Abstract
There is little remedy for the devastating effects resulting from neuronal loss caused by neural injury or neurodegenerative disease. Reconstruction of damaged neural circuitry with stem cell-derived neurons is a promising approach to repair these defects, but controlling differentiation and guiding synaptic integration with existing neurons remain significant unmet challenges. Biomaterial surfaces can present nanoscale topographical cues which influence neuronal differentiation and process outgrowth. By combining these scaffolds with additional molecular biology strategies, synergistic control over cell fate can be achieved. Here, we review recent progress in promoting neuronal fate using techniques at the interface of biomaterial science and genetic engineering. New data demonstrates that combining nanofiber topography with an induced genetic program enhances neuritogenesis in a synergistic fashion. We propose combining patterned biomaterial surface cues with prescribed genetic programs to achieve neuronal cell fates with the desired sublineage specification, neurochemical profile, targeted integration and electrophysiological properties.
Keywords: stem cell, topography, nanofibers, neuron, gene, scaffold
Current challenges in stem cell regeneration of neural circuits
Embryonic stem cells are pluripotent cells capable of self-renewal and differentiation into many cell types1. Given the appropriate cues, these cells may differentiate toward neural cells, making them promising candidates for replacement of neurons lost through injury or degenerative disease2–4. Positive outcomes following stem cell therapy have been reported in a host of neural disease models, including multiple sclerosis, Alzheimer’s and Parkinson’s diseases, and clinical trials are underway3–4. With the advent of induced pluripotent and direct conversion technologies, where somatic cells are reprogrammed to create the characteristics of embryonic stem cells or functional neurons, respectively, a potentially unlimited source of cells which is unfettered by limitations surrounding cell source has emerged5–7.
Despite incredible progress and the untapped potential for stem cell therapy to treat neural injury and disease, integration with, and reconstruction of, native neural circuitry remain elusive goals. Fundamental questions remain regarding the mechanism of stem cell influence and their ability to efficiently, effectively, and safely repair damaged neural circuitry. Tumor formation by uncommitted progenitors raises significant safety concerns, as highlighted by the discovery of a donor tissue-derived brain tumor in a boy four years after he was transplanted with fetal neural stem cells to treat an inheritable neurodegenerative disorder8. Functional integration of grafted cells with host tissue is rarely reported in animal models, and this is cited as a barrier to clinical trials2, 9–11. Often, the degree of functional improvement seems irreconcilable with the number of surviving, integrated cells following transplantation, lending credence to alternative, non-replacement mechanisms of stem cell effects (i.e., neurotrophic factor release or abatement of inflammation)10, 12–14. Even once differentiated and integrated into the host tissue, the ability of stem cell-derived neurons to readily fire action potentials in response to a stimulus is questionable, with some morphologically “mature” neurons reportedly lacking the ability to fire action potentials from their relatively depolarized resting potentials15–18. Sufficient control of neuronal phenotype and function are barriers to progress in the field19. Defining not only a neural lineage from a stem cell, but also the desired neuronal subtype (defined as a neuronal cell with a specific physiological function)20, is a formidable challenge. There are hundreds of distinct neuronal subtypes in the mammalian nervous system according to some estimates, and otherwise morphologically homogenous cells can present diverse biophysical properties necessary for encoding information21–22. Nevertheless, the tools available to tackle these challenges continue to expand and evolve rapidly, and particularly exciting advances in controlling stem cell fate are being made at the interface of materials science and genetic engineering.
Cell fate is progressively restricted as the cell transitions from a stem cell to a committed neural progenitor; extrinsic environmental cues work in concert with intrinsic genetic programs both before and after cell cycle exit to determine neuronal fate and subtype specification23. Remarkable progress has been made in recent years in the use of targeted biomaterial and molecular biology-based approaches to coax stem cells to a desired neuronal fate. Combining strategies from these fields may allow for the creation of prescribed patterns of neuronal fate specification and synapse formation, yielding the de novo creation of neuronal circuits with defined lineages. The convergence of surface engineering and genetic programming techniques creates a new design space for regenerating neural circuits, understanding their development, and directing their integration with existing networks. In this review, we examine the state-of-the art in genetic engineering and materials science in neurodifferentiation of stem cells, and we propose combining strategies from these fields to efficiently achieve patterns of neurons with specified fates.
Surface-mediated effects on gene expression and stem cell fate
Engineering the topography of biomaterial substrates to determine cell fate takes advantage of the natural contact-mediated signaling events that occur in the cellular environment24–27. Within tissues, cells contact and interact with a three-dimensional macromolecular complex known as the extracellular matrix (ECM). ECM is a fibrous acellular matrix comprised of glycosaminoglycans and proteoglycans, fibrillar proteins (collagen and other glycoproteins), and adhesive proteins (fibronectin, laminin) which interact with surface receptors (integrins) in the cell membrane28. ECM was once believed to spontaneously generate life but later came to be viewed as a passive supporting structure following the emergence of cellular theory in the 1800’s29. In recent years, ECM is increasingly conceptualized as a dynamic regulator of the local tissue environment, providing topographical cues, creating patterns of bound growth factors and controlling the content of the soluble milieu that cells interface27–28, 30. ECM plays an essential role in every stage of development beginning with the zona pellucida in fertilization, and an ever-expanding litany of genetic studies demonstrates its role in the formation of a variety of tissue types and developmental processes25, 28. In some sense, the view of ECM has come full circle, as it is now viewed as an active participant with its cellular counterparts in organogenesis.
Beyond its role as a structural support for tissue growth and migration, cues provided by ECM contact mediate cell viability, proliferation, and differentiation in a variety of cell types, and ECM presents chemical and mechanical cues which affect stem cell differentiation25, 31–33. Chemical signals include the presentation of adhesive proteins and bound growth factors and cytokines which have downstream effects on cellular differentiation28. For example, integrin β1 is known to play important roles in neurodevelopment, and scaffold-mediated neuronal differentiation can be inhibited by integrin β1 functional blocking antibodies34–35. Laminin α5 is required for normal differentiation and localization of cranial sensory and trunk sympathetic ganglia, as evidenced by abnormalities in mutant mice36. ECM can bind growth factors, and Nakajima et al. demonstrate that while ECM proteins such as laminin and fibronectin displayed negligible effects on stem cell differentiation, increased neuronal differentiation occurred in a synergistic manner when growth factors were immobilized onto these matrix proteins37. In addition to these types of chemical effects, there is growing evidence for direct mechanical effects of ECM on stem cell fate. Matrix stiffness, as characterized by the elastic modulus of the material, has been linked to lineage specification. This work is based on the fact that the elastic modulus of various tissues in the human body ranges widely from relatively soft materials including brain tissue (~ 1 kPa) to rigid connective tissues exceeding 1 GPa38–39. In their seminal study, Engler et al. demonstrated the sensitivity of differentiating mesenchymal stem cells to the stiffness of the extracellular environment: soft material (0.1–1.0 kPa) promoted neuronal differentiation, stiffer material (~11 kPa) produced muscle cells, and the stiffest material (~34 kPa) was osteogenic39. Furthermore, modulating adhesive protein presentation and matrix architectures can affect the lineage specification of differentiating human embryonic stem cells as shown by the identification of specific ECM-mimetic hydrogel compositions that favored either endothelial or osteogenic differentiation40.
The mechanisms governing the influence of the mechanical and topographical effects of the extracellular environment on stem cell differentiation are only beginning to be understood. While effects are often broadly attributed to integrin clustering, focal adhesion formation, and changes in cellular tension via mechanotransductive signals, recent reports have shed new light on the specific molecular pathways involved. Du et al. demonstrated that increased neuronal differentiation on soft substrates was related to increased internalization of β1 integrins via caveolar/lipid raft-mediated endocytosis41. Blocking this pathway reduced neuronal differentiation on the soft substrates (modulus <1 kPa). Keung et al. reported new mechanisms underlying the effects of substrate stiffness on neuronal differentiation of neural stem cells42. Cells plated on laminin-coated polyacrylamide gels that were relatively soft (100–700 Pa modulus) favored a neuronal lineage while cells on stiffer substrates adopted glial fates. This effect was apparent whether or not the media formulation included retinoic acid (a soluble cue known to promote neurodifferentiation). Atomic force microscopy showed a monotonic increase in cellular stiffness with increasing substrate stiffness, and this effect was derivative of increased activation of the mechanotransductive Rho family of GTPases (specifically, RhoA and CdC42). Neuronal differentiation could be “rescued” on stiffer substrates by inhibiting downstream signaling cascades initiated by Rho GTPases but not by inhibiting other mechanotransductive proteins such as myosin light chain kinase, Src, or focal adhesion kinase. Given the crossover between Rho GTPases and integrin internalization pathways, these results may be related to a common mechanism, perhaps related to the fact that the actin cytoskeleton must be destabilized to enable neuronal differentiation and neuritogenesis43–44.
Additional mechanisms of ECM effects on differentiation involve mechanotransduction of cytoskeletal changes to nuclear events. As a result of integrin clustering, topography cues are transduced to nuclear signaling events via cytoskeletal linkages45–46. In fact, elements of the nuclear scaffold may directly bind transcription and splicing factors, providing a potential direct link between cytoskeletal remodeling and gene expression45. Changes in cell shape increase intranuclear calcium concentration, providing an additional candidate pathway for substrate topography and mechanical properties to alter gene expression47. As understanding of the biological signaling mechanisms responsible for substrate-mediated effects on cell fate expands, the intelligent design of material surfaces to promote neuronal differentiation, as well as subtype specification, from stem cells will improve. Synergy between genetic approaches and substrate design will require the activation of non-redundant, complementary signaling cascades.
By presenting cells with a nanoscale surface architecture similar to ECM, polymer substrates influence stem and progenitor cell fate through multiple mechanisms, yielding an opportune tactic for regenerating neural tissue48. To date, several strategies have been developed to construct polymer scaffolds with defined, nanoscale surface features for tissue engineering applications49. With a growing appreciation for the ability to control stem cell fate through topographical cues, new methods for fabricating a variety of surface features have emerged in recent years, including self-assembly and multiphoton lithography38, 50–52. Moreover, the emerging field of “cell membrane engineering” presents opportunities for improving interactions of individual cells with scaffolds through artificial incorporation of adhesive moieties within the cell membrane53. In this review, we focus on the fabrication and effects of two families of two-dimensional topographies commonly studied in the literature: fibrous scaffolds and pillar arrays. A survey of the effects of these substrates on stem cell fate is presented in Table 1. Methods to modify surface chemistries, substrate stiffness and the presentation of adhesive ligands have been reviewed elsewhere19, 26, 54–55.
Table 1.
Pro-neural effects following presentation of micro- and nano-scale topographies to stem cells.
| Topography | Material | Dimensions | Cells | Outcome | Reference |
|---|---|---|---|---|---|
| Nanofibers | Polyethersulfone | 283 nm 749 nm 1452 nm |
Adult rat hippocampal NSCs | 40% ↑ in oligodendocytes with 283 nm fibers; 20% ↑ in neuronal cells with 749 nm fibers; reduced viability with 1452 nm fibers. | 61 |
| Polycaprolactone | 250 nm | ESCs treated with retinoic acid | Nestin expression in 40% of cells 14 days after plating on fibers. | 63 | |
| Poly-L-lactic acid | 250 nm 1250 nm |
C17.2 stem cell line | Accelerated neurite out-growth for cells on 250 nm fibers compared to 1250 nm fibers. | 62 | |
| Polyurethane | 360 nm | Human ESCs | >80% expression of neuronal markers beyond 2 weeks on fibers compared to predominantly glial cells without fibers. | 68 | |
| Self-assembling scaffolds with pentapeptide epitope IKVAV | 3D network of nanofibers 5-8 nm in diameter | Murine neural progenitors | Rapid differentiation into neurons (35% TUJ1+ after 1 day) in comparison to controls. | 67 | |
| Pillars, pits and grooves | PDMS, fibronectin coat | 600 nm ridge with 600 nm spacing, 600 nm height | Human ESC line | Nanoscale grooves ↓ proliferation, ↑ alignment. | 75 |
| Pillars of silicon sputter-coated with tantalum | 600-2400 nm height; 1000-6000 nm laterally | Murine ESC lines | Pillar spacing of 2000-4000 nm optimal for undifferentiated cells; greater differentiation potential on 600 nm height. | 78 | |
| PDMS, 2-hydroxyethyl methacrylale coat | Pillars with height varying from 35 to 400 nm | Murine ES-derived neural precursors | ↑ neuronal yield by ↑ pillar height; pillars ↑ neuronal yield compared to flat surfaces. | 77 | |
| Polyurethane scrylate | Grooves with 350 nm width and spacing; 500 nm height | Human ESC line | Neuronal differentiation noted within 5 days in the absence of chemical agents. | 72 | |
| PDMS | Grooves with 350 nm, 1 µm, or 10 µm width | Mesenchymal stem cells | ↑ neuronal differentiation on 350 nm widths versus microscale features. | 73 |
Micro and Nanofiber Arrays
Electrospun polymer nanofiber scaffolds are perhaps the most widely studied means of achieving a surface with defined topography in neural tissue engineering to date. The basic procedure is as follows: a polymer solution is fed through a spinneret at the end of a reservoir syringe, and an applied voltage causes the elongation of the solution droplet at the end of the spinneret (the “Taylor cone”) via electrostatic interactions. If the applied electric field exceeds a critical value, the solution erupts from the droplet into a steady stream which travels mid-air to a collector. If the collector is attached to a spinning wheel, various degrees of fiber alignment can be achieved by adjusting the rotational velocity. A long list of natural and synthetic polymer materials have been electrospun into nanofibers, and the list continues to expand56. Nanofibers have been successfully electrospun from numerous materials of synthetic and natural origin: for example, common degradable synthetic polymers such as poly-L-lactic acid (PLLA) and polycaprolactone (PCL), as well as natural materials such as silk fibroin, gelatin and collagen have all been used in electrospinning. The relative simplicity of this method, combined with its versatility, has made it a popular and powerful technique for scaffold fabrication.
Several process variables can be manipulated in the course of nanofiber fabrication to yield different fiber diameters and degrees of fiber alignment, including solution concentration, flow rate, applied voltage, needle size, and the distance between the syringe needle and the collector57–59. Altering surface geometry, in turn, can produce downstream effects on cell morphology and physiology. For example, a variety of process parameters, including solution concentration, strength of the applied electric field, and flow rate all affect fiber diameter57–58, 60. Fiber diameter affects the viability, neurite outgrowth and differentiation status of attached stem cells (Table 1)61–62. Likewise, numerous studies demonstrate alignment of neural process outgrowth with the direction of nanofiber orientation; this phenomenon has been demonstrated in neural stem cell lines and primary neurons62–64. Increasing the rotational velocity not only increases fiber alignment but also improves the bulk material properties of the scaffold (as reflected by an increase in tensile strength)65. Fiber alignment and acceleration of process outgrowth are useful characteristics for guiding integration with the desired target tissues62, 66.
Nanofibers constructed out of a variety of materials have been shown to enhance neuronal differentiation of stem and progenitor cells61–63, 67–68. Additionally, mitosis of undifferentiated neural stem cells is reduced on nanofiber substrates in comparison to tissue culture plastic61. Further, culture on nanofiber substrates has been shown to promote the establishment of polarity of embryonic neurons66, 69. Neural progenitors seeded onto aligned nanofiber arrays show a bipolar morphology with increased neurite length, whereas several shorter processes are observed on randomly oriented fibers62. Nanofiber substrates are a powerful tool for promoting neuronal fate and guiding neurite growth of differentiated stem cells.
Pillar, pit, and groove topographies
Several groups have used lithography techniques, which afford the ability to create micro- and nanoscale features with relative ease and low cost, to create arrays of pillars, pits, and grooves on polymer surfaces70–75. Soft lithography is the most common technique, where the basic fabrication method uses photolithography to create a master pattern on which an elastomeric material is cast (typically, poly(dimethylsiloxane), PDMS)76. Following curing, the elastomer may be used to pattern another surface (as in microcontact printing), create a mold on which another polymer is cast (replication molding), or serve as the cell culture substrate itself. As such, soft lithography is a versatile platform for the creation of a variety of topographies with a high degree of control over surface geometry.
Submicron features created by lithographic methods enhance neuronal differentiation from embryonic stem cells and progenitors in a manner reminiscent of nanofiber topography effects72. Likewise, reduced proliferation and effects on cellular alignment are evident75. Increased expression of neuronal markers in mesenchymal stem cells following plating on grooved PDMS surfaces has been reported73. Similarly to nanofiber substrates, the dimensions of the geometries produce effects on neuronal differentiation61, 73, 77. In a study which systematically evaluated the differentiation of embryonic stem cells on a library of 504 surface architectures created on silicon substrates, the authors concluded that micron-scale lateral dimensions (1–6 µm) of the pillars and the gaps between them yielded optimal maintenance of the undifferentiated state, while submicron-scale vertical heights of the pillars (0.6–1 µm) resulted in spreading thought to favor differentiation78. However, the mechanisms underlying geometric control of neuronal differentiation are still being explored.
Unifying principles of geometric influence on neuronal fate
In a recent paper, Li et al. surmise that, “…the selection of biomaterials for tissue engineering applications is a trial-and-error iterative process without general principle.”79 Indeed, analyzing the broad spectrum of data for global themes in surface cues is complicated by the wide variety of surface chemistries presented by a litany of materials and coatings studied, as well as the entangled interplay between topography and surface chemistry80. Studies of topography effects on stem cell fate typically survey a range of feature sizes and geometries and catalogue observed effects on phenotype. The outcome of such studies is often the identification of an optimal scaffold to produce a particular phenotype within the subset of surfaces sampled. The lack of systematic approaches and continuity between studies make understanding the mechanisms of topography effects extraordinarily difficult, making the rational design of material surfaces to promote a specific stem cell fate a significant challenge. Furthermore, the identification of specific signaling pathways involved in topography effects is necessary to develop synergistic strategies, which ideally target non-redundant pathways.
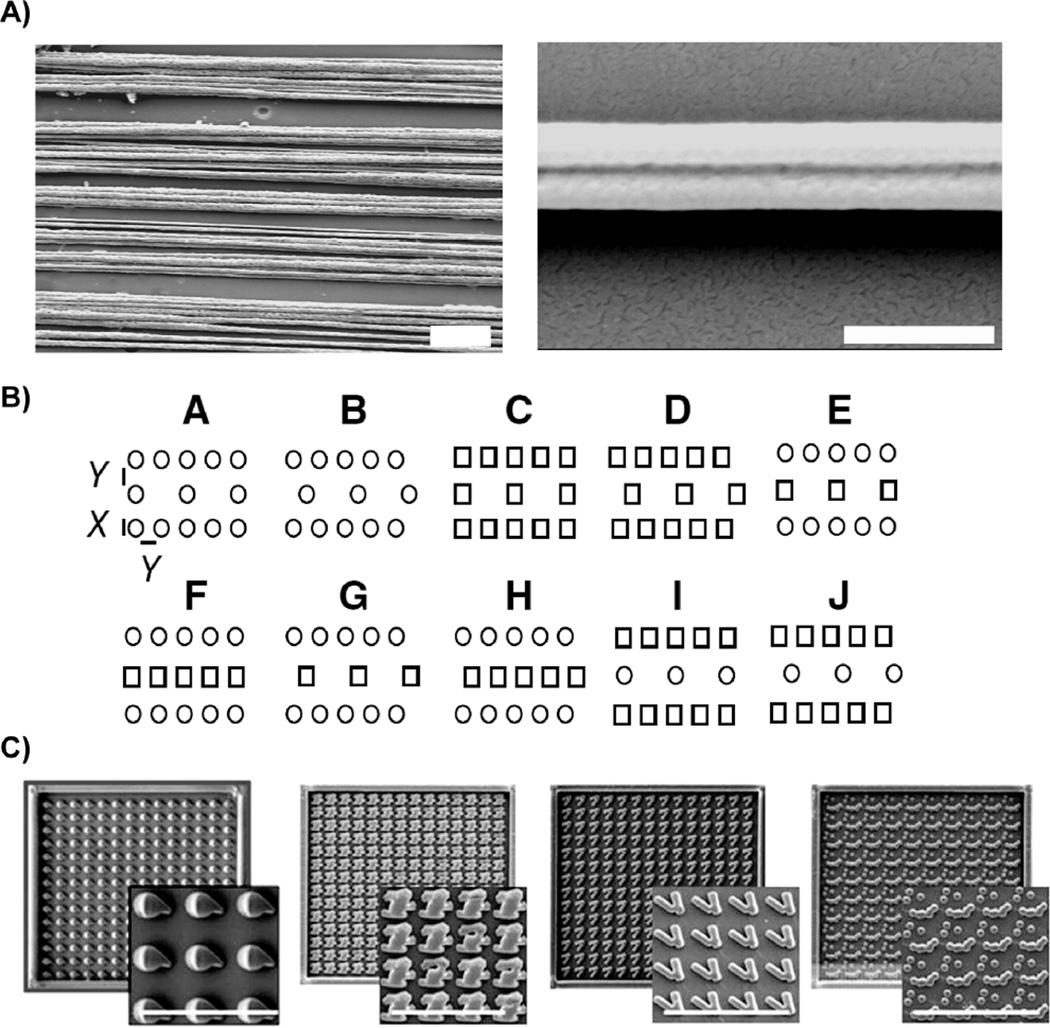
Recent studies approach this issue by screening a vast library of various surface features for effects on a particular cell type78, 81–82. The initial report using this strategy probed the effects of 1,700 polymer chemistries on human embryonic stem cells81. While strategies like this represent an important advance in understanding topography effects, comparison of topographies across two studies which simultaneously analyze hundreds of surface features fails to yield clear unifying principles or mechanisms (Figure 1). Markert et al. surveyed 504 topographies on embryonic stem cell differentiation and proliferation. Analysis indicated that structures with narrow lateral dimensions (“X” = 1 micron) yielded the highest colony numbers for series “A-J” topographies, but this effect was diminished when the vertical height was decreased from 1.6 to 0.6 microns (see Figure 1A). The “K” series tended to have low colony numbers. Unadkat et al. surveyed the effects of 2,176 randomly generated surface geometries on mesenchymal stem cell fate in a “materiomics” approach inspired by screening methods used for drug discovery82. Visual inspection reveals little apparent similarity between the four topographies identified as increasing proliferation (Figure 1B). The authors’ analysis indicated that parameters representing the “fraction of the total energy contained in the feature after applying discrete Fourier transformation that is present in sinusoids with wave number approximately 1.5 and 1” were predictive of increased proliferation. Simple predictive rules for topography design have yet to emerge.
Figure 1.
Topographies which result in a given cellular outcome (increased proliferation) vary widely. Nanofibers similar to those shown in SEMs in (A) tend to favor reduced proliferation (Scale = 10 µm in left panel, 1 µm in higher magnification image in right panel). Markert et al. report that structures with narrow lateral dimensions (“X” = 1 micron) yielded the highest colony numbers for series “A-J” topographies, although the schematics illustrate a variety of feature patterns (B). SEM images of PLA surfaces with the greatest mesenchymal stem cell proliferation show a variety of surface features (C) (Scale = 50 µm). Part (B) is reproduced with permission from ref 78 Copyright 2009 Mary Ann Liebert. Part (C) is reproduced with permission from ref 82 Copyright 2011 National Academy of Sciences.
Nonetheless, certain surface features seem to predict a neuronal fate from stem cells irrespective of the material properties of the underlying substrate. Scaffolds presenting aligned grooves or nanofibers with lateral dimensions on the order of hundreds of nanometers (~250–750 nm) favor cellular elongation in the direction of alignment, reduced proliferation, enhanced expression of markers of neuronal differentiation and acceleration of neurite outgrowth (Table 1)61–63, 68, 72–73, 75. These results are consistent and span mesenchymal stem cells, neural stem cell lines, and embryonic stem cells plated on a variety of polymer materials (Table 1). While mechanisms are unclear, the encouragement of a cell shape which mimics that found in nature, i.e., increased anisotropy or elongation, seems to be a requisite outcome of guiding neuronal lineage commitment in stem cells82–83. It is possible that cell shape affects integrin clustering, turnover and associated Rho GTPases, as well as actin cytoskeleton stability and dynamics. However, a linkage between these mechanisms and underlying topography effects on neural lineage specification of stem cells has yet to be determined.
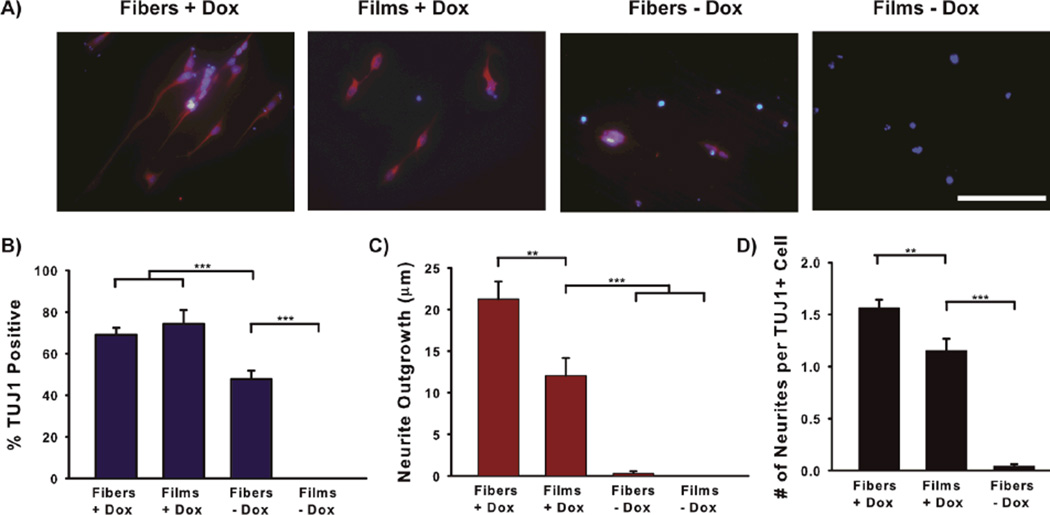
While topography cues are sufficient to enhance expression of neuronal markers from a subpopulation of attached embryonic stem cells, the resulting cell population is still a heterogeneous mixture of undifferentiated and differentiated cells. Xie et al. report that ~20% of retinoic acid-treated embryoid bodies express TUJ1 after plating on nanofibers, whereas expression of this neuronal marker was absent without nanofibers63. Recent data from our laboratory indicates that ~40% of embryonic stem cells attaching to polystyrene nanofibers express a neuronal marker (βIII-tubulin, TUJ1) but do not extend neurites (Figure 2A-C). While aligned, microscale surface topography enhances neuronal differentiation from stem cells, the majority of cells adopt a non-neuronal fate. Recently, a variety of combinatorial strategies have been explored to improve outcomes, where multiple cues are presented to stem cells to promote a neuronal fate in a synergistic fashion.
Figure 2.
Synergistic effects of induced bHLH expression and nanofiber topography on neurite outgrowth from stem cell-derived neurons. Stem cells were plated on polystyrene nanofiber scaffolds or films and either Dox-induced to overexpress Neurog1 or left uninduced. Neuronal differentiation and neuritogenesis were quantified using an automated image analysis program136 to analyze neurites. Dox-induction produces a similar percentage of neuronal cells with or without fiber topography (B), but combining induced Neurog1 with a fiber substrate increased neurite outgrowth in a synergistic manner (C and D). Scale = 100 µm in (A).
Emerging opportunities for synergy
Topography is increasingly combined with additional attributes in order to further fine-tune stem cell fate in a potentially synergistic fashion. Elastic modulus is one potential feature to explore, as there exists compelling evidence for differentiation of mesenchymal stem cells to neurogenic, osteogenic, or myogenic fates by tuning scaffold stiffness to mimic the elasticity of those tissues39. To some degree, fabrication restricts the combination of rigidity and topography achievable; creating micro- and nanoscale features requires material stiffness exceeding a few hundred kPa84. The practical implication is that it is very difficult to mimic both the dimensions and elastic modulus of the extracellular environment of the brain.
A second potential opportunity for synergistic control of stem cell fate is through the release or attachment of neurotrophic factors or other bioactive ligands to a defined material surface. McKay and colleagues have demonstrated that application of specific growth factors can determine the lineage of multipotent stem cells, where platelet-derived growth factor promotes neuronal differentiation, and thyroid hormone and ciliary neurotrophic factor favor glial fates85. The ability to incorporate neurotrophic factors into polymer scaffolds has been demonstrated by several research groups86–88. Attaching brain-derived neurotrophic factor to PCL nanofiber scaffolds increased the neuronal differentiation of attached neural stem cells86. However, very few studies have combined growth factor delivery with topography for the specific purpose of promoting a neuronal fate from stem cells.
A third possibility for synergistic control over neuronal fate involves the conductivity of the scaffold material. Conductive scaffolds retain the ability to differentiate stem cells into neurons through topographical cues while offering electrical interfacing as a complementary mode of influence over stem cell fate. Electrical stimulation is known to promote neuronal differentiation from embryonic stem cells and enhance neurite extension in neuronal cells, and while the mechanisms are still being explored, increased calcium influx appears to play an important role89–90. The nanofiber materials used incorporate carbon nanotubes91–92 or conductive polymers such as polyaniline (PANi)93, polypyrrole (PPy)94, or poly(3,4-ethylenedioxythiophene) (PEDOT)95. Increased neurite outgrowth (>40%) has been observed in pheochromocytoma-12 (PC-12) cells cultured on poly(lactic-co-glycolic acid) (PLGA) films and nanofibers coated with PPy when these scaffolds were electrically stimulated90, 94. A similar report indicates increased neurite extension on electrically stimulated nanofibers for stem cell-derived neurons plated on composites of PANi and PLLA96. Interestingly, reports suggest that the intrinsic electrical conductivity of these materials promotes differentiation and neuronal growth even in the absence of active stimulation97–98. Effects have been proposed to be attributable to alterations in the surrounding ionic environment and membrane transport92.
The combination of microscale topography with additional mechanical, neurotrophic, or electrical cues are all promising strategies for controlling stem cell fate. Meanwhile, the induction of specific genetic programs is perhaps the most effective and efficient method to determine a neuronal fate. Using genetic and surface cues to synergistically control stem cell fate is a promising and comparatively understudied method.
Combined delivery of topographical and genetic cues to promote neuronal fate specification
The combination of surface cues with genetic manipulation presents a broad array of possibilities for engineering stem cell fate decisions. The effects of the surface architecture may be direct, as in the case of vector-releasing substrates, or indirect, as shown by recent reports demonstrating topography effects on transfection efficiency. We explore recent developments in both of these strategic areas, beginning with a primer on genetic engineering strategies.
Strategies for transgene delivery
There are several potential methods of forcing the expression of specific genes in a population cells, and they can be broadly categorized into viral and non-viral delivery systems (“vectors”). Some key assessment criteria when choosing a vector system include: (1) the safety and predictability of the cellular/organism response, (2) the amount of genetic information that can be delivered (“cargo” or “payload” size), (3) the efficiency of transformation in terms of the percentage of the cell population expressing the transgene, (4) the ability to excise the gene delivery system without leaving mutations (“reversibility”), and (5) the duration of the expression. Viral means of transferring genetic material are often highly efficient (yielding transgene expression in 20%–80% of infected embryonic stem cells in some reports), but their limited cargo size restricts the ability to selectively transform cells of specified lineages99–100. Different types of viruses have different properties; i.e., adenoviruses have a larger packaging capacity than lentiviruses and retroviruses, but their use in vivo has raised significant safety concerns arising from early failures and unexpected lethal inflammatory responses in isolated clinical trials100–101. Alternatively, several non-viral means of transferring genetic material to host cells exist, with new and ingenious means of removing any trace of vector material from the transformed cell becoming increasingly available99–100. Non-viral strategies may employ transposons (mobile DNA elements), plasmids (circular, self-replicable double-stranded DNA), short-interfering RNA molecules (siRNAs), cell-penetrating peptides, or bacterial artificial chromosomes (DNA constructs) to deliver a transgene. Non-viral transfection typically employs lipofection or electroporation techniques as the means of encouraging cellular uptake of the vector by endocytosis or the creation of transient pores in the cell membrane (respectively).
Methods of gene delivery are further categorized into reversible and irreversible systems. Irreversible systems leave the vector in the host cell or cause mutations that remain even after the transgene is excised. The clinical applicability of irreversible systems is dubious, given the propensity for unwanted mutagenesis102. Recently, transposon systems have emerged as a promising platform for reversible gene delivery. In these systems, a plasmid delivers a transposon to the cell, which then integrates into the genome in what can be described as a ‘cut-and-paste’ regime. The “Sleeping Beauty” system, so-named for its “awakening” following an extended evolutionary “sleep,” was the first transposon shown to be capable of gene transfer in mammalian cells103. This system was followed by the “PiggyBac” transposon system, which was recently modified to be fully reversible and feature a large payload capacity100. These features are particularly valuable in stem cell applications, where lack of unwanted mutagenesis and control of cell fate are of preeminent importance for safety reasons. A larger cargo capacity enables the inclusion of additional measures of genetic control; for instance, sequences can be included which enable the identification and selection of transformed cells or allow gene expression to be controllably turned on or off. These types of controls are particularly valuable when funneling a heterogenous stem cell population to a neuronal fate via forced gene expression.
Genetic control of neuronal fate
Genetic programming of neuronal fate is initiated by proneural genes of the basic helix-loop-helix (bHLH) class of transcription factors104. These factors promote neuronal fate while inhibiting gliogenesis104–105. Their expression results in a lengthening of the cell cycle, reducing proliferation and favoring differentiation106. The expression of bHLH transcription factors is transient, yielding progenitors that are later terminally differentiated via downstream signaling events. The expression of specific bHLH factors not only gives rise to a generalized neuronal fate but also can determine specific neuronal subtypes. During development, spatiotemporal expression patterns of different combinations of bHLH genes underlies the adoption of distinct neuronal fates at specific locations along the neuraxis107.
That a given bHLH protein may be required for the development of different areas of the nervous system at different stages of development indicates that the resulting subtype depends, in part, on contextual cues108. As an example, the bHLH transcription factor Neurogenin-1 is required for the development of proximal cranial sensory neurons as well as the small diameter fibers of the dorsal root (spinal) ganglia109–110. Likewise, the bHLH transcription factor Atoh1(Math1) is required for the development of a variety of neuronal subtypes but is also necessary for the formation of non-neuronal cells including intestinal secretory cells, touch-sensitive Merkel cells in the skin, and sound-sensing inner ear hair cells108, 111. The expression of bHLH genes is under the control of regulatory pathways which lend themselves to extrinsic manipulation. For instance, expression of a proneural gene in one cell inhibits expression in the neighboring cell in a process known as “lateral inhibition”112. A recent report implicates lateral inhibition and the associated Notch-signaling pathway in the specification of sensory receptor cells and neurons from inner ear stem cells via differential effects on the bHLH factors required for genesis of those cell types113. Thus, neural fate is under the regulatory control of cell-cell contact, a cell density- and Notch-dependent variable which is manipulable by scaffolding conditions114–115. Proneural gene expression may also be affected by growth factors, presenting another potential means of extrinsic control through controlled release116. The regulation of intrinsic genetic programs by context-dependent signaling pathways lends itself to bioengineering strategies to act synergistically with induced genetic cues to promote a specific neural fate. In addition to providing these contextual cues by presenting defined topographical features and surface chemistries, polymer substrates can be used directly for gene delivery, and they can enhance transfection efficiency through topography cues. These substrates can present the combined genetic and environmental signals required for cell fate specification.
Direct gene transfer from polymer substrates
Vector release from a polymer scaffold to cells adhered to its surface is an effective strategy for transforming cells, and the materials used are often amenable to electrospinning or soft lithography techniques117. Not only does this approach allow for localized vector delivery, which may enhance transfection118, but it also enables patterned delivery of a variety of genetic sequences119–121. These traits could be leveraged for creating neuronal circuits from stem cells which recreate the diversity found in native tissue21. Vector release from scaffolds has been achieved by the controlled release of plasmids from commonly used degradable biocompatible polymers such as PLGA or through incorporating genetic material onto a cell culture surface122–125. Reported transfection efficiencies and cellular viabilities tend to be comparable or superior to those offered by traditional, “bolus” approaches to delivering non-viral vectors. For instance, gene transfer from DNA/calcium phosphate nanocomposites on a polystyrene surface achieved transfection efficiencies comparable to a commercially available transfection reagent122. More recently, nanoparticles have been used to deliver siRNAs in order to achieve gene silencing121, 126. Here, the technology enables targeted “knock-down” of the genes of interest, and combination with microprinting methods can be used to deliver siRNAs in a defined pattern121. Likewise, microfluidic systems have been used to create spatial patterns of genetic material onto tissue culture-treated plastic and polymer scaffolds with microscale channels119–120. Scaffold release of siRNA can be used to determine stem cell fate; release of siRNA from three-dimensional scaffolds promoted a mesoderm lineage and downregulated genes associated with an endoderm lineage in human embryonic stem cells127. However, proof-of-concept of these knockdown strategies are typically carried out in model cell lines, and applying this type of technology to determine a neuronal fate from a stem cell population and assessing efficacy in vivo have yet to be accomplished.
Indirect effects of topography on cell fate and transfection efficiency
In addition to providing a depot for vector delivery, the surface features of polymer substrates may directly enhance cellular responsiveness to genetic manipulation. Studies by Kong and colleagues showed that surface stiffness and ligand presentation enhance non-viral transfection of preosteoblast cells seeded in hydrogels128–129. These effects were attributed to increased cellular proliferation associated with increased density of adhesive ligands on the stiffer substrates, where the resulting effects on transfection efficiency were presumably due to enhanced exposure of the nucleus during mitosis. However, recent reports indicate that topography increases transfection efficiency irrespective of effects on proliferation. As a proof-of-concept, attachment to microscale topographies increased transfection efficiency in fibroblasts and mesenchymal stem cells70, 130. Adler et al. created pitted PDMS substrates with a variety of feature dimensions and inter-feature spacings, yielding surfaces that ranged in appearance from relatively smooth to mesh-like substrata70. The mesh-like surfaces increased non-viral transfection efficiency of plated fibroblasts by ~25% in comparison to smoother surfaces after normalizing to cell number. The authors suggest that cell spreading effects on endocytosis underlie these results. Likewise, Teo et al. demonstrated increased expression of green fluorescent protein-encoding plasmids by mesenchymal stem cells when these cells were plated on micro- and nanopillar poly(methyl methacrylate) (PMMA) surfaces130. The accompanying data showing related increases in the uptake of fluorescently labeled dextrans by fibroblasts and mesenchymal stem cells corroborates the idea that topography effects on endocytosis increase non-viral transfection efficiency. While the fact that integrins are a key regulator of endocytosis is well-documented43, 131, the application to controlling stem cell fate has not yet been realized. One can envision using topography cues from a bioengineered scaffold to enhance the uptake of vectors designed to force the expression of neural lineage-determining transcription factors in stem cells. Such a scaffold would promote a defined neuronal fate while using topography cues to enhance and guide neurite outgrowth. Furthermore, it may be possible to deliver a pattern of multiple proneural vectors from a surface to attached stem cells, resulting in a network of neurons with distinct sublineages. With an ability to deliver and enhance uptake of genetic material in prescribed patterns, determine a neuronal fate, and accelerate and guide neuritogenesis, the ability to combine topographic and genetic cues in a single substrate to create diverse neuronal circuits from stem cells is achievable.
Potential synergy between topography and genetic transformation
The duplicity of certain ‘required’ transcription factors in the development of disparate types of neurons in different parts of the body implies that intrinsic genetic programs and extrinsic contextual cues act in a coordinated manner to determine neuronal fate specification108. A combination of genetic engineering and biomaterials-based strategies may be designed to recapitulate these developmental programs in stem cells. This strategy has been used to enhance the differentiation of mesenchymal stem cells into an osteogenic lineage132–133. This approach has yet to be exploited for the creation of neural circuits from stem cells.
Our group recently collected evidence that illustrates the potential to combine topography and gene expression to promote neuronal fate from a population of embryonic stem cells in a synergistic fashion. We plated stem cells with inducible overexpression of the proneural bHLH transcription factor Neurogenin-1 (Neurog1) onto polystyrene nanofibers and assessed the neuronal differentiation and neurite elongation after a three day time period in vitro (Figure 2). This stem cell line, created by Matt Velkey and colleagues, reportedly yields ~60% neuronal differentiation (as indicated by TUJ1 expression) three days after doxycycline induction of Neurog1, and these cells bear many similarities to auditory neurons14, 134–135. We observed a similarly rapid, efficient generation of TUJ1 positive cells following induction in cells plated on either polystyrene nanofibers or polystyrene films (Figure 2B). Meanwhile, synergistic effects on neurite outgrowth were observed (Figure 2C-D). Interestingly, fiber topography elicited TUJ1 expression in cells in the absence of dox (Figure 2B), but these cells did not extend neurites. This indicates that fiber topography is sufficient to promote a marker of neuronal progenitor fate from an embryonic stem cell population but is ineffective in efficiently promoting neuritogenesis in the absence of a proneural genetic program, at least in this relatively short time frame. To the best of our knowledge, these data represent the only report combining genetic and surface cues to influence the neuronal fate of embryonic stem cells to date. The potential for synergy in fate specification and maturation opens new possibilities in neuroengineering, but the way forward will rest on a better understanding of how each factor governs downstream signaling events.
Synergy from combined genetic, chemical, and contact cues offers the exciting possibility that we may one day custom-design neurons with highly specified phenotypic traits. Only then may we reproduce the rich diversity of neuron subtypes found in the human nervous system. To achieve this, we must understand the mechanisms that specify fate and fine-tune physiological traits. Normal neurogenesis is a tightly orchestrated process involving spatial and temporal patterns of gene and protein expression, diffusible signals, and a dynamic extracellular landscape. Driving stem cells to a desired neural subtype will be most effective when a combination of cues work together, but these cues must be chosen based on a priori knowledge of the signaling pathways engaged. Practically speaking, the goal is not to recreate a general cellular niche but to derive a particular cellular phenotype with all of the morphological and physiological traits necessary to promote functional integration and replacement of damaged or missing neural tissue. This approach requires that we move past simple mimicry of basic, first-order properties like neurite extension and spike generation.
Many questions remain if we are to push toward higher-order phenotypic traits. For example, how will combinations of proneural cues modulate the expression of surface receptors, which ultimately guide neurite outgrowth and synaptogenesis? How can these cues be combined to specify axodendritic polarity and neurite arborization? Can surface features enhance or limit myelination? Will combined cues alter spike patterns (phasic, tonic, or bursting) leading to changes in temporal coding of a stimulus? Are certain stem cell sources more suitable for manipulation by combinations of cues than others, and what relative advantages in terms of safety and ease of reprogramming do the differing sources offer (e.g. embryonic stem cells, adult induced pluripotent stem cells, or lineage-restricted neural stem cells)? And, given the achievement of higher-order phenotypic traits in vitro, how will results translate to the in vivo environment, which presents its own complex array of extrinsic soluble and topographical cues? On the last point, what will be the various challenges posed by specific neural circuits within the body, in terms of accessibility, ease of genetic manipulation, permissibility of the extracellular environment, and complexity of the cellular circuits involved? The challenges posed by the in vivo environment are highlighted by mixed reports of transplantation survival and therapeutic efficacy of cell scaffolds implanted in the nervous system137. Ultimately, success will require understanding the interaction of the extracellular environment of the host tissue with the genetic and topographical signaling cues imposed upon the stem cell population by the chosen vector and scaffold.
At the core of many of these questions and challenges lies a more fundamental issue, namely, whether combinations of proneural cues will work synergistically to enhance some traits while diversifying phenotype in other areas. Hints from literature suggest that common, master-controlling pathways are initiated by both nanofibers and chemical neural induction. The combination of retinoic acid and a nanofiber substrate potentiates Wnt/β-catenin signaling, possibly because both of these factors upregulate a key proneural transcription factor, NeuroD1138. NeuroD1 is immediately downstream of several proneural bHLH proteins, including neurogenins139, suggesting that genetic, chemical, and substrate cues could indeed combine to enhance activation of specific signaling pathways. However, Wnt signaling is only one of several major cellular pathways that influence differentiation and physiology. While potentiating Wnt signaling, combined factors may independently activate or inhibit a variety of other pathways leading to desirable or undesirable features. It is clear that systematic investigations of fate specification cannot focus on subsets of downstream events, whether signaling molecules or phenotype traits. One can envision large-scale parametric screens that methodically chip away at the possible combinations, but a matrix that explores major bHLH inducers, diffusible chemical factors, and various substrate topologies and chemistries quickly becomes unwieldy. Instead, we suggest a systems biology approach that incorporates the genes, proteins, and pathways activated by individual induction factors and then combine these factors to maximize expression of key phenotypic markers.
Conclusions and perspectives: synergistic effects of combined approaches to create prescribed networks of neurons from stem cells
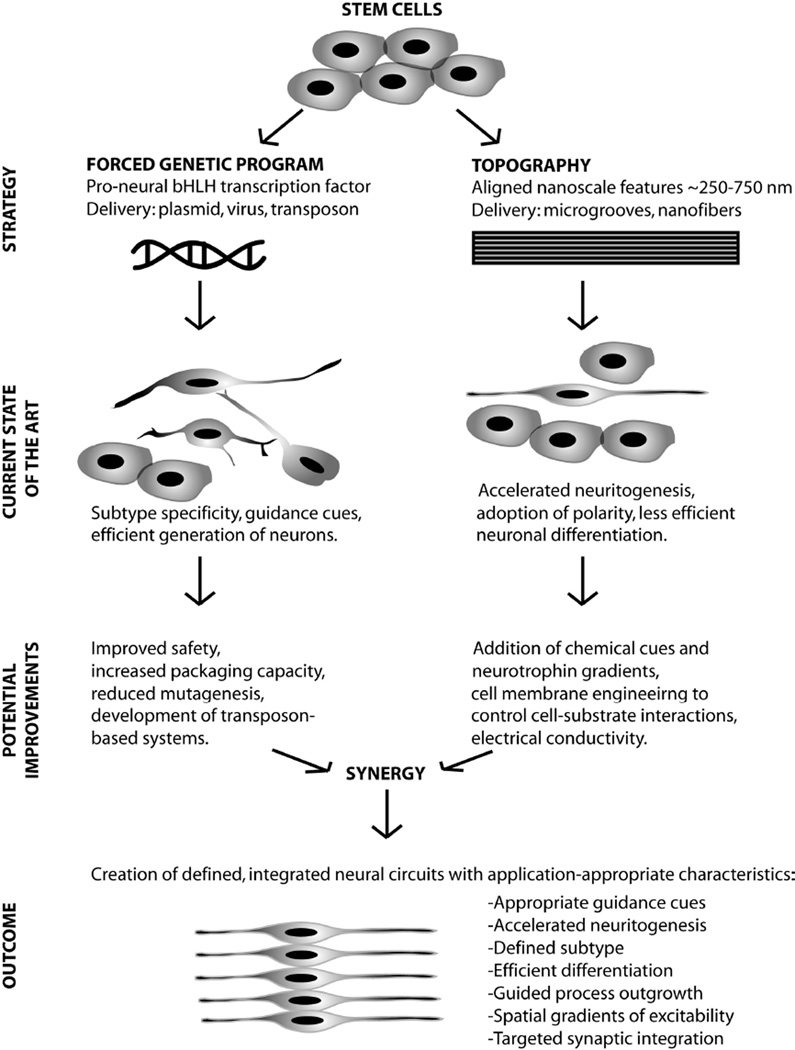
Replacement of damaged neurons with stem cells will require directed differentiation of those cells into neurons of a specific sublineage combined with guided, functional integration of these cells with existing neural circuitry. This is a formidable challenge which demands an exquisite level of control over all aspects of cellular behavior. Native nervous tissue is beautiful in its diversity and complexity, and depending upon the particular peripheral nerve or central circuit of interest, the creation of motor and sensory fibers, excitatory and inhibitory cells, spatial gradients in excitability, unique patterns of dendritic arborization or polarity, and hundreds of synaptic connections on a single cell could be required. We envision rationally designed strategies, inspired by normal developmental processes, which combine intrinsic and extrinsic cues to achieve neuronal cell fates from stem cells with the desired subtype specification, neurotransmitter profile, targeted integration and electrophysiological properties (Figure 3). The ultimate goal is to re-create neural circuitry to a level which is commensurate with the pre-injured state. The combined delivery of fate-determining genes with lineage-defining and process-guiding substrate cues is an emerging strategy for gaining control over stem cell fate. Understanding the mechanisms underlying topographical cues will accelerate the development of approaches to determining stem cell fate. A coherent strategy for unraveling the underlying biological complexity requires comprehensive global profiles of the genes, proteins, and cellular pathways involved in fate specification. The overlap between engineered substrates and stem cell biology must enter the omics era, including transcriptomic, proteomic, and physiomic descriptions of derived neurons. Advances in next-generation sequencing offer unprecedented views into the biology of stem cell-derived neurons, including the complete sequencing of expressed mRNAs, the identification of novel gene products not cataloged on microarray platforms, and information on non-coding RNAs (e.g. microRNAs) that may act on large families of target genes and pathways140–142. Even so, a systems biology approach cannot stop here. Some neural traits will be more specific to protein levels than transcript levels, since the relationship between these is not always straightforward or predictive143. As a consequence, transcriptomic views of fate specification must be combined with global profiles that give proteomic and physiological insights into the biology of derived neurons and ultimately the biology of cell systems once introduced into host tissue. Such an approach will require the assimilation of a broad spectrum of observations analyzed with sophisticated data-mining tools, necessitating combined expertise in stem cell biology, neurophysiology, bioengineering, and bioinformatics. Armed with improved methods of gene delivery and scaffold fabrication and state-of-the-art tools for dissecting the cell system, the reconstruction of neural circuitry is a realistic and attainable goal.
Figure 3.
Future vision for the de novo creation of neuronal networks from stem cells using biomaterial topography and forced genetic programs. Forced genetic programs are advantageous for efficiently yielding neurons of a prescribed sublineage. Aligned nanoscale topographical features accelerate and guide neurogenesis, while less efficiently upregulating the expression of neuronal markers. Combining the guidance properties of patterned substrates with improved techniques for delivering genetic material to cells will result in the ability to create neural circuits with the profile and pattern of physiological properties appropriate for a given regeneration application.
Acknowledgments
Funding Sources
This work was supported by a research grant from the Hearing Health Foundation and a National Institutes of Health training grant (NIDCD T32 DC00011).
Footnotes
Author Contributions
The manuscript was written through contributions of all authors. All authors have given approval to the final version of the manuscript.
References
- 1.Evans MJ, Kaufman MH. Nature. 1981;292(5819):154–156. doi: 10.1038/292154a0. [DOI] [PubMed] [Google Scholar]
- 2.Lindvall O, Kokaia Z. Nature. 2006;441(7097):1094–1096. doi: 10.1038/nature04960. [DOI] [PubMed] [Google Scholar]
- 3.Lunn JS, Sakowski SA, Hur J, Feldman EL. Ann Neurol. 2011;70(3):353–361. doi: 10.1002/ana.22487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Aboody K, Capela A, Niazi N, Stern JH, Temple S. Neuron. 2011;70(4):597–613. doi: 10.1016/j.neuron.2011.05.007. [DOI] [PubMed] [Google Scholar]
- 5.Pang ZP, Yang N, Vierbuchen T, Ostermeier A, Fuentes DR, Yang TQ, Citri A, Sebastiano V, Marro S, Sudhof TC, Wernig M. Nature. 2011;476(7359):220–223. doi: 10.1038/nature10202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vierbuchen T, Ostermeier A, Pang ZP, Kokubu Y, Sudhof TC, Wernig M. Nature. 2010;463(7284):1035–1041. doi: 10.1038/nature08797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Takahashi K, Yamanaka S. Cell. 2006;126(4):663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 8.Amariglio N, Hirshberg A, Scheithauer BW, Cohen Y, Loewenthal R, Trakhtenbrot L, Paz N, Koren-Michowitz M, Waldman D, Leider-Trejo L, Toren A, Constantini S, Rechavi G. PLoS Med. 2009;6(2):e1000029. doi: 10.1371/journal.pmed.1000029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Martino G, Pluchino S. Nat Rev Neurosci. 2006;7(5):395–406. doi: 10.1038/nrn1908. [DOI] [PubMed] [Google Scholar]
- 10.Lindvall O, Kokaia Z, Martinez-Serrano A. Nat Med. 2004;10(Suppl):S42–S50. doi: 10.1038/nm1064. [DOI] [PubMed] [Google Scholar]
- 11.Gaillard A, Prestoz L, Dumartin B, Cantereau A, Morel F, Roger M, Jaber M. Nat Neurosci. 2007;10(10):1294–1299. doi: 10.1038/nn1970. [DOI] [PubMed] [Google Scholar]
- 12.Ourednik J, Ourednik V, Lynch WP, Schachner M, Snyder EY. Nat Biotechnol. 2002;20(11):1103–1110. doi: 10.1038/nbt750. [DOI] [PubMed] [Google Scholar]
- 13.Pluchino S, Zanotti L, Rossi B, Brambilla E, Ottoboni L, Salani G, Martinello M, Cattalini A, Bergami A, Furlan R, Comi G, Constantin G, Martino G. Nature. 2005;436(7048):266–271. doi: 10.1038/nature03889. [DOI] [PubMed] [Google Scholar]
- 14.Hill GW, Purcell EK, Liu LQ, Velkey JM, Altschuler RA, Duncan RK. Stem Cells Dev. 2012 doi: 10.1089/scd.2011.0437. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tong M, Hernandez JL, Purcell EK, Altschuler RA, Duncan RK. Am J Physiol Cell Physiol. 2010;299(6):C1335–C1344. doi: 10.1152/ajpcell.00207.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Balasubramaniyan V, de Haas AH, Bakels R, Koper A, Boddeke HW, Copray JC. Neurosci Res. 2004;49(2):261–265. doi: 10.1016/j.neures.2004.02.010. [DOI] [PubMed] [Google Scholar]
- 17.Carpenter MK, Inokuma MS, Denham J, Mujtaba T, Chiu CP, Rao MS. Exp Neurol. 2001;172(2):383–397. doi: 10.1006/exnr.2001.7832. [DOI] [PubMed] [Google Scholar]
- 18.Martinez-Monedero R, Yi E, Oshima K, Glowatzki E, Edge AS. Dev Neurobiol. 2008;68(5):669–684. doi: 10.1002/dneu.20616. [DOI] [PubMed] [Google Scholar]
- 19.Discher DE, Mooney DJ, Zandstra PW. Science. 2009;324(5935):1673–1677. doi: 10.1126/science.1171643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Teichert RW, Smith NJ, Raghuraman S, Yoshikami D, Light AR, Olivera BM. Proc Natl Acad Sci U S A. 2012;109(5):1388–1395. doi: 10.1073/pnas.1118833109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stevens CF. Curr Biol. 1998;8(20):R708–R710. doi: 10.1016/s0960-9822(98)70454-3. [DOI] [PubMed] [Google Scholar]
- 22.Padmanabhan K, Urban NN. Nat Neurosci. 2010;13(10):1276–1282. doi: 10.1038/nn.2630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Edlund T, Jessell TM. Cell. 1999;96(2):211–224. doi: 10.1016/s0092-8674(00)80561-9. [DOI] [PubMed] [Google Scholar]
- 24.Mitragotri S, Lahann J. Nat Mater. 2009;8(1):15–23. doi: 10.1038/nmat2344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tsang KY, Cheung MC, Chan D, Cheah KS. Cell Tissue Res. 2010;339(1):93–110. doi: 10.1007/s00441-009-0893-8. [DOI] [PubMed] [Google Scholar]
- 26.Guilak F, Cohen DM, Estes BT, Gimble JM, Liedtke W, Chen CS. Cell Stem Cell. 2009;5(1):17–26. doi: 10.1016/j.stem.2009.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hynes RO. Science. 2009;326(5957):1216–1219. doi: 10.1126/science.1176009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rozario T, DeSimone DW. Dev Biol. 2010;341(1):126–140. doi: 10.1016/j.ydbio.2009.10.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Piez KA. Matrix Biol. 1997;16(3):85–92. doi: 10.1016/s0945-053x(97)90037-8. [DOI] [PubMed] [Google Scholar]
- 30.Wickstrom SA, Radovanac K, Fassler R. Cold Spring Harb Perspect Biol. 2011;3(2) doi: 10.1101/cshperspect.a005116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tatulian SA. Biophys J. 2001;80(2):789–800. doi: 10.1016/S0006-3495(01)76058-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Leone DP, Relvas JB, Campos LS, Hemmi S, Brakebusch C, Fassler R, Ffrench-Constant C, Suter U. J Cell Sci. 2005;118(Pt 12):2589–2599. doi: 10.1242/jcs.02396. [DOI] [PubMed] [Google Scholar]
- 33.Flanagan LA, Rebaza LM, Derzic S, Schwartz PH, Monuki ES. J Neurosci Res. 2006;83(5):845–856. doi: 10.1002/jnr.20778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Campos LS. Bioessays. 2005;27(7):698–707. doi: 10.1002/bies.20256. [DOI] [PubMed] [Google Scholar]
- 35.Chen YC, Lee DC, Tsai TY, Hsiao CY, Liu JW, Kao CY, Lin HK, Chen HC, Palathinkal TJ, Pong WF, Tai NH, Lin IN, Chiu IM. Biomaterials. 2010;31(21):5575–5587. doi: 10.1016/j.biomaterials.2010.03.061. [DOI] [PubMed] [Google Scholar]
- 36.Coles EG, Gammill LS, Miner JH, Bronner-Fraser M. Dev Biol. 2006;289(1):218–228. doi: 10.1016/j.ydbio.2005.10.031. [DOI] [PubMed] [Google Scholar]
- 37.Nakajima M, Ishimuro T, Kato K, Ko IK, Hirata I, Arima Y, Iwata H. Biomaterials. 2007;28(6):1048–1060. doi: 10.1016/j.biomaterials.2006.10.004. [DOI] [PubMed] [Google Scholar]
- 38.Kim HN, Kang DH, Kim MS, Jiao A, Kim DH, Suh KY. Ann Biomed Eng. 2012;40(6):1339–1355. doi: 10.1007/s10439-012-0510-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Engler AJ, Sen S, Sweeney HL, Discher DE. Cell. 2006;126(4):677–689. doi: 10.1016/j.cell.2006.06.044. [DOI] [PubMed] [Google Scholar]
- 40.Yang F, Cho SW, Son SM, Hudson SP, Bogatyrev S, Keung L, Kohane DS, Langer R, Anderson DG. Biomacromolecules. 2010;11(8):1909–1914. doi: 10.1021/bm100357t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Du J, Chen X, Liang X, Zhang G, Xu J, He L, Zhan Q, Feng XQ, Chien S, Yang C. Proc Natl Acad Sci U S A. 2011;108(23):9466–9471. doi: 10.1073/pnas.1106467108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Keung AJ, de Juan-Pardo EM, Schaffer DV, Kumar S. Stem Cells. 2011;29(11):1886–1897. doi: 10.1002/stem.746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Caswell PT, Vadrevu S, Norman JC. Nat Rev Mol Cell Biol. 2009;10(12):843–853. doi: 10.1038/nrm2799. [DOI] [PubMed] [Google Scholar]
- 44.Loubet D, Dakowski C, Pietri M, Pradines E, Bernard S, Callebert J, Ardila-Osorio H, Mouillet-Richard S, Launay JM, Kellermann O, Schneider B. FASEB J. 2012;26(2):678–690. doi: 10.1096/fj.11-185579. [DOI] [PubMed] [Google Scholar]
- 45.Wang N, Tytell JD, Ingber DE. Nat Rev Mol Cell Biol. 2009;10(1):75–82. doi: 10.1038/nrm2594. [DOI] [PubMed] [Google Scholar]
- 46.Maniotis AJ, Chen CS, Ingber DE. Proc Natl Acad Sci U S A. 1997;94(3):849–854. doi: 10.1073/pnas.94.3.849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Itano N, Okamoto S, Zhang D, Lipton SA, Ruoslahti E. Proc Natl Acad Sci U S A. 2003;100(9):5181–5186. doi: 10.1073/pnas.0531397100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hoffman-Kim D, Mitchel JA, Bellamkonda RV. Annu Rev Biomed Eng. 2010;12:203–231. doi: 10.1146/annurev-bioeng-070909-105351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Norman JJ, Desai TA. Ann Biomed Eng. 2006;34(1):89–101. doi: 10.1007/s10439-005-9005-4. [DOI] [PubMed] [Google Scholar]
- 50.Nielson R, Kaehr B, Shear JB. Small. 2009;5(1):120–125. doi: 10.1002/smll.200801084. [DOI] [PubMed] [Google Scholar]
- 51.Bakota EL, Wang Y, Danesh FR, Hartgerink JD. Biomacromolecules. 2011;12(5):1651–1657. doi: 10.1021/bm200035r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Spivey EC, Khaing ZZ, Shear JB, Schmidt CE. Biomaterials. 2012;33(17):4264–4276. doi: 10.1016/j.biomaterials.2012.02.043. [DOI] [PubMed] [Google Scholar]
- 53.Cheng H, Byrska-Bishop M, Zhang CT, Kastrup CJ, Hwang NS, Tai AK, Lee WW, Xu X, Nahrendorf M, Langer R, Anderson DG. Biomaterials. 2012;33(20):5004–5012. doi: 10.1016/j.biomaterials.2012.03.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lutolf MP, Gilbert PM, Blau HM. Nature. 2009;462(7272):433–441. doi: 10.1038/nature08602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chew SY, Low WC. J Biomed Mater Res A. 2011;97(3):355–374. doi: 10.1002/jbm.a.33064. [DOI] [PubMed] [Google Scholar]
- 56.Khadka DB, Haynie DT. Nanomedicine. 2012 doi: 10.1016/j.nano.2012.02.013. in press. [DOI] [PubMed] [Google Scholar]
- 57.Sill TJ, von Recum HA. Biomaterials. 2008;29(13):1989–2006. doi: 10.1016/j.biomaterials.2008.01.011. [DOI] [PubMed] [Google Scholar]
- 58.Murugan R, Ramakrishna S. Tissue Eng. 2006;12(3):435–447. doi: 10.1089/ten.2006.12.435. [DOI] [PubMed] [Google Scholar]
- 59.Leach MK, Feng ZQ, Gertz CC, Tuck SJ, Regan TM, Naim Y, Vincent AM, Corey JM. J Vis Exp. 2011;(48):e2389. doi: 10.3791/2389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhang F, Zuo B, Fan Z, Xie Z, Lu Q, Zhang X, Kaplan DL. Biomacromolecules. 2012;13(3):798–804. doi: 10.1021/bm201719s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Christopherson GT, Song H, Mao HQ. Biomaterials. 2009;30(4):556–564. doi: 10.1016/j.biomaterials.2008.10.004. [DOI] [PubMed] [Google Scholar]
- 62.Yang F, Murugan R, Wang S, Ramakrishna S. Biomaterials. 2005;26(15):2603–2610. doi: 10.1016/j.biomaterials.2004.06.051. [DOI] [PubMed] [Google Scholar]
- 63.Xie J, Willerth SM, Li X, Macewan MR, Rader A, Sakiyama-Elbert SE, Xia Y. Biomaterials. 2009;30(3):354–362. doi: 10.1016/j.biomaterials.2008.09.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Corey JM, Lin DY, Mycek KB, Chen Q, Samuel S, Feldman EL, Martin DC. J Biomed Mater Res A. 2007;83(3):636–645. doi: 10.1002/jbm.a.31285. [DOI] [PubMed] [Google Scholar]
- 65.Thomas V, Jose MV, Chowdhury S, Sullivan JF, Dean DR, Vohra YK. J Biomater Sci Polym Ed. 2006;17(9):969–984. doi: 10.1163/156856206778366022. [DOI] [PubMed] [Google Scholar]
- 66.Gertz CC, Leach MK, Birrell LK, Martin DC, Feldman EL, Corey JM. Dev Neurobiol. 2010;70(8):589–603. doi: 10.1002/dneu.20792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Silva GA, Czeisler C, Niece KL, Beniash E, Harrington DA, Kessler JA, Stupp SI. Science. 2004;303(5662):1352–1355. doi: 10.1126/science.1093783. [DOI] [PubMed] [Google Scholar]
- 68.Carlberg B, Axell MZ, Nannmark U, Liu J, Kuhn HG. Biomed Mater. 2009;4(4):045004. doi: 10.1088/1748-6041/4/4/045004. [DOI] [PubMed] [Google Scholar]
- 69.Lee JY, Bashur CA, Gomez N, Goldstein AS, Schmidt CE. J Biomed Mater Res A. 2010;92(4):1398–1406. doi: 10.1002/jbm.a.32471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Adler AF, Speidel AT, Christoforou N, Kolind K, Foss M, Leong KW. Biomaterials. 2011;32(14):3611–3619. doi: 10.1016/j.biomaterials.2011.01.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Milner KR, Siedlecki CA. J Biomed Mater Res A. 2007;82(1):80–91. doi: 10.1002/jbm.a.31049. [DOI] [PubMed] [Google Scholar]
- 72.Lee MR, Kwon KW, Jung H, Kim HN, Suh KY, Kim K, Kim KS. Biomaterials. 2010;31(15):4360–4366. doi: 10.1016/j.biomaterials.2010.02.012. [DOI] [PubMed] [Google Scholar]
- 73.Yim EK, Pang SW, Leong KW. Exp Cell Res. 2007;313(9):1820–1829. doi: 10.1016/j.yexcr.2007.02.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Yim EK, Darling EM, Kulangara K, Guilak F, Leong KW. Biomaterials. 2010;31(6):1299–1306. doi: 10.1016/j.biomaterials.2009.10.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Gerecht S, Bettinger CJ, Zhang Z, Borenstein JT, Vunjak-Novakovic G, Langer R. Biomaterials. 2007;28(28):4068–4077. doi: 10.1016/j.biomaterials.2007.05.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Xia YN, Whitesides GM. Angew Chem Int Ed Engl. 1998;37:551–575. doi: 10.1002/(SICI)1521-3773(19980316)37:5<550::AID-ANIE550>3.0.CO;2-G. [DOI] [PubMed] [Google Scholar]
- 77.Migliorini E, Grenci G, Ban J, Pozzato A, Tormen M, Lazzarino M, Torre V, Ruaro ME. Biotechnol Bioeng. 2011;108(11):2736–2746. doi: 10.1002/bit.23232. [DOI] [PubMed] [Google Scholar]
- 78.Markert LD, Lovmand J, Foss M, Lauridsen RH, Lovmand M, Fuchtbauer EM, Fuchtbauer A, Wertz K, Besenbacher F, Pedersen FS, Duch M. Stem Cells Dev. 2009;18(9):1331–1342. doi: 10.1089/scd.2009.0114. [DOI] [PubMed] [Google Scholar]
- 79.Li YC, Lin YC, Young TH. Acta Biomater. 2012;8(8):3035–3048. doi: 10.1016/j.actbio.2012.04.036. [DOI] [PubMed] [Google Scholar]
- 80.Assender H, Bliznyuk V, Porfyrakis K. Science. 2002;297(5583):973–976. doi: 10.1126/science.1074955. [DOI] [PubMed] [Google Scholar]
- 81.Anderson DG, Levenberg S, Langer R. Nat Biotechnol. 2004;22(7):863–866. doi: 10.1038/nbt981. [DOI] [PubMed] [Google Scholar]
- 82.Unadkat HV, Hulsman M, Cornelissen K, Papenburg BJ, Truckenmuller RK, Post GF, Uetz M, Reinders MJ, Stamatialis D, van Blitterswijk CA, de Boer J. Proc Natl Acad Sci U S A. 2011;108(40):16565–16570. doi: 10.1073/pnas.1109861108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Peng R, Yao X, Ding J. Biomaterials. 2011;32(32):8048–8057. doi: 10.1016/j.biomaterials.2011.07.035. [DOI] [PubMed] [Google Scholar]
- 84.Gordan OD, Persson BN, Cesa CM, Mayer D, Hoffmann B, Dieluweit S, Merkel R. Langmuir. 2008;24(13):6636–6639. doi: 10.1021/la800728x. [DOI] [PubMed] [Google Scholar]
- 85.Johe KK, Hazel TG, Muller T, Dugich-Djordjevic MM, McKay RD. Genes Dev. 1996;10(24):3129–3140. doi: 10.1101/gad.10.24.3129. [DOI] [PubMed] [Google Scholar]
- 86.Horne MK, Nisbet DR, Forsythe JS, Parish CL. Stem Cells Dev. 2010;19(6):843–852. doi: 10.1089/scd.2009.0158. [DOI] [PubMed] [Google Scholar]
- 87.Madduri S, Papaloizos M, Gander B. Biomaterials. 2010;31(8):2323–2334. doi: 10.1016/j.biomaterials.2009.11.073. [DOI] [PubMed] [Google Scholar]
- 88.Patel S, Kurpinski K, Quigley R, Gao H, Hsiao BS, Poo MM, Li S. Nano Lett. 2007;7(7):2122–2128. doi: 10.1021/nl071182z. [DOI] [PubMed] [Google Scholar]
- 89.Yamada M, Tanemura K, Okada S, Iwanami A, Nakamura M, Mizuno H, Ozawa M, Ohyama-Goto R, Kitamura N, Kawano M, Tan-Takeuchi K, Ohtsuka C, Miyawaki A, Takashima A, Ogawa M, Toyama Y, Okano H, Kondo T. Stem Cells. 2007;25(3):562–570. doi: 10.1634/stemcells.2006-0011. [DOI] [PubMed] [Google Scholar]
- 90.Schmidt CE, Shastri VR, Vacanti JP, Langer R. Proc Natl Acad Sci U S A. 1997;94(17):8948–8953. doi: 10.1073/pnas.94.17.8948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Inagaki M, Yang Y, Kang F. Adv Mater. 2012;24(19):2547–2566. doi: 10.1002/adma.201104940. [DOI] [PubMed] [Google Scholar]
- 92.Kabiri M, Soleimani M, Shabani I, Futrega K, Ghaemi N, Ahvaz HH, Elahi E, Doran MR. Biotechnol Lett. 2012;34(7):1357–1365. doi: 10.1007/s10529-012-0889-4. [DOI] [PubMed] [Google Scholar]
- 93.Li M, Guo Y, Wei Y, MacDiarmid AG, Lelkes PI. Biomaterials. 2006;27(13):2705–2715. doi: 10.1016/j.biomaterials.2005.11.037. [DOI] [PubMed] [Google Scholar]
- 94.Lee JY, Bashur CA, Goldstein AS, Schmidt CE. Biomaterials. 2009;30(26):4325–4335. doi: 10.1016/j.biomaterials.2009.04.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Bolin MH, Svennersten K, Wang XJ, Chronakis IS, Richter-Dahlfors A, Jager EWH, Berggren M. Sensor Actuat B-Chem. 2009;142(2):451–456. [Google Scholar]
- 96.Prabhakaran MP, Nair AS, Kai D, Ramakrishna S. Biopolymers. 2012;97(7):529–538. doi: 10.1002/bip.22035. [DOI] [PubMed] [Google Scholar]
- 97.George PM, Lyckman AW, LaVan DA, Hegde A, Leung Y, Avasare R, Testa C, Alexander PM, Langer R, Sur M. Biomaterials. 2005;26(17):3511–3519. doi: 10.1016/j.biomaterials.2004.09.037. [DOI] [PubMed] [Google Scholar]
- 98.Jun I, Jeong S, Shin H. Biomaterials. 2009;30(11):2038–2047. doi: 10.1016/j.biomaterials.2008.12.063. [DOI] [PubMed] [Google Scholar]
- 99.Kane NM, McRae S, Denning C, Baker AH. Drug Discovery Today Technologies. 2008;5(4):e107–e115. [Google Scholar]
- 100.Lacoste A, Berenshteyn F, Brivanlou AH. Cell Stem Cell. 2009;5(3):332–342. doi: 10.1016/j.stem.2009.07.011. [DOI] [PubMed] [Google Scholar]
- 101.Marshall E. Science. 2000;288(5468):951–957. [PubMed] [Google Scholar]
- 102.Baum C, von Kalle C, Staal FJ, Li Z, Fehse B, Schmidt M, Weerkamp F, Karlsson S, Wagemaker G, Williams DA. Mol Ther. 2004;9(1):5–13. doi: 10.1016/j.ymthe.2003.10.013. [DOI] [PubMed] [Google Scholar]
- 103.Ivics Z, Hackett PB, Plasterk RH, Izsvak Z. Cell. 1997;91(4):501–510. doi: 10.1016/s0092-8674(00)80436-5. [DOI] [PubMed] [Google Scholar]
- 104.Bertrand N, Castro DS, Guillemot F. Nat Rev Neurosci. 2002;3(7):517–530. doi: 10.1038/nrn874. [DOI] [PubMed] [Google Scholar]
- 105.Sun Y, Nadal-Vicens M, Misono S, Lin MZ, Zubiaga A, Hua X, Fan G, Greenberg ME. Cell. 2001;104(3):365–376. doi: 10.1016/s0092-8674(01)00224-0. [DOI] [PubMed] [Google Scholar]
- 106.Ali F, Hindley C, McDowell G, Deibler R, Jones A, Kirschner M, Guillemot F, Philpott A. Development. 2011;138(19):4267–4277. doi: 10.1242/dev.067900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Guillemot F. Development. 2007;134(21):3771–3780. doi: 10.1242/dev.006379. [DOI] [PubMed] [Google Scholar]
- 108.Powell LM, Jarman AP. Curr Opin Genet Dev. 2008;18(5):411–417. doi: 10.1016/j.gde.2008.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Ma Q, Fode C, Guillemot F, Anderson DJ. Genes Dev. 1999;13(13):1717–1728. doi: 10.1101/gad.13.13.1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Ma Q, Chen Z, del Barco Barrantes I, de la Pompa JL, Anderson DJ. Neuron. 1998;20(3):469–482. doi: 10.1016/s0896-6273(00)80988-5. [DOI] [PubMed] [Google Scholar]
- 111.Bermingham NA, Hassan BA, Price SD, Vollrath MA, Ben-Arie N, Eatock RA, Bellen HJ, Lysakowski A, Zoghbi HY. Science. 1999;284(5421):1837–1841. doi: 10.1126/science.284.5421.1837. [DOI] [PubMed] [Google Scholar]
- 112.Kageyama R, Ohtsuka T, Shimojo H, Imayoshi I. Nat Neurosci. 2008;11(11):1247–1251. doi: 10.1038/nn.2208. [DOI] [PubMed] [Google Scholar]
- 113.Jeon SJ, Fujioka M, Kim SC, Edge AS. J Neurosci. 2011;31(23):8351–8358. doi: 10.1523/JNEUROSCI.6366-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Lee J, Kotov NA. Small. 2009;5(9):1008–1013. doi: 10.1002/smll.200801242. [DOI] [PubMed] [Google Scholar]
- 115.Beckstead BL, Santosa DM, Giachelli CM. J Biomed Mater Res A. 2006;79(1):94–103. doi: 10.1002/jbm.a.30760. [DOI] [PubMed] [Google Scholar]
- 116.zur Lage PI, Powell LM, Prentice DR, McLaughlin P, Jarman AP. Dev Cell. 2004;7(5):687–696. doi: 10.1016/j.devcel.2004.09.015. [DOI] [PubMed] [Google Scholar]
- 117.Putnam D. Nat Mater. 2006;5(6):439–451. doi: 10.1038/nmat1645. [DOI] [PubMed] [Google Scholar]
- 118.De Laporte L, Shea LD. Adv Drug Deliv Rev. 2007;59(4–5):292–307. doi: 10.1016/j.addr.2007.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Houchin-Ray T, Swift LA, Jang JH, Shea LD. Biomaterials. 2007;28(16):2603–2611. doi: 10.1016/j.biomaterials.2007.01.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Houchin-Ray T, Whittlesey KJ, Shea LD. Mol Ther. 2007;15(4):705–712. doi: 10.1038/mt.sj.6300106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Mehrotra S, Lee I, Chan C. Acta Biomater. 2009;5(5):1474–1488. doi: 10.1016/j.actbio.2009.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Shen H, Tan J, Saltzman WM. Nat Mater. 2004;3(8):569–574. doi: 10.1038/nmat1179. [DOI] [PubMed] [Google Scholar]
- 123.Shea LD, Smiley E, Bonadio J, Mooney DJ. Nat Biotechnol. 1999;17(6):551–554. doi: 10.1038/9853. [DOI] [PubMed] [Google Scholar]
- 124.Zhang J, Chua LS, Lynn DM. Langmuir. 2004;20(19):8015–8021. doi: 10.1021/la048888i. [DOI] [PubMed] [Google Scholar]
- 125.Bengali Z, Pannier AK, Segura T, Anderson BC, Jang JH, Mustoe TA, Shea LD. Biotechnol Bioeng. 2005;90(3):290–302. doi: 10.1002/bit.20393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Andersen MO, Lichawska A, Arpanaei A, Rask Jensen SM, Kaur H, Oupicky D, Besenbacher F, Kingshott P, Kjems J, Howard KA. Biomaterials. 2010;31(21):5671–5677. doi: 10.1016/j.biomaterials.2010.03.069. [DOI] [PubMed] [Google Scholar]
- 127.Zoldan J, Lytton-Jean AK, Karagiannis ED, Deiorio-Haggar K, Bellan LM, Langer R, Anderson DG. Biomaterials. 2011;32(31):7793–7800. doi: 10.1016/j.biomaterials.2011.06.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Kong HJ, Liu J, Riddle K, Matsumoto T, Leach K, Mooney DJ. Nat Mater. 2005;4(6):460–464. doi: 10.1038/nmat1392. [DOI] [PubMed] [Google Scholar]
- 129.Kong HJ, Hsiong S, Mooney DJ. Nano Lett. 2007;7(1):161–166. doi: 10.1021/nl062485g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Teo BK, Goh SH, Kustandi TS, Loh WW, Low HY, Yim EK. Biomaterials. 2011;32(36):9866–9875. doi: 10.1016/j.biomaterials.2011.08.088. [DOI] [PubMed] [Google Scholar]
- 131.Wickstrom SA, Fassler R. Trends Cell Biol. 2011;21(5):266–273. doi: 10.1016/j.tcb.2011.02.003. [DOI] [PubMed] [Google Scholar]
- 132.You MH, Kwak MK, Kim DH, Kim K, Levchenko A, Kim DY, Suh KY. Biomacromolecules. 2010;11(7):1856–1862. doi: 10.1021/bm100374n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Kaur G, Wang C, Sun J, Wang Q. Biomaterials. 2010;31(22):5813–5824. doi: 10.1016/j.biomaterials.2010.04.017. [DOI] [PubMed] [Google Scholar]
- 134.Velkey JM. Lineage differentiation of embryonic stem cells. 2005 p 1 v. [Google Scholar]
- 135.Reyes JH, O'Shea KS, Wys NL, Velkey JM, Prieskorn DM, Wesolowski K, Miller JM, Altschuler RA. J Neurosci. 2008;28(48):12622–12631. doi: 10.1523/JNEUROSCI.0563-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Leach MK, Naim YI, Feng ZQ, Gertz CC, Corey JM. J Neurosci Methods. 2011;199(2):192–198. doi: 10.1016/j.jneumeth.2011.04.033. [DOI] [PubMed] [Google Scholar]
- 137.Cooke MJ, Vulic K, Shoichet MS. Soft Matter. 2010;6(20):4988–4998. [Google Scholar]
- 138.Lim SH, Liu XY, Song H, Yarema KJ, Mao HQ. Biomaterials. 2010;31(34):9031–9039. doi: 10.1016/j.biomaterials.2010.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Seo S, Lim JW, Yellajoshyula D, Chang LW, Kroll KL. EMBO J. 2007;26(24):5093–5108. doi: 10.1038/sj.emboj.7601923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Hurd PJ, Nelson CJ. Brief Funct Genomic Proteomic. 2009;8(3):174–183. doi: 10.1093/bfgp/elp013. [DOI] [PubMed] [Google Scholar]
- 141.Meaburn E, Schulz R. Semin Cell Dev Biol. 2012;23(2):192–199. doi: 10.1016/j.semcdb.2011.10.010. [DOI] [PubMed] [Google Scholar]
- 142.Cullum R, Alder O, Hoodless PA. Respirology. 2011;16(2):210–222. doi: 10.1111/j.1440-1843.2010.01899.x. [DOI] [PubMed] [Google Scholar]
- 143.Ghazalpour A, Bennett B, Petyuk VA, Orozco L, Hagopian R, Mungrue IN, Farber CR, Sinsheimer J, Kang HM, Furlotte N, Park CC, Wen PZ, Brewer H, Weitz K, Pan C, Yordanova R, Neuhaus I, Tilford C, Siemers N, Gargalovic P, Eskin E, Kirchgessner T, Smith DJ, Smith RD, Lusis AJ. PLoS Genet. 2011;7(6):e1001393. doi: 10.1371/journal.pgen.1001393. [DOI] [PMC free article] [PubMed] [Google Scholar]