Significance
We present a novel vaccine that elicits protective immune responses against many different influenza viruses belonging to both group 1 and 2 lineages. The vaccine uses a live vaccinia virus that expresses multiple H5N1 influenza viral proteins and the cytokine IL-15 to stimulate the immune system. The vaccine was able to induce T-cell immune responses that recognize different influenza viruses and these immune responses were augmented when exposed to a challenge virus, resulting in protection against a lethal disease. Vaccine-induced CD4+ T cells that coordinate immune responses were found to be more important than CD8+ T cells in conferring protection. Our vaccine provides a promising strategy for universal protection against novel and emerging influenza viruses.
Keywords: universal vaccine, cell-mediated immunity
Abstract
Current influenza vaccines are ineffective against novel viruses and the source or the strain of the next outbreak of influenza is unpredictable; therefore, establishing universal immunity by vaccination to limit the impact of influenza remains a high priority. To meet this challenge, a novel vaccine has been developed using the immunogenic live vaccinia virus as a vaccine vector, expressing multiple H5N1 viral proteins (HA, NA, M1, M2, and NP) together with IL-15 as a molecular adjuvant. Previously, this vaccine demonstrated robust sterile cross-clade protection in mice against H5 influenza viruses, and herein its use has been extended to mediate heterosubtypic immunity toward viruses from both group 1 and 2 HA lineages. The vaccine protected mice against lethal challenge by increasing survival and significantly reducing lung viral loads against the most recent human H7N9, seasonal H3N2, pandemic-2009 H1N1, and highly pathogenic H7N7 influenza A viruses. Influenza-specific antibodies elicited by the vaccine failed to neutralize heterologous viruses and were unable to confer protection by passive transfer. Importantly, heterologous influenza-specific CD4+ and CD8+ T-cell responses that were elicited by the vaccine were effectively recalled and amplified following viral challenge in the lungs and periphery. Selective depletion of T-cell subsets in the immunized mice revealed an important role for CD4+ T cells in heterosubtypic protection, despite low sequence conservation among known MHC-II restricted epitopes across different influenza viruses. This study illustrates the potential utility of our multivalent Wyeth/IL-15/5Flu as a universal influenza vaccine with a correlate of protective immunity that is independent of neutralizing antibodies.
Influenza causes widespread infection during seasonal epidemics and occasional worldwide pandemics despite available vaccines. The subtype of future outbreaks or pandemic influenza strains is unpredictable as is its source, evident from the most recent H7N9 outbreak from poultry in China, the variant H3N2 outbreak from swine in the United States in 2012, the H1N1 worldwide pandemic of 2009, and the highly pathogenic avian influenza (HPAI) H5N1 in 1997 from domestic poultry. Therefore, the development of a successful universal vaccination strategy is urgently needed.
Universal protection requires heterosubtypic immunity (HSI), whereby vaccination against one influenza virus cross-protects against novel and emerging strains that could potentially be mediated by multiple adaptive immune mechanisms. T cells are potent mediators of HSI, because these cells typically recognize peptide epitopes derived from internal proteins of influenza virus, which are naturally more conserved than surface HA and NA across different strains and even serologically distinct viral subtypes. Because of sequence conservation of the majority of T-cell epitopes between different influenza viruses, cross-reactive T-cell responses have been detected in healthy seronegative individuals against H5N1 and pandemic H1N1-2009 viruses (1, 2).
Previously, we generated a multivalent vaccinia virus-based H5N1 influenza vaccine, which demonstrated effective cross-clade immunity against lethal H5N1 challenges. This vaccine expresses five H5N1-derived influenza proteins (HA, NA, M1, M2, and NP), in combination with the immune stimulatory cytokine IL-15 (3) that increases long-term memory responses, along with enhanced T-cell, B-cell, and NK cell functions, including cytokine production and survival. Despite the use of live vaccine vectors being generally constrained in immune-compromised individuals, the vaccine vector (Wyeth/IL-15) has been proven safe and effective in primates and immune-deficient mice (4).
A cell-culture–derived, live vaccine vector that encodes full-length influenza proteins with inherent capacity to access MHC-I and II processing pathways to establish robust influenza-specific T-cell responses is an excellent approach to overcome issues related to population-wide MHC polymorphism and egg-based production methods for HPAI vaccines. Herein, we extend the use of Wyeth/IL-15/5Flu vaccine against multiple human influenza viruses of different HA subtypes, including the highly pathogenic H7N7, pandemic H1N1-2009, seasonal H3N2, and the most recent human H7N9 viruses. The vaccine proved effective against all of the heterologous strains tested, and the immunological mechanisms of protection were investigated to decipher correlates of immunity.
Results
Vaccinia-Based H5N1 Vaccine Wyeth/IL-15/5Flu Protects Against Group 1 and 2 Influenza Viruses.
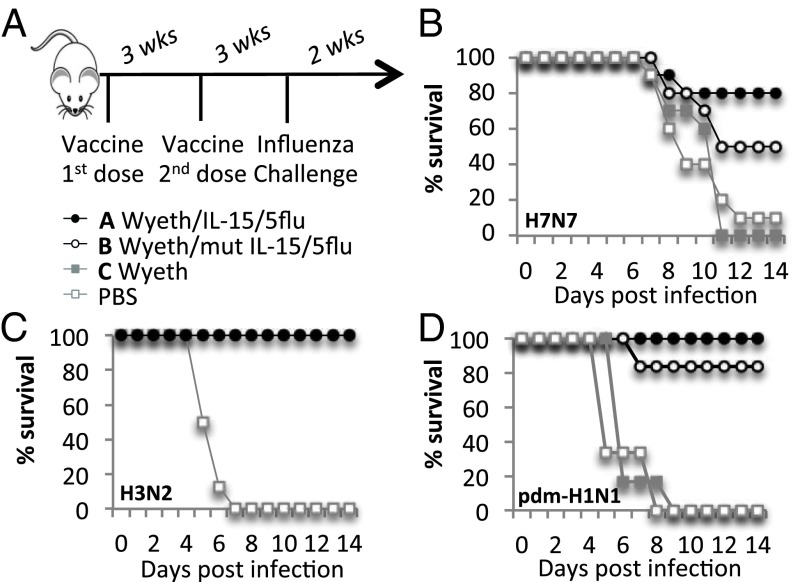
Group 1 (H1N1) and group 2 (H3N2, HPAI H7N7, H7N9) human influenza viruses were used for viral challenges to probe the H5N1-based vaccine for heterosubtypic protection (Fig. 1 and Fig. S1). The multivalent vaccinia virus-based H5N1 influenza vaccine (Wyeth/IL-15/5Flu) enhanced recovery from lethal heterosubtypic influenza infections by improving survival (Fig. 1 B–D), viral clearance (Fig. S1 A–D) (>1 log for H7N7 and H7N9, and total clearance for H1N1 and H3N2 in vaccinated mice), clinical morbidity, and by minimizing weight loss (Fig. S1 E–G). The addition of the molecular adjuvant (IL-15) enhanced survival from 50% (Wyeth/mutIL-15/5Flu) to 80% (Wyeth/IL-15/5Flu) against a lethal HPAI H7N7 challenge (Fig. 1B). The addition of IL-15 in the vaccine also increased survival against H3N2 and H1N1 challenges, and appeared to reduce the duration and severity of symptoms seen with an H1N1 challenge (Fig. S1 E–G).
Fig. 1.
Vaccinia-H5N1 vaccine protects against group 1 and group 2 viruses. (A) BALB/c mice were vaccinated subcutaneously twice, 3 wk apart with 107 pfu of indicated vaccines (Wyeth/IL-15/5Flu, Wyeth/mut-IL-15/5Flu, wild-type Wyeth, or PBS). Mice were challenged 3 wk later with influenza viruses, (B) H7N7 (10 LD50, 1 × 103TCID50), (C) H3N2 (40 LD50, 4.72 × 105 TCID50), or (D) H1N1 (40 LD50, 9.5 × 105 TCID50). Mice were monitored for survival for 14 d following infection (n = 5–11 mice). Experiments were repeated at least twice.
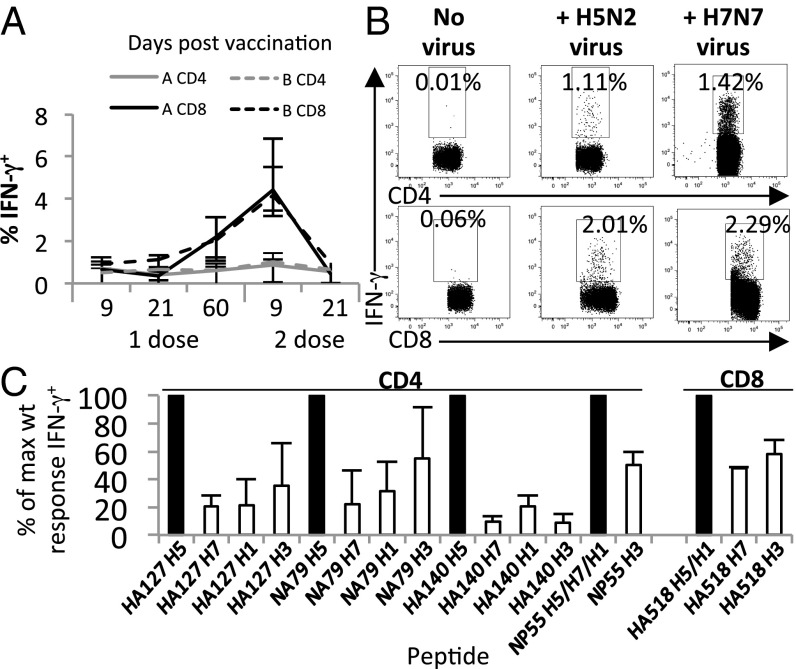
The vaccine also provided heterosubtypic protection in another mouse strain (C57BL6), with improved viral clearance at day 7 after H3N2 challenge (Fig. S2A). Furthermore, a single dose of vaccine was also effective against a lethal HPAI H7N7 challenge (Fig. S3 A and C). However, a two-dose regimen improved the durability of the vaccine-induced protective immunity (Fig. S3 C vs. D) because the time lapsed between priming and challenge did not reduce the protective efficacy with this regimen as opposed to single-dose vaccinations (3 wk vs. 16 wk) (Fig. S3 B vs. D). Therefore, a two-dose vaccination schedule was used for subsequent experiments. Furthermore, the two-dose vaccine regimen also boosted influenza-specific CD8+ T-cell responses (Fig. 2A) and H5-specific antibody titers, as reported previously (3).
Fig. 2.
Vaccine-induced T-cell responses cross-react with heterologous influenza viruses and peptides. (A) Kinetics of IFN-γ+ CD4+, and CD8+ T-cell responses elicited by the vaccine (with and without IL-15) as assessed by ex vivo H5N2 stimulation. Wyeth/IL-15/5Flu vaccinated group is denoted by “A” and Wyeth/mut-IL-15/5Flu vaccinated group is denoted by “B.” (B) Representative FACS plots for IFN-γ production following ex vivo stimulation with H5N2 or H7N7 virus at day 9 after Wyeth/IL-15/5Flu vaccination (splenocytes of BALB/c mice gated on FSC/SSC, CD4+ or CD8+). (C) T-cell responses were determined at day 9 after two doses of Wyeth/IL-15/5Flu. Splenocytes were stimulated with heterologous peptides having >1 aa difference with the cognate vaccine-peptide (Tables 2 and 3). Data shown in B and C represent mean ± SD from five mice; experiments were repeated at least twice.
Antibody Responses Are Nonneutralizing and Nonprotective.
High titer antibody responses were detected by an ELISA against the whole virus both before (day 0, equivalent to 21 d after the second dose of vaccine) and after H3N2 challenge (day 28) (Fig. S4A) (3). Having demonstrated the ability of our vaccine to induce influenza-specific binding antibodies, we next examined whether vaccine-induced antibodies were capable of neutralizing infectious virus by probing against an array of viral subtypes. No neutralizing activity was detected against H1, H3, H5, H7, or H9 viruses with day 0 serum (Fig. S4B). However, neutralizing activity was detectable in the sera 28 d after H3N2 challenge but was limited to homologous H3N2 virus, and was comparable between the vaccine and control groups.
A yeast surface-expression system displaying a library of H3-derived HA fragments (H3-HA-YSD) was used to screen immune sera (Fig. S4 C and D). No H3-HA-YSD binding activity was detected in prechallenged sera from the vaccinated mice. However, the postchallenged sera collected 28 d from both vaccinated and PBS groups showed similar binding patterns against the H3 hemagglutinin (10-230 aa major, 350–550 aa minor epitope). Therefore, the H5N1-vaccine did not appreciably modulate the antibody profiles against heterologous hemagglutinins.
High doses of immune sera have previously been proven effective for heterologous protection mediated by NP-antibodies (5, 6). In multiple experiments, the passive transfer of sera from vaccinated mice extending from –3 d to +1 d (0.5–2 mL) postchallenge showed no protection against either H3N2 or H7N7 challenges, resulting in 100% mortality of recipient mice (Fig. S5).
Amino Acid Sequence Conservation Among Vaccine Antigens and Challenge Viruses.
T-cell–mediated HSI is often dependent on the sequence conservation of the immunogenic peptides (1, 7). Among the challenge viruses used in the present study, surface HA and NA proteins had a conservation rate of 38–87% (Table 1), whereas the internal polypeptides NP, M1, and M2 displayed a conservation rate of 87–99% (Table 1). The majority of known MHC-I–restricted peptides are derived from the NP protein (three of four peptides), whereas the majority known MHC-II–restricted peptides are derived from the surface HA and NA proteins (five of six peptides) (Tables 2 and 3) (8, 9). It is important to note the existence of a greater level of sequence conservation for known MHC-I peptides (Table 2).
Table 1.
Amino acid conservation among vaccine-expressed H5N1 antigens and heterologous influenza viruses used in challenge studies
| HA | NA | NP | M1 | M2 | |
| H1N1 | 62 | 87 | 94 | 96 | 92 |
| H7N7 | 38 | 40 | 99 | 96 | 94 |
| H7N9 | 38 | 41 | 99 | 93 | 88 |
| H3N2 | 38 | 37 | 92 | 95 | 87 |
Protein sequences were aligned using CLUSTALW to identify amino acid homology. Conservation among the entire polypeptides is shown.
Table 2.
Amino acid conservation among vaccine-expressed H5N1 antigens and heterologous influenza viruses used in challenge studies: H2d CD8 peptides
| NP39–47 | NP147–155 | NP218–226 | HA518–526 | % Conservation | |
| H5N1 | FYIQMCTEL | TYQRTRALV | AYERMCNIL | IYSTVASSL | |
| H1N1 | ––––––––– | ––––––––– | ––––––––– | ––––––––– | 100 ± 0 |
| H7N7 | ––––––––– | ––––––––– | ––––––––– | WF––G––CF | 86 ± 28 |
| H7N9 | ––––––––– | ––––––––– | ––––––––– | WF–FG––CF | 83 ± 33 |
| H3N2 | ––––––––– | ––––––––– | ––––––––– | LR–L––––G | 86 ± 28 |
Protein sequences were aligned using CLUSTALW to identify amino acid homology. An en-dash (–) denotes a conserved residue between vaccine encoded epitope and the corresponding epitope of heterologous virus polypeptide. Known BALB/c MHC-I epitopes.
Table 3.
Amino acid conservation among vaccine-expressed H5N1 antigens and heterologous influenza viruses used in challenge studies: H2-IAb CD4 peptides
| HA127–137 | HA177–199 | HA140–154 | NA79–93 | NA191–201 | NP55–69 | % Conservation | |
| H5N1 | HFEKIQIIPKS | TIKRSYNNTNQE | KSSFFRNVVWLIKKN | INGWAVYSKDNSIR | LKYNGIITDTI | RLIQNSITIERMVLS | |
| H1N1 | S––RFEIF–KT | KLSK––I–DKGK | AK––YK–LI––V––E | –––––––––––––– | ––––––––––– | ––––––––––––––– | 67 ± 38 |
| H7N7 | GID–ETMGFTY | QMTK––K––RKD | G–––YAEMK––LSNT | VE––V–IA–––AV– | VY––RRL–T–– | ––––––––––––––– | 45 ± 30 |
| H7N9 | GID–EAMGFTY | QMTK––K––RKS | G–––YAEMK––LSNT | ––S–H––G–––A–– | VW––RRPVAE– | ––––––––––––––– | 46 ± 34 |
| H3N2 | SSGTLEF–TEG | VLNVTMP––D–F | G–G––SRLN––T–SG | –T–F–PF––––––– | FI–D–RLV–S– | ––––––L–––––––– | 46 ± 31 |
Protein sequences were aligned using CLUSTALW to identify amino acid homology. An en-dash (–) denotes a conserved residue between vaccine encoded epitope and the corresponding epitope of heterologous virus polypeptide. Known BALB/c MHC-II epitopes.
Vaccination Establishes Heterologous T-Cell Responses.
Vaccination-induced, influenza-specific T-cell responses were detectable from day 9 by intracellular cytokine staining [ICS; using low pathogenic avian influenza (LPAI) H5N2 with high homology to vaccine H5N1 proteins for stimulation] and remained stable up to 60 d postvaccination, with little contraction of influenza-specific T-cell responses (Fig. 2A). After two doses of the vaccine, influenza-specific CD8+ T-cell responses were significantly boosted. Importantly, in mice vaccinated with Wyeth/IL-15/5Flu that expresses H5N1 antigens, H7N7 virus elicited robust IFN-γ production in both CD4+ and CD8+ T-cell subsets 9 d postvaccination, validating the potential of our vaccine to induce cross-reactive T-cell responses against heterologous influenza viruses (Fig. 2B).
The highest influenza-specific IFN-γ+ CD8+ T-cell response was directed toward the HA518 peptide and the highest IFN-γ+ CD4+ T-cell response was directed toward the NP55 peptide. Low-level IL-4 production (independent of IFN-γ) was detected for CD4+ T-cell responses, which was above no peptide controls yet far below IFN-γ production (IL-4: 0.05%, IFN-γ: 0.8%). Therefore, the vaccine maintains a TH1-type cytokine bias required for effective anti-influenza immunity.
The cross reactivity of the vaccine-induced T-cell responses was further evaluated with heterologous peptides (Tables 2 and 3) by ICS assay (Fig. 2C). The low levels of IFN-γ production relative to the wild-type vaccine-derived peptides paralleled the low sequence homology of known CD4+ epitopes (Tables 1–3) (8–55% of maximum wild-type–specific IFN-γ+). The variable HA518 CD8+ T-cell peptide showed modest cross-recognition of ∼50% of the wild-type response for the H7- and H3-derived peptides.
Comparisons were also made for the role of IL-15 in the vaccine (Wyeth/IL-15/5Flu vs. Wyeth/mutIL-15/5Flu) for influenza and peptide-specific CD4+ and CD8+ T-cell responses (Fig. 2A). Although there were no significant differences in the magnitude of vaccine responses (Fig. 2A), an advantage for the inclusion of IL-15 was clearly apparent after viral challenge (Fig. 1 and Fig. S1).
Robust Recall of T-Cell Responses After Virus Challenge.
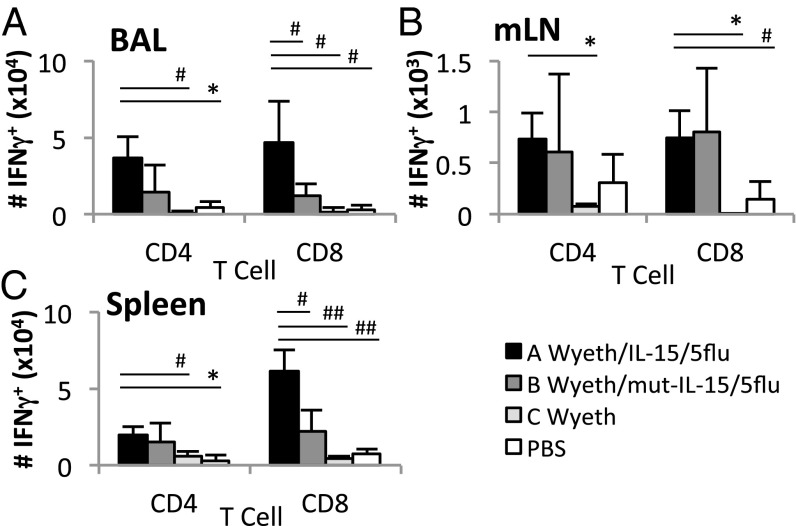
Infection of vaccinated mice with multiple strains of influenza viruses showed increased T-cell responses in vaccinated mice (Fig. 3 and Fig. S6). After H3N2 challenge, total influenza-specific (using H5N2 for stimulation) IFN-γ+ CD4+, and CD8+ T-cell responses were significantly higher (P < 0.05) than unvaccinated controls at the site of infection (bronchoalveolar lavage, BAL), draining lymph nodes (mediastinal lymph nodes, mLN), and periphery (spleen) (Fig. 3). Furthermore, immunization with the IL-15–expressing vaccine significantly increased T-cell responses, especially the CD8+ T-cell responses (P < 0.05) (Fig. 3). Similarly, increased T-cell responses were observed in vaccinated mice after H7N9 infection [activated and proliferating (Fig. S6A), H1N1 infection by peptide ICS (Fig. S6 B–E), and H3N2 infection of C57BL6 mice (Fig. S2)].
Fig. 3.
Vaccination recalls influenza virus-specific CD4+ and CD8+ T-cell responses after H3N2 challenge. Groups of BALB/c mice were vaccinated (two doses) with the respective vaccine agents or PBS and challenged with 40 LD50 H3N2 virus 2 wk later. At day 7 postchallenge, influenza-specific CD4+ and CD8+ T-cell responses were measured by IFN-γ ICS assay after ex vivo stimulation with H5N2 virus. T-cell responses in BAL (A), mLN (B), and spleen (C) are shown. Cells were gated on FSC/SSC, CD4+ or CD8+, IFN-γ+. Data represent mean ± SD; n = 3–5; #P < 0.05, *P < 0.01, ##P < 0.005, **P < 0.001; experiments were repeated at least twice.
The quality of T-cell responses was determined by polyfunctional cytokine production (10). IFN-γ+ CD4+ T-cell responses (Fig. S7 B and C) but not IFN-γ+ CD8+ T cells (Fig. S7 D and E) had improved polyfunctional cytokine production of TNF-α and IL-2, despite CD8+ T-cell responses being quantitatively greater. Additionally, vaccination induced higher TH2-type IL-4 production in IFN-γ− CD4+ T cells after H3N2 challenge in the spleen upon coculture with NP55 peptide (Fig. S7A).
Heterologous Protection Requires Influenza-Specific CD4+ T Cells.
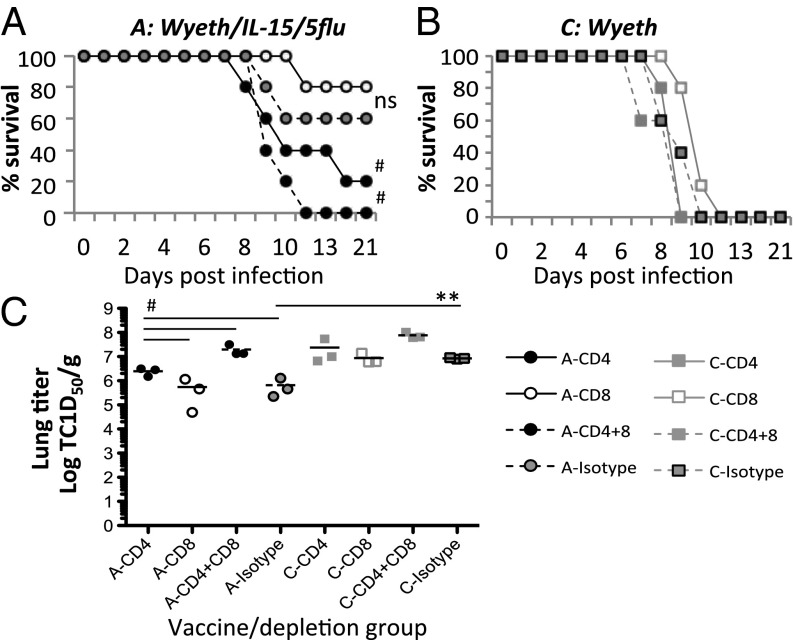
Previous studies using T-cell–depletion experiments have established that both influenza-specific CD4+ T-cell responses and CD8+, especially the memory CD8+ T-cell responses, are important in the protection against heterologous influenza virus infections in mice (ref. 11 and references therein). Therefore, to gain insights as to how Wyeth/IL-15/5Flu confers protection in the absence of neutralizing antibodies, vaccinated mice (Wyeth/IL-15/5Flu or Wyeth) were depleted of their CD4+, CD8+, or both CD4+ and CD8+ T cells, respectively, before challenging with a lethal HPAI H7N7 virus (Fig. 4 A and B).
Fig. 4.
Vaccine-induced heterosubtypic protection is mediated by influenza-specific CD4+ T-cell responses. BALB/c mice were vaccinated with Wyeth/IL-15/5Flu (A) or control Wyeth vaccine (B), twice 3 wk apart, and then treated with respective monoclonal antibodies to deplete total CD4+, CD8+, combination of both CD4+and CD8+ T cells, or treated with an isotype-matched control antibody at days −4, −2, 0, and +3, and challenged with 100 LD50 HPAI H7N7 virus. Mice were monitored for survival (A and B) for 21 d after HPAI H7N7 challenge (n = 5). Wyeth/IL-15/5Flu groups depleted of CD4+ or both CD4+ and CD8+ T cells were compared with isotype controls by Gehan-Breslow-Wilcoxon test. In C, lung viral loads were measured by a standard TCID50 assay on Madin-Darby canine kidney cells, at day 7 postchallenge; (n = 3); #P < 0.05, **P < 0.001.
As shown in Fig. 4, all mice vaccinated with the control vaccine (Wyeth) succumbed to lethal H7N7 infection by day 12 postinfection, whereas Wyeth/IL-15/5Flu–vaccinated mice displayed significant protection (P < 0.01). Surprisingly, the depletion of CD8+ T cells in these vaccinated mice had no impact on survival from the lethal HPAI H7N7 challenge, with comparable survival rates relative to isotype control mice (80% vs. 60%) (Fig. 4A). Importantly, the depletion of CD4+ T cells in vaccinated mice reduced overall survival to 20% (Fig. 4A). Furthermore, the combined depletion of both CD4+and CD8+ T cells was uniformly lethal by day 12 with 100% mortality (P < 0.05).
Viral titers in the lungs at day 7 postchallenge further delineated survival data (Fig. 4C). The depletion of CD8+ T cells in Wyeth/IL-15/5Flu–vaccinated mice did not alter lung viral load, yielding similar titers compared with isotype control-treated mice. However, the depletion of CD4+ resulted in a fourfold higher viral titer than isotype controls (P < 0.05), underscoring the critical importance of CD4+ T cells for heterologous immunity. Additionally, the combined depletion of both CD4+ and CD8+ T cells in vaccinated mice resulted in a viral titer that was eight times higher than what was seen in CD4+-depleted mice (P < 0.05), suggesting that the combined effect of CD4+ and CD8+ T-cell responses is needed for optimal viral clearance.
Innate and Adaptive Immune Cell Populations Are Skewed by Vaccination.
The profiles of innate and adaptive immune cells were compared in the lungs after H1N1 challenge (Fig. S8). At day 7 after H1N1 challenge, the lungs were dominated by B cells, CD4+, and CD8+ T cells, which tended to be higher in the vaccinated mice (Fig. S8A). Conversely, unvaccinated mice had a higher level of total neutrophils in the lungs (P < 0.05), which is likely because of more prolific inflammation associated with higher viral loads in these mice at day 7.
Immune cell types that are likely to be under the control of CD4+ helper functions were also investigated. There was a significantly higher proportion of germinal center B cells in the mLN in vaccinated mice relative to the controls (P < 0.05) (Fig. S9A). However, no differences were seen with respect to TFH cells between the groups. The influx of alternatively activated macrophages, which are typically associated with inflammation and tissue repair (12), along with TH2-type cytokine profiles, was also assessed at day 7 postinfection in the infected lungs (Fig. S9B). There were significantly higher numbers of alternatively activated macrophages in the control Wyeth vaccinated mice (P < 0.05, Wyeth/IL-15/5Flu vs. Wyeth), which may be attributable to heightened inflammation and delayed viral clearance in the control mice.
Discussion
Influenza is a moving target for vaccine development. In this study, broad protection against influenza challenge viruses was achieved using a live vaccine vector, with the vaccinia virus as a backbone incorporating multiple antigenic influenza proteins. Immunization with the live multivalent-influenza vaccine resulted in increased survival, reduced weight loss, reduced symptoms and duration of illness, and accelerated viral clearance. The Wyeth/IL-15/5Flu vaccine induced heterologous influenza-specific CD4+ and CD8+ T-cell immunity, which produced antiviral cytokines, following the stimulation with LPAI H5N2 and H7N7 viruses. Vaccine-induced CD4+ and CD8+ T-cell responses also partially recognized peptide variants derived from H3, H1, and H7 heterologous viruses. Vaccination resulted in significantly larger magnitudes of CD4+ and CD8+ T-cell responses upon influenza challenge in the spleen, lungs, and draining lymph nodes. After influenza challenge, the vaccinated mice also had increased germinal center B cells and, alternatively activated macrophages in the infected lungs while numbers of neutrophils were much reduced.
The importance of memory CD4+ T cells was evident from T-cell–depletion experiments of vaccinated mice before a lethal HPAI H7N7 challenge. Vaccinated mice depleted of CD4+ T cells displayed a dramatic reduction in protection (∼20% survival) from the HPAI H7N7 challenge and higher viral loads at day 7 postchallenge, unlike the CD8+ T-cell–depleted mice or isotype controls. However, the observation that depletion of both CD4+ and CD8+ T cells results in zero survivals implies still a probable minor role for CD8+ T cells in the vaccine-mediated protection. Thus, a unique feature of this vaccine regimen is the dependence on CD4+ T-cell immunity, which was unexpected by the comparison of sequence conservation of known MHC-II–restricted peptides from the vaccine and challenge influenza viruses. Mice depleted of CD4+ T cells may succumb to infection because of dysregulated cytokine production and inflammation that is rampant during lethal influenza challenges (13).
The importance of memory CD4+ T cells in the protection against heterosubtypic influenza A viral infections in mice has been well documented (refs. 14 and 15, and references therein). A prevailing view is that these influenza-specific memory CD4+ T cells confer protection through multiple mechanisms that involve inflammation, provision of helper functions to CD8+ T and B cells, IFN-γ production, as well as direct perforin-dependent cytotoxic activity toward influenza-infected cells. Concordant with these observations in mice, influenza virus challenge studies in healthy human volunteers have revealed that higher levels of existing influenza-specific CD4+ T cells correlate with lower virus shedding and disease protection in the absence of detectable neutralizing antibodies (16). The CD4+ T-cell dominance in conferring heterosubtypic protection in Wyeth/IL-15/5Flu–vaccinated mice further adds to the evolving perspective that CD4+ T cells are central to the HSI against influenza. However, whether the mechanistic basis of CD4+ T-cell–mediated heterosubtypic protection in Wyeth/IL-15/5Flu–vaccinated mice is dependent on direct effector activity of these vaccine-induced influenza-specific CD4+ T cells or through the involvement of other immune effector elements remains to be elucidated. The antibody response induced by vaccination was nonneutralizing against multiple heterologous influenza viruses, and the passive transfer of high amounts of vaccine immune sera was unable to mediate protection in this study. Nonneutralizing anti-influenza antibodies can induce antibody-dependent cellular cytotoxicity in cell types, such as NK cells (17), that is FcRγ-dependent and requires memory influenza-specific CD8+ T-cell responses (18). However, this does not appear to be a dominant mechanism of protection here with our Wyeth/IL-15/5Flu vaccine. Furthermore, the H3-HA-YSD epitope mapping studies indicate that our vaccine does not appear to expand the breadth of humoral responses or mediate protection by the induction of HA2 bnAbs, with no difference in the binding profiles between vaccinated and control mice. Thus, the primary mechanism of protection is not likely to be antibody mediated, but rather dependent upon established heterologous T-cell responses.
Vaccinia as a vaccine vector has been a research focus for many years because of its ability as a large DNA virus to incorporate multiple proteins, along with its inherent potent immunogenicity and limited pathogenicity. Previously, a modified vaccinia virus Ankara (MVA) recombinant virus that expresses the H5 hemagglutinin has been assessed for its potential as an H5N1-specific vaccine in various mouse models (reviewed in ref 19) and the protection conferred by this MVA-HA recombinant was dependent on the H5 clade being homologous or related. Breewoo et al. (20) recently reported the use of an MVA encoding the NP from an H5N1 and HA protein from pandemic H1N1. The MVA-HA/NP vaccine showed heterosubtypic protection against H1N1 and H5N1 challenges (both group 1 viruses), and was only partially protective against a H3N2 challenge (group 2 virus). Furthermore, an MVA-NP/M1 vaccine (21) is currently being evaluated in phase II clinical trials (22). The MVA-NP/M1 has proven effective in human challenge studies by reducing the duration of viral shedding and disease severity, and was also effective in adults >65 y old (22). Thus, the use of vaccinia as a vaccine vector for inducing robust anti-influenza immunity is already underway. Our vaccine strategy represents further improvements to the current efforts, as Wyeth is more immunogenic than MVA, and the integrated IL-15 reduces the residual virulence of the Wyeth strain (4). The Wyeth/IL-15/5Flu vaccine is multivalent, thereby broadening the immune responses to both surface and antigenically conserved internal influenza proteins, and is readily produced in cell culture with the added benefit of a vaccine that can be lyophilized and stockpiled to be rapidly deployed should the necessity arise with a global influenza pandemic.
Materials and Methods
Vaccinia Vaccine: Wyeth/IL-15/5Flu.
The vaccine construct Wyeth/IL-15/5Flu has been described in detail by Poon et al. (3). Briefly, the virus expresses the NP, HA, and NA proteins derived from A/Vietnam/1203/2004, the matrix proteins M1 and M2 from A/CK/Indonesia/PA/2003, and human IL-15 cytokine. A derivative with disrupted IL-15 (Wyeth/mut-IL-15/5Flu), wild-type Wyeth vaccinia and PBS were also used in mouse vaccination experiments.
Influenza Virus Challenge of Vaccinated Mice.
Female BALB/c (H-2d) or C57BL/6J (B6, H-2b) mice (6–8 wk of age) were primed twice 3 wk apart via the subcutaneous route with 107 pfu of vaccine agent in 100 μL PBS and challenged with influenza virus 3 wk later. For influenza challenge, mice were anesthetized and infected intranasally with 25 μL of either H7N9 (A/Shanghai/2/2013, 1 × 105TCID50, nonlethal) (23), HPAI H7N7 (A/Netherlands/219/2003, 1 × 103-4, 10–100 LD50), mouse-adapted H3N2 (A/Hong Kong/1/68-MA20C, 4.72 × 105TCID50, 40 LD50; a gift from Earl G. Brown, University of Ottawa, Ottawa, ON, Canada), or pandemic H1N1 (A/California/04/2009, 1.36 × 106TCID50, 40 LD50). All experiments involving H7N9 and HPAI H7N7 viruses were conducted in a biosafety level 3 laboratory. All animal studies were approved by the Committee on the Use of Live Animals in Teaching and Research , Hong Kong University.
Isolation of Mouse Tissues.
Blood was collected in MiniCollect tubes (Greiner BioOne) and sera harvested by centrifugation. To evaluate cellular immunity, cells were sampled from the site of infection by BAL, lungs, mLN, and spleen. Spleens were disrupted and red blood cells removed by RBC lysis buffer (BioLegend). mLN were homogenized using a sieve and syringe plunger. The protein concentration in supernatants of BAL was measured using Pierce BCA Protein Assay kit (Thermo Scientific).
Lungs were chopped and digested with collagenase II (1,000 U) and DNase I (250 U; Worthington) in DMEM [with 10% (vol/vol) FBS, 1% penstrep] for 60 min at 37 °C. Digested lungs were homogenized and red blood cells were lysed.
Alternatively, lungs were mechanically homogenized (Qiagen) in 1 mL of MEM (with 10 μg/mL TPCK trypsin), clarified by centrifugation at maximum speed, and titered by TCID50 assay (24).
ICS of T-Cell Responses.
For the peptide ICS assay, 0.5–1 × 106 cells in MEM (10% FBS, 1% penstrep) were cultured for 6 h at 37 °C, 5% CO2 in the presence of 10 μM peptide (Tables 2 and 3) (Genscript), 10 U/mL rhIL-2 (Roche Diagnostics), 1 μg/mL Golgi-Plug (BD Biosciences) at 0 h for CD8+ responses, or 2 h for CD4+ responses, plus CD49d and CD28 at 0 h (both 3 μg/mL; BD Biosciences). At 6 h, cells were washed with FACS buffer (PBS, 1% FBS, 0.5% NaN3), and stained with anti–CD8-PerCPCy5.5 and CD4-APCCy7 (BioLegend) for 30 min on ice. Fixation with BD Cytoperm Cytofix buffer for 20 min on ice and stained intracellularly with anti–IL-4-PECy7, IFN-γ-FITC, TNF-α-APC, and IL-2-PE for 30 min on ice.
For the virus ICS assay cells were infected with an multiplicity of infection of 2 with LPAI H5N2 virus (A/Eurasian Wigeon/Hong Kong/MPF461/2007) or LPAI H7N7 (A/Northern Shoveller/Hong Kong/MPF518/2008) plus CD49d and CD28 for 8 h before the addition of Golgi-Plug, then incubated for further 12 h at 37 °C. Cells were then stained and fixed as above. Samples were acquired by flow cytometry on a FACS LSRII and analyzed with FlowJo software. Total cytokine production was calculated by subtracting background fluorescence using no peptide and no virus controls.
Immune Cell Profiling.
Cells were subject to Fc receptor blocking (anti-CD16/CD32; BD Bioscience) and then stained with one of five mixtures containing a panel of monoclonal antibodies for innate and adaptive immune cells (all BioLegend, unless otherwise indicated), for 30 min on ice. Mixture 1: F4/80-PE, I-AE-PerCPCy5.5, CD11b-APCy7, Gr1-PECy7, IA8-APC, CD11c-FITC. Mixture 2: CD3-APC, CD4-APCCy7, CD8-PerCPCy5.5, NK1.1/Dx5-FITC, γδT-PE and CD69-PECy7 or B220-PECy7. Mixture 3: F4/80-PECy7, CD11b-APCCy7, I-AE-PerCPCy5.5, MMR-APC, IL-4Rα-PE, iNOS-FITC (BD Bioscience). Mixture 4: CD3-APC, B220-PECy7, CD4-APCCy7, PNA-FITC, CXCR5-PerCPCy5.5, CD38-APC, Bcl6-PE (BD Bioscience). Mixture 5: CD3-APC, CD4-APCCy7, CD8-PerCPCy5.5, CD44-PE, CD69-PECy7, Ki67-FITC (eBioscience).
For intracellular staining, cells were fixed in fixation/permeabilzation buffer (eBioscience) for 30 min on ice, and stained with inducible nitric oxide synthase-FITC, Bcl6-PE, or Ki67-FITC in Perm wash buffer (BD Bioscience). Cells were finally fixed with 100 μL of 4% PFA for 20 min on ice, and then washed and stained with DAPI in PBS for 10 min on ice. Samples were acquired by flow cytometry on a FACS LSRII and analyzed with FlowJo software.
Depletion of T-Cell Subsets in Vaccinated Mice.
Vaccinated mice were depleted of CD4+, CD8+, or both CD4+ and CD8+ T cells before influenza infection. Mice were treated with GK1.5 (anti-CD4) and 2.43 (anti-CD8) or isotype control (IgG2b) monoclonal antibodies (BioXCell) at 100 μg i.p. at days −4, −2, 0, and 3 (11). Depletion was confirmed at day −1 at >98% compared with isotype treated mice and nondepleted mice.
Statistics.
Results represent the mean ± SD of three to five mice per group, unless indicated otherwise. Statistical significance was compared between vaccine group (Wyeth/IL-15/5Flu vaccinated) and controls using a standard Student t test (unless indicated), #P < 0.05, *P < 0.01, ##P < 0.005, **P < 0.001.
Supplementary Material
Acknowledgments
This project was supported by the National Institutes of Health (NIH); National Institute of Allergy and Infectious Diseases Contract HHSN266200700005C; the Food and Health Bureau Control of Infectious Diseases Commissioned Project (L.L.M.P.); Health and Medical Research Fund Grant 13121142 (to S.A.V.); Areas of Excellence Scheme of the University Grants Committee Grant AoE/M-12/06; and by the Intramural Research Program of the National Cancer Institute, Center for Cancer Research, NIH (T.A.W. and L.P.P.). S.A.V. is an Overseas Biomedical Research Fellow of the National Health and Medical Research Council, Australia.
Footnotes
The authors declare no conflict of interest. The US Government has been granted a use patent (No. 8,663,622) for recombinant vaccine viruses expressing IL-15, which includes L.P.P. and T.A.W. as inventors.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1403684111/-/DCSupplemental.
References
- 1.Lee LY, et al. Memory T cells established by seasonal human influenza A infection cross-react with avian influenza A (H5N1) in healthy individuals. J Clin Invest. 2008;118(10):3478–3490. doi: 10.1172/JCI32460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Greenbaum JA, et al. Pre-existing immunity against swine-origin H1N1 influenza viruses in the general human population. Proc Natl Acad Sci USA. 2009;106(48):20365–20370. doi: 10.1073/pnas.0911580106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Poon LL, et al. Vaccinia virus-based multivalent H5N1 avian influenza vaccines adjuvanted with IL-15 confer sterile cross-clade protection in mice. J Immunol. 2009;182(5):3063–3071. doi: 10.4049/jimmunol.0803467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zielinski RJ, et al. Smallpox vaccine with integrated IL-15 demonstrates enhanced in vivo viral clearance in immunodeficient mice and confers long term protection against a lethal monkeypox challenge in cynomolgus monkeys. Vaccine. 2010;28(43):7081–7091. doi: 10.1016/j.vaccine.2010.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.LaMere MW, et al. Contributions of antinucleoprotein IgG to heterosubtypic immunity against influenza virus. J Immunol. 2011;186(7):4331–4339. doi: 10.4049/jimmunol.1003057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fang Y, et al. Seasonal H1N1 influenza virus infection induces cross-protective pandemic H1N1 virus immunity through a CD8-independent, B cell-dependent mechanism. J Virol. 2012;86(4):2229–2238. doi: 10.1128/JVI.05540-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Valkenburg SA, et al. Protective efficacy of cross-reactive CD8+ T cells recognising mutant viral epitopes depends on peptide-MHC-I structural interactions and T cell activation threshold. PLoS Pathog. 2010;6(8):e1001039. doi: 10.1371/journal.ppat.1001039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen W, Antón LC, Bennink JR, Yewdell JW. Dissecting the multifactorial causes of immunodominance in class I-restricted T cell responses to viruses. Immunity. 2000;12(1):83–93. doi: 10.1016/s1074-7613(00)80161-2. [DOI] [PubMed] [Google Scholar]
- 9.Hessel A, et al. Vectors based on modified vaccinia Ankara expressing influenza H5N1 hemagglutinin induce substantial cross-clade protective immunity. PLoS ONE. 2011;6(1):e16247. doi: 10.1371/journal.pone.0016247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Betts MR, et al. HIV nonprogressors preferentially maintain highly functional HIV-specific CD8+ T cells. Blood. 2006;107(12):4781–4789. doi: 10.1182/blood-2005-12-4818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Guo H, Santiago F, Lambert K, Takimoto T, Topham DJ. T cell-mediated protection against lethal 2009 pandemic H1N1 influenza virus infection in a mouse model. J Virol. 2011;85(1):448–455. doi: 10.1128/JVI.01812-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gordon S. Alternative activation of macrophages. Nat Rev Immunol. 2003;3(1):23–35. doi: 10.1038/nri978. [DOI] [PubMed] [Google Scholar]
- 13.de Jong MD, et al. Fatal outcome of human influenza A (H5N1) is associated with high viral load and hypercytokinemia. Nat Med. 2006;12(10):1203–1207. doi: 10.1038/nm1477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brown DM, Dilzer AM, Meents DL, Swain SL. CD4 T cell-mediated protection from lethal influenza: Perforin and antibody-mediated mechanisms give a one-two punch. J Immunol. 2006;177(5):2888–2898. doi: 10.4049/jimmunol.177.5.2888. [DOI] [PubMed] [Google Scholar]
- 15.McKinstry KK, et al. Memory CD4+ T cells protect against influenza through multiple synergizing mechanisms. J Clin Invest. 2012;122(8):2847–2856. doi: 10.1172/JCI63689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wilkinson TM, et al. Preexisting influenza-specific CD4+ T cells correlate with disease protection against influenza challenge in humans. Nat Med. 2012;18(2):274–280. doi: 10.1038/nm.2612. [DOI] [PubMed] [Google Scholar]
- 17.Jegaskanda S, et al. Cross-reactive influenza-specific antibody-dependent cellular cytotoxicity antibodies in the absence of neutralizing antibodies. J Immunol. 2013;190(4):1837–1848. doi: 10.4049/jimmunol.1201574. [DOI] [PubMed] [Google Scholar]
- 18.Laidlaw BJ, et al. Cooperativity between CD8+ T cells, non-neutralizing antibodies, and alveolar macrophages is important for heterosubtypic influenza virus immunity. PLoS Pathog. 2013;9(3):e1003207. doi: 10.1371/journal.ppat.1003207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rimmelzwaan GF, Sutter G. Candidate influenza vaccines based on recombinant modified vaccinia virus Ankara. Expert Rev Vaccines. 2009;8(4):447–454. doi: 10.1586/erv.09.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Brewoo JN, et al. Cross-protective immunity against multiple influenza virus subtypes by a novel modified vaccinia Ankara (MVA) vectored vaccine in mice. Vaccine. 2013;31(14):1848–1855. doi: 10.1016/j.vaccine.2013.01.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Berthoud TK, et al. Potent CD8+ T-cell immunogenicity in humans of a novel heterosubtypic influenza A vaccine, MVA-NP+M1. Clin Infect Dis. 2011;52(1):1–7. doi: 10.1093/cid/ciq015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lillie PJ, et al. Preliminary assessment of the efficacy of a T-cell-based influenza vaccine, MVA-NP+M1, in humans. Clin Infect Dis. 2012;55(1):19–25. doi: 10.1093/cid/cis327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mok CK, et al. Pathogenicity of the novel A/H7N9 influenza virus in mice. MBio. 2013;4(4):e00362-13. doi: 10.1128/mBio.00362-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Webster RG, Cox N, Stöhr K. WHO Manual on Animal Influenza Diagnosis and Surveillance. Geneva, Switzerland: World Health Organization; 2002. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.