Abstract
Our capacity to sequence human genomes has exceeded our ability to interpret genetic variation. Current genomic annotations tend to exploit a single information type (e.g. conservation) and/or are restricted in scope (e.g. to missense changes). Here, we describe Combined Annotation Dependent Depletion (CADD), a framework that objectively integrates many diverse annotations into a single, quantitative score. We implement CADD as a support vector machine trained to differentiate 14.7 million high-frequency human derived alleles from 14.7 million simulated variants. We pre-compute “C-scores” for all 8.6 billion possible human single nucleotide variants and enable scoring of short insertions/deletions. C-scores correlate with allelic diversity, annotations of functionality, pathogenicity, disease severity, experimentally measured regulatory effects, and complex trait associations, and highly rank known pathogenic variants within individual genomes. The ability of CADD to prioritize functional, deleterious, and pathogenic variants across many functional categories, effect sizes and genetic architectures is unmatched by any current annotation.
Technical Report
A strength of genomic approaches to study disease is the replacement of informed but biased hypotheses with unbiased but generic ones, like the “equal treatment” of all genetic variants in genome-wide association studies (GWAS). However, for both rare variants of large effect and common variants of weak effect, the use of prior knowledge can be critical for disease gene discovery1–4. For example, exome sequencing is an effective discovery strategy because it focuses on protein-altering variation, which is enriched for causal effects5.
While many existing annotations are useful for prioritizing causal variants to boost discovery power (e.g. PolyPhen6, SIFT7, and GERP8), current approaches tend to suffer from one or more of four major limitations. First, annotations vary widely with respect to both inputs and outputs. For example, conservation metrics8–10 are defined genome-wide but do not use functional information and are not allele-specific, while protein-based metrics6,7 apply only to coding, and often only to missense, variants, thereby excluding >99% of human genetic variation. Second, each annotation has its own metric and these metrics are rarely comparable, making it difficult to evaluate the relative importance of distinct variant categories or annotations. Third, annotations trained on known pathogenic mutations are subject to major ascertainment biases and may not generalize. Fourth, it is a major practical challenge to obtain, let alone to objectively evaluate or combine, the existing panoply of partially correlated and partially overlapping annotations; this challenge will only magnify as large-scale projects like ENCODE11 continually increase the amount of relevant data available. The net result of these limitations is that many potentially relevant annotations are ignored, while the subset that are used are applied and combined in ad hoc and subjective ways that undermine their utility.
Here, we describe a general framework, Combined Annotation Dependent Depletion (CADD), for integrating diverse genome annotations and scoring any possible human single nucleotide variant (SNV) or small insertion/deletion (indel) event. The basis of CADD is to contrast the annotations of fixed or nearly fixed derived alleles in humans relative to simulated variants. Deleterious variants – that is, variants that reduce organismal fitness – are depleted by natural selection in fixed but not simulated variation. CADD therefore measures deleteriousness, a property that strongly correlates with both molecular functionality and pathogenicity12. Importantly, metrics of deleteriousness, in contrast with pathogenicity or molecular functionality, have major advantages. Whereas the latter are limited in scope to a small set of genetically or experimentally well-characterized mutations and subject to major ascertainment biases, deleteriousness can be measured systematically across the genome assembly (see refs 8, 9, 10 and below). Further, selective constraint on genetic variants is related to the totality of their phenotype-relevant effects rather than any individual molecular or phenotypic consequence. Measures of deleteriousness can therefore provide, in principle, a genome-wide, data-rich, functionally generic, and organismally relevant estimate of variant impact.
We identified differences between human genomes and the inferred human-chimpanzee ancestral genome13 where humans carry a derived allele with a frequency of at least 95% (14.9 million SNVs and 1.7 million indels). Nearly all of these events are fully fixed in the human lineage, with fewer than 5% appearing as nearly fixed polymorphisms in the 1000 Genomes Project14 variant catalog (derived allele frequency (DAF) ≥ 95%). To simulate an equivalent number of de novo mutations, we used an empirical model of sequence evolution with CpG dinucleotide-specific rates and mutation rates locally estimated at a 1 megabase (Mb) scale (Supplementary Note). Mutation rate parameters as well as the size distribution of indels were estimated from six-way primate genome alignments15.
To generate annotations, we used the Ensembl Variant Effect Predictor16 (VEP), data from the ENCODE project11 and information from UCSC genome browser tracks17 (Supplementary Table 1). The annotations span a range of data types including conservation metrics like GERP8, phastCons9, and phyloP10; regulatory information11 like genomic regions of DNase hypersensitivity18 and transcription factor binding19; transcript information like distance to exon-intron boundaries or expression levels in commonly studied cell lines11; and protein-level scores like Grantham20, SIFT7, and PolyPhen6. The resulting variant-by-annotation matrix contained 29.4 million variants (half fixed or nearly fixed human derived alleles (“observed”), half simulated de novo mutations (“simulated”)) and 63 distinct annotations, some of which are composites that summarize many underlying annotations (Supplementary Note, Supplementary Tables 1–2).
We first assessed the validity of our general approach by constructing a series of univariate models that contrast observed and simulated variants using each of the 63 annotations as individual predictors (Supplementary Note). Nearly all models were highly significant (Supplementary Tables 3–5) and consistent with expectation. For example, we find a nearly 20-fold depletion of nonsense variants, a 2-fold depletion of missense variants, and no depletion of intergenic or upstream/downstream variants (Supplementary Table 6). Nonsense and missense mutations that occur near the starts of cDNAs were more depleted than those occurring near the ends (Supplementary Table 7), and variants within 20, and especially within 2, nucleotides of splice junctions were also depleted (Supplementary Fig. 1). The best performing individual annotations were protein-level metrics such as PolyPhen6 and SIFT7, but these evaluated only missense variants (0.63% of all variants in the training data are missense; of these, 88% had defined PolyPhen values and 90% had defined SIFT values). Conservation metrics were the strongest individual genome-wide annotations (Supplementary Table 3).
We also examined correlations between annotations (Supplementary Fig. 2) and the value of adding interaction terms between annotations (Supplementary Fig. 3). Many annotations were correlated and many interactions were statistically significant, but only a handful of interacting pairs meaningfully improved a simple additive model. Overall, these analyses demonstrate that substantial biological differences are present between the observed and simulated variants with respect to the 63 annotations, and that linear models capture much of this information.
We next trained a support vector machine21 (SVM) with a linear kernel on features derived from the 63 annotations, supplemented by a limited number of interaction terms (Supplementary Note, Supplementary Tables 1–2, Supplementary Fig. 4). Ten models, independently trained on observed variants and different samples of simulated variants, were highly correlated (all pairwise Spearman rank correlations >0.99; Supplementary Fig. 5). An average of these models was applied to score all 8.6 billion possible SNVs of the human reference genome (GRCh37). To simplify interpretation in some contexts, we also defined phred-like22 scores (“scaled C-scores”) based on the rank of the C-score of each variant relative to all 8.6 billion possible SNVs, ranging from 1 to 99 (Supplementary Note). For example, substitutions with the highest 10% (10−1) of all scores - that is, least likely to be observed human alleles under our model - were assigned values of 10 or greater (“≥C10”), while variants in the highest 1% (10−2), 0.1% (10−3), etc. were assigned scores ≥C20, ≥C30, etc.
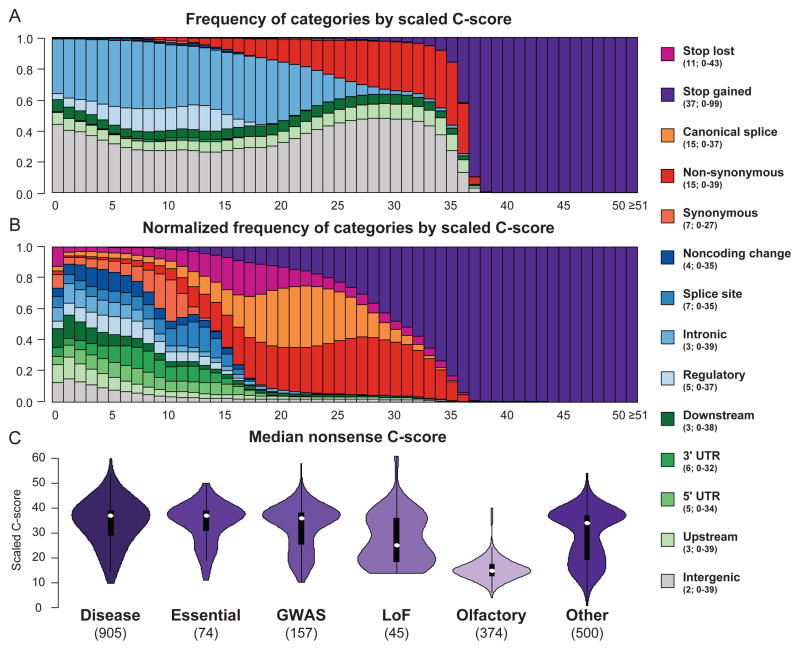
We first calculated the proportion of all possible substitutions with a given scaled C-score having specific functional consequences (Fig. 1; Supplementary Table 8). Although trained solely on the difference between observed and simulated variants, rather than on sets of known disease causing variants that might introduce ascertainment bias, the C-scores of potential nonsense variants are highest (median 37), followed by missense and canonical splice site variants (median 15) and with intergenic variants comprising the bottom of the list (median 2). At the same time, 76% of potential SNVs with ≥C20 are non-coding (i.e. categories other than missense, nonsense, canonical splice or stop loss), while 74% of potential missense and 18% of potential nonsense SNVs are below C20. Further, within each functional class there are distinctions that are biologically relevant and likely predictively useful. For example, potential nonsense variants – often treated as a homogeneous group in disease studies – in olfactory receptors score lower than in other genes, while potential nonsense variants in genes found previously to be “essential”23 score higher (Fig. 1 lower panel, Supplementary Fig. 6). C-scores thus capture considerable information both between and within functional categories. Of note, these same distinctions are absent or muted with other measures, either due to missingness (e.g., for missense-only measures) or lack of functional awareness (e.g., conservation measures cannot distinguish between a nonsense and missense allele at a given position).
Figure 1.
Relationship of scaled C-scores and categorical variant consequences. The upper plot shows the proportion of substitutions with a specific consequence for each scaled C-score bin, while the middle panel shows the proportion of substitutions with a specific consequence after first normalizing by the total number of variants observed in that category. The legend indicates the median and range of scaled C-score values for each category. Consequences are obtained from the Ensembl Variant Effect Predictor16 (Supplementary Note), e.g. “noncoding change” refers to changes in annotated non-coding transcripts. Detailed counts of functional assignments in each C-score bin are in Supplementary Table 8. The lower panel shows violin plots of the median C-scores of potential nonsense (stop-gained) variants for genes that: harbor at least 5 known pathogenic mutations48 (“disease”); are predicted to be “essential”23; harbor variants associated with complex traits41 (“GWAS”); harbor at least 2 loss-of-function mutations in 1000 Genomes49 (“LoF”); encode olfactory receptor proteins; or are in a random selection of 500 genes (“Other”; see Supplementary Note).
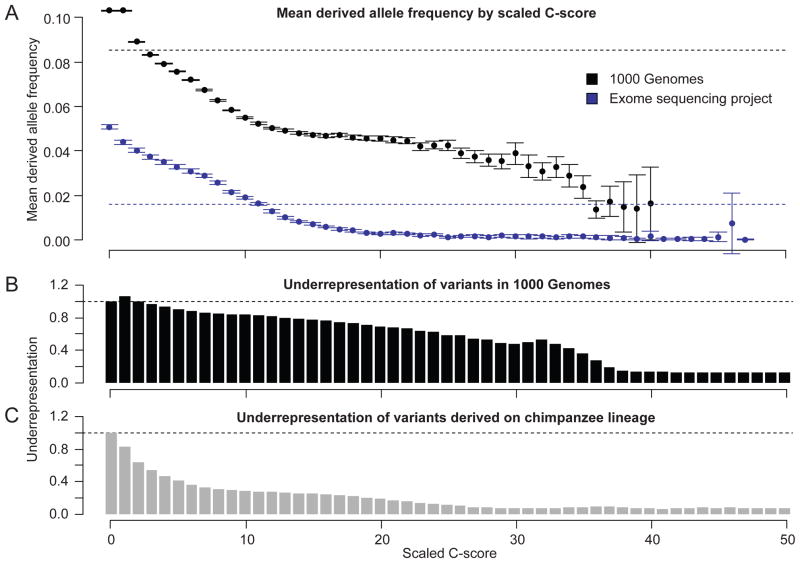
We next compared scaled C-scores with levels of genetic diversity, finding that C-scores are negatively correlated with the DAF of variants identified in the 1000 Genomes Project14 or the Exome Sequencing Project24 (ESP) (Fig. 2a; Supplementary Figs. 7–9), depletion of human genetic variation from the 1000 Genomes Project catalog (Fig. 2b), and depletion of chimp-derived variants (Fig. 2c). Importantly, these validation datasets have minimal overlap with the “observed” subset of the training data, which consists only of fixed or nearly fixed (>95% DAF) human derived alleles. Furthermore, although we cannot fully eliminate confounding by these factors, the negative correlation between C-scores and the DAF of standing variation is robust to controlling for variation in background selection, local GC content, local CpG density, and site-based conservation (Supplementary Fig. 9).
Figure 2.
Relationship between scaled C-scores and: the average derived allele frequency (DAF) of variants identified in the 1000 Genomes Project14 or ESP24 (upper panel); the under-representation of polymorphic sites in 1000 Genomes (middle panel); and chimpanzee lineage derived variants (lower panel). The dashed lines in the upper plot indicate the mean DAF and confidence intervals indicate 1.96x standard errors of the mean (SEM) DAF in each bin. Under-representation is defined as the proportion of 1000 Genomes (middle panel) or chimpanzee-derived (lower panel) variants in a specific scaled C-score bin divided by the frequency with which that scaled C-score is observed for all possible mutations of the human reference assembly (10C-score/−10). The stronger under-representation of chimpanzee-derived variants relative to 1000 Genomes variants is expected given that the former are mostly fixed or high-frequency variants (and have survived many generations of purifying selection) while the latter are mostly low-frequency variants. Depletion values in both panels for C-score bins other than 0 are significantly different from expectation (binomial proportion test, all p-values <10−11).
We next sought to assess the utility of CADD to prioritize functional and disease-relevant variation within five distinct contexts.
First, for MLL2, the gene mutated in Kabuki syndrome, C-scores enable discrimination of a diverse set of disease-associated alleles25 versus rare, likely benign variants from ESP24 (Wilcoxon rank sum test p = 9.9 × 10−94; n = 210/679). Other metrics were markedly inferior in terms of accuracy or comprehensiveness (Supplementary Fig. 10).
Second, for HBB, the gene mutated in beta-thalassemia, C-scores of disease-associated alleles26 – a set of indels (n=93) and SNVs (n=119) with regulatory/upstream (n=54), splicing (n=37), missense (n=22), nonsense (n=18) and other effects – are significantly, and more strongly than other measures, correlated with three levels of phenotypic severity (Kruskal-Wallis rank sum test p = 2.4 × 10−7; n = 48/65/99, Supplementary Fig. 11).
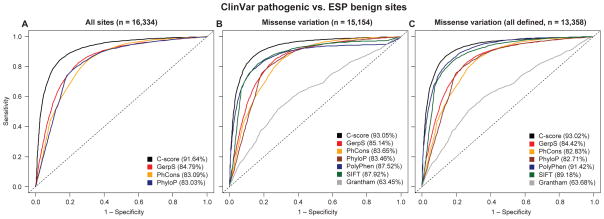
Third, pathogenic variants curated by the NIH ClinVar database27 are well separated from likely benign alleles (ESP24 DAF ≥ 5%) matched to the same categorical consequences (Wilcoxon rank sum test p < 10−300, n = 8174/8174, Fig. 3; Supplementary Figs. 12–16). We note that there is substantial overlap between ClinVar and the training data underlying PolyPhen. When these sites are excluded from the test dataset, or when PolyPhen is excluded as a training feature from CADD, C-scores continue to outperform all or nearly all missense-only metrics and conservation measures (Supplementary Fig. 12).
Figure 3.
Receiver operating characteristics (ROC) for discriminating curated, pathogenic mutations defined by the NIH ClinVar database27 matched to apparently benign ESP alleles (DAF ≥ 5%)24 with the same categorical consequence. The left panel shows genome-wide variants for which GerpS, PhCons, and PhyloP scores are defined (n=16,334), while the middle panel limits the analysis to missense changes (n=15,154), with missing values imputed to an upper value limit of each score, and right panel to missense changes for which PolyPhen, SIFT and Grantham scores are all defined (n=13,358). Versions of the right panel that exclude the overlap between PolyPhen training data and the ClinVar database or use a CADD model trained without PolyPhen as a feature are shown in Supplementary Fig. 12. Area under the curve (AUC) values are provided in the figure legend for each of the scores used.
Fourth, C-scores strongly correlate with the number of observations for somatic cancer mutations in p53 reported to the International Agency for Research on Cancer (Spearman rank correlation 0.38, p = 6 × 10−73, n = 2068, Supplementary Note).
Fifth, we examined two enhancers28 and one promoter29 in which we previously performed saturation mutagenesis. C-scores are significantly correlated, and overall more so than measures of sequence conservation, with the experimentally measured absolute expression fold change of individual variants (Spearman rank correlation of combined data = 0.31, p = 1.9 × 10−65, n = 2847; Supplementary Fig. 17).
Collectively, these analyses demonstrate that CADD is quantitatively predictive of deleteriousness, pathogenicity, and molecular functionality, both protein-altering and regulatory, in a variety of experimental and disease contexts. Within each of these contexts, CADD’s predictive utility is much better than measures of sequence conservation, the only comprehensive type of variant score, and also tends to be better, in most cases substantially so, than function-specific metrics when restricted to the appropriate variant subsets.
We next considered how CADD may be useful in evaluating candidate variation within exome or genome-wide studies.
First, we analyzed de novo exome variants (SNVs and indels) identified in children with autism spectrum disorders30–34 (ASD) and intellectual disability35,36 (ID) along with unaffected siblings or controls, including 88 nonsense, 1,015 missense, 359 synonymous, 32 canonical splice site, and 150 other variants, including indels. Variants in affected children are significantly more deleterious than those in unaffected siblings/controls, considering each disease separately (Supplementary Table 9) or combined (ASD+ID Wilcoxon rank sum test p = 2.0 × 10−4, n = 1130/514). Additionally, de novo variants in ID probands are significantly more deleterious than those of ASD probands (p = 4.7 × 10−5, n=170/960), suggesting a more deleterious global mutation burden in ID, consistent with the observation of increased sizes and numbers of copy number variants in ID relative to ASD37.
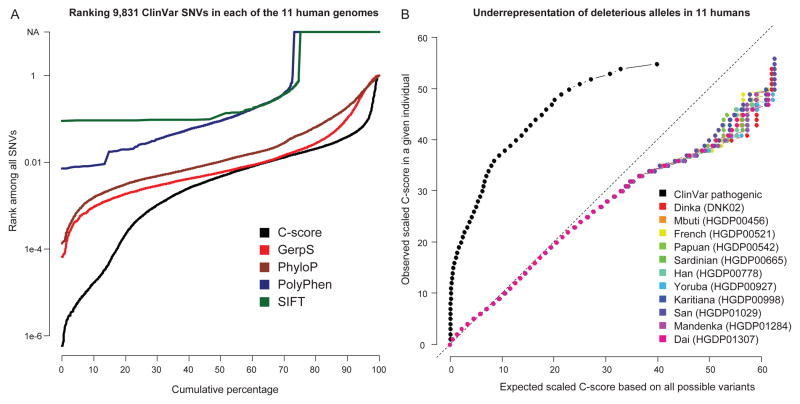
Second, it is well established that annotations like PolyPhen and conservation are valuable in the sequencing-based identification of disease-causal genes by virtue of their ability to highly rank pathogenic variants1,2,38. We therefore examined the distribution of C-scores in the genomes of 11 individuals representing diverse populations39,40, and find that CADD highly ranks known disease-causal variants (ClinVar pathogenic) within the complete spectrum of variation in personal genomes (Fig. 4; Supplementary Fig. 16 and Supplementary Table 10–11). Furthermore, CADD is both more quantitative and comprehensive in this task (e.g., ~27% of pathogenic ClinVar SNVs are not scored by PolyPhen because of missing values or its restriction to missense variation). Given its considerable superiority over the best available protein-based and conservation metrics in terms of ranking known pathogenic variants in the complete spectrum of variation within personal genomes, it is likely that CADD will improve the power of sequence-based disease studies beyond current standard approaches.
Figure 4.
Ranking of pathogenic ClinVar variants among the variants identified by whole genome sequencing of eleven human individuals from diverse populations. Left panel: Cumulative distributions of the ranks of 9,831 pathogenic ClinVar variants when “spiked in” to each of 11 personal genomes. For example, C-scores of ~30% of ClinVar variants rank in the top 0.1% of all variants within a personal genome, and most rank in the top 1%. About 25% of pathogenic ClinVar SNVs are not scored by PolyPhen/SIFT because of missing values or its restriction to missense variation; note also that ranks for PolyPhen/SIFT are computed among missense variants only and are therefore derived from far fewer total variants (see a plot restricted to missense variation in Supplementary Fig. 16). Right panel: A QQ-plot of the C-scores of the SNVs identified from the eleven individuals and pathogenic ClinVar SNVs. For a given scaled C-score observed in an individual, the fraction of that individual’s variants with a C-score at least that large was computed (y-axis). The C-score corresponding to this quantile of the distribution of all possible variants is displayed on the x-axis. High C-scores are underrepresented compared to the set of all possible variants. In contrast, known disease-causal variants from ClinVar have large C-scores relative to the set of all possible variants. This fact can be exploited to prioritize causal variants identified from whole genome sequencing of individual genomes (left panel and Supplementary Tables 10–11).
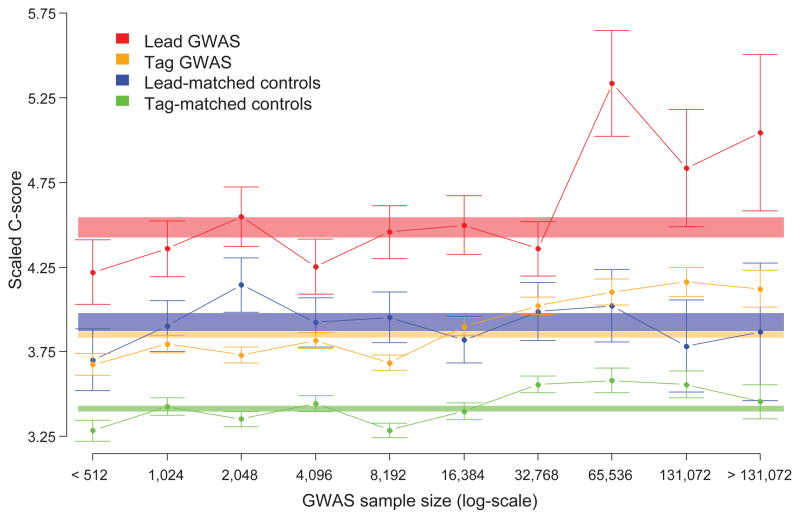
Finally, we analyzed CADD scores for single nucleotide polymorphisms (SNPs) identified by GWAS of complex traits, contrasting them with nearby control SNPs matched for allele frequency and genotyping array availability (Fig. 5, Supplementary Note). We find that lead GWAS SNPs have significantly higher C-scores than control SNPs (one-sided Wilcoxon rank sum test, p-value = 1.3 × 10−12, n = 5498/5498); nearby SNPs in linkage disequilibrium with lead SNPs (“tags”) score lower on average than leads but are also significantly higher than their matched controls (p-value = 5.1 × 10−107). C-score differences remain significant after controlling for properties like gene-body effect, gene expression level, conservation, and regulatory element overlap; each of these are significantly different between associated and control SNPs but none can fully explain the C-score discrepancy (Supplementary Note). C-scores of trait-associated SNPs furthermore correlate with the size of the underlying association study and with statistical significance of the association itself (Fig. 5; Supplementary Figure 16; Supplementary Note), likely due to the increased ability of larger studies and stronger association statistics to enrich for causal variants. While for the most part not causal, our analysis suggests that GWAS-identified SNPs, especially strongly associated lead SNPs from large studies, are enriched for causal variants, consistent with previously observed GWAS enrichments for individual annotations11,41–44.
Figure 5.
C-scores for GWAS SNPs are higher than nearby control SNPs and dependent on study sample size. The average scaled C-score (y-axis) is plotted for each category of SNP, as indicated by color, relative to the sample sizes of the association studies in which the SNPs were identified (x-axis). Sample size bins are log2-scaled and mutually exclusive; for example, the bin labeled “1024” represents all SNPs from studies with between 512 and 1024 samples. Error bars are ±1 standard errors of the mean (SEM). Shaded rectangles represent the overall, i.e. across all sample sizes, scaled C-score means ±1 SEM for each category as indicated by the color.
With CADD, we describe a generic, expandable framework for integrating information contained in diverse annotations of genetic variation to a single score. We demonstrate that in a variety of contexts this approach is better, in some cases modestly but in many cases dramatically, than other widely used annotations at prioritizing functional and pathogenic variants. Further, beyond utility in any one setting, there are practical and conceptual advantages to CADD that should prove of major value to genetic studies of human disease. First, the information content of many individual annotations is objectively merged into a single value, which is far preferable to ad hoc approaches for combining annotations and likely to improve performance, consistent with benefits seen for “consensus” methods in missense-specific annotation45. Second, CADD can readily incorporate expansions to existing annotations and entirely new annotations. The ability to indefinitely and readily integrate new information is crucial in light of projects like ENCODE, which are continuously and rapidly expanding available annotations11. Third, CADD combines the generality of conservation-based metrics with the specificity of subset-relevant functional metrics (e.g. PolyPhen), exploiting the advantages of both while attenuating their respective disadvantages.
CADD also has a number of limitations which may restrict its utility for certain analyses or represent areas for improvement. First, C-scores measure reductions in variation, which correlate with deleteriousness but are also affected by local mutation rate, background selection, biased gene conversion, and other phenomena, potentially limiting accuracy. Second, C-scores reflect the proportion of variants with a given annotation pattern that are visible to selection but may not capture differences in selective intensity; other approaches, such as polymorphism-to-divergence comparisons, may be more accurate for estimating selective coefficients46. Third, there is a strong need for more “gold standard” data, particularly for non-coding regions of the genome, the current paucity of which limits the development of better annotations as well as our ability to validate predictions. Fourth, it is at present not possible to precisely calibrate the relationship between CADD-estimated deleteriousness and the likelihood that a variant is pathogenic. As such, C-scores are best interpreted in terms of “likelihood of deleteriousness” rather than “likelihood of pathogenicity”, e.g. the quantifiable extent of depletion of a given C-score from chimp-derived alleles (Fig. 2c, Supplementary Table 11). Especially for discovering causal variants, CADD should be treated as one piece of information contributing to the totality of evidence for pathogenicity, and evaluated as a supplement, not a replacement, for genetic information.
The “one-stop” nature of CADD is likely to be of great practical and conceptual value to future sequencing studies. It will minimize the scope and diversity of annotations that have to be generated, tracked, and evaluated by a lab or project, and reduce the need for ad hoc combinations of filters, scores, and parameters as is now routinely done. For example, an oft-used approach in exome studies is to merge missense (with or without an annotation of “damage” or given level of conservation), nonsense, and splice-disrupting variants into a single, internally unranked list of “protein-altering” variants prior to genetic analysis5. With CADD, one might avoid arbitrary filters/thresholds altogether, including both coding and non-coding variants on a single, meaningfully ranked list. For example, a recent study of recessive, non-syndromic pancreatic agenesis identified 5 causal non-coding variants that disrupt function of a distal enhancer of PTF1A47. C-scores for these non-coding, disease-causal variants (scaled scores between 23.2 and 24.5) rank them above 99.5% of all possible human SNVs, above 97% of missense SNVs in a typical exome, and higher than 56% of Mendelian pathogenic SNVs in ClinVar27.
Both in research and in the clinic, our capacity to define catalogs of genetic variants exceeds our ability to systematically evaluate their potential impacts. This challenge will deepen as sequencing accelerates, as genomes displace exomes, and as the array of functional categories and annotations expand. A unified, quantitative, and scalable framework capable of exploiting many genomic annotations will be essential to meet this challenge. We anticipate that the model described here and the accompanying freely available pre-computed scores for all possible GRCh37/hg19 SNVs (http://cadd.gs.washington.edu/) will be broadly useful immediately, and improve over time, enabling better interpretation of variants of uncertain significance in a clinical setting and improving discovery power for genetic studies of both Mendelian and complex diseases.
Online Methods
Simulated and observed variants
The basis of the CADD framework is to capture correlates of selective constraint as manifested in differences between simulated variants and observed human derived changes. For the simulated variants, we developed a genome-wide simulator of de novo germline variation. The simulator was motivated by the parameters of the General Time Reversible (GTR) model50, but because the standard GTR does not naturally accommodate asymmetric CpG-specific mutation rates, we use a fully empirical model of sequence evolution with a separate rate for CpG dinucleotides and local adjustment of mutation rates (see Supplementary Note). Simulation parameters were obtained from Ensembl Enredo-Pecan-Ortheus (EPO)13,15 whole genome alignments of six primate species (Ensembl Compara release 66). A custom script and the associated rate matrices underlying the genome-wide simulator are available as Supplementary File 1. We applied these parameters to simulate single nucleotide (SNV) and insertion/deletion (indel) variants based on the human reference sequence (GRCh37).
For observed human derived changes, we extracted sites where the human reference genome differs from the inferred human-chimp ancestral genome from the Ensembl EPO 6 primate alignments defined above, excluding variants in the most recent 1000 Genomes Project14 data (1000G, variant release 3, 20101123) with a frequency of greater than 5%, and including variants where the human reference carries an ancestral allele (i.e. matching the inferred human-chimp ancestor sequence) but where the derived allele is observed with frequency above 95% in the 1000G data. We identified a total of 14,893,290 SNVs, and 627,071 insertions and 1,107,414 deletions (less than 50bp in length).
Variant annotation matrix
We used the Ensembl Variant Effect Predictor (VEP, Ensembl Gene annotation v68)16 to obtain gene model annotation for single nucleotide and indel variants. For single nucleotide variants within coding sequence, we also obtained SIFT7 and PolyPhen-26 scores from VEP. We combined output lines describing MotifFeatures with the other annotation lines, reformatted it to a pure tabular format and reduced the different Consequence output values to 17 levels and implemented a four-level hierarchy in case of overlapping annotations (see Supplementary Note). To the 6 VEP input derived columns (chromosome, start, reference allele, alternative allele, variant type: SNV/INS/DEL, length) and 26 actual VEP output derived columns, we added 56 columns providing diverse annotations (e.g. mapability scores and segmental duplication annotation as distributed by UCSC51,52; PhastCons and phyloP conservation scores53 for three multi-species alignments9 excluding the human reference sequence in score calculation; GERP++ single-nucleotides scores, element scores and p-values54, also defined from alignments with the human reference excluded; background selection score40,55; expression value, H3K27 acetylation, H3K4 methylation, H3K4 trimethylation, nucleosome occupancy and open chromatin tracks provided for ENCODE cell lines in the UCSC super tracks52; genomic segment type assignment from Segway56; predicted transcription factor binding sites and motifs11; overlapping ENCODE ChIP-seq transcription factors11, 1000 Genome variant14 and Exome Sequencing Project57 variant status and frequencies, Grantham scores20 associated with a reported amino acid substitution). The Supplementary Note provides a full description and Supplementary Table 1 lists all columns of the obtained annotation matrix.
Imputation and final training data set
From the annotations described above, some columns are not useful for model training or needed to be excluded from training as they differ between the simulated variants and the human-chimpanzee ancestor differences for technical reasons (see Supplementary Note for a complete list; note that no allele frequency information was used in model training). In order to fit models, we imputed missing values in genome-wide measures by the genome average obtained from the simulated data, or set missing values to 0 where appropriate (Supplementary Table 2). Further, we created an “undefined” category for the categorical annotations in order to accommodate missing values. In order to deal with missing values in annotations that are not defined on a subset of variants (e.g. information only available for protein-coding genes), we set the missing values to zero and also created indicator variables that contain a 1 if the corresponding variant is undefined, and a 0 otherwise. Since insertions and deletions may produce arbitrary length Ref/Alt and nAA/oAA columns (and thus not a fixed number of categorical levels), these values were set to N for Ref/Alt and set to “undefined” for nAA/oAA.
Sites from the simulation were labeled +1 and human derived variants as −1. Only insertions and deletions shorter than 50bp were considered for model training and the Length column was capped at 49 for the prediction of longer events. The ratio of indel events to SNV events obtained for the simulation (1:8.46).
Model training
We generated ten training data sets by sampling an equal number of 13,141,299 SNVs, 627,071 insertions and 926,968 deletions from both the simulated variant and observed variant datasets. In order to train each support vector machine (SVM) model, the processed data was converted to a sparse matrix representation after converting all n-level categorical values to n individual Boolean flags. 1% of sites (~132,000 SNVs, 6,000 insertions and 9,000 deletions each) were randomly selected and used as a test data set. All other sites were used to train linear SVMs using the LIBOCAS v0.96 library21. The SVM model fits a hyperplane as defined below. X1,…,Xn are the 63 annotations described above (which expand to 166 features due to the treatment of categorical annotations), W1,…,W11 are the Boolean features that indicate whether a given feature (out of cDNApos, relcDNApos, CDSpos, relCDSpos, protPos, relProtPos, Grantham, PolyPhenVal, SIFTval, as well as Dst2Splice ACCEPTOR and DONOR) is undefined, 1{A} is an indicator variable for whether the event A holds, and D is the set of bStatistic, cDNApos, CDSpos, Dst2Splice, GerpN, GerpS, mamPhCons, mamPhyloP, minDistTSE, minDistTSS, priPhCons, priPhyloP, protPos, relcDNApos, relCDSpos, relProtPos, verPhCons, and verPhyloP. Due to the coding of categorical values using Boolean variables, the total number of features in this model is 949.
SVM models were trained, using various values for the generalization parameter (C), which assigns the cost of misclassifications. Supplementary Fig. 4 shows the model training convergence in 2000 iterations (~70h) for different settings of C. These results indicate that model training only converges within a reasonable amount of time for C values around 0.0025 and below. We therefore trained models for all ten training data sets with C=0.0025. We determined the average of the model parameters and used the average model.
Model testing and validation
We annotated all 8.6 billion possible substitutions in the human reference genome (GRCh37), and applied the model to score all possible substitutions. When scoring sites with multiple VEP annotation lines, we score all possible annotations first and then report the one with the highest deleteriousness after applying the four hierarchy levels. We mapped the C-scores to a phred-like scale (“scaled C-scores”) ranging from 1 to 99 based on their rank relative to all possible substitutions in the human reference genome, i.e. −10log10(rank/total number of substitutions).
We used several datasets extracted from the literature and public databases to look at the performance of the model scores (see Supplementary Note for details): (1) C-scores in specific gene classes motivated by the analysis performed by Khurana et al.58 (i.e. HGMD48, non-immune essential genes described by Liao et al.23, GWAS genes as available from the Genome.gov catalog, LoF genes from MacArthur et al.49 and olfactory genes from the Ensembl 68 gene build). (2) 210 mutations in MLL2 associated with Kabuki syndrome from Makrythanasis et al.25. We complemented those with 679 putatively benign variants observed in the Exome Sequencing Project (ESP)57. (3) We downloaded a total of 119 SNVs, 30 insertions and 63 deletions (all required to be at most 50nt) within or near HBB that give rise to thalassemia from HbVar26. Disease categories were used as defined by HbVar, except that all types that are not “beta0” or “beta+” were pooled into one category, “other”. (4) We obtained the NCBI ClinVar27 data set (release date June 16 2012) and extracted variants that were marked “pathogenic” or “non-pathogenic (benign)”. We also selected a set of apparently benign (≥5% allele frequency) variants from ESP that were matched to the pathogenic ClinVar sites in terms of their Consequence annotations. In addition, we generated a data set where we matched ESP and ClinVar frequencies to three decimal precisions of the alternative allele frequency. Due to the overlap of ClinVar and ESP variants with the PolyPhen training data set, we trained a separate classifier without the PolyPhen features and we also checked the performance on the subset of ClinVar and ESP variants not used for PolyPhen training. To compare the performance of CADD with other publically available missense annotations not used in model training, we downloaded scores from dbNSFP 2.059. (5) We combined high confidence de novo mutations from five family based autism exome sequencing studies30–34, a total of 948 ASD probands and 590 unaffected siblings. Further, we obtained the coding variants as described above for two family-based intellectual disability (ID) studies35,36, 151 ID and 20 unrelated control families. (6) We obtained the expression fold change for each base substitution in ALDOB and ECR11 from Patwardhan et al.28. This data set contains a total of 777 variants for ALDOB and 1,860 variants for ECR11. Further, we obtained the HBB promoter data of Patwardhan et al.29. The promoter data set contains a total of 210 variants associated with an expression fold change. (7) We obtained a list of 23,788 single nucleotide somatic cancer mutations in p53 which were reported to the International Agency for Research on Cancer (IARC). These mutations correspond to 2,068 distinct variants; we recorded the number of times that each variant was reported. (8) We obtained GATK VCF variant call files for all autosomes and the X chromosome from shotgun sequencing of eleven men originating from diverse human populations40. (9) We obtained the NHGRI genome-wide association study (GWAS) catalog on December 18, 2012, and obtained 9,977 distinct SNP-trait associations spanning 7,531 unique SNPs in 1000 Genomes; these variants are referred to as “lead SNPs”. We used the Genome Variation Server (GVS, http://gvs.gs.washington.edu/GVS137/) to find all SNPs within 100 kb of a lead SNP that have a pairwise correlation of R2 >= 0.8 within Utah residents with ancestry from northern and western Europe (CEU). This resulted in an additional 56,538 unique SNPs, referred to as “tag SNPs”. We also developed “control” SNP sets, selected to match trait-associated SNPs for a variety of features that may bias SNPs found by GWAS in the absence of any causal effects.
Supplementary Material
Acknowledgments
We thank P. Green and members of the Shendure Lab for helpful discussions and suggestions. Our work was supported by National Institutes of Health (N.I.H.) grants U54HG006493 (to J.S. and G.C), DP5OD009145 (to D.W.) and DP1HG007811 (to J.S.).
Footnotes
G.C. and J.S. designed the study; M.K. processed the annotation data and scores, developed and implemented the simulator and scripts required for scoring; P.J. and B.O. prepared and provided data sets and annotations; D.W. and M.K. developed the model and performed model training; D.W. performed the analysis of individual features and interactions; M.K., D.W., G.C., and J.S. analyzed the model’s performance on different data sets; G.C. analyzed the GWAS data; J.S., G.C., M.K. and D.W. wrote the manuscript with input from all authors.
Contributor Information
Martin Kircher, Email: mkircher@uw.edu.
Daniela M. Witten, Email: dwitten@u.washington.edu.
Preti Jain, Email: pjain@hudsonalpha.org.
Brian J. O’Roak, Email: oroak@uw.edu.
Gregory M. Cooper, Email: gcooper@hudsonalpha.org.
Jay Shendure, Email: shendure@uw.edu.
References
- 1.Cooper GM, et al. Single-nucleotide evolutionary constraint scores highlight disease-causing mutations. Nat Methods. 2010;7:250–1. doi: 10.1038/nmeth0410-250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cooper GM, Shendure J. Needles in stacks of needles: finding disease-causal variants in a wealth of genomic data. Nat Rev Genet. 2011;12:628–40. doi: 10.1038/nrg3046. [DOI] [PubMed] [Google Scholar]
- 3.Musunuru K, et al. From noncoding variant to phenotype via SORT1 at the 1p13 cholesterol locus. Nature. 2010;466:714–9. doi: 10.1038/nature09266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ward LD, Kellis M. Interpreting noncoding genetic variation in complex traits and human disease. Nat Biotechnol. 2012;30:1095–106. doi: 10.1038/nbt.2422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ng SB, et al. Targeted capture and massively parallel sequencing of 12 human exomes. Nature. 2009;461:272–6. doi: 10.1038/nature08250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Adzhubei IA, et al. A method and server for predicting damaging missense mutations. Nat Methods. 2010;7:248–9. doi: 10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ng PC, Henikoff S. SIFT: Predicting amino acid changes that affect protein function. Nucleic Acids Res. 2003;31:3812–4. doi: 10.1093/nar/gkg509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cooper GM, et al. Distribution and intensity of constraint in mammalian genomic sequence. Genome Res. 2005;15:901–13. doi: 10.1101/gr.3577405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Siepel A, et al. Evolutionarily conserved elements in vertebrate, insect, worm, and yeast genomes. Genome Res. 2005;15:1034–50. doi: 10.1101/gr.3715005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pollard KS, Hubisz MJ, Rosenbloom KR, Siepel A. Detection of nonneutral substitution rates on mammalian phylogenies. Genome Res. 2010;20:110–21. doi: 10.1101/gr.097857.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.ENCODE Project Consortium et al. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kimura M. The neutral theory of molecular evolution. xv. Cambridge University Press, Cambridge Cambridgeshire; New York: 1983. p. 367. [Google Scholar]
- 13.Paten B, et al. Genome-wide nucleotide-level mammalian ancestor reconstruction. Genome Res. 2008;18:1829–43. doi: 10.1101/gr.076521.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.The 1000 Genomes Project Consortium et al. An integrated map of genetic variation from 1,092 human genomes. Nature. 2012;491:56–65. doi: 10.1038/nature11632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Paten B, Herrero J, Beal K, Fitzgerald S, Birney E. Enredo and Pecan: genome-wide mammalian consistency-based multiple alignment with paralogs. Genome Res. 2008;18:1814–28. doi: 10.1101/gr.076554.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.McLaren W, et al. Deriving the consequences of genomic variants with the Ensembl API and SNP Effect Predictor. Bioinformatics. 2010;26:2069–70. doi: 10.1093/bioinformatics/btq330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Meyer LR, et al. The UCSC Genome Browser database: extensions and updates 2013. Nucleic Acids Res. 2013;41:D64–9. doi: 10.1093/nar/gks1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Boyle AP, et al. High-resolution mapping and characterization of open chromatin across the genome. Cell. 2008;132:311–22. doi: 10.1016/j.cell.2007.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Johnson DS, Mortazavi A, Myers RM, Wold B. Genome-wide mapping of in vivo protein-DNA interactions. Science. 2007;316:1497–502. doi: 10.1126/science.1141319. [DOI] [PubMed] [Google Scholar]
- 20.Grantham R. Amino acid difference formula to help explain protein evolution. Science. 1974;185:862–4. doi: 10.1126/science.185.4154.862. [DOI] [PubMed] [Google Scholar]
- 21.Franc V, Sonnenburg S. Optimized cutting plane algorithm for large-scale risk minimization. The Journal of Machine Learning Research. 2009;10:2157–2192. [Google Scholar]
- 22.Ewing B, Green P. Base-calling of automated sequencer traces using phred. II Error probabilities. Genome Res. 1998;8:186–94. [PubMed] [Google Scholar]
- 23.Liao BY, Zhang J. Null mutations in human and mouse orthologs frequently result in different phenotypes. Proc Natl Acad Sci U S A. 2008;105:6987–92. doi: 10.1073/pnas.0800387105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fu W, et al. Analysis of 6,515 exomes reveals the recent origin of most human protein-coding variants. Nature. 2012 doi: 10.1038/nature11690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Makrythanasis P, et al. MLL2 mutation detection in 86 patients with Kabuki syndrome: a genotype-phenotype study. Clin Genet. 2013 doi: 10.1111/cge.12081. [DOI] [PubMed] [Google Scholar]
- 26.Giardine B, et al. HbVar database of human hemoglobin variants and thalassemia mutations: 2007 update. Hum Mutat. 2007;28:206. doi: 10.1002/humu.9479. [DOI] [PubMed] [Google Scholar]
- 27.Baker M. One-stop shop for disease genes. Nature. 2012;491:171. doi: 10.1038/491171a. [DOI] [PubMed] [Google Scholar]
- 28.Patwardhan RP, et al. Massively parallel functional dissection of mammalian enhancers in vivo. Nat Biotechnol. 2012;30:265–70. doi: 10.1038/nbt.2136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Patwardhan RP, et al. High-resolution analysis of DNA regulatory elements by synthetic saturation mutagenesis. Nat Biotechnol. 2009;27:1173–5. doi: 10.1038/nbt.1589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.O’Roak BJ, et al. Exome sequencing in sporadic autism spectrum disorders identifies severe de novo mutations. Nature genetics. 2011;43:585–9. doi: 10.1038/ng.835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.O’Roak BJ, et al. Sporadic autism exomes reveal a highly interconnected protein network of de novo mutations. Nature. 2012;485:246–50. doi: 10.1038/nature10989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sanders SJ, et al. De novo mutations revealed by whole-exome sequencing are strongly associated with autism. Nature. 2012;485:237–41. doi: 10.1038/nature10945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Neale BM, et al. Patterns and rates of exonic de novo mutations in autism spectrum disorders. Nature. 2012;485:242–5. doi: 10.1038/nature11011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Iossifov I, et al. De novo gene disruptions in children on the autistic spectrum. Neuron. 2012;74:285–99. doi: 10.1016/j.neuron.2012.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rauch A, et al. Range of genetic mutations associated with severe non-syndromic sporadic intellectual disability: an exome sequencing study. Lancet. 2012 doi: 10.1016/S0140-6736(12)61480-9. [DOI] [PubMed] [Google Scholar]
- 36.de Ligt J, et al. Diagnostic Exome Sequencing in Persons with Severe Intellectual Disability. The New England journal of medicine. 2012 doi: 10.1056/NEJMoa1206524. [DOI] [PubMed] [Google Scholar]
- 37.Cooper GM, et al. A copy number variation morbidity map of developmental delay. Nat Genet. 2011;43:838–46. doi: 10.1038/ng.909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ng SB, et al. Exome sequencing identifies MLL2 mutations as a cause of Kabuki syndrome. Nat Genet. 2010;42:790–3. doi: 10.1038/ng.646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rohland N, Reich D. Cost-effective, high-throughput DNA sequencing libraries for multiplexed target capture. Genome Res. 2012;22:939–46. doi: 10.1101/gr.128124.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Meyer M, et al. A high-coverage genome sequence from an archaic Denisovan individual. Science. 2012;338:222–6. doi: 10.1126/science.1224344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hindorff LA, et al. Potential etiologic and functional implications of genome-wide association loci for human diseases and traits. Proc Natl Acad Sci U S A. 2009;106:9362–7. doi: 10.1073/pnas.0903103106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nicolae DL, et al. Trait-associated SNPs are more likely to be eQTLs: annotation to enhance discovery from GWAS. PLoS Genet. 2010;6:e1000888. doi: 10.1371/journal.pgen.1000888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gerstein MB, et al. Architecture of the human regulatory network derived from ENCODE data. Nature. 2012;489:91–100. doi: 10.1038/nature11245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schaub MA, Boyle AP, Kundaje A, Batzoglou S, Snyder M. Linking disease associations with regulatory information in the human genome. Genome Res. 2012;22:1748–59. doi: 10.1101/gr.136127.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gonzalez-Perez A, Lopez-Bigas N. Improving the assessment of the outcome of nonsynonymous SNVs with a consensus deleteriousness score, Condel. Am J Hum Genet. 2011;88:440–9. doi: 10.1016/j.ajhg.2011.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Arbiza L, et al. Genome-wide inference of natural selection on human transcription factor binding sites. Nat Genet. 2013;45:723–9. doi: 10.1038/ng.2658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Weedon MN, et al. Recessive mutations in a distal PTF1A enhancer cause isolated pancreatic agenesis. Nat Genet. 2013;46:61–4. doi: 10.1038/ng.2826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Stenson PD, et al. The Human Gene Mutation Database: 2008 update. Genome Med. 2009;1:13. doi: 10.1186/gm13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.MacArthur DG, et al. A systematic survey of loss-of-function variants in human protein-coding genes. Science. 2012;335:823–8. doi: 10.1126/science.1215040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tavaré S. Some probabilistic and statistical problems in the analysis of DNA sequences. Lect Math Life Sci. 1986;17:57–86. [Google Scholar]
- 51.Fujita PA, et al. The UCSC Genome Browser database: update 2011. Nucleic Acids Res. 2011;39:D876–82. doi: 10.1093/nar/gkq963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rosenbloom KR, et al. ENCODE whole-genome data in the UCSC Genome Browser: update 2012. Nucleic Acids Res. 2012;40:D912–7. doi: 10.1093/nar/gkr1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hubisz MJ, Pollard KS, Siepel A. PHAST and RPHAST: phylogenetic analysis with space/time models. Brief Bioinform. 2011;12:41–51. doi: 10.1093/bib/bbq072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Davydov EV, et al. Identifying a high fraction of the human genome to be under selective constraint using GERP++ PLoS Comput Biol. 2010;6:e1001025. doi: 10.1371/journal.pcbi.1001025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.McVicker G, Gordon D, Davis C, Green P. Widespread genomic signatures of natural selection in hominid evolution. PLoS Genet. 2009;5:e1000471. doi: 10.1371/journal.pgen.1000471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hoffman MM, et al. Unsupervised pattern discovery in human chromatin structure through genomic segmentation. Nat Methods. 2012;9:473–6. doi: 10.1038/nmeth.1937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tennessen JA, et al. Evolution and functional impact of rare coding variation from deep sequencing of human exomes. Science. 2012;337:64–9. doi: 10.1126/science.1219240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Khurana E, Fu Y, Chen J, Gerstein M. Interpretation of genomic variants using a unified biological network approach. PLoS Comput Biol. 2013;9:e1002886. doi: 10.1371/journal.pcbi.1002886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Liu X, Jian X, Boerwinkle E. dbNSFP: a lightweight database of human nonsynonymous SNPs and their functional predictions. Hum Mutat. 2011;32:894–9. doi: 10.1002/humu.21517. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.