Abstract
A meta-analysis in Caucasians was conducted to investigate the possible association of uridine diphosphate glucuronosyltransferase (UGT) 1A1 gene polymorphisms with irinotecan (IRI)-induced neutropenia and diarrhoea in colorectal cancer (CRC). We searched PubMed and Embase until May 2012 to identify eligible studies, extracted data, assessed methodological quality, and performed statistical analysis using REVMAN 5.1 and R software. Subgroups meta-analyses were performed in groups representing different IRI combination regimens and IRI doses. Sixteen trials were included. UGT1A1*28/*28 genotype was associated with more than fourfold (odds ratio (OR)=4.79, 95% confidence intervals (CI): 3.28–7.01; P<0.00001) and threefold (OR=3.44, 95% CI: 2.45–4.82; P<0.00001) increases in the risk of neutropenia when compared with wild type and with at least one UGT1A1*1 allele, respectively. UGT1A1*1/*28 genotype had an OR of 1.90 (95% CI: 1.44–2.51; P<0.00001) for an increased risk of neutropenia. A twofold increase in risk of diarrhoea was associated with UGT1A1*28/*28 genotype (OR=1.84, 95% CI: 1.24–2.72; P=0.002). In subgroup meta-analysis, the higher incidence of diarrhoea in UGT1A1*28/*28 patients was limited to studies where when IRI was given at higher doses (OR=2.37, 95% CI: 1.39–4.04; P=0.002) or combined with 5-fluorouracil (FU or analogue) (OR=1.78, 95% CI: 1.16–2.75; P=0.009). Genotyping of UGT1A1*28 polymorphism before treatment for CRC can tailor IRI therapy and reduce the IRI-related toxicities. IRI-combined 5-FU (or analogue) and a high-dose IRI therapy enhance IRI-induced diarrhoea among patients bearing the UGT1A1*28 allele. Although the toxicity relationships were much stronger with the UGT1A1*28 homozygous variant, associations were also found with the UGT1A1*28 heterozygous variant.
Keywords: colorectal cancer, irinotecan, meta-analysis, polymorphism, toxicities, UGT1A1*28
Introduction
Colorectal cancer (CRC) is the third most common cancer worldwide1 and frequently treated with irinotecan (IRI).2 IRI-based combination therapy demonstrated superiority in overall response and survival as compared with 5-fluorouracil (5-FU)/leucovorin alone.3 However, its use is accompanied by a comparably high incidence of unpredictable severe toxicity.4 The main toxicities of IRI are neutropenia and diarrhoea, resulting in dose reduction, treatment withdrawals or death.3 A number of studies have attempted to explain these toxicities by analyzing candidate genes in the IRI pathways.5
The uridine diphosphate glucuronosyltransferase (UGT) 1A1 is an essential enzyme involved in the complex metabolism of IRI. It inactivates the IRI toxic metabolite 7-ethyl-10-hydroxycamptothecin (SN-38) by biotransforming SN-38 into SN-38 glucuronide (SN-38 G).6 There is a common and well-described polymorphism in the promoter region of UGT1A1 gene where a variable number of TA repeats affects gene transcriptional efficiency.7 A six-repeat allele is the most commonly identified (wild type) form; a seven-repeat allele (designated UGT1A1*28) is associated with dramatically reduced expression of UGT1A1, resulting in lower SN-38 glucuronidation.8
Researchers have investigated the effect of UGT1A1*28 on the IRI-induced toxicities in patients with CRC.9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25, 26 However, results are both conflicting and difficult to interpret because of small sample sizes and associated poor statistical power. Although several meta-analyses demonstrated the association between the UGT1A1*28 polymorphism and IRI-induced neutropenia and diarrhoea,27, 28, 29 they included studies across many different cancers rather than focusing on patients with CRC. This meta-analysis will therefore focus on CRC alone, which will allow an assessment of uniform regimens tied to a single clinical disease site. Moreover, additional CRC studies have been published since these older meta-analyses.9, 10, 11, 12, 13
Materials and methods
Retrieval of published studies
Searches were conducted for papers published before May 2012 by two different authors (XL, WX). PubMed and Embase were surveyed by using the search terms ‘irinotecan', ‘UGT1A1', ‘UGT1A1 polymorphism', ‘UGT1A1*28', ‘colorectal cancer', and ‘toxicity'. Furthermore, we screened titles and abstracts to identify relevant studies. Studies in abstract form or meeting reports, without publication of the full paper, were excluded.
The UGT1A1*28 polymorphism is relatively rare in Asian populations and the prevalence of homozygous UGT1A1*28/*28 genotype is significantly greater in Caucasians than in Asian populations.30, 31, 32 To reduce the heterogeneity among the analyzed studies, ethnic differences were considered. However, available articles in Asian populations were of limited small sample sizes that it was infeasible to perform stratified analysis based on ethnicity. Thus, only studies of primarily Caucasians populations were included in this meta-analysis.
Inclusion and exclusion criteria
Studies were included in the meta-analysis if (1) they were clinical trials or well characterized observational data sets, (2) they explored the association between UGT1A1*28 and IRI-induced toxicities in patients with CRC, (3) numbers of patients with and without IRI-induced neutropenia or diarrhoea (grade III—IV) were provided (or could be calculated), and (4) they were published in English. Exclusion criteria were as follows: (1) case reports; (2) reviews and opinions; (3) allele frequency studies; (4) studies not involving CRC patients; (5) studies that reported general haematological or gastrointestinal toxicity instead of the more specific neutropenia or diarrhoea; and (6) studies conducted only in non-Caucasian populations. When different publications with overlapping subjects were considered eligible, we only included the one with the larger number of patients. Figure 1 summarizes the search methods and inclusion and exclusion steps.
Figure 1.
Flow diagram for study selection in meta-analysis. CRC, colorectal cancer.
Data extraction
The following information was extracted from each eligible publication: name of first author, year of publication, country, race, sample size, age (median or mean), gender, source of population, mutation detection method, IRI dose, chemotherapy regimens, study design, and number of patients with IRI-induced neutropenia or diarrhoea (grade III—IV) in each genotype group (UGT1A1*1/*1, UGT1A1*1/*28, and UGT1A1*28/*28).
Two or three IRI-containing regimens were administered to patients in some studies;12, 15, 16, 20 when possible, we analyzed the patients treated with each regimen as a separate sample. Patients treated with different regimens were analyzed as a single study only if separate data was not available.
Statistical analysis
We followed the PRISMA guidelines.33 All statistical analyses were performed using Review Manager (v5.1; Oxford, England) and R software (R Foundation, Vienna, Austria; http://www.CRAN.R-project.org). Cochran's χ2 test and the inconsistency index (I2) were used to evaluate heterogeneity across the included studies. P-values of >0.05 for the χ2 test indicated a lack of heterogeneity, and the fixed-effects model was then used to calculate the summary odds ratio (OR).34 Otherwise, a random-effects model was applied.35 ORs and their corresponding 95% confidence intervals (CI) were estimated. Z-test was performed to determine the statistical significance of pooled OR, and was considered significant when P<0.05. We assessed potential publication bias by using a funnel plot and Egger's test.36 When the Egger's test was significant (P<0.05), a trim and fill method was used to adjust for publication bias.37 For each outcome (neutropenia and diarrhoea), we compared the following: UGT1A1*28/*28 versus (vs) UGT1A1*1/*1 (equivalent to homozygous variant vs wild type), UGT1A1*1/*28 vs UGT1A1*1/*1 (equivalent to heterozygous variant vs wild type) and UGT1A1*28/*28 vs all others (equivalent to a recessive genetic model).
Previous studies27, 28, 29 showed a dose-dependent relationship between UGT1A1*28 genotypes and IRI-induced toxicities. Additionally, the co-administration of 5-FU, a core component of many IRI-based regiments, might enhance the haematological and digestive tract toxicities.3, 38 Thus, we carried out stratified analyses in two settings: (A) whether 5-FU (or analogue) was combined with IRI; or (B) between high/medium and low doses of IRI. In the former setting, studies were classified into two subgroups, marked +5FU (studies including 5-FU or a 5-FU analogue) and −5FU (no 5-FU or analogue), respectively. For dose intensity analysis, 150 mg m−2 of IRI dose was set as the cutoff value between medium/high (high IRI) and low dose (low IRI). In some studies,11, 25, 26 the patients received different IRI doses at different time points and only combined toxicity-related data were available. The average dose was calculated to classify these studies.
Results
Characteristics of the studies
Figure 1 shows the process of study selection. In total, 32 full-text studies were fully reviewed. Of these, seven did not provide numbers of neutropenia or diarrhoea patients with different genotypes.39, 40, 41, 42, 43, 44, 45 Two studies did not provide data for individuals' genotypes.14, 19 Four studies combined CRC patients with those that had other cancers.46, 47, 48, 49 Three were excluded because they analyzed only Asian populations.50, 51, 52 Thus, only 16 studies were considered eligible for inclusion in our meta-analysis.
Characteristics of the included studies are summarized in Table 1. Methodological components of study designs may be critically important and take priority over aggregate scores in the meta-analyses.53 Thus, we utilized a modified set of criteria to identify methodological soundness when reporting quality of the studies.54 The criteria considered study design, the detection method of the polymorphisms, chemotherapy regimens, and grading systems for toxicity (Table 1).
Table 1. Characteristics and methodological quality of studies included in meta analysis.
| Study (year) | Country, races | Phase of clinical trial | No. of patient (male%) | Agea | Population source | Mutation detection methods | Regimen | IRI dose (mg m−2)/schedule | Grade criteria | Neutropenia grade 3–4 (%) | Diarrhoea grade 3–4 (%) | Study design |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lamas et al9 | Spain, U | U | 100 (63.4) | 67 | U | SPR | FOLFIRI | 180/biweekly | N3 | 18 (18.0) | 12 (12.0) | R |
| Shulman et al10 | Israel, C | I | 214 (46.3) | 63 | M | SPR | TEGAFIRI, XELIRI, FOLFIRI, IFL | U | U | 48 (22.4) | 19 (8.9) | R |
| Martinez et al11 | Spain, C | III | 149 (U) | U | M | Sequencing | FOLFIRI, FUIRI | 80/weekly or 180/biweekly | U | 31 (20.8) | 45 (30.2) | R |
| McLeod et al12 | USA/UK/Canada, mainly C | III | 212 (U) | 61 | M | PYRS | IFL, IROX | 100–125/weekly or 200/every 3 weeks | N2 | Only grade 4: 28 (13.2) | 60 (28.3) | P |
| Glimelius et al13 | Sweden/UK/Norway, mainly C | III | 136 (U) | 62 | M | SPR | FLIRI, Lv5FU-IRI | 180/biweekly | N2 | 18 (13.2) | 10 (7.4) | R |
| Braun et al15 | UK, U | III | 326 (U) | 64 | M | SPR | IrFu, IRI | 300–350/every 3 weeks, 180/biweekly | C2 | 35 (10.7) | 18 (5.5) | P |
| Parodi et al16 | USA, U | III | 110 (52.2) | 60 | M | SPR | FOLFIRI, mIFL, CapeIRI | 125 or 180/ biweekly, 250/every 3 weeks | U | 42 (38.2) | / | P |
| Ferraldeschi et al17 | UK, mainly C | U | 92 (69.0) | 63 | S | SPR | FOLFIRI/ IRI—VEGF inhibitor, CapeIRI, UFT- Lv- IRI- OX | 180/biweekly | N2 | 16 (17.4) | 6 (6.5) | P |
| Toffoli et al18 | Italy, C | I | 250 (64.8) | 61 | M | PYRS | FOLFIRI, mFOLFIRI | 180/biweekly | N2 | 35 (14.0) | 21 (8.4) | P |
| Kweekel et al20 | Netherlands, C | III | 218 (62.8) | 61 | M | PYRS | CapeIRI, IRI | 250 or 350/every 3 weeks | N2 | / | 48 (22.0) | R |
| Ruzzo et al21 | Italy, C | U | 146 (55.6) | 61 | M | SPR | FOLFIRI | 180/biweekly | N2 | 34 (23.0) | / | P |
| Côté et al22 | France, C | III | 89 (U) | U | M | SPR | FOLFIRI | 180/biweekly | N2 | 19 (21.3) | / | P |
| Massacesi et al23 | Italy, C | II | 56 (52.7) | 64 | M | Sequencing | IRI- raltitrexed | 80/weekly | N2 | 4 (7.1) | 10 (17.9) | P |
| Carlini et al24 | USA, mainly C | II | 62 (55.0) | 61 | M | SPR | CapeIRI | 100 or 125/weekly | N2 | 2 (3.3) | 20 (32.3) | P |
| Rouits et al25 | France, C | U | 73 (61.1) | 62 | S | PYRS | FOLFIRI, mFOLFIRI | 85/weekly or 180/biweekly | N2 | 22 (30.1) | 13 (17.8) | R |
| Marcuello et al26 | Spain, C | U | 95 (63.3) | 68 | U | SPR | IRI-Tomudex, IRI-5FU-LV, IRI-5FU, IRI | 80/weekly or 180/biweekly or 350/every 3 weeks | C2 | / | 29 (30.5) | P |
Abbreviations: C, Caucasian; CAPe, capecitabine; C2/3, CTCAEv2/3, Common Terminology Criteria for Adverse Events version 2/3; IRI, irinotecan; LV, leucovorin; M, multicentre; N2/3, NCI-CTC v2/3, National Cancer Institute-Common Cytotoxicity Criteria version 2/3; OX(A), oxaliplatin; P, analysis was planned prospectively; PYRS, pyrosequencing; R, analysis was planned retrospectively; S, single centre; SPR, sizing of PCR products; Sequencing, other DNA sequencing methods; TEGAF, UFT/LV; U, unknown; 5FU, 5-fluorouracil; UFT, uracil/tegafur; VEGF, vascular endothelial growth factor; XEL, xeloda.
median or mean age.
Of the 16 studies, 11 assessed genotype relationships with both neutropenia and diarrhoea,9, 10, 11, 12, 13, 15, 17, 18, 23, 24, 25 two only for diarrhoea,20, 26 and three only for neutropenia.16, 21, 22 There were four studies that did not clearly report the race of the participants9, 15, 16, 23 but they were conducted in Europe or America, and the UGT1A1*28 allele frequencies were similar to Caucasians. Accordingly, the four were assigned as Caucasian studies. Table 1 also showed four ‘mainly Caucasian' studies.12, 13, 17, 24 One13 was reported on the basis of authors' own description, whereas in the other three studies the percentage of Caucasian were 86,12 98,17 and 83%,24 respectively.
For subgroup meta-analysis, in two studies with multiple regimens without separate data,17, 26 only one or several patients received IRI-based chemotherapy without 5-FU (or analogue). Hence, the two studies were classified into the +5FU subgroup. The study by Shulman K et al.10 did not provide the exact IRI-dose, but was included in low IRI subgroup based on the authors' own comments. Meta-analyses of these subsets are presented in Table 2. The results of meta-analysis are summarized in Table 3.
Table 2. Subgroup meta-analysis categorization.
| Sub analyses | Subgroup | Regimens or IRI dose |
No. of identified trials |
|
|---|---|---|---|---|
| Neutropenia | Diarrhoea | |||
| By 5-FU status | +5FU (combined with 5-FU or analogue) | FOLFIRI, mFOLFIRI, IFL, FLIRI, Lv5FU-IR, mIFL, CapeIRI, IrFu, TEGAFIRI, XELIRI, UFT-Lv-IRI-OX, IRI-5FU-LV, IRI-5FU | 139, 10, 11, 12, 13, 15, 16, 17, 18, 21, 22, 24, 25 | 129, 10, 11, 12, 13, 15, 17, 18, 20, 24, 25, 26 |
| −5FU (no 5-FU or analogue) | IROX, IRI-raltitrexed, IRI-alone | 312, 15, 23 | 412, 15, 20, 23 | |
| By IRI dose | High IRI (medium and high dose) | >150 mg m−2 of IRI | 99, 12, 13, 15, 16, 17, 18, 21, 22 | 89, 12, 13, 15, 17, 18, 20, 26 |
| Low IRI (low dose) | <150 mg m−2 of IRI | 710, 11, 12, 16, 23, 24, 25 | 610, 11, 12, 23, 24, 25 | |
Abbreviations: CAPe, capecitabine; IRI, irinotecan; LV, leucovorin; OX(A), oxaliplatin; TEGAF, UFT/LV; UFT, uracil/tegafur; VEGF, vascular endothelial growth factor; XEL, xeloda; 5FU, 5-fluorouracil.
Table 3. Summary of meta-analysis.
| Toxicity | Compared genotype | Group | No. of studies | No. of participants | Odds ratios (ORs) 95% confidence intervals (CI) | P-value |
Test for heterogeneity |
|
|---|---|---|---|---|---|---|---|---|
| P-value | I2(%) | |||||||
| Neutropenia | *28/*28 vs *1/*1 | Total | 13 | 1095 | 4.79 [3.28, 7.01] | <0.00001 | 0.20 | 22 |
| +5FU subgroup | 12 | 932 | 4.67 [3.11, 7.00] | <0.00001 | 0.14 | 30 | ||
| −5FU subgroup | 3 | 163 | 5.87 [1.97, 17.42] | <0.001 | 0.41 | 0 | ||
| High IRI subgroup | 9 | 764 | 4.64 [2.88, 7.17] | <0.00001 | 0.06 | 44 | ||
| Low IRI subgroup | 6 | 331 | 6.37 [2.69, 10.71] | <0.00001 | 0.76 | 0 | ||
| *1/*28 vs 1/*1 | Total | 14 | 1819 | 1.90 [1.44, 2.51] | <0.00001 | 0.98 | 0 | |
| +5FU subgroup | 13 | 1573 | 1.87 [1.39, 2.51] | <0.0001 | 0.93 | 0 | ||
| −5FU subgroup | 3 | 246 | 2.18 [0.91, 5.22] | 0.08 | 0.85 | 0 | ||
| High IRI subgroup | 9 | 1189 | 1.85 [1.32, 2.58] | 0.0003 | 0.98 | 0 | ||
| Low IRI subgroup | 7 | 630 | 2.01 [1.21, 3.34] | 0.007 | 0.61 | 0 | ||
| *28/*28 vs *1/*28 or *1/*1 | Total | 14 | 2015 | 3.44 [2.45, 4.82] | <0.00001 | 0.15 | 26 | |
| +5FU subgroup | 13 | 1740 | 3.40 [2.37, 4.88] | <0.00001 | 0.14 | 29 | ||
| −5FU subgroup | 3 | 275 | 3.70 [1.46, 9.40] | 0.006 | 0.20 | 38 | ||
| High IRI subgroup | 9 | 1311 | 3.34 [2.21, 5.05] | <0.00001 | 0.02 | 54 | ||
| Low IRI subgroup | 7 | 704 | 3.63 [2.02, 6.53] | <0.00001 | 0.96 | 0 | ||
| Diarrhoea | *28/*28 vs *1/*1 | Total | 13 | 1122 | 1.84 [1.24, 2.72] | 0.002 | 0.15 | 27 |
| +5FU subgroup | 12 | 913 | 1.78 [1.16, 2.75] | 0.009 | 0.07 | 41 | ||
| −5FU subgroup | 4 | 209 | 2.09 [0.83, 5.26] | 0.12 | 0.56 | 0 | ||
| High IRI subgroup | 8 | 774 | 2.37 [1.39, 4.04] | 0.002 | 0.31 | 15 | ||
| Low IRI subgroup | 6 | 348 | 1.41 [0.79, 2.51] | 0.24 | 0.12 | 42 | ||
| *1/*28 vs 1/*1 | Total | 13 | 1794 | 1.20 [0.93, 1.56] | 0.16 | 0.55 | 0 | |
| +5FU subgroup | 12 | 1472 | 1.19 [0.89, 1.58] | 0.25 | 0.46 | 0 | ||
| −5FU subgroup | 4 | 322 | 1.28 [0.71, 2.30] | 0.41 | 0.44 | 0 | ||
| High IRI subgroup | 8 | 1201 | 1.39 [0.97, 1.98] | 0.07 | 0.65 | 0 | ||
| Low IRI subgroup | 6 | 593 | 1.02 [0.70, 1.50] | 0.91 | 0.36 | 8 | ||
| *28/*28 vs *1/*28 or *1/*1 | Total | 13 | 1980 | 1.71 [1.18, 2.47] | 0.005 | 0.29 | 14 | |
| +5FU subgroup | 12 | 1626 | 1.67 [1.11, 2.52] | 0.01 | 0.17 | 28 | ||
| −5FU subgroup | 4 | 354 | 1.85 [0.77, 4.43] | 0.17 | 0.53 | 0 | ||
| High IRI subgroup | 8 | 1317 | 2.04 [1.23, 3.38] | 0.006 | 0.41 | 3 | ||
| Low IRI subgroup | 6 | 663 | 1.41 [0.82, 2.43] | 0.21 | 0.20 | 31 | ||
Abbreviations: FU, fluorouracil; IRI, irinotecan.
Association between UGT1A1*28 and severe neutropenia
UGT1A1*28/*28 vs UGT1A1*1/*1
(Presented in Figure 2 and Supplementary Figures 1A and B) Thirteen studies compared the risk of neutropenia between patients with a UGT1A1*28/*28 genotype and those with a wild-type genotype. Pooled data from all studies showed that the risk of neutropenia was higher among UGT1A1*28/*28 patients than among those with a wild-type genotype (OR=4.79, 95% CI: 3.28–7.01; P<0.00001).
Figure 2.
Forest plots of *28/*28 vs *1/*1, outcome: neutropenia. (a) (Stratified analysis based on 5-FU or analogue): +5-FU, received 5-FU or an analogue; −5-FU, did not receive 5-FU or analogue. (b) (Stratified analysis based on IRI-dose): high IRI, received medium or high dose of IRI; low IRI, received low dose of IRI. CI, confidence intervals; FU, fluorouracil; I2, inconsistency index.
In the subgroup analysis, UGT1A1*28/*28 genotype was found to be associated with significantly increased risk of neutropenia in all subgroups (Table 3). No statistical heterogeneity were detected both in the analysis of all studies (I2=22%, P=0.20) and in all subgroup analyses. The funnel plots were symmetric, and the Egger's test was not significant (P=0.75), suggesting little-to-no publication bias.
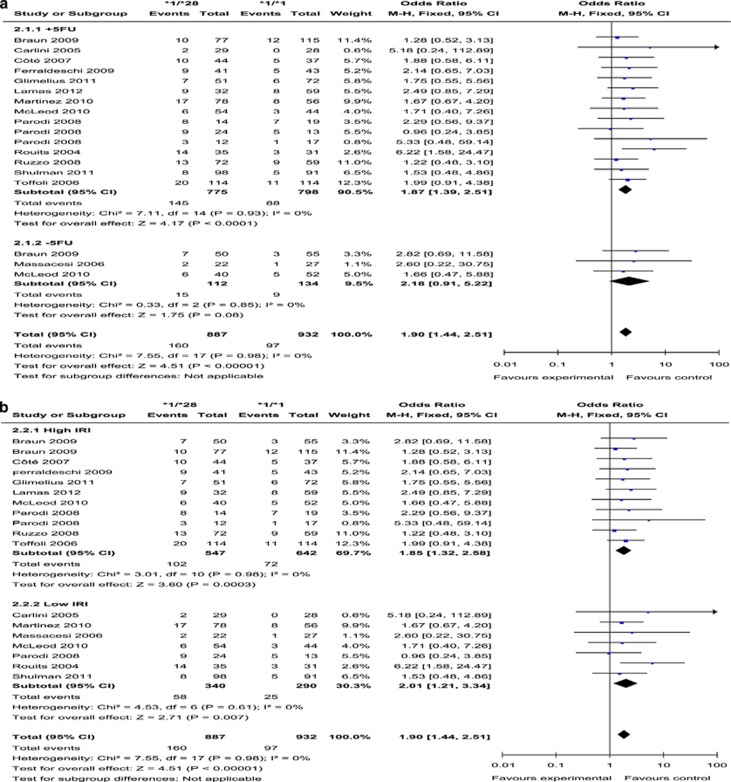
UGT1A1*1/*28 vs UGT1A1*1/*1
(Presented in Figure 3 and Supplementary Figures 1C and D) A total of 14 studies compared the risk of neutropenia among patients with the two different genotypes. The pooled OR was 1.90 (95% CI: 1.44–2.51; P<0.0001) for all studies. Heterogeneity among studies was not detected (I2=0%, P=0.76). In subgroup analysis, there were too few −5FU studies to compare with +5FU subgroups; the OR were qualitatively similar between high and low IRI subgroups. The funnel plot showed some asymmetry, and Egger's test for publication bias was significant (P=0.046). The trim and fill method provided the adjusted estimate of OR=1.68 (95% CI: 1.29–2.19; P<0.0001), which was only a slight change from our original estimate of 1.90.
Figure 3.
Forest plots of *1/*28 vs *1/*1, outcome: neutropenia. (a) (Stratified analysis based on 5-FU or analogue): +5-FU, received 5-FU or an analogue; −5-FU, did not receive 5-FU or analogue. (b) (Stratified analysis based on IRI-dose): high IRI, received medium or high dose of IRI; low IRI, received low dose of IRI. CI, confidence intervals; FU, fluorouracil; I2, inconsistency index.
UGT1A1*28/*28 vs UGT1A1*1/*28 or UGT1A1*1/*1
(Presented in Supplementary Figure 3 and Supplementary Figures 1E and F) Fourteen trials were included for this analysis. No statistical heterogeneity was detected (I2=26%, P=0.15) except in the high IRI subgroup (I2=54%, P=0.02). There was no evidence of publication bias, given the symmetrical distribution of funnel plot and Egger's test (P=0.77). A random-effects model was applied in high IRI subgroup. A pooled OR of 3.42 (95% CI: 1.65–7.09; P=0.0009) was found, which was slightly different from the OR in the fixed-effects model (OR=3.34, 95% CI: 2.21–5.05; P<0.00001). The total and subgroup analyses all suggested an increased risk of neutropenia in UGT1A1*28/*28 patients when compared with patients with at least one UGT1A1*1 allele (Table 3).
Association between UGT1A1*28 and severe diarrhoea
UGT1A1*28/*28 vs UGT1A1*1/*1
(Presented in Figure 4 and Supplementary Figures 2A and B) Relevant data for this comparison were available in 13 trials. Overall, there was an increased risk of diarrhoea associated with the UGT1A1*28/*28 genotype (OR=1.84, 95% CI: 1.24–2.72; P=0.002). The higher incidence of diarrhoea in UGT1A1*28/*28 patients was observed in the +5FU subgroup (OR=1.78, 95% CI: 1.16–2.75; P=0.009) and in the high IRI subgroup (OR =2.37, 95% CI: 1.39–4.04; P=0.002), but not in the other subgroups (Table 3). Heterogeneity was not statistically significant across all studies (I2=27%, P=0.15). There was no evidence of publication bias, given the symmetrical distribution of funnel plot and Egger's test (P=0.84).
Figure 4.
Forest plots of *28/*28 vs *1/*1, outcome: diarrhoea. (a) (Stratified analysis based on 5-FU or analogue): +5-FU, received 5-FU or an analogue; −5-FU, did not receive 5-FU or analogue. (b) (Stratified analysis based on IRI-dose): high IRI, received medium or high dose of IRI; low IRI, received low dose of IRI. CI, confidence intervals; FU, fluorouracil; I2, inconsistency index.
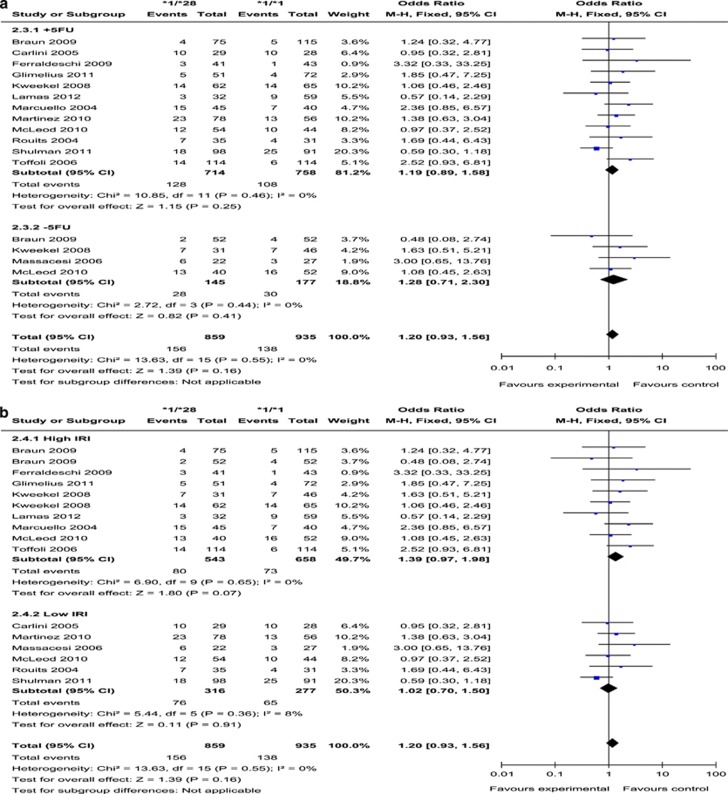
UGT1A1*1/*28 vs UGT1A1*1/*1
(Presented in Figure 5 and Supplementary Figures 2C and D) Thirteen studies were included in the comparison (Table 3). No publication bias was detected in the funnel plot and Egger's test (P=0.18), and there was no heterogeneity noted (I2=0%, P=0.55). Overall analyses showed no statistical difference between UGT1A1*1/*28 and UGT1A1*1/*1 patients for the risk of diarrhoea (OR=1.20, 95% CI: 0.93–1.56; P=0.16). In the high IRI subgroup, there was still a slightly higher risk of diarrhoea in UGT1A1*1/*28 patients (OR=1.39, 95% CI: 0.97–1.98; P=0.07), of borderline significance.
Figure 5.
Forest plots of *1/*28 vs *1/*1, outcome: diarrhoea. (a) (Stratified analysis based on 5-FU or analogue): +5-FU, received 5-FU or an analogue; −5-FU, did not receive 5-FU or analogue. (b) (Stratified analysis based on IRI-dose): high IRI, received medium or high dose of IRI; low IRI, received low dose of IRI. CI, confidence intervals; FU, fluorouracil; I2, inconsistency index.
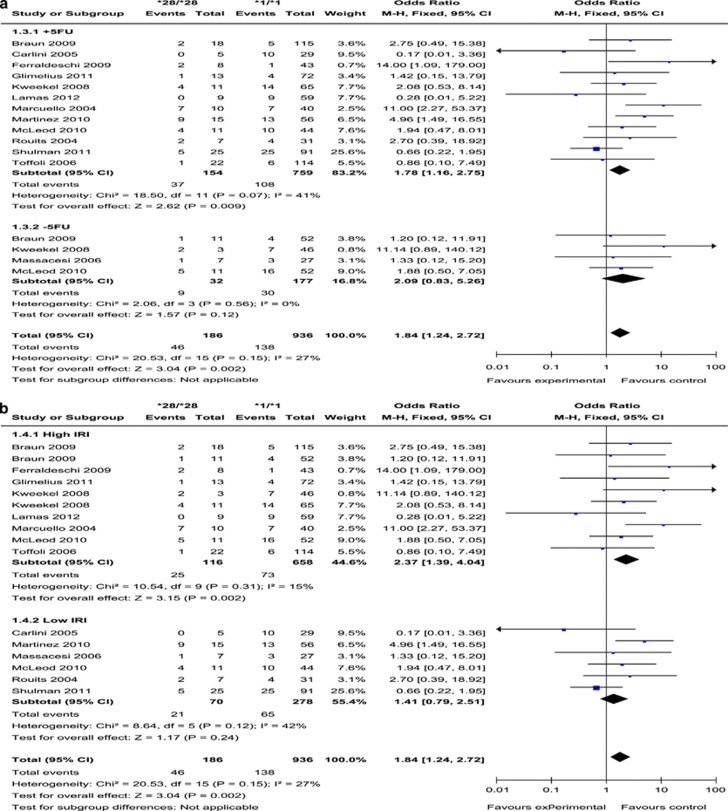
UGT1A1*28/*28 vs UGT1A1*1/*28 or UGT1A1*1/*1
(Presented in Supplementary Figure 4 and Supplementary Figures 2E and F) Thirteen trials provided relevant data for this comparison (Table 3). No statistical heterogeneity were detected (I2=14%, P=0.29). There was no evidence of publication bias, given the symmetrical funnel plot and Egger's test (P=0.34). Although the pooled OR was 1.71 (95% CI: 1.18–2.47; P=0.005) across all studies, the relationship was only seen in the subgroup of studies receiving concurrent 5FU or its analogues (+5FU) and in the subgroup of studies that used a higher dose of IRI (high IRI).
Discussion
Previously reported meta-analyses27, 28, 29 mainly focused on dose-dependent associations between UGT1A1*28 genotype and IRI-induced neutropenia or diarrhoea. However, results from these studies were not completely consistent. This could be a consequence of including studies conducted across various tumour types, as the risk of severe toxicity with IRI can vary by disease site. Thus, the present meta-analysis assessed the association of UGT1A1*28 polymorphisms with IRI-induced neutropenia and diarrhoea in a single cancer site; CRC.
In this meta-analysis, the relationship between UGT1A1*28 genotypes and IRI-induced neutropenia (grade III—IV) was first evaluated; patients with UGT1A1*28 allele in either heterozygote or homozygote form were at an increased risk of neutropenia regardless of the dose of IRI administrated, and regardless of whether 5FU (or analogue) was included in the regimens. The results from our meta-analysis of high IRI and low IRI subgroup are consistent with the meta-analysis of Zhe-Yi H et al,28 where the UGT1A1*28/*28 genotype was associated with an increased risk of neutropenia not only at high doses of IRI but also at low doses in an analysis across various tumour types.
Unlike Zhe-Yi H et al28 we also compared the UGT1A1*28/*28 vs UGT1A1*1/*1 (homozygous model) separately for neutropenia, while the previous meta-analyses focused on various tumour types,27, 28 assessing only the heterozygous and recessive models. An important message of our study is that UGT1A1*28/*28 homozygous patients had more than double the risk than UGT1A1*28 heterozygous patients (OR= 4.79 vs 1.90). Our data support the recommendation of US Food and Drug Administration in 2005 to warn of elevated risk of neutropenia for UGT1A1*28/*28 patients in the IRI product label, but does not identify what the proper clinical management should be for these at-risk patients.
The association between UGT1A1*28 polymorphism and severe diarrhoea, another important toxicity of IRI, was detected in CRC. The UGT1A1*28 genotypes were significantly associated with an increased risk of IRI-induced diarrhoea. We observed that UGT1A1*28/*28 patients were at an increased risk of diarrhoea following medium or high doses of IRI, but not at low doses. A similar result was reported in a previous meta-analysis.29 Our observation did confirm that the IRI dose modulated the association between UGT1A1*28 genotype and IRI-induced diarrhoea. Our results do not suggest that receiving a second diarrhoea-including chemotherapeutic agent (i.e., 5-FU or analogue) modifies the relationship between UGT1A1*28 and diarrhoea. In fact, there was a hint that the relationship was stronger in the no-5-FU (−5FU) subgroup when compared with the +5FU subgroup of patients. Perhaps the greater risk of overall diarrhoea associated with 5-FU overpowered any differential effects of UGT1A1*28 on diarrhoea.
We observed that pooled ORs for diarrhoea were much smaller than the pooled ORs for neutropenia in all comparisons. This is not surprising, as the risk of diarrhoea is modulated by gut-flora-producing enzymes that activate or inactivate SN-38.55 Confounding by gut flora may have attenuated the primary relationship. Another reason may be related to local treatment factors in the gut, such as previous surgery and radiotherapy. For example, local irradiation can worsen diarrhoea independent of the UGT1A1 genotype.26
In both neutropenia and diarrhoea, there were suggestions that the heterozygous variant UGT1A1*1/*28 had an intermediate effect. By assessing the heterozygous variant separately, we are able to suggest that when combined with other clinical factors, even the heterozygous variant may have clinical relevance.
In Caucasian populations, the homozygous variant genotype UGT1A1*28/*28 has been associated with Gilbert's syndrome, which is characterized by intermittent hyperbilirubinaemia.56 With the intention to avoid participants with Gilbert's syndrome, nine studies excluded patients with elevated bilirubin from the trials. The exclusion of patients with markedly pathological laboratory values from the trial may explain why no other correlation was found.57
There are limitations to this analysis. First, there is inherent heterogeneity to all meta-analyses, and in the analyzed studies different combinations of chemotherapy regimens were used, and patients were of varied performance status. Further, there were differences in study design, the source of population, IRI doses, polymorphism detection methods, toxicity grade criteria, stage of CRC, and pretreatment with other regimens. Second, articles included in this meta-analysis were restricted to English language publications. Articles with potentially high-quality data in other languages were excluded because of anticipated difficulties in obtaining accurate medical translation. This may also result in a decreased power during our analysis. Third, not all studies included adequate data for all subgroup comparison analyses. Fourth, the possibility of information and selection biases cannot be completely excluded. For example, some of these studies may have arms where patients are not receiving IRI. We selected patients from subgroup samples receiving IRI.
In summary, this meta-analysis provided evidence for the association between the UGT1A1*28 polymorphism and an increased risk of IRI-induced neutropenia and diarrhoea in CRC. Associations with significant neutropenia were consistent and strong. In contrast, associations with diarrhoea were weaker, and primarily seen when higher doses of IRI were administrated. Clinical validity is only one step of several that determine whether a biomarker is adopted into clinical practice. Assessment of clinical utility is also important. Generally the combination of prolonged severe neutropenia and/or prolonged diarrhoea provides the greatest risk to morbidity and mortality of IRI. Whether UGT1A1*28 rises to the level of clinical utility is still unclear. It is possible that UGT1A1 can be incorporated into a useful risk model of toxicity that includes a panel of clinical and genetic factors. Nonetheless, elucidating the mechanisms through pharmacogenetic association studies will improve our understanding of the biology of drug action and provide the basis upon which personalized medicine can be implemented.
Acknowledgments
This work was sponsored in part by the Alan B Brown Chair in Molecular Genomics, CCO Chair in Experimental Therapeutics and Population studies and Posluns Family Foundation to Professor GL. This work was also supported by the Scholarship Fund Award for Young Teachers Studying Abroad from Hubei University of Chinese Medicine and the Education Department of Hubei Province in PRC Foundation to Dr XL (no. Q20071601).
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies the paper on the The Pharmacogenomics Journal website (http://www.nature.com/tpj)
Supplementary Material
References
- Weitz J, Koch M, Debus J, Hohler T, Galle PR, Buchler MW. Colorectal cancer. Lancet. 2005;365:153–165. doi: 10.1016/S0140-6736(05)17706-X. [DOI] [PubMed] [Google Scholar]
- Pizzolato JF, Saltz LB. The camptothecins. Lancet. 2003;361:2235–2242. doi: 10.1016/S0140-6736(03)13780-4. [DOI] [PubMed] [Google Scholar]
- Douillard JY, Cunningham D, Roth AD, Navarro M, James RD, Karasek P, et al. Irinotecan combined with fluorouracil compared with fluorouracil alone as first-line treatment for metastatic colorectal cancer: a multicentre randomised trial. Lancet. 2000;355:1041–1047. doi: 10.1016/s0140-6736(00)02034-1. [DOI] [PubMed] [Google Scholar]
- Ratain MJ. Irinotecan dosing: does the CPT in CPT-11 stand for ‘‘Can't Predict Toxicity''. J Clin Oncol. 2002;20:7–8. doi: 10.1200/JCO.2002.20.1.7. [DOI] [PubMed] [Google Scholar]
- Funke S, Brenner H, Chang-Claude J. Pharmacogenetics in colorectal cancer: a systematic review. Pharmacogenomics. 2008;9:1079–1099. doi: 10.2217/14622416.9.8.1079. [DOI] [PubMed] [Google Scholar]
- Kawato Y, Aonuma M, Hirota Y, Kuga H, Sato K. Intracellular roles of SN-38, a metabolite of the camptothecin derivative CPT-11, in the antitumor effect of CPT-11. Cancer Res. 1991;51:4187–4191. [PubMed] [Google Scholar]
- Beutler E, Gelbart T, Demina A. Racial variability in the UDP-glucuronosyltransferase 1 (UGT1A1) promoter: a balanced polymorphism for regulation of bilirubin metabolism. Proc Natl Acad Sci USA. 1998;95:8170–8174. doi: 10.1073/pnas.95.14.8170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyer L, Hall D, Das S, Mortell MA, Ramírez J, Kim S, et al. Phenotype-genotype correlation of in vitro SN-38 (active metabolite of irinotecan) and bilirubin glucuronidation in human liver tissue with UGT1A1 promoter polymorphism. Clin Pharmacol Ther. 1999;65:576–582. doi: 10.1016/S0009-9236(99)70078-0. [DOI] [PubMed] [Google Scholar]
- Lamas MJ, Duran G, Balboa E, Bernardez B, Candamio S, Vidal Y, et al. The value of genetic polymorphisms to predict toxicity in metastatic colorectal patients with irinotecan-based regimens. Cancer Chemother Pharmacol. 2012;69:1591–1599. doi: 10.1007/s00280-012-1866-2. [DOI] [PubMed] [Google Scholar]
- Shulman K, Cohen I, Barnett-Griness O, Kuten A, Gruber SB, Lejbkowicz F, et al. Clinical implications of UGT1A1*28 genotype testing in colorectal cancer patients. Cancer. 2011;117:3156–3162. doi: 10.1002/cncr.25735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Balibrea E, Abad A, Martínez-Cardús A, Ginés A, Valladares M, Navarro M, et al. UGT1A and TYMS genetic variants predict toxicity and response of colorectal cancer patients treated with first-line irinotecan and fluorouracil combination therapy. Br J Cancer. 2010;103:581–589. doi: 10.1038/sj.bjc.6605776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLeod HL, Sargent DJ, Marsh S, Green EM, King CR, Fuchs CS, et al. Pharmacogenetic predictors of adverse events and response to chemotherapy in metastatic colorectal cancer: results from North American Gastrointestinal Intergroup Trial N9741. J Clin Oncol. 2010;28:3227–3233. doi: 10.1200/JCO.2009.21.7943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glimelius B, Garmo H, Berglund A, Fredriksson LA, Berglund M, Kohnke H, et al. Prediction of irinotecan and 5-fluorouracil toxicity and response in patients with advanced colorectal cancer. Pharmacogenomics J. 2011;11:61–71. doi: 10.1038/tpj.2010.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulz C, Heinemann V, Schalhorn A, Moosmann N, Zwingers T, Boeck S, et al. UGT1A1 gene polymorphism: impact on toxicity and efficacy of irinotecan-based regimens in metastatic colorectal cancer. World J Gastroenterol. 2009;15:5058–5066. doi: 10.3748/wjg.15.5058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun MS, Richman SD, Thompson L, Daly CL, Meade AM, Adlard JW, et al. Association of molecular markers with toxicity outcomes in a randomized trial of chemotherapy for advanced colorectal cancer: the FOCUS trial. J Clin Oncol. 2009;27:5519–5528. doi: 10.1200/JCO.2008.21.6283. [DOI] [PubMed] [Google Scholar]
- Parodi L, Pickering E, Cisar LA, Lee D, Soufi-Mahjoubi R. Utility of pretreatment bilirubin level and UGT1A1 polymorphisms in multivariate predictive models of neutropenia associated with irinotecan treatment in previously untreated patients with colorectal cancer. Arch Drug Inf. 2008;1:97–106. doi: 10.1111/j.1753-5174.2008.00014.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferraldeschi R, Minchell LJ, Roberts SA, Tobi S, Hadfield KD, Blackhall FH, et al. UGT1A1*28 genotype predicts gastrointestinal toxicity in patients treated with intermediate-dose irinotecan. Pharmacogenomics. 2009;10:733–739. doi: 10.2217/pgs.09.20. [DOI] [PubMed] [Google Scholar]
- Toffoli G, Cecchin E, Corona G, Russo A, Buonadonna A, D'Andrea M, et al. The role of UGT1A1*28 polymorphism in the pharmacodynamics and pharmacokinetics of irinotecan in patients with metastatic colorectal cancer. J Clin Oncol. 2006;24:3061–3068. doi: 10.1200/JCO.2005.05.5400. [DOI] [PubMed] [Google Scholar]
- Rouits E, Charasson V, Pétain A, Boisdron-Celle M, Delord JP, Fonck M, et al. Pharmacokinetic and pharmacogenetic determinants of the activity and toxicity of irinotecan in metastatic colorectal cancer patients. Br J Cancer. 2008;99:1239–1245. doi: 10.1038/sj.bjc.6604673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kweekel DM, Gelderblom H, Van der Straaten T, Antonini NF, Punt CJ, Guchelaar HJ. UGT1A1*28 genotype and irinotecan dosage in patients with metastatic colorectal cancer: a Dutch Colorectal Cancer Group study. Br J Cancer. 2008;99:275–282. doi: 10.1038/sj.bjc.6604461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruzzo A, Graziano F, Loupakis F, Santini D, Catalano V, Bisonni R, et al. Pharmacogenetic profiling in patients with advanced colorectal cancer treated with first-line FOLFIRI chemotherapy. Pharmacogenomics J. 2008;8:278–288. doi: 10.1038/sj.tpj.6500463. [DOI] [PubMed] [Google Scholar]
- Côté JF, Kirzin S, Kramar A, Mosnier JF, Diebold MD, Soubeyran I, et al. UGT1A1 polymorphism can predict hematologic toxicity in patients treated with irinotecan. Clin Cancer Res. 2007;13:3269–3275. doi: 10.1158/1078-0432.CCR-06-2290. [DOI] [PubMed] [Google Scholar]
- Massacesi C, Terrazzino S, Marcucci F, Rocchi MB, Lippe P, Bisonni R, et al. Uridine diphosphate glucuronosyl transferase 1A1 promoter polymorphism predicts the risk of gastrointestinal toxicity and fatigue induced by irinotecan-based chemotherapy. Cancer. 2006;106:1007–1016. doi: 10.1002/cncr.21722. [DOI] [PubMed] [Google Scholar]
- Carlini LE, Meropol NJ, Bever J, Andria ML, Hill T, Gold P, et al. UGT1A7 and UGT1A9 polymorphisms predict response and toxicity in colorectal cancer patients treated with capecitabine/irinotecan. Clin Cancer Res. 2005;11:1226–1236. [PubMed] [Google Scholar]
- Rouits E, Boisdron-Celle M, Dumont A, Guérin O, Morel A, Gamelin E. Relevance of different UGT1A1 polymorphisms in irinotecan-induced toxicity: a molecular and clinical study of 75 patients. Clin Cancer Res. 2004;10:5151–5159. doi: 10.1158/1078-0432.CCR-03-0548. [DOI] [PubMed] [Google Scholar]
- Marcuello E, Altés A, Menoyo A, Del Rio E, Gómez-Pardo M, Baiget M. UGT1A1 gene variations and irinotecan treatment in patients with metastatic colorectal cancer. Br J Cancer. 2004;91:678–682. doi: 10.1038/sj.bjc.6602042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoskins JM, Goldberg RM, Qu P, Ibrahim JG, McLeod HL. UGT1A1*28 genotype and irinotecan-induced neutropenia: dose matters. J Natl Cancer Inst. 2007;99:1290–1295. doi: 10.1093/jnci/djm115. [DOI] [PubMed] [Google Scholar]
- Zhe-Yi H, Qi Y, Pei Q, Guo C. Dose-dependent association between ugt1a1*28 genotype and irinotecan-induced neutropenia: low doses also increase risk. Clin Cancer Res. 2010;16:3832–3842. doi: 10.1158/1078-0432.CCR-10-1122. [DOI] [PubMed] [Google Scholar]
- Hu ZY, Yu Q, Zhao YS. Dose-dependent association between UGT1A1*28 polymorphism and irinotecan-induced diarrhoea: a meta-analysis. Eur J Cancer. 2010;46:1856–1865. doi: 10.1016/j.ejca.2010.02.049. [DOI] [PubMed] [Google Scholar]
- Yong WP, Innocenti F, Ratain MJ. The role of pharmacogenetics in cancer therapeutics. Br J Clin Pharmacol. 2006;62:35–46. doi: 10.1111/j.1365-2125.2006.02591.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall D, Ybazeta G, Destro-Bisol G, Petzl-Erler ML, Di Rienzo A. Variability at the uridine diphosphate glucuronosyltransferase 1A1 promoter in human populations and primates. Pharmacogenetics. 1999;9:591–599. [PubMed] [Google Scholar]
- Premawardhena A, Fisher CA, Liu YT, Verma IC, de Silva S, Arambepola M, et al. The global distribution of length polymorphisms of the promoters of the glucuronosyltransferase 1 gene (UGT1A1): hematologic and evolutionary implications. Blood Cells Mol Dis. 2003;31:98–101. doi: 10.1016/s1079-9796(03)00071-8. [DOI] [PubMed] [Google Scholar]
- Moher D, Liberati A, Tetzlaff J, Altman DG. Preferred reporting items for systematic reviews and meta-analyses: the prisma statement. J Clin Epidemiol. 2009;62:1006–1012. doi: 10.1016/j.jclinepi.2009.06.005. [DOI] [PubMed] [Google Scholar]
- Mantel N, Haenszel W. Statistical aspects of the analysis of data from retrospective studies of disease. J Natl Cancer Inst. 1959;22:719–748. [PubMed] [Google Scholar]
- DerSimonian R, Laird N. Meta-analysis in clinical trials. Control Clin Trials. 1986;7:177–188. doi: 10.1016/0197-2456(86)90046-2. [DOI] [PubMed] [Google Scholar]
- Egger M, Smith GD, Schneider M, Minder C. Bias in meta-analysis detected by a simple, graphical test. BMJ. 1997;315:629–634. doi: 10.1136/bmj.315.7109.629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duval S, Tweedie R. A nonparametric ‘Trim and Fill' method of accounting for publication bias in meta-analysis. Biometrics. 2000;56:455–463. doi: 10.1111/j.0006-341x.2000.00455.x. [DOI] [PubMed] [Google Scholar]
- Kohne CH, van Cutsem E, Wils J, Bokemeyer C, El-Serafi M, Lutz MP, et al. Phase III study of weekly high-dose infusional fluorouracil plus folinic acid with or without irinotecan in patients with metastatic colorectal cancer: European Organisation for Research and Treatment of Cancer Gastrointestinal Group Study 40986. J Clin Oncol. 2005;23:4856–4865. doi: 10.1200/JCO.2005.05.546. [DOI] [PubMed] [Google Scholar]
- Choi YH, Kim TW, Kim KP, Lee SS, Hong YS, Ryu MH, et al. A phase ii study of clinical outcomes of 3-week cycles of irinotecan and s-1 in patients with previously untreated metastatic colorectal cancer: influence of the UGT1A1 and CYP2A6 polymorphisms on clinical activity. Oncology. 2012;82:290–297. doi: 10.1159/000337989. [DOI] [PubMed] [Google Scholar]
- Sunakawa Y, Ichikawa W, Fujita K, Nagashima F, Ishida H, Yamashita K, et al. UGT1A1*1/*28 and *1/*6 genotypes have no effects on the efficacy and toxicity of FOLFIRI in Japanese patients with advanced colorectal cancer. Cancer Chemother Pharmacol. 2011;68:279–284. doi: 10.1007/s00280-010-1485-8. [DOI] [PubMed] [Google Scholar]
- Boige V, Mendiboure J, Pignon JP, Loriot MA, Castaing M, Barrois M, et al. Pharmacogenetic assessment of toxicity and outcome in patients with metastatic colorectal cancer treated with LV5FU2, FOLFOX, and FOLFIRI: FFCD 2000-05. J Clin Oncol. 2010;28:2556–2564. doi: 10.1200/JCO.2009.25.2106. [DOI] [PubMed] [Google Scholar]
- Cecchin E, Innocenti F, D'Andrea M, Corona G, De Mattia E, Biason P, et al. Predictive role of the UGT1A1, UGT1A7, and UGT1A9 genetic variants and their haplotypes on the outcome of metastatic colorectal cancer patients treated with fluorouracil, leucovorin, and irinotecan. J Clin Oncol. 2009;27:2457–2465. doi: 10.1200/JCO.2008.19.0314. [DOI] [PubMed] [Google Scholar]
- Martinez-Balibrea E, Manzano JL, Martinez-Cardus A, Moran T, Cirauqui B, Catot S, et al. Combined analysis of genetic polymorphisms in thymidylate synthase, uridine diphosphate glucoronosyltransferase and X-ray cross complementing factor 1 genes as a prognostic factor in advanced colorectal cancer patients treated with 5-fluorouracil plus oxaliplatin or irinotecan. Oncol Rep. 2007;17:637–645. [PubMed] [Google Scholar]
- Rhodes KE, Zhang W, Yang D, Press OA, Gordon M, Vallböhmer D, et al. ABCB1, SLCO1B1 and UGT1A1 gene polymorphisms are associated with toxicity in metastatic colorectal cancer patients treated with first-line irinotecan. Drug Metab Lett. 2007;1:23–30. doi: 10.2174/187231207779814328. [DOI] [PubMed] [Google Scholar]
- Ishida H, Fujita K, Akiyama Y, Sunakawa Y, Yamashita K, Mizuno K, et al. Regimen selection for first-line FOLFIRI and FOLFOX based on UGT1A1 genotype and physical background is feasible in Japanese patients with advanced colorectal cancer. Jpn J Clin Oncol. 2011;41:617–623. doi: 10.1093/jjco/hyr010. [DOI] [PubMed] [Google Scholar]
- Fakih MG, Ross ME, Starostik P. Increased frequency of uridine diphosphate glucuronosyltransferase 1A1 7/7 in patients experiencing severe irinotecan-induced toxicities. Clin Colorectal Cancer. 2007;6:583–587. doi: 10.3816/CCC.2007.n.026. [DOI] [PubMed] [Google Scholar]
- de Jong FA, Kehrer DF, Mathijssen RH, Creemers GJ, de Bruijn P, van Schaik RH, et al. Prophylaxis of irinotecan-induced diarrhea with neomycin and potential role for UGT1A1*28 genotype screening: a double-blind, randomized, placebo-controlled study. Oncologist. 2006;11:944–954. doi: 10.1634/theoncologist.11-8-944. [DOI] [PubMed] [Google Scholar]
- Park SR, Kong SY, Rhee J, Park YI, Ryu KW, Lee JH, et al. Phase II study of a triplet regimen of S-1 combined with irinotecan and oxaliplatin in patients with metastatic gastric cancer: clinical and pharmacogenetic results. Ann Oncol. 2011;22:890–896. doi: 10.1093/annonc/mdq435. [DOI] [PubMed] [Google Scholar]
- Denlinger CS, Blanchard R, Xu L, Bernaards C, Litwin S, Spittle C, et al. Pharmacokinetic analysis of irinotecan plus bevacizumab in patients with advanced solid tumors. Cancer Chemother Pharmacol. 2009;65:97–105. doi: 10.1007/s00280-009-1008-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okuyama Y, Hazama S, Nozawa H, Kobayashi M, Takahashi K, Fujikawa K, et al. Prospective phase II study of FOLFIRI for mCRC in Japan, including the analysis of UGT1A1 28/6 polymorphisms. Jpn J Clin Oncol. 2011;41:477–482. doi: 10.1093/jjco/hyr001. [DOI] [PubMed] [Google Scholar]
- Hazama S, Nagashima A, Kondo H, Yoshida S, Shimizu R, Araki A, et al. Phase I study of irinotecan and doxifluridine for metastatic colorectal cancer focusing on the UGT1A1*28 polymorphism. Cancer Sci. 2010;101:722–727. doi: 10.1111/j.1349-7006.2009.01428.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu CY, Chen PM, Chiou TJ, Liu JH, Lin JK, Lin TC, et al. UGT1A1*28 polymorphism predicts irinotecan-induced severe toxicities without affecting treatment outcome and survival in patients with metastatic colorectal carcinoma. Cancer. 2008;112:1932–1940. doi: 10.1002/cncr.23370. [DOI] [PubMed] [Google Scholar]
- Stroup DF, Berlin JA, Morton SC, Olkin I, Williamson GD, Rennie D, et al. Meta-analysis of observational studies in epidemiology: a proposal for reporting. Meta-analysis Of Observational Studies in Epidemiology (MOOSE) Group. JAMA. 2000;283:2008–2012. doi: 10.1001/jama.283.15.2008. [DOI] [PubMed] [Google Scholar]
- Jorgensen AL, Williamson PR. Methodological quality of pharmacogenetic studies: issues of concern. Stat Med. 2008;27:6547–6569. doi: 10.1002/sim.3420. [DOI] [PubMed] [Google Scholar]
- Roberts MS, Magnusson BM, Burczynski FJ, Weiss M. Enterohepatic circulation: physiological, pharmacokinetic and clinical implications. Clin Pharmacokinet. 2002;41:751–790. doi: 10.2165/00003088-200241100-00005. [DOI] [PubMed] [Google Scholar]
- Beutler E, Gelbart T, Demina A. Racial variability in the UDP-glucuronosyltransferase 1 (UGT1A1) promoter: a balanced polymorphism for regulation of bilirubin metabolism. Proc Natl Acad Sci USA. 1998;95:8170–8174. doi: 10.1073/pnas.95.14.8170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorbye H, Kohne CH, Sargent DJ, Glimelius B. Patient characteristics and stratification in medical treatment studies for metastatic colorectal cancer: a proposal for standardization of patient characteristic reporting and stratification. Ann Oncol. 2007;18:1666–1672. doi: 10.1093/annonc/mdm267. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.