Abstract
Purpose
To identify and validate copy number aberrations in early-stage primary breast tumors associated with bone or non-bone metastasis.
Patients and Methods
Whole-genome molecular inversion probe arrays were used to evaluate copy number imbalances (CNIs) in breast tumors from 960 early-stage patients with information about site of metastasis. The CoxBoost algorithm was used to select metastasis site-related CNIs and to fit a Cox proportional hazards model.
Results
Gains at 1q41 and 1q42.12 and losses at 1p13.3, 8p22, and Xp11.3 were significantly associated with bone metastasis. Gains at 2p11.2, 3q21.3–22.2, 3q27.1, 10q23.1, and 14q13.2–3 and loss at 7q21.11 were associated with non-bone metastasis. To examine the joint effect of CNIs and clinical predictors, patients were stratified into three risk groups (low, intermediate, and high) based on the sum of predicted linear hazard ratios (HRs). For bone metastasis, the hazard (95% confidence interval) for the low-risk group was 0.32 (0.11–0.92) compared to the intermediate-risk group and 2.99 (1.74–5.11) for the high-risk group. For non-bone metastasis, the hazard for the low-risk group was 0.34 (0.17–0.66) and 2.33 (1.59–3.43) for the high-risk group. The prognostic value of loss at 8p22 for bone metastasis and gains at 10q23.1 for non-bone metastasis, and gain at 11q13.5 for both bone and non-bone metastases were externally validated in 335 breast tumors pooled from four independent cohorts.
Conclusions
Distinct CNIs are independently associated with bone and non-bone metastasis for early-stage breast cancer patients across cohorts. These data warrant consideration for tailoring surveillance and management of metastasis risk.
Keywords: Breast cancer, bone metastasis, non-bone metastasis, copy number imbalances, molecular inversion probe array
Introduction
Breast cancer is the second most common cause of cancer-related death in women, with a yearly toll of more than 40,000 deaths in the United States alone [1]. Primary sites of distant metastasis include bone, lung, liver and brain, with bone being the most common site [2]. The biological mechanisms underlying breast cancer metastasis have been extensively studied since Paget theorized that metastasis is influenced by both, the “seed” (tumor cells) and the “soil” (host environment) [3]. However, to date the driver events of site-specific metastasis in early stage breast cancer remains largely unknown.
Various factors are used to predict risk of metastasis in early stage (I and II) breast cancer patients. These include stage, tumor size, histologic grade, lymph node involvement, hormone receptor status, human epidermal growth factor receptor 2 (HER2) status, and age at diagnosis. Of these, estrogen receptor-positive (ER+) status has the strongest association with bone metastasis [2]. However, other molecular changes are also likely to drive bone-specific metastasis. Kang and colleagues [4] reported that cell lines that are highly metastatic to bone appear to lose a 17-gene metastatic signature set previously described by Ramaswamy et. al., [5]. Kang et al. also identified genes, including DLC1, IL11, CXCR4, and osteopontin that predict bone metastasis [4]. These findings suggest that multiple metastatic competency genes are needed for metastasis, and those alterations in tissue-specific genes may be necessary for tumor cells to grow in a particular “soil”.
Patient’s age and tumor ER-negative (ER−) status [6], amplification of the ERBB2 gene coding for the HER2 receptor [7,8], and triple-negative breast cancer (TNBC) tumor status [9] have emerged as risk factors for breast cancer to metastasize to the brain and/or lung. Although the molecular mechanisms underlying metastasis to the bone have been extensively studied and are partially understood, little information is available on the molecular determinants or molecular drivers of metastasis to non-bone sites. Gene expression studies have examined genetic markers as prognostic factors in breast cancer patients with brain [10] and lung [11] metastasis. However, no comprehensive genomic study characterizing the key aberrations that regulate site-specific bone or non-bone metastasis of breast cancer has been reported yet.
Although metastatic breast cancer is associated with poor prognosis, new treatment strategies, including drugs that target transcription factors and specific cellular pathways, have improved progression-free and overall survival [12]. Somatic alterations, including copy number imbalances (CNIs) and somatic mutations in the primary tumor, may determine the propensity of a tumor to metastasize to a specific site. Importantly, while these targeted genetic abnormalities have a prognostic impact individually or in combination, high-resolution genome profiles show some indications of the prognostic value of CNIs. We hypothesized that evaluation of CNIs provide important insights into the underlying site-specific metastatic propensity of primary early-stage breast cancers, and have potential clinical utility in discriminating between patients who have high versus low risk of developing metastasis. We applied the whole-genome molecular inversion probe (MIP) technique to determine CNIs using DNA from breast tumor tissue from a cohort of patients with early-stage breast cancer for which long-term follow-up data were available. Here, we describe the association between specific CNIs and risk of metastasis to bone or non-bone (lung, liver, brain and others) sites. We then replicated our findings in an external cohort of 335 early-stage breast cancer patients.
Materials and Methods
Patient population and breast tumor specimens
The Early Stage Breast Cancer Repository (ESBCR) at The University of Texas MD Anderson Cancer Center (MDACC) comprised 2,409 women diagnosed with American Joint Committee on Cancer clinical stage I or II breast cancer, and surgically treated at MDACC between 1985 and 2000. Criteria for eligibility and cohort details have been reported previously [13]. From this retrospective cohort study, we identified 1,003 patients with clinical and follow-up data, and adequate tumor DNA from formalin-fixed, paraffin-embedded (FFPE) tissue blocks. Clinical information, primary treatment (i.e., surgery, radiation therapy, chemotherapy, and endocrine therapy), and histopathological information, including patient’s first site of metastasis and other site of metastasis were obtained from medical record [14]. Bone metastasis was defined as the first site of metastasis by bone scan confirmed also by other imaging methods including X-ray, computed tomography (CT), and magnetic resonance imaging (MRI) scans. Non-bone metastasis was defined as the first site of metastasis to lung, liver, brain or others as documented by appropriate CT and MRI scans. The histological type of all tumors was defined according to the WHO classification system. Nuclear grade was defined according to the Black’s nuclear grading system with modification of numbers: 1 represents well-differentiated tumors, and 3 represents poorly differentiated tumors. This study was approved by the Institutional Review Board of the MDACC.
Definition of breast tumor subtypes
The four tumor subtypes of luminal A, luminal B, HER2+, and TNBC were approximated from clinically validated immunohistochemical (IHC) analyses of ER, PR, and Ki67. ER and PR status were obtained from medical records (primary source) and tissue microarray studies (secondary source); the agreement in ER and PR status between the two sources was 84.8% and 76.4%, respectively. ER and PR positivity was defined as ≥ 1% staining. ER-positive/HER2 negative tumors were subclassified using Ki67 and a clinical threshold of ≥17% positivity into LUM A (ER+/Ki67 <17%) and LUM B (ER+/Ki67 ≥17%). HER2+ tumors were defined as a separate subtype, independent of their hormone receptor status, because of the prognostic and predictive significance of HER2 amplification for site of metastasis. Subclassification of HER2+ cases on ER status was explored but did not change the results (data not shown). In this sample set, defining HER2 status using MIP array-based ERBB2 copy number and a threshold of 2.8 for ‘copy number gain’ proved equivalent to IHC area under the curve (AUC) analysis of 0.94 [14]. Copy number thresholds of 2.3, 2.5, and 2.8 changed the frequency of HER2+ tumors in the sample as follows: 26.9%, 21.0%, and 16.3%, respectively.
Molecular inversion probe arrays for copy number analysis
Tumor DNA was extracted from FFPE tissue and copy number data were obtained using the MIP-based, OncoScan FFPE Express (Affymetrix, Santa Clara, CA) as previously described [14]. Data collected from 129 matched normal lymph node samples were used for normalizing the CN data; therefore, common germline CNIs have been normalized by comparing the tumors to this normal set. For each sample, we generated full-genome MIP quantifications (330K MIPs). In order to reduce the data dimension, we computed the running median within groups of 25 consecutive MIPs, yielding 13,175 data points per sample. The Circular Binary Segmentation algorithm was used to convert the data to a list of segments for each sample. CN differences were analyzed with the R package DNAcopy, using thresholds of 2.5 for one copy gained and 1.5 for one copy lost. The parameter alpha (significance level for acceptance of change-points) used in the segmentation algorithm was set to 0.01. We recombined consecutive segments if their gain/loss calls agreed for at least 99.5% of the samples. This procedure yielded 1,593 segments, representing the entire genome. Comparisons of CN patterns across different demographic, clinical, and tumor subtype groups were performed by Fisher’s exact test, chi-square test, or Wilcoxon rank-sum test, as appropriate, with random permutations of the samples to incorporate an FDR adjustment for multiple comparisons. These data are available in the Gene Expression Omnibus (GEO) database accession number GSE31424.
Patient samples with array comparative genomic hybridization for the validation study
For validation, 361 patients with site of metastasis and whole genome copy number data as array comparative genomic hybridization (aCGH) determined on Agilent’s 244K Human Genome CGH collected at different hospitals in Norway and Sweden were pooled from four independent cohorts. After excluding normal samples (n = 20), lymph node metastases samples (n= 1), samples with stage 0 (n = 2), stage IV (n = 2), and a lymph node metastases sample with stage IV (n = 1), we have 335 early-stage breast cancer patients in the analysis: MDG n = 37, MicMa n = 41, FW n = 94, ULL n = 163. This includes 209 cases with complete stage data (MDG n = 32, MicMa n = 40, ULL n = 137) and 126 missing data on stage. All studies have been approved by the local ethical committee and local authorities. These data are available in the GEO database accession numbers GSE20394 and GSE32291.
Cox proportional hazards regression
The primary endpoint was time-to-first metastasis to bone or non-bone site, defined as the time from diagnosis of the primary breast tumor to first documented metastasis to bone or non-bone sites; only the site of first metastasis was considered. First, a univariate Cox proportional hazards regression model was used to calculate a hazard ratio (HR) and 95% confidence interval (CI) for the association between each established clinical and pathological characteristic and risk of metastasis to bone or non-bone sites. Second, we applied the CoxBoost algorithm [15] for fitting a Cox proportional hazards regression model with high-dimensional covariates to select CNIs associated with bone or non-bone metastasis using the R package CoxBoost. Further, we conducted multivariate Cox proportional hazards regression models that included the significant (α < 0.1) clinical, pathological, and CNIs identified in the first and second steps. Finally, using stepwise minimization of the Akaike Information Criterion (AIC), we built the most parsimonious models. To evaluate the predictive accuracy of the model, the concordance-index (C-index) was used to compare the strengths of the models. C-Index can be interpreted as the probability of agreement between what the model predicts and the actual observed risk of breast cancer metastasis. A completely random prediction would have a concordance of 0.5, a perfect rule a concordance of 1. To identify a set of robust factors for time-to-event, we calculated the posterior probability of each covariate using the Bayesian model averaging (BMA) algorithm [16,17]. A higher posterior probability score indicates a stronger effect. To evaluate the cumulative risk, we used the sum of the predicted linear HRs from the final multivariate model to stratify patients into three risk groups: high-risk group (the highest 25% of linear HRs), intermediate-risk group (the middle 50% of linear HRs), or low-risk group (the lowest 25% of linear HRs), denoting poor, intermediate, and good prognosis, respectively. All analyses were performed using R version 2.12.0 (R Foundation for Statistical Computing).
Results
Characteristics of the study populations
For the MDACC discovery cohort of 960 early-stage breast cancer patients, 203 (21%) patients developed a metastasis (Table 1), of which 74 (36%) had first metastasis to the bone, and 129 (64%) had first metastasis to non-bone sites (59 to lung, 35 to liver, 9 to brain, and 26 to others). The median time-to-bone and -non-bone metastasis was 4.6 and 4.7 years, respectively, whereas that for the no-metastasis group was 10.3 years. For the patients with first metastasis to bone, 36 only had one metastasis record, which was to bone. For 38 patients, there were multiple metastasis records for which 29 of them only had one recorded location which was bone. The remaining nine had multiple locations for metastasis as site of first metastasis but were included with first site of metastasis as bone. The median [standard deviation (SD)] period of multiple conversion (solitary-to-multiple metastatic bone lesion development) was 0.68 (1.47) years. The median (SD) time of conversion from solitary metastatic non-bone lesions to multiple lesions was 0.62 (2.00) years for lung, 0.02 (0.62) years for liver, and 0.04 (0.54) years for brain.
Table 1.
Clinical characteristics of breast cancer patients with copy number imbalances
| Characteristic | Discovery dataset (n = 960)
|
Validation dataset (n = 335)
|
||||
|---|---|---|---|---|---|---|
| No n (%)* |
Bone n (%)* |
Non-bone n (%)* |
No n (%)* |
Bone n (%)* |
Non-bone n (%)* |
|
| n | 757 | 74 | 129 | 259 | 26 | 50 |
| Median time to first metastasis, y | — | 4.6 | 4.7 | — | 3.5 | 2.8 |
| Median follow-up time, y | 10.3 | — | — | 9.8 | — | — |
| Age at primary diagnosis, y | ||||||
| Mean (SD) | 55 (13) | 51 (12) | 51 (12) | 62 (14) | 62 (15) | 59 (16) |
| ≤50 | 295 (38.9) | 36 (48.7) | 68 (52.7) | 62 (23.9) | 7 (26.9) | 17 (34.0) |
| >50 | 462 (61.1) | 38 (51.3) | 61 (47.3) | 197 (76.1) | 19 (73.1) | 33 (66.0) |
| Race/ethnicity & | ||||||
| White | 561 (74.1) | 54 (72.9) | 93 (72.1) | — | — | — |
| Black | 95 (12.6) | 11 (14.9) | 17 (13.2) | — | — | — |
| Hispanic | 97 (12.8) | 7 (9.5) | 17 (13.2) | — | — | — |
| Unknown | 4 (0.5) | 2 (2.7) | 2 (1.5) | — | — | — |
| Tumor subtype # | ||||||
| Luminal A | 271 (35.8) | 21 (28.4) | 24 (18.6) | 72 (27.8) | 7 (26.9) | 11 (22.0) |
| Luminal B | 172 (22.7) | 27 (36.5) | 26 (20.2) | 16 (6.2) | 2 (7.7) | 5 (10.0) |
| HER2+ | 112 (14.8) | 14 (18.9) | 31 (24.0) | 26 (10.0) | 2 (7.7) | 5 (10.0) |
| TNBC | 138 (18.2) | 4 (5.4) | 40 (31.0) | 16 (6.2) | 1 (3.9) | 6 (12.0) |
| Normal | — | — | — | 16 (6.2) | 2 (7.7) | 3 (6.0) |
| Unknown | 64 (8.5) | 8 (10.8) | — | 113 (43.6) | 12 (46.1) | 20 (40.0) |
| Stage | ||||||
| I | 278 (36.7) | 12 (16.2) | 14 (10.9) | 56 (21.6) | — | 6 (12.0) |
| II | 479 (63.3) | 62 (83.8) | 115 (89.2) | 76 (29.4) | 10 (38.5) | 16 (32.0) |
| III | — | — | — | 21 (8.1) | 10 (38.5) | 14 (28.0) |
| Unknown | — | — | — | 106 (40.9) | 6 (23.0) | 14 (28.0) |
| Nuclear grade† | ||||||
| I–II | 454 (60.0) | 46 (62.2) | 66 (51.2) | 169 (65.2) | 20 (76.9) | 38 (76.0) |
| III | 255 (33.7) | 19 (25.7) | 56 (43.4) | 59 (22.8) | 6 (23.1) | 11 (22.0) |
| Unknown | 48 (6.3) | 9 (12.1) | 7 (5.4) | 31 (12.0) | — | 1 (2.0) |
| Tumor size | ||||||
| < 2 cm | 477 (63.0) | 30 (40.5) | 56 (43.4) | 126 (48.7) | 6 (23.1) | 18 (36.0) |
| ≥2 cm | 257 (34.0) | 38 (51.4) | 68 (52.7) | 95 (36.7) | 17 (65.4) | 26 (52.0) |
| Infiltrating skin/thoracic wall | — | — | — | 2 (0.8) | 2 (7.7) | 1 (2.0) |
| Carcinoma in situ | — | — | — | 27 (10.4) | — | 1 (2.0) |
| Unknown | 23 (3.0) | 6 (8.1) | 5 (3.9) | 9 (3.4) | 1 (3.8) | 4 (8.0) |
| Lymph node status | ||||||
| Positive | 267 (35.3) | 40 (54.1) | 71 (55.0) | 67 (25.9) | 17 (65.4) | 26 (52.0) |
| Negative | 476 (62.9) | 30 (40.5) | 55 (42.6) | 163 (62.9) | 7 (26.9) | 19 (38.0) |
| Unknown | 14 (1.8) | 4 (5.4) | 3 (2.4) | 29 (11.2) | 2 (7.7) | 5 (10.0) |
| Radiation therapy | ||||||
| Yes | 321 (42.4) | 35 (47.3) | 51 (39.5) | 109 (42.1) | 13 (50.0) | 20 (40.0) |
| No | 418 (55.2) | 36 (48.7) | 75 (58.2) | 112 (43.2) | 12 (46.2) | 24 (48.0) |
| Unknown | 18 (2.4) | 3 (4.1) | 3 (2.3) | 38 (14.7) | 1(3.8) | 6 (12.0) |
| Chemotherapy | ||||||
| Yes | 319 (42.2) | 38 (51.3) | 76 (61.9) | 44 (17.0) | 4 (15.4) | 8 (16.0) |
| No | 397 (52.4) | 31 (41.9) | 47 (36.4) | 209 (80.7) | 21 (80.8) | 41 (82.0) |
| Unknown | 41 (5.4) | 5 (6.8) | 6 (4.7) | 6 (2.3) | 1 (3.8) | 1 (2.0) |
| Endocrine therapy | ||||||
| Yes | 353 (46.6) | 28 (37.8) | 37 (28.7) | 79 (30.5) | 12 (46.2) | 16 (32.0) |
| No | 386 (51.0) | 43 (58.1) | 88 (68.2) | 173 (66.8) | 13 (50.0) | 33 (66.0) |
| Unknown | 18 (2.4) | 3 (4.1) | 4 (3.1) | 7 (2.7) | 1 (3.8) | 1 (2.0) |
Abbreviations: SD, standard deviation; HER2, human epidermal growth factor receptor 2; TNBC, triple-negative breast cancer.
Numbers do not add up because of missing data.
Nearly all of the validation dataset samples are Caucasian; however, some might come from Pakistan or other places.
Tumor subtype was determined using ER, PR, Ki67, and HER2 as defined in Materials and Methods.
Nuclear grade was determined by the modified Black’s method.
For the validation cohort (Norway data), 77 (23%) patients developed metastasis: 27 (35%) to be bone and 50 (65%) had first metastasis to non-bone sites. For the 27 patients were designated as first to bone for which 23 patients only had bone as the first metastasis location, four patients with multiple first metastasis locations, which included bone.
Clinical factors predicting risk of bone or non-bone metastasis
Compared with patients without metastasis in the discovery dataset (n = 757), patients who developed metastasis were more likely to have later stage (II), larger tumor size (≥ 2cm), and positive lymph node status (Table 2). In addition, when compared with the no-metastasis group, patients who developed non-bone metastasis were more likely to be younger than age 50 years at diagnosis, to have higher nuclear grade (grade III), and to have undergone chemotherapy, but not endocrine therapy. Further, risk of developing metastasis to bone or non-bone sites appeared to be subtype-specific. For bone metastasis risk was highest for patients with luminal B tumors (HR, 1.92; 95% CI, 1.08–3.39) and lowest for those with TNBC (HR, 0.38; 95% CI, 0.13–1.11), although is marginally significant. For non-bone metastasis, the risk was highest for patients with TNBC (HR, 3.00; 95% CI, 1.81–4.97).
Table 2.
Univariate Cox proportional hazards models of clinical factors predicting risk of bone or non-bone metastasis in patients with early-stage breast cancer
| Variable | Discovery dataset (n = 960)
|
Validation dataset (n =335)
|
||||||
|---|---|---|---|---|---|---|---|---|
| Bone metastasis
|
Non-bone metastasis
|
Bone metastasis
|
Non-bone metastasis
|
|||||
| HR (95% CI) | P value | HR (95% CI) | P value | HR (95% CI) | P value | HR (95% CI) | P value | |
| Tumor subtype* | 0.0076 | <0.0001 | 0.914 | 0.37 | ||||
| Luminal A | Ref. | Ref. | Ref. | Ref. | ||||
| Luminal B | 1.92 (1.08–3.39) | 1.60 (0.92–2.79) | 1.70 (0.34–8.43) | 2.26 (0.78–6.53) | ||||
| HER2+ | 1.56 (0.79–3.06) | 2.74 (1.61–4.68) | 1.08 (0.22–5.34) | 1.33 (0.46–3.84) | ||||
| TNBC | 0.38 (0.13–1.11) | 3.00 (1.81–4.97) | 0.81 (0.10–6.76) | 2.49 (0.92–6.73) | ||||
| Pathologic stage | 0.0006 | <0.0001 | 0.0036 | 0.0007 | ||||
| I | Ref. | Ref. | — | Ref. | ||||
| II | 2.97 (1.60–5.50) | 4.40 (2.53–7.66) | Ref. | 1.87 (0.73–4.78) | ||||
| III | — | — | 3.71 (1.54–8.98) | 5.40 (2.07–14.08) | ||||
| Tumor size | 0.001 | <0.0001 | 0.0035 | 0.025 | ||||
| <2 cm | Ref. | Ref. | Ref. | Ref. | ||||
| ≥2 cm | 2.23 (1.38–3.61) | 2.07 (1.46–2.96) | 4.43 (1.63–12.0) | 1.99 (1.09–3.64) | ||||
| Lymph node status | 0.0004 | <0.0001 | 0.0002 | <0.0001 | ||||
| Positive | Ref. | Ref. | Ref. | Ref. | ||||
| Negative | 0.43 (0.27–0.69) | 0.47 (0.33–0.67) | 0.18 (0.07–0.45) | 0.31 (0.17–0.56) | ||||
| Age at diagnosis | 0.1809 | 0.015 | 0.7459 | 0.17 | ||||
| ≤50 | Ref. | Ref. | Ref. | Ref. | ||||
| >50 | 0.73 (0.46–1.16) | 0.65 (0.46–0.92) | 0.87 (0.36–2.06) | 0.67 (0.37–1.19) | ||||
| Nuclear grade† | 0.29 | 0.038 | 0.9614 | 0.79 | ||||
| I–II | Ref. | Ref. | Ref. | Ref. | ||||
| III | 0.75 (0.44–1.28) | 1.46 (1.02–2.08) | 1.02 (0.41–2.56) | 0.90 (0.46–1.77) | ||||
| Endocrine therapy | 0.3329 | 0.003 | 0.0667 | 0.50 | ||||
| Yes | Ref. | Ref. | Ref. | Ref. | ||||
| No | 1.27 (0.79–2.04) | 1.80 (1.23–2.66) | 0.48 (0.22–1.05) | 0.82 (0.45–1.47) | ||||
Abbreviations: HR, hazard ratio; CI, confidence interval; HER2, human epidermal growth factor receptor 2; TNBC, triple-negative breast cancer.
Tumor subtype was determined using ER, PR, Ki67, and HER2 as defined in Materials and Methods.
Nuclear grade was determined by the modified Black’s method.
CNIs predicting risk of bone or non-bone metastasis
Supplemental Figure 1 shows the pattern of CNIs for the three subgroups: no metastasis, bone metastasis, and non-bone metastasis. Of the 1,593 segments evaluated (representing the entire genome), we identified five CNIs associated with bone metastasis and six CNIs associated with non-bone metastasis (Table 3). Specifically, we identified gains at 1q41, and 1q42.12 and losses at 1p13.3, 8p22, and Xp11.3 were predictors of bone metastasis, and gains at 2p11.2, 3q21.3-22.2, 3q27.1, 10q23.1, and 14q13.2-3 and loss at 7q21.11 were predictors of non-bone metastasis. Gain at 11q13.5 was selected for both bone and non-bone metastasis suggesting that 11q13.5 is acting as a general risk factor for metastasis independent of site. Adjusting the model for HER2 status by different copy number calling thresholds did not change the association between individual CNIs and first site-of-metastasis (data not shown). Supplemental Figure 2 shows the time-to-bone- or -non-bone-metastasis for each individual CNI marker, and the full list of genes in each region is provided in Supplemental Table 1. Among the non-bone metastasis group, gains at 2p11.2, 3q21.3–22.2, 3q27.1, 10q23.1, and 14q13.2–3 were significantly associated with lung metastasis, whereas gain at 2p11.2, 3q21.3–22.2, 3q27.1, 14q13.2-3 and loss at 7q21.11 were significantly associated with brain metastasis, none of the markers is associated with liver metastasis (Table 4). Because of the small sample size within subgroups, and risk of false discovery, these results are highly exploratory, suggesting a role for selective CNIs in different sites of metastases.
Table 3.
Selected copy number imbalances predicting breast cancer bone or non-bone metastasis, from the CoxBoost analysis
| CNI marker | Start (Mp) | Stop (Mp) | Size (Mb) | Discovery dataset
|
Validation dataset
|
Potential target genes# | ||
|---|---|---|---|---|---|---|---|---|
| HR (95% CI) | P value | HR (95% CI) | P value | |||||
| Bone metastasis | ||||||||
| 1p13.3 loss* | 107.2 | 107.8 | 0.6 | 3.54 (1.82–6.89) | 0.0002 | 2.74 (0.82–9.17) | 0.1021 | PRMT6 |
| 1q41 gain* | 212.6 | 213 | 0.4 | 2.58 (1.63–4.09) | 5.39 × 10−5 | 0.89 (0.39–2.05) | 0.7866 | PTPN14 (associated with metastasis) |
| 1q42.12 gain | 223.9 | 224.4 | 0.5 | 2.77 (1.74–4.41) | 1.64 × 10−5 | 1.28 (0.58–2.78) | 0.5415 | EPHX |
| 8p22 loss | 16.9 | 17.2 | 0.3 | 2.90 (1.75–4.82) | 3.57 × 10−5 | 2.27 (1.01–5.12) | 0.0481 | REAM (associated with metastasis), DLC1 (predictor of bone metastasis) |
| 11q13.5 gain | 76.2 | 76.7 | 0.5 | 3.11 (1.79–5.40) | 6.09 × 10−5 | 2.79 (1.11–46.99) | 0.0291 | OMP (close to metastasis genes EMSY and LRRC32) |
| Xp11.3 loss | 45.9 | 46.8 | 0.9 | 4.36 (2.30–8.28) | 6.75 × 10−6 | 1.49 (0.20–11.04) | 0.6951 | |
|
| ||||||||
| Non-bone metastasis | ||||||||
| 2p11.2 gain | 84.4 | 84.8 | 0.4 | 3.19 (1.56–6.52) | 0.0015 | 4.03 (0.98–16.63) | 0.0541 | SUCLG1 |
| 3q21.3–22.2 gain* | 127.9 | 137.4 | 9.4 | 2.89 (1.56–5.37) | 0.0008 | 1.53 (0.37–6.32) | 0.5542 | EPHB1 (associated with metastasis) |
| 3q27.1 gain | 184.7 | 185.1 | 0.4 | 2.53 (1.55–4.11) | 0.0002 | 0.20 (0.03–1.45) | 0.1109 | PARL |
| 7q21.11 loss* | 77.6 | 77.9 | 0.3 | 3.87 (1.70–8.79) | 0.0012 | 0.98 (0.24–4.02) | 0.9747 | MGC34774 |
| 10q23.1 gain | 82.2 | 82.9 | 0.6 | 3.13 (1.68–5.80) | 0.0003 | 6.39 (2.86–14.27) | 5.97 × 10−6 | SH2D4B |
| 11q13.5 gain | 75.7 | 76.2 | 0.5 | 2.32 (1.41–3.82) | 0.0010 | 2.51 (1.25–5.04) | 0.0098 | EMSY (associated with prognosis in breast and ovarian cancers); LRRC32 (associated with metastasis) |
| 14q13.2-3 gain | 35.3 | 36.2 | 0.9 | 2.54 (1.43–4.51) | 0.0014 | 2.68 (0.96–7.51) | 0.0599 | PAX9 |
Abbreviations: CNI, copy number imbalance; HR, hazard ratio; CI, confidence interval.
Only selected genes with biological plausibility are shown in the table. The full list of genes for each region is given in Supplemental Table 1.
CNI markers identified in this study. The other markers (chromosome region) listed have been previously implicated in predicting poor clinical outcome in breast cancer patients.
Table 4.
Selected copy number imbalances predicting breast cancer site specific non-bone metastasis, from the CoxBoost analysis in the discovery
| CNI marker | Lung metastasis (n = 59)
|
Liver metastasis (n = 35)
|
Brain metastasis (n = 47) #
|
|||
|---|---|---|---|---|---|---|
| HR (95% CI) | P value | HR (95% CI) | P value | HR (95% CI) | P value | |
| 2p11.2 gain | 4.56 (1.83–11.42) | 0.0012 | 1.75 (0.24–11.81) | 0.5811 | 5.43 (2.14–13.75) | 0.0004 |
| 3q21.3–22.2 gain | 4.90 (2.32–10.32) | 2.96 × 10−5 | 0 (0-Infinity) | — | 4.87 (2.06–11.51) | 0.0003 |
| 3q27.1 gain | 3.50 (1.82–6.74) | 0.0002 | 1.00 (0.24–4.18) | 0.9965 | 4.54 (2.31–8.92) | 1.16 × 10−5 |
| 7q21.11 loss | 3.13 (0.76–12.83) | 0.113 | 2.97 (0.41–21.71) | 0.2839 | 4.31 (1.04–17.81) | 0.0435 |
| 10q23.1 gain | 4.03 (1.73–9.37) | 0.0012 | 2.22 (0.53–9.28) | 0.2734 | 2.34 (0.73–7.56) | 0.154 |
| 11q13.5 gain | 1.45 (0.58–3.63) | 0.4281 | 2.40 (0.93–6.2) | 0.07 | 3.64 (1.76–7.56) | 0.0005 |
| 14q13.2–3 gain | 3.94 (1.93–8.01) | 0.0002 | 1.53 (0.37–6.38) | 0.5608 | 3.05 (1.20–7.72) | 0.0187 |
Abbreviations: CNI, copy number imbalance; HR, hazard ratio; CI, confidence interval.
Because there were only 9 patients with first metastasis to brain, we expanded this group to include those with any brain metastasis to improve the power.
Prediction models of bone or non-bone metastasis
Tumor subtype, tumor size, endocrine therapy, lymph node status, and copy number losses at 8p22 and Xp11.3 were significantly associated with bone metastasis, whereas tumor subtype, stage, and copy number loss at 7q21.11 were significantly associated with non-bone metastasis (Table 5). The strongest determinant of bone metastasis was copy number loss at 8p22 (posterior probability = 100%; Table 5). The strongest determinants of non-bone metastasis were tumor subtype and stage (both posterior probabilities = 100%). Among the non-bone metastasis group, tumor subtype, stage, and gains at 3q21.3-22.2, 10q23.1, and 14q13.2-3 and loss at 7q21.11 were significantly associated with lung metastasis; stage was significantly associated with liver metastasis (Table 6).
Table 5.
Multivariate Cox proportional hazards regression model for risk of bone or non-bone metastasis in early-stage breast cancer patients
| Variable | Discovery dataset (n = 960)
|
Validation dataset (n = 335)
|
||||
|---|---|---|---|---|---|---|
| HR (95% CI) | P value | Posterior | HR (95% CI) | P value | Posterior | |
| Bone metastasis | ||||||
| Tumor subtype† | 0.0233 | 30.4 | 0.7136 | 0.4 | ||
| Luminal A | Ref. | Ref. | ||||
| Luminal B | 1.39 (0.75–2.55) | 0.67 (0.08–5.40) | ||||
| HER2+ | 0.85 (0.39–1.82) | 0.56 (0.05–6.46) | ||||
| TNBC | 0.27 (0.09–0.81) | 0.23 (0.02–2.76) | ||||
| Tumor size | 0.0456 | 56.0 | 0.2576 | 43.9 | ||
| < 2 cm | Ref. | Ref. | ||||
| ≥ 2 cm | 1.71 (1.01–2.89) | 2.54 (0.51–12.82) | ||||
| Endocrine | 0.0209 | 51.8 | 0.2210 | 27.5 | ||
| Yes | Ref. | Ref. | ||||
| No | 1.93 (1.11–3.38) | 3.93 (0.44–35.12) | ||||
| Lymph node status | 0.0066 | 94.2 | 0.0371 | 69.7 | ||
| Positive | Ref. | Ref. | ||||
| Negative | 0.48 (0.29–0.82) | 0.10 (0.01–0.87) | ||||
| CNI marker | ||||||
| 8p22 loss | 2.55 (1.44–4.53) | 0.0013 | 100.0 | 5.61 (1.00–31.60) | 0.0503 | 78.8 |
| 11q13.5 gain | 2.09 (1.11–3.94) | 0.0225 | 82.9 | 3.22 (0.51–20.30) | 0.2125 | 44.5 |
| Xp11.3 loss | 2.13 (0.94–4.82) | 0.0684 | 38.7 | 14.08 (0.88–125.26) | 0.0616 | 58.7 |
|
| ||||||
| C-index for clinical factors only | 0.711 | 0.784 | ||||
| C-index for CNIs only | 0.627 | 0.728 | ||||
| C-index for the full model | 0.750 | 0.859 | ||||
|
| ||||||
| Non-bone metastasis | ||||||
| Tumor | 0.0004 | 100 | 0.7804 | 0 | ||
| Luminal A | Ref. | Ref. | ||||
| Luminal B | 1.44 (0.82–2.51) | 1.04 (0.24–4.52) | ||||
| HER2+ | 2.49 (1.46–4.25) | 0.37 (0.05–3.00) | ||||
| TNBC | 2.64 (1.59–4.39) | 1.18 (0.38–3.71) | ||||
| Stage | 1.84 × 10−6 | 100 | 0.1552 | 24.1 | ||
| I | Ref. | Ref. | ||||
| II | 4.07 (2.29–7.24) | 1.37 (0.36–5.24) | ||||
| III | — | 3.49 (0.82–14.80) | ||||
| CNI marker | ||||||
| 7q21.11 loss | 4.15 (1.80–9.54) | 0.0008 | 82.0 | 2.62 (0.54–12.70) | 0.2330 | 28.2 |
| 11q13.5 gain | 2.07 (1.19–3.58) | 0.0098 | 61.1 | 2.96 (1.04–8.41) | 0.0422 | 87.7 |
|
| ||||||
| C-index for clinical factors only | 0.699 | 0.669 | ||||
| C-index for CNIs only | 0.548 | 0.679 | ||||
| C-index for the full model | 0.712 | 0.759 | ||||
Note: The clinical covariates shown were selected by a stepwise model selection procedure that minimizes the Akaike information criterion.
Abbreviations: CNI, copy number imbalance; HR, hazard ratio; CI, confidence interval; HER2, human epidermal growth factor receptor 2; TNBC, triple-negative breast cancer.
The posterior probability was calculated by the Bayesian model averaging algorithm. A higher posterior probability score indicates stronger effect in the model.
Tumor subtype was determined using ER, PR, Ki67, and HER2 as defined in Materials and Methods.
Table 6.
Multivariate Cox proportional hazards regression model for risk of non-bone (lung, liver, brain) metastasis in early-stage breast cancer patients in the discovery
| Variable | Lung metastasis (n = 59) | Liver metastasis (n = 35) | Brain metastasis (n = 47)# | ||||||
|---|---|---|---|---|---|---|---|---|---|
|
| |||||||||
| HR (95% CI) | P value | Posterior probability (%)* | HR (95% CI) | P value | Posterior probability (%)* | HR (95% CI) | P value | Posterior probability (%)* | |
| Tumor Subtype† | 0.0035 | 98.7 | 0.5008 | 0 | 0.0573 | 46.0 | |||
| Luminal A | Ref. | Ref. | Ref. | ||||||
| Luminal B | 2.25 (0.87–5.84) | 1.33 (0.49–3.60) | 2.78 (0.97–7.98) | ||||||
| Her2+ | 3.73 (1.51–9.24) | 1.95 (0.72–5.27) | 3.23 (1.06–9.82) | ||||||
| TNBC | 4.59 (1.92–10.9) | 1.90 (0.70–5.14) | 4.20 (1.49–11.82) | ||||||
| Pathologic stage | 0.0046 | 100 | 0.010 | 99.1 | 0.0675 | 55.7 | |||
| I | Ref. | Ref. | Ref. | ||||||
| II | 3.19 (1.43–7.11) | 4.04 (1.40–11.69) | 2.01 (0.95–4.26) | ||||||
| CNI marker | |||||||||
| 2p11.2 gain | 1.30 (0.45–3.76) | 0.6235 | 10.7 | 1.06 (0.13–8.68) | 0.9550 | 9.7 | 2.08 (0.68–6.39) | 0.1989 | 38.6 |
| 3q21.3–22.2 gain | 3.25 (1.10–9.61) | 0.0334 | 90.1 | 0 (0–Infinity) | 0.9963 | 24.6 | 1.81 (0.49–6.61) | 0.3722 | 27.8 |
| 3q27.1 gain | 1.19 (0.43–3.12) | 0.7355 | 15.0 | 1.32 (0.28–6.20) | 0.7226 | 9.6 | 1.77 (0.62–5.07) | 0.2882 | 73.6 |
| 7q21.11 loss | 4.47 (1.07–18.7) | 0.0401 | 32.6 | 7.09 (0.91–55.1) | 0.0611 | 31.3 | 3.60 (0.72–17.98) | 0.1180 | 29.9 |
| 10q23.1 gain | 3.45 (1.42–8.40) | 0.0064 | 79.2 | 0.93 (0.13–6.98) | 0.9471 | 9.4 | 0.71 (0.14–3.67) | 0.6803 | 5.5 |
| 11q13.5 gain | 0.94 (0.33–2.68) | 0.9024 | 6.4 | 2.69 (1.02–7.10) | 0.0454 | 44.4 | 2.56 (1.08–6.06) | 0.0324 | 52.3 |
| 14q13.2–3 gain | 2.52 (1.14–5.59) | 0.0229 | 61.8 | 1.56 (0.36–6.72) | 0.5520 | 11.3 | 1.63 (0.52–5.10) | 0.3999 | 18.9 |
Note: Clinical covariates shown were selected by a stepwise selection procedure that minimizes Akaike information criterion, except for age at diagnosis.
Abbreviations: CNI, copy number imbalance; HR, hazard ratio; CI, confidence interval; HER2, human epidermal growth factor receptor 2; PP, posterior probability; TNBC, triple-negative breast cancer.
The posterior probability was calculated by the Bayesian model averaging algorithm. A higher posterior probability score indicates stronger effect in the model.
Tumor subtype was determined using ER, PR, Ki67, and HER2 as defined in Materials and Methods.
Because there were only nine patients with first metastasis to brain, we expanded this group to include those with any brain metastasis.
To assess the independent prognostic performance of the CNIs for risk of metastasis, we compared the CNIs only, clinical only, and both clinical and CNIs models. The full model incorporating the clinical and CNIs performed the best, C-index were 0.750 and 0.712, for bone and non-bone metastasis, respectively. For risk of bone metastasis, the clinical only model (C-index = 0.711) is better than the CNIs only model (C-index = 0.627), and similarly, the clinical factors is stronger (C-index = 0.699) than the CNIs only model (C-index = 0.548) for the non-bone metastasis.
Since subtypes are strongly associated with site-specific metastasis, we repeated the same analyses within each subtype. While some of the markers are more common within particular subtypes, CNIs largely occur independently of the defined subtype (Supplemental Table 2). Further, to assess the potential effect of changes in treatment, we conducted sensitivity analyses, stratifying the cohort by time (before/after year 1994). We see no evidence that change in treatment practice modified the association between specific CNIs and risk for first site of metastasis (Supplemental Tables 3 and 4), suggesting that these findings are relevant to contemporary treatment. We also assessed the sensitivity of our models to patients with site of first metastasis versus ‘multiple site of metastasis’ by excluding them from the bone as first as well as including them in the non-bone as first site and found no effect on the performance of either model.
Cumulative risk prediction of bone or non-bone metastasis
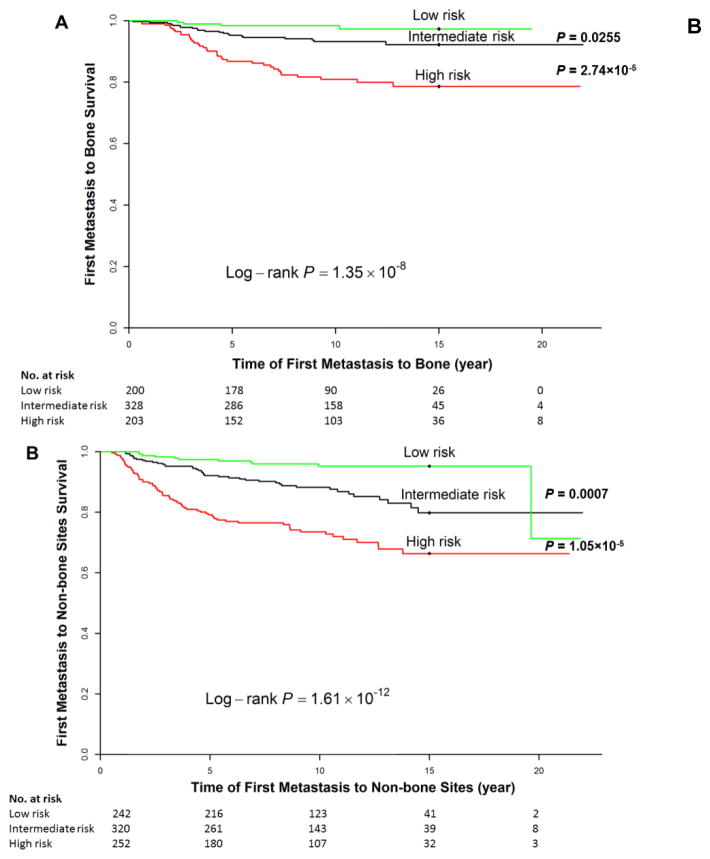
To evaluate joint effects of CNIs and clinical covariates on risk of bone or non-bone metastasis, we used the sum of the predicted linear HRs from the final multivariate model to stratify patients into three risk groups: high-risk group (the highest 25% of linear HRs), intermediate-risk group (the middle 50% of linear HRs), or low-risk group (the lowest 25% of linear HRs), denoting poor, intermediate, and good prognosis, respectively. The difference in risk of bone (Figure 1A) or non-bone metastasis (Figure 1B) among the three groups was highly significant (log-rank test, P = 1.35×10−8 and 1.61×10−12 for bone and non-bone metastasis, respectively). In particular, the 15-year probability of bone metastasis-free survival in the low-, intermediate-, and high-risk group was 97.3%, 92.2%, and 78.6%, respectively; for non-bone, survival probability in the low-, intermediate-, and high-risk group was 95.2%, 79.8%, and 66.4%, respectively. When we used the intermediate-risk group as reference, the HR for bone metastasis was 0.32 (0.11–0.92) for the low-risk group and 2.99 (1.74–5.11) for the high-risk group; for non-bone metastasis, the HR was 0.34 (0.17–0.66) for the low-risk group and 2.33 (1.59–3.43) for the high-risk group.
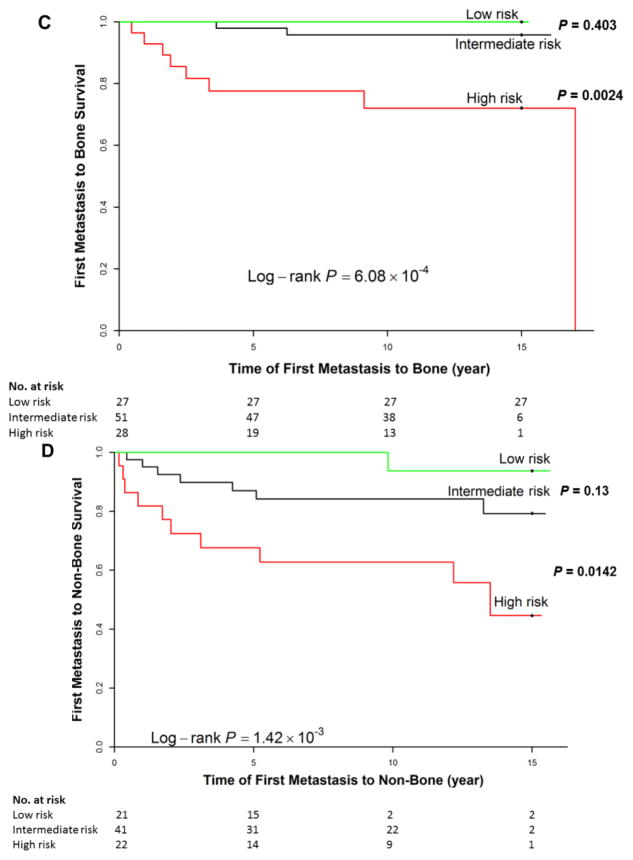
Figure 1. Cumulative risk of bone (A, C) or non-bone (B, D) metastasis according to the full model, including clinical factors and copy number imbalances, in the discovery dataset (A, B) and validation dataset (C, D).
We stratified patients into three risk groups based on the sum of the predicted linear hazard ratios (HRs) from the final multivariate model (see Materials and Methods). Patients with higher predicted linear HRs have a higher risk of developing bone or non-bone metastasis at a given time point. Note that the numbers shown do not add up to the total number of patients in each specific subgroup due to missing data for clinical factors or outcome (time-to-metastasis).
Validation
In the external validation dataset, compared to patients with no metastasis (n = 259), patients who developed metastasis had later stage disease, size ≥ 2 cm, and positive lymph node status (Table 2). Of the 12 CNIs selected in the discovery, and loss at 8p22 (P = 0.048) was significant for bone metastasis, while gains at 10q23.1 (P < 0.001) was significant for non-bone metastasis; and gain at 11q13.5 was significant for both bone (P = 0.029) and non-bone (P = 0.010) metastasis. The C-index of the full models incorporating the clinical and CNIs were 0.859 and 0.759, for bone and non-bone metastasis, respectively (Table 3).
Because of the smaller size of the validation cohort, other CNIs were not significantly associated with metastasis, although the direction and magnitude of risk replicated those observed in the discovery set. Restricting analyses to cases with known stage (n = 209) or those with high-quality aCGH data did not substantially change the associations. For cumulative risk prediction, the risk of bone (Figure 1C) or non-bone (Figure 1D) metastasis was significantly different among the three risk groups (log-rank test, P = 6.08×10−4 and 1.42×10−3 for bone and non-bone metastasis, respectively). Compared with the intermediate-risk group, the HR for the high-risk group was 7.85 (1.63–37.9) for bone metastasis and 3.17 (1.20–8.33) for non-bone metastasis.
Discussion
This study provides one of the first global, high-resolution genome-wide DNA CNI profiles of site-specific metastasis in early-stage breast cancer patients. In the discovery dataset, we found copy number gains at 1q41 and 1q42.12 and losses at 1p13.3, 8p22, and Xp11.3 independently increased risk of bone metastasis. Further, gains at 2p11.2, 3q21.3–22.2, 3q27.1, 10q23.1, and 14q13.2–3 and loss at 7q21.11 were associated with non-bone metastasis. When evaluated in an external dataset, loss at 8p22 acting as a bone-specific marker and gain at 10q23.1 as a non-bone specific marker. Copy number gain at 11q13.5 was significantly associated with risk for both bone and non-bone metastases in the two study populations, which suggest that 11q13.5 gain increases the propensity for metastasis independent of site.
Of the twelve CNIs identified as predictors of bone or non-bone metastasis, gains at 10q23.1 [14], 11q13.5 [18–20,14], and 14q13.2-3 [14] and loss at 8p22 [14,4,21–23] have previously been implicated in metastasis and worse patient outcomes. Putative candidate genes at 11q13.5 include EMSY, which encodes a BRCA2-associated protein, and LRRC32 (leucine rich repeat containing 32), and gains in both have been associated with poor prognosis in a variety of human cancers [19,24–26,20,27,28]. Of interest, amplification on 11q13.5 is a feature of a subset of ER+ breast carcinomas prone to metastasis [20]. Similarly, 8p22 is commonly deleted in metastatic tumors [14,29–31]. Two metastasis suppressor genes have been identified at 8p22: REAM (reduced expression associated with metastasis) [14] and DLC1 (Deleted in Liver Cancer 1) [32,22]. In a clonal model of experimental organ-specific metastasis, DLC1 was down-regulated in breast cancer cells that were highly metastatic to the bone [4]; supporting our observed association between 8p22 loss and bone metastasis in two cohorts.
To our knowledge, this is the first study to identify gain at 1q41 and loss at 1p13.3 for bone metastasis and gain at 3q21.3–22.2 and loss at 7q21.11 for non-bone metastasis. While we failed to replicate theses associations, prior studies support a role for genes in the region and metastasis risk. For example, PTPN14 (protein tyrosine phosphatase non-receptor type 14) at 1q41 has been associated with tumor progression [33] acting via phosphorylation of adherens junctions and facilitating tumor motility and migration [34]. EPHB1 (a member of the Ephrin receptor family) at 3q21.3–22.2 has been correlated with invasion, stage, and metastasis in colorectal cancer [35,36] and shown to influence cell-cell interaction and cell migration in response to environmental signals [29,37].
A number of studies have investigated putative associations between gene expression profiles and metastasis to the bone [4], lung [11], and brain [10]. However, the numbers of patients in these studies is small, and none have explored CNIs as predictors of site of metastasis. The strength of our study is the large sample size and cases from a single treatment center with validated long-term follow-up, including first site of metastasis. There are few studies with site of metastasis and none to our knowledge of this size with the copy number data because of the rarity of the metastasis events and the need to follow patient long term. These two retrospective sample sets (discovery and validation) are unique in that they have collected patient outcomes by site of metastasis. Further, we found strong associations between certain CNIs and site of metastasis that can be replicated across sample sets. Notably, we also found that the metastasis associated CNIs, while not equally distributed, were present across all breast tumor subtypes whether defined by IHC as in the MDACC cohort or by gene expression profiling as in the validation study (Supplemental Table 2). These results suggest that measurement of CNIs may further enhance discrimination of high risk tumors within subtypes.
A limitation of our study is the smaller size of the validation data-set, as well as population heterogeneity (MDACC included non-Hispanic white, Hispanic, and African American patients, whereas the vast majority Swedish/Norwegian patients were non-Hispanic white (only a few immigrants with background from Pakistan or other countries) as well as differences in the resolution of the aCGH and MIP platforms. A further potential limitation is the relevance of outcomes in the context of current treatment protocols. Routine use of an anthracycline with cyclophosphamide (AC) at MDACC began in the 1980s, whereas the first use of AC plus taxane was observed in our cohort in 1986; use of AC plus taxane (now standard of care) was routine by 1994. Among patients receiving chemotherapy, use of AC plus taxane was 5.3% prior to 1994 and 42.3% after 1994. Among patients in our study receiving chemotherapy, 73.7% received a regimen containing an anthracycline and the remainder (26.3%) received both an anthracycline and a taxane. Our sensitivity analyses stratifying the cohort by time (before/after year 1994) shown no evidence that change in treatment practice modified the association between specific CNIs and risk for first site of metastasis.
In summary, we identified specific CNIs that increase risk for bone and/or non-bone metastasis, independently of established risk factors like tumor subtype. Validation of our findings show gain at 11q13.5, loss at 8p22, and gain at 10q23.1, and possibly loss at Xp11.3, are informative as markers of site of first metastasis. These results strongly suggest that these markers, or their underlying genomic drivers, have potential clinical use as biomarkers to tailor post-treatment surveillance and to identify high-risk patients, who would be most likely to benefit from prolonged therapy with treatments given in the adjuvant setting for the purpose of preventing bone or soft tissue metastasis.
Supplementary Material
Acknowledgments
Grant Support: This study was supported by the National Institutes of Health (NIH) through grant R01CA089608. Additional support was provided by Susan G. Komen for the Cure, by the National Breast Cancer Foundation, and by NIH through SPORE P50CA116199, MD Anderson’s Cancer Center Support Grant (CCSG) CA016672, and the Arizona Cancer Center’s CCSG CA023074. Grants to ALBD lab supporting this study: Norwegian Research Council grants 175240/S10, 159188/S10, and 193387/V50, Norwegian Cancer Society grant 138296-PR-2008-0108.
We thank Melissa May, Dr. Jeong Yun Shim, and Dr. Agbe Samuel for retrieving and processing all the tumor specimens used in the study; Wanda Williams, who supervised the medical record abstraction; Phyllis Adatto, who supervised the study staff; and Betsy C. Wertheim for careful review and editing of the manuscript. For providing tumor specimens and clinical data, the authors gratefully acknowledge for the MicMa cohort, Dr. Bjørn Naume and Dr. Marit Synnestvedt; for the ULL cohort, Dr. Anita Langerød; for the MDG cohort, Dr. Vilde D. Haakensen and Dr. Åslaug Helland, all Oslo University Hospital, Norway. For the Uppsala cohort we wish to thank Dr. Johan Botling, Department of Surgery, Uppsala University, Sweden. For technical support performing the aCGH experiments we wish to thank Dr. Hans Kristian M. Vollan, Ida J. Schneider, and Eldri U. Due, Oslo University Hospital, Norway.
Footnotes
Disclosure of Potential Conflicts of Interest: No potential conflicts of interest were disclosed.
References
- 1.American Cancer Society. Breast Cancer Facts & Figures 2009–2010. (Available at: http://www.cancer.org/acs/groups/content/@nho/documents/document/f861009final90809pdf.pdf)
- 2.Coleman RE. Clinical features of metastatic bone disease and risk of skeletal morbidity. Clin Cancer Res. 2006;12 (20 Pt 2):6243s–6249s. doi: 10.1158/1078-0432.CCR-06-0931. 12/20/6243s [pii] [DOI] [PubMed] [Google Scholar]
- 3.Paget S. The distribution of secondary growths in cancer of the breast. 1889. Cancer Metastasis Rev. 1989;8 (2):98–101. [PubMed] [Google Scholar]
- 4.Kang Y, Siegel PM, Shu W, Drobnjak M, Kakonen SM, Cordon-Cardo C, Guise TA, Massague J. A multigenic program mediating breast cancer metastasis to bone. Cancer Cell. 2003;3(6):537–549. doi: 10.1016/s1535-6108(03)00132-6. S1535610803001326 [pii] [DOI] [PubMed] [Google Scholar]
- 5.Ramaswamy S, Ross KN, Lander ES, Golub TR. A molecular signature of metastasis in primary solid tumors. Nat Genet. 2003;33(1):49–54. doi: 10.1038/ng1060. ng1060 [pii] [DOI] [PubMed] [Google Scholar]
- 6.Evans AJ, James JJ, Cornford EJ, Chan SY, Burrell HC, Pinder SE, Gutteridge E, Robertson JF, Hornbuckle J, Cheung KL. Brain metastases from breast cancer: identification of a high-risk group. Clin Oncol (R Coll Radiol) 2004;16 (5):345–349. doi: 10.1016/j.clon.2004.03.012. [DOI] [PubMed] [Google Scholar]
- 7.Duchnowska R, Szczylik C. Central nervous system metastases in breast cancer patients administered trastuzumab. Cancer Treat Rev. 2005;31 (4):312–318. doi: 10.1016/j.ctrv.2005.04.008. S0305-7372(05)00082-4 [pii] [DOI] [PubMed] [Google Scholar]
- 8.Hicks DG, Short SM, Prescott NL, Tarr SM, Coleman KA, Yoder BJ, Crowe JP, Choueiri TK, Dawson AE, Budd GT, Tubbs RR, Casey G, Weil RJ. Breast cancers with brain metastases are more likely to be estrogen receptor negative, express the basal cytokeratin CK5/6, and overexpress HER2 or EGFR. Am J Surg Pathol. 2006;30(9):1097–1104. doi: 10.1097/01.pas.0000213306.05811.b9. 00000478-200609000-00005 [pii] [DOI] [PubMed] [Google Scholar]
- 9.Nam BH, Kim SY, Han HS, Kwon Y, Lee KS, Kim TH, Ro J. Breast cancer subtypes and survival in patients with brain metastases. Breast Cancer Res. 2008;10 (1):R20. doi: 10.1186/bcr1870. bcr1870 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bos PD, Zhang XH, Nadal C, Shu W, Gomis RR, Nguyen DX, Minn AJ, van de Vijver MJ, Gerald WL, Foekens JA, Massague J. Genes that mediate breast cancer metastasis to the brain. Nature. 2009;459 (7249):1005–1009. doi: 10.1038/nature08021. nature08021 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Minn AJ, Gupta GP, Siegel PM, Bos PD, Shu W, Giri DD, Viale A, Olshen AB, Gerald WL, Massague J. Genes that mediate breast cancer metastasis to lung. Nature. 2005;436 (7050):518–524. doi: 10.1038/nature03799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Singletary SE, Walsh G, Vauthey JN, Curley S, Sawaya R, Weber KL, Meric F, Hortobagyi GN. A role for curative surgery in the treatment of selected patients with metastatic breast cancer. Oncologist. 2003;8 (3):241–251. doi: 10.1634/theoncologist.8-3-241. [DOI] [PubMed] [Google Scholar]
- 13.Brewster AM, Do KA, Thompson PA, Hahn KM, Sahin AA, Cao Y, Stewart MM, Murray JL, Hortobagyi GN, Bondy ML. Relationship between epidemiologic risk factors and breast cancer recurrence. J Clin Oncol. 2007;25 (28):4438–4444. doi: 10.1200/JCO.2007.10.6815. JCO.2007.10.6815 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Thompson PA, Brewster AM, Kim-Anh D, Baladandayuthapani V, Broom BM, Edgerton ME, Hahn KM, Murray JL, Sahin A, Tsavachidis S, Wang Y, Zhang L, Hortobagyi GN, Mills GB, Bondy ML. Selective genomic copy number imbalances and probability of recurrence in early-stage breast cancer. PloS one. 2011;6 (8):e23543. doi: 10.1371/journal.pone.0023543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Binder H, Schumacher M. Allowing for mandatory covariates in boosting estimation of sparse high-dimensional survival models. BMC Bioinformatics. 2008;9:14. doi: 10.1186/1471-2105-9-14. 1471-2105-9-14 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hoeting J, Madigan D, Raftery AE, Volinsky C. Bayesian Model Averaging: A Tutorial Statistical Science. 1999;14:382–417. [Google Scholar]
- 17.Volinsky C, Madigan D, Raftery A, Kronmal R. Bayesian model averaging in proportional hazard models: assessing the risk of a stroke. Applied Statistics. 1997;46:433–448. [Google Scholar]
- 18.Rodriguez C, Hughes-Davies L, Valles H, Orsetti B, Cuny M, Ursule L, Kouzarides T, Theillet C. Amplification of the BRCA2 pathway gene EMSY in sporadic breast cancer is related to negative outcome. Clin Cancer Res. 2004;10(17):5785–5791. doi: 10.1158/1078-0432.CCR-03-0410. 10/17/5785 [pii] [DOI] [PubMed] [Google Scholar]
- 19.Szepetowski P, Ollendorff V, Grosgeorge J, Courseaux A, Birnbaum D, Theillet C, Gaudray P. DNA amplification at 11q13.5-q14 in human breast cancer. Oncogene. 1992;7 (12):2513–2517. [PubMed] [Google Scholar]
- 20.Bekri S, Adelaide J, Merscher S, Grosgeorge J, Caroli-Bosc F, Perucca-Lostanlen D, Kelley PM, Pebusque MJ, Theillet C, Birnbaum D, Gaudray P. Detailed map of a region commonly amplified at 11q13-->q14 in human breast carcinoma. Cytogenet Cell Genet. 1997;79 (1–2):125–131. doi: 10.1159/000134699. [DOI] [PubMed] [Google Scholar]
- 21.Hirano A, Emi M, Tsuneizumi M, Utada Y, Yoshimoto M, Kasumi F, Akiyama F, Sakamoto G, Haga S, Kajiwara T, Nakamura Y. Allelic losses of loci at 3p25.1, 8p22, 13q12, 17p13.3, and 22q13 correlate with postoperative recurrence in breast cancer. Clin Cancer Res. 2001;7 (4):876–882. [PubMed] [Google Scholar]
- 22.Utada Y, Haga S, Kajiwara T, Kasumi F, Sakamoto G, Nakamura Y, Emi M. Allelic loss at the 8p22 region as a prognostic factor in large and estrogen receptor negative breast carcinomas. Cancer. 2000;88(6):1410–1416. doi: 10.1002/(SICI)1097-0142(20000315)88:6<1410::AID-CNCR19>3.0.CO;2-X. [pii] [DOI] [PubMed] [Google Scholar]
- 23.Dahmane N, Sanchez P, Gitton Y, Palma V, Sun T, Beyna M, Weiner H, Ruiz i Altaba A. The Sonic Hedgehog-Gli pathway regulates dorsal brain growth and tumorigenesis. Development. 2001;128 (24):5201–5212. doi: 10.1242/dev.128.24.5201. [DOI] [PubMed] [Google Scholar]
- 24.Liu CJ, Lin SC, Chen YJ, Chang KM, Chang KW. Array-comparative genomic hybridization to detect genomewide changes in microdissected primary and metastatic oral squamous cell carcinomas. Mol Carcinog. 2006;45 (10):721–731. doi: 10.1002/mc.20213. [DOI] [PubMed] [Google Scholar]
- 25.Martinez-Cardus A, Martinez-Balibrea E, Bandres E, Malumbres R, Gines A, Manzano JL, Taron M, Garcia-Foncillas J, Abad A. Pharmacogenomic approach for the identification of novel determinants of acquired resistance to oxaliplatin in colorectal cancer. Mol Cancer Ther. 2009;8 (1):194–202. doi: 10.1158/1535-7163.MCT-08-0659. 8/1/194 [pii] [DOI] [PubMed] [Google Scholar]
- 26.Lassmann S, Weis R, Makowiec F, Roth J, Danciu M, Hopt U, Werner M. Array CGH identifies distinct DNA copy number profiles of oncogenes and tumor suppressor genes in chromosomal- and microsatellite-unstable sporadic colorectal carcinomas. J Mol Med (Berl) 2007;85 (3):293–304. doi: 10.1007/s00109-006-0126-5. [DOI] [PubMed] [Google Scholar]
- 27.Maire G, Forus A, Foa C, Bjerkehagen B, Mainguene C, Kresse SH, Myklebost O, Pedeutour F. 11q13 alterations in two cases of hibernoma: large heterozygous deletions and rearrangement breakpoints near GARP in 11q13.5. Genes Chromosomes Cancer. 2003;37 (4):389–395. doi: 10.1002/gcc.10223. [DOI] [PubMed] [Google Scholar]
- 28.Hughes-Davies L, Huntsman D, Ruas M, Fuks F, Bye J, Chin SF, Milner J, Brown LA, Hsu F, Gilks B, Nielsen T, Schulzer M, Chia S, Ragaz J, Cahn A, Linger L, Ozdag H, Cattaneo E, Jordanova ES, Schuuring E, Yu DS, Venkitaraman A, Ponder B, Doherty A, Aparicio S, Bentley D, Theillet C, Ponting CP, Caldas C, Kouzarides T. EMSY links the BRCA2 pathway to sporadic breast and ovarian cancer. Cell. 2003;115(5):523–535. doi: 10.1016/s0092-8674(03)00930-9. S0092867403009309. [pii] [DOI] [PubMed] [Google Scholar]
- 29.Surawska H, Ma PC, Salgia R. The role of ephrins and Eph receptors in cancer. Cytokine & growth factor reviews. 2004;15 (6):419–433. doi: 10.1016/j.cytogfr.2004.09.002. [DOI] [PubMed] [Google Scholar]
- 30.Liu J, Ghanim M, Xue L, Brown CD, Iossifov I, Angeletti C, Hua S, Negre N, Ludwig M, Stricker T, Al-Ahmadie HA, Tretiakova M, Camp RL, Perera-Alberto M, Rimm DL, Xu T, Rzhetsky A, White KP. Analysis of Drosophila segmentation network identifies a JNK pathway factor overexpressed in kidney cancer. Science. 2009;323 (5918):1218–1222. doi: 10.1126/science.1157669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lee J, Sayegh J, Daniel J, Clarke S, Bedford MT. PRMT8, a new membrane-bound tissue-specific member of the protein arginine methyltransferase family. The Journal of biological chemistry. 2005;280 (38):32890–32896. doi: 10.1074/jbc.M506944200. [DOI] [PubMed] [Google Scholar]
- 32.Goodison S, Yuan J, Sloan D, Kim R, Li C, Popescu NC, Urquidi V. The RhoGAP protein DLC-1 functions as a metastasis suppressor in breast cancer cells. Cancer Res. 2005;65 (14):6042–6053. doi: 10.1158/0008-5472.CAN-04-3043. 65/14/6042 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang Z, Shen D, Parsons DW, Bardelli A, Sager J, Szabo S, Ptak J, Silliman N, Peters BA, van der Heijden MS, Parmigiani G, Yan H, Wang TL, Riggins G, Powell SM, Willson JK, Markowitz S, Kinzler KW, Vogelstein B, Velculescu VE. Mutational analysis of the tyrosine phosphatome in colorectal cancers. Science. 2004;304 (5674):1164–1166. doi: 10.1126/science.1096096. [DOI] [PubMed] [Google Scholar]
- 34.Niedergethmann M, Alves F, Neff JK, Heidrich B, Aramin N, Li L, Pilarsky C, Grutzmann R, Allgayer H, Post S, Gretz N. Gene expression profiling of liver metastases and tumour invasion in pancreatic cancer using an orthotopic SCID mouse model. British journal of cancer. 2007;97 (10):1432–1440. doi: 10.1038/sj.bjc.6604031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Batlle E, Bacani J, Begthel H, Jonkheer S, Gregorieff A, van de Born M, Malats N, Sancho E, Boon E, Pawson T, Gallinger S, Pals S, Clevers H. EphB receptor activity suppresses colorectal cancer progression. Nature. 2005;435 (7045):1126–1130. doi: 10.1038/nature03626. nature03626 [pii] [DOI] [PubMed] [Google Scholar]
- 36.Sheng Z, Wang J, Dong Y, Ma H, Zhou H, Sugimura H, Lu G, Zhou X. EphB1 is underexpressed in poorly differentiated colorectal cancers. Pathobiology. 2008;75 (5):274–280. doi: 10.1159/000151707. 000151707 [pii] [DOI] [PubMed] [Google Scholar]
- 37.Nakamoto M, Bergemann AD. Diverse roles for the Eph family of receptor tyrosine kinases in carcinogenesis. Microsc Res Tech. 2002;59 (1):58–67. doi: 10.1002/jemt.10177. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.