The genomes of many species, including man, have been laid bare with hundreds of known and thousands of unknown genes. A major challenge in present day biology is to decipher the molecular function of the unknown genes. Gene knockout by homologous recombination has proven to be very useful but is laborious and expensive. Antisense and ribozyme technology have been useful but are not always reliable or robust. RNA interference has emerged as a novel pathway that offers great hope and promise to study the functions of a vast number of genes. The advent of this technology could not have come at a better time. Briefly, in invertebrates, long double-stranded RNA molecules are processed by the endonuclease dicer into 21- to 23-nt small interfering RNAs (siRNAs), which are then incorporated into RNA-induced silencing complex, a multicomponent nuclease complex that selects and degrades mRNAs that are homologous to the initially delivered double-stranded RNA (1, 2). In mammalian systems, introduction of long double-stranded RNA (>50 bp) results in systemic, nonspecific inhibition of translation due to activation of the PKR response. This formidable obstacle can be overcome by the use of synthetic siRNAs (<30 bp) that can be either delivered exogenously (3) or expressed endogenously from RNA polymerase III promoters, resulting in a powerful tool for achieving specific down-regulation of target mRNAs (4–8). Currently, in mammalian systems, the only way to generate a whole-genome knockdown screening by using RNA interference is to systematically generate synthetic or polymerase III-transcribed siRNA hairpins for every known target gene. Because only 25% of selected target siRNA sequences are functional, several synthetic siRNAs need to be generated and tested for every target gene. Algorithms are being developed to predict effective siRNA sequences for efficient screening of large numbers of genes, but presently they have not been validated and may require automation (9).
siRNA Library
Fortunately, Luo et al. (10), in this issue of PNAS, and two other groups (11, 12) have now described a method for generating an enzymatically synthesized library of siRNA hairpin cassettes obtained from a source of double-stranded (ds)DNA. Such a library will allow the selection of active siRNAs to any target gene, known or unknown, as long as it was represented in the mRNA source of the library. Furthermore, such a library will allow the identification of many new gene functions by using the right selection pressure after introduction of the siRNA library into the cells.
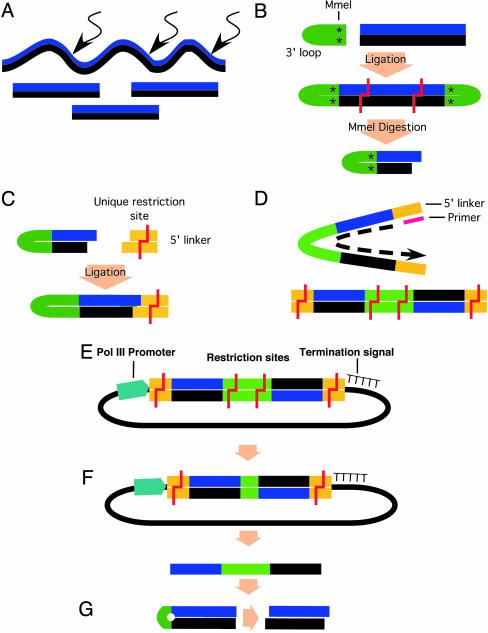
The basic outline of the process (Fig. 1) is as follows. First, dsDNA was synthesized from a source of mRNA or a cloned gene. Then, the dsDNA was converted to small fragments by DNase I treatment (11) or by digestion with a mixture of restriction enzymes that frequently cut and leave an identical overhang (10, 12). Next, the fragmented DNA is ligated to a hairpin linker containing a site for MmeI (a type II restriction endonuclease that cleaves 18 and 20 nt away from its recognition site and leaves a 2-nt 3′ overhang). As a result of the MmeI digestion, the fragmented DNA is converted to a mixture of 20-bp fragments attached to the hairpin linker. Finally, a second linker is ligated to the MmeI-generated termini, and a DNA polymerase converts the short-stranded DNA hairpin structure to a linear dsDNA. These fragments are suitable to generate siRNA expression constructs after cloning them adjacent to an RNA polymerase III-driven promoter. Digestion with a unique restriction enzyme and self-ligation eliminate most of the first linker sequences, leaving a short spacer (8 nt) between the sense and antisense sequences in the final siRNA transcript, which is structurally and functionally identical to most common siRNA transcripts published previously.
Fig. 1.
Outline for enzymatic production of the siRNA library. (A) Double-stranded cDNA is fragmented by DNase I treatment or by a mixture of restriction endonucleases. (B)A3′ loop (green) linker containing an MmeI site (asterisk) and a unique restriction site that will eventually leave an 8-bp spacer is ligated to the fragments, followed by MmeI digestion. (C) MmeI-digested fragments are ligated to a 5′ linker (yellow) containing a unique restriction site. (D) The single-stranded hairpin is denatured and is converted to linear dsDNA after DNA polymerase synthesis of the second strand. (E) The entire siRNA-like sequence, sense and antisense strands (black/blue) and spacer (green), is ligated into a viral expression vector between a polymerase III promoter and a termination signal (five thymidines). (F) The spacer is minimized to 8 bp in length by digestion with restriction endonuclease and self-ligation of the plasmid. (G) Expression from the polymerase III promoter generates a hairpin-shaped siRNA, which is processed by dicer to give rise to a 21-nt sense and antisense functional siRNA.
For users of this approach, it is not yet certain whether fragmentation of the source cDNA (first step) by DNase I treatment is preferable over restriction enzyme mixture. On one hand, DNase I treatment is more homogenous and does not depend on the frequency of restriction enzyme sites. However, DNase I digestion needs fine tuning to prevent under- or overdigestion and may be “user” dependent. Overall, we predict that most researchers would prefer the simple restriction enzymes mixture and might consider using DNase I treatment only when certain mRNAs are underrepresented because of restriction site bias.
Another practical concern is the type of polymerase III promoter used in the siRNA library. Two of the groups favored the U6 promoter (10, 11) over the H1 promoter (12), although there is some evidence that the first guanine in the transcribed RNA is important for the efficiency of that promoter (13). It may be necessary to include an additional guanine just before the siRNA hairpin or to use the H1 promoter.
Because of the nature of the library design, two different orientations of siRNAs are isolated. Half of the clones contain the guide RNA, which is complimentary to the mRNA, on the 3′ strand of the siRNA, but the other half are in the reverse 5′ strand orientation. Sen et al. (12) claim that none of the reverse orientation clones (5′ strand constructs) were functional, thus resulting in 50% loss in complexity of the library. Shirane et al. (11), on the other hand, observe that both orientations give rise to functional clones; however, there was an overall tendency for guide sequences residing in the 3′ side to be more effective. Luo et al. (10) avoided the controversy by using a nondirectional cloning procedure that allows the same MmeI fragment to give rise to both orientations. It has been shown that, for synthetic siRNAs, both strands could function as guide RNA (14); however, it is not clear whether this would also be true for polymerase III-transcribed siRNAs.
All three papers discussed here (10–12) use an integrating retrovirus delivery system, which is crucial when using tissue culture cell lines. However, in primary cells that do not divide in culture, better viral vector systems are needed. Nonintegrating vector systems such as adenovirus and adeno-associated virus viral delivery vectors are known to be able to express siRNAs in terminally differentiated primary cells (15, 16). Lentiviral delivery systems that have a much wider host spectrum (using the VSV-G envelope protein) can express siRNA cassettes in both dividing and nondividing cells and even in transgenic animals (17, 18).
The small interfering RNA library can be used to transduce a pool of cells and isolate a desired phenotype.
Future Applications
The most important application of the use of the siRNA library is the ability to transduce a pool of cells and isolate a desired cellular phenotype resulting from the knockdown of a target gene. An important question is whether a single copy of the siRNA cassette is sufficient to generate a significant phenotype that can be selected from a pool of transduced cells. To address this question, one must design a selectable system efficient enough to discriminate between affected and nonaffected cells and to be able to enrich a single affected cell showing the desired phenotype. Shirane et al. (11), have described a method for enrichment of effective siRNA sequences from a library of GFP siRNAs by fusion of a target gene (i.e., GFP) to a thymidine kinasepuromycin cassette and selecting by using gancyclovir treatment. However, such a method will not be sufficient for selecting from a whole-cell cDNA library because of the high background of cells not eliminated by gancyclovir and the very low number of cells that were actually transduced by an effective siRNA cassette. The manner of selection should match the nature of the desired phenotype; e.g., a phenotype resistant to a lytic viral infection can be directly selected by exposing the siRNA-transduced pool of cells to the virus. If a cellular factor(s), which is crucial for virus propagation, were silenced, one would expect resistant colonies to appear a few days after infection. Analysis and sequencing of the integrated siRNA cassette can identify the silenced cellular factor.
Another utility of the siRNA library is to use high-throughput screening of individual siRNA clones to fish out desired phenotypes (19). Thus, synthesis or cloning of thousands of individual siRNA cassettes will not be necessary, and the question of an effective single-copy siRNA will be avoided. Recently, the issue of modulation of the nontargeted gene has been raised (20); therefore, identifications of multiple siRNA sequences will allow the confirmation of the phenotype elicited by the loss of the target gene product.
Although the approaches of enzymatically generating siRNA libraries appear to be successful, there is still a certain chance of underrepresentation of certain gene transcripts and dependence on robust RNA polymerase III expression. Therefore parallel efforts are under way to generate siRNA libraries that encompass all 30,000 human or mouse genes by generating three to four siRNAs for each gene. Admittedly, this is a laborious and expensive method, but it will ensure siRNAs for every gene. It will also be very helpful for generating knockdown mice when combined with a lentiviral transgenesis approach (17, 18).
See companion article on page 5494.
References
- 1.Fjose, A., Ellingsen, S., Wargelius, A. & Seo, H. C. (2001) Biotechnol. Annu. Rev. 7, 31–57. [DOI] [PubMed] [Google Scholar]
- 2.Hannon, G. J. (2002) Nature 418, 244–251. [DOI] [PubMed] [Google Scholar]
- 3.Elbashir, S. M., Harborth, J., Lendeckel, W., Yalcin, A., Weber, K. & Tuschl, T. (2001) Nature 411, 494–498. [DOI] [PubMed] [Google Scholar]
- 4.Brummelkamp, T. R., Bernards, R. & Agami, R. (2002) Science 296, 550–553. [DOI] [PubMed] [Google Scholar]
- 5.Miyagishi, M. & Taira, K. (2002) Nucleic Acids Res. 2, Suppl., 113–114. [DOI] [PubMed] [Google Scholar]
- 6.Miyagishi, M. & Taira, K. (2002) Nat. Biotechnol. 20, 497–500. [DOI] [PubMed] [Google Scholar]
- 7.Paul, C. P., Good, P. D., Winer, I. & Engelke, D. R. (2002) Nat. Biotechnol. 20, 505–508. [DOI] [PubMed] [Google Scholar]
- 8.Oliveira, D. M. & Goodell, M. A. (2003) Genesis 36, 203–208. [DOI] [PubMed] [Google Scholar]
- 9.Reynolds, A., Leake, D., Boese, Q., Scaringe, S., Marshall, W. S. & Khvorova, A. (2004) Nat. Biotechnol. 22, 326–330. [DOI] [PubMed] [Google Scholar]
- 10.Luo, B., Heard, A. D. & Lodish, H. F. (2004) Proc. Natl. Acad. Sci. USA 101, 5494–5499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shirane, D., Sugao, K., Namiki, S., Tanabe, M., Iino, M. & Hirose, K. (2004) Nat. Genet. 36, 190–196. [DOI] [PubMed] [Google Scholar]
- 12.Sen, G., Wehrman, T. S., Myers, J. W. & Blau, H. M. (2004) Nat. Genet. 36, 183–189. [DOI] [PubMed] [Google Scholar]
- 13.Boden, D., Pusch, O., Lee, F., Tucker, L., Shank, P. R. & Ramratnam, B. (2003) Nucleic Acids Res. 31, 5033–5038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schwarz, D. S., Hutvagner, G., Du, T., Xu, Z., Aronin, N. & Zamore, P. D. (2003) Cell 115, 199–208. [DOI] [PubMed] [Google Scholar]
- 15.Xia, H., Mao, Q., Paulson, H. L. & Davidson, B. L. (2002) Nat. Biotechnol. 20, 1006–1010. [DOI] [PubMed] [Google Scholar]
- 16.Hommel, J. D., Sears, R. M., Georgescu, D., Simmons, D. L. & DiLeone, R. J. (2003) Nat. Med. 9, 1539–1544. [DOI] [PubMed] [Google Scholar]
- 17.Tiscornia, G., Singer, O., Ikawa, M. & Verma, I. M. (2003) Proc. Natl. Acad. Sci. USA 100, 1844–1848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rubinson, D. A., Dillon, C. P., Kwiatkowski, A. V., Sievers, C., Yang, L., Kopinja, J., Rooney, D. L., Ihrig, M. M., McManus, M. T., Gertler, F. B., et al. (2003) Nat. Genet. 33, 401–406. [DOI] [PubMed] [Google Scholar]
- 19.Aza-Blanc, P., Cooper, C. L., Wagner, K., Batalov, S., Deveraux, Q. L. & Cooke, M. P. (2003) Mol. Cell 12, 627–637. [DOI] [PubMed] [Google Scholar]
- 20.Jackson, A. L., Bartz, S. R., Schelter, J., Kobayashi, S. V., Burchard, J., Mao, M., Li, B., Cavet, G. & Linsley, P. S. (2003) Nat. Biotechnol. 21, 635–637. [DOI] [PubMed] [Google Scholar]