Significance
The kinesin-3 family is one of the largest among the kinesin superfamily and its members play important roles in a variety of cellular functions ranging from intracellular transport to mitosis. Defects in kinesin-3 transport have been implicated in a variety of neurodegenerative, developmental, and cancer diseases, yet the molecular mechanisms of kinesin-3 regulation and cargo transport are largely unknown. We show that kinesin-3 motors undergo cargo-mediated dimerization to transport cellular cargoes. We also show that dimerization results in kinesin-3 motors that are fast and superprocessive. Such high processivity has not been observed for any other motor protein and suggests that kinesin-3 motors are evolutionarily adapted to serve as the marathon runners of the cellular world.
Keywords: microtubule, molecular motor, helical plot, intracellular transport
Abstract
The kinesin-3 family is one of the largest among the kinesin superfamily and its members play important roles in a wide range of cellular transport activities, yet the molecular mechanisms of kinesin-3 regulation and cargo transport are largely unknown. We performed a comprehensive analysis of mammalian kinesin-3 motors from three different subfamilies (KIF1, KIF13, and KIF16). Using Forster resonance energy transfer microscopy in live cells, we show for the first time to our knowledge that KIF16B motors undergo cargo-mediated dimerization. The molecular mechanisms that regulate the monomer-to-dimer transition center around the neck coil (NC) segment and its ability to undergo intramolecular interactions in the monomer state versus intermolecular interactions in the dimer state. Regulation of NC dimerization is unique to the kinesin-3 family and in the case of KIF13A and KIF13B requires the release of a proline-induced kink between the NC and subsequent coiled-coil 1 segments. We show that dimerization of kinesin-3 motors results in superprocessive motion, with average run lengths of ∼10 μm, and that this property is intrinsic to the dimeric kinesin-3 motor domain. This finding opens up studies on the mechanistic basis of motor processivity. Such high processivity has not been observed for any other motor protein and suggests that kinesin-3 motors are evolutionarily adapted to serve as the marathon runners of the cellular world.
Molecular motors of the kinesin and dynein superfamilies are responsible for a variety of microtubule-based intracellular functions such as vesicle transport, spindle assembly, and cytoskeletal organization. Several features of the kinesin and dynein mechanochemical cycles have been realized, yet much remains unknown (1, 2). For kinesin motors, much of the current knowledge is based on studies of kinesin-1, the founding member of the kinesin superfamily, where alternating catalysis by the two motor domains of the dimeric molecule results in unidirectional processive movement (the ability to take successive steps along the microtubule). Mechanochemical studies of other members of the kinesin superfamily have revealed interesting evolutionary adaptations to the kinesin motor domain that enable, for example, conversion of ATP hydrolysis into microtubule destabilizing activity (kinesin-8 and kinesin-13 families) (3, 4).
The kinesin-3 family is one of the largest among the kinesin superfamily and consists of five subfamilies in mammals (KIF1, KIF13, KIF14, KIF16, and KIF28) (5). Kinesin-3 family members are characterized by high sequence conservation within their motor domains, a forkhead-associated domain, and in most cases a C-terminal lipid-binding domain such as a pleckstrin homology or Phox homology (PX) domain (5). The founding member of kinesin-3 family, CeUNC-104, was identified in Caenorhabditis elegans based on a mutation that severely affects the transport of synaptic vesicles to the axon terminal (6, 7). Since that time, mammalian kinesin-3 motors have been found to be associated with diverse cellular and physiological functions including vesicle transport (8–12), signaling (13, 14), mitosis (15–17), nuclear migration (18), viral trafficking (19, 20), and development (21). Defects in kinesin-3 transport have been implicated in a wide variety of neurodegenerative, developmental, and cancer diseases (22). However, a mechanistic understanding of this important class of cellular transporters is currently limited.
To date, mechanistic studies have focused on truncated versions of mammalian KIF1A and its homolog CeUNC-104, and these studies have yielded contradictory results (23). For example, murine KIF1A has been proposed to function as a monomer that diffuses along the microtubule surface or as a processive dimer. Furthermore, dimerization has been proposed to occur before cargo binding or on the cargo surface (24–26). Thus, despite their widespread functions and clinical importance, the mechanisms of kinesin-3 motor regulation and motility remain largely enigmatic. Based on extensive characterization at the cellular and single-molecule levels of mammalian kinesin-3 motors from three subfamilies (KIF1, KIF13, and KIF16), we now provide a general model for kinesin-3 motor regulation and motility. We show that kinesin-3 motors are largely regulated by a monomer-to-dimer transition on the cargo surface that results in superprocessive motion for cargo transport.
Results and Discussion
KIF16B Motors Undergo Cargo-Mediated Dimerization to Drive Cargo Transport.
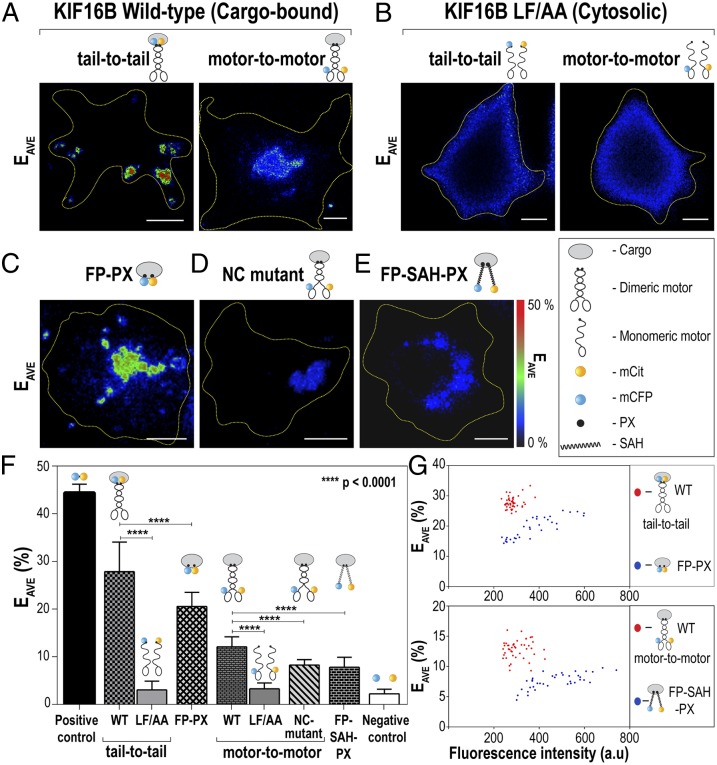
To directly examine the relationship between kinesin-3 motor dimerization and cargo transport, we used the kinesin-3 motor KIF16B, whose molecular and motility properties have not been examined. We took advantage of the fact that wild-type KIF16B motors are entirely cargo-bound (Fig. S1A and ref. 27), whereas introduction of two point mutations (L1248A and F1249A, referred to as LF/AA) into the PX domain abolishes cargo binding (Fig. S1B and ref. 28). We used Forster resonance energy transfer (FRET) analysis of wild-type (cargo-bound) and LF/AA (cytosolic) motors in live cells to assess the monomeric versus dimeric state of these motors. The motors were tagged with monomeric versions of the FRET donor cyan fluorescent protein (mCFP) or the FRET acceptor Citrine (mCit) either at the N terminus to measure the proximity of the motor domains (motor-to-motor FRET) or at the C terminus to measure the proximity of the tail domains (tail-to-tail FRET).
For the cytosolic KIF16B(LF/AA) motors (Fig. S2 B and E), very low levels of FRET were detected (Fig. 1 B and F and Fig. S3 C and F), similar to the FRET levels of coexpressed mCFP and mCit (negative control, Fig. 1F and Fig. S3I), suggesting that non–cargo-bound motors exist in a monomeric state. In contrast, the cargo-bound wild-type KIF16B motors (Fig. S2 A and D) exhibited high FRET efficiencies, consistent with motor dimerization, regardless of whether the FRET pairs were placed on the motor (N-termini) or tail (C-termini) domains of the molecule (Fig. 1 A and F and Fig. S3 B and E). Note that the C-terminally tagged wild-type motors are active motors because their recruitment results in cargo transport to the cell periphery (Fig. S2A), whereas fusion of fluorescent proteins (FPs) to the N terminus of KIF16B seems to hinder its motility because the early endosomal cargoes remain clustered in the cell center (Fig. S2D).
Fig. 1.
Full-length KIF16B motors dimerize on the cargo surface to drive transport. (A–E) Live cell FRET microscopy. COS-7 cells coexpressing (A) wild-type KIF16B tagged with FRET donor (mCFP) and acceptor (mCit) fluorescent proteins or (B) the cargo-binding mutant LF/AA tagged with mCFP and mCit were imaged by FRET microscopy. For tail-to-tail FRET (A and B, Left), FPs were fused to the C termini of the motors, whereas for motor-to-motor FRET (A and B, Right), FPs were fused to the N termini of the motors. (C–E) FRET controls. COS-7 cells coexpressing (C) donor and acceptor FPs targeted to the cargo surface via the monomeric KIF16B PX domain, (D) the N-terminally tagged KIF16B NC mutant, or (E) donor and acceptor FPs extended 30 nm from the cargo surface by a SAH between the FP and PX domains were imaged by FRET microscopy. Representative images of the calculated average FRET efficiency (EAVE) are shown. Yellow dotted lines indicate the outline of the cells. (Scale bars, 10 μm.) (F) Quantification of the FRET efficiencies in live COS-7 cells. For a negative control, the FRET efficiency between mCFP and mCit expressed as separate proteins was measured. For a positive control, the FRET efficiency between linked mCFP and mCit proteins (mCit-16aa-mCFP) was measured. n = 25–40 cells each over three independent experiments. The data are presented as mean ± SD. P values were calculated using the two-tailed t test. (G) Comparison of the FRET efficiency (EAVE) to protein concentration (measured as fluorescence intensity) on a cell-by-cell basis.
To rule out the possibility that the high FRET efficiencies of cargo-bound motors are due to clustering of monomeric motors rather than dimerization, two control experiments were carried out. First, we compared the average FRET efficiency (EAVE) of the C-terminally tagged wild-type motors (tail-to-tail FRET) to that of FRET donor and acceptor molecules targeted to the cargo surface via the monomeric KIF16B-PX domain (aa 1182–1317) (28). mCFP-PX and mCit-PX colocalized on the cargo surface (Fig. S2C) and exhibited relatively high FRET efficiencies (Fig. 1 C and F and Fig. S3D), but the EAVE was significantly lower than that of the C-terminally tagged wild-type KIF16B motors (Fig. 1F, P < 0.0001). Importantly, FRET between monomeric donor (mCFP) and acceptor (mCit) molecules was found to be concentration-dependent, whereas KIF16B tail-to-tail FRET was found to be concentration-independent (Fig. 1G, Upper). Thus, the C termini of KIF16B are closer together owing to dimerization than are monomeric molecules clustered on the same cargo. Second, we compared the average FRET efficiency of the N-terminally tagged wild-type motors (motor-to-motor FRET) to that of a nondimerizing mutant where the hydrophobic residues in the “a” and “d” positions of the neck coil (NC) were changed to charged residues (NC mutant), as described for CeUNC-104 (24). We also compared the motor-to-motor FRET to that of monomeric FPs extended 30 nm from the cargo surface via a single α-helix (SAH) domain (FP-SAH-PX construct) (29). Both the KIF16B NC mutant and the FP-SAH-PX constructs localized to the cargo surface (Fig. S2 F and G). Significantly higher FRET efficiencies were obtained for the N-terminally tagged wild-type KIF16B motor (Fig. 1A, Right and Fig. 1F) than the nondimerizing NC mutant (Fig. 1 D and F and Fig. S3G, P < 0.0001) or the monomeric FPs extended 30 nm from the cargo surface (Fig. 1 E and F and Fig. S3H, P < 0.0001). Importantly, a linear relationship between protein concentration and FRET was observed for the control FP-SAH-PX construct but not for KF16B (Fig. 1G, Lower). These results indicate that the N termini of KIF16B are closer together owing to NC dimerization than are monomeric molecules clustered on the same cargo. Taken together, these results demonstrate that KIF16B motors exist in a dimeric state on the cargo surface to drive cargo transport.
To verify that KIF16B motors exist in a monomeric state when not bound to cargo, we carried out photobleaching analysis at the single-molecule level. To do this, the full-length motor was tagged with three tandem copies of mCit. In our previous work, we used this assay to determine the oligomeric state of KIF1A (26). We thus used KIF1A as a control and also investigated the oligomeric state of two additional kinesin-3 family members, KIF13A and KIF13B, whose molecular mechanisms of motor regulation and cargo transport are unknown. In photobleaching assays, the majority of molecules bleached in two or three steps (Fig. S4C), indicating that the full-length kinesin-3 motors exist primarily in a monomeric state with some dimers in the population. The finding that full-length KIF1A is largely monomeric differs from our previous work (26) where expressed full-length motors were found to be in a predominantly dimeric state. However, the previous photobleaching assays were carried out in the presence of microtubules and adenylylimidodiphosphate and therefore enriched for the subpopulation of dimeric motors (30). Direct comparison of the photobleaching behavior of full-length KIF1A motors in the soluble (adsorbed to the coverslip) versus microtubule-bound state demonstrates that soluble motors are largely monomeric (Fig. S4F). The full-length kinesin-3 motors are also largely inactive for microtubule-based motility based on their behavior in neuronal CAD cells (Fig. S4 A and B) and single-molecule assays (Fig. S4 D and E). These findings are consistent with the general model of kinesin autoinhibition (31), yet these results indicate that for kinesin-3 family motors, the mechanism of regulation involves both cargo binding and a monomer-to-dimer transition.
To our knowledge, this is the first report to directly demonstrate cargo-mediated dimerization for full-length kinesin-3 motors on a cellular cargo. We propose that the monomer-to-dimer transition is a key regulatory mechanism for kinesin-3 motor activity. Although this mechanism of regulation is so far unique among the kinesin superfamily, it is interesting to note that several members of the myosin superfamily have been demonstrated to undergo dimerization and activation upon cargo binding (32–34). Thus, this work extends the parallels between the kinesin and myosin superfamilies.
Regulation of the NC Controls the Dimerization and Activation of Kinesin-3 Motors.
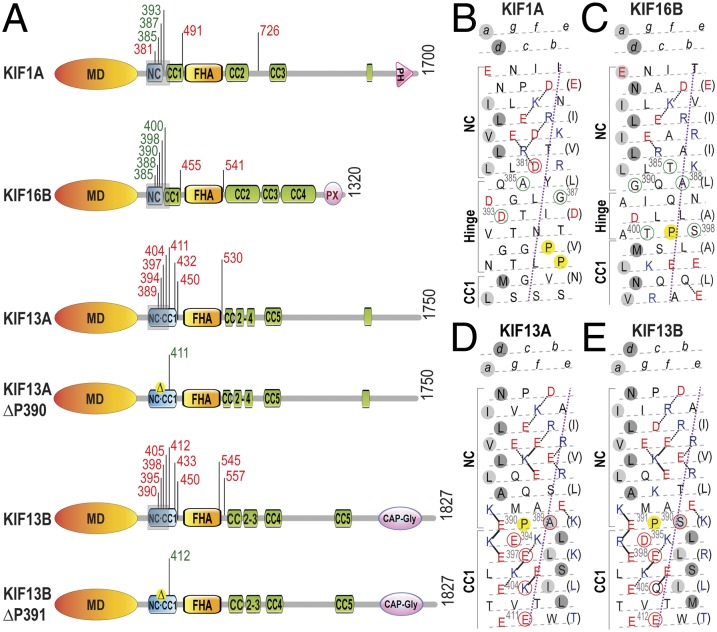
The molecular mechanisms by which kinesin-3 motors are sequestered in a monomeric and inactive state until cargo-mediated dimerization and activation are largely unknown. For MmKIF1A/CeUNC-104, the monomer-to-dimer transition is regulated by the NC segment that immediately follows the motor domain (Fig. 2A). The dimerization potential of the KIF1A NC segment seems to be weak and regulated by the subsequent coiled-coil 1 (CC1) domain such that an intramolecular interaction between the NC and CC1 segments holds the motor in a monomeric state (26, 35). An intermolecular NC–NC interaction on the cargo surface would then result in a dimeric state.
Fig. 2.
The NC is a key regulatory element for kinesin-3 motor dimerization and processive motility. (A) Domain organization of kinesin-3 motors analyzed in this study. Truncations are indicated by black tick marks with the amino acid number. Truncations in green text indicate processive motors, whereas truncations in red text indicate nonprocessive motors based on analysis of motor behavior in CAD cells (Fig. S6) and/or single molecule assays (Fig. 3). CC, coiled-coil; FHA, forkhead-associated; MD, motor domain; NC, neck coil; PH, pleckstrin homology; and PX, Phox homology. (B–E) Heptad net plots of the NC–CC1 regions (gray shaded rectangles in A) of the kinesin-3 motors. The helix has been cut and opened flat to give a 2D representation of the amino acid residues and their potential interactions in the helix. The N terminus is at the top right. Acidic residues are shown in red, basic in blue, and all others in black. Preferred ionic interactions are shown as solid lines and alternative interactions are shown as dotted lines. Every seventh residue is repeated on the right of the plot (in brackets) so that all interactions can be shown. The violet dotted line defines the orientation of the α-helix axis. The light gray circles indicate ‘‘a’’ positions and the dark gray circles indicate ‘‘d’’ positions of the hydrophobic seam in the CC. Residues circled in green indicate truncations that yield processive motors and residues circled in red indicate truncations that yield nonprocessive motors.
We thus tested whether the dimerization potential of the NC segment is a key mechanism that regulates the oligomerization and activity of kinesin-3 family members. We started by analyzing the primary sequence of KIF13A, KIF13B, and KIF16B by the COILS program (Fig. S5) and heptad net plots (36, 37) (Fig. 2 B–E) to identify potential NC segments and then generated several truncations for each motor based on this analysis. To rapidly screen truncated constructs for processive motility, mCit-tagged versions were expressed in neuronal CAD cells (Fig. S6).
COILS analysis of the KIF16B sequence indicated the presence of a short coiled-coil segment that could function as a NC for dimerization (Fig. S5D). A heptad net plot shows that the potential NC segment contains a hydrophobic core (leucine zipper) with a number of flanking ionic interactions (Fig. 2C). Truncations within and following the NC segment resulted in processive motors with strong accumulation at neurite tips, whereas truncations after the subsequent CC1 segment resulted in nonprocessive motors that remained in the cell body (Fig. S6 D and H and Table S1, summarized in Fig. 2 A and C). Analysis of the oligomeric state of the KIF16B constructs showed that nonprocessive motors exist in a monomeric state, whereas processive motors exist in a dimeric state (Fig. S7 and Table S1), similar to previous work on MmKIF1A and CeUNC-104 (26, 35). These results suggest that KIF16B is regulated in a manner similar to KIF1A in which an intramolecular interaction between the NC and CC1 segments inhibits intermolecular motor dimerization via the NC segments.
Analysis of the KIF13A and KIF13B sequences suggested the presence of a NC with strong coiled-coil potential (Fig. S5 B and C); however, truncations within or after the predicted NC resulted in monomeric motors with little to no processive motility in CAD cells or single-molecule assays (Figs. S6 B, C, F, and G and S7 and Table S1, summarized in Fig. 2 A, D, and E). The only exception was KIF13B(1–412), which accumulated at neurite tips in some cells (e.g., Fig. S6C) and showed rare motility events in single-molecule assays, similar to previous work on DmKhc-73, a Drosophila ortholog of KIF13 (38). The inability of truncated KIF13A and KIF13B motors to undergo processive motility was surprising. Further analysis of the secondary structure predictions revealed a CC1 segment immediately following but out of register with the NC, as if the hinge between the NC and CC1 segments has been truncated or deleted (Fig. 2 D and E). Furthermore, a proline residue (P390 and P391 in KIF13A and KIF13B, respectively) is found at the junction of the NC–CC1 segments (highlighted in yellow in Fig. 2 D and E). Proline residues are commonly found at the start of α-helices; when found at internal positions in α-helices and coiled-coil domains, they introduce a helix-destabilizing kink of about 30°. We therefore considered the possibility that the proline residue restricts the conformational mobility of the NC and CC1 segments and thereby prevents dimerization and thus processive motility. To explore this, we created two mutants of the truncated KIF13A(1–411) and KIF13B(1–412) motors in which the proline residue was either mutated to alanine (A) or deleted (Δ). When expressed in CAD cells, the alanine mutants remained monomeric and inactive, identical to the wild-type motors. Surprisingly, deletion of the proline residue resulted in dimeric motors that showed robust processive motility in CAD cells and single-molecule assays (Figs. S6 B, C, F, and G and S7 and Table S1, summarized in Fig. 2 A, D, and E). These data suggest that a proline residue at the NC–CC1 junction plays a critical role in regulating the dimerization potential and processive motility of KIF13A and KIF13B motors.
The proline-induced offset of the NC–CC1 segments that regulates KIF13A and KIF13B motor function is a novel mechanism that has not been observed for any other motor protein. To gain a structural understanding of how this proline residue regulates the dimerization of KIF13A and KIF13B, we used I-TASSER to predict the structural conformation of the NC–CC1 segments (39). The wild-type NC–CC1 sequences are predicted to form anti-parallel coiled-coils with the proline residues responsible for the tight turn between NC and CC1 (Fig. S8 A and D). Mutation of proline to alanine is not predicted to alter this conformation (Fig. S8 B and E), consistent with the mutants remaining monomeric and inactive. In contrast, proline deletion is predicted to result in the formation of a single long coil containing the NC and CC1 segments (Fig. S8 C and F), consistent with the fact that the ΔP mutants are capable of dimerization and processive motility. An important question for the future is to determine how the intramolecular NC–CC1 interactions are relieved upon cargo binding, especially for the proline-induced NC–CC1 interaction. One possibility, based on the fact that proline residues are typically found at the end of pi-helices (40), is that the KIF13 NC forms a pi-helix rather than an α-helix and that the pi-helix is compatible with dimerization. Alternatively, adaptor molecules involved in motor-cargo binding could play a role in isomerizing the proline conformation and allowing the NC and CC1 helices to come in register with each other. Interestingly, isomerization of a key proline residue has been demonstrated to control autoinhibition of the Crk adaptor protein (41, 42).
These findings support two general principles of kinesin motor biochemistry. First, the NC segment is the minimal unit for dimerization of kinesin motors, and second, dimerization is a fundamental requirement for processive motion (1). However, our results indicate that the kinesin-3 motors use a unique mechanism of regulation in which the dimerization potential of the NC is regulated to ensure that cargo binding and processive motility are tightly linked.
Dimeric Kinesin-3 Motors Are Superprocessive.
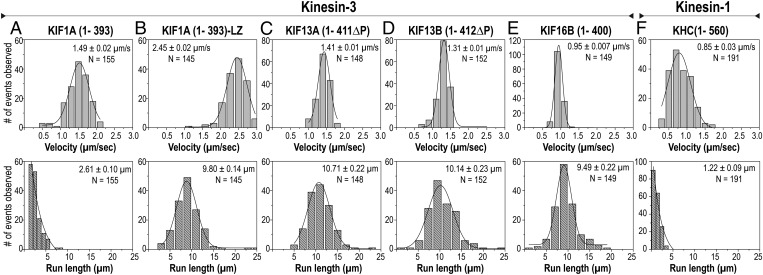
To characterize the motility properties of the dimeric kinesin-3 motors, we performed single-molecule motility assays using total internal reflection fluorescence (TIRF) microscopy. All truncated dimeric kinesin-3 motors underwent diffusion along the microtubule surface in the presence of ADP (Fig. S9 C and D) and processive motility in the presence of ATP (Fig. 3). Surprisingly, the KIF13A, KIF13B, and KIF16 motors were superprocessive, with average run lengths of ∼10 μm (Fig. 3 C–E and Table S2), which is nearly 10 times more processive than the well-characterized kinesin-1 motor (Fig. 3F). Such high processivity was surprising because KIF1A(1–393) motors are only moderately processive with an average run length of ∼2 μm (Fig. 3A, Table S2, and ref. 26). We wondered whether the apparently limited processivity of truncated dimeric forms of KIF1A is due to the presence in the population of both monomeric motors undergoing diffusive motility and weakly dimeric motors undergoing short processive motion, as suggested for DmKhc-73 (38) and the weak propensity of the KIF1A NC to form a stable dimer (Fig. S7 A and F). To test this possibility, we stabilized the NC by fusing the leucine zipper (LZ) of GCN4 to the C terminus of KIF1A(1–393), as was done previously for CeUNC-104 and DmKhc-73 (25, 38). The resulting KIF1A(1–393)-LZ motors exhibited fast and highly processive motion with an average run length of 9.80 ± 0.14 μm (Fig. 3B and Table S2). In addition, strongly dimeric KIF1A(1–393)-LZ motors displayed only processive motion, whereas weakly dimeric KIF1A(1–393) motors displayed both diffusive and processive motion (Fig. S9 A–C). Thus, we conclude that all dimeric kinesin-3 motors are capable of superprocessive motility.
Fig. 3.
Truncated kinesin-3 motors are superprocessive. Truncated kinesin-3 motors tagged with three tandem mCit FPs (3xmCit) at their C termini were expressed in COS-7 cells. Cell lysates were used for single-molecule motility assays. Histograms of the velocities (Upper) and run lengths (Lower) were plotted for each population of motors and fit to a single Gaussian. The peak represents the average velocity and run length of (A) KIF1A(1–393), (B) KIF1A(1–393)-LZ, (C) KIF13A(1–411∆P), (D) KIF13B(1–412∆P), (E) KIF16B(1–400), and (F) KHC(1–560), a well-characterized truncated kinesin-1 motor used as a motility control. Data are the averages from at least two independent experiments. The mean ± SEM and N values for each motor are indicated in the top right corners.
The extremely high processivity of the kinesin-3 motors was surprising. Indeed, these run lengths are the highest reported for any motor protein to date. It is important to note that the ability to undergo highly processive motion is intrinsic to the dimeric kinesin-3 motors because the truncated constructs consist of only the core motor and NC domains. This is important because other motors initially reported to be highly processive [kinesin-7 (CENP-E) and kinesin-8 (mammalian KIF18A and yeast Kip3p)] were later found to contain microtubule-binding sites in their tail domains that prevent dissociation from the microtubule surface (43–46).
What are the molecular mechanisms that enable kinesin-3 motors to undergo superprocessive motion? One mechanism proposed to influence motor processivity is a gating mechanism based on the length of the neck linker that connects the motor and NC domains (47–50). However, this mechanism cannot explain the remarkable processivity of the kinesin-3 motors because their neck linker lengths are similar to that of kinesin-2, a mildly processive motor (run lengths ∼1 μm). An alternative possibility is that the NC engages in electrostatic interactions with the microtubule to increase motor processivity, as shown for kinesin-1 (51). This mechanism also seems unlikely to explain the superprocessivity of kinesin-3 motors because the net charge of their NC regions is predicted to be neutral or net-negative. Nonetheless, to exclude an influence of electrostatic interactions on kinesin-3 processivity, we compared the motility of the motors in low- versus physiological-ionic-strength buffers. Interestingly, higher-ionic-strength buffers had no effect on kinesin-3 or kinesin-1 velocities and run lengths (compare Fig. S10 to Fig. 3) but did cause a decrease in the number of motility events (Fig. S11). Together, these results indicate that the high processivity of kinesin-3 motors is intrinsic to their core motor domains. That kinesin-3 motors are superprocessive was previously unknown and opens the door to understanding how a motor can maintain an interaction with its track for thousands of steps. Clearly, further work is required to determine the molecular, mechanical, and structural features that have evolved to enable kinesin-3 motors to achieve superprocessive motion.
Kinesin-3 Motors Function As the Marathon Runners of the Cellular World.
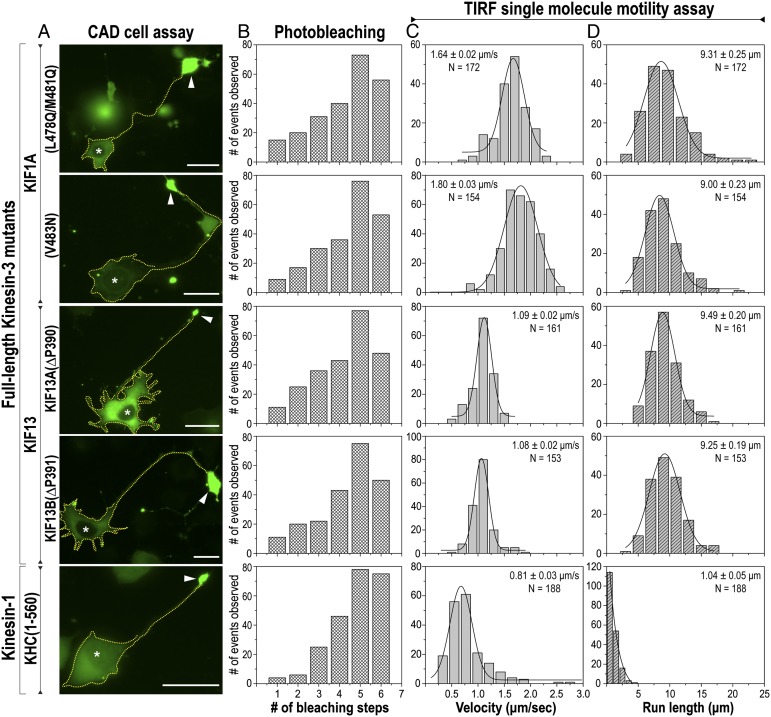
Together, this work indicates that kinesin-3 family motors are regulated by a unique mechanism in which non–cargo-bound motors are monomeric and inactive whereas cargo-bound motors are dimeric and undergo extremely long processive motion for transport. To verify this model, we tested whether the NC/CC1 regulatory mechanism is critical for preventing dimer formation and highly processive motion in the context of the full-length molecules. For KIF1A, recent structural analysis identified key residues at the C terminus of the CC1 segment (L478, M481, and V483) whose mutation results in motor activation in cells (52, 53). For KIF13A and KIF13B, our analysis of the truncated motors identified the proline residue at the NC/CC1 junction as critical for motor regulation. We thus directly compared the oligomeric state and motility properties of the inactive wild-type motors to that of the active mutants for full-length KIF1A, KIF13A, and KIF13B.
In CAD cells, mutation of L478Q/M481Q or V483N in full-length KIF1A or mutation of the proline residues in full-length KIF13A and KIF13B resulted in accumulation at the tips of neurite processes (Fig. 4A and Fig. S12), indicative of processive motility. In single-molecule motility assays, the mutant full-length motors were found to be dimeric and to undergo fast, superprocessive motion with average run lengths of ∼10 μm (Fig. 4 B–D and Table S2). Note that these motility properties are strikingly similar to those of the respective truncated stable dimers KIF1A(1–393)-LZ, KIF13A(1–411ΔP), and KIF13B(1–412ΔP) (Fig. 3 B–D and Table S2), suggesting that mutations that alter the NC–CC1 conformation resulted in the complete activation of full-length motors in cells. Thus, we conclude that kinesin-3 motors are regulated by a monomer-to-dimer transition that occurs upon cargo binding and enables highly processive transport of cellular cargoes. The key mechanism regulating this monomer-to-dimer transition involves regulating whether the NC helix forms an intramolecular interaction with the CC1 segment or an intermolecular interaction with another NC helix (Fig. 5).
Fig. 4.
Dimerization of full-length kinesin-3 motors results in superprocessive motion. The oligomeric state and motility properties of full-length kinesin-3 mutants KIF1A(L478Q/M481Q), KIF1A(V483N), KIF13A(∆P390), and KIF13B(∆P391) were compared with that of an active dimeric kinesin-1 [KHC(1–560)] motor. (A) CAD cell assay to measure motor processivity in cells. Representative images of differentiated CAD cells expressing C-terminal mCit-tagged full-length mutant kinesin-3 motors or KHC(1–560). Expressing cells are outlined with a yellow dotted line. Arrowheads indicate neurite tips. Asterisks indicate nuclei of expressing cells. (Scale bars, 20 μm.) (B) TIRF single-molecule photobleaching assay to measure oligomeric state. The fluorescence intensity over time was measured by TIRF microscopy for individual 3xmCit-tagged motors walking along the microtubule in COS-7 cell lysates. The number of bleaching events per molecule was plotted in a histogram for the population. (C and D) TIRF single-molecule motility assays; 3xmCit-tagged motors in COS-7 cell lysates were added to polymerized microtubules in a flow chamber and observed by TIRF microscopy. The measured (C) velocities and (D) run lengths of each population were plotted in a histogram. The averages of each population are from at least two independent experiments. The mean ± SEM and N values for each motor are indicated in the top right corners.
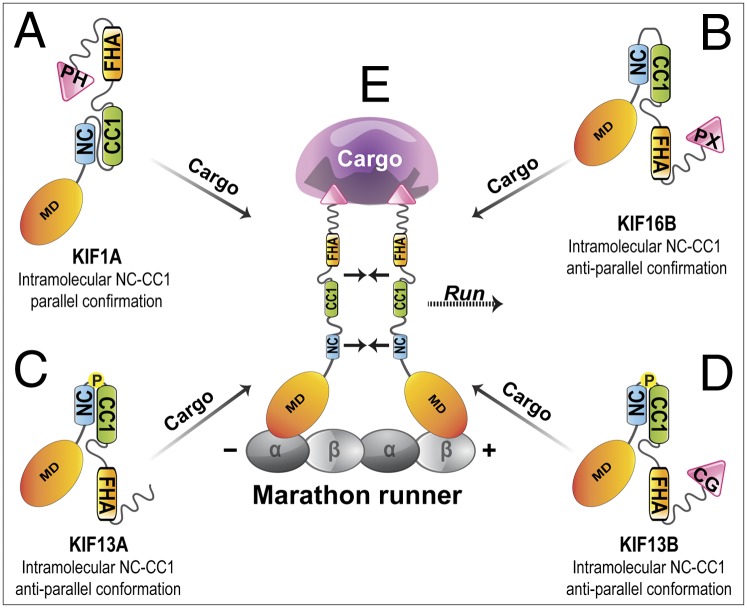
Fig. 5.
Molecular mechanisms of kinesin-3 motor regulation. Kinesin-3 motors use a unique mechanism of regulation in which dimerization and processive motility are tied to cargo binding. Maintaining the monomeric state of cytosolic motors is due to intramolecular interactions between the NC and CC1 segments. (A) For KIF1A, the NC–CC1 region is proposed to form a parallel coiled-coil based on structural studies of CeUNC-104 (35). (B) For KIF16B, the shorter linker between the NC and CC1 helices results in the formation of an anti-parallel coiled-coil. (C and D) For KIF13A and KIF13B, the hinge between NC and CC1 is absent; however, the two segments maintain an anti-parallel coiled-coil conformation owing to the presence of a proline residue at the NC–CC1 junction. (E) Cargo binding increases the effective concentration of the motor such that intermolecular NC–NC interactions are favored, resulting in dimerization and highly processive motility. CG, Cap-Gly; PH, Pleckstrin homology; and PX, phox homology.
In conclusion, we have shown that kinesin-3 motors use a regulatory mechanism unique among the kinesin family in which motor dimerization is prevented by either a self-interaction of NC and CC1 segments (KIF1A and KIF16B) or a proline-mediated shift in the NC–CC1 axis (KIF13A and KIF13B). To our knowledge, we provide the first direct demonstration that cargo binding results in dimerization and subsequent cargo transport. Furthermore, we show that active dimeric kinesin-3 motors are fast and remarkably processive, with run lengths longer than those of any other motor protein. Because these motility properties match the velocities and run lengths of known kinesin-3 cargoes transported in cells (54–57), we suggest that kinesin-3 motors have been evolutionarily adapted to drive long-distance intracellular and axonal transport. Thus, kinesin-3 motors seem to be the marathon runners of the cellular world.
Materials and Methods
A complete description of the materials and methods used is given in SI Materials and Methods.
All single-molecule assays were performed at room temperature using a Nikon Ti-E objective-type TIRF microscope with a 100× 1.49 N.A. CFI APO TIRF objective, and an EMCCD camera (iXon+ DU897; Andor). Motors in cell lysates were tracked on polymerized microtubules at 20 frames per second.
Supplementary Material
Acknowledgments
We thank Sam Straight and the Center for Live Cell Imaging (University of Michigan Medical School) for help with the FRET assays. We thank members of the K.J.V. laboratory for helpful discussions. This work was supported by National Institutes of Health Grant R01GM070862.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1400759111/-/DCSupplemental.
References
- 1.Vale RD. The molecular motor toolbox for intracellular transport. Cell. 2003;112(4):467–480. doi: 10.1016/s0092-8674(03)00111-9. [DOI] [PubMed] [Google Scholar]
- 2.Gennerich A, Vale RD. Walking the walk: How kinesin and dynein coordinate their steps. Curr Opin Cell Biol. 2009;21(1):59–67. doi: 10.1016/j.ceb.2008.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Su X, Ohi R, Pellman D. Move in for the kill: Motile microtubule regulators. Trends Cell Biol. 2012;22(11):567–575. doi: 10.1016/j.tcb.2012.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Moores CA, et al. The role of the kinesin-13 neck in microtubule depolymerization. Cell Cycle. 2006;5(16):1812–1815. doi: 10.4161/cc.5.16.3134. [DOI] [PubMed] [Google Scholar]
- 5.Miki H, Okada Y, Hirokawa N. Analysis of the kinesin superfamily: Insights into structure and function. Trends Cell Biol. 2005;15(9):467–476. doi: 10.1016/j.tcb.2005.07.006. [DOI] [PubMed] [Google Scholar]
- 6.Hall DH, Hedgecock EM. Kinesin-related gene unc-104 is required for axonal transport of synaptic vesicles in C. elegans. Cell. 1991;65(5):837–847. doi: 10.1016/0092-8674(91)90391-b. [DOI] [PubMed] [Google Scholar]
- 7.Otsuka AJ, et al. The C. elegans unc-104 gene encodes a putative kinesin heavy chain-like protein. Neuron. 1991;6(1):113–122. doi: 10.1016/0896-6273(91)90126-k. [DOI] [PubMed] [Google Scholar]
- 8.Horiguchi K, Hanada T, Fukui Y, Chishti AH. Transport of PIP3 by GAKIN, a kinesin-3 family protein, regulates neuronal cell polarity. J Cell Biol. 2006;174(3):425–436. doi: 10.1083/jcb.200604031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jenkins B, Decker H, Bentley M, Luisi J, Banker G. A novel split kinesin assay identifies motor proteins that interact with distinct vesicle populations. J Cell Biol. 2012;198(4):749–761. doi: 10.1083/jcb.201205070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lo KY, Kuzmin A, Unger SM, Petersen JD, Silverman MA. KIF1A is the primary anterograde motor protein required for the axonal transport of dense-core vesicles in cultured hippocampal neurons. Neurosci Lett. 2011;491(3):168–173. doi: 10.1016/j.neulet.2011.01.018. [DOI] [PubMed] [Google Scholar]
- 11.Okada Y, Yamazaki H, Sekine-Aizawa Y, Hirokawa N. The neuron-specific kinesin superfamily protein KIF1A is a unique monomeric motor for anterograde axonal transport of synaptic vesicle precursors. Cell. 1995;81(5):769–780. doi: 10.1016/0092-8674(95)90538-3. [DOI] [PubMed] [Google Scholar]
- 12.Nangaku M, et al. KIF1B, a novel microtubule plus end-directed monomeric motor protein for transport of mitochondria. Cell. 1994;79(7):1209–1220. doi: 10.1016/0092-8674(94)90012-4. [DOI] [PubMed] [Google Scholar]
- 13.Lamason RL, Kupfer A, Pomerantz JL. The dynamic distribution of CARD11 at the immunological synapse is regulated by the inhibitory kinesin GAKIN. Mol Cell. 2010;40(5):798–809. doi: 10.1016/j.molcel.2010.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ahmed SM, et al. KIF14 negatively regulates Rap1a-Radil signaling during breast cancer progression. J Cell Biol. 2012;199(6):951–967. doi: 10.1083/jcb.201206051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sagona AP, et al. PtdIns(3)P controls cytokinesis through KIF13A-mediated recruitment of FYVE-CENT to the midbody. Nat Cell Biol. 2010;12(4):362–371. doi: 10.1038/ncb2036. [DOI] [PubMed] [Google Scholar]
- 16.Torres JZ, et al. The STARD9/Kif16a kinesin associates with mitotic microtubules and regulates spindle pole assembly. Cell. 2011;147(6):1309–1323. doi: 10.1016/j.cell.2011.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Samwer M, et al. The nuclear F-actin interactome of Xenopus oocytes reveals an actin-bundling kinesin that is essential for meiotic cytokinesis. EMBO J. 2013;32(13):1886–1902. doi: 10.1038/emboj.2013.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tsai JW, Lian WN, Kemal S, Kriegstein AR, Vallee RB. Kinesin 3 and cytoplasmic dynein mediate interkinetic nuclear migration in neural stem cells. Nat Neurosci. 2010;13(12):1463–1471. doi: 10.1038/nn.2665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kratchmarov R, et al. Glycoproteins gE and gI are required for efficient KIF1A-dependent anterograde axonal transport of alphaherpesvirus particles in neurons. J Virol. 2013;87(17):9431–9440. doi: 10.1128/JVI.01317-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kramer T, et al. Kinesin-3 mediates axonal sorting and directional transport of alphaherpesvirus particles in neurons. Cell Host Microbe. 2012;12(6):806–814. doi: 10.1016/j.chom.2012.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ueno H, Huang X, Tanaka Y, Hirokawa N. KIF16B/Rab14 molecular motor complex is critical for early embryonic development by transporting FGF receptor. Dev Cell. 2011;20(1):60–71. doi: 10.1016/j.devcel.2010.11.008. [DOI] [PubMed] [Google Scholar]
- 22.Hirokawa N, Niwa S, Tanaka Y. Molecular motors in neurons: Transport mechanisms and roles in brain function, development, and disease. Neuron. 2010;68(4):610–638. doi: 10.1016/j.neuron.2010.09.039. [DOI] [PubMed] [Google Scholar]
- 23.Verhey KJ, Kaul N, Soppina V. Kinesin assembly and movement in cells. Annu Rev Biophys. 2011;40:267–288. doi: 10.1146/annurev-biophys-042910-155310. [DOI] [PubMed] [Google Scholar]
- 24.Klopfenstein DR, Tomishige M, Stuurman N, Vale RD. Role of phosphatidylinositol(4,5)bisphosphate organization in membrane transport by the Unc104 kinesin motor. Cell. 2002;109(3):347–358. doi: 10.1016/s0092-8674(02)00708-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tomishige M, Klopfenstein DR, Vale RD. Conversion of Unc104/KIF1A kinesin into a processive motor after dimerization. Science. 2002;297(5590):2263–2267. doi: 10.1126/science.1073386. [DOI] [PubMed] [Google Scholar]
- 26.Hammond JW, et al. Mammalian kinesin-3 motors are dimeric in vivo and move by processive motility upon release of autoinhibition. PLoS Biol. 2009;7(3):e72. doi: 10.1371/journal.pbio.1000072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hoepfner S, et al. Modulation of receptor recycling and degradation by the endosomal kinesin KIF16B. Cell. 2005;121(3):437–450. doi: 10.1016/j.cell.2005.02.017. [DOI] [PubMed] [Google Scholar]
- 28.Blatner NR, et al. The structural basis of novel endosome anchoring activity of KIF16B kinesin. EMBO J. 2007;26(15):3709–3719. doi: 10.1038/sj.emboj.7601800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sivaramakrishnan S, et al. Combining single-molecule optical trapping and small-angle x-ray scattering measurements to compute the persistence length of a protein ER/K alpha-helix. Biophys J. 2009;97(11):2993–2999. doi: 10.1016/j.bpj.2009.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rashid DJ, Bononi J, Tripet BP, Hodges RS, Pierce DW. Monomeric and dimeric states exhibited by the kinesin-related motor protein KIF1A. J Pept Res. 2005;65(6):538–549. doi: 10.1111/j.1399-3011.2005.00255.x. [DOI] [PubMed] [Google Scholar]
- 31.Verhey KJ, Hammond JW. Traffic control: Regulation of kinesin motors. Nat Rev Mol Cell Biol. 2009;10(11):765–777. doi: 10.1038/nrm2782. [DOI] [PubMed] [Google Scholar]
- 32.Yu C, et al. Myosin VI undergoes cargo-mediated dimerization. Cell. 2009;138(3):537–548. doi: 10.1016/j.cell.2009.05.030. [DOI] [PubMed] [Google Scholar]
- 33.Umeki N, et al. Phospholipid-dependent regulation of the motor activity of myosin X. Nat Struct Mol Biol. 2011;18(7):783–788. doi: 10.1038/nsmb.2065. [DOI] [PubMed] [Google Scholar]
- 34.Phichith D, et al. Cargo binding induces dimerization of myosin VI. Proc Natl Acad Sci USA. 2009;106(41):17320–17324. doi: 10.1073/pnas.0909748106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Al-Bassam J, et al. Distinct conformations of the kinesin Unc104 neck regulate a monomer to dimer motor transition. J Cell Biol. 2003;163(4):743–753. doi: 10.1083/jcb.200308020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Peckham M, Knight PJ. When a predicted coiled coil is really a single α-helix, in myosins and other proteins. Soft Matter. 2009;5:2493–2503. [Google Scholar]
- 37.Baboolal TG, et al. The SAH domain extends the functional length of the myosin lever. Proc Natl Acad Sci USA. 2009;106(52):22193–22198. doi: 10.1073/pnas.0909851106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Huckaba TM, Gennerich A, Wilhelm JE, Chishti AH, Vale RD. Kinesin-73 is a processive motor that localizes to Rab5-containing organelles. J Biol Chem. 2011;286(9):7457–7467. doi: 10.1074/jbc.M110.167023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Roy A, Kucukural A, Zhang Y. I-TASSER: A unified platform for automated protein structure and function prediction. Nat Protoc. 2010;5(4):725–738. doi: 10.1038/nprot.2010.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fodje MN, Al-Karadaghi S. Occurrence, conformational features and amino acid propensities for the pi-helix. Protein Eng. 2002;15(5):353–358. doi: 10.1093/protein/15.5.353. [DOI] [PubMed] [Google Scholar]
- 41.Sarkar P, Reichman C, Saleh T, Birge RB, Kalodimos CG. Proline cis-trans isomerization controls autoinhibition of a signaling protein. Mol Cell. 2007;25(3):413–426. doi: 10.1016/j.molcel.2007.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sarkar P, Saleh T, Tzeng SR, Birge RB, Kalodimos CG. Structural basis for regulation of the Crk signaling protein by a proline switch. Nat Chem Biol. 2011;7(1):51–57. doi: 10.1038/nchembio.494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mayr MI, Storch M, Howard J, Mayer TU. A non-motor microtubule binding site is essential for the high processivity and mitotic function of kinesin-8 Kif18A. PLoS ONE. 2011;6(11):e27471. doi: 10.1371/journal.pone.0027471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Stumpff J, et al. A tethering mechanism controls the processivity and kinetochore-microtubule plus-end enrichment of the kinesin-8 Kif18A. Mol Cell. 2011;43(5):764–775. doi: 10.1016/j.molcel.2011.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Su X, et al. Mechanisms underlying the dual-mode regulation of microtubule dynamics by Kip3/kinesin-8. Mol Cell. 2011;43(5):751–763. doi: 10.1016/j.molcel.2011.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Weaver LN, et al. Kif18A uses a microtubule binding site in the tail for plus-end localization and spindle length regulation. Curr Biol. 2011;21(17):1500–1506. doi: 10.1016/j.cub.2011.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Clancy BE, Behnke-Parks WM, Andreasson JO, Rosenfeld SS, Block SM. A universal pathway for kinesin stepping. Nat Struct Mol Biol. 2011;18(9):1020–1027. doi: 10.1038/nsmb.2104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Düselder A, Thiede C, Schmidt CF, Lakämper S. Neck-linker length dependence of processive Kinesin-5 motility. J Mol Biol. 2012;423(2):159–168. doi: 10.1016/j.jmb.2012.06.043. [DOI] [PubMed] [Google Scholar]
- 49.Yildiz A, Tomishige M, Gennerich A, Vale RD. Intramolecular strain coordinates kinesin stepping behavior along microtubules. Cell. 2008;134(6):1030–1041. doi: 10.1016/j.cell.2008.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shastry S, Hancock WO. Interhead tension determines processivity across diverse N-terminal kinesins. Proc Natl Acad Sci USA. 2011;108(39):16253–16258. doi: 10.1073/pnas.1102628108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Thorn KS, Ubersax JA, Vale RD. Engineering the processive run length of the kinesin motor. J Cell Biol. 2000;151(5):1093–1100. doi: 10.1083/jcb.151.5.1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Huo L, et al. The CC1-FHA tandem as a central hub for controlling the dimerization and activation of kinesin-3 KIF1A. Structure. 2012;20(9):1550–1561. doi: 10.1016/j.str.2012.07.002. [DOI] [PubMed] [Google Scholar]
- 53.Yue Y, et al. The CC1-FHA dimer is essential for KIF1A-mediated axonal transport of synaptic vesicles in C. elegans. Biochem Biophys Res Commun. 2013;435(3):441–446. doi: 10.1016/j.bbrc.2013.05.005. [DOI] [PubMed] [Google Scholar]
- 54.Lee JR, et al. Characterization of the movement of the kinesin motor KIF1A in living cultured neurons. J Biol Chem. 2003;278(4):2624–2629. doi: 10.1074/jbc.M211152200. [DOI] [PubMed] [Google Scholar]
- 55.Soppina V, Rai AK, Ramaiya AJ, Barak P, Mallik R. Tug-of-war between dissimilar teams of microtubule motors regulates transport and fission of endosomes. Proc Natl Acad Sci USA. 2009;106(46):19381–19386. doi: 10.1073/pnas.0906524106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhou HM, Brust-Mascher I, Scholey JM. Direct visualization of the movement of the monomeric axonal transport motor UNC-104 along neuronal processes in living Caenorhabditis elegans. J Neurosci. 2001;21(11):3749–3755. doi: 10.1523/JNEUROSCI.21-11-03749.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Barkus RV, Klyachko O, Horiuchi D, Dickson BJ, Saxton WM. Identification of an axonal kinesin-3 motor for fast anterograde vesicle transport that facilitates retrograde transport of neuropeptides. Mol Biol Cell. 2008;19(1):274–283. doi: 10.1091/mbc.E07-03-0261. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.