Abstract
Obesity is major public health concern worldwide and obese individuals exhibit a higher risk of chronic diseases such as type 2 diabetes. Inflammation plays a significant role in metabolic regulation and mounting evidence highlight the contribution of adipose tissue to systemic inflammatory state. Food extracts with a high content of (-)-epicatechin have been found to exert systemic anti-inflammatory actions, however the anti-inflammatory actions of (-)-epicatechin on adipose tissue remain to be determined. The aim of this study was to investigate the capacity of (-)-epicatechin to prevent tumor necrosis alpha (TNFα)-induced activation of cell signals involved in inflammation and insulin resistance (NF-κB, mitogen-activated protein kinases (MAPKs), AP-1, and peroxisome proliferator activated receptor γ (PPARγ)) in differentiated white adipocytes (3T3-L1). TNFα triggered the activation of transcription factors NF-κB and AP-1, and MAPKs ERK1/2, JNK, and p38. (-)-Epicatechin caused a dose (0.5-10 μM)-dependent decrease in TNFα-mediated JNK, ERK1/2, and p-38 phosphorylation, and nuclear AP-1-DNA binding. (-)-Epicatechin also inhibited TNFα-triggered activation of the NF-κB signaling cascade, preventing TNFα-mediated p65 nuclear transport and nuclear NF-κB-DNA binding. (-)-Epicatechin also attenuated the TNFα-mediated downregulation of PPARγ expression and decreased nuclear DNA binding. Accordingly, (-)-epicatechin inhibited TNFα-mediated altered transcription of genes (MCP-1, interleukin-6, TNFα, resistin, and protein-tyrosine phosphatase 1B) involved in inflammation and insulin signaling. In conclusion, (-)-epicatechin can attenuate TNFα-mediated triggering of signaling cascades involved in inflammation and insulin resistance. These findings could be of relevance in the dietary management of obesity and metabolic syndrome.
Keywords: Adipocytes, NF-κB, MAPK, PPAR, epicatechin, inflammation, insulin resistance
Introduction
Obesity and metabolic syndrome result from excess calorie intake and genetic predisposition, and obese individuals exhibit a higher risk of chronic diseases such as type 2 diabetes [1–3]. Mounting evidence highlight the contribution of adipose tissue to a systemic inflammatory state, which can play a significant role in metabolic regulation [4, 5]. Adiposity promotes the secretion of proinflammatory cytokines, such as tumor necrosis factor alpha (TNFα), interleukin (IL)-6, resistin, monocyte chemoattractant protein-1 (MCP-1), by both adipocytes and infiltrated inflammatory cells, including recruited macrophages. These proinflammatory cytokines contribute to the sustained adipose and systemic inflammation and insulin resistance associated with obesity [6]. A significant role of TNFα on these events was initially suggested by the findings that adipose-secreted TNFα contributed to insulin resistance in rodent models of obesity [7], and that obese mice lacking a functional TNFα are protected from dietary obesity-induced insulin resistance [8]
TNFα is a major player mediating the activation of signaling cascades in adipocytes that are central to inflammation and insulin resistance. In this regard, TNFα triggers adipocyte activation of the mitogen activated kinases (MAPKs) extracellular-signal-regulated kinases (ERK1/2), c-Jun N-terminal kinases (JNK), and p38, and of transcription factors AP-1 and nuclear factor kappa B (NF-κB) [9–11]. These signaling cascades are in part redox regulated, given that binding of TNFα to its receptor results in the activation of NADPH oxidase leading to an increase in oxidant production [12, 13]. Activation of these signaling pathways leads to an increased expression of pro-inflammatory cytokines, including IL-6, IL-8, IL-1β, and MCP-1. NF-κB also drives [14] an increased expression of protein-tyrosine phosphatase 1B (PTP1B), a negative regulator of insulin signaling, which dephosphorylates tyrosine residues of the insulin receptor (IR) and insulin receptor substrate1 (IRS1) [15]. Furthermore, TNFα down regulates the nuclear receptor transcription factor peroxisome proliferator-activator receptor gamma (PPARγ) that plays a major role in the regulation of both adipose glucose and lipid metabolism [14].
Flavonoids are polyphenolic compounds that are widely present in human diets. Epidemiological studies have shown an inverse relationship between consumption of flavonoid-rich foods and pathologies with inflammatory components [16]. The beneficial health effects of flavonoids can be in part attributed to their capacity to regulate oxidant production and pro-inflammatory signals [17]. Recent reports support an anti-inflammatory action of select polyphenols in adipocytes in cultures. Addition of a polyphenol-rich grape powder extract to human adipocytes, inhibits TNFα-triggered activation of MAPKs and NF-κB, and the expression of proteins involved in inflammation and insulin resistance [18]. Similarly, high concentrations of the phenolic compounds p-coumaric acid, quercetin and resveratrol prevent TNFα-induced increase in several parameters of inflammation and oxidative stress, and of decreased insulin sensitivity in 3T3-L1 [19] and human adipocytes [9]. Oligomerized grape seed polyphenols attenuated inflammatory events in co-cultures of adipocytes and macrophages, through mechanisms attributed to their antioxidative properties [20].
(-)-Epicatechin is one of the most abundant flavonoids in human diets, being present in high concentrations in grapes, cocoa, tea, and many other fruits and vegetables. (-)-Epicatechin has a basic chemical structure of two aromatic rings (A and B) linked by an oxygenated heterocycle (C) with a hydroxyl group in position 4. Consumption or supplementation with (-)-epicatechin or (-)-epicatechin-containing foods in humans and experimental animals are associated with the improvement of parameters related to cardiovascular disease, which pathology involves a significant proinflammatory component [21, 22].
In vitro, (-)-epicatechin modulates the production of reactive oxygen species, activation of NF-κB, and production of cytokines in Jurkat T cells and Hodgkin’s lymphoma cells [23, 24]. However, the capacity of (-)-epicatechin to attenuate inflammation in adipocytes has not been yet characterized. This study investigated in 3T3-L1 adipocytes, the capacity of (-)-epicatechin to inhibit TNFα-triggered deregulation of signaling pathways (MAPKs ERK1/2, JNK and p38, and transcription factors NF-κB, AP-1, and PPARγ) that regulate genes involved in inflammation and insulin resistance.
Materials and methods
Materials
3T3-L1 and Raw 264.7 cells were obtained from the American Type Culture Collection (Rockville, MA, USA). Cell culture media and reagents, and TRIzol reagent were obtained from Invitrogen Life Technologies (Carlsbad, CA, USA). Antibodies for ERK (sc-93), p-Erk (sc-7383), heterogeneous nuclear ribonucleoprotein A1 (sc-32301), JNK (sc-572), p-JNK (sc-6254), PPARγ (7273), MCP-1 (1785), TNFα (sc-1351); and the oligonucleotide containing the consensus sequence for PPAR were from Santa Cruz Biotechnology (Santa Cruz, CA, USA). The antibody for p-p38 (9211) was obtained from Cell Signaling Technology (Danvers, MA, USA). The antibody for PTP1B was from Abcam Inc. (Cambridge, MA). PVDF membranes were obtained from BIO-RAD (Hercules, CA, USA) and Chroma Spin-10 columns were from Clontech (Palo Alto, CA, USA). The ECL Western blotting system was from GE Healthcare (Piscataway, NJ, USA). The oligonucleotides containing the consensus sequence for NF-κB, and AP-1, the reagents for the electrophoretic mobility shift assays (EMSA), and cDNA reverse transcriptase were obtained from Promega (Madison, WI, USA). (-)-Epicatechin, TNFα, lipopolysaccharide (LPS), and all other reagents were from the highest quality available and were purchased from Sigma (St. Louis, MO, USA).
Cell culture and incubations
3T3-L1 preadipocytes were maintained in DMEM containing 25 mM glucose, 10% (v/v) new born calf serum, 50 U/ml penicillin, and 50 μg/ml streptomycin. To induce cell differentiation, 3T3-L1 pre-adipocytes were grown to confluence in culture medium containing 10% (v/v) fetal bovine serum. Confluent cells were then switched to differentiation medium (DIFM) containing 20% (v/v) fetal bovine serum, 20 nM insulin and 1 nM triiodothyronine for 48 h. Adipocyte differentiation was induced by treating cells for 48 h in DIFM further supplemented with 0.5 μM dexamethasone, 0.5 mM isobutylmethylxanthine, and 0.125 mM indomethacin (induction media). After induction, cells were returned to DIFM medium, exhibiting at day 12 a fully differentiated phenotype with massive accumulation of multilocular and unilocolar fat droplets. 3T3-L1 adipocytes were treated with 0.5–10 μM (-)-epicatechin during 4 h, and subsequently treated with TNFα (20 ng/ml) for 15 min-24 h depending on the determination.
RAW 264.7 macrophages were cultured in DMEM medium supplemented with 10% (v/v) fetal bovine serum. Cells were pre-incubated for 4 h in the absence or the presence of 1 and 10 μM (-)-epicatechin, and subsequently for 24 h without or with 0.5 μg/ml lypopolisacharide (LPS). TNFα concentration in the cell culture media was determined by ELISA using a commercial kit (BD Biosciences, San Diego, CA) according to the manufacturer's protocol.
Western blot analysis
To prepare total extracts, cells were rinsed with PBS, scraped and centrifuged. The pellet was rinsed with PBS, and resuspended in 200 μL of 50 mM HEPES (pH 7.4), 125 mM KCl contained containing protease inhibitors and 2% (v/v) Igepal. The final concentration of the inhibitors was 0.5 mmol/L PMSF, 1 mg/L leupeptin, 1 mg/L pepstatin, 1.5 mg/L aprotinin, 2 mg/L pestatin and 0.4 mM sodium pervanadate. Samples were exposed to one cycle of freezing and thawing, incubated at 4° C for 30 min and centrifuged at 15,000 x g for 30 min. The supernatant was decanted and protein concentration was measured (Bradford, 1976). Aliquots of total cell lysates containing 25–40 μg protein were denatured with Laemmli buffer, separated by reducing 10–12.5% (w/v) polyacrylamide gel electrophoresis, and electroblotted to PVDF membranes. Membranes were blotted for 2 h in 5% (w/v) non-fat milk, and subsequently incubated in the presence of corresponding primary antibodies (1:1,000 dilution for all the antibodies except for TNFα (1:500) and PTP-1B (1:5,000)) overnight at 4°C. After incubation for 90 min at room temperature in the presence of the secondary antibody (HRP conjugated) (1:10,000 dilution) the conjugates were visualized and quantified by chemiluminescence detection in a Phosphoimager 840 (Amersham Pharmacia Biotech. Inc., Piscataway, NJ).
Electrophoretic mobility shift assay (EMSA)
Nuclear fractions were isolated as previously described [25, 26] with minor modifications [27]. For the EMSA, the oligonucleotides containing the consensus sequences for NF-κB, AP-1 and PPARγ were end-labelled with [γ-32P] ATP using T4 polynucleotide kinase, and purified using Chroma Spin-10 columns. Samples were incubated with the labelled oligonucleotide (20,000-30,000 cpm) for 20 min at room temperature in 1X binding buffer [5X binding buffer: 50 mM Tris-HCl buffer, pH 7.5, containing 20% (v/v) glycerol, 5 mM MgCl2, 2.5 mM EDTA, 2.5 mM DTT, 250 mM NaCl, and 0.25 mg/ml poly(dI-dC)]. The products were separated by electrophoresis in a 6% (w/v) non-denaturing polyacrilamide gel using 0.5 X TBE (Tris/borate 45 mM, EDTA 1mM) as the running buffer. The gels were dried and the radioactivity quantitated in a Phosphoimager 840.
RNA isolation and real-time PCR
RNA was extracted from cells using TRIzol reagent. cDNA was generated using high-capacity cDNA reverse transcriptase. IL-6, MCP-1, resistin and TNFα mRNA levels were assessed by quantitative real-time PCR (iCycler, BioRad) using appropriate primers (Table 1) and normalized to β-actin mRNA content.
Table 1.
Primers used for RT-PCR
| Gene | 5’ 3’ sequence | |
|---|---|---|
| β-Actin | f | CAGGGTGTGAGTTGGGGAATG |
| r | CGCACGATTTCCCTCTCAGCTG | |
| IL-6 | f | ACAACCACGGCCTTCCCTACTT |
| r | CAGGATTTCCCAGCGAACATGTG | |
| MCP-1 | f | GCCCCACTCACCTGCTGCTACT |
| r | CCTGCTGCTGGTGATCCTCTTGT | |
| Resistin | f | CAGAAGGCACAGCAGTCTTG |
| r | GACCGGAGGACATCAGACAT | |
| TNFα | f | AGCCCCCAGTCTGTATCCTT |
| r | CTCCCTTTGCAGAACTCAGG | |
Statistical analysis
Data were analyzed by one-way analysis of variance (ANOVA) using Statview 5.0 (SAS Institute Inc., Cary, NC, USA). Fisher least significance difference test was used to examine differences between group means. A p value < 0.05 was considered statistically significant. Data are shown as mean ± SEM.
Results
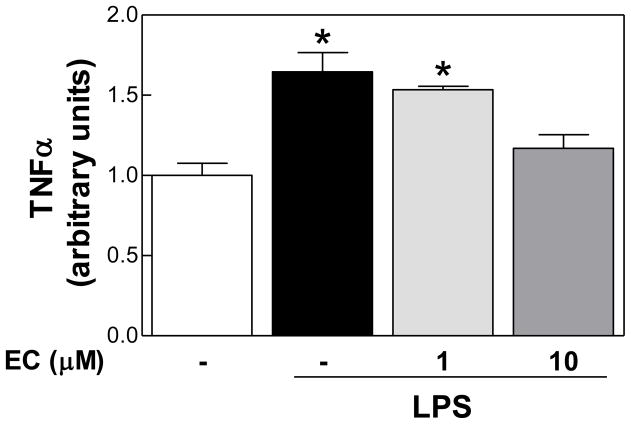
(-)-Epicatechin decreased LPS-induced release of TNFα in RAW 264.7 macrophages
Macrophage infiltration is a key event in sustained adipose tissue inflammation. Thus, the effects of (-)-epicatechin on LPS-induced production of TNFα in RAW 264.7 mouse macrophage cells were initially evaluated. Incubation of RAW 264.7 cells with 0.5 μg/ml LPS caused a 65% increase in TNFα secretion. Notably, simultaneous incubation of cells with LPS and 1 μM (-)-epicatechin had no effect, while with 10 μM (-)-epicatechin the increased release of TNFα to the incubation media triggered by LPS was completely prevented (Fig. 1).
Figure 1. (-)-Epicatechin inhibits LPS-induced TNFα release in RAW 264.7 macrophages.
RAW 264.7 cells were incubated without or with (-)-epicatechin (EC) (1 and 10 μM) for 4 h, and subsequently in the absence or presence of 0.5 μg/ml LPS for further 24 h. TNFα concentration in the media was determined by ELISA and results were referred to untreated cell values (Arbitrary unit=1). Data represent means ± SEM of three independent experiments. *Significantly different compared to control (no additions) (p< 0.05, One way ANOVA test).
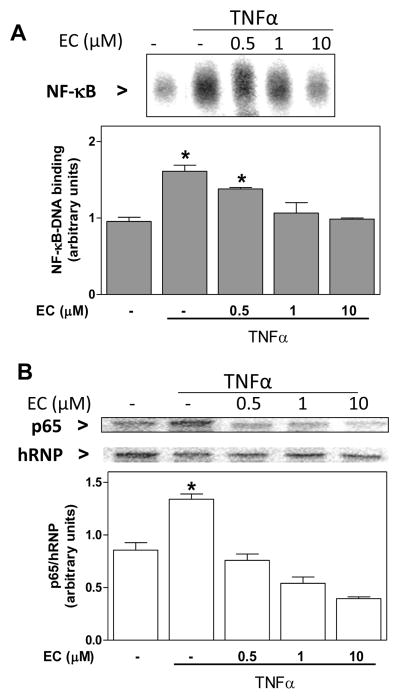
(-)-Epicatechin prevents TNFα-induced NF-κB activation in 3T3-L1 adipocytes
NF-κB is at the core of chronic inflammation. Thus, we investigated the effects of (-)-epicatechin on TNFα (20 ng/ml)-induced NF-κB activation measuring NF-κB-DNA binding in nuclear fractions by EMSA. Adipocytes incubated for 2 h in the presence of TNFα showed a 61% increase in NF-κB-DNA binding compared with controls (Fig. 2A). When cells were incubated with (-)-epicatechin (0.5–10 μM) prior and during the incubation with TNFα, a dose-dependent inhibition in NF-κB-DNA binding was observed. The inhibition was complete at 1 μM (-)-epicatechin. To evaluate if the effects of (-)-epicatechin were due to (-)-epicatechin interactions with NF-κB proteins, or to upstream events, the nuclear content of the NF-κB protein p65 (RelA) was evaluated by Western blot. TNFα caused a 56% increase in p65 nuclear content, and incubation with (-)-epicatechin (0.5–10 μM) prevented this increase (Fig. 2B).
Figure 2. (-)-Epicatechin prevents TNFα-induced NF-κB activation in 3T3-L1 adipocytes.
3T3-L1 adipocytes were incubated without or with (-)-epicatechin (EC) (0.5-10 μM) for 4 h, and subsequently in the absence or presence of 20 ng/ml TNFα for further 2 h. NF-κB activation was evaluated measuring, A- nuclear NF-κB-DNA binding by EMSA; and B- nuclear p65 (Rel A)levels by Western blot. Upper panels: representative images. A,B- Bands were quantified and results were referred to untreated cell values (Arbitrary unit=1). Data represent means ± SEM of three-four independent experiments. p65 levels were referred to the nuclear content of heterogeneous nuclear ribonucleoprotein (hRNP). *Significantly different compared to all other groups (p < 0.05, One way ANOVA test).
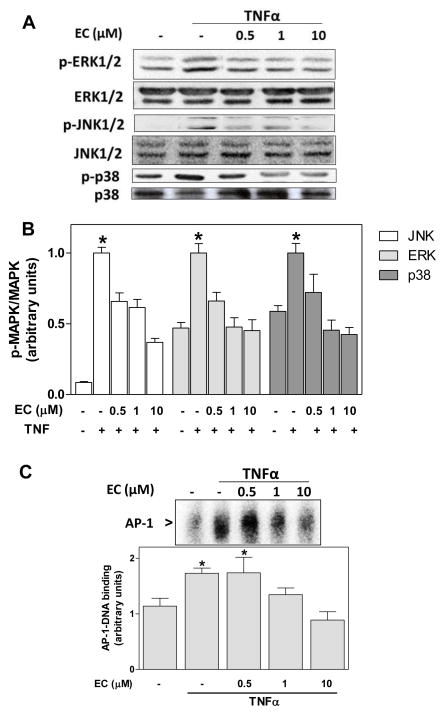
(-)-Epicatechin prevents TNFα-induced MAPKs and AP-1 activation in 3T3-L1 adipocytes
Activation of the MAPKs occurs downstream the binding of TNFα to its receptor. In 3T3-L1 adipocytes, TNFα (20 ng/ml) caused a 12- and 2-fold increase in JNK1/2 (Thr 183, Tyr 185) and ERK1/2 (Tyr 204) phosphorylation, respectively, and a 60% increase in p38 (Thr180/Tyr182) phosphorylation after 15 min incubation (Fig. 3A,B). Preincubation of cells for 4 h in the presence of 0.5–10 μM (-)-epicatechin caused a dose-dependent inhibition of TNFα-induced MAPKs phosphorylation (Fig. 3A,B). At 1 μM (-)-epicatechin, the inhibition was complete for ERK1/2 and p38 phosphorylation, and partial (33%) for JNK1/2.
Figure 3. (-)-Epicatechin prevents TNFα-induced MAPK and AP-1 activation in 3T3-L1 adipocytes.
3T3-L1 adipocytes were incubated without or with (-)-epicatechin (EC) (0.5-10 μM) for 4 h, and subsequently in the absence or presence of 20 ng/ml TNFα for further 15 min (MAPKs) or 2 h (AP-1): A- Representative images and B- quantification of Western blots for phosphorylated and non-phosphorylated JNK1/2, ERK1/2, and p38 levels in total cell extracts; C- representative image and quantification of AP-1-DNA binding in nuclear fractions as determined by EMSA. B-C, bands were quantified and results were referred to untreated cell values (Arbitrary unit=1). For Western blots (B) results were expressed as the ratio phosphorylated/non phosphorylated protein levels. Data represent means ± SEM of three to four independent experiments. *Significantly different compared to all other groups (p < 0.05, One way ANOVA test).
Activation of the transcription factor AP-1, a downstream MAPKs target, was subsequently investigated. Incubation of 3T3-L1 adipocytes in the presence of 20 ng/ml TNFα caused a 52% increase in AP-1-DNA binding (Fig. 3C). Preincubation with 1 and 10 μM (-)-epicatechin prevented this increase.
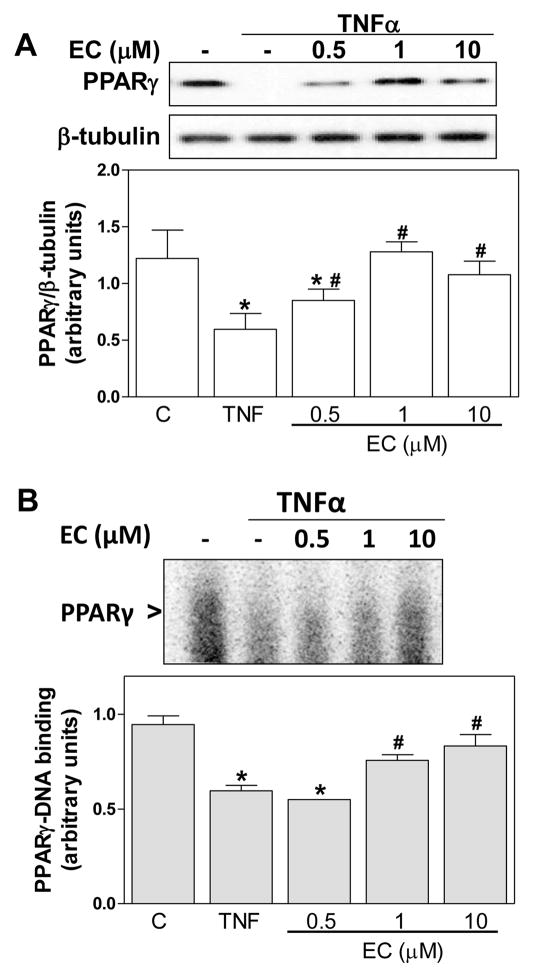
(-)-Epicatechin prevents TNFα-induced PPARγ downregulation in 3T3-L1 adipocytes
TNFα downregulates PPARγ, which is a major modulator of adipogenesis. In 3T3-L1 adipocytes, and after 24 h incubation with TNFα, a 51% decrease in PPARγ protein levels was observed in total cell lysates. (-)-Epicatechin prevented this decrease in a dose (0.5-10μM)-dependent manner (Fig. 4A). Consistent with these findings, the incubation with TNFα for 2 h caused a 37% decrease in PPARγ-DNA binding in nuclear fractions compared with control cells (Fig. 4B). (-)-Epicatechin caused a dose-dependent prevention of this decrease that reached a 50% at 1 μM (-)-epicatechin.
Figure 4. (-)-Epicatechin prevents TNFα-induced PPARγ downregulation in differentiated 3T3-L1 adipocytes.
3T3-L1 adipocytes were incubated without or with (-)-epicatechin (EC) (0.5-10 μM) for 4 h, and subsequently in the absence or presence of 20 ng/ml TNFα for further 24 h (PPARγ protein levels) or 2 h (PPARγ-DNA binding). A- Representative image and quantification of Western blots for PPARγ and β-tubulin in total cell extracts, and B- Representative image and quantification of PPARγ-DNA binding in nuclear fractions as determined by EMSA. A,B- Bands were quantified and results were referred to untreated cell values (Arbitrary unit=1). For Western blots (A), results were expressed as the ratio PPARγ/β-tubulin protein levels. Results are shown as mean ± SEM of 3 independent experiments. Values having different superscripts are significantly different (p<0.05, one way ANOVA test).
(-)-Epicatechin prevents TNFα-induced transcriptional activity involved in inflammation and insulin resistance
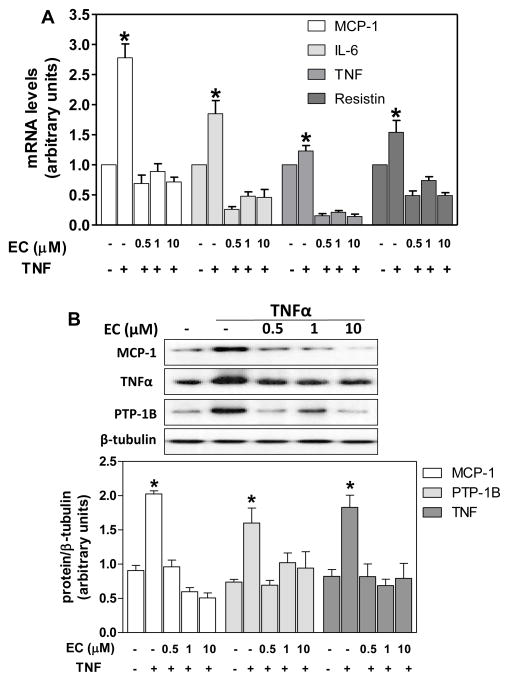
The capacity of (-)-epicatechin to modulate the expression of genes regulated by NF-κB, MAPKs, and AP-1 was next investigated. Incubation of 3T3-L1 adipocytes for 24 h in the presence of TNFα, led to a marked increase in the mRNA levels of the cytokines IL-6, MCP-1, and TNFα, as assessed by RT-PCR (Fig. 5A). At all the concentrations tested (0.5-10 μM), simultaneous incubation with (-)-epicatechin, prevented those increases. Similarly, TNFα caused a 54% increase in the mRNA level of the adipocytokine resistin, and (-)-epicatechin (0.5-10 μM) prevented this increase (Fig. 5A).
Figure 5. (-)-Epicatechin prevents TNFα-induced expression of PTP1B and of proinflammatory genes.
3T3-L1 adipocytes were incubated without or with (-)-epicatechin (EC) (0.5-10 μM) for 4 h, and subsequently in the absence or presence of 20 ng/ml TNFα for further 24 h. A-mRNA levels for MCP-1, IL-6, TNFα and resistin were measured by quantitative real-time PCR, normalized against β-actin and expressed relative to control cells (Arbitrary unit=1). B- MCP-1, PTP1B, and TNFα protein levels were measured by Western blot. Bands were quantified, results expressed as the ratio protein/β-tubulin protein levels, and referred to untreated cell values (1). Data represent means ± SEM of three independent experiments. *Significantly different from all other groups (p<0.05, one way ANOVA test).
Similarly to that observed for the mRNA levels, the incubation of 3T3-L1 adypocytes with TNFα for 24 h caused a 2.2-fold increase in MCP-1 and TNFα protein levels as evaluated by Western blot (Fig. 5B).. At all the tested concentrations, (-)-epicatechin prevented these increases. TNFα also led to a significant increase in PTP1B protein levels, which was prevented by simultaneous incubation with 0.5–10 μM (-)-epicatechin (Fig. 5B).
Discussion
Chronic inflammation is one of the major contributors to white adipose tissue insulin resistance in obesity. This is due to the associated production of pro-inflammatory cytokines (TNFα, IL-6, IL-1β) [28]. These pro-inflammatory cytokines drive the recruitment and activation of macrophages and other immune cells that completes a cycle of inflammation and impaired metabolic function [6]. The current work demonstrates that, at concentrations compatible with those found in human tissues (high nanomolar-low micromolar range) after dietary consumption [29-32] , (-)-epicatechin inhibited TNFα-induced deregulation of adipocyte cell signals (NF-κB, MAPKs, AP-1, PPARγ) and the downstream upregulation of genes involved in inflammation and insulin resistance.
(-)-Epicatechin not only showed a capacity to protect adipocytes from TNFα deleterious effects, but was also effective inhibiting LPS-induced TNFα release by RAW 264.7 macrophages. At higher (-)-epicatechin and catechin concentrations, a similar finding of TNFα release prevention was described in RAW 264.7 macrophages either unstimulated or stimulated with interferon gamma [33]. Thus, the capacity of (-)-epicatechin to prevent obesity-related inflammation could be due to its actions at both, the infiltrating macrophages and adipocytes.
TNFα is a major player in adipose tissue chronic inflammation triggering the activation/inhibition of signaling cascades that perpetuate the inflammatory scenario, cause insulin resistance and hyperlipemia [10, 28]. In this regard, TNFα acts activating MAPKs, and transcription factors AP-1 and NF-κB [10], and downregulating PPARγ. MAPKs, NF-κB and AP-1 activation can act promoting adipocyte inflammation and insulin resistance by: i) antagonizing PPARγ [10], ii) inducing the expression of pro-inflammatory genes and signaling proteins [10], iii) antagonizing the insulin signaling through a direct modulation of IRS-1/2 expression and phosphorylation [10, 34, 35] and by increasing the expression of PTP1B (NF-κB) [36], and iv) modulating the oxidative stress condition associated with inflammation. Even at low concentrations (0.5–1 μM), (-)-epicatechin inhibited partially or totally TNFα-induced MAPKs, NF-κB and AP-1 activation. Accordingly, TNF-α-dependent transcription of pro-inflammatory genes (MCP-1, IL-6, TNFα, resistin) was inhibited by (-)-epicatechin. PTP1B expression was upregulated by TNFα, as previously described in cultured cells and in mice [36]. The expression of this protein, central to the modulation of insulin signaling, is regulated by NF-κB [36]. Significantly, (-)-epicatechin inhibited TNFα-induced PTP1B expression at all concentrations tested. (-)-Epicatechin also prevented TNFα-mediated increase of resistin, a proinflammatory adipocytokine which expression is in part regulated by ERK1/2 and NF-κB [37]. It has been proposed that resistin links adiposity and diabetes [38], being found elevated in conditions of obesity and insulin resistance
PPARγ binds different ligands (e.g. fatty acids) and is the central modulator of adipogenesis, improving adipocyte insulin sensitivity, increasing adipocyte lipid uptake, and regulating the expression of adipokines (upregulation of adiponectin, downregulation of TNFα, MCP-1, resistin) (reviewed in [14]). At the systemic level, PPARγ-induced changes in adipokines cause a decrease in hepatic lipid accumulation and glucose output, increasing fatty acid oxidation. Importantly, PPARγ also changes macrophages from M1 into an anti-inflammatory M2 phenotype, and decreases macrophage adipose tissue infiltration [28]. (-)-Epicatechin acted decreasing TNFα-mediated downregulation of PPARγ in 3T3-L1 adipocytes. This positive effect of (-)-epicatechin on PPARγ could contribute to improve the loss of insulin sensitivity associated with adipose tissue inflammation.
Flavonoids have been previously shown to have beneficial effects in adipocytes. Quercetin reduced visceral adipose inflammation in obese Zucker rats [39]. In vitro, quercetin (10-60 μM) inhibited TNFα-induced MAPK, NF-κB and AP-1 activation and PPARγ inhibition, improving inflammatory and insulin resistance parameters in primary cultures of human adipocytes [9]. Similar effects were observed for a grape extract containing catechins, although the complex nature of the extract does not allow a direct extrapolation to a particular active component [20]. (-)-Catechin, an (-)-epicatechin isomer, activates PPARγ and promotes adipocyte differentiation [40]. In immune (Jurkat T) cells (-)-epicatechin, (-)-catechin, and the B2 epicatechin dimer inhibits phorbol 12-myristate 13-acetate (PMA)-induced NF-κB activation, in part through a decrease in the associated increased cell oxidant production [23]. Furthermore, (-)-epicatechin, (-)-catechin, and dimers B1 and B2 can directly interact with NF-κB proteins preventing its binding to κB DNA sites [23, 41]. Both in PMA-stimulated Jurkat T cells and in Hodgkin’s lymphoma cells, (-)-epicatechin inhibited NF-κB-regulated cytokine production [23, 24]. Accordingly, our current findings demonstrate that (-)-epicatechin also acts inhibiting TNFα-triggered NF-κB activation, which in conjunction with its inhibitory effects on MAPKs and AP-1, leads to a decreased cytokine and PTP1B expression in 3T3-L1 adipocytes.
The mechanisms of (-)-epicatechin protective actions in adipocytes challenged with TNFα cannot be addressed from the current results. It is only evident that the inhibitory actions of (-)-epicatechin on NF-κB activation do not occur at the nucleus, but upstream, given it prevents TNFα-triggered p65 nuclear translocation. Although the concentrations of flavonoids attainable in adipose tissue would not afford a direct antioxidant mechanism of protection [42], the regulation of NADPH oxidase by (-)-epicatechin (EC) and its metabolites [43, 44], could contribute to prevent TNFα-induced and redox regulated MAPK, NF-κB and AP-1 activation.
The obtained results demonstrate that (-)-epicatechin, at concentrations that are attainable in human tissues, prevent the deregulation of key signaling cascades (NF-κB, MAPKs, AP-1, PPARγ) triggered by an inflammatory stimuli (TNFα). These signaling pathways are central to the regulation, among others, of genes required for insulin sensitivity, glycogen and fatty acid synthesis, transcriptional regulation, and cytokine-mediated inflammation [10, 45]. Thus, the obtained results suggest that (-)-epicatechin-containing diets could improve obesity-associated impaired insulin sensitivity and sustained inflammation.
(-)-Epicatechin (EC) inhibits TNFα-triggered inflammatory response in adipocytes.
MAPK, NF-κB, and AP-1 inhibition associates with reduced pro-inflammatory gene transcription.
In parallel, EC prevents PPARγ downregulation, central to energy metabolism.
In conclusion, EC could provide benefits against obesity-associated inflammation.
Acknowledgments
This work was supported by the University of California, Davis; a Research Grant from the Juvenile Diabetes Research Foundation (1-2009-337) and NIH grant R56 DK084317 to F.G.H.; a grant from the University of Buenos Aires (01/1111), Argentina to CGF. CGF and MVP are researchers, and PIO correspondent researcher from CONICET, Argentina. MVP was a recipient of a fellowship from CONICET and from Program I+D 2010, Argentina.
Abbreviations
- DIFM
differentiation medium
- EMSA
electrophoretic mobility shift assay
- ERK
extracellular-signal-regulated kinases
- IL
interleukin
- IR
insulin receptor
- IRS-1
insulin receptor subtrates-1
- JNK
c-Jun N-terminal kinases
- LPS
lipopolysaccharide
- MAPKs
mitogen-activated protein kinases
- MCP-1
monocyte chemoattractant protein 1
- NF-κB
nuclear factor kappa B
- PPARγ
peroxisome proliferator-activator receptor gamma
- PTP1B
protein-tyrosine phosphatase 1B
- TNFα
tumor necrosis factor alpha
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Friedman JM. Nature. 2000;404:632–634. doi: 10.1038/35007504. [DOI] [PubMed] [Google Scholar]
- 2.Spiegelman BM, Flier JS. Cell. 2001;104:531–543. doi: 10.1016/s0092-8674(01)00240-9. [DOI] [PubMed] [Google Scholar]
- 3.Mensah GA, Mokdad AH, Ford E, Narayan KM, Giles WH, Vinicor F, Deedwania PC. Cardiol Clin. 2004;22:485–504. doi: 10.1016/j.ccl.2004.06.005. [DOI] [PubMed] [Google Scholar]
- 4.Hotamisligil GS. Nature. 2006;444:860–867. doi: 10.1038/nature05485. [DOI] [PubMed] [Google Scholar]
- 5.Hotamisligil GS. International journal of obesity 2005. 2008;32(Suppl 7):S52–54. doi: 10.1038/ijo.2008.238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gregor MF, Hotamisligil GS. Annu Rev Immunol. 2011;29:415–445. doi: 10.1146/annurev-immunol-031210-101322. [DOI] [PubMed] [Google Scholar]
- 7.Hotamisligil GS, Shargill NS, Spiegelman BM. Science. 1993;259:87–91. doi: 10.1126/science.7678183. [DOI] [PubMed] [Google Scholar]
- 8.Uysal KT, Wiesbrock SM, Marino MW, Hotamisligil GS. Nature. 1997;389:610–614. doi: 10.1038/39335. [DOI] [PubMed] [Google Scholar]
- 9.Chuang CC, Martinez K, Xie G, Kennedy A, Bumrungpert A, Overman A, Jia W, McIntosh MK. Am J Clin Nutr. 2010;92:1511–1521. doi: 10.3945/ajcn.2010.29807. [DOI] [PubMed] [Google Scholar]
- 10.Ruan H, Hacohen N, Golub TR, Van Parijs L, Lodish HF. Diabetes. 2002;51:1319–1336. doi: 10.2337/diabetes.51.5.1319. [DOI] [PubMed] [Google Scholar]
- 11.Gil A, Maria Aguilera C, Gil-Campos M, Canete R. Br J Nutr. 2007;98(Suppl 1):S121–126. doi: 10.1017/S0007114507838050. [DOI] [PubMed] [Google Scholar]
- 12.Yang B, Rizzo V. Am J Physiol Heart Circ Physiol. 2007;292:H954–962. doi: 10.1152/ajpheart.00758.2006. [DOI] [PubMed] [Google Scholar]
- 13.Jung Y, Kim H, Min SH, Rhee SG, Jeong W. J Biol Chem. 2008;283:23863–23871. doi: 10.1074/jbc.M803072200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tontonoz P, Spiegelman BM. Annu Rev Biochem. 2008;77:289–312. doi: 10.1146/annurev.biochem.77.061307.091829. [DOI] [PubMed] [Google Scholar]
- 15.Nieto-Vazquez I, Fernandez-Veledo S, Kramer DK, Vila-Bedmar R, Garcia-Guerra L, Lorenzo M. Arch Physiol Biochem. 2008;114:183–194. doi: 10.1080/13813450802181047. [DOI] [PubMed] [Google Scholar]
- 16.Mennen LI, Sapinho D, de Bree A, Arnault N, Bertrais S, Galan P, Hercberg S. J Nutr. 2004;134:923–926. doi: 10.1093/jn/134.4.923. [DOI] [PubMed] [Google Scholar]
- 17.Fraga CG, Galleano M, Verstraeten SV, Oteiza PI. Mol Aspects Med. 2010;31:435–445. doi: 10.1016/j.mam.2010.09.006. [DOI] [PubMed] [Google Scholar]
- 18.Chuang CC, Bumrungpert A, Kennedy A, Overman A, West T, Dawson B, McIntosh MK. J Nutr Biochem. 2011;22:89–94. doi: 10.1016/j.jnutbio.2009.12.002. [DOI] [PubMed] [Google Scholar]
- 19.Yen GC, Chen YC, Chang WT, Hsu CL. J Agric Food Chem. 2011;59:546–551. doi: 10.1021/jf1036992. [DOI] [PubMed] [Google Scholar]
- 20.Sakurai T, Kitadate K, Nishioka H, Fujii H, Kizaki T, Kondoh Y, Izawa T, Ishida H, Radak Z, Ohno H. J Nutr Biochem. 2010;21:47–54. doi: 10.1016/j.jnutbio.2008.10.003. [DOI] [PubMed] [Google Scholar]
- 21.Fraga CG, Actis-Goretta L, Ottaviani JI, Carrasquedo F, Lotito SB, Lazarus S, Schmitz HH, Keen CL. Clin Dev Immunol. 2005;12:11–17. doi: 10.1080/10446670410001722159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Galleano M, Oteiza PI, Fraga CG. J Cardiovasc Pharmacol. 2009;54:483–490. doi: 10.1097/FJC.0b013e3181b76787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mackenzie GG, Carrasquedo F, Delfino JM, Keen CL, Fraga CG, Oteiza PI. FASEB J. 2004;18:167–169. doi: 10.1096/fj.03-0402fje. [DOI] [PubMed] [Google Scholar]
- 24.Mackenzie GG, Oteiza PI. Free Radic Res. 2006;40:1086–1094. doi: 10.1080/10715760600788396. [DOI] [PubMed] [Google Scholar]
- 25.Dignam JD, Lebovitz RM, Roeder RG. Nucleic Acids Res. 1983;11:1475–1489. doi: 10.1093/nar/11.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Osborn L, Kunkel S, Nabel GJ. Proc Natl Acad Sci U S A. 1989;86:2336–2340. doi: 10.1073/pnas.86.7.2336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Erlejman AG, Jaggers G, Fraga CG, Oteiza PI. Arch Biochem Biophys. 2008 doi: 10.1016/j.abb.2008.01.024. [DOI] [PubMed] [Google Scholar]
- 28.Wellen KE, Hotamisligil GS. J Clin Invest. 2005;115:1111–1119. doi: 10.1172/JCI25102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ottaviani JI, Momma TY, Kuhnle GK, Keen CL, Schroeter H. Free Radic Biol Med. 2011 doi: 10.1016/j.freeradbiomed.2011.12.010. [DOI] [PubMed] [Google Scholar]
- 30.Mullen W, Borges G, Donovan JL, Edwards CA, Serafini M, Lean ME, Crozier A. Am J Clin Nutr. 2009;89:1784–1791. doi: 10.3945/ajcn.2008.27339. [DOI] [PubMed] [Google Scholar]
- 31.Baba S, Osakabe N, Yasuda A, Natsume M, Takizawa T, Nakamura T, Terao J. Free Radic Res. 2000;33:635–641. doi: 10.1080/10715760000301151. [DOI] [PubMed] [Google Scholar]
- 32.Holt RR, Lazarus SA, Sullards MC, Zhu QY, Schramm DD, Hammerstone JF, Fraga CG, Schmitz HH, Keen CL. 2002;76:4. doi: 10.1093/ajcn/76.4.798. [DOI] [PubMed] [Google Scholar]
- 33.Park YC, Rimbach G, Saliou G, Valacchi G, Packer L. FEBS Letters. 2000;465:93–97. doi: 10.1016/s0014-5793(99)01735-4. [DOI] [PubMed] [Google Scholar]
- 34.Aguirre V, Uchida T, Yenush L, Davis R, White MF. J Biol Chem. 2000;275:9047–9054. doi: 10.1074/jbc.275.12.9047. [DOI] [PubMed] [Google Scholar]
- 35.Fujishiro M, Gotoh Y, Katagiri H, Sakoda H, Ogihara T, Anai M, Onishi Y, Ono H, Abe M, Shojima N, Fukushima Y, Kikuchi M, Oka Y, Asano T. Mol Endocrinol. 2003;17:487–497. doi: 10.1210/me.2002-0131. [DOI] [PubMed] [Google Scholar]
- 36.Zabolotny JM, Kim YB, Welsh LA, Kershaw EE, Neel BG, Kahn BB. J Biol Chem. 2008;283:14230–14241. doi: 10.1074/jbc.M800061200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang BW, Hung HF, Chang H, Kuan P, Shyu KG. Am J Physiol Heart Circ Physiol. 2007;293:H2305–2312. doi: 10.1152/ajpheart.00361.2007. [DOI] [PubMed] [Google Scholar]
- 38.Steppan CM, Bailey ST, Bhat S, Brown EJ, Banerjee RR, Wright CM, Patel HR, Ahima RS, Lazar MA. Nature. 2001;409:307–312. doi: 10.1038/35053000. [DOI] [PubMed] [Google Scholar]
- 39.Rivera L, Moron R, Sanchez M, Zarzuelo A, Galisteo M. Obesity (Silver Spring) 2008;16:2081–2087. doi: 10.1038/oby.2008.315. [DOI] [PubMed] [Google Scholar]
- 40.Shin DW, Kim SN, Lee SM, Lee W, Song MJ, Park SM, Lee TR, Baik JH, Kim HK, Hong JH, Noh M. Biochem Pharmacol. 2009;77:125–133. doi: 10.1016/j.bcp.2008.09.033. [DOI] [PubMed] [Google Scholar]
- 41.Mackenzie GG, Delfino JM, Keen CL, Fraga CG, Oteiza PI. Biochem Pharmacol. 2009;78:1252–1262. doi: 10.1016/j.bcp.2009.06.111. [DOI] [PubMed] [Google Scholar]
- 42.Fraga CG, Oteiza PI. Free Radic Biol Med. 2011;51:813–823. doi: 10.1016/j.freeradbiomed.2011.06.002. [DOI] [PubMed] [Google Scholar]
- 43.Steffen Y, Gruber C, Schewe T, Sies H. Arch Biochem Biophys. 2008;469:209–219. doi: 10.1016/j.abb.2007.10.012. [DOI] [PubMed] [Google Scholar]
- 44.Steffen Y, Schewe T, Sies H. Biochem Biophys Res Commun. 2007;359:828–833. doi: 10.1016/j.bbrc.2007.05.200. [DOI] [PubMed] [Google Scholar]
- 45.Stephens JM, Lee J, Pilch PF. J Biol Chem. 1997;272:971–976. doi: 10.1074/jbc.272.2.971. [DOI] [PubMed] [Google Scholar]