Abstract
The modulation of vascular smooth muscle cell (VSMC) phenotype is an essential element to fabricate engineered conduits of clinical relevance. In vivo, owing to their close proximity, endothelial cells (ECs) play a role in VSMC phenotype switching. Although considerable progress has been made in vascular tissue engineering, significant knowledge gaps exist on how the contractile VSMC phenotype is induced at the conclusion of the tissue fabrication process. The objectives of this study were as follows: (1) to establish ligand presentation modes on transcriptional activation of VSMC-specific genes, (2) to develop a three-dimensional (3D) coculture model using human coronary artery smooth muscle cells (HCASMCs) and human coronary artery endothelial cells (HCAECs) on porous synthetic scaffolds and, (3) to investigate EC-mediated Notch signaling in 3D cultures and the induction of the HCASMC contractile phenotype. Whereas transcriptional activation of VSMC-specific genes was not induced by presenting soluble Jagged1 and Jagged1 bound to protein G beads, a direct link between HCAEC-bound Jagged1 and HCASMC differentiation genes was observed. Our 3D culture results showed that HCASMCs seeded to scaffolds and cultured for up to 16 days readily attached, infiltrated the scaffold, proliferated, and formed dense confluent layers. HCAECs, seeded on top of an HCASMC layer, formed a distinct, separate monolayer with cell-type partitioning, suggesting that HCAEC growth was contact inhibited. While we observed EC monolayer formation with 200,000 HCAECs/scaffold, seeding 400,000 HCAECs/scaffold revealed the formation of cord-like structures akin to angiogenesis. Western blot analyses showed that 3D coculture induced an upregulation of Notch3 receptor in HCASMCs and its ligand Jagged1 in HCAECs. This was accompanied by a corresponding induction of the contractile HCASMC phenotype as demonstrated by increased expression of smooth muscle-α-actin (SM-α-actin) and calponin. Knockdown of Jagged1 with siRNA showed a reduction in SM-α-actin and calponin in cocultures, identifying a link between Jagged1 and the expression of contractile proteins in 3D cocultures. We therefore conclude that the Notch3 signaling pathway is an important regulator of VSMC phenotype and could be targeted when fabricating engineered vascular tissues.
Introduction
Despite the urgent need for a tissue-engineered vascular substitute, several unresolved challenges such as the regulation of the vascular smooth muscle cell (VSMC) phenotype in three-dimensional (3D) cultures hinder progress.1,2 VSMCs are known to exhibit remarkable phenotypic plasticity at different developmental stages and in response to changing local environmental cues.2–4 During vasculogenesis, VSMCs are in a synthetic phenotype characterized by high proliferative and migratory indices and lay down an abundance of extracellular matrix (ECM) components, such as collagen and elastin.3 On the other hand, VSMCs in mature vessels are largely in a contractile phenotype with a principal function of contraction. The distinctive phenotypes stated just now are, however, not limited to developmental events and to the matured vasculature. VSMC phenotype plasticity is also observed during vascular disease, injury, and repair, such as atherosclerosis and postangioplasty restenosis.3 After vascular injury and in response to changes in local environmental cues, VSMCs rapidly undergo phenotypic modulation, which results in suppression of genes that define the contractile phenotype, while upregulating genes required for proliferation and migration to lay down a new ECM and repair the vessel wall. This response is required for vascular repair and is beneficial,4 if cells revert back to the more contractile phenotype once the injury has been stabilized by reinducing VSMC contractile differentiation marker genes. Unimpeded, however, this response could be detrimental due to neointimal hyperplasia and luminal narrowing that is life threatening.4 The synthetic phenotype of mature SMCs is somewhat akin to the embryonic and fetal vascular smooth muscle phenotype.5,6
The phenotype shifting property of VSMCs is considered to be of paramount importance in the engineering of a functional vasculature.7,8 Initially, the synthetic phenotype is desirable to enable cells to expand rapidly in culture. This would, in turn, promote infiltration and population of scaffolds by VSMCs to create dense cellular layers. This proliferation phase would also result in the synthesis and secretion of appropriate ECM components that provide the tissue-engineered vessel with sufficient mechanical properties, and a substrate for endothelial cell (EC) adhesion. Once the desired population of cells and ECM components have been put in place, VSMCs must then shift to a quiescent contractile phenotype to impart the contractile property found in native blood vessels. Failure of the VSMCs to achieve this shift would, upon implantation of the engineered tissue, result in the onset of intimal hyperplasia and restenosis and, could lead to graft failure. In view of this, induction of a contractile VSMC phenotype after tissue fabrication is imperative. Although there are a number of reported strategies to induce the contractile VSMC phenotype following culture,7,9 none are as relevant as using ECs.10 In an intact vasculature, ECs reside in close proximity to SMCs and play a crucial role by interacting with the underlying SMCs through direct cell–cell contact or through the synthesis and release of mediators into the surrounding medium.11 Despite early reports that suggest that ECs promote the contractile phenotype of VSMCs,12–15 some of the molecular mechanisms, such EC activation of VSMC PI 3-kinase/Akt pathway16 and protein kinase A pathway via prostacyclin receptor,17 only started to emerge mainly from two-dimensional (2D) coculture experiments.
One additional pathway that has recently been shown to be an important regulator of EC-induced contractile VSMC phenotype switching during development is Notch signaling.18 This pathway is evolutionarily conserved and dictates cell fate through the control of proliferation, differentiation, and apoptosis.18,19 Components of the Notch signaling pathway include the Notch family of transmembrane receptors (Notch1–4), the Delta, Serrate/Jagged, Lag-2 family of transmembrane ligands (Delta like1, 3, and 4 and Jagged1 and 2), and the various effectors from the hairy enhancer of split (HES) and HES-related repressor protein (HERP) family.19,20 Tissue distribution of Notch signaling components varies significantly; however, several of these are confined to the vasculature, including the ligands Delta like4; Jagged1 and 2; the receptors Notch1, 3, and 4; and the effectors HERP1, 2, and 3.19 Interaction of the extracellular domain of the Notch receptors with their ligands on neighboring cells leads to proteolytic cleavage of the receptor thereby freeing the Notch intracellular domain (ICD). Once the ICD translocates to the nucleus, it associates with C-promoter-binding factor-1 to form a multiprotein complex that initiates DNA transcription of the Notch effector genes (HES and HERP).21 In VSMCs in particular, Notch activity regulates cell differentiation, proliferation, migration, and survival.20,22,23 Notch3 is the primary receptor that is expressed by VSMCs and its ligand Jagged1 is predominantly expressed by vascular ECs.19,24,25 Notwithstanding a number of in-vivo-gene-knockout experiments that demonstrate the role of EC-mediated Notch signaling during vascular development and maturity,26–28 it is unknown whether Notch signaling is activated in vitro in 3D cultures to induce the VSMC contractile phenotype.29 The objectives of the current work were, therefore, threefold: (1) to establish ligand presentation modes on the transcriptional activation of VSMC-specific genes, (2) to develop a 3D coculture model using human coronary artery smooth muscle cell (HCASMCs) and human coronary artery endothelial cell (HCAECs) on porous synthetic scaffolds, and (3) to investigate EC-mediated Notch signaling pathway in 3D cultures and induction of HCASMC contractile phenotype.
Materials and Methods
Scaffold fabrication and preparation for cell culture
Three-dimensional scaffolds were fabricated from poly(carbonate urethane) (PCU; Bionate® 55D; DSM Biomedical) using a pressure differential solvent casting and particulate leaching method as established in our laboratory.30 Briefly, ground NH4Cl porogen particles (180–210 μm) were packed into the cylindrical glass tube and compressed using air pressure to achieve high packing density and uniformity. About 20% (w/v) PCU solution in dimethylformamide was poured over the porogen bed and infiltrated by the application of a pressure gradient. Following solvent evaporation, the porogen particles were leached out using water and the scaffolds were dried and sectioned into 0.5-mm-thick discs using a rotary blade prior to use in cell culture studies. Scaffold morphology was visualized using a scanning electron microscope (S-2600N; Hitachi).
Jagged1/Fc protein immobilization to protein G Dynabeads
Protein G Dynabeads were washed three times with phosphate-buffered saline (PBS; pH 7.4, 0.02% Tween) and mixed with 5 μg of human Jagged1/Fc chimera protein (R&D Systems) in the original bead volume. The mixture was incubated for 10 min under rotation at room temperature and the Jagged1-immobilized beads were washed three times with PBS. As a control for Jagged1/Fc chimeric protein, Protein G beads were incubated with human immunoglobulin G (IgG) solution (5 μg/mL) at the same conditions. This control addresses the effect of the Fc fragment of Jagged1 for any possible nonspecific effects. Beads were added to cell cultures at a concentration of 3.5×105 beads per well corresponding to 200 beads/cell at a seeding density of 1.7×104 cells per well.
Mono- and cocultures of cells
Primary HCASMCs and primary HCAECs purchased from Lonza Walkersville, Inc., were cultured in smooth muscle growth media (SmGM; SmGM®-2 BulletKit) and endothelial cell growth media (EGM; EGM®-2 Bullet Kit), respectively, according to the supplier's instruction. Both media were supplemented with 100 U/mL penicillin G and 100 μg/mL streptomycin sulfate. Cell cultures were maintained in a humidified incubator at 5% CO2 and 37°C and were used between passages 5 and 9. For 2D cell culture studies, HCASMCs were seeded at a density of 1.7×104 cells/well and cultured for 48 h with the addition of the following: 5 μg/mL of soluble Jagged1 protein or IgG protein (Invitrogen), 3.5×105 Dynabeads (Invitrogen), and IgG or Jagged1-immobilized 3.5×105 Dynabeads. HCASMCs cultured alone served as controls. For cocultures of smooth muscle and ECs, HCASMCs were seeded at a density of 1.7×104 cells/well and cultured for 48 h in SmGM. Equal number of HCAECs were then seeded over the HCASMC layer and cultured for an additional 48 h in coculture media (one part EGM and one part SmGM) determined in screening experiments. For 3D cultures, HCASMCs were seeded onto the scaffolds at varying initial densities depending on the experiment and allowed to attach in a 37°C and 5% CO2 incubator for 3 h and cultured in a 24-well culture plate with 2 mL of SmGM for prescribed times. For 3D cocultures, varying numbers of HCAECs were seeded onto scaffolds containing HCASMCs and cultured for an additional 48 h in the presence of 1:1 EGM/SmGM.
Transfection of HCAECs with Jagged1 siRNA
Prior to transfection, HCAECs were passaged in antibiotic-free growth media such that they would be at 50% confluence at the time of transfection. Two hundred picomoles of human Jagged1 siRNA or scrambled control nontargeting siRNA (ON-TARGETplus; Thermo Scientific Dharmacon®) was diluted in 1 mL of Opti-MEM reduced serum medium. Each of these solutions was then mixed with another 1 mL of Opti-MEM reduced serum medium containing 20 μL of Lipofectamine™ RNAiMAX. Solutions were incubated at room temperature for 20 min and added to a culture dish with 50% confluent HCAECs. Following culture for 24 h, HCAECs were trypsinized and transferred to scaffolds that had been previously seeded with HCASMCs and cultured. The cocultures were maintained for 48 h before cell harvesting and lysis to test the transfection efficiency and protein expression levels.
Separation of HCAECs from coculture
To examine target protein expression in response to coculture in each cell type separately, anti-PECAM conjugated Dynabeads (Invitrogen; 25 μL corresponding to 107 beads for 105 HCAECs) were employed to separate the HCAECs from the HCASMCs. First, cells were recovered from scaffolds or culture plates by incubating in a 0.25% Trypsin/ethylenediaminetetraaceticacid (EDTA) solution at 37°C for 5 min. This method has proven effective in the past for cell recovery from PCU scaffolds.31 Scaffolds or culture plates were then rinsed several times with a low serum-content buffer (5% fetal bovine serum in 1×PBS) to neutralize the trypsin activity. The trypsinized cell suspension was centrifuged for 5 min at room temperature and the pellet was resuspended in 0.1% bovine serum albumin (BSA)/PBS. Washed anti-PECAM conjugated Dynabeads were mixed with the cell suspension and rotated at 4°C for 20 min to facilitate attachment to HCAECs. Following incubation, samples were placed in a magnet to separate the bead-bound HCAECs from the supernatant (i.e., HCASMCs). The supernatant was collected and the bead-bound HCAECs were rinsed and magnetized three times to increase the separation efficiency.
RNA isolation and quantitative real-time polymerase chain reaction analysis
Real-time polymerase chain reaction (PCR) combined with reverse transcription was used to quantify messenger RNA of Notch3, smooth muscle-α-actin (SM-α-actin), and calponin in HCASMCs. Total RNA from HCASMCs was isolated using TRIzol® reagent following the manufacturer's protocol. Complementary DNA was synthesized using 1 μg of total RNA primed with oligo(dT)12–18 as described in SuperScript™. Conventional reverse transcription PCR was used to test primer specificity by running PCR for 40 cycles at 95°C for 20 s and 52°C for 1 min. Quantitative real-time PCR was conducted in 10 μL reaction volumes, using a Chromo4 Real-time Thermal Cycler, and gene expression of human Notch3, SM-α-actin, calponin, and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was then determined with iQ™ SYBR® Green Supermix according to the recommended protocol of the manufacturer. Notch3 forward primer, 5′-CCT AGA CCT GGT GGA CAA G-3′, and reverse primer, 5′-ACA CAG TCG TAG CGG TTG-3′; SM-α-actin forward primer, 5′-CAA GTG ATC ACC ATC GGA AAT G-3′, and reverse primer, 5′-GAC TCC ATC CCG ATG AAG GA-3′; calponin forward primer, 5′-TGA AGC CCC ACG ACA TTT TT-3′, and reverse primer, 5′-GGG TGG ACT GCA CCT GTG TA-3′; and GAPDH forward primer, GGT GGT CTC CTC TGA CTT CAA CA, and reverse primer, GTT GCT GTA GCC AAA TTC GTT GT, were used.32 Cycling parameters were optimized as follows: denaturation 95°C (10 s), gradient annealing 50°C/65°C (10 s), extension 72°C (30 s), and running for 39 cycles. Notch3, SM-α-actin, and calponin gene expression in HCASMCs was normalized to GAPDH with at least three repeats per experimental group and expressed as relative ratios using the Gene Expression Macro analysis software.
Immunofluorescence staining and laser scanning confocal microscopy
For some 3D coculture experiments, HCASMCs and HCAECs were live-stained using CellTracker™ green (CTG) and CellTracker™ red (CTR), respectively, at a concentration of 10 μM in serum-free media for 45 min. Following two 30-min incubations in SmGM or EGM to rinse out the unincorporated CTG and CTR, respectively, the respective cells were seeded and cultured for a prescribed time. Cells were then fixed using a 4% solution of paraformaldehyde, permeabilized in 0.5% Triton X-100, and washed three times with 1× PBS. Cocultured cell–scaffold constructs were incubated in 1% BSA/PBS with vascular endothelial (VE)-cadherin antibody (1:50 dilution; Santa Cruz, Inc.) for 1 h followed by three washes in 1× PBS. Scaffolds were then incubated in Alexa Fluor® 568-conjugated secondary antibody for 1 h, washed three times with 1× PBS, and incubated with Hoechst 33342 (10 μg/mL; Sigma-Aldrich) for 5 min to label the nuclei. Samples were mounted on glass microscope slides in a mounting medium composed of glycerol and water (glycerol:water, 9:1 v/v) and sealed using nail enamel. A Zeiss LSM 410 confocal microscope (Carl Zeiss) equipped with argon/neon and UV lasers was used for imaging the samples. Images were captured by taking serial optical slices at regular increments through the sample depths. Where applicable, long-term HCASMC monocultures were stained using Alexa Fluor® 488 Phalloidin (1:50 dilution) in 1% BSA/PBS for 1 h.
Protein extraction and western blot
Expression levels of Jagged1, Notch3, SM-α-actin, and calponin in 3D cultures were evaluated using western blotting. Scaffolds were incubated in 0.25% Trypsin/EDTA for 5 min at 37°C. The trypsin solution was pipetted repeatedly into the scaffolds to recover cells after which the cells were centrifuged and collected. Cells from cocultures were separated using anti-PECAM conjugated Dynabeads (as described in the “Separation of HCAECs from coculture” section) before they are recovered using elution buffer and lysed. Lysates were microcentrifuged and total protein concentrations were determined using 660 nm colorimetric protein assay (Pierce™ 660-nm Protein Assay Kit) according to the manufacturer's instructions. Twenty micrograms per well of protein was loaded and separated by 10% sodium dodecyl sulfate–polyacrylamide gel electrophoresis for 50 min and then subsequently transferred onto a nitrocellulose membrane. Ponceau S stain was used to verify proper transfer. Membranes were then blocked with 5% nonfat dry milk in 1× PBS for 1 h and incubated in primary antibodies (diluted in 5% nonfat dry milk in 1× PBS): anti-SM α-actin (1:1000 dilution), anti-calponin (1:1000 dilution), anti-Jagged1 (1:200 dilution), anti-Notch3 (1:200 dilution), and anti-GAPDH (1:2000 dilution) all from Santa Cruz, Inc., for 2 h. After incubation with horseradish peroxidase-conjugated secondary antibodies for 45 min, membranes were incubated for 5 min in SuperSignal®West Pico Chemiluminescent substrate. Bio-Rad's ChemiDoc™ XRS+System was used to image the membranes and blots were quantified using Image Lab™ software.
Statistical analysis
Quantified data for RNA and protein expression levels were plotted and analyzed using GraphPad Prism 5. Values were normalized against GAPDH and are presented as mean±standard deviation from at least three independent experiments. Statistical analyses were conducted by one-way analysis of variance followed by Tukey's post hoc test to compare differences between two groups. Values of p<0.05 were considered to be statistically significant.
Results
Jagged1-induced Notch3, SM-α-actin, and calponin gene expression levels in HCASMCs
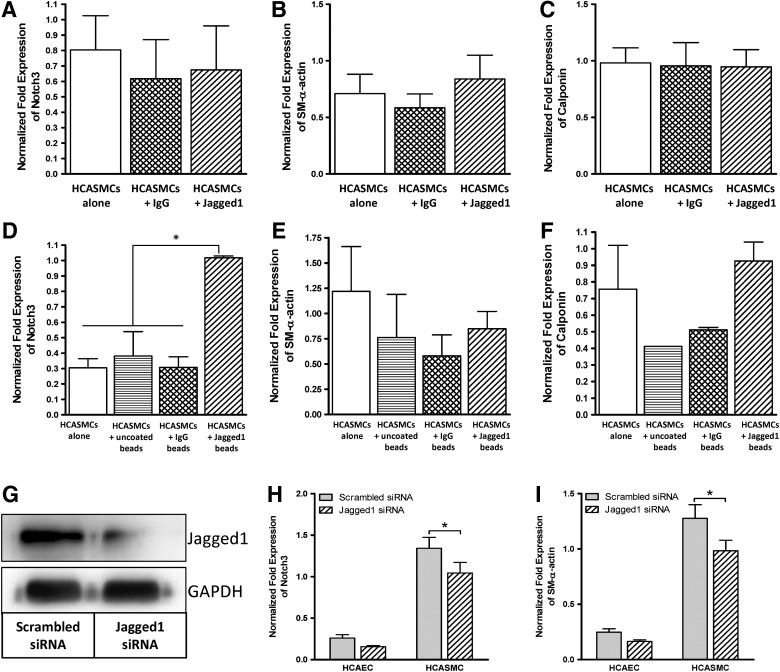
Although Notch3-dependent protein synthesis in HCASMCs has been reported previously in 2D cultures,33 it was not clear whether the differential protein synthesis was the result of transcriptional repression/activation or mRNA stability/instability. Therefore, one of the objectives of this study was to examine Notch-induced differential SM-α-actin and calponin gene expression of HCASMCs in response to Jagged1 presentation strategies using quantitative real time (RT)-PCR. For our targeted gene analysis, Notch3 was chosen because it is the primary receptor for Jagged1 in arterial SMCs. SM-α-actin and calponin were targeted as these are early-to-mid stage SMC differentiation markers34,35 and direct targets for Notch activity.21,32,36 Quantitative RT-PCR (Fig. 1A–C) indicated that soluble Jagged1 was unable to produce any significant changes of Notch transcriptional activity and HCASMC marker gene expression compared with the controls in which HCASMCs were cultured either in SmGM or in IgG-supplemented SmGM (p>0.05), suggesting that soluble Jagged1 does not stimulate Notch3 at the transcriptional level. The IgG control accounts for any nonspecific effects of the Fc-fragment found on the Jagged1 protein. Since Notch3 signaling is generally thought to be paracrine, we conjugated Jagged1 on the surface of protein G beads such that the protein G beads could potentially mimic a signaling cell surface presenting the Jagged1 ligand to adjacent HCASMCs. Results shown in Figure 1D–F demonstrated that, although Jagged-immobilized beads were able to significantly upregulate Notch3 gene expression in HCASMCs (p<0.05), a corresponding increase in contractile gene expression was not detected (p>0.05), suggesting the possibility of posttranscriptional mRNA transcript instability.37 This also suggests that other biochemical or structural cues—which are not offered by the Jagged1-immobilized beads—may be necessary to activate the signaling cascade that results in HCASMC differentiation. Given that both soluble and bead-bound Jagged1 failed to upregulate SM-α-actin and calponin that are downstream targets of Notch3 signaling, we tested the possibility that HCAEC-expressed Jagged1 may be required. To do this, we used a combination of direct coculture system and human Jagged1 siRNA. As shown in Figure 1G, Jagged1 in HCAECs was effectively knocked down by siRNA compared with the control scrambled siRNA. As a result of Jagged1 knockdown in HCAECs, both Notch3 and SM-α-actin genes were significantly downregulated in HCASMCs compared with the scrambled control siRNA (Fig. 1H, I; p<0.05), demonstrating a direct link between HCAEC-bound Jagged1 and HCASMC differentiation. Unlike bead-immobilized Jagged1, which was able to upregulate the expression of Notch3 in HCASMCs but failed to upregulate contractile marker gene expression, HCAEC-bound Jagged1 appears to have a direct effect on the expression of Notch3 and SM-α-actin in HCASMCs. The data collectively presented in Figure 1 for 2D cultures underscore the need for a direct coculture system for studying the modulation of HCASMC phenotype.
FIG. 1.
Human coronary artery endothelial cell (HCAEC)–bound Jagged1 in a direct coculture of human coronary artery smooth muscle cells (HCASMCs) and HCAECs upregulates mRNA expression of Notch3 and modulates phenotypic marker genes in HCASMCs. HCASMCs were seeded on two-dimensional (2D) culture dishes and cultured for 4 days in smooth muscle growth media (SmGM) with or without soluble Jagged1 (A–C), or in SmGM supplemented with or without Jagged1-immobilized beads (D–F). Quantitative real-time polymerase chain reaction (PCR) analysis revealed that soluble Jagged1 has no effect on Notch3 transcriptional activity and smooth muscle cell (SMC) contractile marker genes. Conjugated Jagged1 beads were able to upregulate the expression of Notch3 but failed to upregulate SMC contractile marker genes. (G) HCAECs were transiently transfected with Jagged1 siRNA or scrambled siRNA (as a control) for 2 days prior to coculture with HCASMCs on 2D surfaces. Western blot analysis revealed that transfection was successful and the corresponding Jagged1 expression in HCAECs was significantly reduced. (H, I) Following 2 days of HCASMC and HCAEC coculture, quantitative real-time PCR analysis revealed that direct coculture of HCASMCs and HCAECs significantly downregulates when Jagged1 was knocked down in HCAECs using siRNA. Data are represented as mean±standard deviation (SD) from three independent experiments (*p<0.05).
The effect of porous and interconnected 3D scaffolds on cocultured HCASMCs and HCAECs
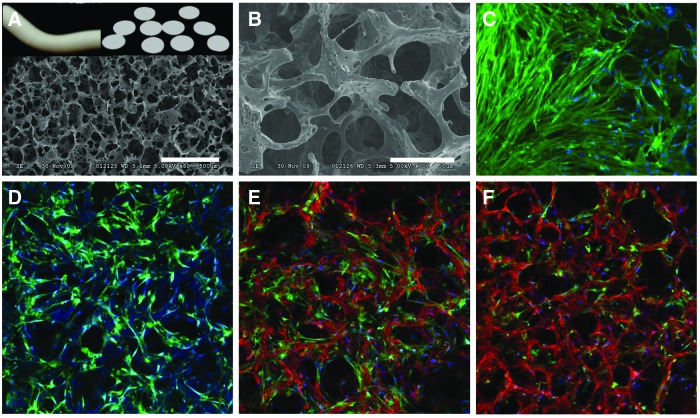
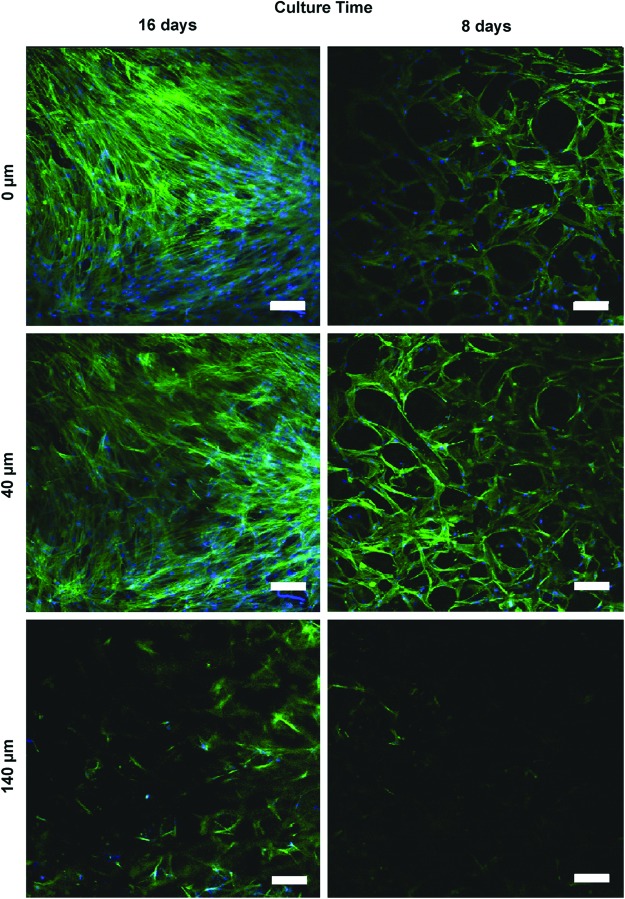
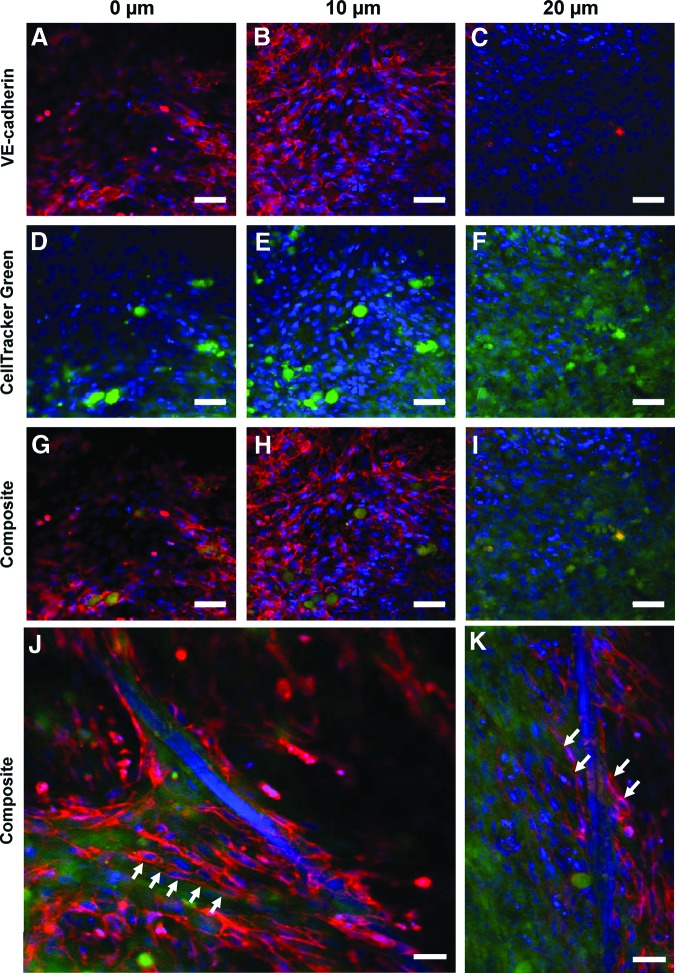
To evaluate Notch-induced HCASMC marker protein expression mediated by HCAECs, we established a 3D coculture system. This was accomplished by fabricating 3D PCU scaffolds that are highly porous with interconnected channels between adjacent pores (Fig. 2A, B). In addition to the macropores (∼180 μm), micropores (∼10 μm) are also evident (Fig. 2B) on the scaffold struts that could potentially alter surface topography and also facilitate mass transport within the construct thus enhancing intercellular interactions.38,39 HCASMCs seeded at 750,000 cells/scaffold and cultured for 16 days were densely packed and expressed abundant and aligned F-actin that covered the scaffold surface (Fig. 2C). To gain insight into the interaction of HCAECs with HCASMCs in 3D cultures, HCASMCs were first seeded onto the scaffolds at a density of 10,000 cells/scaffold and were cultured for 14 days. The initial low seeding density was chosen since it may allow visualization of how HCAECs interact with the less-crowded CTG-stained HCASMCs cultured on 3D scaffolds (Fig. 2D). After the 14 days of culture, 6000 HCAECs labeled with CTR were seeded onto the HCASMC layer. Both cell types were cocultured on the scaffolds for 24 and 48 h. As shown in Figure 2E and F, the HCAECs were located on top of the underlying HCASMC layer. By 48 h, the relative number of CTR-positive cells encountered increased compared with 24 h, suggesting the proliferation of the ECs. Although HCASMCs were densely packed (Fig. 2C) it was important to ascertain whether these cells were actively migrating into the cross-section of the 3D scaffolds with increasing culture time. Serial confocal images of HCASMCs on the scaffolds (Fig. 3) revealed that cells were, in fact, both migrating and proliferating as the culture time increased. Contrary to the need for active migration and proliferation of HCASMCs on the 3D scaffold cross-section, ECs should form only a monolayer when seeded on the SMC layer. This is important when mimicking vascular physiology since ECs only form monolayers with contact-inhibited growth characteristics. Serial confocal analysis (Fig. 4) revealed cell-type partitioning where the top 10 μm of the scaffold was a pure HCAEC population whereas at depths beyond 10 μm pure SMC populations were evident, suggesting that the ECs were forming a monolayer. Since the whole-cell height for an EC is 10–11 μm,40 our data suggest that our 3D coculture model is feasible in forming an EC monolayer. It is also interesting to point out that the seeding density of ECs appeared to make a considerable difference on EC morphogenesis. While we were able to observe EC monolayer formation with 200,000 HCAECs/scaffold (Fig. 4A, B, G, H), scaffolds seeded with 400,000 HCAECs/scaffold revealed morphogenesis of the HCAECs into cord-like structures (Fig. 4J, K).
FIG. 2.
Coculture of HCASMCs and HCAECs on porous and interconnected three-dimensional (3D) scaffolds. (A) Images (top) of rod-like and sectioned polyurethane scaffold discs (0.5 mm), and scanning electron micrograph (SEM) images (bottom) of porous scaffolds using NH4Cl (180–210 μm). Scale bar=500 μm. (B) SEM of porous 3D polyurethane scaffolds under higher magnification. Scale bar=50 μm. HCASMCs were seeded into 3D porous scaffolds and cultured for 16 days. Confocal microscopy after staining for F-actin [(C), green; magnification=40×] showed that HCASMCs were densely packed and covered the scaffold surface. Nuclei are labeled blue. (D) HCASMCs were sparsely seeded into 3D scaffold and were cultured for 14 days prior to coculture. Confocal microscopy showed that HCASMCs were labeled by CellTracker Green. Following CellTracker Red-labeled HCAEC seeding and cocultured for 24 h (E) and 48 h (F), the cells were imaged. Nuclei are labeled blue. More red-stained cells were encountered by 48 h compared with 24 h, suggesting proliferation of the endothelial cells. Magnification=40×. Color images available online at www.liebertpub.com/tea
FIG. 3.
HCASMC-infiltrated 3D polycarbonate urethane (PCU) scaffolds in long-term cultures. HCASMCs were seeded on 3D porous scaffolds and were cultured for 8 and 16 days. Images by confocal microscopy taken at different focal planes after staining for F-actin (green) showed that cells were both migrating and proliferating as the culture time increased. Nuclei are labeled blue. Scale bar=100 μm. Color images available online at www.liebertpub.com/tea
FIG. 4.
In 3D coculture, HCAECs organize into a monolayer on top of underlying HCASMCs. HCASMCs (750,000 cells/scaffolds) were cultured on 3D scaffolds for 14 days prior to coculture for additional 2 days with 200,000 HCAECs. Confocal microscopy images taken at different focal planes show HCASMCs and HCAECs stained with a cytoplasmic stain (CellTracker Green) and VE-cadherin (red) antibody, respectively. Images are shown split into their component red (A–C) and green (D–F) channels as well as composite images (G–I). HCAECs were only found within the top 10 μm of the scaffold surface (A, B, G, H) whereas HCASMCs could be seen at lower levels (F, I), suggesting the formation of two distinct cell layers. When 400,000 HCAECs were seeded on top of the smooth muscle cells, morphogenesis of the cell monolayers into cord-like structures akin to capillaries (arrow indicated) was observed (J, K). Scale bar=20 μm. Color images available online at www.liebertpub.com/tea
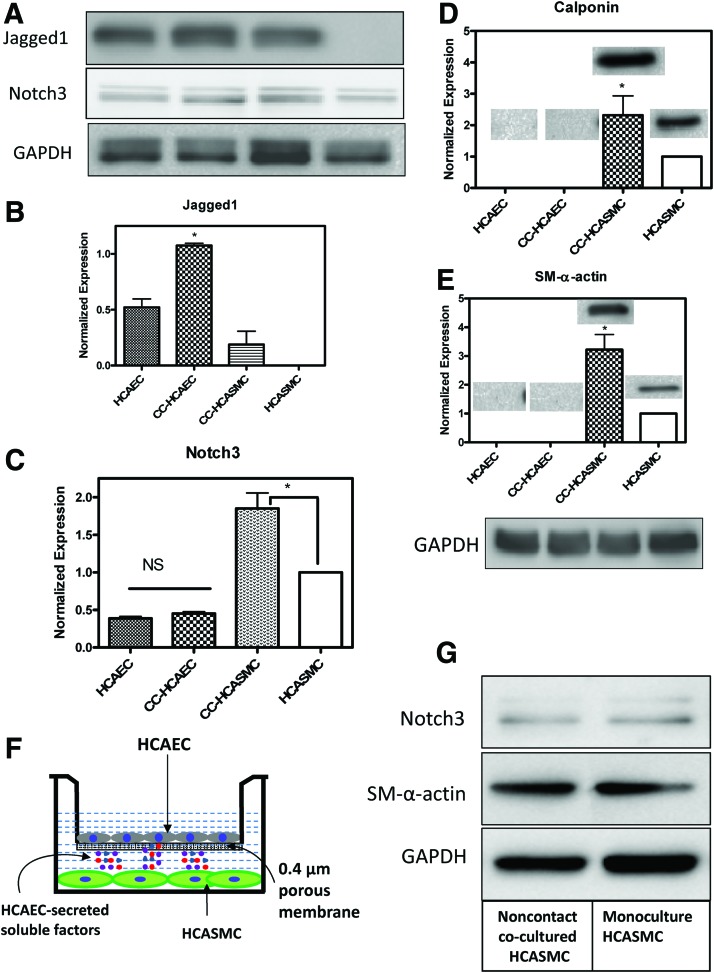
The effect of 3D cocultures on Notch- and SMC-specific protein expression levels
As stated in the “Introduction” section, induction of a contractile VSMC phenotype is an important stage in engineering vascular tissues and is required for tissue functionality. ECs, in addition to providing a nonthromobogenic surface, are thought to regulate the contractile VSMC phenotype. To explore this, we first seeded 750,000 HCASMCs on 3D PCU scaffolds and cultured them for 14 days, after which HCAECs were seeded and cultured for an additional 2 days while making direct contact with the SMCs. Western blotting analyses demonstrated that both Jagged1 in HCAECs and Notch3 in HCASMCs were significantly upregulated in the cocultures compared with monocultures (Fig. 5A–C, p<0.001). This increase in the cocultures was nearly twofold for both Jagged1 and Notch3. Concomitant with the increased expression of Jagged1 and Notch3 in the cocultures, the contractile phenotype marker proteins SM-α-actin and calponin in HCASMCs were also significantly upregulated when cocultured with HCAECs (Fig. 5D, E, p<0.001), suggesting that in the absence of ECs, HCASMCs were in a more synthetic phenotype when cultured on 3D PCU scaffolds. The finding that HCASMCs lacked their contractile marker protein expression in 3D cultures indicates that topographical effects may have been in play. We noted that both SM-α-actin and calponin were not detected in the separated EC blots, demonstrating that the separation protocol using PECAM-incubated magnetic beads was selective to HCAECs. It is worth mentioning that following coculture with HCAECs and cell separation, Jagged1 was detected in the HCASMC fraction (Fig. 5A, B). The detection of Jagged1 in HCASMCs is not related to EC contamination caused by separation inefficiency but rather attributed to Jagged1 expression in VSMCs following a physical association with HCAECs.32 Interaction of Jagged1-expressing ECs with VSMCs that express Notch3 receptors promotes Notch3 upregulation, smooth muscle differentiation, and initiates expression of Jagged1 in the newly differentiated VSMCs thus initiating a feed-forward pathway.41 We also observed that HCAEC-expressed Notch3 was not affected by coculturing (Fig. 5C). Although Notch3 expression is thought to be restricted to arterial SMCs, our finding is consistent with emerging data that suggest Notch3 expression in other cell types, including HCAECs.32,33,42 Given that we cocultured the two cell types in our scaffolds such that they make direct contact with each other, it is plausible to postulate that other soluble factors secreted by HCAECs could also play a role in upregulating Notch3 and subsequent VSMC differentiation marker expression. To test this possibility, we adopted a 2D transwell coculture where the two cell types are not making physical contacts while HUAEC-secreted molecules could freely diffuse into the HCASMC layer (see Fig. 5F for the setup). Western blotting showed that both Notch3 and SM-α-actin expression was unchanged in the noncontact cocultured HCASMCs compared with the monocultured HCASMCs (Fig. 5G), suggesting that soluble factors had no effect in Notch3 activation and SM-α-actin expression in HCASMCs.
FIG. 5.
Upregulation of Jagged1 in HCAECs and Notch3 in HCASMCs enhances SMC-specific marker protein SM-α-actin and calponin expression in 3D coculture. HCASMCs were seeded on 3D scaffolds for 14 days to achieve a dense cell population and extracellular matrix components, after which HCAECs were seeded and cocultured for an additional 2 days. Following separation of cells, western blot analysis demonstrated that both Jagged1 in HCAECs and Notch3 in HCASMCs were significantly upregulated in the cocultures compared with the monocultures (A–C). The SMC contractile phenotype marker proteins SM-α-actin and calponin in HCASMCs were significantly upregulated in HCASMCs cocultured with HCAECs (D, E). These upregulations were dependent on cell–cell contact since noncontact coculture model did not have any effect (F, G). Data are represented as mean±SD from three independent experiments (*p<0.05). NS, not significant. Color images available online at www.liebertpub.com/tea
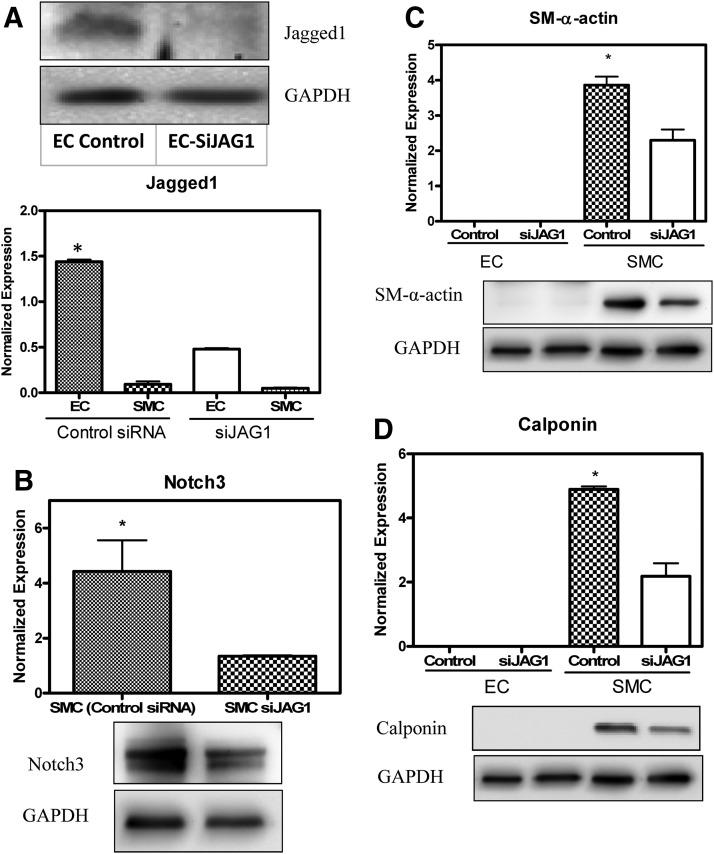
Effect of siRNA knockdown of Jagged1 on protein expression levels in 3D cocultures
Although the data presented in Figure 5 indicated that SM-α-actin and calponin upregulation in response to 3D cocultures was concomitant with Jagged1 and Notch3 activity, they do not provide direct evidence that Jagged1-selective Notch3 signaling pathway is the reason for the observation. To establish causative link, we knocked down the expression of Jagged1 in HCAECs using siRNA and transfection efficiency was tested to ensure sufficient knockdown of Jagged1 in HCAECs (Fig. 6A, B; p<0.0001). Jagged1 expression in HCAECs in the 3D coculture system was significantly higher when treated with control siRNA than with Jagged1-specific siRNA. A corresponding Notch3 downregulation was observed in cocultured HCASMCs with siRNA-treated HCAECs thus linking Jagged1 to its target receptor Notch3. As expected, both SM-α-actin and calponin were significantly downregulated (Fig. 6C, D; p<0.001), suggesting that Notch3 signaling is responsible for contractile VSMC phenotype regulation in 3D cocultures.
FIG. 6.
siRNA-mediated knockdown of Jagged1 reduces the expression levels of SMC-specific marker proteins associated with downregulation of Notch3 in 3D coculture. HCASMCs were seeded on 3D scaffolds and were cultured prior to coculture. HCAECs were transiently transfected with Jagged1 siRNA or scrambled siRNA (as a control), seeded on the HCASMC layer, and cocultured for 2 days. Following separation of cells, Western blot analysis revealed that transfection was successful and the corresponding Jagged1 expression in HCAECs was significantly reduced (A). Notch3 was significantly downregulated in HCASMCs by siRNA-mediated knockdown of Jagged1 as determined by western blot analysis (B). Both SM-α-actin and calponin were significantly downregulated, suggesting that Notch3 signaling is responsible for contractile VSMC phenotype regulation in 3D cultures (C, D). Data are represented as mean±SD from three independent experiments (*p<0.05).
Discussion
In this study we have developed and evaluated 3D coculture systems that utilizes HCASMCs and HCAECs and demonstrated Notch signaling to be responsible for inducing the contractile phenotype of HCASMCs in 3D that otherwise have predominantly a synthetic phenotype. Previously, several in vitro coculture models have been developed to study EC–SMC interactions. These are cocultures of ECs and SMCs on opposite sides of a porous membrane,43 microcarrier techniques,44 direct 2D coculture12,45,46 where these cells are separated by a layer of either collagen47 or fibronectin,48 and a coculture system consisting of SMCs within a 3D collagen and gelatin gel and a confluent monolayer of ECs on the gel surface.49,50 While these models have helped to elucidate some important interactions between the two cell types, most of them were performed only in 2D cultures often evaluating how SMCs influence EC behavior. Thee-dimensional coculture approaches such as microcarriers are limited to aggregates of cellular spheroids whereas the collagen gel system appeared to promote the synthetic phenotype that is undesirable in later stages of tissue engineering. To recapitulate the cellular microenvironment that more closely mimics the one observed in vivo, it is desirable that VSMC phenotype be studied in 3D engineered vascular tissue models rather than in conventional 2D culture systems. Although 2D coculture methods have been used to study Notch3 signaling,32,33 they do not replicate the 3D microenvironment found in tissues. For instance, the 2D culture systems involve a single layer of SMCs directly in contact with the signal-initiating ECs whereas a 3D culture system has multiple SMC layers that allow studies on signal propagation through these multiple layers thereby mimicking a typical arterial wall. Our data presented in Figures 2 and 4 showed that HCASMCs and HCAECs can be cocultured on highly porous 3D polyurethane scaffolds with distinctive layers forming for each cell type. Remarkably, the thickness of the EC layer in our study was found to be similar to that observed in vivo, suggesting that HCAECs only formed a monolayer on our scaffolds. Williams and Wicks51 and Pullens et al.52 seeded VSMCs and ECs onto a 3D porous poly(glycolic acid) tubular scaffolds and studied the effect of ECs on SMC function. In these studies, however, the goal was exploring collagen and proteoglycan deposition within the 3D scaffold mesh. Whether or not the two cell types partitioned into separate layers was not studied. Partitioning of the two cell types is often achieved artificially by separating them using coating with ECM components, with porous membranes, or by encapsulating the VSMCs.48,50 Unlike previous studies, we believe that our work demonstrated that a dense layer of HCASMCs can effectively support a monolayer of HCAEC formation as shown by the VE-cadherin staining (Fig. 4). The number of HCAECs seeded on a dense HCASMC layer also played a role since higher EC numbers formed corded and tube-like structures akin to a capillary network presumably stimulating morphogenesis of ECs into branching networks of cord structures.53
Unlike cardiac and skeletal muscle cells that are terminally differentiated and hence cannot revert to an earlier developmental stage, VSMCs are able to modulate their contractile or synthetic phenotypes in response to changing environmental cues. Understanding the regulating pathways in response to local environmental cues required for phenotype modulation is imperative for engineering living vascular substitutes. When seeded onto a 3D scaffold template, VSMCs must adopt a synthetic phenotype to facilitate cellular proliferation and ECM secretion. Studies from our laboratory demonstrated that 3D porous polyurethane scaffolds promote the synthetic VSMC phenotype in response to fibronectin conjugation54,55 and promoted elastin synthesis via Ras-dependent signaling pathway.56 Following tissue maturation, VSMCs must switch to a quiescent and contractile phenotype to mimic the functional properties of the native blood vessel, an event likely to be influenced by the presence of ECs. Emerging in vivo data on vascular development and maturation strongly suggest Notch signaling as a modulator of VSMC plasticity toward maturation and contraction. However, it was unknown whether the in vivo observation can be recapitulated in vitro using a synthetic 3D tissue engineering scaffold. In arterial SMCs, Notch3 is the predominant receptor while Jagged-1 is the principal ligand expressed by ECs. Given such unique arterial tissue distribution, the present study investigated Jagged1/Notch3 signaling. Indeed, our findings collectively presented in Figures 5 and 6 revealed that coculture significantly upregulated the expression of Notch3 and Jagged1 in HCASMCs and HCAECs, respectively. Concomitantly, SM-α-actin and calponin that are components of the contractile apparatus in VSMCs were upregulated. The absence of SM-α-actin and calponin from HCAECs in coculture indicates that the separation protocol used was highly efficient. siRNA knockdown of Jagged1 in HCASMCs abrogated the expression of the contractile apparatus in VSMCs, establishing a link between Notch signaling and VSMC maturation.
Notwithstanding a number of in vivo studies that documented the requirement of EC-expressed Notch ligand Jagged1 for vascular smooth muscle development and maturation,26,28,32 in vitro studies yielded conflicting data (reviewed in Boucher et al.57). Further, it is not clear whether Jagged1 presented to VSMCs without ECs will be able to activate Notch signaling and subsequent contractile phenotype. While soluble,58 protein G bead bound,59 and alginate and (polystyrene)-bound Jagged1 were able to induce mesenchymal stem cell differentiation toward cardiomyocytes and osteogenic lineages, respectively, it seems that arterial VSMC phenotype switching requires Jagged1 ligand expressed by ECs. With respect to the role of material-bound Jagged1 on the differentiation and maturation of VSMCs, both transcriptional activation60 and repression61 effects have been reported. Such discrepancies may be a result of a highly context-dependent nature and tight spatiotemporal regulation of Notch pathway components during development, in the adult vasculature, and in response to physiological changes in vivo.57 Although our gene expression data with Jagged1 bound to protein G beads showed Notch3 upregulation, neither SM-α-actin nor calponin was activated at the transcriptional level (Fig. 1). To the contrary, coculture of HCASMCs with HCAECs upregulated Notch3 and correspondingly SMC contractile markers (Figs. 5 and 6), suggesting the possibility that Notch3 expression may be a necessary but not a sufficient condition for SMC differentiation. One unique feature of the Notch signaling pathway is the requirement of ligand internalization by the signal-sending cells,62 which leads to receptor activation by pulling on the receptor. This pulling helps to generate the physical forces needed to dissociate and activate the receptor, and downstream target genes.63–65 This reliance on endocytosis by signal-sending cells is believed to be an important step in activating Notch downstream target genes and may explain why Notch signaling requires contact between the signal-sending cell and the signal-receiving cell.66,67 Given that ligand internalization in the case of Jagged1-bound beads is not a likely process (note that the signal-sending cell is replaced by Jagged-1-bound beads), it is reasonable to expect that presenting Jagged1 bound to beads may not be a sufficient condition for effective Notch activation and downstream target gene expression in HCASMCs, which underscores the need for cell–cell contact in our culture system. Finally, although our study focused on the role of Jagged1 in upregulating Notch3, we are not ruling out the possibility that Notch expression may also be regulated by other signaling pathways. Taken together, our work demonstrates an approach by which a contractile SMC phenotype may be induced following vascular tissue engineering.
Conclusions
In this study we developed a 3D coculture system for HCASMCs and HCAECs and demonstrated that the two cell types formed separate layers whereby the top 10-μm thickness was attributed to ECs. Targeted SMC contractile gene and protein expression studies showed that upregulations of SM-α-actin and calponin were dependent on Notch3 receptor expression on HCASMCs and Jagged1 expression on the HCAEC surface. Knockdown of Jagged1 using siRNA attenuated both Notch3 and the contractile phenotype marker proteins SM-α-actin and calponin. Collectively, our studies showed the importance of mechanistic studies to identify signaling events in 3D cultures.
Acknowledgment
This work is funded by the Heart and Stroke Foundation of Canada (HSFC; grant No. T7262).
Disclosure Statement
No competing financial interests exist.
References
- 1.Duncan D.R., and Breuer C.K.Challenges in translating vascular tissue engineering to the pediatric clinic. Vasc Cell 3,23, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chan-Park M.B., Shen J.Y., Cao Y., Xiong Y., Liu Y., Rayatpisheh S., et al. . Biomimetic control of vascular smooth muscle cell morphology and phenotype for functional tissue-engineered small-diameter blood vessels. J Biomed Mater Res A 88,1104, 2009 [DOI] [PubMed] [Google Scholar]
- 3.Owens G.K., Kumar M.S., and Wamhoff B.R.Molecular regulation of vascular smooth muscle cell differentiation in development and disease. Physiol Rev 84,767, 2004 [DOI] [PubMed] [Google Scholar]
- 4.Wamhoff B.R., Bowles D.K., and Owens G.K.Excitation-transcription coupling in arterial smooth muscle. Circ Res 98,868, 2006 [DOI] [PubMed] [Google Scholar]
- 5.Bacakova L., Wilhelm J., Herget J., Novotna J., and Eckhart A.Oxidized collagen stimulates proliferation of vascular smooth muscle cells. Exp Mol Pathol 64,185, 1997 [DOI] [PubMed] [Google Scholar]
- 6.Owens G.K., Geisterfer A.A., Yang Y.W., and Komoriya A.Transforming growth factor-beta-induced growth inhibition and cellular hypertrophy in cultured vascular smooth muscle cells. J Cell Biol 107,771, 1988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stegemann J.P., Hong H., and Nerem R.M.Mechanical, biochemical, and extracellular matrix effects on vascular smooth muscle cell phenotype. J Appl Physiol 98,2321, 2005 [DOI] [PubMed] [Google Scholar]
- 8.Louis S.F., and Zahradka P.Vascular smooth muscle cell motility: from migration to invasion. Exp Clin Cardiol 15,e75, 2010 [PMC free article] [PubMed] [Google Scholar]
- 9.Han M., Wen J.K., Zheng B., Cheng Y., and Zhang C.Serum deprivation results in redifferentiation of human umbilical vascular smooth muscle cells. Am J Physiol Cell Physiol 291,C50, 2006 [DOI] [PubMed] [Google Scholar]
- 10.Lacolley P., Regnault V., Nicoletti A., Li Z., and Michel J.B.The vascular smooth muscle cell in arterial pathology: a cell that can take on multiple roles. Cardiovasc Res 95,194, 2012 [DOI] [PubMed] [Google Scholar]
- 11.Heydarkhan-Hagvall S., Chien S., Nelander S., Li Y.C., Yuan S., Lao J., et al. . DNA microarray study on gene expression profiles in co-cultured endothelial and smooth muscle cells in response to 4- and 24-h shear stress. Mol Cell Biochem 281,1, 2006 [DOI] [PubMed] [Google Scholar]
- 12.Hirschi K.K., Rohovsky S.A., and D'Amore P.A.PDGF, TGF-beta, and heterotypic cell-cell interactions mediate endothelial cell-induced recruitment of 10T1/2 cells and their differentiation to a smooth muscle fate. J Cell Biol 141,805, 1998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Powell R.J., Bhargava J., Basson M.D., and Sumpio B.E.Coculture conditions alter endothelial modulation of TGF-beta 1 activation and smooth muscle growth morphology. Am J Physiol 274,H642, 1998 [DOI] [PubMed] [Google Scholar]
- 14.Powell R.J., Cronenwett J.L., Fillinger M.F., Wagner R.J., and Sampson L.N.Endothelial cell modulation of smooth muscle cell morphology and organizational growth pattern. Ann Vasc Surg 10,4, 1996 [DOI] [PubMed] [Google Scholar]
- 15.Powell R.J., Hydowski J., Frank O., Bhargava J., and Sumpio B.E.Endothelial cell effect on smooth muscle cell collagen synthesis. J Surg Res 69,113, 1997 [DOI] [PubMed] [Google Scholar]
- 16.Brown D.J., Rzucidlo E.M., Merenick B.L., Wagner R.J., Martin K.A., and Powell R.J.Endothelial cell activation of the smooth muscle cell phosphoinositide 3-kinase/Akt pathway promotes differentiation. J Vasc Surg 41,509, 2005 [DOI] [PubMed] [Google Scholar]
- 17.Fetalvero K.M., Shyu M., Nomikos A.P., Chiu Y.F., Wagner R.J., Powell R.J., et al. . The prostacyclin receptor induces human vascular smooth muscle cell differentiation via the protein kinase A pathway. Am J Physiol Heart Circ Physiol 290,H1337, 2006 [DOI] [PubMed] [Google Scholar]
- 18.Hofmann J.J., and Iruela-Arispe M.L.Notch signaling in blood vessels: who is talking to whom about what? Circ Res 100,1556, 2007 [DOI] [PubMed] [Google Scholar]
- 19.Iso T., Hamamori Y., and Kedes L.Notch signaling in vascular development. Arterioscler Thromb Vasc Biol 23,543, 2003 [DOI] [PubMed] [Google Scholar]
- 20.Shawber C.J., and Kitajewski J.Notch function in the vasculature: insights from zebrafish, mouse and man. Bioessays 26,225, 2004 [DOI] [PubMed] [Google Scholar]
- 21.Tang Y., Urs S., and Liaw L.Hairy-related transcription factors inhibit Notch-induced smooth muscle alpha-actin expression by interfering with Notch intracellular domain/CBF-1 complex interaction with the CBF-1-binding site. Circ Res 102,661, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Doi H., Iso T, Shiba Y., Sato H., Yamazaki M., Oyama Y., et al. . Notch signaling regulates the differentiation of bone marrow-derived cells into smooth muscle-like cells during arterial lesion formation. Biochem Biophys Res Commun 381,654, 2009 [DOI] [PubMed] [Google Scholar]
- 23.Weber D.S.A novel mechanism of vascular smooth muscle cell regulation by Notch: platelet-derived growth factor receptor-beta expression? Circ Res 102,1448, 2008 [DOI] [PubMed] [Google Scholar]
- 24.Ghosh S., Paez-Cortez J.R., Boppidi K., Vasconcelos M., Roy M., Cardoso W., et al. . Activation dynamics and signaling properties of Notch3 receptor in the developing pulmonary artery. J Biol Chem 286,22678, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Villa N., Walker L., Lindsell C.E., Gasson J., Iruela-Arispe M.L., and Weinmaster G.Vascular expression of Notch pathway receptors and ligands is restricted to arterial vessels. Mech Develop 108,161, 2001 [DOI] [PubMed] [Google Scholar]
- 26.Domenga V., Fardoux P., Lacombe P., Monet M., Maciazek J., Krebs L.T., et al. . Notch3 is required for arterial identity and maturation of vascular smooth muscle cells. Genes Dev 18,2730, 2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang T., Baron M., and Trump D.An overview of Notch3 function in vascular smooth muscle cells. Prog Biophys Mol Biol 96,499, 2008 [DOI] [PubMed] [Google Scholar]
- 28.High F.A., Lu M.M., Pear W.S., Loomes K.M., Kaestner K.H., and Epstein J.A.Endothelial expression of the Notch ligand Jagged1 is required for vascular smooth muscle development. Proc Natl Acad Sci U S A 105,1955, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Carlson M.E., O'Connor M.S., Hsu M., and Conboy I.M.Notch signaling pathway and tissue engineering. Front Biosci 12,5143, 2007 [DOI] [PubMed] [Google Scholar]
- 30.Grenier S., Sandig M., and Mequanint K.Polyurethane biomaterials for fabricating 3D porous scaffolds and supporting vascular cells. J Biomed Mater Res A 82A,802, 2007 [DOI] [PubMed] [Google Scholar]
- 31.Grenier S., Sandig M., and Mequanint K.Smooth muscle alpha-actin and calponin expression and extracellular matrix production of human coronary artery smooth muscle cells in 3D scaffolds. Tissue Eng Part A 15,3001, 2009 [DOI] [PubMed] [Google Scholar]
- 32.Liu H., Kennard S., and Lilly B.NOTCH3 expression is induced in mural cells through an autoregulatory loop that requires endothelial-expressed JAGGED1. Circ Res 104,466, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Xia Y., Bhattacharyya A., Roszell E.E., Sandig M., and Mequanint K.The role of endothelial cell-bound Jagged1 in Notch3-induced human coronary artery smooth muscle cell differentiation. Biomaterials 33,2462, 2012 [DOI] [PubMed] [Google Scholar]
- 34.Miano J.M., and Olson E.N.Expression of the smooth muscle cell calponin gene marks the early cardiac and smooth muscle cell lineages during mouse embryogenesis. J Biol Chem 271,7095, 1996 [DOI] [PubMed] [Google Scholar]
- 35.Gong Z., and Niklason L.E.Small-diameter human vessel wall engineered from bone marrow-derived mesenchymal stem cells (hMSCs). FASEB J 22,1635, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Noseda M., Fu Y., Niessen K., Wong F., Chang L., McLean G., et al. . Smooth muscle alpha-actin is a direct target of Notch/CSL. Circ Res 98,1468, 2006 [DOI] [PubMed] [Google Scholar]
- 37.Cisneros E., Latasa M.J., Garcia-Flores M., and Frade J.M.Instability of Notch1 and Delta1 mRNAs and reduced Notch activity in vertebrate neuroepithelial cells undergoing S-phase. Mol Cell Neurosci 37,820, 2008 [DOI] [PubMed] [Google Scholar]
- 38.Gao J., Crapo P.M., and Wang Y.Macroporous elastomeric scaffolds with extensive micropores for soft tissue engineering. Tissue Eng 12,917, 2006 [DOI] [PubMed] [Google Scholar]
- 39.Ilagan B.G., and Amsden B.G.Macroporous photocrosslinked elastomer scaffolds containing microposity: preparation and in vitro degradation properties. J Biomed Mater Res A 93,211, 2010 [DOI] [PubMed] [Google Scholar]
- 40.Stamatas G.N., and McIntire L.V.Rapid flow-induced responses in endothelial cells. Biotechnol Prog 17,383, 2001 [DOI] [PubMed] [Google Scholar]
- 41.Feng X., Krebs L.T., and Gridley T.Patent ductus arteriosus in mice with smooth muscle-specific Jag1 deletion. Development 137,4191, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jin S., Hansson E.M., Tikka S., Lanner F., Sahlgren C., Farnebo F., et al. . Notch signaling regulates platelet-derived growth factor receptor-beta expression in vascular smooth muscle cells. Circ Res 102,1483, 2008 [DOI] [PubMed] [Google Scholar]
- 43.Rose S.L., and Babensee J.E.Complimentary endothelial cell/smooth muscle cell co-culture systems with alternate smooth muscle cell phenotypes. Ann Biomed Eng 35,1382, 2007 [DOI] [PubMed] [Google Scholar]
- 44.Korff T., Kimmina S., Martiny-Baron G., and Augustin H.G.Blood vessel maturation in a 3-dimensional spheroidal coculture model: direct contact with smooth muscle cells regulates endothelial cell quiescence and abrogates VEGF responsiveness. FASEB J 15,447, 2001 [DOI] [PubMed] [Google Scholar]
- 45.Niwa K., Kado T., Sakai J., and Karino T.The effects of a shear flow on the uptake of LDL and acetylated LDL by an EC monoculture and an EC-SMC coculture. Ann Biomed Eng 32,537, 2004 [DOI] [PubMed] [Google Scholar]
- 46.Chaterji S., Park K., and Panitch A.Scaffold-free in vitro arterial mimetics: the importance of smooth muscle-endothelium contact. Tissue Eng Part A 16,1901, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sakamoto N., Kiuchi T., and Sato M.Development of an endothelial-smooth muscle cell coculture model using phenotype-controlled smooth muscle cells. Ann Biomed Eng 39,2750, 2011 [DOI] [PubMed] [Google Scholar]
- 48.Lavender M.D., Pang Z., Wallace C.S., Niklason L.E., and Truskey G.A.A system for the direct co-culture of endothelium on smooth muscle cells. Biomaterials 26,4642, 2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Imberti B., Seliktar D., Nerem R.M., and Remuzzi A.The response of endothelial cells to fluid shear stress using a co-culture model of the arterial wall. Endothelium 9,11, 2002 [DOI] [PubMed] [Google Scholar]
- 50.Liu Y., Rayatpisheh S., Chew S.Y., and Chan-Park M.B.Impact of endothelial cells on 3D cultured smooth muscle cells in a biomimetic hydrogel. ACS Appl Mater Interfaces 4,1378, 2012 [DOI] [PubMed] [Google Scholar]
- 51.Williams C., and Wick T.M.Endothelial cell-smooth muscle cell co-culture in a perfusion bioreactor system. Ann Biomed Eng 33,920, 2005 [DOI] [PubMed] [Google Scholar]
- 52.Pullens R.A, Stekelenburg M., Baaijens F.P., and Post M.J.The influence of endothelial cells on the ECM composition of 3D engineered cardiovascular constructs. J Tissue Eng Regen Med 3,11, 2009 [DOI] [PubMed] [Google Scholar]
- 53.Kunz-Schughart L.A., Schroeder J.A., Wondrak M., van Rey F., Lehle K., Hofstaedter F., et al. . Potential of fibroblasts to regulate the formation of three-dimensional vessel-like structures from endothelial cells in vitro. Am J Physiol Cell Physiol 290,C1385, 2006 [DOI] [PubMed] [Google Scholar]
- 54.Dubey G., and Mequanint K.Conjugation of fibronectin onto three-dimensional porous scaffolds for vascular tissue engineering applications. Acta Biomater 7,1114, 2011 [DOI] [PubMed] [Google Scholar]
- 55.Lin S., Sandig M., and Mequanint K.Three-dimensional topography of synthetic scaffolds induces elastin synthesis by human coronary artery smooth muscle cells. Tissue Eng Part A 17,1561, 2011 [DOI] [PubMed] [Google Scholar]
- 56.Lin S., and Mequanint K.The role of Ras-ERK-IL-1beta signaling pathway in upregulation of elastin expression by human coronary artery smooth muscle cells cultured in 3D scaffolds. Biomaterials 33,7047, 2012 [DOI] [PubMed] [Google Scholar]
- 57.Boucher J., Gridley T., and Liaw L.Molecular pathways of notch signaling in vascular smooth muscle cells. Front Physiol 3,81, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Li H., Yu B., Zhang Y., Pan Z., and Xu W.Jagged1 protein enhances the differentiation of mesenchymal stem cells into cardiomyocytes. Biochem Biophys Res Commun 341,320, 2006 [DOI] [PubMed] [Google Scholar]
- 59.Osathanon T., Ritprajak P., Nowwarote N., Manokawinchoke J., Giachelli C., and Pavasant P.Surface-bound orientated Jagged-1 enhances osteogenic differentiation of human periodontal ligament-derived mesenchymal stem cells. J Biomed Mater Res A 101,358, 2013 [DOI] [PubMed] [Google Scholar]
- 60.Boucher J.M., Peterson S.M., Urs S., Zhang C., and Liaw L.The miR-143/145 cluster is a novel transcriptional target of Jagged-1/Notch signaling in vascular smooth muscle cells. J Biol Chem 286,28312, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Li X., Zhang X., Leathers R., Makino A., Huang C., Parsa P., et al. . Notch3 signaling promotes the development of pulmonary arterial hypertension. Nat Med 15,1289, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Le Borgne R., Bardin A., and Schweisguth F.The roles of receptor and ligand endocytosis in regulating Notch signaling. Development 132,1751, 2005 [DOI] [PubMed] [Google Scholar]
- 63.Wang X., and Ha T.Defining single molecular forces required to activate integrin and notch signaling. Science 340,991, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Shergill B., Meloty-Kapella L., Musse A.A., Weinmaster G., and Botvinick E.Optical tweezers studies on Notch: single-molecule interaction strength is independent of ligand endocytosis. Dev Cell 22,1313, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Meloty-Kapella L., Shergill B., Kuon J., Botvinick E., and Weinmaster G.Notch ligand endocytosis generates mechanical pulling force dependent on dynamin, epsins, and actin. Dev Cell 22,1299, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Fortini M.E., and Bilder D.Endocytic regulation of Notch signaling. Curr Opin Genet Dev 19,323, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Pratt E.B., Wentzell J.S., Maxson J.E., Courter L., Hazelett D., and Christian J.L.The cell giveth and the cell taketh away: an overview of Notch pathway activation by endocytic trafficking of ligands and receptors. Acta Histochem 113,248, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]