Abstract
Introduction:
Leishmania infantum is the causative agent of autochthonous cutaneous and visceral cases of leishmaniasis and transmitted by female sandflies. The dogs are considered the main reservoir hosts; however, there are the reports on Leishmania infection in other animals. In this study, occurrence types of L. infantum isolates have been analyzed by polymerase chain reaction-restriction fragment length polymorphism (PCR-RFLP) technique.
Materials and Methods:
In this experimental study, 77 samples were cultured and prepared for microscopic study and examined through PCR-RFLP. The samples were used for both deoxyribonucleic acid (DNA) and smear-slide preparations. The DNAs were amplified by PCR for the detection of Leishmania subgenus and PCR products were restricted with HaeIII for the species differentiation.
Results:
The visceral Leishmania parasites were genotyped as L. infantum. It was also determined sensitivity in PCR (100%) was higher than microscopic examination.
Conclusion:
PCR-RFLP technique appears to be most sensitive for the detection and differentiation of L. infantum. There exists a relationship between genetic heterogeneousness and clinical manifestation and geographical regions of this disease in human.
Keywords: Detection, Leishmania infantum, polymerase chain reaction-restriction fragment length polymorphism, visceral leishmaniasis
INTRODUCTION
Leishmaniasis is a vector-borne infection that is present in the Americas, Africa, Eastern Europe, Western and Central Asia, India and Australia. In some parts of World, cutaneous leishmaniasis (CL) is usually caused by Leishmania major, Leishmania tropica, or Leishmania aethiopica; mucocutenous leishmaniasis by L. aethiopica; and visceral leishmaniasis (VL) by Leishmania donovani or L. donovani, Leishmania infantum. CL and VL has been an important health problem. L. infantum is the causative agents of zoonotic VL, a fatal disease if left untreated. Dogs represent the main reservoir of this parasite.[1]
The diagnosis of leishmaniasis, is performed by direct visualization of amastigotes using microscopic examination of stained material, by isolation of the parasite in culture and by detection parasite using serological methods. Microscopic examination of stained-Giemsa slides, though rapid and low-cost, has limited sensitivity. In vitro culture techniques are susceptible to microbiological contamination and are hampered by the particular growth requirements of different strains.[2,3,4] Polymerase chain reaction (PCR)-based methods are highly sensitive and specific, compared with standard methods and are considered valuable for diagnosis. No single molecular test could serve as a “gold standard” against to the different assays could be evaluated. The PCR-restriction fragment length polymorphism (RFLP)-based genotyping assay has proven to be a reliable test for the detection of Leishmania in a wide range of clinical samples. The primary goal of this work was to detect and identify the Leishmania species specially L. infantum.[4,5,6]
MATERIALS AND METHODS
A total of 77 isolates obtained from humans were analyzed. Parasites were isolated from patients by bone marrow aspirates and were cultivated in liver infusion tryptose medium supplemented with 10% of heat inactivated fetal calf serum and grown at 24-26°C until the parasite concentration reached ~107 parasites/ml. Furthermore, the smears were prepared and stained with Giemsa stain and examined microscopically for the presence of amastigotes. The promastigote culture was washed in phosphate-buffered saline (pH 7.2) and deoxyribonucleic acid (DNA) extraction was performed using a commercial kit (DNG™ -Plus; CinnaGen, Tehran-Iran). PCR amplification was performed by specific primers. These included homologous 5'-CCC AAA CTT TTC TGG TCC TTC G-3', positioned at 24-45 and complementary 5'- CCA CGA CGC ATC CAA TCC AA-3', positioned at 360-341. PCR amplification carried out in a 25 μl reaction volume containing ×1.0 PCR buffer, 2.0 mM MgCl2 , 0.2 mM dNTP's, 0.5 mM each primers, 1-2 units of recombinant Taq DNA polymerase (CinnaGen, Tehran, Iran) and 5 μl of template. The reaction conditions included lid temperature of 105°C along with 4 min of initial denaturation at 95°C followed by 35 cycles of 95°C (30 s), 65°C (30 s) and 72°C (1.0 min). The reactions were ended by additional extension at 72°C for 10 min. PCR products were separated by horizontal electrophoresis in 1.5% agarose gel, along with a molecular weight marker. After staining the gel in ethidium bromide solution, the photograph was taken under ultraviolet illumination. 5 μl PCR product was digested with five Units of HaeIII (Fermentas) restriction endonuclease overnight at 37°C in a 20-μl of reaction volume. The digested PCR product was then separated on 2% agarose gel and analyzed for the presence of DNA ladder. DNA fragments on gel according to previously published procedure were analyzed.
Ethical approval
This study was reviewed and approved by the Ethics Committees of Iran University of Medical Sciences, Iran.
RESULTS
The samples of VL were analyzed by using the PCR-RFLP assay and microscopic examination. Microscopic observations was resulted as positive in 58 of the 77 samples and found to be less sensitive than PCR [Figure 1]. Leishmania subgenus was detected by the primers selected. The expected DNA bands were created by PCR-RFLP technique and a single band (250-bp) was observed in patients with VL [Figure 2]. These parasites were genotyped as a type of L. infantum, when compared with previously published studies and band patterns.
Figure 1.
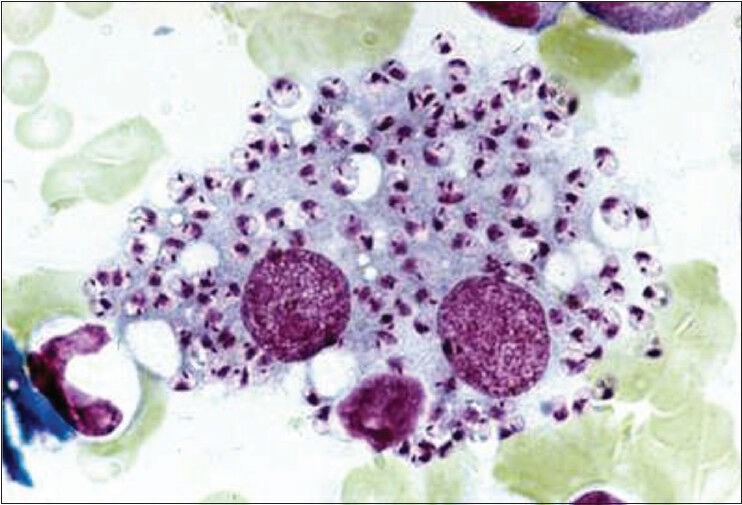
The images of amastigotes in human macrophages (×100, Giemsa stain)
Figure 2.
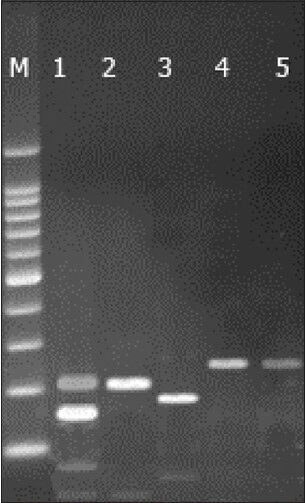
Polymerase chain reaction-restriction fragment length polymorphism analysis of selected strains of Leishmania pecies using Hae III restriction enzyme. Lanes: M–100 bp ladder (Fermentas); (1) Leishmania major (MHOM/IR/75/ER); (2) Leishmania tropica (MHOM/IR/03/Mash-878); (3) Leishmania donovani (IPER/IR/2007/HS10); (4) Leishmania infantum (positive control); (5) Obtained isolate from patient. Standard species were taken from reference laboratory of the Parasitology Department, Tehran University of Medical Sciences
The analytical sensitivity of our assay was tested by using serial dilutions of parasite DNA extracted from a known number of parasites. Six 10-fold serial dilutions of the extracted DNA were performed and each dilution sample separately underwent amplification. The detection limit of the assay was determined to be 10 pg.
DISCUSSION
Characterization of Leishmania species in clinical infections is important, as different species may require distinct treatment regimens.[4,6] Most of PCR assays are based on highly repetitive genomic gene loci or extrachromosomal kinetoplast DNA sequences, but their use is confined to the detection of the parasite at the genus level. Our aim of this study was to optimize and evaluate PCR-RFLP-based assay for Leishmania species. When the amplicon is digested with restriction enzymes, it is possible to identify almost all pathogenic Leishmania species by RFLP, allowing direct, rapid characterization and identification of the infecting parasite.[6,7,8,9] the PCR-RFLP method was used for the detection and the differentiation Leishmania species. We analyzed other several species of Leishmania by PCR-RFLP such as L. tropica, L. major, L. donovani respectively by 215-bp and 35-bp bands, 215, 155, 95, 35, about 180-bp and 70-bp [Figure 2].
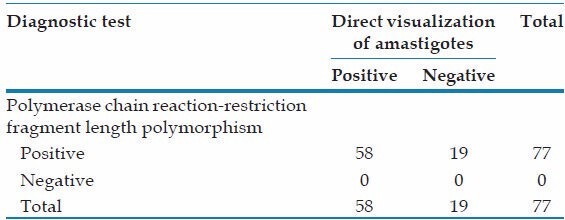
We also compared two diagnostic procedures, direct visualization of amastigotes and PCR-RFLP, in patients with VL. PCR resulted in a higher sensitivity (100%) than microscopy, detecting an additional 75-76% of the positive cases and identified the species [Table 1].[10]
Table 1.
Positivity of diagnostic procedures in patients with visceral leishmaniasis
Footnotes
Source of Support: Nil.
Conflict of Interest: None declared.
REFERENCES
- 1.Uzun S, Uslular C, Yücel A, Acar MA, Ozpoyraz M, Memişoğlu HR. Cutaneous leishmaniasis: Evaluation of 3,074 cases in the Cukurova region of Turkey. Br J Dermatol. 1999;140:347–50. doi: 10.1046/j.1365-2133.1999.02673.x. [DOI] [PubMed] [Google Scholar]
- 2.Andresen K, Gaafar A, El-Hassan AM, Ismail A, Dafalla M, Theander TG, et al. Evaluation of the polymerase chain reaction in the diagnosis of cutaneous leishmaniasis due to Leishmania major: A comparison with direct microscopy of smears and sections from lesions. Trans R Soc Trop Med Hyg. 1996;90:133–5. doi: 10.1016/s0035-9203(96)90112-1. [DOI] [PubMed] [Google Scholar]
- 3.Culha G, Uzun S, Ozcan K, Memisoglu HR, Chang KP. Comparison of conventional and polymerase chain reaction diagnostic techniques for leishmaniasis in the endemic region of Adana, Turkey. Int J Dermatol. 2006;45:569–72. doi: 10.1111/j.1365-4632.2006.02695.x. [DOI] [PubMed] [Google Scholar]
- 4.Chargui N, Bastien P, Kallel K, Haouas N, Akrout FM, Masmoudi A, et al. Usefulness of PCR in the diagnosis of cutaneous leishmaniasis in Tunisia. Trans R Soc Trop Med Hyg. 2005;99:762–8. doi: 10.1016/j.trstmh.2005.06.002. [DOI] [PubMed] [Google Scholar]
- 5.Minodier P, Piarroux R, Gambarelli F, Joblet C, Dumon H. Rapid identification of causative species in patients with Old World leishmaniasis. J Clin Microbiol. 1997;35:2551–5. doi: 10.1128/jcm.35.10.2551-2555.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schönian G, Nasereddin A, Dinse N, Schweynoch C, Schallig HD, Presber W, et al. PCR diagnosis and characterization of Leishmania in local and imported clinical samples. Diagn Microbiol Infect Dis. 2003;47:349–58. doi: 10.1016/s0732-8893(03)00093-2. [DOI] [PubMed] [Google Scholar]
- 7.Serin MS, Daglioglu K, Bagirova M, Allahverdiyev A, Uzun S, Vural Z, et al. Rapid diagnosis and genotyping of Leishmania isolates from cutaneous and visceral leishmaniasis by microcapillary cultivation and polymerase chain reaction-restriction fragment length polymorphism of miniexon region. Diagn Microbiol Infect Dis. 2005;53:209–14. doi: 10.1016/j.diagmicrobio.2005.05.007. [DOI] [PubMed] [Google Scholar]
- 8.Singh B. Molecular methods for diagnosis and epidemiological studies of parasitic infections. Int J Parasitol. 1997;27:1135–45. doi: 10.1016/s0020-7519(97)00111-2. [DOI] [PubMed] [Google Scholar]
- 9.Marfurt J, Nasereddin A, Niederwieser I, Jaffe CL, Beck HP, Felger I. Identification and differentiation of Leishmania species in clinical samples by PCR amplification of the miniexon sequence and subsequent restriction fragment length polymorphism analysis. J Clin Microbiol. 2003;41:3147–53. doi: 10.1128/JCM.41.7.3147-3153.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dilmec F, Matur A, Alhan E, Uzun S, Karakas M, Alabaz D, et al. Investigation of Leishmania parasites from clinical samples using polymerase chain reaction technique in Cukurova Region, Turkey. Journal of Harran University Medical Faculty. 2008;5:10–4. [Google Scholar]


