light-driven proton pump for neural silencing. J Biol Chem. 2013;288(28):20624–20632. doi: 10.1074/jbc.M113.475533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang W, et al. Tuning the electronic absorption of protein-embedded all-trans-retinal. Science. 2012;338(6112):1340–1343. doi: 10.1126/science.1226135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Possart A, Fleck C, Hiltbrunner A. Shedding (far-red) light on phytochrome mechanisms and responses in land plants. Plant Sci. 2014;217–218:36–46. doi: 10.1016/j.plantsci.2013.11.013. [DOI] [PubMed] [Google Scholar]
- 14.Rockwell NC, Martin SS, Feoktistova K, Lagarias JC. Diverse two-cysteine photocycles in phytochromes and cyanobacteriochromes. Proc Natl Acad Sci USA. 2011;108(29):11854–11859. doi: 10.1073/pnas.1107844108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kidd DG, Lagarias JC. Phytochrome from the green alga Mesotaenium caldariorum. Purification and preliminary characterization. J Biol Chem. 1990;265(12):7029–7035. [PubMed] [Google Scholar]
- 16.Jorissen HJMM, Braslavsky SE, Wagner G, Gärtner W. Heterologous expression and characterization of recombinant phytochrome from the green alga Mougeotia scalaris. Photochem Photobiol. 2002;76(4):457–461. doi: 10.1562/0031-8655(2002)076<0457:heacor>2.0.co;2. [DOI] [PubMed] [Google Scholar]
- 17.Wang W-J, Wang F-J, Sun X-T, Liu F-L, Liang Z-R. Comparison of transcriptome under red and blue light culture of Saccharina japonica (Phaeophyceae) Planta. 2013;237(4):1123–1133. doi: 10.1007/s00425-012-1831-7. [DOI] [PubMed] [Google Scholar]
- 18.Bhattacharya D, et al. Genome of the red alga Porphyridium purpureum. Nat Commun. 2013;4:1941. doi: 10.1038/ncomms2931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Merchant SS, et al. The Chlamydomonas genome reveals the evolution of key animal and plant functions. Science. 2007;318(5848):245–250. doi: 10.1126/science.1143609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cock JM, et al. The Ectocarpus genome and the independent evolution of multicellularity in brown algae. Nature. 2010;465(7298):617–621. doi: 10.1038/nature09016. [DOI] [PubMed] [Google Scholar]