Abstract
Nematode anthelminthic resistance is widespread for the 3 major drug classes commonly used in agriculture: benzamidazoles, macrocyclic lactones, and nicotinic agonists e.g. levamisole. In parasitic nematodes the genetics of resistance are unknown other than to the benzimidazoles which primarily involve a single gene. In previous work with a levamisole resistant Oesophagostomum dentatum isolate, the nicotinic acetylcholine receptor (nAChR) exhibited decreased levamisole sensitivity. Here, using a transcriptomic approach on the same isolate, we investigate whether that decreased nAChR sensitivity is achieved via a 1-gene mechanism involving 1 of 27 nAChR pathway genes. 3 nAChR receptor subunit genes exhibited ≥ 2-fold change in transcript abundance: acr-21 and acr-25 increased, and unc-63 decreased. 4 SNPs having a ≥ 2-fold change in frequency were also identified. These data suggest that resistance is likely polygenic, involving modulated abundance of multiple subunits comprising the heteropentameric nAChR, and is not due to a simple 1-gene mechanism.
Keywords: resistance, levamisole, nAChR, nodular worm
Treatment of intestinal nematode infection is largely afforded by drug therapy using one of 3 major classes of anthelminthic compounds, benzimidazoles, macrocyclic lactones, or nicotinic agonists, that respectively interact with nematode beta-tubulin, glutamate-gated chloride channels, and nicotinic acetylcholine receptors (nAChR) [1]. Resistance to anthelminthics has been reported in agricultural use of all 3 major drug classes [2].
Use of the anthelminthic levamisole, a selective nicotinic agonist that induces spastic paralysis due to somatic muscle contraction, has been critical in allowing economically feasible treatment of animals throughout the world given its broad spectrum of activity and effectiveness against many larval stages. Levamisole resistance has become an increasing problem [3] since its first report in 1979. Investigations of laboratory-induced levamisole resistance in the free-living Clade V nematode Caenorhabditis elegans have identified a number of resistance-associated phenotypes and genes (e.g. [4]) including those involving the levamisole sensitive nAChR, a heteropentameric transmembrane protein located at the neuromuscular junction that is critical to muscle contraction (Fig. 1).
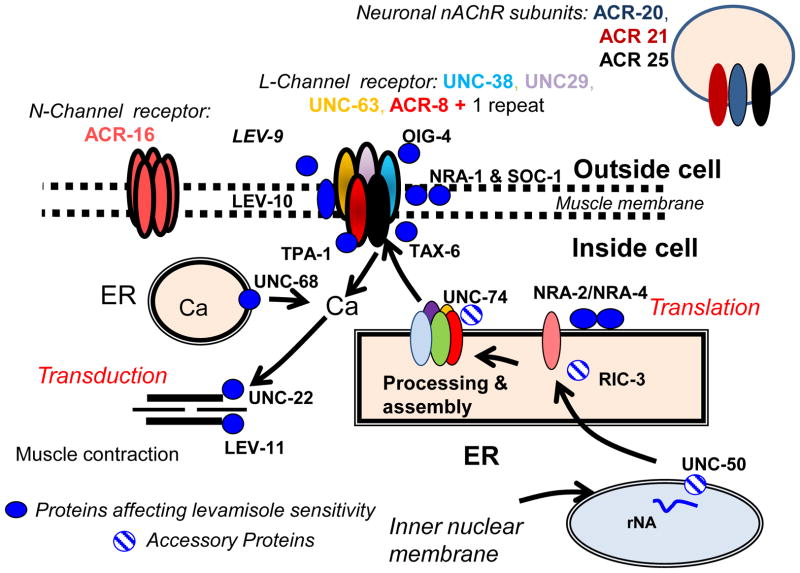
Fig. 1. Putative assembly and function of levamisole (L-channel or L-nAChRs) and nicotine (N-channel or N-nAChRs) nAChR of O. dentatum based on the C. elegans model modified using information from H. contortus.
The N-channel receptor is more sensitive to nicotine and composed of a homooligomer of ACR-16 subunits. The levamisole sensitive nAChR channels in the muscle membrane are heteropentamers formed from 3 alpha subunits (UNC-38, blue; UNC-63, yellow; and ACR-8, red), 1 non-alpha subunit (UNC-29, purple), and 1 additional subunit (black) that is a repeat of 1 of the same 4 subunits. It is not known which subunit is repeated, but if it were UNC-29 it would allow two adjacent alpha:non-alpha junctions as agonist binding sites. The normal function of the receptor is supported by OIG-4, NRA-1, NRA-2, NRA-4, SOC-1, TAX-6, TPA-1, LEV-9 and LEV-10 proteins. Once the levamisole nAChR channel opens, calcium enters and its signal is increased by the ryanodine receptor (UNC-68); the increase in cytoplasmic calcium then initiates contraction, requiring the proteins UNC-22 and LEV-11. The expression of the levamisole receptor subunits requires RNAs encoding the channel subunits together with three ancillary proteins involved in nAChR assembly (RIC-3), folding (UNC-74) and trafficking (UNC-50). Note: the information within this figure should be considered general in nature due to the significant nAChR heterogeneity among the nematodes, as is indicated from functional receptor reconstitution experiments and from analyses seeking to identify orthologous genes, e.g. [17, 18]. We note that a number of the nAChR pathway genes are involved in other pathways, e.g. TAX-6 [19]. Abbreviation: ER, endoplasmic reticulum; SR sarcoplasmic reticulum.
Although the genetics of levamisole resistance have not been established for any parasitic nematode, the nAChR pathway has been implicated in resistance in a number of parasitic nematodes including O. dentatum, Haemonchus contortus, Teledorsagia circumcincta and Trichostrongylus colubriformis [5]. Previous work with levamisole-sensitive (SENS) versus - resistant (LEVR) O. dentatum isolates determined that LEVR nAChR exhibited decreased sensitivity to levamisole [6]. The present study investigates the simplest genetic explanation for the resistance- and nAChR-properties of the LEVR isolate, that they result from a specific change in abundance or sequence of a single nAChR receptor subunit, in an analogous fashion to the 1-gene resistance mechanism that is causal to nematode resistance to the benzimidazoles (e.g. [7]). Even for an organism like O. dentatum lacking genome information, such a hypothesis is amenable to testing using the techniques of in silico sequencing (to determine sequences for nAChR-pathway target genes) and comparative transcriptomics.
A first requirement was to determine the sequences for genes of the nAChR pathway, both those encoding receptor subunits as well as genes encoding non-subunit proteins involved in the pathway by which nAChR is expressed (Fig. 1). Of note is the complex repertoire of genes that encode nAChR receptor subunits; subunits can be encoded by any of more than 25 different genes grouped within 5 gene families (Fig. 1, reviewed in [8]). Based upon the nAChR pathway as characterized primarily in C. elegans (Fig. 1), 27 genes were targeted for in silico sequencing and study in O. dentatum. This gene set included 11 subunit and 16 non-subunit genes, and of these the O. dentatum coding sequence was unknown for 21 genes (Table S1). The sequences of all 27 target nAChR pathway genes were determined in silico from RNA-Seq datasets using an optimized method we recently developed [9]. Briefly, RNA-Seq library datasets for a single isolate were assembled into contigs, the contigs searched by protein BLAST against target nAChR pathway sequences of H. contortus and Caenorhabditis genera to identify best match contigs having a BLAST expect value ≤ 1E−10, and then those best-match contigs were optimized for length and similarity through an iterative process involving additional read mapping and contig reassembly. Detailed methodology on experimental design, RNA-Seq library construction, and data analyses are provided within the Fig. 2 Legend.
Fig. 2. Differential expression of nAChR pathway genes.
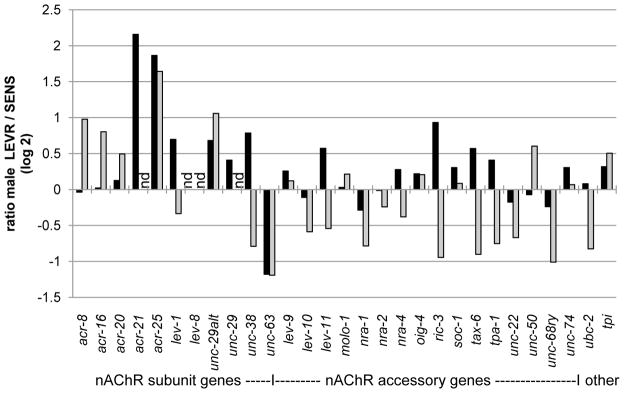
For GAII (grey bars) and HiSeq (black bars) RNA-Seq libraries, the graph represents the log 2 transformation of the ratio of LEVR/SENS DESeq normalized read counts (Tables S2-S4); “nd” indicates not determined because of low read-counts (≤ 10). Gene sequences determined in silico and deposited at the GenBank™ database: acr-8 GAAS01000001, acr-16 GAAS01000002, acr-20 GAAS01000003, acr-21 GAAS01000004, acr-25 GAAS01000005, lev-1 GAAR01000001, lev-8 GAAS01000006, unc-38 GAAR01000011, unc-63 GAAR01000012, lev-9 GAAR01000003, lev-10 GAAR01000002, lev-11 GAAS01000007, nra-1 GAAS01000008, nra-2 GAAR01000004, nra-4 GAAS01000009, oig-4 GAAR01000005, ric-3 GAAR01000006, soc-1 GAAR01000007, tax-6 GAAR01000008, tpa-1 GAAR01000009, unc-22 GAAR01000010, unc-50 GAAS01000010, unc-68ry GAAR01000013, unc-74 GAAS01000011. Two non-nAChR gene sequences, for ubc-2 (ubiquitin conjugating enzyme 2, AM181594.1) and tpi (triosephosphate isomerase: AF344333.1), were included for reference. RNA-Seq library reads (from which in silico sequences were assembled) are deposited at GenBank™: HiSeq SENS male SRX290901 and female SRX290923, HiSeq LEVR male SRX290893 and female SRX290897, GAII SENS male SRX113576 and female SRX113577, GAII LEVR male SRX113574 and female SRX113575. RNA sources and RNA-Seq library: Workup of RNA, including extraction, DNase I treatment, and quality assessment were as described [9]. From the first of 2 biological replicates, RNA was extracted from 4 separate pools of SENS (68 male, 69 female) and LEVR (58 male, 141 female) adults which had been isolated from 2 infected pigs, and was used for Illumina GAII sequencing. For the 2nd biological replicate, using worms isolated from different pigs that had been SENS- and LEVR-infected only after the first replicate was completed through the sequencing step, RNA was extracted from 4 separate pools of SENS (70 male, 36 female) and LEVR (40 male, 114 female) adults, and used for Illumina HiSeq sequencing. Construction of indexed, non-normalized, paired-end RNA-Seq libraries, and subsequent 75- or 100-cycle pyrosequencing on Illumina GAIIx or HiSeq 2000 platforms (respectively), were performed by the DNA Facility, Iowa State University, using 4–5 ug total RNA (per sample) having an RNA Integrity Number ≥ 7.2. The number of reads per library ranged from 4.2* 107 to 16.4 * 107 for HiSeq and from 2.0* 107 to 2.2 107 for GAII. Quantitation: A single reference transcriptome was assembled comprised of all contigs ≥ 300 bases assembled from the GAIIx SENS male library using Velvet (with Velvet kmer set to 31), less those with identity to the nAChR pathway genes, plus the O. dentatum nAChR pathway gene coding sequences. Removal of redundant contig sequences from the transcriptome utilized cd-hit-est [20]. Samtools [21] version 0.1.18 was used to count reads mapping to each contig. Normalization: Normalization of reads was performed using upper quartile [22] or DESeq [23] methods. Relative RNA expression profiles were obtained by (i) constructing a single reference transcriptome comprised all assembled contigs ≥ 300 bases, minus those contigs having high identity to the nAChR pathway genes, plus the O. dentatum nAChR pathway gene coding sequences (Table S1), with redundant sequences finally removed, (ii) using BWA [24] version 0.6.2-r126 to map male library reads onto that transcriptome, then (iii) normalizing mapped gene read values within libraries using either upper quartile or DESeq methodologies. BWA [24] version 0.6.2-r126 was used to map reads for SNP/indel and expression analyses. We note that the experimental replicates were performed more than 8 months apart, and, due to upgrades in the Illumina sequencing platform that occurred during that time, the first experiment yielded data via the GAIIx system and the second yielded data via HiSeq; consequently, the ratio of the expression levels for the target genes were calculated within each replicate but not among the replicates (Table S4).
The gene names and accession numbers for nAChR gene sequences determined in silico are included in Fig. 2 Legend, while information derived from comparison of the in silico gene sequences against the homologous sequences of H. contortus or C. elegans used initially to BLAST-query the contigs is listed in Table S1. Overall, there was high identity between the deduced protein sequence of the in silico-determined genes and their nearest homolog, with only 3 instances of identity lower that 50 % (Table S1). As might be expected based upon nematode Clade V phylogeny [10], each of the 5 O. dentatum genes for which H. contortus was the comparator (acr-16, lev-1, ric-3, unc-50 and -74) exhibited higher deduced protein identity to H. contortus than to C. elegans homologs (Table S1).
Next, the relative mRNA expression of the 27 nAChR pathway genes was assessed in each of the 2 SENS vs LEVR biological replicates, but only for libraries representing males. Female libraries were not used for expression assessment because, being gravid, they contained a combination of adult and egg transcripts. Reads from each of the 4 RNA-Seq male libraries were mapped, counted, and then the counts were normalized using DESeq (Fig. 2 Legend and Tables S2–4). The read expression ratios identified several nAChR-pathway genes whose mRNAs were differentially expressed in LEVR versus SENS males using a threshold of 2- fold change in expression, i.e. those genes shown in Fig. 2 having log base 2 ratios of ≥ 1 or ≤ −1. Receptor subunit mRNA levels for unc-63 were lower, and for acr-25 were higher, in LEVR males in both biological replicates (i.e. for both GAII and HiSeq analyses). Receptor subunit mRNA levels for acr-21 were also higher in LEVR males in the HiSeq analysis, but were not calculated for the GAII dataset due to low read counts of ≤ 10 (Table S2). Receptor subunit gene unc-29alt exhibited ≥ 2-fold change in the GAII experiment, but its ratio in the HiSeq dataset of 1.61 (Table S4) was below the threshold. No accessory target gene exhibited ≥ 2-fold differential expression in both biological replicates, but 1 (unc-68ry) reached the threshold in the GAII experiment.
The differential expression data suggest that levamisole resistance in the LEVR isolate is polygenic, i.e. due to changing the mix of subunits that comprise nAChR receptors on muscle by increasing ACR-21 and -25 while decreasing UNC-63 concentrations. Such a polygenic mechanism would allow nAChR channels to be maintained at levels required to control body muscle, albeit with a modified mix of subunit proteins that effectively decrease sensitivity to levamisole. If so, and given the relatively large number of genes that can encode nAChR subunits, one consequence of a polygenic resistance mechanism may be an increased likelihood for resistance to arise via different gene-expression patterns in isolated parasite populations.
A number of the gene targets, but especially of the accessory non-subunit mRNAs, exhibited expressions ratios that were up in 1 replicate but down in the other, e.g. ric-3 (Fig. 2). One possible explanation for this is that some accessory proteins (e.g. RIC-3) function in more pathways than just that of the nAChR, and consequently the observed differences may reflect gene expression plasticity in pathways not involved in levamisole resistance. In any event, assuming the differences between replicates in expression patterns of individual genes relate to biological plasticity within parasite populations of the same isolate, then such changes would be hallmarks of genes that are not singly-required to achieve levamisole resistance. As already indicated, the data were normalized using DESeq to compensate for differences in read numbers that invariably arise when sequencing different RNA-Seq libraries (Table S2–3). The data were also normalized using the Upper Quartile method (Table S2–3). Neither DESeq nor Upper Quartile algorithms utilize control “housekeeping” genes assumed to be stably expressed, and both methods tend to out-perform other methods that do incorporate such controls, e.g. that normalize to the total number of mapped reads per library or to the reads per kilobase per million mapped reads [11]. That said, Fig. 2 shows two non-nAChR-pathway genes (ubc-2 and tpi) that are unlikely to be involved in levamisole resistance and that, as expected, exhibit expression ratios below the 2-fold threshold of significance.
SNP calling from RNA-Seq was used to identify non-synonymous nAChR genes changes that exhibited ≥ 2- fold LEVR vs SENS frequency change in both the HiSeq and GAII male libraries. SNP calls were filtered to retain high-confidence calls by using published criteria [12]: briefly, these criteria included observation within a single RNA-Seq library of (i) ≥ 10 different reads per SNP position, (ii) a SNP frequency of ≥ 20% relative to the reference nucleotide base frequency, (iii) ≥ 3 different library reads containing the alternate allele, and (iv) ≥ 50 SNP-quality score (Tables S5–6). Predictably, only a small number of target gene SNPS were identified. Out of 3000 HiSeq library and 2277 GAII library high confidence SNPs identified, 64 GAII and 78 HiSeq SNPS were non-synonymous of which 52 were present in both HiSeq and GAII libraries (Table S5–6). Of the 52 non-synonymous SNPS, 25 (HiSeq) and 24 (GAII) exhibited ≥ 2- fold change in frequency in LEVR vs SENS, but only 4 SNPs exhibited ≥ 2- fold change in both experiments: SNPs that encode NRA-2 (H219N) and UNC-74 (V133I) substitutions increased ≥ 2- fold in LEVR vs SENS, while SNPs that encode UNC-38 (E2G) and UNC-22 (I4063V) substitutions increased ≥ 2- fold in SENS vs LEVR libraries.
Although several insertions/deletions were detected in the target genes, none of these exhibited a ≥ 2-fold change in LEVR vs SENS in either GAII or HiSeq library sets. Only one coding-sequence insertion or deletion was identified in the GAII libraries, a 6 base deletion in nra-1 mRNA beginning at coding sequence base 772. The HiSeq libraries contained the same nra-1 deletion along with a single base insertion (“A”) at lev-10 mRNA base 602 that resulted in a frame-shift that introduced a downstream stop codon and a truncated protein of 236 residues (versus the 902 residue full length LEV-10 listed in Table S1).
Interestingly, there were no SNP or insertions/deletions that indicated the existence of RNAs that would encode truncated forms of UNC-63 as have been reported in levamisole-resistant isolates of 3 species of trichostrongylid nematodes [13]; this may represent a basic difference between the repertoire of resistance mechanisms utilized by different nematode families. In conclusion, the gene differential expression (Fig. 2, Table S4), SNP (Tables S5–6), and indel analyses failed to identify a gene whose characteristics of expression or sequence exhibited the high differential prevalence within LEVR vs SENS isolates that would indicate that gene’s central ability to (individually) confer levamisole resistance. Therefore the data fail to support the simplest explanation for resistance, that being that a single gene is responsible for the levamisole resistance of this O. dentatum isolate (as is the case in a number of nematode genera for resistance to the benzimidazoles, e.g. [7]). As such, the data support the alternative possibility, that resistance is polygenic and involves changing the mix of subunits that comprise nAChR receptors on muscle (by increasing ACR-21 and -25, and decreasing UNC-63) and by accumulating specific SNPs. Such a polygenic mechanism of resistance would be similar to that proposed for Ancylostoma caninum which has basis in a study that associates decreased expression of UNC-29, -38, and -63 with pyrantel resistance [14]. Given a polygenic mechanism, further characterization using the tools of transcriptomics/genomics will require an increased number of biological replicates. One caveat applicable to all transcriptomic assessments is the midlevel correlation between mRNA transcript abundance and protein abundance (reviewed in [15]). Consequently, it is important that these findings be extended not just with additional transcriptomic assessments but also with functional assays, e.g. electrophysical characterization of functional recombinant nAChR receptors expressed in Xenopus laevis eggs [16].
Supplementary Material
Highlights.
27 nAChR pathway genes in levamisole resistant versus sensitive O. dentatum were analyzed by transcriptomics
nAChR-subunit acr-21 and acr-25 exhibited increased mRNA in resistant isolate
nAChR-subunit unc-63 mRNA decreased in resistant isolate
by SNPs analysis, NRA-2 (H219N) and UNC-74 (V133I) increased in resistant isolate
by SNPs analysis, UNC-38 (E2G) and UNC-22 (I4063V) decreased in resistant isolate
Acknowledgments
We thank from Iowa State University Hui-Hsien Chou, Andrew Severin, and James Koltes for assistance with (respectively) general bioinformatics, RNA-Seq normalization, and GATK. We thank Claude Charvet (Institut de Biologie de l’École Normale Supérieure, Paris, France) for the use of non-public sequence information (JX429921 and JX429919).
Research funding was by The Hatch Act, State of Iowa, and by NIH grant R01 AI047194 National Institute of Allergy and Infectious Diseases. The funding agencies had no role in the design, execution or publication of this study.
Abbreviations
- nAChR
nicotinic acetylcholine receptor
- SNP
single nucleotide polymorphism
Footnotes
Conflict of interest statement
The authors have no financial or personal conflicts of interest or competing interests that could influence (bias) the work reported within this publication.
The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institute of Allergy and Infectious Diseases.
Animal use in this study was approved by the Iowa State University Animal Care and Use Committee.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Robertson AP, Buxton SK, Puttachary S, Williamson SM, Wolstenholme AJ, Neveu C, et al. Antinematodal Drugs- Modes of Action and Resistance: And Worms Will not come to Thee. In: Caffrey ER, editor. Parasitic Helminths, Targets, Screens, Drugs and Vaccines. Wiley-Blackwell; 2012. pp. 233–49. [Google Scholar]
- 2.Kaplan RM. Drug resistance in nematodes of veterinary importance: a status report. Trends Parasitol. 2004;20:477–81. doi: 10.1016/j.pt.2004.08.001. [DOI] [PubMed] [Google Scholar]
- 3.James CE, Hudson AL, Davey MW. Drug resistance mechanisms in helminths: is it survival of the fittest? Trends Parasitol. 2009;25:328–35. doi: 10.1016/j.pt.2009.04.004. [DOI] [PubMed] [Google Scholar]
- 4.Jones AK, Sattelle DB. The cys-loop ligand-gated ion channel gene superfamily of the nematode, Caenorhabditis elegans. Invertebrate Neuroscience. 2008;8:41–7. doi: 10.1007/s10158-008-0068-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.WHO. Preventive chemotherapy in human helminthiasis: coordinated use of anthelminthic drugs in control interventions: a manual for health professionals and programme managers. 2006 [Google Scholar]
- 6.Robertson AP, Bjorn HE, Martin RJ. Resistance to levamisole resolved at the single-channel level. FASEB J. 1999;13:749–60. doi: 10.1096/fasebj.13.6.749. [DOI] [PubMed] [Google Scholar]
- 7.Beech RN, Prichard RK, Scott ME. Genetic variability of the beta-tubulin genes in benzimidazole-susceptible and -resistant strains of Haemonchus contortus. Genetics. 1994;138:103–10. doi: 10.1093/genetics/138.1.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Martin RJ, Robertson AP, Buxton SK, Beech RN, Charvet CL, Neveu C. Levamisole receptors: a second awakening. Trends Parasitol. 2012;28:289–96. doi: 10.1016/j.pt.2012.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Romine NM, Martin RJ, Beetham JK. Computational cloning of drug target genes of a parasitic nematode, Oesophagostomum dentatum. BMC Genet. 2013;14:55. doi: 10.1186/1471-2156-14-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Blaxter ML, De Ley P, Garey JR, Liu LX, Scheldeman P, Vierstraete A, et al. A molecular evolutionary framework for the phylum Nematoda. Nature. 1998;392:71–5. doi: 10.1038/32160. [DOI] [PubMed] [Google Scholar]
- 11.Dillies MA, Rau A, Aubert J, Hennequet-Antier C, Jeanmougin M, Servant N, et al. A comprehensive evaluation of normalization methods for Illumina high-throughput RNA sequencing data analysis. Brief Bioinfor. 2012 doi: 10.1093/bib/bbs046. [DOI] [PubMed] [Google Scholar]
- 12.Koepke T, Schaeffer S, Krishnan V, Jiwan D, Harper A, Whiting M, et al. Rapid gene-based SNP and haplotype marker development in non-model eukaryotes using 3′UTR sequencing. BMC Genomics. 2012;13:18. doi: 10.1186/1471-2164-13-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Neveu C, Charvet CL, Fauvin A, Cortet J, Beech RN, Cabaret J. Genetic diversity of levamisole receptor subunits in parasitic nematode species and abbreviated transcripts associated with resistance. Pharmacogenet Genom. 2010;20:414–25. doi: 10.1097/FPC.0b013e328338ac8c. [DOI] [PubMed] [Google Scholar]
- 14.Kopp SR, Coleman GT, Traub RJ, McCarthy JS, Kotze AC. Acetylcholine receptor subunit genes from Ancylostoma caninum: altered transcription patterns associated with pyrantel resistance. Int J Parasitol. 2009;39:435–41. doi: 10.1016/j.ijpara.2008.08.005. [DOI] [PubMed] [Google Scholar]
- 15.Vogel C, Marcotte EM. Insights into the regulation of protein abundance from proteomic and transcriptomic analyses. Nat Rev Genet. 2012;13:227–32. doi: 10.1038/nrg3185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Williamson SM, Robertson AP, Brown L, Williams T, Woods DJ, Martin RJ, et al. The nicotinic acetylcholine receptors of the parasitic nematode Ascaris suum: formation of two distinct drug targets by varying the relative expression levels of two subunits. PLOS Pathog. 2009;5:e1000517. doi: 10.1371/journal.ppat.1000517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Boulin T, Fauvin A, Charvet CL, Cortet J, Cabaret J, Bessereau JL, et al. Functional reconstitution of Haemonchus contortus acetylcholine receptors in Xenopus oocytes provides mechanistic insights into levamisole resistance. Brit J Pharmacol. 2011;164:1421–32. doi: 10.1111/j.1476-5381.2011.01420.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hernando G, Berge I, Rayes D, Bouzat C. Contribution of subunits to Caenorhabditis elegans levamisole-sensitive nicotinic receptor function. Mol Pharmacol. 2012;82:550–60. doi: 10.1124/mol.112.079962. [DOI] [PubMed] [Google Scholar]
- 19.Lee JI, Mukherjee S, Yoon KH, Dwivedi M, Bandyopadhyay J. The multiple faces of calcineurin signaling in Caenorhabditis elegans: Development, behaviour and aging. J Biosciences. 2013;38:417–31. doi: 10.1007/s12038-013-9319-6. [DOI] [PubMed] [Google Scholar]
- 20.Wu S, Zhu Z, Fu L, Niu B, Li W. WebMGA: a customizable web server for fast metagenomic sequence analysis. BMC Genomics. 2011;12:444. doi: 10.1186/1471-2164-12-444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics (Oxford, England) 2009;25:2078–9. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bullard JH, Purdom E, Hansen KD, Dudoit S. Evaluation of statistical methods for normalization and differential expression in mRNA-Seq experiments. BMC Bioinformatics. 2010;11:94. doi: 10.1186/1471-2105-11-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Anders S, Huber W. Differential expression analysis for sequence count data. Genome Biolog. 2010;11:R106. doi: 10.1186/gb-2010-11-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics (Oxford, England) 2009;25:1754–60. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.