Significance
The diverse T-cell receptor (TCR) repertoire is generated by selection of T cells that have undergone TCR-gene recombination during intrathymic development. This process is precisely regulated to prevent DNA damage and to minimize the escape of self-reactive T cells. Peripheral T cells with self-reactive TCRs can be neutralized by undergoing further TCR-gene rearrangements, through a process known as TCR revision. Although a potentially useful source of new TCR specificities, revision incurs the risk of off-target DNA damage. Our work demonstrates that TCR revision occurs in the germinal center, a distinct microenvironment comprising specialized cells that engage in specific interactions. Confinement to this well-regulated environment may explain how a potentially risky process can occur safely.
Keywords: Rag-mediated recombination, T-cell tolerance
Abstract
Peripheral CD4 T cells in Vβ5 transgenic (Tg) C57BL/6J mice undergo tolerance to an endogenous superantigen encoded by mouse mammary tumor virus 8 (Mtv-8) by either deletion or T-cell receptor (TCR) revision. Revision is a process by which surface expression of the Vβ5+ TCR is down-regulated in response to Mtv-8 and recombination activating genes are expressed to drive rearrangement of the endogenous TCRβ locus, effecting cell rescue through the expression of a newly generated, non–self-reactive TCR. In an effort to identify the microenvironment in which revision takes place, we show here that the proportion of T follicular helper cells (Tfh) and production of high-affinity antibody during a primary response are increased in Vβ5 Tg mice in an Mtv-8–dependent manner. Revising T cells have a Tfh-like surface phenotype and transcription factor profile, with elevated expression of B-cell leukemia/lymphoma 6 (Bcl-6), CXC chemokine receptor 5, programmed death-1, and other Tfh-associated markers. Efficient revision requires Bcl-6 and is inhibited by B lymphocyte-induced maturation protein-1. Revision completes less efficiently in the absence of signaling lymphocytic activation molecule-associated protein although initiation proceeds normally. These data indicate that Tfh formation is required for the initiation of revision and germinal-center interactions for its completion. The germinal center is known to provide a confined space in which B-cell antigen receptors undergo selection. Our data extend the impact of this selective microenvironment into the arena of T cells, suggesting that this fluid structure also provides a regulatory environment in which TCR revision can safely take place.
During T-cell development in the thymus, conventional T cells undergo recombination-activating gene (Rag)-mediated rearrangement of the gene clusters encoding T-cell receptor (TCR) α and β chains (1). Developing T cells are subjected to sequential selection processes survived by 1–5% of cells, such that the bulk of T cells exiting the thymus express TCRs that are both useful and safe (2). Rag-mediated rearrangement can induce off-target mutations, creating a potential risk of oncogenesis, a danger that is mitigated by precise regulation of Rag expression during T-cell development (1). It is currently unclear what regulatory processes are in place to dampen the risks incurred by postthymic TCR rearrangement, or TCR revision, a process known to occur in both mice (3–11) and humans (12–15).
TCR revision has been well-studied in Vβ5 transgenic (Tg) C57BL/6J (B6) mice, in which all T cells exit the thymus with Vβ5 paired to endogenous TCRα chains (16). Vβ5+ TCRs interact with an extrathymic superantigen (superAg) encoded by mouse mammary tumor virus 8 (Mtv-8), a defective retrovirus (17, 18). Mtv-8 is very poorly expressed and only weakly stimulates T cells (17–19). Most Mtv-8–reactive Vβ5+ CD4 T cells become anergic and are deleted, leading to an age-dependent decline in the CD4:CD8 T-cell ratio in Vβ5 Tg B6 mice (16, 20). Fewer cells undergo TCR revision, in which interaction of the Vβ5+ TCR with Mtv-8 leads to down-regulation of TCR surface expression, induction of Rag and terminal deoxynucleotidyl transferase (TdT) expression, and rearrangement of endogenous TCRβ-chain genes (21, 22). The newly generated TCRβ chain is expressed on the cell surface, driving age-dependent accumulation of Vβ5−TCRβ+ CD4 T cells (20). This accumulation of postrevision T cells is prevented by deletion of Rag in peripheral T cells (23), demonstrating that revision depends on extrathymic Rag expression.
TCR revision is an effective tolerance process, as revised TCRs are no longer responsive to Mtv-8 and replicate the endogenous TCR repertoire (24, 25). Postrevision T cells respond to homeostatic signals and generate MHC-restricted antigen (Ag)-specific responses (25). Given that revision generates a functional and self-tolerant TCR, the revising T cell is likely subjected to some form of selection. Indeed, the frequency of revising T cells is increased in the absence of the proapoptotic molecule Bcl-2–interacting mediator of cell death (26), and the accumulation of postrevision T cells is enhanced in the absence of the death receptor Fas (27). These results suggest that apoptosis plays a role in the selection of the postrevision T-cell repertoire.
Formulating a rational hypothesis for the regulation of TCR revision requires an understanding of secondary Ag receptor rearrangement in generative compartments. TCR editing in the thymus and B-cell receptor (BCR) editing in the bone marrow are regulated by their confinement to specialized environments (28, 29). The potential requirement for a confined microenvironment raises the possibility that TCR revision occurs in the germinal center (GC), a site in which B cells and CD4 T cells interact, thereby driving B-cell differentiation into high-affinity antibody-secreting plasma cells or memory B cells (30). In line with this notion, TCR revision in most models excludes CD8 T cells (3) and, unlike deletion, requires B cells, inducible T-cell costimulator (ICOS), and CD28 (27). In addition, immunohistochemistry of revising T cells, identified in Rag2p-GFP Tg mice in which GFP is expressed under the control of the Rag2 promoter (22), suggests that revising T cells localize predominantly in or near splenic GCs (31).
Using these prior studies as a foundation, we hypothesized that revising T cells are follicular helper T cells (Tfh), the subset of CD4 T cells interacting with B cells in the GC (32). Because the generation of Tfh requires specific cell interactions and the specialized GC microenvironment, we investigated whether revising T cells share these features to help determine whether they are Tfh. We demonstrate here that revising T cells have a Tfh-like surface phenotype and transcription factor profile and that TCR revision is regulated by many of the same factors known to control Tfh differentiation. We now propose that revision occurs in three distinctly localized stages: first, down-regulation of Vβ5 and expression of Rag at the T cell–B cell boundary of the B-cell follicle, followed by surface expression of an endogenous TCRβ in the GC, and, finally, exit from the GC after revision is complete. Our work indicates that GCs are required for revision and suggests that GCs may provide the confined regulatory microenvironment needed to mitigate the risks inherent in extrathymic Rag expression and gene rearrangement.
Results
The Proportion of GC Tfh Increases in Vβ5 Tg Mice in an Mtv-8–Dependent Manner.
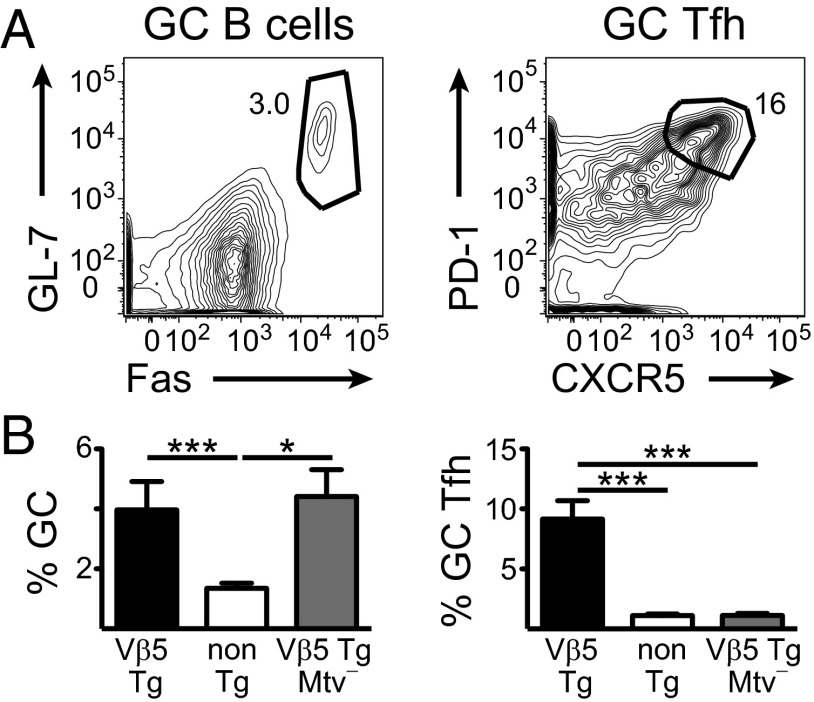
GC B-cell and Tfh populations (defined as shown in Fig. 1A) are elevated as a percent of total B or T cells in Vβ5 Tg mice compared with their TCR non-Tg littermates (Fig. 1B). Analysis of the same phenotype in Vβ5 Tg Mtv− mice indicates that the increased proportion of GC Tfh is Mtv-8–dependent, but the GC B-cell phenotype is not (Fig. 1B).
Fig. 1.
The proportion of GC Tfh increases in Vβ5 Tg mice in an Mtv-8–dependent manner. (A) Representative flow-cytometric plots show GC gating on mLN cells from a 22-wk-old Vβ5 Tg mouse. B cells are gated as CD4− and CD19+ or B220+ and T cells are gated as CD4+ and CD19− or B220−. Numbers next to gates represent the percent of cells in that gate. (B) mLN cells from Vβ5 Tg Mtv+ (black), TCR non-Tg Mtv+ (white), and Vβ5 Tg Mtv− (gray) mice ranging from 20 to 29 wk of age were analyzed for % GC of B cells and % GC Tfh of CD4 T cells. Bars indicate mean ± SEM from 4 to 12 mice per group in two to six independent experiments. *P < 0.05, ***P < 0.005.
Mtv+ Vβ5 Tg Mice Have Elevated Memory B-Cell Formation and High-Affinity Ab Production During a Primary Ab Response.
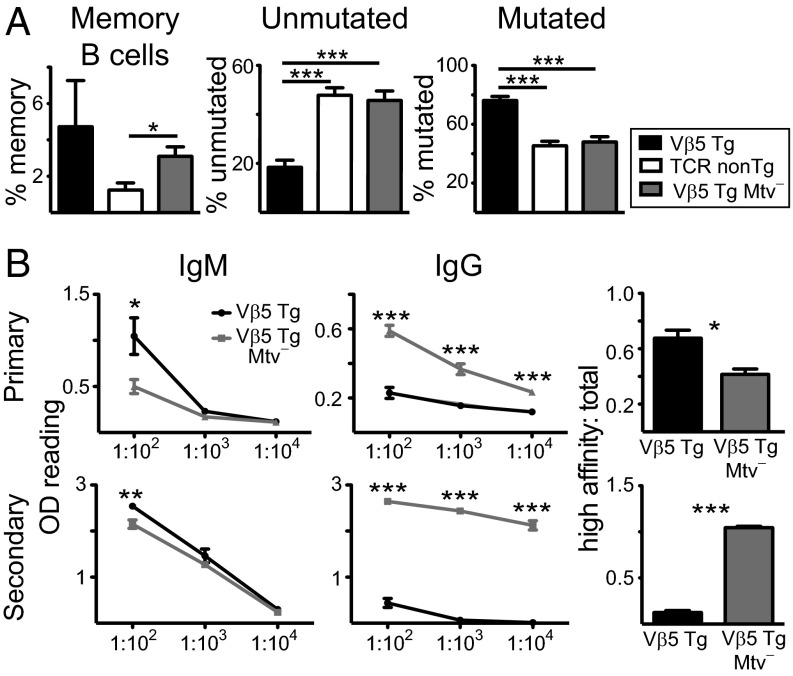
CD73+CD80+ memory B cells constitute an elevated percentage of B cells in Vβ5 Tg Mtv+ B6 mice relative to TCR non-Tg and Vβ5 Tg Mtv− mice (Fig. 2A). CD35 expression can be used to distinguish memory B cells with mutated (CD35−) and unmutated (CD35+) Ig genes (33). Memory B cells from Vβ5 Tg Mtv+ mice have a higher frequency of mutated than unmutated Ig whereas TCR non-Tg and Vβ5 Tg Mtv− mice have the reverse phenotype (Fig. 2A). Three weeks after primary immunization with 4-hydroxy-4-nitrophenyl acetyl-keyhole limpet hemocyanin (NP-KLH), Vβ5 Tg Mtv− mice produce lower levels of NP-specific IgM and higher levels of total NP-specific IgG compared with Vβ5 Tg Mtv+ mice. Interestingly, of this total NP-specific IgG, high-affinity IgG constitutes a lower proportion in Vβ5 Tg Mtv− mice (Fig. 2B). However, 7 d after secondary immunization with NP-chicken gamma globulin (CGG), in addition to higher total NP-specific IgG, Vβ5 Tg Mtv− mice produce higher levels of high-affinity IgG (Fig. 2B). Thus, the Mtv-8–dependent increase in GC Tfh in Vβ5 Tg mice is correlated with increases in Ab affinity maturation in primary but not in secondary Ab responses.
Fig. 2.
Mtv+ Vβ5 Tg mice have elevated memory B-cell formation and high-affinity Ab production during a primary Ab response. (A, Left to Right) The % memory (CD62L+MHC class II+CD73+CD80+), % memory with unmutated Ig (CD35+), and % memory with mutated Ig (CD35−) of B220+ B cells in mLN from Vβ5 Tg Mtv+ (black), TCR non-Tg Mtv+ (white), and Vβ5 Tg Mtv− (gray) mice ranging from 28 to 45 wk of age. (B, Left and Center) Titrations of NP-specific IgM and IgG Ab. (Right) Ratio of high-affinity IgG to total IgG (1:100 dilution) in sera from Vβ5 Tg Mtv+ (black) and Mtv− (gray) mice ranging from 15 to 17 wk of age 21 d after primary immunization with NP-KLH (Upper) or 7 d after secondary immunization with NP-CGG (Lower). Bars indicate mean ± SEM from three to seven mice per group in one to five independent experiments. *P < 0.05, **P < 0.01, ***P < 0.005.
Increasing the Frequency of Activated B Cells Does Not Enhance TCR Revision.
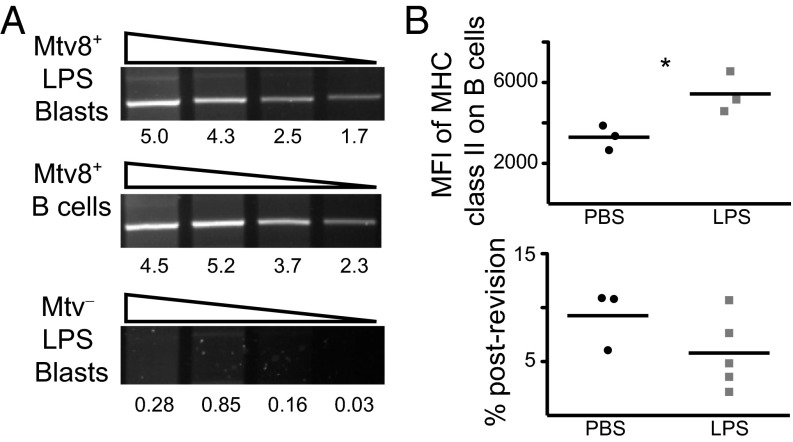
Mtv surface expression increases along with MHC class II expression on activated B cells (34). It is therefore possible that TCR revision is selectively induced by Mtvhigh activated B cells and that the apparent GC requirement for TCR revision simply reflects an activated B-cell requirement. To address this possibility, expression of RNA specific for the Mtv-8 superAg was analyzed in nonactivated splenic B cells and lipopolysaccharide (LPS)-activated blasts from Mtv-8+ and Mtv− mice. Mtv-8+ B cells and blasts express similar levels of Mtv-8 RNA (Fig. 3A). As a proxy for Mtv-8 surface expression, mice were injected with LPS to identify the effect of elevated B-cell MHC class II expression on accumulation of postrevision T cells (defined as Vβ5−TCRβ+ in Fig. S2A). Two weeks after a second injection, MHC class II expression was higher on a per-cell basis on B cells from LPS-injected than from PBS-injected mice (Fig. 3B). However, there was no difference in accumulation of postrevision T cells between LPS- and PBS-injected mice 8 wk after a third injection (Fig. 3B), indicating that B-cell activation is not sufficient to enhance TCR revision.
Fig. 3.
Increasing the frequency of activated B cells does not enhance TCR revision. (A) RNA was isolated from Mtv-8+ and Mtv− B cells and LPS blasts. Semiquantitative PCR was performed on serial threefold dilutions of cDNA for expression of Mtv-8, with hypoxanthine guanine phosphoribosyltransferase (HPRT) used as a control for quantification (Fig. S1). Numbers below gel images represent quantification of band intensity. Data are representative of three experiments. (B) Mice were injected i.p. with LPS or PBS at 3-wk intervals. (Upper) Two weeks after the second injection, median fluorescence intensity (MFI) of MHC class II was measured on CD19+ B cells from mLN. (Lower) Eight weeks after the third injection, % postrevision of CD4 T cells in mLN was measured. Data are from three to five mice, representative of two independent experiments. *P < 0.05.
Revising T Cells Have a Tfh-Like Localization, Surface Phenotype, and Transcription Factor Expression Pattern.
The correlation between Mtv-8 expression and GC function indicates that GC interactions may be required for revision, in which case, revising T cells should have a phenotype similar to that of Tfh. Revising T cells were identified as GFP+Vβ5+ peripheral CD4 T cells in thymectomized (Tx) Rag2p-GFP Tg Vβ5 Tg mice, cells that have been shown to express Rag (22), and postrevision T cells were identified as GFP−Vβ5−TCRβ+ peripheral CD4 T cells (Fig. S2A). CCR7 is a chemokine receptor expressed by cells in the T-cell zone of secondary lymphoid organs whereas CXCR5 is expressed by cells that migrate into the B-cell follicle, the site of GCs (reviewed in ref. 35). Intermediate levels of these receptors suggest localization at the T cell–B cell boundary (Fig. S2B). Although naive CD4 T cells are primarily CCR7+, revising and postrevision T cells express a range of CCR7 and CXCR5 levels (Fig. S2C). Therefore, revising T cells likely localize to the T cell–B cell boundary of follicles.
Revising T cells display a Tfh phenotype (Fig. 4A) for the markers programmed death-1 (PD-1), B- and T-lymphocyte attenuator (BTLA), ICOS, OX40, and IL-6 receptor (IL-6R) α (32, 36). Postrevision T cells also express high levels of PD-1 and ICOS (Fig. 4A). Expression of IL-6Rα on naive T cells (Fig. 4A) is consistent with previous data (37). Production of the cytokine IL-4 by Tfh promotes Ab class switch (32). Revising T cells express IL-4 RNA at a higher level than naive T cells, but at a lower level than Th2-skewed T cells (Fig. 4B). Tfh differentiation requires the transcriptional repressor B-cell leukemia/lymphoma 6 (Bcl-6) and is inhibited by B lymphocyte-induced maturation protein-1 (Blimp-1), its mutually antagonistic repressor (38). Revising T cells and Tfh have similar high Bcl-6 and low Blimp-1 RNA expression patterns (38) whereas postrevision T cells have lower Bcl-6 and higher Blimp-1 expression (Fig. 4B). In line with this RNA expression, revising T cells express Bcl-6 protein (Fig. 4B). Thus, revising T cells have a phenotype resembling that of Tfh.
Fig. 4.
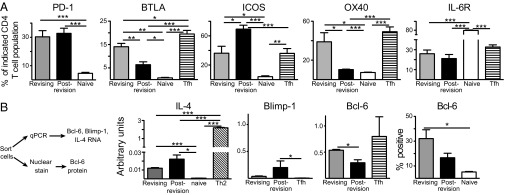
Revising T cells have a Tfh-like phenotype. (A) Shown are % revising (gray), postrevision (black), naive (white), or Tfh (striped) CD4 T cells expressing the indicated Tfh-associated markers. Naive CD4 T cells were defined as CD44lo Vβ5+ CD4 T cells from 5- to 8-wk-old Vβ5 Tg mice. Tfh control was GC Tfh from mLN of Vβ5 Tg mice 7 d after NP-CGG immunization i.p. (B, Left) Experimental scheme. (Center) IL-4, Blimp-1, and Bcl-6 RNA levels were measured using qPCR. The 2−ΔCT values were calculated for triplicate samples and normalized to HPRT. Tfh control is OT-II T cells ∼7 d after NP-ovalbumin immunization in vivo. (Right) The graph depicts protein expression of Bcl-6. Naive control (white) was splenic CD44lo CD4 T cells from TCR non-Tg mice. Bars indicate mean ± SEM from pools of two to nine mice per group in two to four independent experiments. *P < 0.05, **P < 0.01, ***P < 0.005.
Bcl-6 Is Required for Efficient Revision Whereas Blimp-1 Inhibits It.
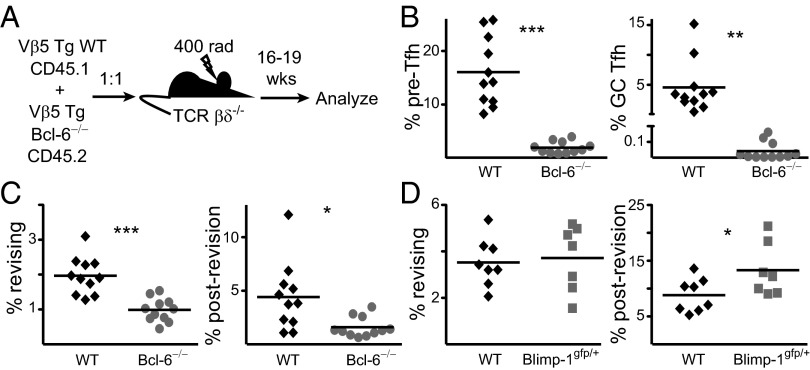
Given the antagonistic relationship between Bcl-6 and Blimp-1 and the impact of these transcription factors on Tfh formation, revision was studied in the absence of both proteins. Because Bcl-6−/− B6 embryos die late in gestation (39), recipients were reconstituted with a mixture of congenically marked Vβ5 Tg WT and Bcl-6−/− fetal liver (Fig. 5A). The kinetics of revision are impacted by high levels of irradiation (27) so chimeras were made in sublethally irradiated T cell-deficient (TCR βδ−/−) hosts. To evaluate the effect of Bcl-6 deficiency on Tfh formation, two stages of Tfh differentiation were analyzed: pre-Tfh (PD-1intCXCR5int) localize to the T cell–B cell boundary, and more differentiated GC Tfh (PD-1+CXCR5+) are found in the B-cell follicle (32). In mixed fetal-liver chimeras, both populations are dramatically reduced in the absence of Bcl-6 (Fig. 5B).
Fig. 5.
Bcl-6 and Blimp-1 inversely regulate revision. (A) Vβ5 Tg WT (black diamonds) and Bcl-6−/− (gray circles) CD4 T cells from mLN of mixed fetal-liver chimeras were analyzed 16–19 wk postreconstitution for (B) % pre-Tfh and GC Tfh and (C) % revising and postrevision T cells. Data are representative of two independent groups of chimeras, totaling 11 mice. (D) mLN CD4 T cells from 15- to 24-wk-old Vβ5 Tg WT (black diamonds) and Blimp-1gfp/+ (gray squares) mice were analyzed for % revising and postrevision T cells. Data are from seven to eight mice per group in five independent experiments. *P < 0.05, **P < 0.01, ***P < 0.005.
To analyze revision in the absence of Bcl-6, we took advantage of the fact that Vβ5loTCRβ+ cells are revision intermediates that become postrevision T cells in adoptive hosts (26). Thus, we analyzed Vβ5loTCRβ+ revising cells and Vβ5−TCRβ+ postrevision T cells, which are subpopulations confined to CD4 T cells from Mtv+ mice (Fig. S3). Both the revising and postrevision T-cell populations are decreased in the absence of Bcl-6 (Fig. 5C), indicating that efficient initiation and completion of revision require this transcription factor.
The effect of Blimp-1 on revision was analyzed in Vβ5 Tg Blimp-1gfp/+ mice (40). Revising T cells are not affected, but there is an increase in postrevision T cells in Blimp-1gfp/+ mice (Fig. 5D). Consistent with the congruence between Tfh and revising T cells, Bcl-6 and Blimp-1 regulate TCR revision antagonistically.
SAP Is Required for Efficient Completion, but Not Initiation, of Revision.
SAP is an intracellular adaptor protein that interacts with SLAM receptors to stabilize interactions between B and T cells in the GC (41). Mice deficient in SAP have reduced Tfh formation and a severely diminished GC Tfh compartment (42). Analysis of Vβ5 Tg WT and SAP−/− mice confirms the reduction in both Tfh populations (Fig. 6A). The postrevision T-cell population is diminished in the absence of SAP (Fig. 6B), indicating that the completion of revision requires long-lived interactions between B cells and T cells.
Fig. 6.
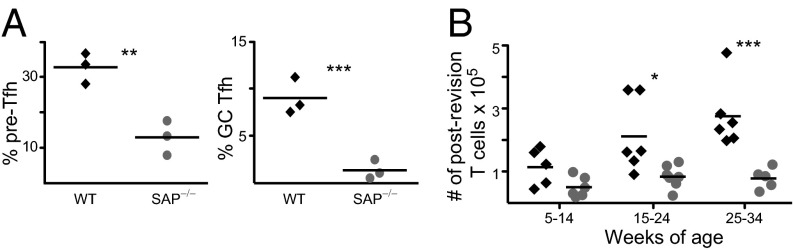
SAP is required for efficient completion of revision. (A) mLN CD4 T cells from 20- to 29-wk-old Vβ5 Tg WT (black diamonds) and SAP−/− (gray circles) mice were analyzed for % pre-Tfh and GC Tfh. (B) mLN CD4 T cells from 5- to 34-wk-old Vβ5 Tg WT and SAP−/− mice were analyzed for total number of postrevision T cells. Data are from three to seven mice per group in five to nine independent experiments. *P < 0.05, **P < 0.01, ***P < 0.005.
Irradiated recipients were reconstituted with congenically marked Vβ5 Tg WT and SAP−/− bone marrow (Fig. S4A). Vβ5+ CD4 T cells become activated (CD44hi) by exposure to Mtv-8 and can then be deleted or undergo revision (Fig. S4B). The proportion of Vβ5+ CD4 T cells that are CD44hi is increased in the absence of SAP (Fig. S4 B and C), and the revising T-cell population is not affected (Fig. S4D), indicating that initiation of revision proceeds normally in the absence of SAP.
Discussion
This work demonstrates that revising T cells congregate at the T cell–B cell boundary of B-cell follicles, have a Tfh-like phenotype, and can function as Tfh. Efficient revision requires Bcl-6 and SAP but is inhibited by Blimp-1. These data localize TCR revision to the GC, a microenvironment in which this tolerance process can be regulated, preventing DNA damage and promoting the generation of cells expressing TCRs that are both useful and safe.
TCR revision is not the only example of Ag receptor modification after initial receptor gene rearrangement. There is evidence of secondary TCRα rearrangement or TCR editing in the thymus after encounter with a cortical Ag recognized by the TCR (28). Immature B cells undergo a similar BCR editing process, confined to the bone marrow (29). The BCR evolves further through somatic hypermutation, which also creates some risk of genomic instability, and is precisely regulated in the GC through activation-induced deaminase (AID) expression (reviewed in ref. 43). These processes are examples of Ag receptor alteration that can reduce autoreactivity or fine-tune a response. The potential dangers of Ag-receptor manipulation can be diminished by limiting the expression of Rag, TdT, and AID to a controlled microenvironment. Similarly, the GC localization and Bcl-6 and SAP requirements of TCR revision suggest that revision is induced by GC-specific B cell–T cell interactions, providing a means for regulating Rag expression and imposing selection for the newly expressed TCR.
An alternate explanation for the GC requirements would suggest that TCR revision is induced selectively by activated B cells, given that surface expression of Mtvs is increased upon LPS stimulation, along with MHC class II expression (34). However, our data indicate that activating B cells to up-regulate Mtv-8 surface expression does not enhance TCR revision, demonstrating that the GC localization we observe is not simply the result of a requirement for activated B cells. Our results showing no effect of LPS stimulation on Mtv-8 superAg RNA expression are surprising in light of evidence that LPS stimulation enhances expression of Mtv-8 envelope RNA (44). However, superAg and envelope are encoded by different portions of the Mtv-8 locus and are differentially regulated (19, 45).
When taken together, the GC and Mtv-8 requirements for TCR revision could suggest that revision is induced only by B cell-expressed antigens. However, although superAg-induced revision models predominate, revision can be driven by conventional self Ag as well (3, 6, 7, 11). The demonstration that TCR revision can be induced by conventional Ag indicates that this process is limited neither to Ag expressed by B cells nor to superAg presented outside the peptide-binding groove of MHC class II (46).
Consistent with the Tfh-like phenotype of revising T cells, the enhanced proportion of Tfh in Vβ5 Tg mice is Mtv-8–dependent. The Mtv-8 independence of the enriched GC B-cell population was surprising. However, the memory B-cell phenotype and high-affinity antibody levels indicate that Mtv-8 does influence GC function. The combination of enhanced primary response and diminished secondary response in Vβ5 Tg Mtv+ mice suggests that the T cells activated in the primary response either are deleted or undergo revision and consequently no longer provide Ag-specific help for a secondary Ab response. Vβ5 Tg Mtv− mice undergo normal secondary responses, presumably because their CD4 T cells neither delete nor revise. Our data on IL-4 and IL-6R expression by revising T cells also suggest that these cells function as Tfh. GC Tfh express IL-6R, and IL-6 produced by Ag-presenting cells contributes to Bcl-6 expression and differentiation and maintenance of Tfh (32, 35). IL-4 production by GC Tfh promotes Ab class-switch, GC B-cell survival, and GC maintenance (32, 36). Overall, these data indicate that revising T cells are functional Tfh that promote affinity maturation, plasma-cell formation, and GC maintenance.
The requirement of Bcl-6 for both initiation and completion of revision and SAP only for completion is consistent with previous research on the differentiation of Tfh. Bcl-6 expression is up-regulated during the initial stages of Tfh differentiation (47) whereas SAP is required only for GC Tfh formation (42). Analysis of revision in SAP−/− mice is complicated by the fact that Mtv-8–dependent deletion of CD4 T cells (16) is reduced in the absence of SAP (Fig. S4C). The elevated CD44 expression by SAP−/− Vβ5+ CD4 T cells indicates that Mtv-8–induced T-cell activation is not affected, but the deletion defect complicates direct comparison of the proportion of WT and SAP−/− postrevision T cells in the same mouse. However, our data do suggest that Bcl-6+ pre-Tfh initiate revision and that the transition from revising to postrevision T cell requires sustained SAP-mediated interactions in the GC.
Although revising T cells have a distinctive Tfh phenotype, cells that complete revision become functional memory T cells. They undergo lymphopenia-induced proliferation and respond to microbial Ag in chronically lymphopenic mice (25). In addition, they recognize Ag in a self-MHC restricted manner and become effectors after pathogen challenge (25). However, postrevision T cells are unusual in that they are skewed to a Th17 phenotype, do not become regulatory T cells (48), and have elevated steady-state proliferation compared with other memory T cells (25). Our data are consistent with the memory phenotype of postrevision T cells and show that they have a phenotype distinct from that of revising T cells. In particular, chemokine-receptor expression suggests that they exit the GC and traffic to other tissues, consistent with previous findings (16, 25).
The existence of postrevision T cells in SAP−/− mice raises the question of whether cells that revise in the absence of SAP have the same phenotype as their WT counterparts. SAP−/− postrevision T cells express reduced CCR7 and elevated CXCR5 levels (Fig. S5A), indicating that they are likely to remain in the B-cell follicle. However, the chemokine receptor phenotype (Fig. S5A) and Bcl-6 and Blimp-1 RNA levels (Fig. S5B) of SAP−/− revising T cells mirror those of their WT counterparts. These data indicate that revising T cells acquire a pre-Tfh phenotype and localize normally in the absence of SAP, but that homing of cells after revision is affected. GC defects in the absence of SAP are T cell-intrinsic (49), and SAP contributes to signaling within T cells (50), raising the possibility that this homing phenotype may result from changes in T-cell signaling in the absence of SAP.
TCR revision has been depicted as a two-step process. First, interaction of TCRβ with Mtv-8 causes down-regulation of the TCR and up-regulation of Rag expression. Then, transcription and translation of the new endogenous TCRβ leads to surface expression of the TCR and transition into a postrevision T cell (51). This model is consistent with in vitro data (52) and studies on TCR editing (28) showing that surface TCR down-regulation promotes Rag expression. Our current data indicate that the first step occurs at the T cell–B cell boundary of the B-cell follicle and is Bcl-6–dependent but SAP-independent (Fig. S6A). The second step, occurring in the B-cell follicle, is dependent on both Bcl-6 and SAP and inhibited by Blimp-1 (Fig. S6B). The SAP−/− Tfh phenotype data suggest that exit from the B-cell follicle is a separate step usually completed by postrevision T cells (Fig. S6C). Overall, these results indicate that revision is a three-step process affecting cells with a Tfh-like phenotype, with each stage occurring in a distinct microenvironment, identifying the GC as the site of Ag receptor modification for T cells as well as B cells. Overall, results from our laboratory and others demonstrate that these processes have evolved to occur in confined microenvironments to preserve the potential benefits while minimizing the risks associated with Ag receptor modification.
Materials and Methods
Mice.
WT B6 (B6 CD45.2+), B6.SJL-PtprcaPepcb/BoyJ CD45.1+ (B6 CD45.1+), and B6.129P2-Tcrbtm1MomTcrdtm1Mom/J (TCR βδ−/−) mice were purchased from The Jackson Laboratory or bred under specific pathogen-free conditions at the University of Washington. Vβ5 Tg and OVA-specific OT-II Tg B6 mice were bred in-house and maintained as heterozygotes. Rag2p-GFP Tg mice (53) have been backcrossed at least 12 generations on the B6 background in our laboratory. Mtv-8+ and Mtv− mice were derived by intercross/backcross breeding of Vβ5 Tg mice to a male WLC-0 mouse provided by D. Morris (University of California, Irvine, CA). WLC-0 mice are wild-derived and Mtv− whereas B6 mice express Mtvs -8, -9, -17, and -30 (16). Bcl-6+/− B6 (54) mice were provided by A. Dent (Indiana University, Indianapolis, IN). Blimp-1gfp/+ B6 mice, in which a knock-in allele expresses GFP and nonfunctional Blimp-1 protein (40), were provided by P. Greenberg (University of Washington, Seattle, WA). SAP−/− B6 (41) mice were provided by P. Stein (Northwestern University, Chicago, IL). Tx were performed as described previously (31) and verified upon euthanizing by staining for CD4 and CD8 expression in cells from any tissue remaining in the thymic region. All experiments were conducted in accordance with the University of Washington Institutional Animal Care and Use Committee.
Mixed Radiation Chimeras.
Bone marrow was isolated from donors, dispersed into a single-cell suspension, and T cell-depleted by incubation with Ab to Thy1.2 (13.4.6), CD8 (3.168.6), and CD4 (RL172K) followed by Low-Tox-M rabbit complement (CedarLane). For fetal-liver chimeras, Vβ5 Tg CD45.1+ Bcl-6+/+ control embryos and those from Vβ5 Tg CD45.2+ Bcl-6+/− intercross matings were collected on approximately embryonic day 16. Livers were teased into single-cell suspension and resuspended in HBSS. Tail DNA was isolated using the Gentra Puregene Tissue Kit (Qiagen), and Vβ5 and Bcl-6 genotypes were identified by PCR (SI Materials and Methods, PCR Protocols and Primers for Vβ5 and Bcl-6 Genotyping). For both bone-marrow and fetal-liver chimeras, Vβ5 Tg WT and null samples were mixed at the indicated ratios and resuspended in HBSS. Then, 5 × 106 cells were injected into the lateral tail vein of sublethally irradiated (400 rad) CD45.2+ TCR βδ−/− B6 mice.
Cell Preparation, Flow Cytometry, and Sorting.
Single-cell suspensions were prepared from spleen and peripheral (axial, brachial, inguinal) and mesenteric (mLN) lymph nodes. RBCs were removed from spleens by water lysis. For flow cytometry, Fc receptors were blocked with anti-CD16/32 (2.4G2; BD Pharmingen). Cells were surface-stained in HBSS containing 1% BSA using fluorochrome-conjugated or biotinylated Abs, all purchased from BD Biosciences, BioLegend, or eBioscience. Abs were specific for mouse CD4 (RM4-5), CD8α (53-6.7), CD19 (1D3, MB19.1), CD35 (8C12), CD44 (IM7), CD45R/B220 (RA3-6B2), CD45.1 (A20), CD45.2 (104), CD62L (MEL-14), CD73 (TY/11.8), CD80 (16-10A1), CD95 (Jo2), GL-7 (GL7), PD-1 (J43), CCR7 (4B12), CXCR5 (2G8), IL-6Rα (D7715A7), BTLA (8F4), ICOS (7E.17G9), MHC class II (M5/115.15.2), OX40 (OX-86), panTCRβ (H57-597.13), and Vβ5 (MR9-4). Staining with biotinylated Abs was followed by FITC-conjugated (eBioscience), allophycocyanin-conjugated (eBioscience), or Brilliant Violet 421-conjugated (BioLegend) streptavidin. Abs recognizing Vβ5 and TCRβ do not cross-block. For CXCR5 and CCR7 only, cells were stained for 30 min at 37 °C. For detection of Bcl-6, surface-stained cells were stained with Alexa Fluor 647-conjugated anti–Bcl-6 (K112-91) using the Foxp3 Staining Buffer Set and protocol (eBioscience). Alexa Fluor 647-conjugated IgG1κ (BD Biosciences) was used as an isotype control. Flow cytometry data were collected on a FACSCanto or LSRII (Becton Dickinson) and analyzed using FlowJo software (Tree Star). For cell sorting, enriched CD4 T cells were obtained using an EasySep Negative Selection Mouse CD4+ T-cell Enrichment Kit (Stem Cell Technologies), surface stained, and sorted using a FACSAria II (Becton Dickinson). Th2-differentiated CD4 T cells were prepared as described previously (55).
Immunizations and Ab Quantification.
Vβ5 Tg Mtv+ B6 and Mtv− mice were immunized i.p. with 100 μg of NP-KLH or NP-CGG (Biosearch Technologies) in a 1:1 solution with Imject Alum (Thermo Scientific). Secondary Ab responses were induced by immunization with 20 μg of NP-CGG 21 d later. B6 recipients of 1–3 × 106 OT-II T cells were immunized with 100 μg of NP-ovalbumin in alum at the base of the tail, and responding OT-II T cells sorted from the draining inguinal nodes 6.5–7.5 d later. Then, 96-well ELISA plates (Thermo Scientific) were coated overnight with NP(4)-BSA and NP(23)-BSA (Biosearch Technologies) and blocked overnight with PBS/1% BSA. Sera from day 21 post primary immunization or day 7 post secondary immunization were added to plates in triplicate at dilutions of 1:100, 1:1,000, and 1:10,000. Anti-NP Abs were detected with goat anti-mouse IgG and IgM (Southern Biotech) conjugated to HRP. TMB substrate reagent (Becton Dickinson) was used for detection, and plates were read at OD450 and OD570 on an iMark microplate reader (Bio-Rad).
LPS Blast Formation.
Vβ5 Tg mice were injected i.p. with 50 μg of LPS (Calbiochem) or 200 μL of PBS. Injection was repeated 3 and 6 wk later. For in vitro blast formation, B cells were enriched from Mtv-8+ and Mtv− splenocytes using an EasySep Negative Selection Mouse B-cell Isolation Kit (Stem Cell Technologies) and cultured for 3 d in complete RPMI medium 1640 containing 10% (vol/vol) FBS, 10 mM Hepes, 4 mM l-Gln, and 50 μM 2-mercaptoethanol at 37 °C in the presence of 50 μg/mL LPS.
Quantitative PCR and Semiquantitative PCR.
Total RNA was extracted from purified cells using the RNeasy Micro kit (Qiagen), and first-strand cDNA synthesis was performed with oligo(dT) primers using the SuperScript III Reverse Transcriptase kit and protocol (Invitrogen). Quantitative PCR (qPCR) was performed using an ABI 7300 Real Time PCR System (Applied Biosystems) and Power SYBR Green PCR Master Mix (Applied Biosystems) using the primers and settings indicated (SI Materials and Methods, Quantitative PCR Protocol and Primers). Semiquantitative PCR was performed on serial threefold dilutions of cDNA starting with 150 ng cDNA using the primers and settings described (SI Materials and Methods, Semiquantitative PCR Protocol and Primers).
Statistics.
P values were calculated using an unpaired or paired (mixed chimeras in Fig. 5 and Fig. S4 only) Student t test.
Supplementary Material
Acknowledgments
We thank Drs. A. Dent, P. Greenberg, and P. Stein for mice. This work was supported by National Institutes of Health Grants T32CA009537 and T32GM007270 (to L.E.H.) and R01 AG13078 and R21 AI097950 (to P.J.F.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1321803111/-/DCSupplemental.
References
- 1.Schlissel MS. Regulating antigen-receptor gene assembly. Nat Rev Immunol. 2003;3(11):890–899. doi: 10.1038/nri1225. [DOI] [PubMed] [Google Scholar]
- 2.Starr TK, Jameson SC, Hogquist KA. Positive and negative selection of T cells. Annu Rev Immunol. 2003;21:139–176. doi: 10.1146/annurev.immunol.21.120601.141107. [DOI] [PubMed] [Google Scholar]
- 3.Hale JS, Fink PJ. T-cell receptor revision: Friend or foe? Immunology. 2010;129(4):467–473. doi: 10.1111/j.1365-2567.2010.03250.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wagner DH., Jr Re-shaping the T cell repertoire: TCR editing and TCR revision for good and for bad. Clin Immunol. 2007;123(1):1–6. doi: 10.1016/j.clim.2006.08.006. [DOI] [PubMed] [Google Scholar]
- 5.Dash P, et al. Paired analysis of TCRα and TCRβ chains at the single-cell level in mice. J Clin Invest. 2011;121(1):288–295. doi: 10.1172/JCI44752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Takase M, Kanagawa EM, Kanagawa O. Age-dependent TCR revision mediated by interaction between alphabeta TCR and self-antigens. J Immunol. 2007;179(4):2163–2169. doi: 10.4049/jimmunol.179.4.2163. [DOI] [PubMed] [Google Scholar]
- 7.Huang CY, Golub R, Wu GE, Kanagawa O. Superantigen-induced TCR α locus secondary rearrangement: Role in tolerance induction. J Immunol. 2002;168(7):3259–3265. doi: 10.4049/jimmunol.168.7.3259. [DOI] [PubMed] [Google Scholar]
- 8.Serra P, et al. RAG-dependent peripheral T cell receptor diversification in CD8+ T lymphocytes. Proc Natl Acad Sci USA. 2002;99(24):15566–15571. doi: 10.1073/pnas.242321099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jones SC, et al. Impact of post-thymic cellular longevity on the development of age-associated CD4+ T cell defects. J Immunol. 2008;180(7):4465–4475. doi: 10.4049/jimmunol.180.7.4465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vaitaitis GM, Poulin M, Sanderson RJ, Haskins K, Wagner DH., Jr Cutting edge: CD40-induced expression of recombination activating gene (RAG) 1 and RAG2: a mechanism for the generation of autoaggressive T cells in the periphery. J Immunol. 2003;170(7):3455–3459. doi: 10.4049/jimmunol.170.7.3455. [DOI] [PubMed] [Google Scholar]
- 11.Bynoe MS, Viret C, Flavell RA, Janeway CA., Jr T cells from epicutaneously immunized mice are prone to T cell receptor revision. Proc Natl Acad Sci USA. 2005;102(8):2898–2903. doi: 10.1073/pnas.0409880102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li TT, Han S, Cubbage M, Zheng B. Continued expression of recombination-activating genes and TCR gene recombination in human peripheral T cells. Eur J Immunol. 2002;32(10):2792–2799. doi: 10.1002/1521-4141(2002010)32:10<2792::AID-IMMU2792>3.0.CO;2-I. [DOI] [PubMed] [Google Scholar]
- 13.Lantelme E, et al. Cutting edge: Recombinase-activating gene expression and V(D)J recombination in CD4+CD3low mature T lymphocytes. J Immunol. 2000;164(7):3455–3459. doi: 10.4049/jimmunol.164.7.3455. [DOI] [PubMed] [Google Scholar]
- 14.Lantelme E, et al. Increased frequency of RAG-expressing, CD4(+)CD3(low) peripheral T lymphocytes in patients with defective responses to DNA damage. Eur J Immunol. 2000;30(5):1520–1525. doi: 10.1002/(SICI)1521-4141(200005)30:5<1520::AID-IMMU1520>3.0.CO;2-D. [DOI] [PubMed] [Google Scholar]
- 15.Lantelme E, et al. Analysis of secondary V(D)J rearrangements in mature, peripheral T cells of ataxia-telangiectasia heterozygotes. Lab Invest. 2003;83(10):1467–1475. doi: 10.1097/01.lab.0000092228.51605.6a. [DOI] [PubMed] [Google Scholar]
- 16.Blish CA, et al. Chronic modulation of the TCR repertoire in the lymphoid periphery. J Immunol. 1999;162(6):3131–3140. [PubMed] [Google Scholar]
- 17.Günzburg WH, Hynes NE, Groner B. The methylation pattern of endogenous mouse mammary tumor virus proviral genes is tissue specific and stably inherited. Virology. 1984;138(2):212–224. doi: 10.1016/0042-6822(84)90346-5. [DOI] [PubMed] [Google Scholar]
- 18.Scherer MT, Ignatowicz L, Pullen A, Kappler J, Marrack P. The use of mammary tumor virus (Mtv)-negative and single-Mtv mice to evaluate the effects of endogenous viral superantigens on the T cell repertoire. J Exp Med. 1995;182(5):1493–1504. doi: 10.1084/jem.182.5.1493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Waanders GA, Lees RK, Held W, MacDonald HR. Quantitation of endogenous mouse mammary tumor virus superantigen expression by lymphocyte subsets. Eur J Immunol. 1995;25(9):2632–2637. doi: 10.1002/eji.1830250934. [DOI] [PubMed] [Google Scholar]
- 20.Fink PJ, Swan K, Turk G, Moore MW, Carbone FR. Both intrathymic and peripheral selection modulate the differential expression of Vβ5 among CD4+ and CD8+ T cells. J Exp Med. 1992;176(6):1733–1738. doi: 10.1084/jem.176.6.1733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McMahan CJ, Fink PJ. RAG reexpression and DNA recombination at T cell receptor loci in peripheral CD4+ T cells. Immunity. 1998;9(5):637–647. doi: 10.1016/s1074-7613(00)80661-5. [DOI] [PubMed] [Google Scholar]
- 22.Cooper CJ, Orr MT, McMahan CJ, Fink PJ. T cell receptor revision does not solely target recent thymic emigrants. J Immunol. 2003;171(1):226–233. doi: 10.4049/jimmunol.171.1.226. [DOI] [PubMed] [Google Scholar]
- 23.Hale JS, Ames KT, Boursalian TE, Fink PJ. Cutting Edge: Rag deletion in peripheral T cells blocks TCR revision. J Immunol. 2010;184(11):5964–5968. doi: 10.4049/jimmunol.1000876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McMahan CJ, Fink PJ. Receptor revision in peripheral T cells creates a diverse Vβ repertoire. J Immunol. 2000;165(12):6902–6907. doi: 10.4049/jimmunol.165.12.6902. [DOI] [PubMed] [Google Scholar]
- 25.Hale JS, Wubeshet M, Fink PJ. TCR revision generates functional CD4+ T cells. J Immunol. 2010;185(11):6528–6534. doi: 10.4049/jimmunol.1002696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hale JS, Nelson LT, Simmons KB, Fink PJ. Bcl-2-interacting mediator of cell death influences autoantigen-driven deletion and TCR revision. J Immunol. 2011;186(2):799–806. doi: 10.4049/jimmunol.1002933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ali M, Weinreich M, Balcaitis S, Cooper CJ, Fink PJ. Differential regulation of peripheral CD4+ T cell tolerance induced by deletion and TCR revision. J Immunol. 2003;171(11):6290–6296. doi: 10.4049/jimmunol.171.11.6290. [DOI] [PubMed] [Google Scholar]
- 28.McGargill MA, Derbinski JM, Hogquist KA. Receptor editing in developing T cells. Nat Immunol. 2000;1(4):336–341. doi: 10.1038/79790. [DOI] [PubMed] [Google Scholar]
- 29.Sandel PC, Monroe JG. Negative selection of immature B cells by receptor editing or deletion is determined by site of antigen encounter. Immunity. 1999;10(3):289–299. doi: 10.1016/s1074-7613(00)80029-1. [DOI] [PubMed] [Google Scholar]
- 30.Allen CDC, Okada T, Cyster JG. Germinal-center organization and cellular dynamics. Immunity. 2007;27(2):190–202. doi: 10.1016/j.immuni.2007.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cooper CJ, Turk GL, Sun M, Farr AG, Fink PJ. Cutting edge: TCR revision occurs in germinal centers. J Immunol. 2004;173(11):6532–6536. doi: 10.4049/jimmunol.173.11.6532. [DOI] [PubMed] [Google Scholar]
- 32.Crotty S. Follicular helper CD4 T cells (TFH) Annu Rev Immunol. 2011;29:621–663. doi: 10.1146/annurev-immunol-031210-101400. [DOI] [PubMed] [Google Scholar]
- 33.Anderson SM, Tomayko MM, Ahuja A, Haberman AM, Shlomchik MJ. New markers for murine memory B cells that define mutated and unmutated subsets. J Exp Med. 2007;204(9):2103–2114. doi: 10.1084/jem.20062571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.King LB, Lund FE, White DA, Sharma S, Corley RB. Molecular events in B lymphocyte differentiation: Inducible expression of the endogenous mouse mammary tumor proviral gene, Mtv-9. J Immunol. 1990;144(8):3218–3227. [PubMed] [Google Scholar]
- 35.Vinuesa CG, Cyster JG. How T cells earn the follicular rite of passage. Immunity. 2011;35(5):671–680. doi: 10.1016/j.immuni.2011.11.001. [DOI] [PubMed] [Google Scholar]
- 36.King C, Tangye SG, Mackay CR. T follicular helper (TFH) cells in normal and dysregulated immune responses. Annu Rev Immunol. 2008;26:741–766. doi: 10.1146/annurev.immunol.26.021607.090344. [DOI] [PubMed] [Google Scholar]
- 37.Jones GW, et al. Loss of CD4+ T cell IL-6R expression during inflammation underlines a role for IL-6 trans signaling in the local maintenance of Th17 cells. J Immunol. 2010;184(4):2130–2139. doi: 10.4049/jimmunol.0901528. [DOI] [PubMed] [Google Scholar]
- 38.Johnston RJ, et al. Bcl6 and Blimp-1 are reciprocal and antagonistic regulators of T follicular helper cell differentiation. Science. 2009;325(5943):1006–1010. doi: 10.1126/science.1175870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yu D, et al. The transcriptional repressor Bcl-6 directs T follicular helper cell lineage commitment. Immunity. 2009;31(3):457–468. doi: 10.1016/j.immuni.2009.07.002. [DOI] [PubMed] [Google Scholar]
- 40.Kallies A, et al. Plasma cell ontogeny defined by quantitative changes in Blimp-1 expression. J Exp Med. 2004;200(8):967–977. doi: 10.1084/jem.20040973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Qi H, Cannons JL, Klauschen F, Schwartzberg PL, Germain RN. SAP-controlled T-B cell interactions underlie germinal centre formation. Nature. 2008;455(7214):764–769. doi: 10.1038/nature07345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yusuf I, et al. Germinal center T follicular helper cell IL-4 production is dependent on signaling lymphocytic activation molecule receptor (CD150) J Immunol. 2010;185(1):190–202. doi: 10.4049/jimmunol.0903505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Keim C, Kazadi D, Rothschild G, Basu U. Regulation of AID, the B-cell genome mutator. Genes Dev. 2013;27(1):1–17. doi: 10.1101/gad.200014.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lund FE, Corley RB. Regulated expression of mouse mammary tumor proviral genes in cells of the B lineage. J Exp Med. 1991;174(6):1439–1450. doi: 10.1084/jem.174.6.1439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Günzburg WH, et al. Endogenous superantigen expression controlled by a novel promoter in the MMTV long terminal repeat. Nature. 1993;364(6433):154–158. doi: 10.1038/364154a0. [DOI] [PubMed] [Google Scholar]
- 46.Zamoyska R. Superantigens: Supersignalers? Sci STKE. 2006;2006(358):pe45. doi: 10.1126/stke.3582006pe45. [DOI] [PubMed] [Google Scholar]
- 47.Baumjohann D, Okada T, Ansel KM. Cutting Edge: Distinct waves of BCL6 expression during T follicular helper cell development. J Immunol. 2011;187(5):2089–2092. doi: 10.4049/jimmunol.1101393. [DOI] [PubMed] [Google Scholar]
- 48.Zehn D, Bevan MJ, Fink PJ. Cutting edge: TCR revision affects predominantly Foxp3− cells and skews them toward the Th17 lineage. J Immunol. 2007;179(9):5653–5657. doi: 10.4049/jimmunol.179.9.5653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Veillette A, et al. SAP expression in T cells, not in B cells, is required for humoral immunity. Proc Natl Acad Sci USA. 2008;105(4):1273–1278. doi: 10.1073/pnas.0710698105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schwartzberg PL, Mueller KL, Qi H, Cannons JL. SLAM receptors and SAP influence lymphocyte interactions, development and function. Nat Rev Immunol. 2009;9(1):39–46. doi: 10.1038/nri2456. [DOI] [PubMed] [Google Scholar]
- 51.Simmons KB, et al. Modulation of TCRβ surface expression during TCR revision. Cell Immunol. 2012;272(2):124–129. doi: 10.1016/j.cellimm.2011.10.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Roose JP, et al. T cell receptor-independent basal signaling via Erk and Abl kinases suppresses RAG gene expression. PLoS Biol. 2003;1(2):E53. doi: 10.1371/journal.pbio.0000053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yu W, et al. Continued RAG expression in late stages of B cell development and no apparent re-induction after immunization. Nature. 1999;400(6745):682–687. doi: 10.1038/23287. [DOI] [PubMed] [Google Scholar]
- 54.Dent AL, Shaffer AL, Yu X, Allman D, Staudt LM. Control of inflammation, cytokine expression, and germinal center formation by BCL-6. Science. 1997;276(5312):589–592. doi: 10.1126/science.276.5312.589. [DOI] [PubMed] [Google Scholar]
- 55.Hendricks DW, Fink PJ. Recent thymic emigrants are biased against the T-helper type 1 and toward the T-helper type 2 effector lineage. Blood. 2011;117(4):1239–1249. doi: 10.1182/blood-2010-07-299263. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.