Abstract
The tobacco hornworm Manduca sexta has served as a model for insect biochemical and physiological research for decades. However, knowledge of the posttranscriptional regulation of gene expression by microRNAs is still rudimentary in this species. Our previous study (Zhang et al., 2012) identified 163 conserved and 13 novel microRNAs in M. sexta, most of which were present at low levels in pupae. To identify additional M. sexta microRNAs and more importantly to examine their possible roles in the expression regulation of immunity-related genes, we constructed four small RNA libraries using fat body and hemocytes from naïve or bacteria-injected larvae and obtained 32.9 million reads of 18-31 nucleotides by Illumina sequencing. Mse-miR-929 and mse-miR-1b (antisense microRNA of mse-miR-1) were predicted in the previous study and now found to be conserved microRNAs in the tissue samples. We also found four novel microRNAs, two of which result from a gene cluster. Mse-miR-281-star, mse-miR-965-star, mse-miR-31-star, and mse-miR-9b-star were present at higher levels than their respective mature strands. Abundance changes of microRNAs were observed after the immune challenge. Based on the quantitative data of mRNA levels in control and induced fat body and hemocytes as well as the results of microRNA target site prediction, we suggest that certain microRNAs and microRNA*s regulate gene expression for pattern recognition, prophenoloxidase activation, cellular responses, antimicrobial peptide synthesis, and conserved intracellular signal transduction (Toll, IMD, JAK-STAT, MAPK-JNK-p38, and small interfering RNA pathways). In summary, this study has enriched our knowledge on M. sexta microRNAs and how some of them may participate in the expression regulation of immunity-related genes.
Keywords: posttranscriptional regulation, Illumina sequencing, Lepidoptera, insect immunity, target site prediction
1. Introduction
MicroRNAs (miRNAs) are non-coding RNAs, generally between 20 and 22 nucleotides (nt) in length. Their precursors, from either primary transcripts or intron lariats, are transported into the cytoplasm for processing (Asgari, 2011). The RNase III-type enzyme Dicer 1 trims the loop to generate miRNA:miRNA* duplexes, of which the mature miRNA strands are usually incorporated into the RNA-induced silencing complex (RISC) to initiate target mRNA translational repression or degradation, mostly binding to 3’-untranslated regions (3’-UTRs) of the mRNAs. The passenger strands (miRNA*s) are usually disposed of rapidly and detected at lower levels by high-throughput sequencing. In some cases, however, miRNA*s are maintained at high levels (Jagadeeswaran et al., 2010; Kato et al., 2009; Zhang et al., 2012). The dominant usage of 5’ or 3’ arms of miRNA precursors is considered to be a possible mechanism for insect miRNA evolution (Marco et al., 2010). After their discovery, miRNAs were found to regulate diverse physiological processes, including insect development, host defense, and metabolism (Asgari, 2011; Baker and Thummel, 2007; Chawla and Sokol, 2011).
Insects only possess innate immunity. As a lepidopteran model species, Manduca sexta has contributed significantly to biochemical research on insect antimicrobial defense (Jiang et al., 2010). Hemocytes and fat body are major sources of plasma proteins. Upon exposure to bacteria and fungi, various recognition proteins interact with pathogen-associated molecular patterns to stimulate cellular and humoral immune responses. Phagocytosis, nodule formation, and encapsulation are early hemocyte responses aimed at eliminating the invading pathogens. Pathogen recognition initiates a serine proteinase cascade to activate prophenoloxidase (PPO) for melanization, pro-Spätzle for Toll pathway activation, and paralytic peptide precursor for plasmatocyte spreading. Melanization entraps and kills pathogens (Cerenius et al., 2008; Nappi and Christensen, 2005). A superfamily of plasma serine proteinase inhibitors (serpins) modulates the serine proteinase cascade by specifically inhibiting various pathway members (Jiang et al., 2010). The Toll pathway, together with the immune deficiency (Imd) pathway, is important for induced production of antimicrobial peptides (AMPs) (Lemaitre and Hoffmann, 2007). Highly conserved JNK, JAK-STAT, and MAPK pathways in the insect cells also assist in host defense against pathogens (Bond and Foley, 2009; Goto et al., 2010; Ragab et al., 2011).
Although miRNAs extensively modulate insect immunity against viruses and apicomplexan parasites (Asgari, 2011; Fullaondo and Lee, 2012b; Hakimi and Cannella, 2011), knowledge is limited on miRNA-regulated reactions against pathogenic bacteria and fungi. As detected by microarray using 455 arthropod mature miRNAs as probes, abundances of 59 miRNAs in Tribolium castaneum changed after injection of peptidoglycan (PG) from Micrococcus luteus (Freitak et al., 2012). Out of the 59, fourteen were previously identified in T. castanuem and the others are either conserved or novel miRNAs in other arthropods. While peptidoglycans initiate strong immune responses, differences exist in PGs from Gram-positive (G+) and Gram-negative (G-) bacteria, and PGs induced somewhat different responses as compared with whole bacteria (Sumathipala and Jiang, 2010). In Drosophila melanogaster, let-7 directly interacts with the 3’-UTR of an AMP gene diptericin and miR-8 negatively regulates the basal expression of diptericin and drosomycin without pathogen stimulation (Choi and Hyun, 2012; Garbuzov and Tatar, 2010). An in silico screening method was developed to predict miRNAs which may regulate D. melanogaster immune responses (Fullaondo and Lee, 2012a). However, there are no miRNA expression profiles presented and their abundances, based on the premise of expression co-regulation, were deduced from the microarray expression data of their adjacent genes. Differential regulation of D. melanogaster AMP genes in S2 and Sf9 cell lines implied that intracellular immune signaling pathways involve species-specific regulators (Rao et al., 2011). Upon encountering Serratia marcescens or M. luteus, Apis mellifera workers mounted immune responses and, among the thirteen miRNAs predicted to regulate immunity in the honeybee, only two exhibited significant changes at 6 h after S. marcescens infection (Lourenco et al., 2013). This result also suggests some miRNAs act differently in various insects and experimental data on levels of miRNAs and transcripts of their putative target genes are both needed to establish regulatory relationships. In the transcriptome analysis (Zhang et al., 2011; Gunaratna and Jiang, 2013), we determined the transcript levels of 232 putative immunity-related genes in M. sexta, which increased or decreased in fat body and/or hemocytes 24 h after injection of a mixture of bacteria and curdlan into the 5th instar larvae. Nevertheless, there is no report on related miRNA level changes and more efforts are needed to explore the expression regulation of M. sexta immunity-related genes by miRNAs.
In this work, we used the same total RNA samples from fat body (F) and hemocytes (H) of control (C) and bacteria-induced (I) M. sexta 5th instar larvae (Zhang et al., 2011) to prepare four small RNA libraries (CF, IF, CH and IH) for Illumina sequencing. Due to their spatiotemporal expression specificity, we were able to identify additional miRNAs identified from four developmental stages of M. sexta (Zhang et al., 2012). Numbers of miRNA reads were normalized and compared (CF vs. IF; CH vs. IH) to assess miRNA regulation upon pathogen invasion. We predicted miRNA target sites in 3’-UTRs of the 232 mRNAs that encode pathogen recognition proteins, hemolymph proteinases (HPs), serpins, AMPs, and members of the Toll, Imd, JNK, JAK-STAT and MAPK pathways. By correlating the miRNA and corresponding transcript levels (Zhang et al., 2011; Gunaratna and Jiang, 2013), we explored possible regulatory pairs of miRNA:mRNA for future research on M. sexta miRNA functions.
2. Materials and methods
2.1. Pathogen injection, total RNA extraction, and small RNA library construction
The same four total RNA samples (CF, IF, CH, IH) as used previously (Zhang et al., 2011) were used for small RNA library construction. Briefly, a mixture of E. coli, M. luteus, and curdlan was injected into day 2, 5th instar larvae (60) to induce immune responses. After 24 h, hemolymph was collected for hemocyte preparation and RNA isolation. Fat body was dissected from the induced larvae for RNA isolation. Similarly, hemocytes and fat body tissue were collected from day 3, 5th instar naïve larvae (60) for preparing control hemocyte and fat body RNA. The small RNA libraries were constructed for Illumina sequencing at National Center for Genome Resources (Santa Fe, NM) as described previously (Zhang et al., 2012).
2.2. Sequence analysis and identification of microRNAs
The analysis procedures were described previously (Zhang et al., 2012). Briefly, reads were first removed if they had no perfect match to 3’-adaptor sequence. Repeats, known noncoding RNAs (rRNAs, tRNAs, snRNAs, snoRNAs, etc.), mitochondrial nucleotide sequences were filtered out according to respective online databases. Compared to the M. sexta hemocyte-fat body EST dataset (http://ftp.genome.ou.edu/pub/for_haobo/manduca/fourlibrariesassembly/), M. sexta midgut EST dataset (http://rfc.ex.ac.uk/iceblast/iceblast.php) and M. sexta Cufflink RNA-Seq Assembly 1.0 (http://agripestbase.org/manduca/), possible degradation products of mRNAs were eliminated. The remaining sequences were aligned to miRBase (v20, http://www.miRBase.org/) to obtain conserved miRNAs and their frequencies. M. sexta Genome Assembly 1.0 (http://agripestbase.org/manduca/) was searched using the mature miRNA sequences to locate corresponding precursors and genomic loci, in which the precursors have at least 18 matched base pairs, only one central loop, and folding energy lower than −18 kCal/mol (http://mfold.rna.albany.edu/?q=mfold/RNA-Folding-Form) (Zuker, 2003). The other small RNA reads were designated as novel M. sexta mature miRNAs if they had fewer than 5 genomic loci, low-energy fold-back precursor structures, highest abundance among reads mapped to the respective precursors, and existence of predicted corresponding miRNA*s in the dataset. The ones without accompanying miRNA* sequences were named as novel miRNA candidates. Frequencies of conserved miRNAs and miRNA*s, novel miRNAs and miRNA*s, and novel miRNA candidates were calculated based on read numbers and library sizes.
2.3. Prediction of M. sexta miRNA targets
Quality of the 232 immunity-related transcripts (Gunaratna and Jiang, 2013) was improved using the hemocyte and fat body EST contigs (Zhang et al., 2012), 52 RNA-Seq datasets (Cao et al., unpublished data), and sequences in M. sexta Genome Assembly 1.0. While the open reading frames encode pattern recognition proteins, serine proteinases, serpins, intracellular immune signaling pathway members, and AMPs, their 3’-UTRs were retrieved for miRNA target site analysis using Hitsensor (Zheng and Zhang, 2010).
3. Results
3.1. Overview of the small RNA dataset
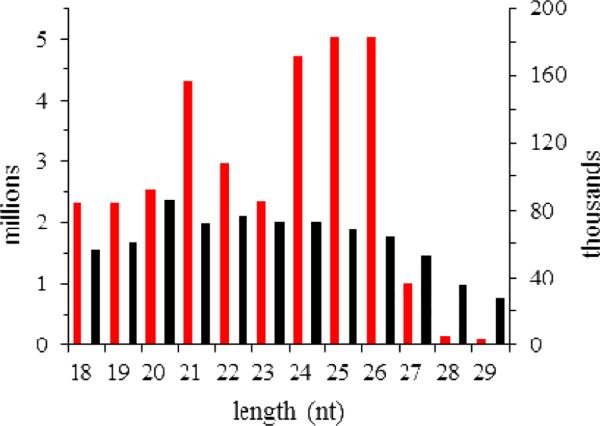
A total of 32.9 million reads were obtained by sequencing four independent small RNA libraries generated from CF, IF, CH and IH (Table 1). Length distribution of the total reads exhibited two peaks somewhat similar to those of the distinct M. sexta developmental stages (Zhang et al., 2012), whereas the unique read distribution did not have a peak between 26 and 28 nt (Fig. 1). As discussed before, the peak in the unique read distribution represented the robust diversity but low levels of piRNAs, which commonly function in germline development. Thus, the fat body and hemocytes small RNA libraries contained less piRNA size small RNAs than those of the whole insects. The M. sexta developmental series had 0.05% total reads matching the silkworm mRNAs (Zhang et al., 2012). Our current dataset had 3.07% total and 20.89% unique reads matching M. sexta Cufflinks 1.0 transcripts (Table 1). The higher percentages (3.76% total and 35.51% unique reads) of match with the smaller dataset of hemocyte, fat body, and midgut EST contigs indirectly reflected high redundancy of Cufflinks 1.0, which compromised its higher gene coverage. Given the large size of these tissue libraries, the sequence match greatly reduced the workload of data analysis. When compared with the 176 identified M. sexta miRNAs (Zhang et al., 2012), the number of unique reads matching miRBase precursors was much larger (Table 1). Many of them represented mature miRNAs, miRNA*s, or degradation products of the precursors, while others can be novel or conserved miRNAs not found before.
Table 1.
Absolute read numbers in different RNA categories in the small RNA libraries*
| Category | CF | IF | CH | IH | ||||
|---|---|---|---|---|---|---|---|---|
| total | unique | total | unique | total | unique | total | unique | |
| Noncoding RNAs | 357,010 | 46,860 | 259,482 | 40,753 | 1,160,076 | 106,739 | 395,502 | 32,400 |
| miRBase precursors | 157,866 | 2,813 | 153,771 | 2,995 | 438,471 | 5,179 | 299,117 | 3,777 |
| Hemocyte, fat body, and midgut ESTs | 246,461 | 55,507 | 138,235 | 47,007 | 888,465 | 134,125 | 86,099 | 29,799 |
| Cufflinks transcripts | 206,609 | 30,535 | 157,336 | 26,935 | 511,945 | 75,048 | 237,804 | 24,220 |
| Repeats | 68,889 | 16,289 | 41,293 | 13,937 | 294,669 | 36,415 | 42,546 | 10,978 |
| Genome Assembly 1.0 | 306,216 | 41,301 | 239,380 | 36,791 | 883,999 | 97,738 | 375,741 | 34,812 |
| Total | 3,333,930 | 138,886 | 11,810,233 | 162,065 | 8,686,565 | 295,841 | 9,070,983 | 153,632 |
CF, IF, CH, and IH: control (C) and induced (I) fat body (F) and hemocytes (H). The unique read numbers are the counts after the removal of redundant reads.
Fig. 1.
Size distributions of numbers of the total (red bars, left y-axis) and unique (black bars columns, right y-axis) reads in the four libraries combined.
3.2. New novel and conserved miRNAs
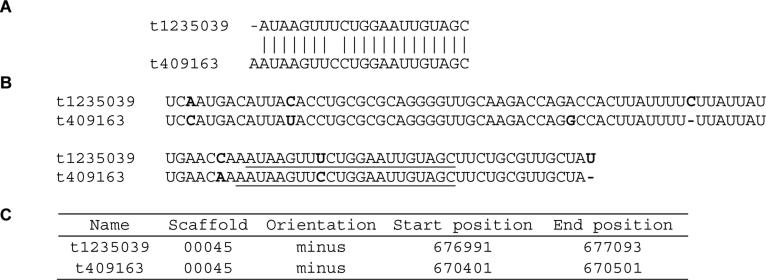
We indeed identified four novel miRNAs and respective star strands, all of which had low read numbers (Table 2). Their predicted precursors had low-energy fold-back structures (Fig. 2). Their minimal free energy was all below −18 kCal/mol: −40.0, −38.3, −37.7 and −39.6 for t3040582, t1306847, t1235039 and t409163, respectively. We also found a cluster of two miRNA (t1235039 & t409163) residing closely in the genome (Fig. 3). Their precursor and mature strands are highly similar, suggesting they arose from recent gene duplication. Since their seed region (nucleotides 2-7), which is critical for target recognition, is identical, t1235039 and t409163 may regulate similar genes.
Table 2.
Novel miRNAs identified in the four libraries*
| Name | Mature miRNA sequence | CF | IF | CH | IH | ||||
|---|---|---|---|---|---|---|---|---|---|
| miR | miR* | miR | miR* | miR | miR* | miR | miR* | ||
| t3040582 | GAAUUGACCAAUUGUAGGGAGUC | 0 | 0 | 0 | 1 | 1 | 0 | 0 | 0 |
| t1306847 | AUAUAUUGAUUCCGAUACACAAG | 0 | 1 | 1 | 0 | 0 | 0 | 0 | 1 |
| t1235039 | AUAAGUUUCUGGAAUUGUAGC | 0 | 1 | 0 | 1 | 0 | 0 | 1 | 2 |
| t409163 | AAUAAGUUCCUGGAAUUGUAGC | 1 | 4 | 0 | 1 | 1 | 13 | 2 | 5 |
Read numbers are absolute values from each of the control (C) and induced (I) fat body (F) and hemocyte (H) libraries.
Fig. 2.
Predicted stem-loop structures of novel M. sexta miRNAs. The precursor sequences are retrieved from the genome based on the loci of mature and star strands (Section 2.2), with the mature ones shown in bold red capital letters.
Fig. 3.
One cluster of novel miRNAs. A) Alignment of the mature miRNA sequences with identical residues labeled “│”. B) alignment of the miRNA precursor sequences with different residues shown in bold red font and mature miRNA sequences underlined. C) Genomic loci of the miRNA precursors in M. sexta Genome Assembly 1.0.
The presence of a corresponding star strand is an essential criterion to validate a novel miRNA (Section 2.2). Because the star strands are degraded rapidly in most cases, failure to detect them by deep sequencing leads to lists of novel miRNA candidates (Table S2 in Zhang et al., 2012; Table S1). Among the 28 new ones, five have more than one gene copy and constitute two potential miRNA families: t1480635 (3) and t6233600 (2). Interestingly, t6233600a and t6233600b reside in the same genomic location but use opposite DNA strands as templates and, hence, may act as antisense miRNAs. Likewise, t454580 and t4479723 are also putative antisense miRNAs.
We predicted six conserved miRNAs based on M. sexta Genome Assembly 1.0 but did not detect them in the developmental series (Zhang et al., 2012). Here, mature strands of mse-miR-1b and mse-miR-929 are found (Table 3). mse-miR-1b is the antisense miRNA of mse-miR-1 and, due to differences in the seed regions, they are hypothesized to regulate different genes, including those involved in M. sexta immunity (Table 4).
Table 3.
Abundance of miRNAs with precursors identified*
| name | miRNA | miRNA* | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CF | IF | CH | IH | IF/CF | IH/CH | CF | IF | CH | IH | IF/CF | IH/CH | |
| mse-miR-1 | 48891 | 5310 | 3012 | 1293 | 0.11 | 0.43 | ||||||
| mse-miR-1b | 6 | 3 | 8 | |||||||||
| mse-miR-2a | 897 | 318 | 1234 | 482 | 0.35 | 0.39 | 456 | 107 | 404 | 216 | 0.23 | 0.53 |
| mse-miR-2b | 243 | 82 | 367 | 160 | 0.34 | 0.44 | 84 | 19 | 50 | 23 | 0.23 | 0.46 |
| mse-miR-7 | 90 | 16 | 205 | 118 | 0.18 | 0.58 | 1 | |||||
| mse-miR-8 | 34494 | 24829 | 7961 | 8698 | 0.72 | 1.09 | 10939 | 1469 | 4233 | 2888 | 0.13 | 0.68 |
| mse-miR-9a | 660 | 264 | 715 | 354 | 0.40 | 0.50 | 63 | 64 | 77 | 22 | 1.02 | 0.29 |
| mse-miR-9b | 117 | 14 | 10 | 4 | 0.12 | 0.40 | 201 | 14 | 17 | 2 | 0.07 | 0.12 |
| mse-miR-10a | 771 | 88 | 130 | 114 | 0.11 | 0.88 | 9 | 3 | 2 | |||
| mse-miR-11 | 1830 | 563 | 2065 | 2937 | 0.31 | 1.42 | 1 | |||||
| mse-miR-12 | 828 | 541 | 140 | 247 | 0.65 | 1.76 | ||||||
| mse-miR-14 | 195 | 221 | 579 | 72 | 1.13 | 0.12 | 33 | 29 | 39 | 20 | 0.88 | 0.51 |
| mse-miR-31 | 3 | 5 | 2 | 110299 | 10207 | 71413 | 34506 | 0.09 | 0.48 | |||
| mse-miR-33 | 180 | 80 | 97 | 47 | 0.44 | 0.48 | 1 | 3 | 2 | |||
| mse-miR-34 | 309 | 179 | 585 | 677 | 0.58 | 1.16 | 11 | 12 | 20 | 1.67 | ||
| mse-miR-71 | 732 | 229 | 877 | 993 | 0.31 | 1.13 | 3 | 28 | 56 | 20 | 9.33 | 0.36 |
| mse-miR-79 | 336 | 306 | 361 | 191 | 0.91 | 0.53 | 12 | 10 | 35 | 25 | 0.83 | 0.71 |
| mse-miR-87 | 243 | 76 | 300 | 349 | 0.31 | 1.16 | 2 | 3 | 1 | |||
| mse-miR-92a | 30 | 30 | 63 | 77 | 1.00 | 1.22 | 3 | |||||
| mse-miR-92b | 6 | 10 | 3 | 40 | 1.67 | 13.33 | 1 | |||||
| mse-miR-100 | 33 | 26 | 28 | 2 | 0.79 | 0.07 | ||||||
| mse-miR-124 | 2 | |||||||||||
| mse-miR-133 | 9 | 1 | ||||||||||
| mse-miR-184 | 82770 | 22483 | 67853 | 105093 | 0.27 | 1.55 | 1 | |||||
| mse-miR-190 | 78 | 14 | 56 | 108 | 0.18 | 1.93 | 1 | 7 | 3 | |||
| mse-miR-252 | 2715 | 482 | 433 | 513 | 0.18 | 1.18 | ||||||
| mse-miR-263a | 1377 | 137 | 280 | 180 | 0.10 | 0.64 | 3 | 2 | ||||
| mse-miR-263b | 3 | |||||||||||
| mse-miR-275 | 1515 | 524 | 684 | 853 | 0.35 | 1.25 | 3 | 2 | 1 | |||
| mse-miR-276 | 2460 | 1898 | 2916 | 3564 | 0.77 | 1.22 | 330 | 167 | 547 | 345 | 0.51 | 0.63 |
| mse-miR-277 | 219 | 429 | 181 | 417 | 1.96 | 2.30 | 3 | 9 | 1 | |||
| mse-miR-278 | 27 | 26 | 30 | 30 | 0.96 | 1.00 | 6 | 2 | ||||
| mse-miR-279a | 780 | 207 | 832 | 298 | 0.27 | 0.36 | 9 | 2 | 15 | 13 | 0.22 | 0.87 |
| mse-miR-279b | 1266 | 668 | 800 | 340 | 0.53 | 0.43 | 2 | 3 | ||||
| mse-miR-279c | 210 | 130 | 100 | 67 | 0.62 | 0.67 | 2 | 3 | ||||
| mse-miR-279d | 14214 | 3779 | 12756 | 3939 | 0.27 | 0.31 | 2 | 3 | 4 | |||
| mse-miR-281 | 6 | 4 | 1 | 1 | 2595 | 950 | 262 | 299 | 0.37 | 1.14 | ||
| mse-miR-282 | 318 | 42 | 45 | 24 | 0.13 | 0.53 | 3 | |||||
| mse-miR-283 | 66 | 110 | 26 | 112 | 1.67 | 4.31 | ||||||
| mse-miR-306 | 9997 | 2065 | 3447 | 406 | 0.21 | 0.12 | ||||||
| mse-miR-307 | 32115 | 10677 | 128054 | 22303 | 0.33 | 0.17 | 1 | 2 | ||||
| mse-miR-308 | 927 | 179 | 328 | 128 | 0.19 | 0.39 | 4 | 9 | 4 | |||
| mse-miR-316 | 57 | 3 | 5 | 3 | 0.05 | 0.60 | 1 | |||||
| mse-miR-317 | 282 | 319 | 611 | 335 | 1.13 | 0.55 | 3 | 3 | 1 | |||
| mse-miR-745 | 786 | 292 | 1791 | 627 | 0.37 | 0.35 | ||||||
| mse-miR-750 | 15 | 3 | 6 | 1 | 0.20 | 0.17 | 2 | |||||
| mse-miR-929 | 3 | |||||||||||
| mse-miR-932 | 3 | 6 | 7 | 3 | 1 | |||||||
| mse-miR-965 | 105 | 25 | 66 | 6 | 0.24 | 0.09 | 228 | 43 | 135 | 172 | 0.19 | 1.27 |
| mse-miR-970 | 16353 | 8984 | 58144 | 52406 | 0.55 | 0.90 | 1 | 3 | 6 | |||
| mse-miR-989 | 30 | 3 | 17 | 7 | 0.10 | 0.41 | ||||||
| mse-miR-998 | 30 | 25 | 111 | 165 | 0.83 | 1.49 | ||||||
| mse-miR-2755 | 696 | 682 | 1325 | 848 | 0.98 | 0.64 | 36 | 22 | 35 | 29 | 0.61 | 0.83 |
| mse-miR-2763 | 42 | 6 | 7 | 4 | 0.14 | 0.57 | ||||||
| mse-miR-2766 | 10960 | 4959 | 17985 | 9178 | 0.45 | 0.51 | 399 | 131 | 381 | 127 | 0.33 | 0.33 |
| mse-miR-2767 | 5387 | 3710 | 14360 | 7845 | 0.69 | 0.55 | ||||||
| mse-miR-2768 | 9 | 4 | ||||||||||
| mse-miR-2779 | 96 | 21 | 36 | 44 | 0.22 | 1.22 | ||||||
| mse-miR-2796 | 1 | |||||||||||
| mse-miR-3286 | 1 | |||||||||||
| mse-miR-6100 | 426 | 46 | 247 | 169 | 0.11 | 0.68 | ||||||
| mse-bantam | 549 | 329 | 498 | 419 | 0.60 | 0.84 | 1 | 2 | ||||
| mse-miR-iab-4 | 30 | 47 | 6 | 6 | 1.57 | 1.00 | ||||||
| mse-let-7a | 10300 | 4919 | 19444 | 10340 | 0.48 | 0.53 | 3 | 1 | ||||
Abundance is shown with normalized read numbers (reads per million) in the control (C) and induced (I) fat body (F) and hemocyte (H) libraries. Read numbers are shown as blank for those either non-detectable or whose values are below 0.5 after normalization. Up (I/C >1.25) and down (I/C <0.80) regulated ones are shaded orange and green, respectively.
Table 4.
Putative immunity-related target genes for M. sexta miRNAs
| Name | Putative Targets |
|---|---|
| mse-miR-1 | ANKRD54, cecropin-like, Draper, Hdd13, Tab2, tetraspanin |
| mse-miR-1b | Aminoacylase, PPBP1, MASK |
| mse-miR-2a | Aop, Atg6, Domeless, galectin-2, Hdd23, Pelle, PI-like, salivary cysteine-rich peptide |
| mse-miR-2b | Aop, Atg6, Domeless, galectin-2, Hdd23, Pelle, PI-like, salivary cysteine-rich peptide |
| mse-miR-7 | Aop, ERK, focal adhesion kinase, galectin-4, Hdd13, HP12, JAK/Hopscotch, MyD88, serpin4, tyrosine protein kinase |
| mse-miR-8 | aPKC, cdc42, ERK, focal adhesion kinase, HP17, HP17s, integrin linked protein kinase, Jra, MLK1, p38, PPBP2, Pelle, PPO2, protein phosphatase type 2c, salivary cysteine-rich peptide, Serrate, Tollip |
| mse-miR-9a | Brahma, Cactus, cecropin-like, CTL10, Domeless, Eiger, FADD, GTP/GDP exchange factor, Hdd1, HP17, HP17s, HP21, HP5, integrin β1, moricin, PAP2, PSP, Punch, Rac1, thioredoxin peroxidase-3, transferrin, tyrosine protein kinase |
| mse-miR-9b | Alk, Domeless, Draper, HP22, HP7, Imd, p38, protein phosphatase type 2c, PVR, serpin4 |
| mse-miR-10a | aPKC, βGRP3, cecropin-like, Domeless, PGRP-L2, Phe hydroxylase, PI6, Spz1A, Spz1B, STAT, Tab2, TAK1 |
| mse-miR-11 | Aop, attacin1, Brahma, Domeless, Dsor1, FADD, HP12, HP14, HP8, IKKβ, integrin related 1, JAK/Hopscotch, lectin, Misshapen, PPBP2, PGRP-L2, protein phosphatase type 2c, PPO1, PSP, Ref2P, serpin4, serpin6, Tab2, tetraspanin |
| mse-miR-12 | Aminoacylase, βGRP2, cecropin B, HP5, HP6, IKKβ, MLK1, PAP3, PPBP2, serpin3, tetraspanin, thioredoxin peroxidase-3 |
| mse-miR-14 | ANKRD54, Dicer-2, HP7, JAK/Hopscotch, MEKK1, PGRP-L5, Stam, tetraspanin, thioredoxin peroxidase-3 |
| mse-miR-31 | ECSIT, MEKK1, Rel2B |
| mse-miR-33 | Alk, JAK/Hopscotch, Notch |
| mse-miR-34 | cdc42, Draper, ERK, Hdd13, Hem, integrin β1, integrin related-1, JNK, MKK4, nuclear transport factor 2, Serrate, tetraspanin, Ubc13/ben |
| mse-miR-71 | Atg4LP, Domeless, Dscam, gallerimycin, Lesswright, MKK4, MLK1, MyD88, PAP2, PGRP-L2, PGRP-L5, Punch, Rel2A |
| mse-miR-79 | Alk, Domeless, Draper, HP2, p38, protein phosphatase type 2c, serpin4 |
| mse-miR-87 | Draper, FADD, HP13, IML3, Spz1A, Spz1B, Tab2, tetraspanin, Tollip |
| mse-miR-92b | Dsor1, HP13, HP2, JAK/Hopscotch, JNK, leureptin 1, MLK1, MyD88, neuroglian, Rel2A, STAT |
| mse-miR-100 | Argonaute-1, MyD88 |
| mse-miR-184 | HP12, p38 |
| mse-miR-190 | Dicer-2, focal adhesion kinase, PGRP-L2, PPO1 |
| mse-miR-252 | Cecropin-like, Dicer-2, Domeless, Hdd1, Hdd13, IKKβ, integrin β1, Jra, PAP2, Ref2P, scolexinB |
| mse-miR-263a | ANKRD54, Atg4, Cactus, Draper, ECSIT, Eiger, HP14, integrin linked protein kinase, Licrone/MKK3, protein phosphatase type 2c, PSP precursor, serpin3, serpin4, tetraspanin |
| mse-miR-275 | HP12, integrin β1, PIAS, protein phosphatase type 2c, Rac1, Spz1A, Spz1B, tetraspanin, tyrosine protein kinase, Uba2 |
| mse-miR-276 | Domeless, Dsor1, IML3, leureptin 1, secreted peptide 30, Tab2 |
| mse-miR-277 | Aop, Atg3, Domeless, Draper, Dscam, Dsor1, ERK, focal adhesion kinase, galectin-2, HP12, HP13, HP6, HP8, IAP, JAK/Hopscotch, Lesswright, MEKK1, MKK4, p38, protein phosphatase type 2c, Rac1, serpin2, serpin4, Sickie, SPH2, tetraspanin, tyrosine protein kinase |
| mse-miR-279a | GTP/GDP exchange factor, HP12, integrin related 1, integrin linked protein kinase, protein phosphatase type 2c, Rac1, tetraspanin, Ubc13/ben |
| mse-miR-279b | Atg4, GTP/GDP exchange factor, HP12, integrin related 1, integrin linked protein kinase, protein phosphatase type 2c, Rac1, Ubc13/ben |
| mse-miR-279c | GTP/GDP exchange factor, IKKγ, integrin related 1, integrin linked protein kinase, protein phosphatase type 2c, tetraspanin, Ubc13/ben |
| mse-miR-279d | Atg4, GTP/GDP exchange factor, Hdd1, HP12, HP6, IKKγ, integrin related 1, integrin linked protein kinase, protein phosphatase type 2c, scolexinB, tetraspanin, Ubc13/ben |
| mse-miR-281 | aPKC, Dsor1, PPBP2 |
| mse-miR-282 | galectin-2, Hdd1, PGRP-L2 |
| mse-miR-283 | Atg3, Atg4, Atg4LP, attacin1, attacin3, Domeless, galectin-4, GTP/GDP exchange factor, Hdd1, IKKβ, Imd, integrin related 1, integrin linked protein kinase, JAK/Hopscotch, lebocin B, Misshapen, MLK1, Notch, Nuclear transport factor 2, PPBP2, PGRP-L2, protein phosphatase type 2c, PSP, Punch, PVR, Rel2A, Tab2, tyrosine hydroxylase |
| mse-miR-306 | HP12, PIAS, Rac1, Spz1A, Spz1B, Uba2 |
| mse-miR-307 | aPKC, HP19, lysozyme, MLK1, MASK, scolexinB |
| mse-miR-308 | Aop, Atg4LP, βGRP2, Dsor1, HP12, HP8, IKKβ, JAK/Hopscotch, Misshapen, PPBP3, PGRP-L2, PPO1, PI-like, PSP, Rac1, Rel2A, salivary cysteine-rich peptide, serpin4, serpin7, SPH1b, Tab2, TEP1, TEP2, tetraspanin |
| mse-miR-316 | Aop, Argonaute-1, Atg2, aPKC, Domeless, focal adhesion kinase, galectin-2, Hem, HP7, IAP, Lacunin, MEKK1, Rac1, tetraspanin |
| mse-miR-317 | Atg4LP, Draper, Licrone/MKK3, PGRP-L2, PGRP-L5, Ral GTP/GDP exchange factor, MASK, STAT, thioredoxin peroxidase-2 |
| mse-miR-745 | Atg3, IAP, IKKβ, MLK1, Tube |
| mse-miR-750 | CTL10, Ref2P |
| mse-miR-965 | Alk, GTP/GDP exchange factor, MLK1, STAT, TEP1 |
| mse-miR-970 | FADD, hemicentin-2, serpin3, tetraspanin, Tollip |
| mse-miR-989 | Atg4LP, aPKC, nuclear transport factor 2, serpin4, Spz1A, Spz1B, transferrin |
| mse-miR-998 | galectin-2, GTP/GDP exchange factor, p38, protein phosphatase type 2c |
| mse-miR-2755 | Aop, Eiger, PGRP-L2, TEP1 |
| mse-miR-2763 | Alk, ANKRD54, Atg2, Atg3, Atg4, Atg4LP, βGRP2, Domeless, FADD, Hdd13, hemicentin-1, HP14, JAK/Hopscotch, Licrone/MKK3, p38, PAP3, PGRP-L2, PGRP-L5, Phe hydroxylase, PVR, Rel2A, Rel2B, STAT, tetraspanin |
| mse-miR-2766 | ANKRD54, antileukoproteinase, Atg3, Dicer-2, dipeptidyl peptidase, FADD, galectin-4, Hem, hemocyte-specific integrin α2, HP6, HP7, JAK/Hopscotch, Kazal type PI, Lesswright, Licrone/MKK3, MEKK1, Notch, PGRP-L5, PIAS, protein phosphatase type 2c, TEP2, tetraspanin |
| mse-miR-2767 | Aos1, Atg6, cdc42, Draper, Eiger, FADD, HAIP, HP14, HP7, IKKγ, Imd, MEKK1, tyrosine protein kinase |
| mse-miR-2779 | Hdd13, TEP2 |
| mse-miR-6100 | Dopa decarboxylase, MLK1, Punch |
| mse-bantam | galectin-2, p38, PVR, Ubc13/ben |
| mse-miR-iab-4 | Domeless, Dsor1, Hdd13, IAP, lebocinD, MEKK1, PGRP-L5, PSP, Sickie, thioredoxin peroxidase-3 |
| mse-let-7a | cecropin B, Dsor1, leureptin 1, MLK1, serpin4 |
| mse-miR-2a* | Alk, ANKRD54, Atg4, cdc42, Dscam, Imd, PGRP-L2, Ral GTP/GDP exchange factor, SOCS, STAT, thioredoxin peroxidase-2 |
| mse-miR-2b* | Alk, Atg4LP, Atg6, Dredd/Caspase 6, Dsor1, FADD, galectin-4, Hdd1, hemocyte specific integrin α1, HP14, HP4, HP5, IKKp, Integrin β1, JAK/Hopscotch, Jra, leureptin-1, PPBP2, Pelle, PGRP-L5, PGRP-SA, Ras85D, Serrate, tetraspanin, Tollip |
| mse-miR-8* | ANKRD54, Atg2, hemolectin, HP14, HP19, integrin related 1, TAK1, tyrosine hydroxylase, Vrille |
| mse-miR-9a* | Dscam, ECSIT, ERK, Hdd1, IML3, JAK/Hopscotch, Nimrod A, Pellino, PGRP-L2, protein phosphatase type 2c, Rac1, Rel2A, Rel2B |
| mse-miR-9b* | ERK, HP22, JAK/Hopscotch, Nimrod A, Pellino, PGRP-L2, protein phosphatase type 2c, Rac1, Rel2A, Rel2B |
| mse-miR-14* | serpin4 |
| mse-miR-31* | Aos1, βGRP2, Domeless, HP6, HP8, leureptin-1, PGRP-L2, secreted peptide 30 |
| mse-miR-34* | Atg12, Dsor1, ERK, hemocyte specific integrin α1, integrin related 1, PPBP2, Pelle, PI6, MASK, Rel2B, tetraspanin, Tube |
| mse-miR-71* | Domeless, IML3, lebocinD, Licrone/MKK3, MLK1, MASK, Sickie, tetraspanin |
| mse-miR-79* | HP12, JNK, Rac1, serpin6 |
| mse-miR-279a* | Caspar, cdc42, lysozyme, serpin4, Serrate |
| mse-miR-281* | Domeless, hemolectin, Misshapen, Nuclear transport factor 2, serpin4 |
| mse-miR-965* | Atg4LP, hemocyte specific integrin α2, JNK, Rel2B, tyrosine protein kinase |
| mse-miR-2755* | Atg4LP, Dscam, ERK, FADD, Hdd13, HP6, serpin6, SPH4 |
| mse-miR-2766* | Dsor1, Licrone/MKK3 |
3.3. Abundance changes of miRNAs after the immune challenge
Read numbers were normalized against total read numbers of respective small RNA libraries. Normalized read counts for miRNAs with and without identified precursors (Tables 3 & S2) allowed us to omit those with normalized abundances <10 in all four libraries and calculate IF/CF and IH/CH to reveal up- and down-regulation after the immune challenge. We found 77, 14 and 11 of the 102 qualified miRNAs fell into the IF/CF value ranges of 0 to 0.80, 0.80 to 1.25, and above 1.25, respectively. Similarly, 64, 21 and 20 of the 105 miRNAs had IH/CH ratios in the categories of down, no, and up regulation. Due to incomplete coverage of the M. sexta genome assembly and lack of identified precursors for miRNA variants, we focused on those with precursors (Table 3). Thirty-four miRNAs including mse-miR-1, -2a, -2a*, -2b, -2b*, -7, -8*, -9a, -9b, -9b*, -31*, -33, -100, -263a, -276*, -279a, -279b, -279c, -279d, -282, -306, -307, -308, -316, -745, -750, -965, -989, -2763, -2766, -2766*, -2767, -6100 and -let-7a were down-regulated in both fat body and hemocytes. In contrast, mse-miR-92b, -277, and -283 levels increased in both tissues after the immune challenge. There were 14 miRNAs down-regulated in fat body and unchanged in hemocytes: mse-miR-8, -10a, -34, -71, -87, -252, -275, -276, -279a*, -281*, -970, -2755*, -2779, and -bantam. Levels of mse-miR-11, -12, -184, -190 and -965* became lower in fat body but higher in hemocytes. Seven miRNAs (mse-miR-9a*, -14, -14*, -79, -79*, -317, and -2755) were down-regulated in hemocytes and unchanged in fat body. Besides, mse-miR-998 level increased in hemocytes but did not change in fat body; mse-miR-iab-4 level increased in fat body but did not change in hemocytes.
3.4. miRNA*s maintained at high levels
We also examined levels of miRNA-miRNA* pairs and found several miRNA*s were comparable to their mature strands. Mse-miR-281*, mse-miR-965* and mse-miR-31* were more abundant than mse-miR-281, mse-miR-965 and mse-miR-31 in all four samples (Table 3). The three miRNA*s exhibited the same preference over the developmental course (Zhang et al., 2012), implying the dominant usage of their passenger arms of the miRNA precursors. During development, mse-miR-9a*, mse-miR-9b* and mse-miR-2a* in whole animals were present at similar levels to the respective miRNAs. However, in the fat body and hemocyte small RNA libraries, only mse-miR-9b* showed a similar pattern (Table 3). Members of the same miRNA family could have distinct preferences in terms of star strand maintenance, as is the case with mse-miR-9a and mse-miR-9b. The differences in miRNA* abundances, between the whole insect development series and the tissue-immunity series, may reflect variations in spatiotemporal regulation of miRNA expression and maturation.
3.5. Target sites in the immunity-related genes
For miRNA target analysis, we focused on the 232 immunity-related genes expressed in the fat body and hemocytes (Zhang et al., 2011; Gunaratna and Jiang, 2013) and collected their 3’-UTRs. We disregarded mse-miR-92a and mse-miR-278, whose levels remained similar in both tissues after the bacterial injection, and the ones with no normalized read numbers exceeding 10 in all the tissue samples except for mse-miR-1b, -31 and -281. The putative targets (Table 4) include pattern recognition proteins, AMPs, and members of the PPO system, cellular immunity, and conserved intracellular signaling pathways such as Toll, Imd, JAK-STAT, MAPK-JNK-p38, and small-interfering RNA.
4. Discussion
4.1. miRNAs and corresponding miRNA*s may regulate different genes
miRNAs and miRNA*s differed in abundance maintenance and some miRNA*s exhibited different patterns of level changes in comparison to respective miRNAs (e.g. miR-276 and -276*; miR-279a and -279a*; miR-965 and -965*) (Table 3). Induction or suppression of miRNA*s was observed in Lymantria dispar and Plutella xylostella after being parasitized by Glyptapanteles flavicoxis and Diadegma semiclausum, respectively (Etebari et al., 2013; Gundersen-Rindal and Pedroni, 2010), supporting that miRNA*s may play substantial roles in the regulation of immunity against bacteria and fungi. Comparison of potential targets of miRNAs and miRNA*s showed, while certain miRNA:miRNA* pairs (e.g. miR-2b and -2b*, miR-9a and -9a*) shared a few putative targets, the entire target lists cover diverse immunity genes (Table 4). Notably, out of the four miRNA*s discussed in Section 3.4, miR-9b:9b* shared two potential targets (HP22 and protein phosphatase type 2c), while the other three pairs may regulate different targets. Conserved miR-8 was validated to substantially regulate AMP production in fat body of D. melanogaster and P. xylostella (Choi and Hyun, 2012; Etebari and Asgari, 2013). Our data supported that by showing mse-miR-8 was down-regulated only in fat body. Intriguingly, with similar high levels of mature strands (Table 3), only mse-miR-8* was maintained at a relatively high level (around 1/3 to 1/2 compared to mature levels in CF, CH and IH) and the ratio of IF/CF was 0.13. Thus, mse-miR-8* is likely involved in regulating immunity-related genes in fat body. To date, there has been no validation of biological functions of miRNA*s in insects; future work is necessary to confirm the regulation of miRNA mature and star strands in an immune responsive M. sexta cell line.
4.2. Possible functional pairs of miRNA:mRNA in M. sexta immunity
Compared with other target prediction algorithms, Hitsensor exhibited superior performance with a testing pool of the validated miRNA:mRNA pairs (Zheng and Zhang, 2010). Nevertheless, we cannot exclude the possibility of false-positive predictions. Our quantitative analyses of the same RNA samples revealed changes in immunity-related transcript levels in M. sexta hemocytes and fat body (Gunaratna and Jiang, 2013; Zhang et al., 2011). While their expression can be regulated at the transcription level, we looked for negative correlations between miRNA/miRNA* levels and their putative target mRNA levels, which suggest contribution of miRNAs in post-transcriptional regulation of gene expression.
4.2.1. Pattern recognition receptors (PRRs)
PRRs are proteins that recognize molecular patterns on the surface of microbes, such as peptidoglycans (PGs), lipopolysaccharide (LPS), and β-1,3-glucan. PGRPs bind bacterial PGs and βGRPs bind fungal β-1,3-glucans. M. sexta βGRP2 mRNA level drastically increased upon immune challenge in both fat body and hemocytes while βGRP3 was only up-regulated in fat body. Only mse-miR-10a was predicted to target βGRP3 and mse-miR-10a was only down-regulated in fat body. Thus, mse-miR-10a had the potential to modulate the level of βGRP3 mRNA. Three miRNAs (mse-miR-308, -2763, and -31*) became less abundant in hemocytes and fat body after the immune challenge. Interestingly they all seem to target βGRP2 mRNA and their disregulation may have contributed to the up-regulation of βGRP2 expression. Mse-miR-12 may also regulate the βGRP2 transcript level in fat body. M. sexta immulectins (IMLs) recognize LPS on G- bacteria and IML3 was predicted as a potential target of mse-miR-87, -276, -9a* and -71*. IML3 up-regulation in fat body concurred with the mse-miR-87 and -276 down-regulation in the same tissue. Also in fat body, the increase in M. sexta C-type lectin-10 (CTL10) mRNA level concurred with the abundance decrease in mse-miR-9a and -750, putative regulators of the CTL. Two LPS-binding leureptins were found in M. sexta and leureptin-1 mRNA was highly induced in hemocytes and less so in fat body. Among the five miRNAs targeting leureptin-1, mse-let-7a, -2b* and -31* levels decreased in both tissues. The increases in Ig domain-containing hemicentin-1 and -2 mRNA levels in fat body were accompanied by abundance decreases of their potential regulatory miRNAs (mse-miR-2763 for hemicentin-1 and mse-miR-970 for hemicentin-2) after the immune challenge. In summary, transcripts of one PRR may be regulated by one or more miRNAs; mse-miR-2763 and -31* could control multiple PRRs by targeting their transcripts.
4.2.2. Extracellular signal transduction and melanization
We found miRNAs may regulate the mRNAs of 13 HPs, 2 PPO-activating proteinases (PAPs), 3 serine proteinase homologs (SPHs), and 5 serpins that mediate and modulate the extracellular signal transduction for immune responses (Table 4). They may also contribute to fine tuning of the gene expression for melanization, involving Phe and Tyr hydroxylases, Punch, dopa decarboxylase, and PPOs. After examination of the miRNA and mRNA levels, we identified the putative regulatory pairs if they displayed opposite trends of change after the bacterial injection in either fat body or hemocytes (Table 5). HP6 activates proHP8 and HP8 activates pro-Spätzle to induce the Toll pathway (An et al., 2010). Mse-miR-31* seems to regulate levels of both HP6 and HP8 transcripts, which is crucial to the Toll pathway activation. HP14, HP21, HP6, PAPs and SPHs form a PPO activation system to generate active compounds and melanin to kill and sequestrate pathogens. HP21 cleaves proPAP2/3 and mse-miR-9a potentially regulates HP21 and PAP2 mRNA levels. Mse-miR-2763 may affect HP14 and PAP3 transcript abundances. The 3’-UTR of HP22 contains putative recognition sites of both strands of the mse-miR-9b duplex. Serpins inhibit cognate serine proteinases to modulate the extracellular signaling pathways. Serpin-3, -4, -6, and -7 inhibit three (HP8, PAP1, PAP3), three (HP1, 6, 21), three (HP8, PAP1, PAP3) and one (PAP3) proteinases, respectively (Christen et al., 2012; Jiang, 2008; Suwanchaichinda et al., 2013). Notably, mse-miR-11, -263a, and -308 may down-regulate the transcripts of serpin-4 & 6, serpin-3 & 4, and serpin-4 & 7, respectively. While mse-miR-12 may regulate the expression of a serpin-proteinase pair (i.e. serpin-3 and PAP3), mRNA levels of serpins and target serine proteinases seem to be modulated by different miRNAs in most cases.
Table 5.
miRNA:mRNA pairs with reverse profiles involved in extracellular signal transduction and melanization
| Target gene | Putative regulatory miRNA |
|---|---|
| HP2 | mse-miR-79 |
| HP5 | mse-miR-9a, -2b* |
| HP6 | mse-miR-12, -279d, -2766, -31* |
| HP7 | mse-miR-9b, -316, -2766, -2767 |
| HP8 | mse-miR-308, -31* |
| HP14 | mse-miR-263a, -2763, -2767, -2b*, -8* |
| HP17 | mse-miR-8, -9a |
| HP19 | mse-miR-307, -8* |
| HP21 | mse-miR-9a |
| HP22 | mse-miR-9b, -9b* |
| PAP2 | mse-miR-9a, -71, -252 |
| PAP3 | mse-miR-12, -2763 |
| SPH1 | mse-miR-308 |
| Serpin3 | mse-miR-12, -263a, -970 |
| Serpin4 | mse-miR-7, -9b, -11, -79, -263a, -308, -989, -let-7a, -281* |
| Serpin6 | mse-miR-11, -79* |
| Serpin7 | mse-miR-308 |
| tyrosine hydroxylase | mse-miR-8* |
| dopa decarboxylase | mse-miR-6100 |
| Phe hydroxylase | mse-miR-10a, -2763 |
| Punch | mse-miR-9a, -71, -6100 |
During melanogenesis, Phe and Tyr hydroxylases, Punch, and dopa decarboxylase were highly induced in fat body and/or hemocytes (Gunaratna and Jiang, 2013). Intriguingly, M. sexta novel miRNA mse-miR-6100 may modulate mRNA levels of punch and dopa decarboxylase to affect melanization. Although mRNA levels of PPOs did not change much in hemocytes after the immune challenge, active POs play important roles in various insect physiological processes. Further studies are needed to test if PPO1 and PPO2 transcripts are regulated by mse-miR-11, -190, 308, and/or -8.
4.2.3. Antimicrobial proteins/peptides (AMPs)
Antimicrobial effectors kill invading pathogens and their synthesis is highly induced in fat body as well as hemocytes. Nineteen miRNAs probably recognize 3’-UTRs of the transcripts of attacin-1 and -3, lebocins B and D, cecropin B and cecropin-like peptide, moricin, lysozyme, transferrin, gallerimycin, salivary Cys-rich peptide, and antileukoproteinase (Table 4). Except for mse-miR-283, -iab-4 and -71*, all these miRNAs became more scarce in fat body after the immune challenge. While the level of mse-miR-283 increased in both fat body and hemocytes, it recognizes the 3’-UTRs of the three AMP transcripts. Of the 16 depressed miRNAs, only mse-miR-9a was predicted to target more than one antimicrobial effecter, cecropin-like peptide, moricin, and transferin.
4.2.4. Intracellular immune signaling pathways
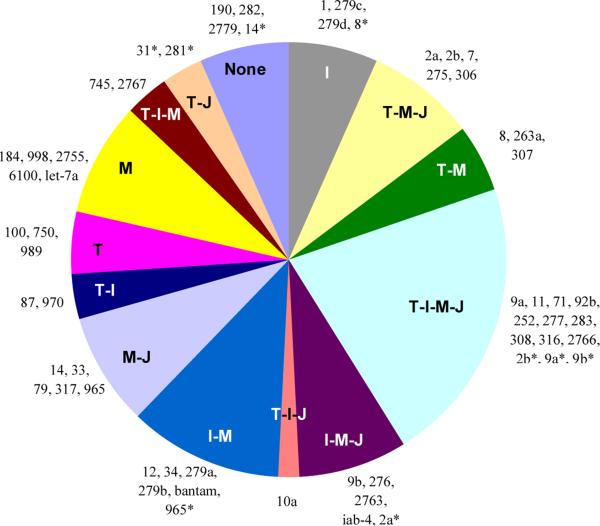
The Toll, IMD, MAPK-JNK-p38 and JAK-STAT pathways transduce signals of bacterial, fungal, viral and parasite infection. Predicted interactions form a complicated network between miRNAs and immune pathways in honeybees and Drosophila (Fullaondo and Lee, 2012a; Lourenco et al., 2013). One miRNA recognizes multiple genes and one 3’-UTR is recognized by several miRNAs. Since most transcript levels for the four pathways remained similar after the immune challenge in M. sexta (Gunaratna and Jiang, 2013), we focused on the interactions based on miRNA target predictions. From Table 4, we omitted those of mse-miR-34*, -71*, -79*, -279a*, -2755*, -2766*, -31, and -281 as they appeared to be incompletely processed miRNA duplex passenger strands, and mse-miR-1b as it existed at an extremely low level (Table 3). For the other 60 miRNAs or miRNA*s, their interactions with the immune signaling pathways were summarized (Fig. 4). The different categories provide the guidance for specific research aims in future studies. Four miRNAs are predicted to have no targets in the four pathways and twelve may only regulate one immune pathway. The twelve miRNAs can serve as candidates only to modify one immune pathway. For the other miRNAs, if their expression levels were to be modulated by genetic modifications, cautions should be taken as they may affect at least two immune pathways. The Toll and Imd pathways operate mainly against bacterial pathogens while JAK-STAT acts for antiviral defense. Thus, mse-miR-87, -970 and -10a are good candidates to modify both Toll and Imd pathways in defense against bacteria. Intriguingly, strands of some miRNA:miRNA* duplexes (mse-miRs 8, 2b, 9b, 2a and 965) fell into different categories
Fig. 4.
Summary of miRNA-targeted immune pathways, Toll (T), Imd (I), MAPK-JNK-p38 (M), and JAK-STAT (J). Each slice represents the group of miRNAs that may regulate member(s) of one or more of the four pathways, with miRNA names listed outside.
5. Conclusions
With the small RNA libraries of naïve and induced fat body and hemocytes analyzed, we extended the conserved and novel M. sexta miRNA lists and found, through examination of miRNA level changes accompanying the immune challenge, that some of them may regulate innate immunity to some extent. Opposite level changes in their respective target mRNAs, tethered by miRNA target site prediction, suggest they likely form pairs in the regulation of gene expression. With the miRNA and predicted targets both available, we are in the unique position to systematically investigate a network of putative posttranscriptional regulators of the major immune pathways in a model species. Further research using immune responsive cell lines of M. sexta is anticipated to greatly enrich our knowledge of the post-transcriptional regulation of insect innate immunity.
Supplementary Material
Highlights.
Identification of additional conserved and novel microRNAs in Manduca sexta;
Examination of microRNA levels in hemocytes and fat body after immune challenge;
Prediction of microRNA targets in the 232 M. sexta immunity-related genes;
Finding negatively correlated changes in the microRNA and putative target mRNA levels;
Possible regulation of intracellular immune signaling pathways by microRNAs.
Acknowledgments
We appreciate the comments on the manuscript from Dr. Ulrich Melcher at Oklahoma State University. The research was supported by National Institutes of Health Grant GM58634 (to H.J.), and a start-up fund from Kunming University of Science and Technology (to Y.Z.). This article was approved for publication by the Director of the Oklahoma Agricultural Experiment Station and supported in part under projects OKLO2450 (to H.J.) and OKLO2611 (to R.S.). We also thank Manduca Genome Project for Genome Assembly 1.0, funded by Defense Advanced Research Projects Agency (Gary Blissard, Boyce Thompson Institute) and National Institutes of Health (Michael Kanost, Kansas State University).
Abbreviations
- Alk
anaplastic lymphoma kinase
- AMP
antimicrobial peptides
- ANKRD54
ankyrin repeat domain 54
- Aop
anterior open
- aPKC
atypical protein kinase C
- AtgX
autophagy-related protein X
- βGRP
β-1,3-glucan recognition protein
- CF, IF, CH and IH
control (C) and induced (I) fat body (F) and hemocytes (H)
- CTL
C-type lectin
- Dscam
Downs syndrome cell adhesion molecule
- ECSIT
evolutionarily conserved intermediate in Toll pathway
- ERK
extracellular signal regulated kinase
- HAIP
hemocyte aggregation inhibitor protein
- Hem
hemipterous
- HP
hemolymph proteinase
- IAP
inhibitor of apoptosis
- IKK
IκB kinase
- IMD
immune deficiency
- IML
immulectin
- JAK-STAT
Janus kinase-signal transducer and activator of transcription
- JNK
Jun N-terminal kinase
- Jra
Jun related antigen
- MAPK
mitogen-activated protein kinase
- MASK
multiple ankyrin repeats single KH domain
- MEKK
MEK kinase
- MKK
MAP kinase kinase
- MLK
mixed-lineage kinase
- PAP
prophenoloxidase activating proteinase
- PGRP
peptidoglycan recognition protein
- PIAS
protein inhibitor of activated STAT
- PO and PPO
phenoloxidase and its precursor
- PPBP
paralytic peptide binding protein
- PRR
pattern recognition receptors
- PSP
plasmatocyte spreading peptide
- Pvr
PDGF/VEGF receptor
- serpin
serine proteinase inhibitor
- SOCS
suppressor of cytokine signaling
- SPH
serine proteinase homolog
- Spz
spatzle
- TAK
transforming growth factor β-activated kinase
- TEP
thioester-containing protein
- Tollip
Toll interacting protein
- Ubc
ubiquitin-conjugating domain
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- An CJ, Jiang H, Kanost MR. Proteolytic activation and function of the cytokine Spatzle in the innate immune response of a lepidopteran insect, Manduca sexta. FEBS J. 2010;277:148–162. doi: 10.1111/j.1742-4658.2009.07465.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asgari S. Role of microRNAs in insect host-microorganism interactions. Front Physiol. 2011;2:48. doi: 10.3389/fphys.2011.00048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker KD, Thummel CS. Diabetic larvae and obese flies-emerging studies of metabolism in Drosophila. Cell Metab. 2007;6:257–266. doi: 10.1016/j.cmet.2007.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bond D, Foley E. A quantitative RNAi screen for JNK modifiers identifies Pvr as a novel regulator of Drosophila immune signaling. PLoS Pathog. 2009;5:e1000655. doi: 10.1371/journal.ppat.1000655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerenius L, Lee BL, Soderhall K. The proPO-system: pros and cons for its role in invertebrate immunity. Trends Immunol. 2008;29:263–271. doi: 10.1016/j.it.2008.02.009. [DOI] [PubMed] [Google Scholar]
- Chawla G, Sokol NS. MicroRNAs in Drosophila development. Int Rev Cell Mol Biol. 2011;286:1–65. doi: 10.1016/B978-0-12-385859-7.00001-X. [DOI] [PubMed] [Google Scholar]
- Choi IK, Hyun S. Conserved microRNA miR-8 in fat body regulates innate immune homeostasis in Drosophila. Dev Comp Immunol. 2012;37:50–54. doi: 10.1016/j.dci.2011.12.008. [DOI] [PubMed] [Google Scholar]
- Christen JM, Hiromasa Y, An C, Kanost MR. Identification of plasma proteinase complexes with serpin-3 in Manduca sexta. Insect Biochem Mol Biol. 2012;42:946–955. doi: 10.1016/j.ibmb.2012.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunn PE, Drake DR. Fate of bacteria injected into naive and immunized larvae of the tobacco hornworm Manduca sexta. J Invertebr Pathol. 1983;41:77–85. [Google Scholar]
- Etebari K, Asgari S. Conserved microRNA miR-8 blocks activation of the Toll pathway by upregulating serpin 27 transcripts. RNA Biol. 2013;10:1356–1364. doi: 10.4161/rna.25481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Etebari K, Hussain M, Asgari S. Identification of microRNAs from Plutella xylostella larvae associated with parasitization by Diadegma semiclausum. Insect Biochem Mol Biol. 2013;43:309–318. doi: 10.1016/j.ibmb.2013.01.004. [DOI] [PubMed] [Google Scholar]
- Freitak D, Knorr E, Vogel H, Vilcinskas A. Gender- and stressor-specific microRNA expression in Tribolium castaneum. Biol Lett. 2012 doi: 10.1098/rsbl.2012.0273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fullaondo A, Lee SY. Identification of putative miRNA involved in Drosophila melanogaster immune response. Dev Comp Immunol. 2012a;36:267–273. doi: 10.1016/j.dci.2011.03.034. [DOI] [PubMed] [Google Scholar]
- Fullaondo A, Lee SY. Regulation of Drosophila-virus interaction. Dev Comp Immunol. 2012b;36:262–266. doi: 10.1016/j.dci.2011.08.007. [DOI] [PubMed] [Google Scholar]
- Garbuzov A, Tatar M. Hormonal regulation of Drosophila microRNA let-7 and miR-125 that target innate immunity. Fly. 2010;4:306–311. doi: 10.4161/fly.4.4.13008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goto A, Yano T, Terashima J, Iwashita S, Oshima Y, Kurata S. Cooperative regulation of the induction of the novel antibacterial Listericin by peptidoglycan recognition protein LE and the JAK-STAT pathway. J Biol Chem. 2010;285:15731–15738. doi: 10.1074/jbc.M109.082115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunaratna RT, Jiang H. A comprehensive analysis of the Manduca sexta immunotranscriptome. Dev Comp Immunol. 2013;39:388–398. doi: 10.1016/j.dci.2012.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gundersen-Rindal DE, Pedroni MJ. Larval stage Lymantria dispar microRNAs differentially expressed in response to parasitization by Glyptapanteles flavicoxis parasitoid. Arch Virol. 2010;155:783–787. doi: 10.1007/s00705-010-0616-1. [DOI] [PubMed] [Google Scholar]
- Hakimi MA, Cannella D. Apicomplexan parasites and subversion of the host cell microRNA pathway. Trends Parasitol. 2011;27:481–486. doi: 10.1016/j.pt.2011.07.001. [DOI] [PubMed] [Google Scholar]
- Jagadeeswaran G, Zheng Y, Sumathipala N, Jiang H, Arrese EL, Soulages JL, Zhang W, Sunkar R. Deep sequencing of small RNA libraries reveals dynamic regulation of conserved and novel microRNAs and microRNA-stars during silkworm development. BMC Genomics. 2010;11:52. doi: 10.1186/1471-2164-11-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang H, Vilcinskas A, Kanost MR. Immunity in lepidopteran insects. Adv Exp Med Biol. 2010;708:181–204. doi: 10.1007/978-1-4419-8059-5_10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang H. The biochemical basis of antimicrobial responses in Manduca sexta. Insect Sci. 2008;15:53–66. [Google Scholar]
- Kato M, de Lencastre A, Pincus Z, Slack FJ. Dynamic expression of small non-coding RNAs, including novel microRNAs and piRNAs/21U-RNAs, during Caenorhabditis elegans development. Genome Biol. 2009;10:R54. doi: 10.1186/gb-2009-10-5-r54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemaitre B, Hoffmann J. The host defense of Drosophila melanogaster. Annu Rev Immunol. 2007;25:697–743. doi: 10.1146/annurev.immunol.25.022106.141615. [DOI] [PubMed] [Google Scholar]
- Lourenco AP, Guidugli-Lazzarini KR, Freitas FC, Bitondi MM, Simoes ZL. Bacterial infection activates the immune system response and dysregulates microRNA expression in honey bees. Insect Biochem Mol Biol. 2013;43:474–482. doi: 10.1016/j.ibmb.2013.03.001. [DOI] [PubMed] [Google Scholar]
- Marco A, Hui JH, Ronshaugen M, Griffiths-Jones S. Functional shifts in insect microRNA evolution. Genome Biol Evol. 2010;2:686–696. doi: 10.1093/gbe/evq053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nappi AJ, Christensen BM. Melanogenesis and associated cytotoxic reactions: applications to insect innate immunity. Insect Biochem Mol Biol. 2005;35:443–459. doi: 10.1016/j.ibmb.2005.01.014. [DOI] [PubMed] [Google Scholar]
- Ragab A, Buechling T, Gesellchen V, Spirohn K, Boettcher AL, Boutros M. Drosophila Ras/MAPK signalling regulates innate immune responses in immune and intestinal stem cells. EMBO J. 2011;30:1123–1136. doi: 10.1038/emboj.2011.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao XJ, Xu XX, Yu XQ. Manduca sexta moricin promoter elements can increase promoter activities of Drosophila melanogaster antimicrobial peptide genes. Insect Biochem Mol Biol. 2011;41:982–992. doi: 10.1016/j.ibmb.2011.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sumathipala N, Jiang H. Involvement of Manduca sexta peptidoglycan recognition protein-1 in the recognition of bacteria and activation of prophenoloxidase system. Insect Biochem Mol Biol. 2010;40:487–495. doi: 10.1016/j.ibmb.2010.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suwanchaichinda C, Ochieng R, Zhuang S, Kanost MR. Manduca sexta serpin-7, a putative regulator of hemolymph prophenoloxidase activation. Insect Biochem Mol Biol. 2013;43:555–561. doi: 10.1016/j.ibmb.2013.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S, Gunaratna RT, Zhang X, Najar F, Wang Y, Roe B, Jiang H. Pyrosequencing-based expression profiling and identification of differentially regulated genes from Manduca sexta, a lepidopteran model insect. Insect Biochem Mol Biol. 2011;41:733–746. doi: 10.1016/j.ibmb.2011.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Zheng Y, Jagadeeswaran G, Ren R, Sunkar R, Jiang H. Identification and developmental profiling of conserved and novel microRNAs in Manduca sexta. Insect Biochem Mol Biol. 2012;42:381–395. doi: 10.1016/j.ibmb.2012.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Y, Zhang W. Animal microRNA target prediction using diverse sequence-specific determinants. J Bioinform Comput Biol. 2010;8:763–788. [Google Scholar]
- Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.