Abstract
SUMMARY
Over the past few decades, nucleic acid-based methods have been developed for the diagnosis of intestinal parasitic infections. Advantages of nucleic acid-based methods are numerous; typically, these include increased sensitivity and specificity and simpler standardization of diagnostic procedures. DNA samples can also be stored and used for genetic characterization and molecular typing, providing a valuable tool for surveys and surveillance studies. A variety of technologies have been applied, and some specific and general pitfalls and limitations have been identified. This review provides an overview of the multitude of methods that have been reported for the detection of intestinal parasites and offers some guidance in applying these methods in the clinical laboratory and in epidemiological studies.
INTRODUCTION
Soon after the first publications on the in vitro amplification of DNA by PCR (1, 2), it was predicted that this new technology would lead to a breakthrough in molecular parasitology and in the diagnosis of parasitic infections (3, 4). Until that time, specific DNA probes had been used in basic research and also in diagnostic applications despite being hampered by the limited sensitivity of such direct hybridization assays without a preceding amplification step. It was expected that this could be overcome by the specific amplification of minute amounts of DNA using PCR. In 1995, a large number of papers on DNA-based methods for the detection and identification of a range of parasitic infections were reviewed by J. B. Weiss (5). At that time, the use of PCR was still limited, and most research had been limited to malaria, Leishmania, trypanosome, and Toxoplasma parasites, all of which are tissue parasites. Moreover, except for toxoplasmosis, the experience of using PCR on DNA isolated directly from patient material was limited.
Over the last 10 to 15 years, many clinical microbiology laboratories have been provided with facilities for performing molecular diagnostics. Moreover, technical advances, especially the introduction of real-time PCR, have overcome many drawbacks of PCR from the early years, such as contamination risk from amplified products. It also became possible to combine more than one target in a multiplex assay relatively simply. In addition, the implementation of automated DNA/RNA isolation methods has made it possible to use nucleic acid-based detection techniques in a high-throughput format.
Molecular detection, differentiation, and genotyping methods for a large number of parasites have been described and implemented in both diagnostic and research settings. In this review, we focus primarily on molecular diagnostics and the molecular epidemiology of intestinal parasites and parasites that reside elsewhere in the body but whose DNA can be detected in fecal samples (e.g., Schistosoma and Paragonimus).
TECHNIQUES
PCR
First described in 1985, PCR allows the in vitro amplification of a specific DNA fragment in a cyclic process of denaturation, hybridization of primers, and elongation of the DNA strand using a thermostable DNA polymerase (1, 2). In nested PCR, amplicons from a PCR are used as the template in a second PCR using one primer (“seminested” or “heminested”) or two primers different from those used in the initial PCR and located within the sequence amplified by the first primer set. Nested PCRs are used to increase sensitivity and specificity but have been more or less abandoned in diagnostic laboratories due to the risk of contamination by PCR products. The amplification products in a conventional PCR are usually visualized with ethidium bromide or alternative, less mutagenic dyes after agarose gel electrophoresis. Specificity is based on the expected size of the PCR product. Multiplex PCRs, combining PCRs for different DNA targets, can be achieved by choosing primers for each target in such a way that different-sized amplicons for each target are produced. Optimization of such conventional multiplex PCRs is difficult, as the efficiency of the PCR is correlated with the size of the amplicon, which can result in preferential amplification of smaller products.
Reverse Transcriptase PCR
In reverse transcriptase PCR (RT-PCR), cDNA copies are made of RNA, followed by normal PCR amplification of the desired target.
Real-Time PCR
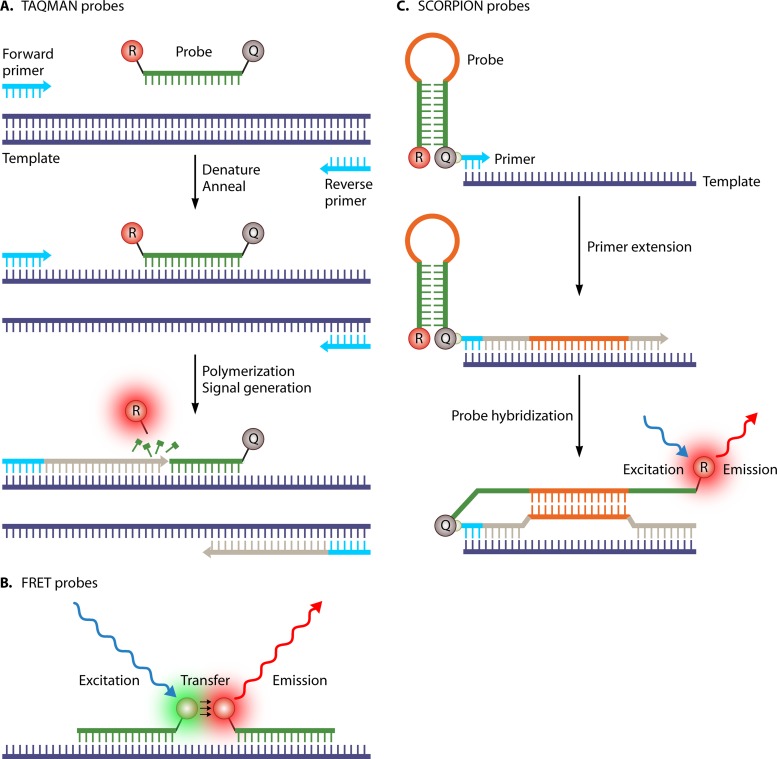
In real-time PCR, the production of amplicons is measured in “real time” during the amplification process (6). Numerous methods have been described, ranging from the use of nonspecific staining of double-stranded DNA using intercalating dyes to the use of fluorescence-labeled DNA probes (7). One of the most commonly used probe-based chemistries is the use of hydrolysis or TaqMan probes (Fig. 1A), in which the 5′-to-3′ exonuclease activity of Taq polymerase cleaves the hybridized probe during the elongation phase of the amplification reaction. In this process, the fluorescent molecule at the 5′ end of the probe is separated from the quencher molecule at the 3′ end of the probe, resulting in a fluorescent signal that can be measured after each amplification cycle. Amplification can also be detected by using fluorescence resonance energy transfer (FRET) probes (Fig. 1B), which comprise two adjacent hybridizing probes that are labeled at the 3′ and 5′ ends of the probe with a donor and an acceptor fluorescent molecule, respectively. An additional melt curve analysis (see below) can be used for the detection of point mutations in the probe sites, which can provide additional differentiation of the sequence detected. Another example of probe-based real-time PCR chemistry is scorpion probes (or primers), which are composed of a primer region with a covalently linked probe and a self-complementary stem sequence with a 5′ fluorophore and a 3′ quencher (Fig. 1C). In the amplification process, the loop sequence hybridizes with the complementary internal target sequence, separating the reporter from the quencher and allowing the reporter to fluoresce.
FIG 1.
Three examples of probe-based real-time PCR chemistry. (A) Hydrolysis or TaqMan probes; (B) fluorescence resonance energy transfer (FRET) probes; (C) scorpion probes.
The amplification cycle at which the level of fluorescent signal exceeds the background fluorescence (threshold cycle [CT] value) is directly correlated with the initial amount of target DNA in the sample, making quantification possible. The absence of a postamplification process reduces the risk of contamination, labor time, and reagent costs. Separate measurement of probes with different fluorophores emitting fluorescence at different wavelengths enables the implementation of multiplex PCRs of similar-sized DNA fragments with the same efficiency.
High-Resolution Melt Curve Analysis
In high-resolution melt curve (HRM) analysis, the decrease of fluorescence of an intercalating dye is measured in the process of the separation of double-stranded DNA by a gradual increase in temperature. Differences in the melting temperature (Tm) of an amplicon reflect differences in the nucleotide sequence.
PCR-Restriction Fragment Length Polymorphism
In PCR-restriction fragment length polymorphism (PCR-RFLP) analysis, PCR products are digested with restriction enzymes to produce different numbers and sizes of fragments depending on differences in the number and location of restriction sites in the amplicon. Target genes and restriction enzymes are chosen in a way that, for example, different species within a genus produce the same-sized amplicon but reveal different banding patterns by gel electrophoresis after digestion.
Random Amplified Polymorphic DNA
Random amplification of polymorphic DNA (RAPD) is performed by using single primers with arbitrarily chosen short nucleotide sequences to amplify products from genomic DNA. After optimization, genus-, species-, or strain-specific banding patterns representing different DNA regions throughout the whole genome can be obtained. The use of these nonspecific primers requires DNA from a pure isolate without contamination by DNA from other organisms, which makes it impossible to use this method on genomic DNA isolated from clinical samples. Specific DNA products of interest can be isolated, sequenced, and used for the development of specific assays targeting these products (8).
Amplification Fragment Length Polymorphism
The amplification fragment length polymorphism (AFLP) technique is based on selective amplification of restriction fragments derived from digested genomic DNA. DNA is digested with restriction enzymes, and oligonucleotide adapters are ligated to the restriction fragments. Thereafter, PCR is used for selective amplification of the restriction fragments, and the amplified fragments are separated by gel or capillary electrophoresis. Similar to the case for RAPD, genus-, species-, or strain-specific banding patterns can be obtained provided that pure isolates are available.
Single-Strand Conformation Polymorphism
Single-strand conformation polymorphism (SSCP) is a mutation-scanning method based on the differential migration in gel electrophoresis of single-stranded DNA molecules of the same size but with different conformations due to differences in the nucleotide sequences.
Multiplex Ligation-Dependent Probe Amplification
Multiplex ligation-dependent probe amplification (MLPA) uses two target-specific oligonucleotides (probes) for each target of interest with a universal primer sequence on the 5′ end of one probe and a sequence of a known length assigned to the target of interest (stuffer sequence) followed by a universal primer sequence on the 3′ end of the second probe. The two hybridized probes are joined together through ligation using a DNA ligase enzyme. The resulting product can be amplified by using primers targeting the universal primer sequences on both ends of the product. The PCR products can be separated on the basis of the different lengths of the unique stuffer sequences using gel or capillary electrophoresis, enabling multiplex detection of a large number of targets (9).
PCR-Reverse Line Blot Assay
In PCR-reverse line blot (RLB) analysis, biotin-labeled primers are used to produce a biotin-labeled PCR product and probes that are covalently bound to a membrane. The biotin-labeled PCR product is hybridized to the probes on the membrane, and the hybridization product is visualized on an X-ray film by a biotin-streptavidin-peroxidase-mediated chemiluminescence reaction.
Loop-Mediated Isothermal Amplification
Loop-mediated isothermal amplification (LAMP) typically uses four target-specific oligonucleotides to amplify DNA under isothermal conditions (10). The amplified DNA product can be detected by the naked eye as a white precipitate in the reaction tube or under UV light after the addition of a fluorescent intercalating dye.
Multilocus Sequence Typing
For multilocus sequence typing (MLST), depending on the degree of discrimination required, a number of housekeeping genes of an isolate are amplified and sequenced on both strands. Each sequence variant within each gene is assigned to a distinct allele, and the combination of alleles within an isolate defines its allelic profile or sequence type (SQT).
Basic Local Alignment Search Tool
The Basic Local Alignment Search Tool (BLAST) is an online algorithm that can be used to compare DNA sequences of interest against several DNA sequence databases.
DNA ISOLATION
Without an appropriate nucleic acid isolation method, DNA amplification techniques will not be reliable. Basically, there are two essential points that must be kept in mind. First, will the isolation method used be able to release the nucleic acids from the parasitic stage (e.g., cysts, spores, or eggs), which is to be expected in the clinical sample? Second, will the nucleic acids that are isolated be free of substances that may interfere with or inhibit the amplification reaction? It is not hard to imagine that the latter is especially important in the isolation of parasite DNA from a complex matrix such as feces (11). Heating of the stool specimen and the addition of absorbent substances such as polyvinyl polypyrrolidone during the DNA isolation procedure or the addition of inhibition factor-binding substances, such as bovine serum albumin (BSA), or inhibitor-resistant DNA polymerases in the PCR mixture can be used to prevent inhibition of the amplification reaction (12–15). However, it remains important to include an internal inhibition control in each reaction mixture. For example, phocin herpesvirus (PhHV) is frequently used and is added to each sample for the isolation procedure, after which a PhHV-specific PCR within the multiplex of the respective targets is performed (16). Because the same amount of virus is added to each sample, it is expected that the fluorescent signal of the PhHV-specific probe will cross the threshold at the same amplification cycle for each sample (CT value). If a higher or no CT value is found in a sample, it suggests inhibition of the amplification reaction. In such cases, DNA isolation and PCR should be repeated using a diluted sample. With regard to the first aspect, it appears that the efficient release of nucleic acids depends on a balanced combination of actions in the DNA isolation procedure. For example, some papers mention specifically that additional mechanical disruption is needed for the efficient isolation of DNA from Trichuris eggs or Entamoeba cysts, while others use a standard isolation protocol without mention of additional rigorous steps to break down the egg shell or cyst wall (15, 17–19). Another example is the negative effect of preserving feces in formalin or sodium acetate-acetic acid-formalin (SAF) on the specific amplification of Entamoeba, Giardia, and Cryptosporidium DNAs; this effect increases with the duration of fixation (20–23). Species-specific DNA extraction using magnetic beads which capture oligonucleotides specific for Cryptosporidium and Giardia improved (lowered) real-time PCR CT values by averages of 10.7 and 9.7 cycles, respectively (24). Nowadays, more and more commercially available DNA isolation kits and protocols for automated DNA isolation systems are becoming available for DNA isolation from feces (25–27). The efficiency of DNA isolation for each target in the expected variety of clinical samples should be verified when such systems are introduced.
ASSAY DEVELOPMENT AND VALIDATION
Extensive knowledge of genetic variation across species and genera is paramount in efforts to tailor DNA-based assays to relevant organisms. Alignments of target sequences from related and unrelated organisms are used to aid the design of organism-specific detection assays, and a choice is made of a target sequence that is specific for the organism of interest and does not show sequence variation within the organism. For example, it is important to know that Cryptosporidium assays based on the DnaJ-like sequence, which are quite commonly used in diagnostic settings, will most likely detect C. hominis and C. parvum only, and as a consequence, other rarer Cryptosporidium species will remain undetected. This can be overcome by using an assay based on the small-subunit (SSU) rRNA gene, but it should be taken into account that the SSU rRNA gene is highly conserved among some apicomplexan genera. Thus, other related species, such as Cyclospora and Cystoisospora, could be detected as well. Obviously, in the design of genotype-specific detection assays, target sequences that show genotype-specific variation within the organism are used to design genotype-specific primers and probes (28).
Sequence data that are used for the design and evaluation of diagnostic primers and probes using BLAST searches are limited mainly to data available in GenBank (28). Moreover, as pointed out by Stensvold et al. (28) and Burnet et al. (29), the nomenclature in GenBank does not always follow the changing taxonomy, and there are several falsely annotated sequences, so care should be taken when designing primers and interpreting PCR results, whether in silico (i.e., using software to analyze and predict PCR outcomes) or in clinical situations.
Such evaluations should not be performed only when a diagnostic assay is introduced but have to be maintained through continuous monitoring of the literature. While in silico evaluation of primer/probe specificity and sensitivity is a key component in the development, integration, and validation of diagnostic assays, extensive laboratory testing using panels of control DNA samples of individual worms, cultured or purified organisms, and isolated fecal genomic DNAs may further challenge the sensitivity and specificity of any given assay. For instance, the SSU rRNA gene is highly conserved among some apicomplexan genera, but the lack of specificity is somewhat balanced by the fact that many of these genera are parasites of nonhuman hosts (28).
PROTISTS
More than 15 protistan genera are known to parasitize the human intestine. For some genera, only one species has been found in humans, for instance, Balantidium, while humans are natural hosts of several species of Entamoeba, for instance. Human intestinal protists each belong to one of six biologically very different groups: amoebozoans, metamonads, ciliates, apicomplexans, microsporidia, and stramenopiles. Microsporidia and the Apicomplexa are all obligate intracellular parasites and as such are directly responsible for tissue damage that often leads to symptoms in the infected host. Asymptomatic carriage is not uncommon for most parasitic infections; however, treatment of even asymptomatic carriers of, for instance, Entamoeba histolytica and Giardia is important to reduce transmission and episodes of recrudescence. In this review, only species that have been positively linked to disease are included. A positive link means that the parasite may cause dysentery (e.g., Entamoeba histolytica and Balantidium coli) and/or has been identified as a recurring cause of outbreaks of diarrhea (e.g., Cryptosporidium, Cyclospora, and Giardia) or multiple sporadic cases of diarrhea, for instance, in immunocompromised individuals (e.g., microsporidia and Cystoisospora belli).
In addition to these organisms, Blastocystis and Dientamoeba fragilis have been included, although the pathogenicity of these organisms is the subject of ongoing debate. However, without the routine use of permanently stained fecal smears, these parasites are notoriously difficult to detect, and the use of nucleic acid-based techniques has largely facilitated both detection and differentiation. Thanks to these techniques, we are starting to build a picture of their epidemiology and clinical significance, which is why we have chosen to include them.
AMOEBOZOA
Entamoeba
Entamoeba histolytica is the causative agent of amoebic dysentery and is able to penetrate the gut wall and reach the liver, where it can cause severe damage by lysing liver tissue (30). Worldwide, an estimated 50 million people are infected with E. histolytica, and 40,000 people die annually from the consequences of this infection (31).
In 1997, the existence of E. histolytica and Entamoeba dispar as two distinct but morphologically identical species was officially acknowledged, with only the former causing disease and the latter being regarded as a harmless intestinal commensal (30, 32). The global epidemiology of E. histolytica and E. dispar has been reviewed quite extensively, showing large variations in the ratios between E. histolytica and E. dispar in different regions (33, 34). Studies of travelers and immigrants performed in laboratories situated in countries where these organisms are not endemic have shown a 1:10 E. histolytica/E. dispar ratio (35–38). which is in agreement with the assumed worldwide ratio (39).
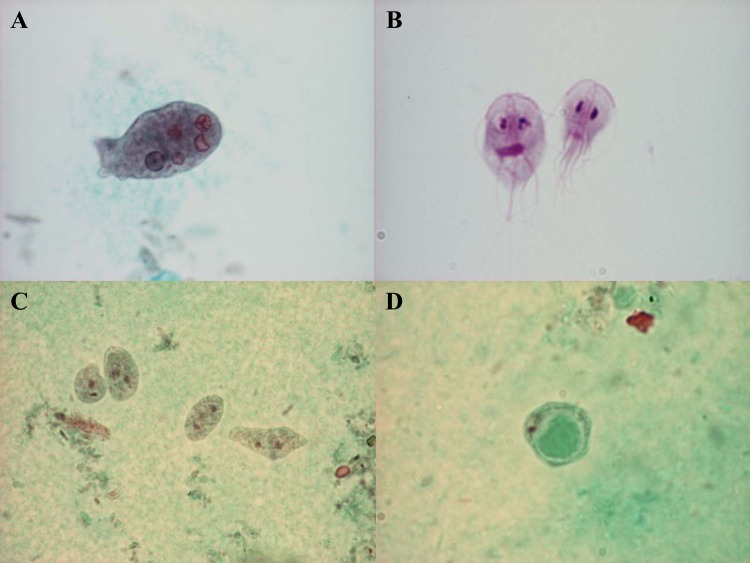
In nondiarrheic stool samples, Entamoeba cysts may be identified by, e.g., microscopy of fecal concentrates. Several species of Entamoeba (e.g., E. histolytica, E. dispar, E. moshkovskii, E. coli, E. hartmanni, and E. polecki) are capable of establishing infection in the human intestine, and these species can be separated in part based on morphological analysis of cysts. Mature cysts from Entamoeba species infecting humans are mainly uni-, quadri-, or octanucleated. E. histolytica produces quadrinucleated cysts, which are generally morphologically indistinguishable from cysts produced by some nonpathogenic species of Entamoeba. Demonstration of erythrocytes within trophozoites of E. histolytica (Fig. 2A) is diagnostic but requires the availability of highly specialized personnel and access to freshly passed dysenteric stool samples or the use of fecal fixatives for specimen submission (40). Stool antigen assays appear to be specific and sensitive for the detection of E. histolytica infections in areas of endemicity (41) but lack sensitivity in settings where the disease is not endemic (42, 43).
FIG 2.
Trichrome staining of an Entamoeba histolytica trophozoite with ingested red blood cells (A), Dientamoeba fragilis trophozoites (C), and the Blastocystis vacuolar stage (D). (B) Giemsa staining of Giardia lamblia trophozoites. (Parasite images courtesy of Marianne Lebbad; reprinted with permission.)
Apart from E. histolytica and E. dispar, two additional species producing quadrinucleated cysts, E. moshkovskii and E. bangladeshi, have been identified in humans. E. moshkovskii has been detected frequently by using nested PCRs in mixed infections with E. histolytica and/or E. dispar (Table 1). Although E. moshkovskii is regarded mostly as nonpathogenic, it has been associated with gastrointestinal complaints in some studies (44–47). Little is yet known about the genetic presence of virulence factors or the expression thereof in comparison with E. histolytica. The biology, diagnosis, epidemiology, and clinical aspects of infections with E. moshkovskii were recently reviewed (48).
TABLE 1.
PCR-based studies that have included PCR detection of E. moshkovskii rDNAh
| Country | Method | No. of patients | Sample selection criterion | No. of samples tested | % positive samples |
Reference(s) | |||
|---|---|---|---|---|---|---|---|---|---|
| E. histolyticaa | E. dispara | E. moshkovskiia | E. moshkovskii only | ||||||
| Bangladesh | Nested PCR | 109 | 109 | 16 | 36 | 21 | 6 | 427 | |
| Ghana | PCR-RLB | 246 | 20 | 0 | 45 | 0 | 428 | ||
| India | Nested PCR-RFLPc | 746 | Microscopy positive | 68 | 19 | 97 | 25 | 1 | 429 |
| Thailand | Multiplex PCR | Microscopy positive | 30 | 13 | 20 | 0 | 0 | 430 | |
| Iran | Nested PCRd | 1,037 | Microscopy positive | 88 | 0 | 100 | 1 | 1 | 431 |
| India | Nested PCR-RFLPd | 1,720 | Microscopy positive/culture positive | 202 | 30 | 79 | 16 | 1 | 432 |
| India | Nested multiplex PCR | 1,720 | Microscopy positive/culture positive | 202 | 35 | 84 | 18 | 0 | 433 |
| Turkey | Nested PCR-RFLPd | 100 | Diarrhea cases | 100 | 23c | NA | 2 | NA | 434 |
| Australia | Nested PCRd | 1,246 | MSM/microscopy positive | 54 | 5 | 65 | 31 | 15 | 44 |
| Australia | Nested PCRd | 5,921 | Microscopy positive | 110 | 5 | 57 | 50 | 24 | 435, 436 |
| Tunisia | Nested PCRd | Microscopy positive | 27 | 0 | 89 | 7 | 0 | 437 | |
| India | Nested multiplex PCRe | Microscopy positive | 202 | 35 | 85 | 18 | NA | 438 | |
| Tanzania | Nested PCRd | 136 | HIV positive | 136 | 4c | 5 | 13 | 12 | 439 |
| 622 | Controls | 0 | 0 | 0 | 0 | ||||
| Thailand | Multiplex real-time (FRET) PCR | Microscopy positive | 33 | 6 | 85 | 1 | 0 | 61 | |
| India | Nested multiplex PCRe | 246 | Microscopy positive | 49 | 12 | 29 | 8 | NA | 440 |
| NAi | PCR-pyrosequencing | NA | Microscopy positive/E. histolytica-E. dispar PCR positive | 102 | 17 | 86 | 0 | 0 | 64 |
| Iran | PCRf | 3,825 | Microscopy positive | 58 | 3 | 93 | 5 | 2 | 441 |
| Brazil | PCR-sequencing | Microscopy positive/culture positive | 29 | 14 | 66 | 3b | 3b | 63 | |
| Ecuador | PCR-RLBg | 674 | Microscopy positive | 101 | 0 | 8 | 0 | 0 | 442 |
| Pakistan | Nested multiplex PCRe | 129 | Diarrhea cases | 129 | 9 | 19 | 19 | NA | 45 |
| 151 | Controls | 151 | 1 | 27 | 4 | NA | |||
| Malaysia | PCRf | 500 | 500 | 6 | 17 | 3 | 1 | 46, 443–445 | |
| Malaysia | Nested multiplex PCRf | 426 | 426 | 9 | 4 | 1 | 1 | 446 | |
Including mixed infections.
DNA isolated from culture.
Antigen detection only.
PCR primers are described in reference 427.
PCR primers are described in reference 433.
PCR primers are described in reference 430.
PCR primers are described in reference 428.
Abbreviations: RLB, reverse line blot; RFLP, restriction fragment length polymorphism; FRET, fluorescence resonance energy transfer; MSM, men who have sex with men; NA, not available.
Samples from the indicated study were from patients that were tested in Sweden, Denmark, and The Netherlands.
Recently, E. bangladeshi was identified in stool samples from children in Bangladesh (49). A genus-specific primer pair (50) was used to analyze fecal DNAs from Bangladeshi children with and without diarrhea who were microscopy positive for quadrinucleated cysts but PCR negative for E. histolytica, E. dispar, and E. moshkovskii. Sequencing of PCR products produced evidence of a novel species, named E. bangladeshi. The morphology of cysts and trophozoite stages of E. bangladeshi appears similar to that of E. histolytica (49, 51). Phylogenetic analysis of the relationship between E. bangladeshi and other Entamoeba parasites reveals that, although distinct, E. bangladeshi clearly groups with the clade of Entamoeba parasites infecting humans, which includes E. histolytica, E. dispar, and E. moshkovskii; this “complex” moreover contains two additional species that have not been found in humans so far, namely, E. ecuadoriensis (sewage) and E. nuttalli (nonhuman primates [NHPs]) (52, 53). Of note, E. nuttalli differs from E. histolytica by 2 to 3 bp precisely in the DNA sequence of the E. histolytica detection probe of a widely used real-time PCR based on SSU rRNA genes of E. histolytica and E. dispar (28). As a result, DNA amplification will occur but will probably not be detected by this assay. Whereas E. nuttalli appears to be virulent in nonhuman primates, at present, it remains unclear whether E. nuttalli can infect and cause disease in humans (53–57).
The genetic universe of Entamoeba is currently rapidly expanding, which is due mainly to the recent application of sequencing of PCR products amplified from DNA extracted directly from feces. This has resulted in the discovery of a large number of novel Entamoeba lineages and subtypes (STs) from human and nonhuman hosts. To date, at least 29 distinct lineages have been identified (50, 58). The search for and ongoing discovery of such new lineages warrant substantial additional sampling in order to identify the clinical significance of each lineage and to enable further intrageneric analysis of Entamoeba.
The absence of morphologically apparent differences has made it necessary to develop new diagnostic techniques both for diagnosis and for an understanding of the epidemiology of E. histolytica infections. Laboratory techniques for the diagnosis of E. histolytica infections have been reviewed in detail (59, 60). A range of DNA targets, single- and multicopy genes, have been used in single-round and nested conventional PCRs and real-time PCRs. More recent assays (35, 61–64) using (multiplex) real-time PCR or (pyro)sequencing have been based on the SSU rRNA gene, which appears to be the best diagnostic target in terms of sensitivity and specificity compared to real-time PCR assays targeting non-SSU rRNA genes (65). A method for the detection and differentiation of five species of Entamoeba, including E. histolytica, based on Luminex technology was recently developed (66). One of the species targeted was Entamoeba coli, which exhibits remarkable intraspecies variation, and currently, two subtypes are known (ST1 and ST2) (50). Meanwhile, the probes used for detection of E. coli by Santos et al. (66) were identical only to ST1 and might not detect E. coli ST2, which is currently represented in GenBank under accession numbers AF149914 and AB444953. While the approach is appealing, the challenges and limitations of basing diagnostics exclusively on detection probes are evident.
Genetic variation within E. histolytica in protein-coding genes, noncoding genes, and short tandem-repeat (STR) loci was reviewed by Ali et al. (33). Although one study in Bangladesh has shown a link between genotypes and symptoms, it appears that most strains of E. histolytica have the ability to invade host tissue and cause disease. While the overall genetic diversity of E. histolytica based on single-nucleotide polymorphisms (SNPs) appears to be low, there is substantial genetic variability in highly repetitive DNA regions and across SNPs in some coding genes. A multilocus sequence typing (MLST) system was recently developed based on 16 polymorphic loci identified by next-generation sequencing of E. histolytica genomes and verified by Sanger sequencing (67). As this system is based on SNPs in protein-coding genes potentially involved in pathogenesis rather than on surrogate markers such as tRNA STR patterns, this method may hold promise for further clinical and epidemiological investigations. However, it remains unclear whether the extensive genetic diversity seen among the isolates analyzed reflects a high rate of evolution by recombination or reassortment events that may drive any observed differences between E. histolytica genotypes in samples isolated from the same geographical area. Hence, although a couple of SNPs in the cyclin-2 locus appeared to be associated with disease, it is not yet clear whether differences in SNPs are associated with differences in clinical outcomes or geographical differences. In summary, genotyping currently has very limited clinical significance and is not used in a diagnostic setting but for epidemiology research only.
PCRs for the detection and differentiation of E. histolytica and E. dispar were probably some of the first parasite PCRs to be widely introduced into routine diagnostic laboratories and are now typically used in situations where quadrinucleated cysts have been found in stool samples for species identification, sometimes in addition to serology testing. PCR technology also facilitates efforts toward identification of E. histolytica in samples other than stool, e.g., aspirates from liver abscesses, cerebrospinal fluid (CSF), and urine (51, 68–70). The wider availability of real-time PCR platforms has paved the way for the routine use of PCRs for this and other parasite targets as first-line diagnostic methods. Indeed, E. histolytica-specific gene targets have been integrated into several recent high-throughput multiplex assays aiming to detect a steadily increasing number of enteropathogens (Table 2).
TABLE 2.
Examples of multiplex pathogen detection approaches including parasitic targetsm
| Assay format | Combined assay targets | Sample selection criterion | No. of samples tested | Commercial availability | Description | Reference(s) |
|---|---|---|---|---|---|---|
| Multiplex real–time PCRa | E. histolytica, C. parvum sensu lato,l G. lamblia, and PhHV | Microscopy–positive stool samples | 112 | In–house | First multiplex real–time PCR described for detection of diarrhea–causing protozoa; has been adapted in many diagnostic laboratories since then | 79 |
| Multiplex PCR array | E. histolytica, E. dispar, Cryptosporidium, and G. lamblia | Control DNA only | In-house | Simultaneous detection, species differentiation, and genotyping after hybridization of a multiplex PCR in a microarray format | 447 | |
| Multiplex real-time PCRb | G. lamblia, Cryptosporidium, and E. histolytica | Antigen- and PCR-positive stool samples | 129 | In-house | Primers designed from the COWP gene should be able to detect C. parvum, C. hominis, and C. meleagridis but not other species of Cryptosporidium | 448 |
| Multiplex real-time PCRc | E. histolytica, C. parvum sensu lato, G. lamblia, and PhHV | Gastrointestinal complaints | 956 | In-house | Higher detection rates found for Giardia and Cryptosporidium with PCR, and no additional parasites found with microscopy in a general practitioner patient population | 104 |
| Multiplex real-time PCR | N. americanus, A. duodenale, O. bifurcum, and PhHV | No selection, population based | 339 | In-house | Good correlation found between DNA load and egg/larval counts | 328 |
| Multiplex real-time PCRd | E. histolytica, C. parvum sensu lato, G. lamblia, and PhHV; S. stercoralis and PhHV | Stool samples from travelers | 2,591 | In-house | PCR for the targets used outperformed expert microscopy; strikingly, even in travelers, not many additional parasites were found by microscopy | 107 |
| Multiplex real-time PCRd,e | E. histolytica, C. parvum sensu lato, G. lamblia, and PhHV; D. fragilis and PhHV | Stool samples from patients with gastrointestinal complaints | 397 | In-house | Increased detection rate of D. fragilis with PCR (31%) compared to that with microscopy (17%) | 27 |
| Multiplex real-time PCRb | G. lamblia, Salmonella enterica, Campylobacter jejuni, and PhHV | Stool samples from patients with gastrointestinal complaints | 13,974 | In-house | Implementation of Giardia PCR in a routine diagnostic laboratory | 106 |
| Multiplex real-time PCRf,g | A. duodenale, N. americanus, A. lumbricoides, S. stercoralis, and PhHV | No selection, population based | 1,312 | In-house | Large albendazole placebo-controlled trial on the effect of STH infections on allergy, atherosclerosis, and malaria; effect of treatment was monitored by quantitative real-time PCR | 319, 321, 333, 449 |
| Multiplex nested PCR-RFLP | Microsporidia, Cyclospora, and Cryptosporidium | Controls | In-house | Conventional and nested PCR are not very practical in a routine setting | 450 | |
| Multiplex tandem real-time PCR | E. histolytica, Cryptosporidium, G. lamblia, and D. fragilis | Stool samples from patients with gastrointestinal complaints | 472 | AusDiagnostics, Beaconsfield, Australia | Automated system of multiplex preamplification followed by target-specific real-time PCR | 451 |
| Multiplex PCR using Luminex beadsb,f,g,h,i | E. histolytica, Cryptosporidium, G. lamblia, A. duodenale, N. americanus, A. lumbricoides, and S. stercoralis | Microscopy- and/or PCR-positive stool samples | 319 | In-house | Luminex detection of multiplex PCR products using primers and probes that were described previously as real-time TaqMan-based assays | 322 |
| Multiplex PCR using Luminex beads | C. cayetanensis, C. belli, E. bieneusi, and E. intestinalis | Microscopy- and/or PCR-positive stool samples | 234 | In-house | Luminex detection of multiplex PCR products using primers and probes that were described previously as real-time TaqMan-based assays | 193 |
| Multiplex real-time PCRf,g,h | A. duodenale, N. americanus, A. lumbricoides, S. stercoralis, and PhHV | Stool samples from patients with gastrointestinal complaints | 78 | In-house | Application of multiplex real-time PCR in Malaysia for STH infection, finding higher detection rates with PCR, especially for A. lumbricoides and S. stercoralis | 320 |
| Multiplex real-time PCRc,e,j | E. histolytica, E. dispar, C. parvum sensu lato, and G. lamblia; E. bieneusi and Encephalitozoon spp.; D. fragilis; Blastocystis | HIV positive | 96 | In-house | Although a high prevalence of intestinal parasites was detected, there was no association between any of the parasites and the presence of diarrhea | 160 |
| Multiplex tandem real-time PCR | Campylobacter, Clostridium difficile, Salmonella, Cryptosporidium, G. lamblia, Shigella, adenovirus 40/41, and norovirus | Cryptosporidium-positive samples and negative-control samples | 267 | AusDiagnostics | Extension of the no. of targetsn | 452 |
| Multiplex real-time PCRc,f,g,h | E. histolytica, Cryptosporidium, and G. lamblia; A. duodenale, N. americanus, A. lumbricoides, and S. stercoralis | Stool samples from patients with gastrointestinal complaints | 229 | In-house | Application of multiplex real-time PCR in Malaysia for STHs and diarrhea-causing protozoa | 161 |
| Multiplex PCR | E. histolytica, astrovirus, calicivirus, and EIEC | Stool samples from patients with gastrointestinal complaints | 103 | In-house | Conventional PCR is not very practical in a routine setting with large numbers of samples | 453 |
| Multiplex real-time PCRc,e | E. histolytica, C. parvum sensu lato, and G. lamblia; E. dispar; D. fragilis | Stool samples from patients with gastrointestinal complaints | 889 | In-house | PCR showed higher detection rates of the parasites targeted; no additional pathogens were found by FECT-microscopy | 162 |
| TaqMan open array | Cryptosporidium, G. lamblia, Campylobacter spp., Clostridium difficile, Salmonella, Vibrio parahaemolyticus, diarrheagenic Escherichia coli (EHEC), Shigella, Yersinia enterocolitica, Listeria monocytogenes, adenovirus, astrovirus, norovirus GI, norovirus GII, rotavirus, and sapovirus | Stool samples from patients with gastrointestinal complaints | 86 | In-house | A DNA sample with PCR mix is distributed by hydrophilic forces into 64 through-holes, in which primers and a probe are spotted by the manufacturer; in this way, up to 64 PCRs per sample can be performed for 48 samples simultaneously | 87, 454 |
| Multiplex real-time PCRd | E. histolytica, C. parvum sensu lato, G. lamblia, and PhHV | Stool samples from patients with gastrointestinal complaints | 396 | In-house | Microscopy showed low sensitivity and specificity compared to real-time PCR, especially for detection of E. histolytica | 87 |
| TaqMan array card | Cryptosporidium, G. lamblia, Ascaris lumbricoides, Trichuris trichiura Campylobacter jejuni-C. coli, Clostridium difficile, Salmonella, Vibrio cholerae, diarrheagenic Escherichia coli (EAEC, ETEC, EPEC, and STEC), Shigella/EIEC, adenovirus, astrovirus, norovirus GII, rotavirus, sapovirus, and PhHV | Microscopy-, culture-, and/or immunoassay-positive stool samples | 109 | In-house | A DNA sample with PCR mix is distributed by centrifugation into 42 wells of a microfluidic card in which primers and probe are spotted by the manufacturer | 309 |
| TaqMan panelf,i | E. histolytica, Cryptosporidium, G. lamblia, A. duodenale, N. americanus, A. lumbricoides, S. stercoralis, and T. trichiura | No selection, population based | 525 | In-house | Parallel testing of 8 singleplex PCR mixtures that were prealiquoted and stored frozen until use; good correlation between egg counts and DNA load; all Ascaris PCR-positive cases tested negative 3 wk after treatment | 18 |
| Multiplex real-time PCR | E. histolytica, Cryptosporidium, and G. lamblia; Campylobacter coli-C. jejuni, E. coli (VTEC), Salmonella spp., Shigella spp., E. coli (EIEC), Y. enterocolitica, and C. difficile; norovirus GI, norovirus GII, and IC; adenovirus, astrovirus, and rotavirus | Stool samples from patients with gastrointestinal complaints | 1,758 | CE labeled (Fast-Track Diagnostics, Luxembourg) | Enhanced rate of detection of diarrhea-causing pathogens using a broad approach due to more sensitive detection of pathogens and detection of pathogens that were not requested | 413 |
| Multiplex PCR using Luminex beads | E. histolytica, Cryptosporidium, G. lamblia, Salmonella spp., Shigella spp., Campylobacter spp., E. coli O157, ETEC, STEC, C. difficile, Y. enterocolitica, V. cholerae, norovirus GI, norovirus GII, adenovirus 40/41, and rotavirus A | Stool samples from patients with gastrointestinal complaints | 901 | FDA cleared, CE labeled (Luminex) | Multicenter study comparing standard routine procedures including culture, antigen tests, and real-time PCR with Luminex detection of multiplex PCR products of 15 viral, bacterial, and parasitic pathogens (Luminex xTAG GPP) | 423 |
| Multiplex PCR using Luminex beads | E. histolytica, Cryptosporidium, G. lamblia, Salmonella spp., Shigella spp., Campylobacter spp., E. coli O157, ETEC, STEC, C. difficile, Y. enterocolitica, V. cholerae, norovirus GI, norovirus GII, adenovirus 40/41, and rotavirus A | Stool samples from diarrheic patients | 440 | FDA cleared, CE labeled (Luminex) | Luminex xTAG GPP was compared with standard routine procedures which did not include real-time PCR; no further analysis of discrepant results | 422 |
| Multiplex PCR using Luminex beads | E. histolytica, Cryptosporidium, G. lamblia, Salmonella spp., Shigella spp., Campylobacter spp., E. coli O157, ETEC, STEC, C. difficile, Y. enterocolitica, V. cholerae, norovirus GI, norovirus GII, adenovirus 40/41, rotavirus A | Stool samples from patients with gastrointestinal complaints | 393 | FDA cleared, CE labeled (Luminex) | Luminex xTAG GPP was compared to standard routine procedures including culture, antigen tests, and real-time PCR; 5 of 6 E. histolytica-positive results by xTAG GPP could not be confirmed | 455 |
| Multiplex real-time PCRf,g,j,k | A. duodenale, N. americanus, A. lumbricoides, S. stercoralis, and PhHV; Schistosoma and PhHV | Stool samples from HIV-positive patients | 153 | In-house | Improved detection of helminth infections using real-time PCR; discrepancies in risk factor analysis between qualitative data from PCR and microscopy; this was resolved by using quantitative data from PCR | 456 |
PCR primers and probe for Cryptosporidium are described in reference 158.
PCR primers and probe for Giardia are described in reference 79.
PCR primers and probes for E. histolytica, Cryptosporidium, and Giardia are described in reference 79.
PCR primers and probes for E. histolytica and Giardia are described in reference 79.
PCR primers and probe for D. fragilis are described in reference 128.
PCR primers and probes for N. americanus and A. duodenale are described in reference 328.
PCR primers and probe for S. stercoralis are described in reference 354.
PCR primers and probe for A. lumbricoides are described in reference 319.
PCR primers and probe for E. histolytica are described in reference 448.
PCR primers and probes for E. bieneusi and Encephalitozoon spp. are described in reference 457.
PCR primers and probe for Schistosoma are described in reference 458.
Nomenclature of the sequence from GenBank used for the design of primers and probes was for C. parvum, which was later separated into C. hominis and C. parvum.
Abbreviations: RFLP, restriction fragment length polymorphism; SSU, small subunit; ITS, internal transcribed spacer; STH, soil-transmitted helminth; GPP, gastrointestinal pathogen panel; COWP, Cryptosporidium oocyst wall protein; FECT, formol ethyl-acetate concentration technique; EIEC, enteroinvasive Escherichia coli; EHEC, enterohemorrhagic E. coli; EAEC, enteroaggregative E. coli; ETEC, enterotoxigenic E. coli; EPEC enteropathogenic E. coli; STEC, Shiga-toxigenic E. coli; VTEC, verotoxin-producing Escherichia coli; IC, internal control; CE, European Community; PhHV, phocine herpesvirus.
As described in reference 451.
METAMONADS
Giardia
The genus Giardia comprises multiple species whose taxonomy is in a state of flux. Giardia lamblia (synonyms, G. duodenalis and G. intestinalis) (Fig. 2B) is one of the most common intestinal parasites of humans (71); in some cohorts, for instance, Cuban children, the prevalence may exceed 50% (72). Transmission is by the fecal-oral route, and the parasite has a simple life cycle comprising excystation in the duodenum; colonization by rapidly multiplying, noninvasive trophozoites on the mucosal surface of the small intestine; and, eventually, the production of environmentally resistant cysts that are shed with host feces. Food- and waterborne outbreaks have frequently been reported (73). Symptoms may include late-onset and persistent diarrhea (rather than acute diarrhea), abdominal cramps, bloating, steatorrhea, malabsorption, weight loss, and stunting (74–76). Stools from symptomatic carriers may be mushy, greenish, and foul-smelling.
Persistent symptoms due to, or following, Giardia infection take a significant toll on human health. Reappearance of symptoms, including abdominal symptoms and fatigue, can result from reinfection, recrudescence, perturbation of the gut flora, or postinfection syndromes (77). In developed countries, such sequelae may have a vast impact on quality of life; in developing countries, particularly in children, they add yet another burden to already disadvantaged populations (77).
Several conventional and real-time PCRs for the primary diagnosis of giardiasis that target the SSU ribosomal DNA (rDNA), β-giardin, triosephosphate isomerase (TPI), and intergenic spacer (IGS) regions have been reported (21, 78–81). Many targets are being used for further genotyping directly on DNA from fecal samples or from Giardia isolates. SSU rRNA, glutamate dehydrogenase (GDH), TPI, β-giardin, IGS region, and elongation factor 1-alpha (EF1-alpha) gene amplification products are used for direct sequencing or RFLP in post-PCR analysis (82–85). Recently, novel sequence information was used to identify new assemblage A- and B-specific loci. Two assemblage-specific PCRs based on these loci showed an excellent performance when used on DNAs extracted from feces (86, 87). The molecular epidemiology of Giardia and genotyping methods have recently been reviewed (88, 89). Genotypes of Giardia are traditionally named “assemblages,” which are identified by analysis of single or multiple loci such as SSU rDNA, β-giardin, TPI, and GDH; the SSU rRNA and GDH genes appear to be the genes that are most easily amplified (84, 90). Humans host mainly assemblage B, while assemblage A is less common; both assemblages are shared with many other mammals (91). Monis and coworkers have suggested using G. duodenalis for assemblage A and renaming assemblage B Giardia enterica. Assemblages C and D are found mainly in canids and have been referred to as Giardia canis; assemblage E, found in livestock, has been referred to as G. bovis; assemblage F, found in cats, has been referred to as G. cati, and assemblage G, found in rodents, has been referred to as G. simondi, but for assemblage H in marine vertebrates, no species name has yet been proposed (88, 89, 92–95). The zoonotic potential of both assemblages A and B is evident when studied at the levels of assemblages and subassemblages and even at each single locus. However, when genotypes are defined by using a multilocus sequence typing scheme, only 2 of 84 multilocus genotypes (MLGs) of assemblage A and none (n = 99) of assemblage B appear to have zoonotic potential (88, 96, 97).
While molecular markers for assemblage A appear to produce robust and easy-to-read sequences, the allelic heterozygosity shown to exist at the single-cell level in assemblage B isolates and sometimes further complicated by other coinfecting assemblage B subgenotypes (91) makes precise identification impossible. Therefore, development of alternative genotyping methods appears to be relevant.
Findings on the clinical significance of different assemblages have been contradictory; assemblage B was recently shown to be associated with flatulence in children, and assemblage B appears to be more common in patients with suspected treatment failure (98). Homan and Mank found that assemblage A isolates were detected solely in patients with intermittent diarrhea, while assemblage B isolates were present in patients with persistent diarrhea (99). In Saudi children, a strong correlation between the presence of assemblage B and symptoms was found, while assemblage A was found mainly in cases of asymptomatic giardiasis (85). In contrast, a study in Australia found assemblage A to be associated with diarrhea, while assemblage B was found mainly in asymptomatic children (100). In a study among Rwandan children, assemblage A was associated with vomiting and abdominal pain (101). There are also examples of studies where clinical differences associated with the two assemblages were not identified (102, 103).
As for other parasites, the introduction of PCR-based diagnostic assays for Giardia took place about a decade ago. Until then, state-of-the-art diagnosis included mainly microscopy of fecal concentrates (cysts), permanent staining of fixed fecal smears (trophozoites), and antigen detection by using enzyme-linked immunosorbent assays (ELISAs) or direct fluorescent-antibody (DFA) tests typically integrated into an assay also enabling the detection of Cryptosporidium. DNA amplification techniques have excellent sensitivity and specificity compared with microscopy and antigen detection (27, 78, 79, 81, 101, 104–108). Giardia-specific DNA detection (including all assemblages) is increasingly being incorporated into multiplex assays, which are listed in Table 2.
Dientamoeba fragilis
Dientamoeba fragilis (Fig. 2C) was first described in 1918 as an amoeba of the intestinal tract of humans (109). Later, by means of antigen and ultrastructural studies and analysis of rRNA, however, the organism was reclassified as a trichomonad flagellate although lacking external flagella (110–112). Since its discovery, the pathogenicity of this organism has remained controversial. Although in recent years, several authors have reported the clinical importance of D. fragilis as a cause of gastrointestinal symptoms (113, 114), a consensus on its pathogenicity is lacking, mainly because so many D. fragilis infections remain asymptomatic (115–118). D. fragilis appears to be extremely common and may have a cosmopolitan distribution, although there are large variations in prevalence. D. fragilis has been linked to intestinal symptoms, especially in children (119, 120). Some studies report a higher prevalence in patients with intestinal symptoms than in healthy individuals (121), while others report the reverse situation (122). Dientamoeba infections are potentially chronic (123), which is one of the reasons why dientamoebiasis has been speculated to be a neglected differential diagnosis of irritable bowel syndrome (IBS) (124). It was found that metronidazole was capable of eradicating D. fragilis in 60% of 25 positive patients fulfilling the Rome III criteria for IBS; however, microbiological and clinical cures were not associated, and the study did not support a hypothesis of a simple association between D. fragilis and IBS.
The SSU rRNA gene of D. fragilis was amplified and completely sequenced for the first time in 1996 (110). Phylogenetic analysis of housekeeping genes such as the SSU rRNA, EF1-alpha, and actin genes consistently reveals Histomonas, a potentially invasive parasite causing blackhead disease in birds, as the closest relative (110, 125). Possibly due to insurmountable methodological challenges, such as obtaining sufficient amounts of DNA from axenic cultures, genomic data from Dientamoeba are not yet available, and this precludes studies aiming to predict the existence of virulence factors and other effector proteins.
Although one recent paper describes the existence of a cyst stage of D. fragilis (112), the apparent absence of a cyst stage has always meant to date that microscopy of fecal concentrates was not applicable, and traditional parasitological detection of D. fragilis relies on morphological detection of trophozoites by light microscopy of fixed and permanently stained fecal smears. The sensitivity of a single examination is not high because the day-to-day variation of D. fragilis trophozoites in feces seems to be even more irregular than that observed in other intestinal protozoan infections such as G. lamblia and E. histolytica (113, 126).
A number of PCRs have been developed for diagnosis of and research into Dientamoeba (Table 3). Conventional PCRs have been used mainly for confirmation of microscopy results and subsequent characterization of the Dientamoeba ribosomal genes in human fecal samples. A real-time PCR targeting 77 bp of the SSU rRNA gene was the first molecular assay developed as a screening tool, potentially replacing expensive and time-consuming parasitological diagnosis (127). Interestingly, with a real-time PCR targeting the 5.8S ribosomal gene, which also included an internal process control, it was shown that no reduction in amplification efficiency could be detected when comparing fresh material with material that had been stored unpreserved at 4°C for 8 weeks (128).
TABLE 3.
Primer and probe sequences for PCR-based assays for detection and molecular characterization of Dientamoeba fragilisa
| Target | Method | Amplicon size (bp) | Primer or probe (sequence) | Reference(s) |
|---|---|---|---|---|
| Detection | ||||
| SSU rDNA | PCR | 887 | DF400 (5′-TATCGGAGGTGGTAATGACC-3′) | 130, 459, 460 |
| DF1250 (5′-CATCTTCCTCCTGCTTAGACG-3′) | ||||
| SSU rDNA | Real-time (TaqMan) PCR | 77 | DF3 (5′-GTTGAATACGTCCCTGCCCTTT-3′) | 127, 138, 451, 460, 461 |
| DF4 (5′-TGATCCAATGATTTCACCGAGTCA-3′) | ||||
| Probe (5′-FAM-CACACCGCCCGTCGCTCCTACCG-TAMRA-3′) | ||||
| rDNA (5.8S) | Real-time (TaqMan) PCR | 98 | Df-124F (5′-CAACGGATGTCTTGGCTCTTTA-3′) | 27, 128, 135, 162, 462 |
| Df-221R (5′-TGCATTCAAAGATCGAACTTATCAC-3′) | ||||
| Df-172revT (5′-FAM-CAATTCTAGCCGCTTAT-MGB-3′) | ||||
| Molecular characterization/genotyping | ||||
| SSU rDNA | PCR-RFLP | 887 | DF400 (5′-TATCGGAGGTGGTAATGACC-3′) | 130, 459, 460 |
| DF1250 (5′-CATCTTCCTCCTGCTTAGACG-3′) | ||||
| ITS1-5.8S-ITS2 | PCR-sequencing | ≈440 | TFR1 (5′-TGCTTCAGTTCAGCGGGTCTTCC-3′) | 133 |
| TFR2 (5′-CGGTAGGTGAACCTGCCGTTGG-3′) | ||||
| SSU rDNA | PCR | 1,700 | TRD5 (5′-GATACTTGGTTGATCCTGCCAAGG-3′) | 110, 129–131 |
| TRD3 (5′-GATCCAACGGCAGGTTCACCTACC-3′) | ||||
| SSU rDNA | PCR-RFLP | 662 | DF1 (5′-CTCATAATCTACTTGGAACCAATT-3′) | 85, 90, 131, 463 |
| DF4 (5′-CCCCGATTATTCTCTTTGATATT-3′) | ||||
| SSU rDNA | PCR-HRM | 662 | DF1 (5′-CTCATAATCTACTTGGAACCAATT-3′) | 85 |
| DF4 (5′-CCCCGATTATTCTCTTTGATATT-3′) | ||||
| SSU rDNA | Nested PCR-sequencing | 366 | DF1 (5′-CTCATAATCTACTTGGAACCAATT-3′) | 90 |
| DF4 (5′-CCCCGATTATTCTCTTTGATATT-3′) | ||||
| DF322For (5′-GAGAAGGCGCCTGAGAGATA-3′) | ||||
| DF687Rev (5′-TTCATACTGCGCTAAATCATT-3′) | ||||
| ITS1-5.8S-ITS2 | PCR-sequencing | 500 | ssu2 (5′-GGAATCCCTTGTAAATGCGT-3′) | 134 |
| lsu1 (5′-AGTTCAGCGGGTCTTCCTG-3′) | ||||
| ITS1 | PCR-sequencing | ≈100 | ssu2 (5′-GGAATCCCTTGTAAATGCGT-3′) | 134 |
| 5.8s1 (5′-TGTGAGGAGCCAAGACATCC-3′) | ||||
| ITS1 | Nested PCR-sequencing | 366 | SSU2 (5′-GGAATCCCTTGTAAATGCGT-3′) | 90 |
| Df-ITSRev (5′-GCGGGTCTTCCTATATAAACAAGAACC-3′) | ||||
| Df-ITSnesFor (5′-ATACGTCCCTGCCCTTTGTA-3′) | ||||
| Df-ITSnesRev (5′-GCAATGTGCATTCAAAGATCGAAC-3′) | ||||
| SSU rDNA | PCR-pyrosequencing | 129 | D.FRAGILISpyroF (5′-CGGAGGTGGTAATGACCAGTTAT-3′) | 132 |
| D.FRAGILISpyroR (5′-[biotin-C6]-TTGCAGAGCTGGAATTACCG-3′) | ||||
| D.FRAGILISpyroS (5′-TGGTAATGACCAGTTATAA-3′) | ||||
| SSU rDNA | PCR-sequencing | 364 | DFpn_1f (5′-GCCAAGGAAGCACACTATGG-3′) | 136 |
| DFpn_364r (5′-GTAAGTTTCGCGCCTGCT-3′) | ||||
| Actin | PCR-sequencing | 134–840 | DF ACTIN_3f (5′-CCACACATTCTACAACGAATTAC-3′) | 136 |
| DF_ACTIN_157f (5′-TTCTTTCACTTTACTCATCAGGTC-3′) | ||||
| DF_ACTIN_291r (5′-GACCAGCAAGGTTGAGTCTC-3′) | ||||
| DF_ACTIN_843r (5′-TGGACCAGCTTCATTGTATTC-3′) | ||||
| EF-1α | PCR-sequencing | 99–836 | DF_EF_1f (5′-CTCACTTTGGAAGTTCGAATC-3′) | 136 |
| DF_EF_265f (5′-TCAAAGGCTCGTTATGATGAAATC-3′) | ||||
| DF_EF_364r (5′-GAAACCTGAGATTGGAACAAAC-3′) | ||||
| DF_EF_836r (5′-CTGTGTGGCAATCGAAAAC-3′) |
Abbreviations: SSU, small subunit; ITS, internal transcribed spacer; EF-1α, elongation factor 1α; RFLP, restriction fragment length polymorphism; HRM, high-resolution melting; TAMRA, 6-carboxytetramethylrhodamine; FAM, 6-carboxyfluorescein; MGB, minor groove binder.
RFLP analysis has been used to distinguish between the two genotypes currently known and which differ by 2% across the SSU rRNA gene (129–131). Genotyping has also been performed by analysis of SNPs detected by PCR and pyrosequencing (132). The Bi/PA strain (GenBank accession no. U37461) is commonly acknowledged as a representative of genotype 2. Although there are still few studies of genotypes, genotype 1 appears to account for the vast majority of cases (130, 131, 133). The value of sequencing of the internal transcribed spacer (ITS) region for typing studies of D. fragilis is limited due to intrastrain genetic heterogeneity (133). A profiling method using the variability within ITS1 of D. fragilis was developed by Bart et al. (134) as a means of extracting useful data from sequenced ITS clones, but so far, little is known regarding its applicability and epidemiological relevance.
Studies of other housekeeping genes, such as EF1-alpha and actin, may prove useful in terms of obtaining a higher resolution than can be obtained by studies of SSU rRNA genes alone, as demonstrated for other metamonads (e.g., Giardia and Trichomonas) (125). Given the high prevalence of the parasite (135) and the recently discovered potential for zoonotic transmission (90), high-resolution markers for distinguishing between strains are warranted. However, a preliminary study of D. fragilis in 40 patients revealed that the EF1-alpha and actin genes appear to be remarkably conserved among patient isolates (136), but the clinical and epidemiological utility of multilocus sequencing of housekeeping genes can be fully determined only after comparing D. fragilis isolates from symptomatic cases to those from asymptomatic carriers. The clinical significance of the two known genotypes of D. fragilis needs further investigation. Is the rarer genotype more virulent than the other, and could this possibly contribute to the differences in clinical perceptions regarding the organism's pathogenicity?
Until a few years ago, humans were the only known hosts of Dientamoeba, but recently, the parasite was discovered in nonhuman primates (gorillas) and pigs as well (90, 137, 138). Unfortunately, no comparisons were made between sequences obtained from humans and those obtained from gorillas, while analysis of Dientamoeba ribosomal sequences from pigs showed that these sequences were identical to those of genotype 1 commonly found in humans (90).
Microscopic examination of permanent stains of fixed fecal smears is insensitive compared to nucleic acid-based techniques: in a study comparing microscopy and real-time PCR, Bruijnesteijn van Coppenraet et al. showed D. fragilis prevalences of 17% and 31%, respectively (27). Reported prevalence figures may reflect differences in diagnostic modalities as well as geographical variation or age variation in study cohorts (135). Whether or not routine detection of D. fragilis should be part of an overall parasitological screen for patients suspected of having intestinal parasitic disease is a matter of contentious debate, mainly due to the predicament that guidelines as to when to try and eradicate the parasite are still to be defined. However, the fact that D. fragilis detection can now be easily integrated into multiplex nucleic acid-based detection techniques (Table 2) means that it is relatively inexpensive and straightforward to implement D. fragilis PCR in a routine diagnostic panel. First and foremost, this can provide accurate data on possible differences in prevalence and infection intensity between symptomatic and asymptomatic carriers, which can also be exploited in randomized controlled treatment studies for evaluation of treatment efficacy. Moreover, positive DNAs can be stored and used for epidemiological analyses of the prevalence and significance of the two genotypes.
APICOMPLEXA
Some apicomplexan parasites belonging to the suborder Eimeriorina can complete their life cycles in the human intestinal tract and hence can be found in human feces; these parasites include Cryptosporidium (Cryptosporidiidae), Cyclospora (Eimeriidae), Cystoisospora, and Sarcocystis (Sarcocystidae).
Cryptosporidium
Cryptosporidium has emerged as an important cause of diarrheal illness worldwide, particularly in young children (<5 years old) and immunocompromised patients (139). At least 6,000 Cryptosporidium-caused cases of gastroenteritis occur annually in the United Kingdom, where Cryptosporidium is the most common protozoan agent involved in acute gastroenteritis (140). Infections in immunocompetent individuals are self-limiting but may last for 1 to 2 weeks; asymptomatic shedding of oocysts may be common (141). Cryptosporidiosis may be chronic and particularly debilitating in patients with T-cell immune deficiencies, with complications such as sclerosing cholangitis and, rarely, biliary cirrhosis and pancreatitis (140).
Transmission is by the fecal-oral route by accidental ingestion of mature oocysts containing infectious sporozoites (140). As the oocysts are immediately infectious, unlike those of Cyclospora, infection may result from direct exposure to mammalian (including human) feces, but food and water contaminated by oocysts may also represent a significant vehicle of transmission. Food- and waterborne outbreaks are not uncommon (142) but may be identified only by chance and after ruling out other causes (143).
While treatment options remain limited, nitazoxanide, which is subject to availability and often requires a special license/approval, may reduce the severity of symptoms, which may include watery diarrhea, abdominal cramps, vomiting, mild fever, and loss of appetite (144). Affected children in developing countries may suffer from malnourishment, and in some nonindustrialized countries, cryptosporidiosis may be a significant cause of morbidity and mortality (145, 146). The introduction of highly active antiretroviral treatment (HAART) for immune reconstruction has dramatically reduced the incidence and severity of cryptosporidiosis in patients with HIV/AIDS (140).
In 2010, 6,605 laboratory-confirmed cases of cryptosporidiosis were reported by 21 European Union/European Economic Area countries; however, 4 countries reported zero cases, and 9 countries failed to report (http://www.ecdc.europa.eu/en/publications/Publications/Annual-Epidemiological-Report-2012.pdf). It is likely that in countries where Cryptosporidium infections are not notifiable, diagnostic methods are far from standardized. Individual national prevalence estimates can be difficult to obtain, and even when they are available, they should be interpreted carefully due to variability in diagnostic modalities. In other countries, surveillance systems include sub- and genotyping of laboratory-confirmed cases, which is useful for epidemiological and outbreak investigations (147).
The genus Cryptosporidium comprises over 20 established species (Table 4), of which the morphologically indistinguishable species C. parvum and C. hominis (previously C. parvum genotype H or genotype 1) account for most human cases (for a list of loci used to discriminate the two species, see reference 148). However, geographical variation may be seen, and both immunocompromised and immunocompetent individuals may be infected by unusual species and genotypes (149). Cryptosporidium meleagridis, in particular, appears to be an emerging pathogen and was found at a rate of 12% in a large study of Peruvian HIV-infected cryptosporidiosis patients (150). In some countries, around 10% of all human cryptosporidiosis cases are due to species other than C. parvum, C. hominis, and C. meleagridis (150, 151). C. parvum may be seen more commonly in mixed infections in humans than C. hominis (152). Human infections due to C. canis, C. cuniculus, C. felis, C. ubiquitum, and C. viatorum, as well as other species and unusual genotypes, are also emerging (150, 153–155; M. Lebbad, personal communication). It appears that these genotypes are found more frequently in asymptomatic carriers than in patients with symptoms, which suggests that some “unusual” genotypes may be more common than thought (140). The risk of C. parvum infections is higher during spring, while C. hominis infections peak in late summer and autumn (140). In contrast to human infection by C. parvum, infections due to C. hominis may result not only in diarrhea but also in nausea, vomiting, malaise, and nonintestinal sequelae (156).
TABLE 4.
Species and genotypes of Cryptosporidium found in humans, listed according to frequency of reportinga
| Report frequency and species or genotype | Host reservoir(s) | GenBank accession no. |
||||
|---|---|---|---|---|---|---|
| Complete SSU rDNA sequence available (∼1.75 kbp) (examples) | COWP sequence (∼550 bp) (examples) | LIB13 | DnaJ-like protein (HSP40) | ITS2 | ||
| Common | ||||||
| C. hominis | Humans | AF093489/L16997 | GU904404, GU904390, GU904388, GU904389, GQ983374, GQ983372, DQ388389, EU186155 | AF190627 | AF400132 | AF093012 |
| C. parvum | Humans, ruminants | AF093490, AF161856 | DQ187314, DQ060433, DQ062120, JX547011, GU904402, GU904400, GU904398 | B78618 | AF400131 | AF093008 |
| C. meleagridis | Birds, mammals (including humans) | AF112574 | EU310392, DQ116568, JX568159, GU904403, AB471654, AY166840, AF248742 | NA | AF400133 | AF381169 |
| Less common | ||||||
| C. canis | Dog | AF112576 | AF266274 | NA | NA | NA |
| C. cuniculus (previously rabbit genotype) | Rabbit | NA | EU437411, GU327782, GU904391, GU904394 | NA | NA | NA |
| C. felis | Cat | AF112575 | AF266263 | NA | NA | AF093013 |
| C. ubiquitum | Various mammals | AF442484 | JX861404, JX861396, JX861405 | NA | NA | NA |
| C. viatorum | Humans | NA | JX984441 | NA | NA | NA |
| Rare | ||||||
| C. andersoni | Cattle | AB089285, AY954885, AF093496 | DQ060431, AB089289, AB514043, AB514044 | NA | NA | NA |
| C. bovis | Cattle | EF514234 | NA | NA | NA | NA |
| C. fayeri | Red kangaroo | AF112570 | AF266269 | NA | NA | NA |
| C. muris | Rodents | AF093498 | DQ060430, AB089287 | NA | NA | AF381167 |
| C. scrofarum | Pig | NA | NA | NA | NA | NA |
| C. suis | Pig | AF108861 | AF266270 | NA | NA | NA |
| C. tyzzeri (previously, mouse genotype I) | Mouse | AF112571 | NA | NA | NA | NA |
| Chipmunk genotype I | Chipmunk, possibly other sciuridae | NA | JX984442 | NA | NA | NA |
| Horse genotype | Horse | NA | EU437416 | NA | NA | NA |
| Monkey genotype | Monkey | AF112569 | NA | NA | NA | NA |
| Skunk genotype | Skunk, possibly other mustelids | NA | NA | NA | NA | NA |
Selected information on nucleotide sequences for ribosomal genes (SSU rRNA and ITS2), Cryptosporidium oocyst wall protein (COWP), LIB13, and DnaJ-like proteins (or heat shock protein 40 [HSP40]) currently available in GenBank is also shown. LIB13 is a Cryptosporidium-specific gene with unknown function. Abbreviations: SSU, small subunit; ITS, internal transcribed spacer; NA, not available.
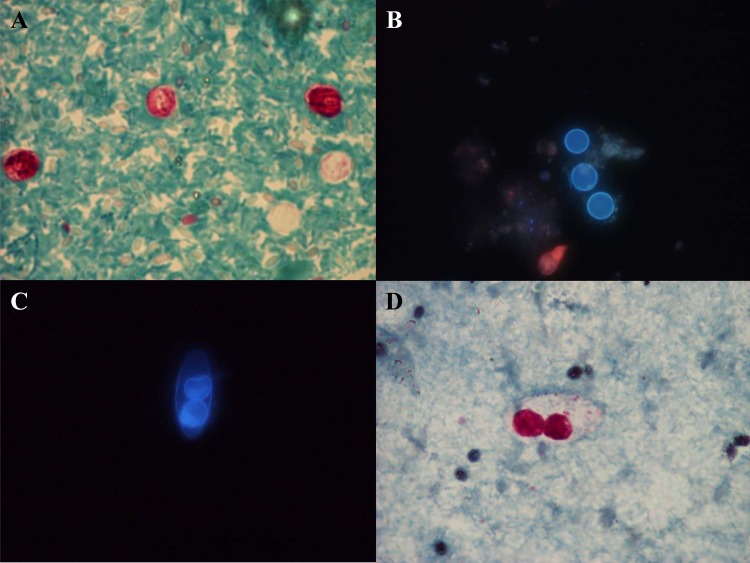
Similar to many other types of intestinal parasitic disease, symptoms due to cryptosporidiosis are nonpathognomonic, and diagnosis should be confirmed by laboratory tests. Cryptosporidiosis should be suspected in any patient with acute gastroenteritis, particularly in young children and if symptoms are prolonged (140). As a consequence, cryptosporidiosis should be a differential diagnosis to other causes of gastroenteritis, including Giardia, Cyclospora, Cystoisospora, microsporidia, noro- and rotaviruses, Campylobacter, Salmonella, Shigella, and enterohemorrhagic E. coli, such as E. coli O157 (141). However, in many laboratories, Cryptosporidium is not traditionally included in test panels for gastroenteritis. Clinical samples appropriate for laboratory diagnosis of cryptosporidiosis were recently reviewed by Davies and Chalmers (140) and include biopsy specimens (jejunal/gastric), bile (obtained by endoscopic retrograde cholangiopancreatography [ERCP]), sputum samples (if respiratory symptoms are present), and antral washouts (in high-risk patients with unexplained sinusitis), in addition to stool samples. Traditional diagnosis relies on microscopy of modified acid-fast-stained fecal concentrates (Fig. 3A) or auramine-phenol staining and/or antigen detection by DFA or immunochromatographic assays.
FIG 3.
(A and D) Modified Ziehl-Neelsen acid-fast staining of Cryptosporidium parvum/C. hominis oocysts (A) and a Cystoisospora belli oocyst (D). (B and C) Unstained wet mounts for UV fluorescence microscopy showing autofluorescence of Cyclospora cayetanensis oocysts (B) and a Cystoisospora belli oocyst (C). (Parasite images courtesy of Marianne Lebbad; reprinted with permission.)
The vast number of species reported to infect humans makes a genus-specific PCR assay the most appropriate diagnostic approach in routine clinical laboratories. DNA samples can be stored for later epidemiological analysis in research and surveillance laboratories. The main targets for diagnostic PCRs typically include the SSU rRNA gene, the Cryptosporidium oocyst wall protein (COWP) gene, or the DnaJ-like protein gene (157–159). While partial SSU rRNA gene sequences are available for all species of Cryptosporidium known to infect humans, surprisingly, only a fraction of these species are currently represented by complete SSU rDNA sequences in GenBank (Table 4). COWP sequences are readily available, but for some other loci, for instance, the DnaJ-like protein used in widely used diagnostic assays (27, 79, 87, 104, 107, 158, 160–162), sequence data are not available for most species other than C. parvum and C. hominis, and other Cryptosporidium species may go undetected by this assay.
Because there is little genetic variation across Cryptosporidium SSU rRNA genes, the design of primers/probes targeting the entire genus is relatively straightforward, but there are limited targets for designing species-specific primers/probes. Therefore, other loci have been targeted, such as COWP and the LIB13 locus (a coding region of unknown function) (Table 4), for the differentiation of Cryptosporidium species. A few published assays include both genus- and species-specific primers, enabling not only sensitive screening but also real-time identification of at least some species. One such example is a PCR targeting 125 bp of the SSU rRNA gene designed to amplify and partially differentiate Cryptosporidium species pathogenic to humans (152). Scorpion probes were designed to enable differentiation between C. parvum, C. meleagridis, and other species. The assay had a detection limit of 500 to 5,000 oocysts/g feces and was validated against microscopy and antigen detection; species identification by scorpion probes was validated by using RFLP analysis of amplicons, using VspI digestion specific for C. hominis. A generic TaqMan assay targeting the SSU rRNA gene and additional TaqMan assays for subsequent distinction between species infecting humans and those infecting cattle were reported recently by Burnet et al. (29).
Some of the challenges associated with developing standardized, “one-size-fits-all,” nucleic acid-based tests are exemplified in an interesting paper by Hadfield et al., who introduced a PCR assay based on two duplex reactions. One reaction targeted the entire genus by amplification of the SSU rRNA gene coupled with a reaction targeting the C. parvum-specific LIB13 locus (163). The second reaction targeted the C. hominis-specific LIB13 locus and also included an internal process control. Hence, the assay allowed direct detection of C. hominis and C. parvum, and in the event of a positive genus-specific (SSU rDNA) result in the absence of a positive result from either of the two LIB loci, the 300-bp-long SSU rRNA gene product could be sequenced for identification of the species present. It has been argued that SSU rDNA PCR is compromised in its ability to detect mixed species due to preferential amplification of the predominant species in a sample (153, 163, 164). Therefore, the setup developed by Hadfield et al. does not circumvent this potential problem, since this genus-specific PCR is based on SSU rRNA gene amplification. Interestingly, detection and differentiation of C. hominis, C. parvum, and C. meleagridis in human fecal samples can be performed by using high-resolution melting curve analysis of amplicons of the ITS2 region (165). Using this assay, C. hominis, C. parvum, and C. meleagridis were detected in 97, 44, and 2 samples, respectively, of 143 Cryptosporidium oocyst DNA samples originating from Australians with clinical cryptosporidiosis, and the results were in agreement with results previously obtained by single-strand conformation polymorphism analysis. Melting curve analyses in assays using intercalating dyes to distinguish between C. parvum and C. hominis were also reported by Tanriverdi et al. (166, 167). Fluorescence resonance energy transfer (FRET) probes (Fig. 1B) have also been used to distinguish between C. parvum and C. hominis (168), but there are issues regarding diagnostic sensitivity and potentially impaired performance in terms of species resolution in mixed infections.
For phylogenetic analysis and molecular epidemiological and outbreak investigations, PCR-RFLP or PCR-sequencing analysis of various loci, including SSU rDNA, COWP, GP60, heat shock protein 70 (HSP70), actin, thrombospondin-related adhesive protein, and many other gene targets, has been useful (169, 170). Although GP60 remains the locus most widely targeted, the ideal combination of loci for molecular epidemiological purposes remains to be identified (169, 171). Surprisingly, there are a number of species and genotypes for which complete SSU rRNA gene sequences are not available (Table 4), including the recently described species C. cuniculus and C. viatorum. This may be due to the fact that phylogenetic analysis of Cryptosporidium isolates are often carried out by using only partial SSU rRNA genes (despite the fact that variation is seen across the entire gene) and sometimes in conjunction with phylogenetic analysis of, for example, HSP70 and actin genes (172, 173).
PCR is slowly gaining a foothold in clinical microbiology laboratories, and the main approach has been the application of generic primers/probes used alone or multiplexed in assays targeting other relevant parasites; typically, these are used as a first-line screening tool as an alternative to traditional diagnostics (Table 2). DNA-based diagnostics offer improved diagnostic sensitivity, as shown by Morgan et al. in 1998 (12) and in a multitude of other studies. One example is seen in a study by Amar et al., who found that PCR resulted in a 22-fold increase in the detection of Cryptosporidium and Giardia versus conventional microscopy (174). An increased detection rate was found by using a DnaJ-like gene-based TaqMan assay (158) compared to commonly used commercial kits such as Merifluor Cryptosporidium/Giardia (Meridian Bioscience) and the ImmunCard STAT! Crypto/Giardia Rapid assay (Meridian Bioscience) (175). In a study by Stensvold and Nielsen (162), the multiplex assay was remarkably more sensitive than modified Ziehl-Neelsen staining of fecal concentrates, strongly supporting the implementation of a molecular screening platform for cryptosporidiosis in low-endemicity Denmark: 16/889 samples were positive for Cryptosporidium by real-time PCR, compared to none by modified Ziehl-Neelsen microscopy. Along these lines, Chalmers et al. (176) recently showed that the sensitivity of modified Ziehl-Neelsen microscopy was 75.4% compared to the real-time PCR developed by Hadfield et al. (163). Conversely, comparably high sensitivities were reported for auramine-phenol microscopy and commercial kits based on immunofluorescence microscopy and enzyme immunoassays (176). The validation study was carried out by using Cryptosporidium samples, 97% of which were either C. parvum or C. hominis, and therefore, the diagnostic sensitivity reported there must be interpreted in this light.
Cyclospora
Species of Cyclospora are obligate intracellular apicomplexan parasites infecting primates and a number of nonprimate hosts, including other mammals, reptiles, and arthropods. Cyclospora cayetanensis is the only species so far known to infect humans and is presumably host specific. Oocysts are excreted in feces, and thereafter, it takes >1 week for oocysts to sporulate in the environment. Infection is due to the ingestion of sporulated oocysts. Sporozoites released from ingested oocysts can infect the duodenum and jejunum. Oocysts are highly resistant to disinfectants used in the food industry (177).
Areas where cyclosporiasis is endemic include the Americas, the Middle East, Southeast and South Asia, South Africa, and southern Europe, but the parasite may even be seen in outbreaks in areas where the disease is not endemic, mainly due to the distribution of contaminated food produce such as raspberries, basil, lettuce, sugar snap peas, or other vegetables (178–183) or outbreaks in groups of individuals from areas where disease is not endemic who travel to areas of endemicity (184–186) or as sporadic cases after travel to areas of endemicity (186). Asymptomatic presentation is not uncommon in areas where the disease is endemic (177), but symptoms related to Cyclospora infection may include low-grade fever, anorexia, nausea, diarrhea, and weight loss and may be seen primarily in children and HIV/AIDS patients in areas of endemicity (177).
Recently, a study on population-based active surveillance for Cyclospora infection by the U.S. Foodborne Disease Active Surveillance Network concluded that clinicians should include Cyclospora infection in the differential diagnosis of prolonged or relapsing diarrheal illness and should explicitly request stool examinations for this parasite (187).
Traditional diagnostic methods include mainly autofluorescence (Fig. 3B) or, perhaps more commonly, modified Ziehl-Neelsen staining of fecal concentrates in which oocysts of Cyclospora (size, 8 to 10 μm) can be differentiated from those of Cryptosporidium (4 to 6 μm) and Cystoisospora (25 to 30 μm) by morphological characteristics.
PCR methods in various formats have been used for direct detection (screening) and confirmation of microscopy results. While the ITS region may offer higher resolution and therefore may be better for molecular epidemiological purposes (188, 189), the SSU rRNA gene has been the primary target for PCR-based diagnosis of Cyclospora in human fecal samples (Table 5).
TABLE 5.
PCR-based assays for detection and molecular characterization of Cyclosporaf
| Species | Amplification method | Detection method | Target | Sample type(s) | Reference |
|---|---|---|---|---|---|
| Cyclospora, Eimeria | Nested PCR | Gel electrophoresis | SSU rDNA | Purified oocysts | 464 |
| Cyclospora, Eimeria | Nested PCRa | Gel electrophoresis | SSU rDNA | Stool | 465 |
| Cyclospora, Eimeria | Nested PCR-RFLPa | Gel electrophoresis | SSU rDNA | Raspberries | 466 |
| Cyclospora, Eimeria | Nested PCRa | Oligonucleotide ligation | SSU rDNA | Controls | 467 |
| Cyclospora, Eimeria | Nested PCRa | Sequencing | SSU rDNA | Stool | 468 |
| Cyclospora | PCR | Gel electrophoresis | ITS1 | 188 | |
| Cyclospora | Real-time PCR | Hydrolysis probe | SSU rDNA | Controls | 190 |
| Cyclospora | Real-time PCR | Hydrolysis probe | SSU rDNA | Stool | 191 |
| Cyclospora, Eimeria | Nested PCR-RFLP | Gel electrophoresis | SSU rDNA | Water | 469 |
| C. cayetanensis, C. colobi, and Eimeria | Multiplex nested PCRb | Gel electrophoresis | SSU rDNA | Stool | 470 |
| Cyclospora, Eimeria | Nested PCRa | Gel electrophoresis | SSU rDNA | Sputum | 471 |
| Cyclospora, Eimeria | Nested PCRa | Gel electrophoresis | SSU rDNA | Stool | 472 |
| Cyclospora | Real-time PCRe | Hydrolysis probe | SSU rDNA | Stool | 473 |
| C. cayetanensis, C. colobi, and Eimeria | Multiplex Nested PCRd | Gel electrophoresis | SSU rDNA | Stool | 194 |
| Cyclospora, Eimeria | Nested PCR-RFLPa | Gel electrophoresis | SSU rDNA | Stool | 192 |
| C. cayetanensis | PCR | Gel electrophoresis | ITS2 | Spiked samples | 474 |
| Cyclospora | PCRc | Gel electrophoresis | ITS1 | Stool | 189 |
| Nested PCR | Gel electrophoresis | SSU rDNA | Controls | 450 | |
| Cyclospora, Cryptosporidium, Toxoplasma, and Sarcocystis | Real-time PCR | SYBR green MCA | SSU rDNA | Control samples | 219 |
| C. cayetanensis, C. colobi, and Eimeria | Multiplex PCRd | Luminex | SSU rDNA | Stool | 193 |
| C. cayetanensis, C. colobi, and Eimeria | Multiplex nested PCRd | Gel electrophoresis | SSU rDNA | Stool | 184 |
| Cyclospora | Nested PCR | Sequencing | SSU rDNA | Stool | 475 |
| Cyclospora | Nested PCR | Sequencing | HSP70 | Stool | 195 |
| C. cayetanensis, C. colobi, and Eimeria | Nested PCRd | Electrophoresis | SSU rDNA | Raspberry, basil, pesto | 476 |
| C. cayetanensis, C. cercopitheci | Real-time PCR | Hydrolysis probe | SSU rDNA | Raspberry, basil, pesto | |
| C. cayetanensis | Real-time PCR | Hydrolysis probe | HSP70 | Raspberry, basil, pesto |
PCR primers are described in reference 464.
First-round PCR primers are described in reference 464.
PCR primers are described in reference 188.
PCR primers are described in reference 470.
PCR primers are described in reference 190.
Abbreviations: SSU, small subunit; ITS, internal transcribed spacer; HSP, heat shock protein; RFLP, restriction fragment length polymorphism; MCA, melt curve analysis.
In 2003, the first TaqMan assays were reported (190, 191), enabling the incorporation of Cyclospora into multiplex screening assays to detect and differentiate multiple genera and species of apicomplexa and microsporidia, with the key driving forces being dramatically increased sensitivity compared to that of microscopy (191, 192) and the fact that these parasites are rarely encountered in clinical samples in areas where the disease is not endemic. A multiplex PCR method to detect Cyclospora, Cystoisospora, and microsporidia in stool samples based on Luminex technology was recently reported (193). While the probe and reverse primer appear to be genus specific, the forward primer might be species specific for C. cayetanensis. The assay could possibly be modified toward the development of a Cyclospora genus-specific PCR. It appears relevant to combine diagnostics for intestinal sporozoa (Cystoisospora, Cyclospora, Cryptosporidium, and microsporidia) in multiplex PCR assays, as these parasites are often waterborne, most can be found in outbreaks, and there may be significant overlap in risk factors and symptoms caused by these parasites.
Unfortunately, little has been published on the use of nucleic acid-based tests for the diagnosis of Cyclospora infections in a clinical diagnostic setting. In a small study in Peru, nested PCR detected 57.1% positive results in diarrheal cases (20/35) versus only 18.9% positive results in controls (3/15), compared to only 18.9% and 6.7% positive samples in cases and controls, respectively, by microscopy (192). In Egypt, Cyclospora infections were detected in 35 of 140 (25%) diarrheic children by using nested PCR, compared to 17.8% and 22.2% with modified Kinyoun stain and autofluorescence microscopy, respectively (194). In The Netherlands, 100 stool specimens from returning travelers with diarrhea were screened for the presence of C. cayetanensis by using fluorescence microscopy and real-time PCR. C. cayetanensis was found in five cases by PCR, one of which (20%) could be confirmed by microscopy only after examination of several additional slides; C. cayetanensis was the most frequent parasitic cause of diarrhea after G. lamblia (191).
Genotyping has been performed for the differentiation of Cyclospora species by using the ITS region and the SSU rRNA gene. Very few other genes have been explored, and no sequence variation was found in the HSP70 genes of Cyclospora cayetanensis isolates from Nepal, Mexico, and Peru (195).
Cystoisospora
The apicomplexan genus Cystoisospora (Sarcocystidae; formerly Isospora) comprises species that infect humans, dogs, cats, and other mammals (196, 197). While cystoisosporiasis may be common in nonhuman hosts, human infection by Cystoisospora appears to be relatively rare, and symptomatic infections are probably seen mostly in immunocompromised individuals, including patients with HIV/AIDS or lymphoproliferative disorders, in whom infections are often chronic and severe (198–201). Superinfection of the small bowel during administration of systemic corticosteroids for eosinophilic gastroenteritis has been documented (202).
Regions of endemicity include tropical Africa, Southeast Asia, and Central and South America; in Western countries, human infections may largely be imported from travel to or origins in countries where the disease is endemic (203).
Infection is probably self-limiting in immunocompetent hosts. Symptoms may include diarrhea, steatorrhea, abdominal pain, fever, malaise, nausea, vomiting, weight loss, dehydration, and cachexia (197). Bile duct infection may be seen, while disseminated disease is probably rare but has been reported for AIDS patients. Recurrences are common, and the chronic nature of the illness contributes to morbidity and mortality among these patients (204–206). Sulfamethoxazole-trimethoprim treatment provides a good clinical response and is the drug of choice (207).
Mature oocysts are infective and contain two sporocysts, each containing four sporozoites. Sporozoites released into the small bowel by ingested oocysts invade the intestinal epithelium, and once intracellular, the parasites undergo schizogony with merozoite, trophozoite, schizont, gametocyte, and oocyst formation (196, 197). Tissue cysts (lamina propria) may be seen by endoscopy in the absence of fecal shedding of oocysts; tissue cysts may be unizoite (208, 209). Oocysts are oblong and characterized by a thin, transparent shell surrounding the sporocysts and are usually detected in the laboratory by Ziehl-Neelsen staining of fecal concentrates (Fig. 3D) or by phase-contrast or fluorescence microscopy of unstained wet mounts (Fig. 3C). Flotation techniques are sometimes used.
There is some indication that oocyst morphology differs from host to host (210). Very few molecular studies have been undertaken to characterize Cystoisospora from humans and other animals, so host specificity and the level of genetic diversity are incompletely known; however, Cystoisospora belli (formerly Isospora belli) is the species that has been reported in humans.
While Murphy et al. diligently described how Cystoisospora can be detected by using a broad-specificity primer approach (211), such an approach is not feasible in general practice and on routine screening platforms due to potential requirements such as cloning and low cost-effectiveness. Reports of targeted PCR assays are scarce but include an assay based on nested PCR (212). This nested PCR used an outer primer pair, IsoFO (5′-GTGCCTCTTCCTCTGGAAGG-3′) and IsoRO (5′-GCACTCCACCCAGTTAAGTGC-3′), and an inner primer pair, IsoFI (5′-CGATGGATCATTCAAGTTTC-3′) and IsoRI (5′-ACCACGTACACACCCCTAAG-3′), followed by probe hybridization (IsoOP [5′-GT{T/A}ATGGCTTCGGCCGGCGATGGA-3′]). While this assay could potentially be implemented in a TaqMan format, the primers have not been validated on fecal DNA. A real-time PCR with internal process control was described by ten Hove et al. (213). It uses primers Ib-40F (5′-ATATTCCCTGCAGCATGTCTGTTT-3′) and Ib-129R (5′-CCACACGCGTATTCCAGAGA-3′) and amplifies an 89-bp fragment of the ITS2 region, which is detected by the double-labeled probe Ib-81Taq (5′-FAM [6-carboxyfluorescein]-CAAGTTCTGCTCACGCGCTTCTGG-BHQ1-3′). The assay was shown to be specific for 147 bacterial, parasitic, and fecal control DNA samples and detected C. belli-specific amplification in 21 microscopy-positive stool samples. This assay may be specific for C. belli; at least the probe is located in a region where C. ohioensis exhibits a vast degree of divergence, including an insertion of 3 bases. C. ohioensis is one of several species of Cystoisospora identified in synanthropic carnivores (214). However, for several species of Cystoisospora, no sequence data are available for this particular region, so the specificity of the probe is difficult to determine at present.
A multiplex PCR with 4 primer sets to amplify C. cayetanensis, C. belli, Enterocytozoon bieneusi, and Encephalitozoon intestinalis was developed, in which detection of amplicons occurs by specific probes coupled to Luminex beads (193) (Table 2). For C. belli, primers were based on C. belli 5.8S rRNA and ITS2 and amplify a 213-bp sequence. These primers may enable the detection of species of Cystoisospora other than C. belli; currently, only data for C. ohioensis are available in GenBank for this region, and only 1 to 2 mismatches are seen across the primers.
The SSU rRNA gene and ITS1 sequences are used to determine the taxonomic status of Cystoisospora isolates (210, 214–216).
Sarcocystis
Humans may serve as both intermediate and definitive hosts of Sarcocystis; only intestinal Sarcocystis infections are briefly discussed here. Although Sarcocystis may include distinct species pathogenic to humans, very little is known regarding host specificity, taxonomy, and overall epidemiology (217). Considering the fairly high prevalence in various cohorts across the globe, as reviewed by Fayer (217), it is surprising that reports on Sarcocystis infections of humans are so scarce. While DNA-based confirmation is still pending, it is possible that intestinal infection by Sarcocystis in humans may be caused by both Sarcocystis hominis and Sarcocystis suihominis and acquired through the consumption of tissue cysts in undercooked beef and pork, respectively (217).
Symptoms related to intestinal infection include nausea, loss of appetite, vomiting, stomach ache, bloating, diarrhea, dyspnea, and tachycardia; they appear only a few hours after ingestion of tissue cysts and may last for about 36 h. The prepatent period is 5 to 12 days, and patency may last for at least 120 days. Importantly, symptoms overlap those characteristic of acute gastroenteritis (217).
Seminested PCR combined with RFLP analysis has been used to detect and distinguish S. hominis, S. fusiformis, and S. cruzi-like organisms (218). Sarcocystis cruzi was included in a real-time PCR assay targeting various coccidia of animal health and zoonotic importance, where organisms were detected and differentiated based on melting curve analysis (219).
CILIATES
One genus of ciliates is known to be able to infect humans: Balantidium.
Balantidium
Balantidium has a cosmopolitan distribution and is present generally wherever pigs are present (220). Both B. coli and B. suis infections of pigs have been reported, but molecular studies still remain to be performed to settle whether B. coli and B. suis are in fact separate species. Recent data, however, suggest that B. coli has low host specificity (221). A study from Denmark revealed a prevalence of 100% in some pig cohorts, but so far, cases of human balantidiosis acquired in Denmark remain to be reported. It has been suggested that Balantidium does not readily produce patent infections in humans (220). While asymptomatic infections may be common, dysentery and invasion of the colon have been reported (220, 222). Bowel perforation caused by B. coli, leading to severe peritonitis, was described in France (223). Extraintestinal spread to the peritoneal cavity, genitourinary tract, and lungs has been reported (224–227).
Transmission occurs by ingestion of cysts contaminating food or drink. Risk factors include contact with pigs and pig excreta (224, 228). In humans, B. coli is the species reported; the same species is known to infect pigs. It is the only ciliate and the largest protozoon known to infect humans, and while human infections may be rare, Balantidium may be extremely common in other hosts, including pigs, causing asymptomatic infections. In fact, a number of ciliates are known to colonize ruminants, to which hosts these parasites are often beneficial, assisting the host in food digestion and carbohydrate metabolism (229).
DNA or cysts of Balantidium have been detected in fecal samples from diverse hosts such as humans and nonhuman primates, pigs, ostriches, rats, guinea pigs, salamanders, frogs, and fish (230–234) and even in nonfecal human specimens such as bronchoalveolar lavage (BAL) fluid (224–227).
Because the cysts measure 40 to 60 μm and trophozoites might be as large as 200 μm and are easily overstained in iodine-stained wet mounts, they may easily be missed when focusing on finding protozoan cysts by routine microscopy.
So far, PCR has been used mainly to characterize Balantidium from various hosts, including humans (221, 232–234). Very few Balantidium sequences are currently available in GenBank, and most represent the SSU rDNA or ITS regions of B. coli. Diagnostic PCRs remain to be reported.
Application of general, broad-specificity primers targeting nonhuman eukaryotic SSU rDNA may be of significant utility in efforts to cost-effectively screen for Balantidium in clinical samples other than feces (e.g., biopsy specimens and BAL fluid, etc.).
MICROSPORIDIA
Microsporidia are obligate, intracellular, single-celled fungi that infect a wide variety of vertebrate and invertebrate hosts, comprising >160 genera representing over 1,500 species, at least 14 of which are known to be able to infect humans. Intestinal microsporidiosis in humans is due mainly to Enterocytozoon bieneusi and Encephalitozoon species, mainly E. intestinalis and, probably to a lesser extent, E. cuniculi and E. hellem (235–238). E. bieneusi is generally the species associated most frequently with human infections, although a recent study on Russian HIV-positive patients found that E. intestinalis was much more common than E. bieneusi in this particular cohort (239). In humans, opportunistic infections associated with persistent diarrhea and weight loss may be seen, especially in individuals with HIV/AIDS and organ transplant patients, while in immunocompetent individuals, symptomatic, self-limiting infection may occur (240–245). In HIV-infected patients, a CD4+ T-cell count of <100 cells per μl appears to be a significant risk factor for microsporidiosis (246–248), although a study of HIV patients in Nigeria reported microsporidiosis associated with a CD4+ T-cell count of <200 cells per μl (249); in a study of Danish HIV patients with unexplained diarrhea, most of whom were treated with HAART and had CD4+ T-cell counts of >100 cells/μl, no cases of microsporidiosis were detected by PCR for Enterocytozoon and Encephalitozoon (160). Infections due to Encephalitozoon may also cause rhinosinusitis, keratoconjunctivitis, nephritis, hepatitis, and systemic infections.
Probably because of the increased attention to microsporidiosis during the AIDS epidemic and the improvement of diagnostic techniques, microsporidiosis is now increasingly being diagnosed in transplant patients, children, the elderly, and travelers (241). Although reports of outbreaks of microsporidia are scarce, both food- and waterborne outbreaks have been reported (250, 251). Most infections in immunocompetent individuals are self-limiting; in immunocompromised patients, primary treatment may include chemotherapeutic intervention with the aim of restoring immunocompetence, such as administration of granulocyte colony-stimulating factor in combination with the use of antibiotics such as nitazoxanide, albendazole, or fumagillin (252, 253).
The traditional gold standard for the detection of microsporidia relies on the demonstration of 1- to 2-μm spores by transmission electron microscopy, but this is an insensitive method for finding spores, since the specimen analyzed is relatively small. Detection of microsporidia in stool samples often includes the identification of spores in fecal smears by nonspecific histochemical chemofluorescent agent stains or trichrome stain as well as monoclonal antibody immunofluorescence assays (246, 254–256). Even using these staining techniques, detection and identification of microsporidial spores are difficult, and expertise is required. PCR has been shown to be a valuable technique in the diagnosis of these important opportunistic pathogens (238). Moreover, as distinct treatment options are available for different genera, identification to the genus and species levels is clinically important (252).
Molecular diagnostic tests for the detection of microsporidia have been reviewed extensively (238, 257, 258). PCR diagnosis for E. bieneusi was developed as early as 1993 (259), and since then, a variety of conventional PCRs for detection and species identification have been described. Meanwhile, relatively few real-time PCR methods have been reported (Table 6). A PCR followed by detection of E. bieneusi, E. cuniculi, E. hellem, and E. intestinalis by microarray has been developed (260). The assay developed by Taniuchi et al. included a multiplex PCR setup along with primers and probes for Cyclospora and Cystoisospora (193). This appears to be a relevant option for the detection of diarrheagenic parasites that are difficult to detect by conventional methods, particularly in susceptible individuals such as HIV/AIDS patients and recipients of organ transplants.
TABLE 6.
PCR-based assays for detection and molecular characterization of microsporidiaf
| Species | Amplification method | Detection method(s) | Target | Sample type(s) | Reference |
|---|---|---|---|---|---|
| E. intestinalis, E. cuniculi, E. hellem | Real-time PCRe | Hydrolysis probe | SSU rDNA | Controls | 477 |
| E. intestinalis, Encephalitozoon species | Real-time PCR | FRET probes and MCA | SSU rDNA | Stool (spiked) | 478 |
| E. intestinalis | Real-time PCR | Hydrolysis probe | SSU rDNA | Stool, blood, urine, tissue biopsy specimens, and bronchopulmonary specimens | 479 |
| E. bieneusi | Real-time PCR | Hydrolysis probe | SSU rDNA | Stool | 480 |
| E. bieneusi, Encephalitozoon species | PCR-hybridization | Chemiluminescence | SSU rDNA | Stool, urine (spiked) | 481 |
| E. bieneusi | Real-time PCR | Hydrolysis probe | SSU rDNA | Stool | 482 |
| E. bieneusi, E. cuniculi, E. hellem, and E. intestinalis | PCR | Microarray | SSU rDNA | Stool | 260 |
| E. bieneusi, E. intestinalis | Real-time PCRa,b | Hydrolysis probe | SSU rDNA | Stool | 266 |
| E. bieneusi, E. intestinalis | PCR | Gel electrophoresis | SSU rDNA | Stool | 483 |
| E. bieneusi | Multiplex real-time PCR | Hydrolysis probe | ITS | Positive stool | 457 |
| Encephalitozoon species | SSU rDNA | 457 | |||
| E. bieneusi | PCR | Sequencing | SSU rDNA | Positive stool | 37 |
| E. bieneusi, E. intestinalis | Multiplex PCRc | Luminex | SSU rDNA | Stool | 193 |
| E. bieneusi, Encephalitozoon species | Real-time PCR | SYBR green and MCA | SSU rDNA | Stool | 484 |
| E. bieneusi, E. intestinalis | Real-time PCRb,d,e | Hydrolysis probe | SSU rDNA | Stool | 485 |
E. bieneusi primers and probe are described in reference 482.
E. intestinalis primers and probe are described in reference 479.
E. intestinalis primers and probe are described in reference 457.
E. bieneusi primers and probe are described in reference 480.
Singleplex PCRs in parallel.
Abbreviations: SSU, small subunit; ITS, internal transcribed spacer; HSP, heat shock protein; RFLP, restriction fragment length polymorphism; MCA, melt curve analysis; FRET, fluorescent resonance energy transfer.
Based on the ITS nucleotide sequence of E. bieneusi recovered from feces of infected humans and animals, E. bieneusi comprises a perplexing array of genotypes, several of which have been found in humans. New genotypes keep emerging, and thus, the number of E. bieneusi genotypes today may be well over 100 (37, 249, 256, 261, 262). Some of these genotypes have been recognized as being host specific, while others have been found to infect both humans and other animals, supporting the likelihood of zoonotic transmission (235). Interestingly, pigeons appear to constitute a reservoir for species of microsporidia seen in humans; however, little is known about the potential overlap of genotypes between humans and pigeons (263).
Genotype B was the only genotype identified in samples from patients with HIV in Australia and was also the predominant genotype in France and The Netherlands (37, 248, 264). In contrast, a variety of genotypes were identified in HIV-infected individuals in Portugal, Peru, Thailand, Niger, Nigeria, Vietnam, and Gabon and in unselected individuals in Cameroon (247, 249, 265–267). Genotype D was recently reported for two Spanish transplant patients (242); in two other studies of organ transplant recipients in Europe, genotype C was the predominant genotype (37, 264). Genotype C was also found to be responsible for a food-borne outbreak in Sweden (250).
E. cuniculi infection in humans is rarely reported, while birds, canids, rabbits, and rodents may be common hosts (236, 268). At least four genotypes are known (269), among which genotype II has not been detected in humans to date to our knowledge.
While genetic variation in E. intestinalis remains to be described, E. hellem comprises at least 3 genotypes with some intragenotypic variation (269–274). Interestingly, analysis of four E. hellem isolates from humans revealed that the C-terminal regions of the spore wall proteins EhSWP1a and EhSWP1b are polymorphic, which is of interest for epidemiological studies (275).
While microsporidia may not currently qualify as part of a routine test panel for intestinal pathogens, PCR-based detection appears to be relevant in cases of HIV-related diarrhea, diarrhea in patients undergoing organ transplantation, and unexplained diarrhea in otherwise immunocompromised patients. Genotyping has immediate relevance to outbreak investigations but has also proven useful in surveillance studies of microsporidia in humans and other animals to identify transmission patterns and other aspects of epidemiology. It appears that there is a plethora of genotypes for E. bieneusi, and while ITS sequence analysis may prove valuable for further exploration of the complex epidemiology of microsporidia, it may be relevant to identify alternatives to ITS sequence analysis for a more clinically relevant strain identification approach.
Of note, few diagnostic PCRs have been developed for species that potentially may cause systemic infections, and validation studies on DNA extracted from clinical samples other than stool, such as urine and pulmonary samples, remain limited.
STRAMENOPILES
Stramenopiles encompass a range of very diverse organisms, most of which are free-living. Blastocystis is remarkable in that it has adapted to a strictly anaerobic parasitic life-style and is capable of colonizing a vast variety of mammalian, avian, reptilian, amphibian, and arthropod hosts. Only one other stramenopile is known to be able to cause parasitic infections in humans, namely, Pythium, which is a rare cause of skin disorders and systemic granulomatous disease. Proteromonas lacertae, which is closely related to Blastocystis, can be found in reptiles.
Blastocystis
Blastocystis (Fig. 2D) is a common, strictly anaerobic, unicellular intestinal parasitic protist of humans and a wide variety of nonhuman hosts (276–278). Despite the fact that it is one of the most common microbial eukaryotes to colonize the human intestine and its clinical significance is largely unknown, it remains a relatively little-studied parasite. This may be due in part to a variety of unrelated predicaments: the inconspicuousness of the parasite, which makes diagnosis based on morphology difficult; the difficulties associated with isolation in axenic culture; the resilience of infections, which may be chronic and difficult to eradicate; and the frequency with which the parasite presents itself along with other intestinal microbial eukaryotes, such as Dientamoeba, making it extremely difficult to know whether potential symptoms are due to Blastocystis or another organism. It is not known how to eradicate the parasite, and Blastocystis is not even remotely genetically related to other microeukaryotes that colonize or infect the human intestine, such as other protists and yeasts. Although all the stages involved in the life cycle of Blastocystis have not been fully clarified, transmission is by the fecal-oral route and possibly mostly involves the accidental ingestion of the cyst stage (278).
While asymptomatic carriage is common, Blastocystis has been linked to disease in a variety of case reports (279). The parasite exhibits remarkable genetic diversity, and to date, nine genetically distinct lineages, so-called subtypes (STs) (arguably species), have been found in humans, of which ST1 to ST4 account for >90% of human Blastocystis carriage (277). Efforts continue to identify potential associations between subtypes and clinical outcomes of colonization; so far, results are pointing in different directions.
Microscopy of fecal concentrates has low diagnostic sensitivity, which has most likely led to substantial underreporting of the parasite (280–282), whereas the use of permanently stained smears of preserved stool specimens will increase the rate of detection of Blastocystis (283). The use of molecular tools in Blastocystis research and routine diagnostics has had a crucial impact on our understanding of Blastocystis epidemiology and transmission. The first report of a diagnostic PCR for Blastocystis, in 2006, was partly inspired by the tendency toward screening of fecal DNAs for common intestinal protozoa by PCR as a supplement to or even as a substitute for microscopy of fecal concentrates (284). Primer design was based on sequences available in the NCBI database at that particular time. Analysis of newer data leads to the conclusion that this PCR assay may exhibit preferential amplification of some STs over others due to sequence variation in primer annealing sites, which may impair its use in epidemiological studies. Indeed, the extensive intrageneric diversity of Blastocystis has made the design of a diagnostic genus-specific PCR applicable to fecal DNA templates challenging.
Three diagnostic real-time PCR assays have been reported. A real-time PCR based on an unknown Blastocystis gene using FRET probes was validated against ST1, ST3, and ST4 (285). A SYBR green real-time PCR was designed based on the SSU rRNA gene for the detection of Blastocystis-specific DNA and subsequent subtyping by melting curve analysis (286). Low sensitivity can be expected due to the relatively large PCR product (320 to 342 bp, depending on the subtype), and the specificity of the assay is only 95%. The third real-time assay, using a hydrolysis probe based on the SSU rRNA gene, was characterized by 100% specificity (287). The use of real-time PCR in large-scale surveys will assist in identifying whether the development of symptoms is related to infection intensity by simple analysis of threshold cycle (CT) values for individual samples.
Xenic in vitro culture (XIVC) sensitivity ranged between 52 and 79% compared to these real-time PCR assays (286, 287). Previously, XIVC was found to have a sensitivity of 89% compared to conventional PCR (280). Conventional PCR relies on visual evaluation of PCR results, and this PCR was based on primers that amplify a relatively large PCR product (∼550 to 585 bp), which was suitable for sequencing and subtype identification but too large to be relevant for diagnostic PCR, especially in situations where fecal DNAs are of suboptimal purity. An estimate of the number of rRNA gene copies in one Blastocystis cell is not available, although it may lie somewhere between 20 and 100 (287, 288).
Generally, two methods have been employed for the genetic characterization (subtyping) of Blastocystis, namely, barcoding (289) and sequence-tagged-site (STS) PCR (290). These two methods were recently evaluated by Stensvold (291), who concluded that barcoding is the method of choice, enabling the detection of novel subtypes and further scrutiny of genetic diversity, including analysis of SSU rRNA alleles, since the barcode region has been validated as a marker of overall genetic diversity of Blastocystis (289, 292).
Barcode primers (RD5/BhRDr) amplify 600 bp of the 5′ end of the SSU rRNA gene, and phylogenetic analysis has demonstrated that this region is a valid surrogate genetic marker for complete SSU rRNA gene sequences and even for markers in the genome of the mitochondrion-like organelle (289, 292). The drawbacks compared to the STS method are that sequencing is needed and that mixed-subtype colonization may be difficult to decipher. On the other hand, barcoding enables a more subtle analysis, namely, SSU rDNA allele analysis. A public database is available (http://pubmlst.org/blastocystis/), which includes a sequence depository for barcode sequences and sequences obtained by multilocus sequence typing (MLST) (see below). It also has a BLAST facility, where individual or bulk fasta files can be uploaded and analyzed for quick identification of subtype number, hence obviating the need for phylogenetic analysis (292, 293). SSU rDNA allele analysis is a useful indicator of intrasubtype genetic variation (292), and to date, >35 SSU rDNA alleles for ST3 have been identified, whereas the numbers of SSU rDNA alleles for ST4 and some other subtypes remain much more limited. However, some of the allelic variation in databases is the result of cloning and sequencing of individual genes from strains rather than sequences based on PCR products produced from the whole genome; intragenomic SSU rDNA polymorphism has been reported (288).
Subtyping has revealed significant differences in Blastocystis epidemiology: ST4 appears to be common in Europe, while it is generally rare in most other regions. ST6 and ST7 account for about 20% of the cases in Africa, while only sporadic cases are seen in other regions. Independent data from Denmark (294) and Spain (295) show associations between ST4 and diarrhea, and ST4 is also common in United Kingdom patients suffering from IBS (277). Geographical differences in the distribution of subtypes may hamper attempts to identify subtypes specifically linked to disease.
“Genotype” has been used by some authors interchangeably with “subtype.” Presuming that Blastocystis subtypes are equivalent to separate species, subtype allele analysis should be regarded as the equivalent of genotyping in other organisms. At this level, there is currently very little information on Blastocystis. However, it is likely that the introduction of the SSU rDNA allele database at PubMLST (http://pubmlst.org/blastocystis/) will greatly facilitate studies of both subtypes and SSU rDNA alleles, enabling rapid analysis of fasta files obtained by barcoding (289). Indeed, allele analysis is likely to be an essential tool in future analyses of the potential zoonotic transmission of Blastocystis.
MLST analysis of Blastocystis is currently based on analysis of loci in the mitochondrion-like genome (292); mitochondrial DNA (mtDNA) is especially useful in MLST analyses due to its haploid structure, hence bypassing problems related to sequence heterozygosity, as seen, for example, in some Giardia MLST loci (91). So far, MLST systems are available for ST3 and ST4, and similar assays for ST1 and ST2 will follow. Analysis of 132 ST3 and ST4 isolates from humans and nonhuman primates recently revealed dramatic differences in intrasubtype diversity. No fewer than 58 sequence types (SQTs) were detected among 81 ST3 samples, while only 5 SQTs were found among 50 ST4 samples belonging to the common genotype (215). ST4 samples obtained from Denmark, England, and Nigeria shared the same SQT.
While MLST therefore appears to be a very useful tool for investigating patterns of transmission, at least for ST3, the information obtained by the much simpler SSU rDNA allele analysis can be an extremely cost-effective tool. A recent study of Blastocystis in NHPs representing 30 genera showed that NHPs typically host the same subtypes as humans, apart from the fact that ST4 is rare in NHPs while ST5 and ST8 are rare in humans. However, despite ST1 and ST3 being common, it was noted that many of the ST3 alleles found in NHPs are not found in humans, and the same holds true for ST1 (296). A large overlap, however, appears to be present for ST2 SSU rDNA alleles, but only MLST can confirm whether or not NHP ST2 isolates are indeed identical to human ST2 isolates.
Due to limited knowledge of the clinical significance of Blastocystis, implementation of molecular diagnostics in the routine clinical setting may be considered premature. However, since large-scale epidemiological studies of different cohorts represent a simple pathway to knowledge in Blastocystis research, the use of DNA detection methods will most likely prove pivotal in distinguishing between carriers and noncarriers; barcoding can subsequently be used for subtyping and allele identification with no particular a priori knowledge by using the sequence query facilities at the PubMLST website (http://pubmlst.org/blastocystis/) (277, 292, 293). In the event that differences in the clinical outcomes of Blastocystis infections reflect differences in subtypes or strains, nucleic acid-based methods will most likely be implemented as first-line diagnostics (for recent reviews and updates on Blastocystis research, see references 276, 297, and 298).
SOIL-TRANSMITTED HELMINTHS
Eggs of Trichuris trichiura, Ascaris lumbricoides, Necator americanus, and Ancylostoma duodenale develop into infective eggs or larvae when excreted onto soil, hence the collective name soil-transmitted helminths (STHs) (Fig. 4). New infections are acquired by ingestion of eggs or penetration of larvae through the skin. With a global estimate that hundreds of millions of people are infected, especially in developing countries, these infections are the most common but also the most neglected infections worldwide (299–302). Although Strongyloides stercoralis has a similar infection route, it is usually mentioned only as an aside under the STH heading and is therefore perhaps even more neglected (303, 304), which is strange considering that, as a result of autoinfection, this very chronic disease may suddenly derail, for example, through the use of corticosteroids, leading to a fatal outcome (305–307).
FIG 4.
Wet mounts of a Trichuris trichiura egg (A), an Ascaris lumbricoides egg (B), a hookworm egg (C), and Strongyloides stercoralis larva (D). (Parasite images courtesy of Marianne Lebbad; reprinted with permission.)
Diagnosis appears to be simple and quite straightforward compared to the difficult microscopic diagnosis of protist infections. However, in Kato-Katz slides, which are often used in epidemiological studies, hookworm eggs will disappear if slides are not read within 30 to 60 min. The diagnosis of S. stercoralis infection is notoriously difficult due to the often very small numbers of larvae in chronic infections; multiple stools have to be tested by using Baermann and coproculture techniques to achieve adequate sensitivity (308). Conventional and real-time PCRs for the detection and quantification of soil-transmitted helminths have been developed but are still used mainly for epidemiological studies in areas of endemicity (Table 7).
TABLE 7.
PCR-based assays for detection and molecular characterization of intestinal nematodesh
| Species | Amplification method | Detection method(s) | Target(s) | Sample type(s) (no. of samples) | Reference |
|---|---|---|---|---|---|
| O. bifurcum | Nested PCR | Gel electrophoresis | ITS2 | Stool (119) | 486 |
| N. americanus, O. bifurcum | Nested PCRa | Gel electrophoresis | ITS2 | Stool (262) | 13 |
| A. duodenale, N. americanus | Nested PCRb | Gel electrophoresis | ITS2 | Stool (503) | 487 |
| Ancylostoma, N. americanus, O. bifurcum | Multiplex real-time PCR | Hydrolysis probe | ITS2 | Stool (339) | 328 |
| A. lumbricoides | Nested PCR | Gel electrophoresis and sequencing | Cytb | Coprolites (9) | 488 |
| A. lumbricoides | Nested PCR | Gel electrophoresis and sequencing | ITS1, Cytb | Stool (13) | 318 |
| S. stercoralis | Real-time PCR | Hydrolysis probe | SSU rDNA | Stool (370) | 354 |
| A. duodenale, N. americanus, Trichostrongylus | Nested PCR | Gel electrophoresis | ITS1-5.8S-ITS2 | Stool (203) | 489 |
| Trichuris vulpis, T. trichiura | Nested PCR | Gel electrophoresis | SSU rDNA | Human stool (80), dog stool (79) | 311 |
| Ancylostoma, N. americanus, A. lumbricoides, S. stercoralis | Multiplex real-time PCRc,d | Hydrolysis probe | ITS2 (Ancylostoma, N. americanus), ITS1 (A. lumbricoides), SSU rDNA (S. stercoralis) | Stool (1,312) | 319, 321, 333 |
| Ancylostoma, N. americanus, A. lumbricoides, S. stercoralis | Multiplex real-time PCRe | Hydrolysis probe | ITS2 (Ancylostoma, N. americanus), ITS1 (A. lumbricoides), SSU rDNA (S. stercoralis) | Stool (78, 229) | 161, 320 |
| S. stercoralis | Real-time PCR | Hydrolysis probe | 28S | Spiked stool | 353 |
| S. stercoralis (and O. viverrini) | Real-time multiplex PCR | FRET probes-MCA | SSU rDNA | Stool | 356 |
| Ancylostoma, N. americanus, A. lumbricoides, S. stercoralis | Multiplex PCRe | Luminex detection | ITS2 (Ancylostoma, N. americanus), ITS1 (A. lumbricoides), SSU rDNA (S. stercoralis) | Stool (319) | 322 |
| Ancylostoma, N. americanus, A. lumbricoides, S. stercoralis, T. trichiura | Real-time PCRc,d | Hydrolysis probe | ITS2 (Ancylostoma, N. americanus), ITS1 (A. lumbricoides, T. trichiura), SSU rDNA (S. stercoralis) | Stool (525) | 18 |
| T. trichiura, A. lumbricoides | Real-time PCRf | Hydrolysis probe | SSU rDNA (T. trichiura), ITS1 (A. lumbricoides) | Stool | 309 |
| N. americanus | Real-time PCR | SYBR green | ITS2 | Stool (216) | 332 |
| A. duodenale, N. americanus | Nested PCRg | Gel Electrophoresis | ITS2 | Stool (634) | 490 |
| N. americanus, A duodenale, A. ceylanicum, A. caninum, A. braziliense | Real-time PCR | SYBR green-HRM | ITS2 | Stool (634) | 330 |
| S. stercoralis | Real-time PCRd | Hydrolysis probe | SSU rDNA | Stool (160) | 355 |
| Ancylostoma, N. americanus | Real-time PCRc | Hydrolysis probe | ITS2 | Stool (195) | 329 |
O. bifurcum primers are described in reference 486.
N. americanus PCR primers and probes are described in reference 13.
A. duodenale and N. americanus PCR primers and probes are described in reference 328.
S. stercoralis PCR primers and probe are described in reference 354.
PCR primers and probes are described in reference 319.
A. lumbricoides PCR primers are described in reference 319.
PCR primers are described in reference 487.
Abbreviations: SSU, small subunit; ITS, internal transcribed spacer; MCA, melt curve analysis; HRM, high-resolution melt curve analysis; Cytb, cytochrome b; FRET, fluorescent resonance energy transfer.
Trichuris trichiura
To date, only two papers have been published on the use of PCR for the detection of T. trichiura-specific DNA with an intended use as a first-line diagnostic tool. This might be due to difficulties in the isolation of parasite DNA from the very robust Trichuris eggs (17, 161). The first PCR targeting the T. trichiura SSU rRNA gene for detection and quantification of T. trichiura infections was incorporated into a real-time PCR in a TaqMan array card format that detected 19 enteropathogens (309). By testing analytical performance using spiked stool samples, the assay showed 100% sensitivity and specificity. Clinical samples from Tanzania and Bangladesh with a variety of known pathogens detected by conventional assays showed T. trichiura-specific DNA amplification in 8 of 8 microscopy-positive samples, and an additional infection was detected in 1 of 80 microscopy-negative samples. In a recent paper, a T. trichiura-specific singleplex real-time PCR designed from the ITS1 sequence was performed as part of a screen of stool samples collected from schoolchildren in rural Ecuador in parallel with seven additional singleplex PCRs for other intestinal parasitic infections (18). PCR detected T. trichiura-specific DNA in 12 of 400 samples, whereas direct wet mount slides identified 3 of 400 samples with T. trichiura eggs only. There was a significant correlation between egg counts measured by the Kato-Katz method and the T. trichiura-specific DNA load quantified by PCR.
Phylogenetic analysis of Trichuris SSU, ITS1, 5.8S, ITS2, and mitochondrial genes derived from worms and eggs isolated from different host species has been performed. Although T. trichiura and T. suis cannot be differentiated by using morphological and biometric measurements, the ITS1 and ITS2 sequences of T. trichiura and T. suis isolated from nonhuman primates and different porcine hosts revealed clear differences between the two Trichuris species (310). Phylogenetic analysis of the ITS1, ITS2, and SSU rDNA sequences from various Trichuris species showed that T. trichiura and T. suis are closely related but genetically distinct species (310–312). A detailed analysis of the complete mitochondrial genome also confirmed that T. trichiura and T. suis are separate species (313). Sequence analysis of the ITS2 region of Trichuris species collected from pigs and from humans in the Kabale district of Uganda showed that cross-infections of humans with T. suis occurs but that cross-infections of pigs with T. trichiura were not found. The genetic differentiation and the presence of the two species in humans were confirmed by sequence analysis of a part of the β-tubulin gene. In three cases, worms in humans showed both the T. suis and T. trichiura ITS2 genotypes, suggesting the existence of heterozygous worms (314). Combining ITS1-5.8S-ITS2 sequences from adult Trichuris sp. worms isolated from baboons with GenBank records for Trichuris isolated from other humans, other nonhuman primates, and pigs revealed two distinct Trichuris genotypes separated from T. suis infecting baboons and human patients from Cameroon (315). The zoonotic potential of T. suis and different Trichuris genotypes is yet not fully understood, and further studies on these sympatric Trichuris infections are needed.
Pyrosequencing assays have been developed for the detection of single-nucleotide polymorphisms (SNPs) in the β-tubulin gene that are associated with benzimidazole resistance. The finding of a benzimidazole resistance-associated codon 200 TAC SNP in T. trichiura might explain the reported ineffectiveness of benzimidazole anthelmintics against Trichuris (316). In Guatemala, by using nested PCR and sequence analysis, nine T. trichiura-positive fecal samples revealed the TTC codon in the β-tubulin gene, which is associated with benzimidazole-sensitive parasites (317).
Ascaris lumbricoides
Conventional PCR assays targeting the ITS1 region and the cytochrome b gene using gel electrophoresis for the detection of the PCR product were used to detect and genotype Ascaris DNA from coprolites found in pre-Columbian South American archaeological sites and in fecal samples from patients attending a health center in Rio de Janeiro, Brazil (318). Another PCR for the detection of A. lumbricoides DNA designed from the ITS1 sequence (319) was used in different platforms as part of panels or multiplex assays for the simultaneous detection of several linked pathogens. A multiplex real-time PCR using hydrolysis (TaqMan) probes (18) for the detection and quantification of A. lumbricoides, N. americanus, A. duodenale, and S. stercoralis with an internal control was validated against a large panel of control samples and was used in Malaysia and Indonesia (161, 319–321). In two studies in Malaysia, Ascaris was detected by PCR in 6/77 and 12/225 cases, respectively, compared to detection in 3/77 and 7/225 cases by microscopy. Microscopy-negative samples showed higher median CT values (i.e., lower DNA loads) than did microscopy-positive samples. In Indonesia, in a large placebo-controlled study on the effect of albendazole treatment on malarial parasitemia and allergy, Ascaris prevalence and DNA load measured by PCR showed little decrease in the placebo group (n = 466), and although the decline in the albendazole group (n = 423) was much greater, there were still positive cases after treatment 6 times at 3-month intervals. This ITS1-based Ascaris PCR was also validated for use in a TaqMan array card for the simultaneous detection of 19 enteropathogens, including viruses, bacteria, protozoa, and helminths (A. lumbricoides and T. trichiura) (309). The same primer-and-probe design for the detection of A. lumbricoides DNA was incorporated into a multiplex PCR for the detection of three intestinal protozoa and four helminths using probe-based detection with Luminex beads (322). TaqMan probe chemistry with another primer-and-probe design from the ITS1 sequence was used in a study in which an Ascaris PCR was performed in parallel with singleplex PCRs for seven additional parasitic targets (18). Compared to microscopy, only a small number of additional cases were detected by PCR (28/400 versus 22/400). The microscopy-positive samples showed a much higher DNA load than did the PCR-positive/microscopy-negative samples. DNA loads showed an excellent correlation with the egg counts measured by the Kato-Katz method. In an additional group of 125 children who were tested before and after anthelmintic treatment, all initial Ascaris PCR-positive cases tested negative 21 days after treatment.
Whole-genome fingerprinting by amplified fragment length polymorphism (AFLP) analysis, RFLP analysis, and sequencing of the ITS1 and mitochondrial genes and microsatellites have been used as genotyping methods to study the epidemiology and zoonotic potential of Ascaris. These studies have been reviewed from different angles to elucidate the taxonomic status of A. lumbricoides and A. suum and their zoonotic potential (323–325). Several studies using different markers showed zoonotic potential of Ascaris, as humans and pigs were found to share common haplotypes. Although some researchers leave the taxonomic status undefined, Leles et al. concluded that based on nematode cross-infections between humans and pigs, the hybridization between A. lumbricoides and A. suum, and the high levels of genetic similarity between the complete mtDNA genomes, there is a single interbreeding population of Ascaris, and those authors recommended synonymizing the two species with the name A. lumbricoides Linnaeus 1758 (326).
The codon 200 TAC SNP in the β-tubulin gene was not found in any of the A. lumbricoides samples (n = 38) tested by using a pyrosequencing assay for the detection of SNPs in this gene that are associated with benzimidazole resistance (316). In Guatemala, sequence analysis of the β-tubulin gene of microscopy-positive A. lumbricoides samples showed the TCC codon that is associated with benzimidazole-sensitive parasites in all samples (317).
Hookworm
As reviewed by Gasser et al., species-specific markers defined in the ribosomal and mitochondrial genes were validated for species identification of adult hookworms and were later employed in epidemiological studies using conventional PCR for the detection of species-specific hookworm DNA in fecal samples (327). Primers and probes designed from the ITS2 sequences of N. americanus and A. duodenale showed high specificity (100%) and sensitivity (98.5% to 100%) using a range of controls and field samples from northern Ghana in a multiplex real-time PCR for the detection of N. americanus, Ancylostoma, Oesophagostomum bifurcum, and an inhibition control (328). Moreover, there was a good correlation between egg counts and parasite-specific DNA load. In an area of low N. americanus endemicity in southern Ghana, the detection rate by PCR for a single fecal sample almost equaled that of Kato examination of three fecal samples. Additionally, N. americanus infections were detected in 8 of 31 microscopy-negative cases by PCR using three consecutive fecal samples (329). The same primer-and-probe design from the ITS2 sequence was used in multiplex TaqMan-based assays targeting different helminth infections, in singleplex assays tested in parallel with other targets, and in a multiplex PCR combined with probe-based detection with Luminex beads (Table 7). Considerable numbers of additional N. americanus and Ancylostoma cases were detected with a helminth pentaplex real-time PCR targeting the ITS2 sequences of N. americanus and A. duodenale in microscopy-negative fecal samples in two studies in Malaysia (161, 320). N. americanus DNA was detected in 21/76 and 20/225 microscopy-negative samples, and Ancylostoma DNA was detected in 11/76 and 14/225 samples. Because of the identity between ITS2 sequences of the different Ancylostoma species, the exact identification of the Ancylostoma species involved in these studies is as yet unclear. Real-time PCR targeting the ITS2 sequence followed by HRM analysis enabled the detection and differentiation of N. americanus, A. duodenale, A. ceylanicum, and A. caninum (330). Moreover, this PCR assay was more sensitive for the detection of N. americanus than was nested PCR for testing of 634 samples from Malaysia. In a study in Cambodia, microscopy was compared with these N. americanus and Ancylostoma PCRs in a singleplex format. Although hookworm-specific amplification was detected in an additional 35 of 166 microscopy-negative fecal samples, both PCRs remained negative for 11 of 52 samples in which hookworm-like eggs were seen by microscopy. Moreover, there was no difference in the PCR CT values (or DNA load) detected in microscopy-negative and microscopy-positive samples (331). The possibility of a less optimal DNA isolation procedure or the misidentification of hookworm-like eggs (e.g., Trichostrongylus) can therefore not be excluded. A SYBR green-based real-time PCR targeting the ITS2 sequence of N. americanus detected one egg in 200 mg of fecal material and detected N. americanus-specific DNA amplification in stool samples from 49 of 261 subjects, compared to 30 cases detected by microscopy (332). PCR-based detection showed large reductions in the percentage of N. americanus-positive subjects as well as in the intensity, measured as N. americanus-specific DNA loads, in stool samples following intensive albendazole treatment compared to placebo in a study in Indonesia (333). Unexpectedly, PCR revealed that A. duodenale and not N. americanus was the most prevalent hookworm infection, and A. duodenale was shown to be a key factor in severe anemia and iron deficiency in Malawian children (334). Infections with moderate and high A. duodenale DNA loads were positively associated with severe anemia, with odds ratios (ORs) of 2.49 (95% confidence interval [CI], 1.16 to 5.33) and 9.04 (95% CI, 2.52 to 32.47), respectively. Iron deficiency measured in bone marrow was positively associated with an increasing A. duodenale DNA load (ORs of 3.63 [95% CI, 1.18 to 11.20], 16.98 [95% CI, 3.88 to 74.35], and 44.91 [95% CI, 5.23 to 385.77] for low, moderate, and high loads, respectively).
Genetic diversity within hookworm species investigated by whole-genome fingerprinting methods such as random amplified polymorphic DNA (RAPD) and AFLP analyses, different mutation scanning methods (e.g., SSCP), and sequencing of selected protein-coding mitochondrial genes was reviewed by Gasser et al. (327, 335). Population substructuring was revealed within A. duodenale in China by using PCR-SSCP to detect genetic diversity in the cox1 gene (336). Using adult worms from Africa, Asia, and South America, multiple genetically distinct groups of N. americanus were shown by using AFLP (337), confirming previous findings of ribosomal and mitochondrial genetic variation (327).
Single-nucleotide polymorphisms in the β-tubulin gene at codons 167 and 200 have been associated with resistance to benzimidazole anthelmintics in several strongylid nematodes of livestock. A real-time PCR for the detection of these polymorphisms in A. caninum, A. duodenale, and N. americanus was performed on hookworm specimens from schoolchildren in Uganda for whom a reduced response to treatment with mebendazole was observed. However, polymorphisms in codons 167 and 200 of the β-tubulin gene were not detected in any of these samples (338). Amplification and sequencing primers for pyrosequencing were designed to detect and identify benzimidazole resistance-associated SNPs in the β-tubulin genes of N. americanus, A. lumbricoides, and T. trichiura (339). Recently, new-generation sequencing and analysis of the N. americanus transcriptome revealed 18 predicted drug targets, which did not have homologues in the human host (340).
Strongyloides stercoralis
Strongyloides stercoralis is endemic to many tropical and subtropical regions; it is estimated that 30 million to 100 million people worldwide are infected, with prevalence rates in some cohorts being as high as 50% (299, 303, 341). S. stercoralis behaves differently than other intestinal nematodes; rhabditiform larvae are excreted in the stool but can already develop into infective filariform larvae in the gut and then penetrate the intestinal mucosa or perianal skin, resulting in a prolonged cycle of autoinfection, which can continue for many decades (342–344). Chronic strongyloidiasis may result in hyperinfection or systemic strongyloidiasis and is often seen in immunocompromised hosts such as transplant patients, patients receiving chemotherapy, and patients treated with corticosteroids (303, 305, 345–347).
Laboratory diagnosis is based mainly on serology and the detection of S. stercoralis larvae by microscopic examination of fecal samples. Microscopy is labor-intensive and, especially in chronic infections, the sensitivity is low due to the often very small number of larvae, and even after formalin-ether concentration, the use of the Baermann method, or coproculture, multiple samples have to be examined to achieve ample sensitivity (348–352).
Recently, a S. stercoralis real-time PCR with primers and FRET probes designed from the 28S gene was described and showed high specificity and specificity using controls and spiked samples, but it was not evaluated further on clinical samples (353). Another real-time PCR with primers and a hydrolysis probe based on the SSU rRNA gene sequence of S. stercoralis was 10 to 100 times more sensitive than assays designed from cox1 and a S. stercoralis-specific sequence. The assay showed 100% specificity when used on a large panel of DNAs from fecal samples and controls. When applied to stool samples (n = 212) from northern Ghana, the sensitivity of the SSU rRNA PCR compared to the Baermann test or coproculture was 86% or 91%, respectively. In samples with positive Baermann and coproculture results, PCR showed 100% sensitivity, but lower sensitivity was achieved in samples with discrepant results by the Baermann and culture methods (354). A lower sensitivity of PCR for patients with lower larval counts was also found in a study in Bangladesh using the same PCR method. A positive PCR result was found in all Harada-Mori culture-positive samples with high and moderate larval loads, whereas only 15% of the samples with a low larval load were found to be positive by PCR (355). The PCR design was evaluated in asymptomatic schoolchildren in Cambodia (n = 218) and showed 88.9% sensitivity and 92.7% specificity compared to the combination of the Baermann method and Koga agar culture as the gold standard; a lower sensitivity was found for samples with discrepant results by conventional methods. Additional PCR-positive samples, however, were found in Baermann- and Koga agar-negative samples as well (331). The SSU rDNA primer-and-probe set (354) has been incorporated into multiplex assays on a variety of different platforms (18, 309, 319, 320, 322) (Table 2). In two studies in Malaysia, S. stercoralis DNA was detected in 21/76 and 28/225 microscopy-negative samples (161, 320). The SSU rRNA gene was also used as a target in a duplex real-time FRET PCR combined with melting-curve analysis for the simultaneous detection of Opisthorchis viverrini and S. stercoralis (356) and showed 100% sensitivity and specificity compared to the agar plate culture method, detecting S. stercoralis in 32 of 66 stool samples and 30 controls (356). A conventional PCR designed from the 5.8S rRNA gene amplified S. stercoralis-specific DNA in all 16 agar plate-positive samples and in 5 of 30 microscopy-negative samples in a survey of 782 individuals in Iran (357).
An early study using a PCR-RFLP approach targeting the ITS and a partial 23S rRNA gene using larval DNA revealed species-specific amplicon sizes for S. stercoralis, S. fuelleborni, and S. ratti. RFLP showed identical patterns for human isolates of S. stercoralis from different parts of the world and differentiated these from a dog isolate of S. stercoralis (358). Sequencing of the SSU rRNA gene of larvae isolated from fecal material collected from orangutans and a human working with orangutans identified S. stercoralis and S. fuelleborni. The ITS1 sequences of 17 S. fuelleborni isolates showed high variability, falling into two clusters, without differentiating the orangutan and human isolates (359).
Considering the rather laborious and time-consuming concentration and stool culture methods that have to be applied on multiple fresh stool samples to achieve ample sensitivity in the diagnosis of S. stercoralis infections, which are relatively rare in the Western clinical setting, molecular diagnostics appear to be a worthwhile alternative screening method. In a study using stool samples from returning travelers (n = 2,591), real-time S. stercoralis SSU rDNA PCR detected 21 positive cases, compared to only 3 cases detected by using microscopy of Baermann sediments (107). The higher sensitivity of the PCR could be explained by the detection of DNA from larvae that died before arrival of the samples by regular mail, whereas Baermann tests or coprocultures are dependent on the presence of live larvae. In a small study in Argentina, using 17 positive stool samples from strongyloidiasis patients that were confirmed by agar plate culture of the first, second, or third additional sample collected at 1-week intervals, a PCR using the same primers and a PCR with a novel primer set designed from the SSU rRNA gene performed on the first sample showed positive results in all 17 cases (360).
FOOD-BORNE TREMATODES
Food-borne trematode infections are zoonotic infections and are caused by >80 different trematode species (361). The species considered of public health importance are the liver flukes Clonorchis sinensis, Opisthorchis spp., and Fasciola spp.; the lung flukes Paragonimus spp.; and a number of intestinal flukes. The small liver flukes C. sinensis, O. viverrini, and O. felineus are endemic to East and Southeast Asia and central northern Eurasia, and western Eurasia (362). Increasing inland fish production causes a local increase in the number of food-borne trematode infections, and increasing international travel, human migration, food trade, and changing eating habits may cause an increase in the number of cases diagnosed in countries where the disease is not endemic (363–365). Infections with liver and lung flukes are rarely fatal, but the burden of disease is considerable, and moreover, C. sinensis and O. viverrini infections may lead to cholangiocarcinoma (366, 367). Although Fasciola spp., Paragonimus spp., and intestinal flukes are found worldwide, higher rates of transmission are known for certain areas, such as the Andean region, Africa, and Asia (366).
Detection and differential diagnosis based on microscopic detection of eggs in feces and sputum (only for lung flukes) are notoriously difficult because eggs of different species resemble each other and are often present in very small numbers (362, 366). DNA-based methods for the detection and differentiation of food-borne trematodes have been used, including PCR, nested PCR, real-time PCR, and LAMP, designed based on the O. viverrini-specific repetitive DNA fragment, mitochondrial DNA, ITS1, ITS2, and the SSU rRNA gene (Table 8). A real-time PCR with primers and a hydrolysis probe based on the O. viverrini-specific repetitive DNA fragment showed positive results in liver tissue samples from Thai patients with hepatocellular carcinoma and cholangiocarcinoma. O. viverrini-specific DNA was not detected in 7 control liver specimens from nonprimary liver cancers (368). The ITS2 sequence of C. sinensis was used to develop a TaqMan real-time PCR to detect C. sinensis-specific DNA in fecal samples. Beside cross-reacting with O. felineus, the assay showed high specificity and 100% sensitivity using samples (n = 74) with >100 eggs per gram of feces (epg) and 91.4% sensitivity using samples (n = 70) with ≤100 epg. Three additional positive samples were detected out of 26 samples in which no eggs were found by microscopy. The CT values were strongly correlated with the intensity of infections, as determined by egg counts in Kato-Katz smears (369). A similar approach with primers and a hydrolysis probe designed from the C. sinensis ITS1 sequence showed specific (with the exception of O. viverrini) and sensitive detection of C. sinensis in a small number of fecal samples and fish tissue samples (370). A multiplex real-time PCR based on the 2 DNA sequences of C. sinensis and O. viverrini was developed by using FRET probes and melting curve analysis. C. sinensis-specific DNA and O. viverrini-specific DNA were amplified and detected in 8 and 30 C. sinensis- and O. viverrini-positive stool samples, respectively. Surprisingly, no correlation was found between the CT values and the intensity of infection determined by C. sinensis and O. viverrini egg counts in stool samples (371). An interesting approach, using MLPA, was used for the identification of C. sinensis, O. viverrini, and O. felineus and differentiation from other closely related species with three probe pairs derived from the ITS1 sequence. The assay was validated by testing DNA samples from adult C. sinensis flukes and fecal samples from C. sinensis-infected rats (372). The performance of this technique has not yet been validated on clinical samples from areas where C. sinensis, O. viverrini, and O. felineus are endemic.
TABLE 8.
PCR-based assays for detection and molecular characterization of food-borne trematodesc
| Species | Amplification method | Detection method(s) | Target | Sample type(s) | Reference |
|---|---|---|---|---|---|
| O. viverrini | PCR | Gel electrophoresis | O. viverrini-specific repetitive DNA fragment | Stool | 491 |
| O. viverrini | PCRa | Gel electrophoresis | O. viverrini-specific repetitive DNA fragment | Stool | 492 |
| O. viverrini, C. sinensis | Multiplex PCR | Gel electrophoresis | Mitochondrial DNA | Worms, metacercariae, eggs | 493 |
| Opisthorchiidae | PCR | Gel electrophoresis | ITS2 | Stool | 494 |
| O. viverrini | PCRa | Gel electrophoresis | O. viverrini-specific repetitive DNA fragment | Stool | 495 |
| O. viverrini | Real-time PCR | Hydrolysis probe | O. viverrini-specific repetitive DNA fragment | Tumor tissue | 368 |
| O. viverrini | PCR | Gel electrophoresis | O. viverrini genomic DNA clone | Stool | 496 |
| O. viverrini, C. sinensis, Haplorchis taichui | PCR-RFLP | Gel electrophoresis | ITS2 | Stool | 497 |
| O. viverrini, H. taichui | Nested PCR | Gel electrophoresis | cox1 | Stool | 498 |
| C. sinensis, O. felineus, O. viverrini | Real-time PCR | Hydrolysis probe | ITS2 | Stool | 369 |
| O. viverrini | Real-time PCR | FRET probes-MCA | O. viverrini-specific repetitive DNA fragment | Stool | 499 |
| O. viverrini, H. taichui | PCR | Gel electrophoresis | ITS1 | Stool | 489 |
| O. viverrini, H. taichui | PCR | Gel electrophoresis | ITS2 | Stool | |
| C. sinensis, O. viverrini, O. felineus | MLPA | Gel electrophoresis, capillary sequencer | ITS1 | Worm | 372 |
| O. viverrini (and S. stercoralis) | Real-time multiplex PCRb | FRET probes-MCA | O. viverrini-specific repetitive DNA fragment | Stool | 356 |
| O. viverrini, H. taichui | HAT-RAPD PCR | Gel electrophoresis | Genomic | Microscopy-positive stool | 500 |
| O. viverrini | LAMP | Visual | nad1 | Stool, fish | 374 |
| O. viverrini | LAMP | Real time, visual | ITS1 | Stool | 373 |
| O. viverrini, C. sinensis | Real-time multiplex PCRb | FRET probes-MCA | nad2 | Stool | 371 |
| C. sinensis | Real-time PCR | Hydrolysis probe | ITS1 | Stool, fish | 370 |
| F. hepatica, F. gigantica, intermediate Fasciola | Sequence-related amplified polymorphism | Gel electrophoresis | Random nucleotide sequence | Worms | 501 |
| F. hepatica, F. gigantica | PCR-RFLP | Gel electrophoresis | SSU rDNA | Worms | 502 |
| F. hepatica, F. gigantica, intermediate Fasciola | PCR-RFLP | Gel electrophoresis | ITS2 | Worms | 503 |
| F. hepatica, F. gigantica, intermediate Fasciola | PCR | Gel electrophoresis | ITS2 | Worms | 504 |
| F. hepatica, F. gigantica | LAMP | Visual | IGS | Worms, cercariae, eggs | 505 |
| F. hepatica, F. gigantica | Multiplex PCR | Gel electrophoresis | Mitochondrial DNA | Worms, miracidia, eggs, stools | 506 |
| Paragonimus | PCR sequencing | Sequence | ITS2 | Eggs | 507 |
| P. westermani | LAMP | Visual | ITS2 | Sputum, pleural fluid, crabs, crayfish | 380 |
| P. westermani, Fasciolopsis buski, F. gigantica | PCR | Gel electrophoresis | ITS2 | Worms | 508 |
| Paragonimus heterotremus, P. westermani, P. macrorchis, P. siamensis, P. harinasutai, and P. bangkokensis | Real-time PCR | FRET probes and MCA | ITS2 | Feces of experimentally infected cats | 509 |
| Paragonimus bangkokensis, P. harinasutai, P. heterotremus, P. macrorchis, P. siamensis, P. westermani | PCR pyrosequencing | Sequence | ITS2 | Metacercariae | 510 |
PCR primers are described in reference 491.
PCR primers are described in reference 499.
Abbreviations: RFLP, restriction fragment length polymorphism; MCA, melt curve analysis; HRM, high-resolution melt curve analysis; SSU, small subunit; ITS, internal transcribed spacer; HSP, heat shock protein; IGS, intergenic spacer region; cox1, mitochondrial cytochrome c oxidase subunit I gene; nad1, NADH dehydrogenase subunit 1; nad2, NADH dehydrogenase subunit 2; MLPA, multiple ligation-dependent probe amplification; HAT-RAPD, high-annealing-temperature random amplified polymorphic DNA; LAMP, loop-mediated isothermal amplification; FRET, fluorescence resonance energy transfer.
Primer sets for the detection of O. viverrini DNA in stool samples using LAMP were designed from the mitochondrial nad1 sequence and from the ribosomal ITS1 sequence of O. viverrini (373, 374). The ITS1-based LAMP assay showed a 100% sensitivity compared to microscopy when tested on stool samples (n = 50) from schoolchildren from an area of endemicity in Thailand, compared to a remarkably low sensitivity of only 24% using conventional PCR on the same target (373).
The phylogenetic relationship between C. sinensis and Opisthorchis is not yet clear. There are only small differences between C. sinensis and Opisthorchis across the SSU rDNA, ITS2, and 28S sequences. Based on the ITS1 sequence, O. viverrini and O. felineus group together and are closely related to C. sinensis, whereas analysis of ITS2 and cox1 sequences suggests that O. felineus is more closely related to C. sinensis than to O. viverrini. In contrast, the ninth intron of the paramyosin gene (Pm-int9), which is more variable than nuclear markers, showed a closer relationship between O. viverrini and C. sinensis (375, 376). Microsatellite DNA analysis of O. viverrini in Thailand using 12 microsatellite loci revealed the existence of genetic diversity and population substructuring in O. viverrini, which can be a useful tool to study transmission dynamics and control (377, 378).
Molecular tools for the detection, differentiation, and genetic characterization of Fasciola spp. have been reviewed recently (379). Molecular approaches based on ITS1, ITS2, nad1, and cox1 have been used mainly for the differentiation of adult flukes of F. hepatica, F. gigantica, and the novel “intermediate Fasciola” (i.e., hybrid form between F. hepatica and F. gigantica) and to detect genetic variation among Fasciola spp. The assays have not yet been fully validated in population-based studies or for patient diagnosis. DNA-based methods are not widely used for the diagnosis of paragonimiasis in human cases; PCR and PCR followed by (pyro)sequencing, usually targeting the ITS2 sequence, are used primarily for the detection and identification of Paragonimus species from metacercariae, crabs, and experimentally infected animals or for confirmation of eggs found in sputum or stool samples (Table 8). A LAMP assay using primers targeting the ITS2 sequence detected Paragonimus westermani-specific DNA in 17 microscopy-positive pleura and sputum samples, and no P. westermani-specific DNA was detected in the negative-control sputum samples (380).
Recent outbreaks of opisthorchiasis after eating raw fillets of tench (Tinca tinca) in Italy, with cases found in Italy, Austria, and The Netherlands (381), are only one example that urges the need for specific and sensitive diagnostic tools in a clinical diagnostic setting. In such cases, usually only very small numbers of the very small eggs are present, which makes molecular diagnostic tools such as real-time PCR a very appealing and worthwhile approach to confirm the clinical diagnosis.
SCHISTOSOMA
Although adult schistosome worms reside, depending on the species, in the blood vessels around the intestine or urinary bladder and are therefore strictly blood parasites, the eggs of Schistosoma mansoni are excreted in feces through the intestinal wall, and the eggs of Schistosoma haematobium pass through the bladder wall into the urine. The number of eggs, especially for travelers, is often very small, and therefore, large quantities of feces and/or urine are concentrated to improve the sensitivity of microscopic examination. Due to the often very light infections in settings where the disease is not endemic, serology is much more sensitive than microscopy for the diagnosis of schistosomiasis in travelers (382). Recent studies have shown that the detection of Schistosoma-specific DNA can achieve a higher sensitivity for the diagnosis of acute schistosomiasis than serology (383, 384).
The S. mansoni and S. haematobium tandem-repeat sequences described by Hamburger et al. (385, 386) have been used in a large number of studies using conventional and real-time PCR for the detection of Schistosoma-specific DNA in stool, urine, and serum samples as well as the SSU rDNA, 28S, and ITS sequences and mitochondrial genes (Table 9). In areas of endemicity, Schistosoma-specific PCR performed on DNA isolated from a small amount of urine or feces has been shown to be more sensitive than microscopic examination (387). In an area of low endemicity in Brazil (n = 149), the detection rate of a conventional PCR designed from the S. mansoni 121-bp tandem-repeat sequence and performed on DNA isolated from one stool sample (38.1%) was higher than the detection rate found after microscopic examination of Kato-Katz thick smears of three stool samples (30.9%) (388). Although the sensitivity of a genus-specific real-time PCR targeting the Schistosoma ITS2 sequence using DNA isolated from 200 μl of urine from schoolchildren in Ghana with ≤50 eggs/10 ml was only 85.2%, the overall detection rate found for PCR was 20.8%, compared to 15.4% for microscopy with 10 ml urine. The higher sensitivity of the real-time PCR resulted in much higher negative predictive values than for microscopy (389). A higher sensitivity of PCR in samples with low egg counts might be achieved by the use of larger volumes of urine samples and filter-based DNA isolation methods (390–393). Latent class analysis of hematuria, microscopy, and PCR targeting the S. haematobium-specific DraI repeat sequence using DNA isolated from one-quarter of a filter paper after filtration of 50 ml of urine showed sensitivities of 87%, 70%, and 100%, respectively (392). ten Hove et al. performed a multiplex real-time PCR targeting the cox1 genes of S. mansoni and S. haematobium on a selection of duplicate stool samples from 88 subjects in northern Senegal. S. mansoni-specific DNA and S. haematobium-specific DNA were amplified, and DNA loads showed a significant correlation with microscopic egg counts for S. mansoni in stool and S. haematobium in urine; however, the sensitivity of PCR was slightly lower than that of microscopy (394). Real-time PCR targeting the Schistosoma ITS2 sequence performed on vaginal lavage samples proved to be a promising tool in the notoriously difficult diagnosis of genital schistosomiasis (395, 396). S. haematobium DNA in vaginal lavage samples was highly associated with genital mucosal manifestations typical of female genital schistosomiasis (395).
TABLE 9.
PCR-based assays for detection and molecular characterization of Schistosoma infectionsf
| Species | Amplification method | Detection method | Target(s) | Sample type(s) | Reference |
|---|---|---|---|---|---|
| S. mansoni | PCR | Gel electrophoresis | S. mansoni tandem repeat | Adult worm | 385 |
| S. haematobium | PCR | Gel electrophoresis | S. haematobium tandem repeat | Adult worm | 386 |
| S. mansoni | PCR | Gel electrophoresis | S. mansoni tandem repeat | Stool, serum | 387 |
| S. mansoni | PCRa | Gel electrophoresis | S. mansoni tandem repeat | Stool | 388 |
| S. mansoni | PCR | Gel electrophoresis | Mitochondrial DNA | Stool | 511 |
| S. japonicum | PCR | Gel electrophoresis | Mitochondrial DNA | Stool | 511 |
| Schistosoma genus | PCR | Gel electrophoresis | 28S | Urine | 512 |
| S. mansoni | PCR | Gel electrophoresis | 28S | Urine | 512 |
| S. japonicum | PCR | Gel electrophoresis | 28S | Urine | 512 |
| S. haematobium, S. bovis, S. intercalatum | PCR | Gel electrophoresis | ITS | Urine | 512 |
| S. mansoni | Real-time PCR | SYBR green | SSU rDNA | 513 | |
| S. japonicum | Real-time PCR | SYBR green | nad1 | Stool | 514 |
| S. mansoni | Multiplex real-time PCR | Hydrolysis probe | cox1 | Stool | 394 |
| S. haematobium | Multiplex real-time PCR | Hydrolysis probe | cox1 | Stool | 394 |
| Schistosoma genus | Real-time PCR | Hydrolysis probe | ITS2 | Urine | 458 |
| S. japonicum | Real-time PCR | Hydrolysis probe | nad1 | Stool | 515 |
| S. mansoni | PCRa | Gel electrophoresis | S. mansoni tandem repeat | Stool | 516 |
| Schistosoma genus | Real-time PCR | Hydrolysis probe | S. mansoni tandem repeat | Plasma | 397 |
| Schistosoma genus | Real-time PCRc | Hydrolysis probe | ITS2 | Vaginal lavage | 395 |
| S. japonicum | LAMP | Visual | Retrotransposon SjR2 repeat | Serum | 517 |
| S. mansoni | PCRa | Gel electrophoresis | S. mansoni tandem repeat | Stool | 518 |
| Schistosoma genus | PCR-ELISAa | ELISA | S. mansoni tandem repeat | Stool | 519 |
| S. mansoni | PCRa,b | Gel electrophoresis | S. mansoni tandem repeat and 28S | Stool | 520 |
| S. haematobium | PCRe | Gel electrophoresis | S. haematobium tandem repeat | Cerebrospinal fluid | 521 |
| Schistosoma genus | Real-time PCRd | Hydrolysis probe | S. mansoni tandem repeat | Serum | 383 |
| S. haematobium | PCRe | Gel electrophoresis | S. haematobium tandem repeat | Urine | 391 |
| S. haematobium | PCRe | Gel electrophoresis | S. haematobium tandem repeat | Urine | 392 |
| S. japonicum | Nested PCR | Gel electrophoresis | S. japonicum repeat sequence | Serum | 522 |
| S. mansoni | PCRa | Gel electrophoresis | S. mansoni tandem repeat | Urine | 523 |
| S. japonicum | PCR | Gel electrophoresis | Retrotransposon SjR2 repeat | Stool | 524 |
| S. mansoni | PCRa | Gel electrophoresis | S. mansoni tandem repeat | Stool | 525 |
| S. mansoni | Nested PCRa | Gel electrophoresis | S. mansoni tandem repeat | Spiked stool | 526 |
| S. haematobium | PCR | Gel electrophoresis | nad1 and cox1 | Urine | 393 |
| Schistosoma genus | Real-time PCR | Hydrolysis probe | 28S | Stool, urine, serum | 398 |
| Schistosoma genus | Real-time PCRd | Hydrolysis probe | S. mansoni tandem repeat | Plasma | 384 |
| Schistosoma genus | Real-time PCRc | Hydrolysis probe | ITS2 | Urine | 389 |
| S. mansoni | PCRa | Gel electrophoresis | S. mansoni tandem repeat | Urine | 527 |
| Schistosoma genus | Real-time PCRc | Hydrolysis probe | ITS2 | Urine, vaginal lavage | 396 |
PCR primers are described in reference 387.
PCR primers are described in reference 512.
PCR primers and probe are described in reference 458.
PCR primers and probe are described in reference 397.
PCR primers are described in reference 386.
Abbreviations: RFLP, restriction fragment length polymorphism; SSU, small subunit; ITS, internal transcribed spacer; cox1, mitochondrial cytochrome c oxidase subunit I gene; nad1, NADH dehydrogenase subunit 1.
Recently, there has been increased interest in the detection of Schistosoma DNA in serum samples for the (early) diagnosis of schistosomiasis. Already in 2002, amplification of the S. mansoni tandem-repeat sequence using conventional PCR was shown in sera from two schistosomiasis patients (387). High sensitivities for the detection of Schistosoma DNA using a real-time PCR targeting the S. mansoni tandem-repeat sequence have been reported, using DNA isolated from 10 ml plasma from patients with chronic disease infected with S. mansoni, S. haematobium, and S. japonicum and from patients with Katayama syndrome. Amplification of the S. mansoni tandem-repeat sequence was shown up to 58 weeks after treatment (397). In a cluster of travelers (n = 13) from Rwanda, all presenting with acute schistosomiasis, the same PCR approach using 2 ml of serum detected Schistosoma DNA in all 13 serum samples, whereas eggs of S. mansoni were found in 9 stool samples, and antischistosome antibodies were detected in 10 patients (383). In a multicenter study, the Schistosoma tandem-repeat sequence was amplified in 35 of 38 patients (92%) with acute schistosomiasis, compared to sensitivities of 70% and 24% using serology and microscopy, respectively (384). However, for patients (n = 140) who presented at an outpatient clinic in Antwerp, Belgium, using a real-time PCR developed to target the 28S rDNA sequence, Schistosoma-specific DNA was detected in all microscopy-positive stool (n = 67) and urine (n = 4) samples and additionally in microscopy-negative stool (n = 9) and urine (n = 1) samples, whereas the Schistosoma 28S target was amplified in only 2 of 38 serum samples of patients with confirmed schistosomiasis (398).
CESTODES
The impressive length of tapeworms has made them among the most well-known and evocative parasites to the general public. Although the pathology of intestinal cestodes is usually minor, if metacestode stages occur in human tissues, pathology and morbidity can be severe. Several species are known to infect humans, with Taenia solium (pork tapeworm), which can cause neurocysticercosis when metacestode stages develop in the brain after infection with eggs; Taenia saginata (beef tapeworm); Hymenolepis nana (dwarf tapeworm); and Diphyllobothrium latum (broad or fish tapeworm) being the most common (399).
Frequently, the diagnosis has already been made when patients notice the presence of proglottids in their feces. Parasitological differentiation based on morphological criteria might be difficult due to degeneration or the juvenile stage of the proglottids found. Microscopic detection of eggs in feces from patients with Taenia infections is known to be insensitive, as eggs expelled from proglottids are not equally distributed in the feces. Immunodiagnostic tests for the detection of Taenia antigens in stool samples have been used mainly in epidemiological studies, and DNA-based methods are used mainly for species discrimination (Table 10) (400, 401). Primarily, conventional (multiplex) PCR and PCR-RFLP designed based on the HDP1 repeat, HDP2, mitochondrial 12S rDNA, cox1, the pTsol9 repeat, and the Tso31 sequence have been used for species discrimination using DNA isolated from proglottids and have been tested only on small numbers of usually known positive and negative stool samples (Table 10). Recently, Bayesian modeling was used to estimate and compare the performances of microscopy, coproantigen ELISA, and multiplex real-time PCR for the detection of Taenia carriers by using stool samples (n = 871) collected in two Zambian communities where the disease is endemic. Specificities were 99.9%, 92.0%, and 99.0% and sensitivities were 82.5%, 84.5%, and 82.7% for microscopy, ELISA, and PCR, respectively (402). In a study comparing LAMP and a conventional multiplex PCR based on the cox1 sequence using 43 known positive fecal samples, LAMP was more sensitive than PCR, 88.4% and 37.2%, respectively (403).
TABLE 10.
PCR-based assays for detection and molecular characterization of cestode infectionsg
| Genus or species | Amplification method | Detection method | Target(s) | Sample type(s) (no. of samples) | Reference(s) |
|---|---|---|---|---|---|
| Diphyllobothrium genus | Multiplex PCR | Gel electrophoresis | cox1 | Proglottids, eggs | 407 |
| T. saginata | PCR | Gel electrophoresis | HDP1 | Proglottids | 528, 529 |
| T. solium, T. saginata | Multiplex PCR | Gel electrophoresis | HDP2 | Proglottids | |
| T. solium, T. saginata | Multiplex PCR-RFLPe | Gel electrophoresis | HDP2 | Proglottids | 530 |
| T. solium, T. saginata, T. asiatica | PCR-RFLP | Gel electrophoresis | Mitochondrial 12S rDNA | Proglottids | 531 |
| T. solium, T. saginata | Multiplex PCRe | Gel electrophoresis | HDP2 | Stool | 532 |
| T. solium, T. saginata, T. asiatica | Multiplex PCR | Gel electrophoresis | cox1 | Proglottids, stool (6) | 533 |
| T. saginata, T. asiatica | Multiplex PCR-RFLP | Gel electrophoresis | HDP2 | Proglottids | 534 |
| T. solium, T. saginata, T. asiatica | Multiplex PCRa | Gel electrophoresis | cox1 | Proglottids, stool (25) | 535 |
| T. solium, T. saginata | PCR-RFLP | Gel electrophoresis | cox1 | Stool (12) | 536 |
| T. solium | PCR | Gel electrophoresis | Tsol9 repeat sequence | Cerebrospinal fluid | 404 |
| T. solium, T. saginata, T. asiatica | PCR-RFLPb | Gel electrophoresis | Mitochondrial 12S rDNA | Proglottids | 537, 538 |
| T. solium, T. saginata, T. asiatica | Multiplex PCRa | Gel electrophoresis | cox1 | Proglottids, stool (19) | 539 |
| T. solium, T. saginata, T. asiatica | Multiplex PCRa | Gel electrophoresis | cox1 | Proglottids | 540 |
| T. solium, T. saginata, T. asiatica | Multiplex PCRa | Gel electrophoresis | cox1 | Proglottids | 541 |
| T. solium | Nested PCR | Gel electrophoresis | Tsol31 | Stool (155) | 542 |
| T. solium | Nested PCR | Gel electrophoresis | HDP2 | Cerebrospinal fluid | 543 |
| Taenia genus | PCR | Sequencing | ITS1 | Cerebrospinal fluid | 425 |
| T. solium, T. saginata, T. asiatica | Multiplex PCR | Gel electrophoresis | cox1 | Proglottids | 544 |
| T. solium, T. saginata, T. asiatica | LAMP | Gel electrophoresis | Clp, cox1 | Proglottids, cysticerci, stool (6) | 545 |
| T. solium, T. saginata, T. asiatica | LAMPf | Gel electrophoresis | Clp, cox1 | Stool (43) | 403 |
| T. solium, T. saginata, T. asiatica | PCR | Gel electrophoresis | HDP2 | Proglottids | 546 |
| T. solium, T. saginata, T. asiatica | Multiplex PCRa | Gel electrophoresis | cox1 | Proglottids | 547 |
| T. solium | PCRd | Gel electrophoresis | Tsol9 repeat sequence | Cerebrospinal fluid | 405 |
| T. solium | Real-time PCR | LNA hydrolysis probe | Tsol9 repeat sequence | Cerebrospinal fluid | 406 |
| T. solium, T. saginata, T. asiatica | Multiplex PCRc | Gel electrophoresis | cox1 | Proglottids, eggs | 548 |
| T. solium, T. saginata, T. asiatica | PCR-RFLPb | Gel electrophoresis | Mitochondrial 12S rDNA | Proglottids | 537 |
| T. solium, T. saginata, T. asiatica | PCR-RFLPb | Gel electrophoresis | Mitochondrial 12S rDNA | Proglottids | 549 |
| T. solium, T. saginata, T. asiatica | Multiplex PCRc | Gel electrophoresis | cox1 | Proglottids | 550 |
| T. solium, T. saginata | Real-time multiplex PCR | Hydrolysis probe | ITS1 | Stool (817) | 402 |
PCR primers are described in reference 533.
PCR primers are described in reference 531.
PCR primers are described in reference 544.
PCR primers are described in reference 404.
LAMP primers were described previously (545).
Abbreviations: RFLP, restriction fragment length polymorphism; LNA, locked nucleic acid; SSU, small subunit; ITS, internal transcribed spacer; cox1, mitochondrial cytochrome c oxidase subunit I gene; Clp, cathepsin L-like cysteine peptidase; HDP1, T. saginata-specific repetitive sequence; HDP2, cestode-specific sequence.
Molecular diagnostics have proven to be extremely valuable in the diagnosis of neurocysticercosis (404–406). In a group of 121 patients with confirmed neurocysticercosis, PCR targeting the pTsol9 repetitive element performed on DNA isolated from CSF samples showed the highest sensitivity (95.9%) compared to antibody detection by ELISA or immunoblotting and HP10 antigen detection by ELISA, with sensitivities of 90.1%, 53.7%, and 81%, respectively (405).
Species-specific PCRs for non-Taenia tapeworm species have not been widely used. A multiplex PCR targeting the cox1 sequence with a general reverse primer and four species-specific forward primers was used for the specific amplification of Diphyllobothrium latum, D. dendriticum, D. pacificum, and D. nihonkaiense DNAs from adult worms, plerocercoid larvae, and eggs (407).
PCR followed by sequencing using universal primers to amplify the ribosomal ITS1, ITS2, 5.8S, and SSU rDNA sequences or mitochondrial genes is used to identify and confirm the species identity of tapeworms found in individual cases as well as interspecies variation to construct phylogenetic relationships (408–411). Sequence analysis of the ITS1, cox1, and paramyosin gene sequences from H. nana isolates from northwest Australia provided genetic support that the life cycle involves mainly human-to-human transmission (412).
CONCLUDING REMARKS
A large number of studies using a variety of nucleic acid-based techniques have contributed to our understanding of the genetic diversity, epidemiology, and clinical relevance of intestinal parasites. For obvious reasons, in a routine setting, standardization and harmonization of protocols are essential for cost-effective implementation of these new techniques. A syndrome-driven approach that includes the detection of the most likely pathogens in a particular setting using a single overall technique can be facilitated by the use of multiplex real-time PCR. Such an approach offers a highly sensitive and specific diagnostic alternative to labor-intensive microscopy in clinical laboratory practice for the diagnosis of intestinal pathogens. As molecular diagnostic facilities in general microbiology laboratories are already widely available, an increasing number of laboratories have implemented real-time PCR for the first-line diagnosis of intestinal parasitic infections. The implementation of nucleic acid-based tests is particularly useful for diarrhea-causing protists when used in combination with additional panels for the detection of bacterial and viral enteritis agents (106, 413). In comparison with microscopic examination, the rate of detection of parasitic infections when the targets are included in a multiplex PCR is considerably high (27, 87, 104, 107, 162, 414). However, parasites that are not included as targets in the multiplex PCR will not be detected, which is one of the arguments that is frequently raised against the use of molecular diagnostics in parasitology. Although true, it is good to realize that, diarrhea-causing protists aside, the prevalence of other parasitic pathogens detected by microscopy of stool samples submitted to most general microbiology laboratories in industrialized countries is extremely low (27, 162, 328, 413, 415–418). Even in travelers, additional parasites are found by microscopy in only a small minority of cases, with the highest detection rates in travelers to high-risk areas (107, 419). The use of additional diagnostic methods for the detection of those parasitic infections that are not included in a standard fecal PCR panel may therefore be limited to a select group of patients. For some laboratories, this would probably mean that there are only a few cases in which additional tests are needed, whereas, for example, in laboratories located in large metropolitan areas with large immigrant populations and serving many travelers, it would be more efficient to employ a more elaborate PCR panel and/or to perform additional microscopy on all samples submitted. This approach is similar to algorithms that are used where PCR is not a method of choice for the routine diagnosis of parasitic infections. Decisions on a standard screening technique to employ are based on the general patient population profile of the laboratory, including travel (immigration and adoption), immune status, eosinophilia, larva currens, persistent symptoms, and so on, with or without additional techniques for the diagnosis of other infections (27, 417, 420). For example, in immunocompromised patients with diarrhea, an additional method for the detection of opportunistic pathogens such as microsporidia and C. belli may be appropriate. This could be performed by optical white staining and acid-fast staining, respectively, or by using a multiplex PCR for the detection of both pathogens.
Molecular techniques are ideally suited for automation, and following the rapid growth of nucleic acid-based techniques for a large variety of targets, automation of the diagnostic process is currently being implemented in many laboratories. Automation of nucleic acid isolation, PCR setup, and PCR, when integrated through middleware with a laboratory information management system (LIMS), facilitates a rapid sample-to-answer process. This could be seen by some as another step in the separation of the laboratory, the clinician, and the individual patient. However, in this process, primary screening through a general algorithm can be followed by additional techniques decided upon during medical authorization based on individual clinical records and patient history.
Obviously, just as quality assessment schemes are common practice for microscopy-based diagnosis, the application of molecular diagnostics requires quality assurance as well. Initiatives to this end have been taken (421). Since 2012, Quality Control for Molecular Diagnostics (QCMD) (http://www.qcmd.org/) provides a yearly quality assessment scheme for gastrointestinal diseases, including a panel for intestinal protozoa. The Dutch Foundation for Quality Assessment in Medical Laboratories (SKML) (http://www.skml.nl/) has started an international scheme for molecular diagnosis of intestinal protozoa as well.
FUTURE DIRECTIONS
Throughout the world, including developing countries, real-time PCR is available in an increasing number of research centers, and molecular diagnosis of parasitic infections can be applied to large-scale epidemiological and more fundamental research. One major advantage is the possibility of an integrated high-throughput approach for the detection of a range of parasitic, bacterial, and viral targets using the same technique and for which samples can be collected and transported to central laboratories. Moreover, samples can be stored and used later for testing of additional targets when new research questions arise or, for example, for detection of mutations that are associated with drug resistance. Although, at first glance, PCR is more expensive than conventional microscopy-based techniques, this must be weighed against the complexities of organizing the prolonged stay of a large team of technicians and support staff in the field, sometimes under primitive conditions.
In a number of European countries, (automated) multiplex PCRs, which are usually in-house tests, are routinely used in clinical diagnostic laboratories. Commercial multiplex PCRs for the detection of a range of enteropathogens, which are already optimized and validated for a variety of instruments, as well as kits to be used on more extended multiplex platforms, such as Luminex, and variations on microarrays are all available (413, 422–424). This trend will make it easier for laboratories that are just starting to use molecular diagnostics to catch up with the rapid developments in this field. The first FDA-approved multiplex gastroenteric panel will lead to the more widespread use of nucleic acid-based tests in the United States as well (Table 2). Test formats that combine sample processing and DNA amplification of enteropathogens for single or multiple samples into one device have recently been launched or are still under development (e.g., BDmax [Becton, Dickinson and Company] and FilmArray [Biofire Diagnostics]) and will make it feasible for smaller laboratories to implement nucleic acid-based diagnostics. The cost-effectiveness of such an approach compared to in-house or commercially available high-throughput platforms, however, needs to be investigated. Another interesting approach is the use of an extended-range PCR screening strategy for the detection of bacterial, viral, fungal, and parasitic organisms in complex cases with a broad differential diagnosis; this has been reported in a case of cystoisosporiasis and in a case of neurocysticercosis (211, 425).
While there is no doubt that molecular diagnostic methods will continue to rely on the detection of genus- or species-specific DNA or RNA, new-generation sequencing platforms, which as yet are used primarily for research, will become more user-friendly and will be applied in a diagnostic setting. This will enable, for example, screening of fecal DNAs using broad-specificity primers for in-depth analysis of the microbially diverse populations present in such samples (426). In addition to targeting microorganisms, the host factors potentially involved in pathogenesis can also be explored.
ACKNOWLEDGMENTS
We thank Marianne Lebbad, Public Health Agency of Sweden, Solna, Sweden, for providing the parasite pictures in Fig. 2 to 4. We greatly acknowledge Graham Clark, London School of Hygiene and Tropical Medicine, for his critical comments on the manuscript.
Biographies

Jaco J. Verweij earned his Ph.D. at Leiden University, The Netherlands, on molecular tools in the diagnosis of intestinal parasitic infections. For more than 20 years, he has worked in the department of Parasitology of the Leiden University Medical Center on the development, validation, and implementation of new diagnostic tools, molecular methods in particular. He has a passion for teaching and is a strong advocate for technology transfer and capacity building in the countries suffering the most from parasitic diseases. Since 2012, he has held a position as a medical molecular microbiologist and parasitologist in the Laboratory for Medical Microbiology and Immunology of St. Elisabeth Hospital, Tilburg, The Netherlands.

C. Rune Stensvold, B.Med.Sc., M.Sc., Ph.D., is a diagnostic parasitologist and senior scientist at the Department of Microbiology and Infection Control, Statens Serum Institut, Copenhagen, Denmark. He did his postdoc training at the Faculty of Infectious and Tropical Diseases, London School of Hygiene and Tropical Medicine. His chief academic areas include the development and optimization of molecular diagnostics and typing methods for parasitic infections. Dr. Stensvold moreover takes a vast research interest in exploring the genetic diversity and host specificity of intestinal parasitic protists and in the role of common intestinal microbial eukaryotes in human health and disease, with special emphasis on Blastocystis, Entamoeba, and Dientamoeba. In 2013, Dr. Stensvold was awarded the Fritz Kauffmann Prize for his contribution to clinical microbiology in Denmark.
REFERENCES
- 1.Saiki RK, Scharf S, Faloona F, Mullis KB, Horn GT, Erlich HA, Arnheim N. 1985. Enzymatic amplification of beta-globin genomic sequences and restriction site analysis for diagnosis of sickle cell anemia. Science 230:1350–1354. 10.1126/science.2999980 [DOI] [PubMed] [Google Scholar]
- 2.Saiki RK, Gelfand DH, Stoffel S, Scharf SJ, Higuchi R, Horn GT, Mullis KB, Erlich HA. 1988. Primer-directed enzymatic amplification of DNA with a thermostable DNA polymerase. Science 239:487–491. 10.1126/science.2448875 [DOI] [PubMed] [Google Scholar]
- 3.de Bruijn MH. 1988. Diagnostic DNA amplification no respite for the elusive parasite. Parasitol. Today 4:293–295. 10.1016/0169-4758(88)90028-2 [DOI] [PubMed] [Google Scholar]
- 4.Barker RH., Jr 1990. DNA probe diagnosis of parasitic infections. Exp. Parasitol. 70:494–499. 10.1016/0014-4894(90)90136-Z [DOI] [PubMed] [Google Scholar]
- 5.Weiss JB. 1995. DNA probes and PCR for diagnosis of parasitic infections. Clin. Microbiol. Rev. 8:113–130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Klein D. 2002. Quantification using real-time PCR technology: applications and limitations. Trends Mol. Med. 8:257–260. 10.1016/S1471-4914(02)02355-9 [DOI] [PubMed] [Google Scholar]
- 7.Espy MJ, Uhl JR, Sloan LM, Buckwalter SP, Jones MF, Vetter EA, Yao JD, Wengenack NL, Rosenblatt JE, Cockerill FR, III, Smith TF. 2006. Real-time PCR in clinical microbiology: applications for routine laboratory testing. Clin. Microbiol. Rev. 19:165–256. 10.1128/CMR.19.1.165-256.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Williams JG, Kubelik AR, Livak KJ, Rafalski JA, Tingey SV. 1990. DNA polymorphisms amplified by arbitrary primers are useful as genetic markers. Nucleic Acids Res. 18:6531–6535. 10.1093/nar/18.22.6531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schouten JP, McElgunn CJ, Waaijer R, Zwijnenburg D, Diepvens F, Pals G. 2002. Relative quantification of 40 nucleic acid sequences by multiplex ligation-dependent probe amplification. Nucleic Acids Res. 30:e57. 10.1093/nar/gnf056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Notomi T, Okayama H, Masubuchi H, Yonekawa T, Watanabe K, Amino N, Hase T. 2000. Loop-mediated isothermal amplification of DNA. Nucleic Acids Res. 28:e63. 10.1093/nar/28.12.e63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Monteiro L, Bonnemaison D, Vekris A, Petry KG, Bonnet J, Vidal R, Cabrita J, Mégraud F. 1997. Complex polysaccharides as PCR inhibitors in feces: Helicobacter pylori model. J. Clin. Microbiol. 35:995–998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Morgan UM, Pallant L, Dwyer BW, Forbes DA, Rich G, Thompson RC. 1998. Comparison of PCR and microscopy for detection of Cryptosporidium parvum in human fecal specimens: clinical trial. J. Clin. Microbiol. 36:995–998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Verweij JJ, Pit DS, van Lieshout L, Baeta SM, Dery GD, Gasser RB, Polderman AM. 2001. Determining the prevalence of Oesophagostomum bifurcum and Necator americanus infections using specific PCR amplification of DNA from faecal samples. Trop. Med. Int. Health 6:726–731. 10.1046/j.1365-3156.2001.00770.x [DOI] [PubMed] [Google Scholar]
- 14.Kreader CA. 1996. Relief of amplification inhibition in PCR with bovine serum albumin or T4 gene 32 protein. Appl. Environ. Microbiol. 62:1102–1106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Demeler J, Ramunke S, Wolken S, Ianiello D, Rinaldi L, Gahutu JB, Cringoli G, von Samson-Himmelstjerna G, Krucken J. 2013. Discrimination of gastrointestinal nematode eggs from crude fecal egg preparations by inhibitor-resistant conventional and real-time PCR. PLoS One 8:e61285. 10.1371/journal.pone.0061285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Niesters HG. 2002. Clinical virology in real time. J. Clin. Virol. 25:S3–S12. 10.1016/S1386-6532(02)00197-X [DOI] [PubMed] [Google Scholar]
- 17.Andersen LO, Röser D, Nejsum P, Nielsen HV, Stensvold CR. 2013. Is supplementary bead beating for DNA extraction from nematode eggs using the NucliSENS easyMag protocol necessary? J. Clin. Microbiol. 51:1345–1347. 10.1128/JCM.03353-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mejia R, Vicuna Y, Broncano N, Sandoval C, Vaca M, Chico M, Cooper PJ, Nutman TB. 2013. A novel, multi-parallel, real-time polymerase chain reaction approach for eight gastrointestinal parasites provides improved diagnostic capabilities to resource-limited at-risk populations. Am. J. Trop. Med. Hyg. 88:1041–1047. 10.4269/ajtmh.12-0726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cnops L, Esbroeck MV. 2010. Freezing of stool samples improves real-time PCR detection of Entamoeba dispar and Entamoeba histolytica. J. Microbiol. Methods 80:310–312. 10.1016/j.mimet.2010.01.001 [DOI] [PubMed] [Google Scholar]
- 20.Troll H, Marti H, Weiss N. 1997. Simple differential detection of Entamoeba histolytica and Entamoeba dispar in fresh stool specimens by sodium acetate-acetic acid-formalin concentration and PCR. J. Clin. Microbiol. 35:1701–1705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kuk S, Yazar S, Cetinkaya U. 2012. Stool sample storage conditions for the preservation of Giardia intestinalis DNA. Mem. Inst. Oswaldo Cruz 107:965–968. 10.1590/S0074-02762012000800001 [DOI] [PubMed] [Google Scholar]
- 22.Higgins JA, Fayer R, Trout JM, Xiao L, Lal AA, Kerby S, Jenkins MC. 2001. Real-time PCR for the detection of Cryptosporidium parvum. J. Microbiol. Methods 47:323–337. 10.1016/S0167-7012(01)00339-6 [DOI] [PubMed] [Google Scholar]
- 23.Ramos F, Zurabian R, Moran P, Ramiro M, Gomez A, Clark CG, Melendro EI, Garcia G, Ximenez C. 1999. The effect of formalin fixation on the polymerase chain reaction characterization of Entamoeba histolytica. Trans. R. Soc. Trop. Med. Hyg. 93:335–336. 10.1016/S0035-9203(99)90045-7 [DOI] [PubMed] [Google Scholar]
- 24.Stroup S, Tongjai S, Swai N, Maro A, Kibiki G, Houpt ER. 2012. Dual probe DNA capture for sensitive real-time PCR detection of Cryptosporidium and Giardia. Mol. Cell. Probes 26:104–106. 10.1016/j.mcp.2011.12.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Persson S, de Boer RF, Kooistra-Smid AM, Olsen KE. 2011. Five commercial DNA extraction systems tested and compared on a stool sample collection. Diagn. Microbiol. Infect. Dis. 69:240–244. 10.1016/j.diagmicrobio.2010.09.023 [DOI] [PubMed] [Google Scholar]
- 26.van Zanten E, Wisselink GJ, de Boer W, Stoll S, Alvarez R, Kooistra-Smid AM. 2011. Comparison of the QIAsymphony automated nucleic acid extraction and PCR setup platforms with NucliSens easyMAG and Corbett CAS-1200 liquid handling station for the detection of enteric pathogens in fecal samples. J. Microbiol. Methods 84:335–340. 10.1016/j.mimet.2010.12.024 [DOI] [PubMed] [Google Scholar]
- 27.Bruijnesteijn van Coppenraet LE, Wallinga JA, Ruijs GJ, Bruins MJ, Verweij JJ. 2009. Parasitological diagnosis combining an internally controlled real-time PCR assay for the detection of four protozoa in stool samples with a testing algorithm for microscopy. Clin. Microbiol. Infect. 15:869–874. 10.1111/j.1469-0691.2009.02894.x [DOI] [PubMed] [Google Scholar]
- 28.Stensvold CR, Lebbad M, Verweij JJ. 2011. The impact of genetic diversity in protozoa on molecular diagnostics. Trends Parasitol. 27:53–58. 10.1016/j.pt.2010.11.005 [DOI] [PubMed] [Google Scholar]
- 29.Burnet JB, Ogorzaly L, Tissier A, Penny C, Cauchie HM. 2013. Novel quantitative TaqMan real-time PCR assays for detection of Cryptosporidium at the genus level and genotyping of major human and cattle-infecting species. J. Appl. Microbiol. 114:1211–1222. 10.1111/jam.12103 [DOI] [PubMed] [Google Scholar]
- 30.Stanley SL. 2003. Amoebiasis. Lancet 361:1025–1034. 10.1016/S0140-6736(03)12830-9 [DOI] [PubMed] [Google Scholar]
- 31.Anonymous. 1997. WHO/PAHO/UNESCO report. A consultation with experts on amoebiasis. Mexico City, Mexico 28-29 January, 1997. Epidemiol. Bull. 18:13–14 [PubMed] [Google Scholar]
- 32.Pritt BS, Clark CG. 2008. Amebiasis. Mayo Clin. Proc. 83:1154–1159; quiz 1159–1160. 10.4065/83.10.1154 [DOI] [PubMed] [Google Scholar]
- 33.Ali IK, Clark CG, Petri WA. 2008. Molecular epidemiology of amebiasis. Infect. Genet. Evol. 8:698–707. 10.1016/j.meegid.2008.05.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ximenez C, Moran P, Rojas L, Valadez A, Gomez A. 2009. Reassessment of the epidemiology of amebiasis: state of the art. Infect. Genet. Evol. 9:1023–1032. 10.1016/j.meegid.2009.06.008 [DOI] [PubMed] [Google Scholar]
- 35.Gutiérrez-Cisneros MJ, Cogollos R, López-Vélez R, Martín-Rabadán P, Martínez-Ruiz R, Subirats M, Merino FJ, Fuentes I. 2010. Application of real-time PCR for the differentiation of Entamoeba histolytica and E. dispar in cyst-positive faecal samples from 130 immigrants living in spain. Ann. Trop. Med. Parasitol. 104:145–149. 10.1179/136485910X12607012373759 [DOI] [PubMed] [Google Scholar]
- 36.Herbinger KH, Fleischmann E, Weber C, Perona P, Loscher T, Bretzel G. 2011. Epidemiological, clinical, and diagnostic data on intestinal infections with Entamoeba histolytica and Entamoeba dispar among returning travelers. Infection 39:527–535. 10.1007/s15010-011-0155-z [DOI] [PubMed] [Google Scholar]
- 37.ten Hove RJ, Van Lieshout L, Beadsworth MB, Perez MA, Spee K, Claas EC, Verweij JJ. 2009. Characterization of genotypes of Enterocytozoon bieneusi in immunosuppressed and immunocompetent patient groups. J. Eukaryot. Microbiol. 56:388–393. 10.1111/j.1550-7408.2009.00393.x [DOI] [PubMed] [Google Scholar]
- 38.Verweij JJ, van Lieshout L, Blotkamp C, Brienen EA, van Duivenvoorden S, van Esbroeck M, Polderman AM. 2000. Differentiation of Entamoeba histolytica and Entamoeba dispar using PCR-SHELA and comparison of antibody response. Arch. Med. Res. 31:S44–S46. 10.1016/S0188-4409(00)00221-6 [DOI] [PubMed] [Google Scholar]
- 39.Jackson TF. 1998. Entamoeba histolytica and Entamoeba dispar are distinct species; clinical, epidemiological and serological evidence. Int. J. Parasitol. 28:181–186. 10.1016/S0020-7519(97)00177-X [DOI] [PubMed] [Google Scholar]
- 40.Kebede A, Verweij JJ, Petros B, Polderman AM. 2004. Short communication: misleading microscopy in amoebiasis. Trop. Med. Int. Health 9:651–652. 10.1111/j.1365-3156.2004.01236.x [DOI] [PubMed] [Google Scholar]
- 41.Haque R, Petri WA., Jr 2006. Diagnosis of amebiasis in Bangladesh. Arch. Med. Res. 37:273–276. 10.1016/j.arcmed.2005.09.001 [DOI] [PubMed] [Google Scholar]
- 42.Visser LG, Verweij JJ, Van Esbroeck M, Edeling WM, Clerinx J, Polderman AM. 2006. Diagnostic methods for differentiation of Entamoeba histolytica and Entamoeba dispar in carriers: performance and clinical implications in a non-endemic setting. Int. J. Med. Microbiol. 296:397–403. 10.1016/j.ijmm.2006.03.001 [DOI] [PubMed] [Google Scholar]
- 43.Stark D, van Hal S, Fotedar R, Butcher A, Marriott D, Ellis J, Harkness J. 2008. Comparison of stool antigen detection kits to PCR for diagnosis of amebiasis. J. Clin. Microbiol. 46:1678–1681. 10.1128/JCM.02261-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Stark D, Fotedar R, van Hal S, Beebe N, Marriott D, Ellis JT, Harkness J. 2007. Prevalence of enteric protozoa in human immunodeficiency virus (HIV)-positive and HIV-negative men who have sex with men from Sydney, Australia. Am. J. Trop. Med. Hyg. 76:549–552 http://www.ajtmh.org/content/76/3/549.long [PubMed] [Google Scholar]
- 45.Yakoob J, Abbas Z, Beg MA, Naz S, Khan R, Jafri W. 2012. Entamoeba species associated with chronic diarrhoea in Pakistan. Epidemiol. Infect. 140:323–328. 10.1017/S0950268811000215 [DOI] [PubMed] [Google Scholar]
- 46.Anuar TS, Al-Mekhlafi HM, Ghani MK, Azreen SN, Salleh FM, Ghazali N, Bernadus M, Moktar N. 2012. First molecular identification of Entamoeba moshkovskii in Malaysia. Parasitology 139:1521–1525. 10.1017/S0031182012001485 [DOI] [PubMed] [Google Scholar]
- 47.Shimokawa C, Kabir M, Taniuchi M, Mondal D, Kobayashi S, Ali IK, Sobuz SU, Senba M, Houpt E, Haque R, Petri WA, Jr, Hamano S. 2012. Entamoeba moshkovskii is associated with diarrhea in infants and causes diarrhea and colitis in mice. J. Infect. Dis. 206:744–751. 10.1093/infdis/jis414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Heredia RD, Fonseca JA, López MC. 2012. Entamoeba moshkovskii perspectives of a new agent to be considered in the diagnosis of amebiasis. Acta Trop. 123:139–145. 10.1016/j.actatropica.2012.05.012 [DOI] [PubMed] [Google Scholar]
- 49.Royer TL, Gilchrist C, Kabir M, Arju T, Ralston KS, Haque R, Clark CG, Petri WA. 2012. Entamoeba bangladeshi nov. sp., Bangladesh. Emerg. Infect. Dis. 18:1543–1545. 10.3201/eid1809.120122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Stensvold CR, Lebbad M, Victory EL, Verweij JJ, Tannich E, Alfellani M, Legarraga P, Clark CG. 2011. Increased sampling reveals novel lineages of Entamoeba: consequences of genetic diversity and host specificity for taxonomy and molecular detection. Protist 162:525–541. 10.1016/j.protis.2010.11.002 [DOI] [PubMed] [Google Scholar]
- 51.Haque R, Kabir M, Noor Z, Rahman SM, Mondal D, Alam F, Rahman I, Al Mahmood A, Ahmed N, Petri WA., Jr 2010. Diagnosis of amebic liver abscess and amebic colitis by detection of Entamoeba histolytica DNA in blood, urine, and saliva by a real-time PCR assay. J. Clin. Microbiol. 48:2798–2801. 10.1128/JCM.00152-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Clark CG, Kaffashian F, Tawari B, Windsor JJ, Twigg-Flesner A, Davies-Morel M, Blessmann J, Ebert F, Peschel B, Le Van A, Jackson CJ, Macfarlane L, Tannich E. 2006. New insights into the phylogeny of Entamoeba species provided by analysis of four new small-subunit rRNA genes. Int. J. Syst. Evol. Microbiol. 56:2235–2239. 10.1099/ijs.0.64208-0 [DOI] [PubMed] [Google Scholar]
- 53.Tachibana H, Yanagi T, Pandey K, Cheng XJ, Kobayashi S, Sherchand JB, Kanbara H. 2007. An Entamoeba sp. strain isolated from rhesus monkey is virulent but genetically different from Entamoeba histolytica. Mol. Biochem. Parasitol. 153:107–114. 10.1016/j.molbiopara.2007.02.006 [DOI] [PubMed] [Google Scholar]
- 54.Levecke B, Dreesen L, Dorny P, Verweij JJ, Vercammen F, Casaert S, Vercruysse J, Geldhof P. 2010. Molecular identification of Entamoeba spp. in captive nonhuman primates. J. Clin. Microbiol. 48:2988–2990. 10.1128/JCM.00013-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tachibana H, Yanagi T, Akatsuka A, Kobayashi S, Kanbara H, Tsutsumi V. 2009. Isolation and characterization of a potentially virulent species Entamoeba nuttalli from captive Japanese macaques. Parasitology 136:1169–1177. 10.1017/S0031182009990576 [DOI] [PubMed] [Google Scholar]
- 56.Takano J, Narita T, Tachibana H, Terao K, Fujimoto K. 2007. Comparison of Entamoeba histolytica DNA isolated from a cynomolgus monkey with human isolates. Parasitol. Res. 101:539–546. 10.1007/s00436-007-0510-2 [DOI] [PubMed] [Google Scholar]
- 57.Takano J, Tachibana H, Kato M, Narita T, Yanagi T, Yasutomi Y, Fujimoto K. 2009. DNA characterization of simian Entamoeba histolytica-like strains to differentiate them from Entamoeba histolytica. Parasitol. Res. 105:929–937. 10.1007/s00436-009-1480-3 [DOI] [PubMed] [Google Scholar]
- 58.Stensvold CR, Lebbad M, Clark CG. 2010. Genetic characterisation of uninucleated cyst-producing Entamoeba spp. from ruminants. Int. J. Parasitol. 40:775–778. 10.1016/j.ijpara.2010.03.003 [DOI] [PubMed] [Google Scholar]
- 59.Tanyuksel M, Petri WA., Jr 2003. Laboratory diagnosis of amebiasis. Clin. Microbiol. Rev. 16:713–729. 10.1128/CMR.16.4.713-729.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Fotedar R, Stark D, Beebe N, Marriott D, Ellis J, Harkness J. 2007. Laboratory diagnostic techniques for Entamoeba species. Clin. Microbiol. Rev. 20:511–532. 10.1128/CMR.00004-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hamzah Z, Petmitr S, Mungthin M, Leelayoova S, Chavalitshewinkoon-Petmitr P. 2010. Development of multiplex real-time polymerase chain reaction for detection of Entamoeba histolytica, Entamoeba dispar, and Entamoeba moshkovskii in clinical specimens. Am. J. Trop. Med. Hyg. 83:909–913. 10.4269/ajtmh.2010.10-0050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Liang SY, Hsia KT, Chan YH, Fan CK, Jiang DD, Landt O, Ji DD. 2010. Evaluation of a new single-tube multiprobe real-time PCR for diagnosis of Entamoeba histolytica and Entamoeba dispar. J. Parasitol. 96:793–797. 10.1645/GE-2373.1 [DOI] [PubMed] [Google Scholar]
- 63.Santos HL, Bandea R, Martins LA, de Macedo HW, Peralta RH, Peralta JM, Ndubuisi MI, da Silva AJ. 2010. Differential identification of Entamoeba spp. based on the analysis of 18S rRNA. Parasitol. Res. 106:883–888. 10.1007/s00436-010-1728-y [DOI] [PubMed] [Google Scholar]
- 64.Stensvold CR, Lebbad M, Verweij JJ, Jespersgaard C, von Samson-Himmelstjerna G, Nielsen SS, Nielsen HV. 2010. Identification and delineation of members of the Entamoeba complex by pyrosequencing. Mol. Cell. Probes 24:403–406. 10.1016/j.mcp.2010.07.008 [DOI] [PubMed] [Google Scholar]
- 65.Qvarnstrom Y, James C, Xayavong M, Holloway BP, Visvesvara GS, Sriram R, da Silva A. 2005. Comparison of real-time PCR protocols for differential laboratory diagnosis of amebiasis. J. Clin. Microbiol. 43:5491–5497. 10.1128/JCM.43.11.5491-5497.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Santos HL, Bandyopadhyay K, Bandea R, Peralta RH, Peralta JM, Da Silva AJ. 2013. LUMINEX: a new technology for the simultaneous identification of five Entamoeba spp. commonly found in human stools. Parasit. Vectors 6:69. 10.1186/1756-3305-6-69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Gilchrist CA, Ali IK, Kabir M, Alam F, Scherbakova S, Ferlanti E, Weedall GD, Hall N, Haque R, Petri WA, Caler E. 2012. A multilocus sequence typing system (MLST) reveals a high level of diversity and a genetic component to Entamoeba histolytica virulence. BMC Microbiol. 12:151. 10.1186/1471-2180-12-151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Parija SC, Khairnar K. 2007. Detection of excretory Entamoeba histolytica DNA in the urine, and detection of E. histolytica DNA and lectin antigen in the liver abscess pus for the diagnosis of amoebic liver abscess. BMC Microbiol. 7:41. 10.1186/1471-2180-7-41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Solaymani-Mohammadi S, Lam MM, Zunt JR, Petri WA., Jr 2007. Entamoeba histolytica encephalitis diagnosed by PCR of cerebrospinal fluid. Trans. R. Soc. Trop. Med. Hyg. 101:311–313. 10.1016/j.trstmh.2006.05.004 [DOI] [PubMed] [Google Scholar]
- 70.Othman N, Mohamed Z, Verweij JJ, Huat LB, Olivos-Garcia A, Yeng C, Noordin R. 2010. Application of real-time polymerase chain reaction in detection of Entamoeba histolytica in pus aspirates of liver abscess patients. Foodborne Pathog. Dis. 7:637–641. 10.1089/fpd.2009.0427 [DOI] [PubMed] [Google Scholar]
- 71.Guerrant RL, Hughes JM, Lima NL, Crane J. 1990. Diarrhea in developed and developing countries: magnitude, special settings, and etiologies. Rev. Infect. Dis. 12:S41–S50. 10.1093/clinids/12.Supplement_1.S41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Canete R, Diaz MM, Avalos Garcia R, Laud Martinez PM, Manuel Ponce F. 2012. Intestinal parasites in children from a day care centre in Matanzas City, Cuba. PLoS One 7:e51394. 10.1371/journal.pone.0051394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Baldursson S, Karanis P. 2011. Waterborne transmission of protozoan parasites: review of worldwide outbreaks—an update 2004-2010. Water Res. 45:6603–6614. 10.1016/j.watres.2011.10.013 [DOI] [PubMed] [Google Scholar]
- 74.Nematian J, Gholamrezanezhad A, Nematian E. 2008. Giardiasis and other intestinal parasitic infections in relation to anthropometric indicators of malnutrition: a large, population-based survey of schoolchildren in Tehran. Ann. Trop. Med. Parasitol. 102:209–214. 10.1179/136485908X267876 [DOI] [PubMed] [Google Scholar]
- 75.Boeke CE, Mora-Plazas M, Forero Y, Villamor E. 2010. Intestinal protozoan infections in relation to nutritional status and gastrointestinal morbidity in Colombian school children. J. Trop. Pediatr. 56:299–306. 10.1093/tropej/fmp136 [DOI] [PubMed] [Google Scholar]
- 76.Ali SA, Hill DR. 2003. Giardia intestinalis. Curr. Opin. Infect. Dis. 16:453–460. 10.1097/00001432-200310000-00012 [DOI] [PubMed] [Google Scholar]
- 77.Robertson LJ, Hanevik K, Escobedo AA, Morch K, Langeland N. 2010. Giardiasis—why do the symptoms sometimes never stop? Trends Parasitol. 26:75–82. 10.1016/j.pt.2009.11.010 [DOI] [PubMed] [Google Scholar]
- 78.Verweij JJ, Schinkel J, Laeijendecker D, van Rooyen MA, van Lieshout L, Polderman AM. 2003. Real-time PCR for the detection of Giardia lamblia. Mol. Cell. Probes 17:223–225. 10.1016/S0890-8508(03)00057-4 [DOI] [PubMed] [Google Scholar]
- 79.Verweij JJ, Blange RA, Templeton K, Schinkel J, Brienen EA, van Rooyen MA, van Lieshout L, Polderman AM. 2004. Simultaneous detection of Entamoeba histolytica, Giardia lamblia, and Cryptosporidium parvum in fecal samples by using multiplex real-time PCR. J. Clin. Microbiol. 42:1220–1223. 10.1128/JCM.42.3.1220-1223.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Amar CF, Dear PH, McLauchlin J. 2003. Detection and genotyping by real-time PCR/RFLP analyses of Giardia duodenalis from human faeces. J. Med. Microbiol. 52:681–683. 10.1099/jmm.0.05193-0 [DOI] [PubMed] [Google Scholar]
- 81.Ghosh S, Debnath A, Sil A, De S, Chattopadhyay DJ, Das P. 2000. PCR detection of Giardia lamblia in stool: targeting intergenic spacer region of multicopy rRNA gene. Mol. Cell. Probes 14:181–189. 10.1006/mcpr.2000.0302 [DOI] [PubMed] [Google Scholar]
- 82.Zhang P, Liu Y, Alsarakibi M, Li J, Liu T, Li Y, Li G. 2012. Application of HRM assays with EvaGreen dye for genotyping Giardia duodenalis zoonotic assemblages. Parasitol. Res. 111:2157–2163. 10.1007/s00436-012-3064-x [DOI] [PubMed] [Google Scholar]
- 83.Siripattanapipong S, Leelayoova S, Mungthin M, Thompson RC, Boontanom P, Saksirisamphant W, Tan-Ariya P. 2011. Determination of discriminatory power of genetic markers used for genotyping Giardia duodenalis. Southeast Asian J. Trop. Med. Public Health 42:764–771 [PubMed] [Google Scholar]
- 84.Asher AJ, Waldron LS, Power ML. 2012. Evaluation of a PCR protocol for sensitive detection of Giardia intestinalis in human faeces. Parasitol. Res. 110:853–858. 10.1007/s00436-011-2565-3 [DOI] [PubMed] [Google Scholar]
- 85.Hussein EM, Al-Mohammed H, Hussein AM. 2009. Genetic diversity of Dientamoeba fragilis isolates of irritable bowel syndrome patients by high-resolution melting-curve (HRM) analysis. Parasitol. Res. 105:1053–1060. 10.1007/s00436-009-1515-9 [DOI] [PubMed] [Google Scholar]
- 86.Vanni I, Cacciò SM, van Lith L, Lebbad M, Svard SG, Pozio E, Tosini F. 2012. Detection of Giardia duodenalis assemblages A and B in human feces by simple, assemblage-specific PCR assays. PLoS Negl. Trop. Dis. 6:e1776. 10.1371/journal.pntd.0001776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Nazeer JT, El Sayed Khalifa K, von Thien H, El-Sibaei M, Abdel-Hamid M, Tawfik RA, Tannich E. 2013. Use of multiplex real-time PCR for detection of common diarrhea causing protozoan parasites in Egypt. Parasitol. Res. 112:595–601. 10.1007/s00436-012-3171-8 [DOI] [PubMed] [Google Scholar]
- 88.Ryan U, Cacciò SM. 2013. Zoonotic potential of Giardia. Int. J. Parasitol. 43:943–956. 10.1016/j.ijpara.2013.06.001 [DOI] [PubMed] [Google Scholar]
- 89.Feng Y, Xiao L. 2011. Zoonotic potential and molecular epidemiology of Giardia species and giardiasis. Clin. Microbiol. Rev. 24:110–140. 10.1128/CMR.00033-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Cacciò SM, Sannella AR, Manuali E, Tosini F, Sensi M, Crotti D, Pozio E. 2012. Pigs as natural hosts of Dientamoeba fragilis genotypes found in humans. Emerg. Infect. Dis. 18:838–841. 10.3201/eid1805.111093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Ankarklev S, Svard SG, Lebbad M. 2012. Allelic sequence heterozygosity in single Giardia parasites. BMC Microbiol. 12:65. 10.1186/1471-2180-12-65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Monis PT, Cacciò SM, Thompson RC. 2009. Variation in Giardia: towards a taxonomic revision of the genus. Trends Parasitol. 25:93–100. 10.1016/j.pt.2008.11.006 [DOI] [PubMed] [Google Scholar]
- 93.Thompson RC, Monis P. 2012. Giardia—from genome to proteome. Adv. Parasitol. 78:57–95. 10.1016/B978-0-12-394303-3.00003-7 [DOI] [PubMed] [Google Scholar]
- 94.Lasek-Nesselquist E, Welch DM, Sogin ML. 2010. The identification of a new Giardia duodenalis assemblage in marine vertebrates and a preliminary analysis of G. duodenalis population biology in marine systems. Int. J. Parasitol. 40:1063–1074. 10.1016/j.ijpara.2010.02.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Cacciò SM, Ryan U. 2008. Molecular epidemiology of giardiasis. Mol. Biochem. Parasitol. 160:75–80. 10.1016/j.molbiopara.2008.04.006 [DOI] [PubMed] [Google Scholar]
- 96.Sprong H, Cacciò SM, van der Giessen JW, ZOOPNET Network and Partners 2009. Identification of zoonotic genotypes of Giardia duodenalis. PLoS Negl. Trop. Dis. 3:e558. 10.1371/journal.pntd.0000558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Takumi K, Swart A, Mank T, Lasek-Nesselquist E, Lebbad M, Cacciò SM, Sprong H. 2012. Population-based analyses of Giardia duodenalis is consistent with the clonal assemblage structure. Parasit. Vectors 5:168. 10.1186/1756-3305-5-168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Lebbad M, Petersson I, Karlsson L, Botero-Kleiven S, Andersson JO, Svenungsson B, Svärd SG. 2011. Multilocus genotyping of human Giardia isolates suggests limited zoonotic transmission and association between assemblage B and flatulence in children. PLoS Negl. Trop. Dis. 5:e1262. 10.1371/journal.pntd.0001262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Homan WL, Mank TG. 2001. Human giardiasis: genotype linked differences in clinical symptomatology. Int. J. Parasitol. 31:822–826. 10.1016/S0020-7519(01)00183-7 [DOI] [PubMed] [Google Scholar]
- 100.Read C, Walters J, Robertson ID, Thompson RC. 2002. Correlation between genotype of Giardia duodenalis and diarrhoea. Int. J. Parasitol. 32:229–231. 10.1016/S0020-7519(01)00340-X [DOI] [PubMed] [Google Scholar]
- 101.Ignatius R, Gahutu JB, Klotz C, Steininger C, Shyirambere C, Lyng M, Musemakweri A, Aebischer T, Martus P, Harms G, Mockenhaupt FP. 2012. High prevalence of Giardia duodenalis assemblage B infection and association with underweight in Rwandan children. PLoS Negl. Trop. Dis. 6:e1677. 10.1371/journal.pntd.0001677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Bonhomme J, Le Goff L, Lemée V, Gargala G, Ballet JJ, Favennec L. 2011. Limitations of tpi and bg genes sub-genotyping for characterization of human Giardia duodenalis isolates. Parasitol. Int. 60:327–330. 10.1016/j.parint.2011.05.004 [DOI] [PubMed] [Google Scholar]
- 103.Breathnach AS, McHugh TD, Butcher PD. 2010. Prevalence and clinical correlations of genetic subtypes of Giardia lamblia in an urban setting. Epidemiol. Infect. 138:1459–1467. 10.1017/S0950268810000208 [DOI] [PubMed] [Google Scholar]
- 104.ten Hove R, Schuurman T, Kooistra M, Moller L, van Lieshout L, Verweij JJ. 2007. Detection of diarrhoea-causing protozoa in general practice patients in the Netherlands by multiplex real-time PCR. Clin. Microbiol. Infect. 13:1001–1007. 10.1111/j.1469-0691.2007.01788.x [DOI] [PubMed] [Google Scholar]
- 105.Yakoob J, Abbas Z, Beg MA, Naz S, Khan R, Islam M, Jafri W. 2010. Prevalences of Giardia lamblia and Cryptosporidium parvum infection in adults presenting with chronic diarrhoea. Ann. Trop. Med. Parasitol. 104:505–510. 10.1179/136485910X12786389891209 [DOI] [PubMed] [Google Scholar]
- 106.de Boer RF, Ott A, Kesztyüs B, Kooistra-Smid AM. 2010. Improved detection of five major gastrointestinal pathogens by use of a molecular screening approach. J. Clin. Microbiol. 48:4140–4146. 10.1128/JCM.01124-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.ten Hove RJ, van Esbroeck M, Vervoort T, van den Ende J, van Lieshout L, Verweij JJ. 2009. Molecular diagnostics of intestinal parasites in returning travellers. Eur. J. Clin. Microbiol. Infect. Dis. 28:1045–1053. 10.1007/s10096-009-0745-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Calderaro A, Gorrini C, Montecchini S, Peruzzi S, Piccolo G, Rossi S, Gargiulo F, Manca N, Dettori G, Chezzi C. 2010. Evaluation of a real-time polymerase chain reaction assay for the laboratory diagnosis of giardiasis. Diagn. Microbiol. Infect. Dis. 66:261–267. 10.1016/j.diagmicrobio.2009.10.004 [DOI] [PubMed] [Google Scholar]
- 109.Jepps MW, Dobell C. 1918. Dientamoeba fragilis n.g., n. sp., new intestinal amoeba from man. Parasitology 10:352–367. 10.1017/S0031182000003929 [DOI] [Google Scholar]
- 110.Silberman JD, Clark CG, Sogin ML. 1996. Dientamoeba fragilis shares a recent common evolutionary history with the trichomonads. Mol. Biochem. Parasitol. 76:311–314. 10.1016/0166-6851(95)02516-2 [DOI] [PubMed] [Google Scholar]
- 111.Camp RR, Mattern CF, Honigberg BM. 1974. Study of Dientamoeba fragilis Jepps & Dobell. I. Electronmicroscopic observations of the binucleate stages. II. Taxonomic position and revision of the genus. J. Protozool. 21:69–82 [DOI] [PubMed] [Google Scholar]
- 112.Munasinghe VS, Vella NG, Ellis JT, Windsor PA, Stark D. 2013. Cyst formation and faecal-oral transmission of Dientamoeba fragilis—the missing link in the life cycle of an emerging pathogen. Int. J. Parasitol. 43:879–883. 10.1016/j.ijpara.2013.06.003 [DOI] [PubMed] [Google Scholar]
- 113.Johnson EH, Windsor JJ, Clark CG. 2004. Emerging from obscurity: biological, clinical, and diagnostic aspects of Dientamoeba fragilis. Clin. Microbiol. Rev. 17:553–570. 10.1128/CMR.17.3.553-570.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Stark D, Barratt J, Roberts T, Marriott D, Harkness J, Ellis J. 2010. A review of the clinical presentation of dientamoebiasis. Am. J. Trop. Med. Hyg. 82:614–619. 10.4269/ajtmh.2010.09-0478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Farthing MJ. 2006. Treatment options for the eradication of intestinal protozoa. Nat. Clin. Pract. Gastroenterol. Hepatol. 3:436–445. 10.1038/ncpgasthep0557 [DOI] [PubMed] [Google Scholar]
- 116.Okhuysen PC, White AC. 1999. Parasitic infections of the intestines. Curr. Opin. Infect. Dis. 12:467–472. 10.1097/00001432-199910000-00009 [DOI] [PubMed] [Google Scholar]
- 117.Okhuysen PC. 2001. Traveler's diarrhea due to intestinal protozoa. Clin. Infect. Dis. 33:110–114. 10.1086/320894 [DOI] [PubMed] [Google Scholar]
- 118.Gendrel D, Treluyer JM, Richard-Lenoble D. 2003. Parasitic diarrhea in normal and malnourished children. Fundam. Clin. Pharmacol. 17:189–197. 10.1046/j.1472-8206.2003.00169.x [DOI] [PubMed] [Google Scholar]
- 119.Bosman DK, Benninga MA, van de Berg P, Kooijman GC, van Gool T. 2004. Dientamoeba fragilis: possibly an important cause of persistent abdominal pain in children. Ned. Tijdschr. Geneeskd. 148:575–579 (In Dutch.) [PubMed] [Google Scholar]
- 120.Preiss U, Ockert G, Brömme S, Otto A. 1990. Dientamoeba fragilis infection, a cause of gastrointestinal symptoms in childhood. Klin. Padiatr. 202:120–123. 10.1055/s-2007-1025503 [DOI] [PubMed] [Google Scholar]
- 121.Girginkardeşler N, Coşkun S, Cüneyt Balcioğlu I, Ertan P, Ok UZ. 2003. Dientamoeba fragilis, a neglected cause of diarrhea, successfully treated with secnidazole. Clin. Microbiol. Infect. 9:110–113. 10.1046/j.1469-0691.2003.00504.x [DOI] [PubMed] [Google Scholar]
- 122.de Wit MA, Koopmans MP, Kortbeek LM, van Leeuwen NJ, Vinjé J, van Duynhoven YT. 2001. Etiology of gastroenteritis in sentinel general practices in the Netherlands. Clin. Infect. Dis. 33:280–288. 10.1086/321875 [DOI] [PubMed] [Google Scholar]
- 123.Windsor JJ, Rafay AM, Shenoy AK, Johnson EH. 1998. Incidence of Dientamoeba fragilis in faecal samples submitted for routine microbiological analysis. Br. J. Biomed. Sci. 55:172–175 [PubMed] [Google Scholar]
- 124.Engsbro AL, Stensvold CR, Nielsen HV, Bytzer P. 2012. Treatment of Dientamoeba fragilis in patients with irritable bowel syndrome. Am. J. Trop. Med. Hyg. 87:1046–1052. 10.4269/ajtmh.2012.11-0761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Noda S, Mantini C, Meloni D, Inoue J, Kitade O, Viscogliosi E, Ohkuma M. 2012. Molecular phylogeny and evolution of parabasalia with improved taxon sampling and new protein markers of actin and elongation factor-1α. PLoS One 7:e29938. 10.1371/journal.pone.0029938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.van Gool T, Weijts R, Lommerse E, Mank TG. 2003. Triple faeces test: an effective tool for detection of intestinal parasites in routine clinical practice. Eur. J. Clin. Microbiol. Infect. Dis. 22:284–290. 10.1007/s10096-003-0919-1 [DOI] [PubMed] [Google Scholar]
- 127.Stark D, Beebe N, Marriott D, Ellis J, Harkness J. 2006. Evaluation of three diagnostic methods, including real-time PCR, for detection of Dientamoeba fragilis in stool specimens. J. Clin. Microbiol. 44:232–235. 10.1128/JCM.44.1.232-235.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Verweij JJ, Mulder B, Poell B, van Middelkoop D, Brienen EA, van Lieshout L. 2007. Real-time PCR for the detection of Dientamoeba fragilis in fecal samples. Mol. Cell. Probes 21:400–404. 10.1016/j.mcp.2007.05.006 [DOI] [PubMed] [Google Scholar]
- 129.Johnson JA, Clark CG. 2000. Cryptic genetic diversity in Dientamoeba fragilis. J. Clin. Microbiol. 38:4653–4654 http://jcm.asm.org/content/38/12/4653.long [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Stark D, Beebe N, Marriott D, Ellis J, Harkness J. 2005. Prospective study of the prevalence, genotyping, and clinical relevance of Dientamoeba fragilis infections in an Australian population. J. Clin. Microbiol. 43:2718–2723. 10.1128/JCM.43.6.2718-2723.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Peek R, Reedeker FR, van Gool T. 2004. Direct amplification and genotyping of Dientamoeba fragilis from human stool specimens. J. Clin. Microbiol. 42:631–635. 10.1128/JCM.42.2.631-635.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Stensvold CR, Jespersgaard C, Dinesen K, Sorensen JF, Molbak K, Nielsen HV. 2007. Prevalence of Dientamoeba fragilis genotypes in different Danish populations as assessed by PCR and SNP genotyping. Trop. Med. Int. Health 12:182. 10.1111/j.1365-3156.2007.01867.x [DOI] [Google Scholar]
- 133.Windsor JJ, Macfarlane L, Clark CG. 2006. Internal transcribed spacer dimorphism and diversity in Dientamoeba fragilis. J. Eukaryot. Microbiol. 53:188–192. 10.1111/j.1550-7408.2006.00092.x [DOI] [PubMed] [Google Scholar]
- 134.Bart A, van der Heijden HM, Greve S, Speijer D, Landman WJ, van Gool T. 2008. Intragenomic variation in the internal transcribed spacer 1 region of Dientamoeba fragilis as a molecular epidemiological marker. J. Clin. Microbiol. 46:3270–3275. 10.1128/JCM.00680-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Röser D, Simonsen J, Nielsen HV, Stensvold CR, Mølbak K. 2013. Dientamoeba fragilis in Denmark: epidemiological experience derived from four years of routine real-time PCR. Eur. J. Clin. Microbiol. Infect. Dis. 32:1303–1310. 10.1007/s10096-013-1880-2 [DOI] [PubMed] [Google Scholar]
- 136.Stensvold CR, Clark CG, Röser D. 2013. Limited intra-genetic diversity in Dientamoeba fragilis housekeeping genes. Infect. Genet. Evol. 18:284–286. 10.1016/j.meegid.2013.05.003 [DOI] [PubMed] [Google Scholar]
- 137.Lankester F, Kiyang JA, Bailey W, Unwin S. 2010. Dientamoeba fragilis: initial evidence of pathogenicity in the western lowland gorilla (Gorilla gorilla gorilla). J. Zoo Wildl. Med. 41:350–352. 10.1638/2009-0190.1 [DOI] [PubMed] [Google Scholar]
- 138.Stark D, Phillips O, Peckett D, Munro U, Marriott D, Harkness J, Ellis J. 2008. Gorillas are a host for Dientamoeba fragilis: an update on the life cycle and host distribution. Vet. Parasitol. 151:21–26. 10.1016/j.vetpar.2007.10.002 [DOI] [PubMed] [Google Scholar]
- 139.Kotloff KL, Nataro JP, Blackwelder WC, Nasrin D, Farag TH, Panchalingam S, Wu Y, Sow SO, Sur D, Breiman RF, Faruque AS, Zaidi AK, Saha D, Alonso PL, Tamboura B, Sanogo D, Onwuchekwa U, Manna B, Ramamurthy T, Kanungo S, Ochieng JB, Omore R, Oundo JO, Hossain A, Das SK, Ahmed S, Qureshi S, Quadri F, Adegbola RA, Antonio M, Hossain MJ, Akinsola A, Mandomando I, Nhampossa T, Acácio S, Biswas K, O'Reilly CE, Mintz ED, Berkeley LY, Muhsen K, Sommerfelt H, Robins-Browne RM, Levine MM. 2013. Burden and aetiology of diarrhoeal disease in infants and young children in developing countries (the Global Enteric Multicenter Study, GEMS): a prospective, case-control study. Lancet 382:209–222. 10.1016/S0140-6736(13)60844-2 [DOI] [PubMed] [Google Scholar]
- 140.Davies AP, Chalmers RM. 2009. Cryptosporidiosis. BMJ 339:b4168. 10.1136/bmj.b4168 [DOI] [PubMed] [Google Scholar]
- 141.Chalmers RM, Davies AP. 2010. Clinical cryptosporidiosis. Exp. Parasitol. 124:138–146. 10.1016/j.exppara.2009.02.003 [DOI] [PubMed] [Google Scholar]
- 142.Chalmers RM, Giles M. 2010. Zoonotic cryptosporidiosis in the UK—challenges for control. J. Appl. Microbiol. 109:1487–1497. 10.1111/j.1365-2672.2010.04764.x [DOI] [PubMed] [Google Scholar]
- 143.Ethelberg S, Lisby M, Vestergaard LS, Enemark HL, Olsen KE, Stensvold CR, Nielsen HV, Porsbo LJ, Plesner AM, Mølbak K. 2009. A foodborne outbreak of Cryptosporidium hominis infection. Epidemiol. Infect. 137:348–356. 10.1017/S0950268808001817 [DOI] [PubMed] [Google Scholar]
- 144.Hunter PR, Hughes S, Woodhouse S, Raj N, Syed Q, Chalmers RM, Verlander NQ, Goodacre J. 2004. Health sequelae of human cryptosporidiosis in immunocompetent patients. Clin. Infect. Dis. 39:504–510. 10.1086/422649 [DOI] [PubMed] [Google Scholar]
- 145.Molbak K, Hojlyng N, Gottschau A, Sa JC, Ingholt L, da Silva AP, Aaby P. 1993. Cryptosporidiosis in infancy and childhood mortality in Guinea Bissau, West Africa. BMJ 307:417–420. 10.1136/bmj.307.6901.417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Molbak K, Wested N, Hojlyng N, Scheutz F, Gottschau A, Aaby P, da Silva AP. 1994. The etiology of early childhood diarrhea: a community study from Guinea-Bissau. J. Infect. Dis. 169:581–587. 10.1093/infdis/169.3.581 [DOI] [PubMed] [Google Scholar]
- 147.Fournet N, Deege MP, Urbanus AT, Nichols G, Rosner BM, Chalmers RM, Gorton R, Pollock KG, van der Giessen JW, Wever PC, Dorigo-Zetsma JW, Mulder B, Mank TG, Overdevest I, Kusters JG, van Pelt W, Kortbeek LM. 2013. Simultaneous increase of Cryptosporidium infections in the Netherlands, the United Kingdom and Germany in late summer season, 2012 Euro Surveill. 18(2): pii=20348 http://www.eurosurveillance.org/ViewArticle.aspx?ArticleId=20348 [PubMed] [Google Scholar]
- 148.Morgan-Ryan U, Fall A, Ward LA, Hijjawi N, Sulaiman I, Fayer R, Thompson RC, Olson M, Lal A, Xiao L. 2002. Cryptosporidium hominis n. sp. (Apicomplexa: Cryptosporidiidae) from Homo sapiens. J. Eukaryot. Microbiol. 49:433–440. 10.1111/j.1550-7408.2002.tb00224.x [DOI] [PubMed] [Google Scholar]
- 149.Cama VA, Ross JM, Crawford S, Kawai V, Chavez-Valdez R, Vargas D, Vivar A, Ticona E, Navincopa M, Williamson J, Ortega Y, Gilman RH, Bern C, Xiao L. 2007. Differences in clinical manifestations among Cryptosporidium species and subtypes in HIV-infected persons. J. Infect. Dis. 196:684–691. 10.1086/519842 [DOI] [PubMed] [Google Scholar]
- 150.Cama VA, Bern C, Sulaiman IM, Gilman RH, Ticona E, Vivar A, Kawai V, Vargas D, Zhou L, Xiao L. 2003. Cryptosporidium species and genotypes in HIV-positive patients in Lima, Peru. J. Eukaryot. Microbiol. 50(Suppl):531–533. 10.1111/j.1550-7408.2003.tb00620.x [DOI] [PubMed] [Google Scholar]
- 151.Insulander M, Silverlas C, Lebbad M, Karlsson L, Mattsson JG, Svenungsson B. 2013. Molecular epidemiology and clinical manifestations of human cryptosporidiosis in Sweden. Epidemiol. Infect. 141:1009–1020. 10.1017/S0950268812001665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Stroup SE, Roy S, Mchele J, Maro V, Ntabaguzi S, Siddique A, Kang G, Guerrant RL, Kirkpatrick BD, Fayer R, Herbein J, Ward H, Haque R, Houpt ER. 2006. Real-time PCR detection and speciation of Cryptosporidium infection using scorpion probes. J. Med. Microbiol. 55:1217–1222. 10.1099/jmm.0.46678-0 [DOI] [PubMed] [Google Scholar]
- 153.Cama V, Gilman RH, Vivar A, Ticona E, Ortega Y, Bern C, Xiao L. 2006. Mixed Cryptosporidium infections and HIV. Emerg. Infect. Dis. 12:1025–1028. 10.3201/eid1206.060015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Chalmers RM. 2012. Waterborne outbreaks of cryptosporidiosis. Ann. Ist. Super. Sanita 48:429–446. 10.4415/ANN_12_04_10 [DOI] [PubMed] [Google Scholar]
- 155.Xiao L. 2010. Molecular epidemiology of cryptosporidiosis: an update. Exp. Parasitol. 124:80–89. 10.1016/j.exppara.2009.03.018 [DOI] [PubMed] [Google Scholar]
- 156.Bouzid M, Hunter PR, Chalmers RM, Tyler KM. 2013. Cryptosporidium pathogenicity and virulence. Clin. Microbiol. Rev. 26:115–134. 10.1128/CMR.00076-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Guy RA, Payment P, Krull UJ, Horgen PA. 2003. Real-time PCR for quantification of Giardia and Cryptosporidium in environmental water samples and sewage. Appl. Environ. Microbiol. 69:5178–5185. 10.1128/AEM.69.9.5178-5185.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Fontaine M, Guillot E. 2002. Development of a TaqMan quantitative PCR assay specific for Cryptosporidium parvum. FEMS Microbiol. Lett. 214:13–17. 10.1111/j.1574-6968.2002.tb11318.x [DOI] [PubMed] [Google Scholar]
- 159.Jothikumar N, da Silva AJ, Moura I, Qvarnstrom Y, Hill VR. 2008. Detection and differentiation of Cryptosporidium hominis and Cryptosporidium parvum by dual TaqMan assays. J. Med. Microbiol. 57:1099–1105. 10.1099/jmm.0.2008/001461-0 [DOI] [PubMed] [Google Scholar]
- 160.Stensvold CR, Nielsen SD, Badsberg JH, Engberg J, Friis-Møller N, Nielsen SS, Nielsen HV, Friis-Møller A. 2011. The prevalence and clinical significance of intestinal parasites in HIV-infected patients in Denmark. Scand. J. Infect. Dis. 43:129–135. 10.3109/00365548.2010.524659 [DOI] [PubMed] [Google Scholar]
- 161.Basuni M, Mohamed Z, Ahmad M, Zakaria NZ, Noordin R. 2012. Detection of selected intestinal helminths and protozoa at Hospital Universiti Sains Malaysia using multiplex real-time PCR. Trop. Biomed. 29:434–442 http://www.msptm.org/files/434_-_442_Rahmah_Noordin.pdf [PubMed] [Google Scholar]
- 162.Stensvold CR, Nielsen HV. 2012. Comparison of microscopy and PCR for detection of intestinal parasites in Danish patients supports an incentive for molecular screening platforms. J. Clin. Microbiol. 50:540–541. 10.1128/JCM.06012-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Hadfield SJ, Robinson G, Elwin K, Chalmers RM. 2011. Detection and differentiation of Cryptosporidium spp. in human clinical samples by use of real-time PCR. J. Clin. Microbiol. 49:918–924. 10.1128/JCM.01733-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Reed C, Sturbaum GD, Hoover PJ, Sterling CR. 2002. Cryptosporidium parvum mixed genotypes detected by PCR-restriction fragment length polymorphism analysis. Appl. Environ. Microbiol. 68:427–429. 10.1128/AEM.68.1.427-429.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Pangasa A, Jex AR, Campbell BE, Bott NJ, Whipp M, Hogg G, Stevens MA, Gasser RB. 2009. High resolution melting-curve (HRM) analysis for the diagnosis of cryptosporidiosis in humans. Mol. Cell. Probes 23:10–15. 10.1016/j.mcp.2008.10.003 [DOI] [PubMed] [Google Scholar]
- 166.Tanriverdi S, Tanyeli A, Baslamisli F, Köksal F, Kilinc Y, Feng X, Batzer G, Tzipori S, Widmer G. 2002. Detection and genotyping of oocysts of Cryptosporidium parvum by real-time PCR and melting curve analysis. J. Clin. Microbiol. 40:3237–3244. 10.1128/JCM.40.9.3237-3244.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Tanriverdi S, Arslan MO, Akiyoshi DE, Tzipori S, Widmer G. 2003. Identification of genotypically mixed Cryptosporidium parvum populations in humans and calves. Mol. Biochem. Parasitol. 130:13–22. 10.1016/S0166-6851(03)00138-5 [DOI] [PubMed] [Google Scholar]
- 168.Limor JR, Lal AA, Xiao L. 2002. Detection and differentiation of Cryptosporidium parasites that are pathogenic for humans by real-time PCR. J. Clin. Microbiol. 40:2335–2338. 10.1128/JCM.40.7.2335-2338.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Robinson G, Chalmers RM. 2012. Assessment of polymorphic genetic markers for multi-locus typing of Cryptosporidium parvum and Cryptosporidium hominis. Exp. Parasitol. 132:200–215. 10.1016/j.exppara.2012.06.016 [DOI] [PubMed] [Google Scholar]
- 170.Jex AR, Gasser RB. 2009. Diagnostic and analytical mutation scanning of Cryptosporidium: utility and advantages. Expert Rev. Mol. Diagn. 9:179–185. 10.1586/14737159.9.2.179 [DOI] [PubMed] [Google Scholar]
- 171.Widmer G, Lee Y. 2010. Comparison of single- and multilocus genetic diversity in the protozoan parasites Cryptosporidium parvum and C. hominis. Appl. Environ. Microbiol. 76:6639–6644. 10.1128/AEM.01268-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Elwin K, Hadfield SJ, Robinson G, Crouch ND, Chalmers RM. 2012. Cryptosporidium viatorum n. sp. (Apicomplexa: Cryptosporidiidae) among travellers returning to Great Britain from the Indian subcontinent, 2007-2011. Int. J. Parasitol. 42:675–682. 10.1016/j.ijpara.2012.04.016 [DOI] [PubMed] [Google Scholar]
- 173.Nemejc K, Sak B, Kvetonova D, Kernerova N, Rost M, Cama VA, Kvac M. 2013. Occurrence of Cryptosporidium suis and Cryptosporidium scrofarum on commercial swine farms in the Czech Republic and its associations with age and husbandry practices. Parasitol. Res. 112:1143–1154. 10.1007/s00436-012-3244-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Amar CF, East C, Maclure E, McLauchlin J, Jenkins C, Duncanson P, Wareing DR. 2004. Blinded application of microscopy, bacteriological culture, immunoassays and PCR to detect gastrointestinal pathogens from faecal samples of patients with community-acquired diarrhoea. Eur. J. Clin. Microbiol. Infect. Dis. 23:529–534. 10.1007/s10096-004-1149-x [DOI] [PubMed] [Google Scholar]
- 175.Calderaro A, Montecchini S, Gorrini C, Dettori G, Chezzi C. 2011. Similar diagnostic performances of antigen detection and nucleic acid detection of Cryptosporidium spp. in a low-prevalence setting. Diagn. Microbiol. Infect. Dis. 70:72–77. 10.1016/j.diagmicrobio.2010.11.017 [DOI] [PubMed] [Google Scholar]
- 176.Chalmers RM, Campbell BM, Crouch N, Charlett A, Davies AP. 2011. Comparison of diagnostic sensitivity and specificity of seven Cryptosporidium assays used in the UK. J. Med. Microbiol. 60:1598–1604. 10.1099/jmm.0.034181-0 [DOI] [PubMed] [Google Scholar]
- 177.Ortega YR, Sanchez R. 2010. Update on Cyclospora cayetanensis, a food-borne and waterborne parasite. Clin. Microbiol. Rev. 23:218–234. 10.1128/CMR.00026-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Insulander M, Svenungsson B, Lebbad M, Karlsson L, de Jong B. 2010. A foodborne outbreak of Cyclospora infection in Stockholm, Sweden. Foodborne Pathog. Dis. 7:1585–1587. 10.1089/fpd.2010.0628 [DOI] [PubMed] [Google Scholar]
- 179.Hoang LM, Fyfe M, Ong C, Harb J, Champagne S, Dixon B, Isaac-Renton J. 2005. Outbreak of cyclosporiasis in British Columbia associated with imported Thai basil. Epidemiol. Infect. 133:23–27. 10.1017/S0950268804003176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Shah L, MacDougall L, Ellis A, Ong C, Shyng S, LeBlanc L, British Columbia Cyclospora Investigation Team 2009. Challenges of investigating community outbreaks of cyclosporiasis, British Columbia, Canada. Emerg. Infect. Dis. 15:1286–1288. 10.3201/eid1508.081585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Milord F, Lampron-Goulet E, St-Amour M, Levac E, Ramsay D. 2012. Cyclospora cayetanensis: a description of clinical aspects of an outbreak in Quebec, Canada. Epidemiol. Infect. 140:626–632. 10.1017/S095026881100121X [DOI] [PubMed] [Google Scholar]
- 182.Chacín-Bonilla L. 2010. Epidemiology of Cyclospora cayetanensis: a review focusing in endemic areas. Acta Trop. 115:181–193. 10.1016/j.actatropica.2010.04.001 [DOI] [PubMed] [Google Scholar]
- 183.Samie A, Guerrant RL, Barrett L, Bessong PO, Igumbor EO, Obi CL. 2009. Prevalence of intestinal parasitic and bacterial pathogens in diarrhoeal and non-diarroeal human stools from Vhembe district, South Africa. J. Health Popul. Nutr. 27:739–745 http://www.ncbi.nlm.nih.gov/pmc/articles/PMC2928113/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Kasper MR, Lescano AG, Lucas C, Gilles D, Biese BJ, Stolovitz G, Reaves EJ. 2012. Diarrhea outbreak during U.S. military training in El Salvador. PLoS One 7:e40404. 10.1371/journal.pone.0040404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Blans MC, Ridwan BU, Verweij JJ, Rozenberg-Arska M, Verhoef J. 2005. Cyclosporiasis outbreak, Indonesia. Emerg. Infect. Dis. 11:1453–1455. 10.3201/eid1109.040947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Paschke C, Apelt N, Fleischmann E, Perona P, Walentiny C, Locher T, Herbinger KH. 2011. Controlled study on enteropathogens in travellers returning from the tropics with and without diarrhoea. Clin. Microbiol. Infect. 17:1194–1200. 10.1111/j.1469-0691.2010.03414.x [DOI] [PubMed] [Google Scholar]
- 187.Hall RL, Jones JL, Hurd S, Smith G, Mahon BE, Herwaldt BL. 2012. Population-based active surveillance for Cyclospora infection—United States, Foodborne Diseases Active Surveillance Network (FoodNet), 1997-2009. Clin. Infect. Dis. 54(Suppl 5):S411–S417. 10.1093/cid/cis049 [DOI] [PubMed] [Google Scholar]
- 188.Olivier C, van de Pas S, Lepp PW, Yoder K, Relman DA. 2001. Sequence variability in the first internal transcribed spacer region within and among Cyclospora species is consistent with polyparasitism. Int. J. Parasitol. 31:1475–1487. 10.1016/S0020-7519(01)00283-1 [DOI] [PubMed] [Google Scholar]
- 189.Ozdamar M, Hakko E, Turkoglu S. 2010. High occurrence of cyclosporiasis in Istanbul, Turkey, during a dry and warm summer. Parasit. Vectors 3:39. 10.1186/1756-3305-3-39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Varma M, Hester JD, Schaefer FW, Ware MW, Lindquist HD. 2003. Detection of Cyclospora cayetanensis using a quantitative real-time PCR assay. J. Microbiol. Methods 53:27–36. 10.1016/S0167-7012(02)00209-9 [DOI] [PubMed] [Google Scholar]
- 191.Verweij JJ, Laeijendecker D, Brienen EA, van Lieshout L, Polderman AM. 2003. Detection of Cyclospora cayetanensis in travellers returning from the tropics and subtropics using microscopy and real-time PCR. Int. J. Med. Microbiol. 293:199–202. 10.1078/1438-4221-00252 [DOI] [PubMed] [Google Scholar]
- 192.Mundaca CC, Torres-Slimming P, Araujo-Castillo R, Morán M, Bacon DJ, Ortega Y, Gilman RH, Blazes DL. 2008. Use of PCR to improve diagnostic yield in an outbreak of cyclosporiasis in Lima, Peru. Trans. R. Soc. Trop. Med. Hyg. 102:712–717. 10.1016/j.trstmh.2008.03.003 [DOI] [PubMed] [Google Scholar]
- 193.Taniuchi M, Verweij JJ, Sethabutr O, Bodhidatta L, Garcia L, Maro A, Kumburu H, Gratz J, Kibiki G, Houpt ER. 2011. Multiplex polymerase chain reaction method to detect Cyclospora, Cystoisospora, and microsporidia in stool samples. Diagn. Microbiol. Infect. Dis. 71:386–390. 10.1016/j.diagmicrobio.2011.08.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Hussein EM. 2007. Molecular identification of Cycospora spp. using multiplex PCR from diarrheic children compared to others conventional methods. J. Egypt. Soc. Parasitol. 37:585–598 [PubMed] [Google Scholar]
- 195.Sulaiman IM, Torres P, Simpson S, Kerdahi K, Ortega Y. 2013. Sequence characterization of heat shock protein gene of Cyclospora cayetanensis isolates from Nepal, Mexico, and Peru. J. Parasitol. 99:379–382. 10.1645/GE-3114.1 [DOI] [PubMed] [Google Scholar]
- 196.Barta JR, Schrenzel MD, Carreno R, Rideout BA. 2005. The genus Atoxoplasma (Garnham 1950) as a junior objective synonym of the genus Isospora (Schneider 1881) species infecting birds and resurrection of Cystoisospora (Frenkel 1977) as the correct genus for Isospora species infecting mammals. J. Parasitol. 91:726–727. 10.1645/GE-3341.1 [DOI] [PubMed] [Google Scholar]
- 197.Lindsay DS, Dubey JP, Blagburn BL. 1997. Biology of Isospora spp. from humans, nonhuman primates, and domestic animals. Clin. Microbiol. Rev. 10:19–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Ud Din N, Torka P, Hutchison RE, Riddell SW, Wright J, Gajra A. 2012. Severe Isospora (Cystoisospora) belli diarrhea preceding the diagnosis of human T-cell-leukemia-virus-1-associated T-cell lymphoma. Case Rep. Infect. Dis. 2012:640104. 10.1155/2012/640104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Atambay M, Bayraktar MR, Kayabas U, Yilmaz S, Bayindir Y. 2007. A rare diarrheic parasite in a liver transplant patient: Isospora belli. Transplant. Proc. 39:1693–1695. 10.1016/j.transproceed.2007.02.080 [DOI] [PubMed] [Google Scholar]
- 200.Ferreira MS. 2000. Infections by protozoa in immunocompromised hosts. Mem. Inst. Oswaldo Cruz 95(Suppl 1):159–162. 10.1590/S0074-02762000000700026 [DOI] [PubMed] [Google Scholar]
- 201.Lewthwaite P, Gill GV, Hart CA, Beeching NJ. 2005. Gastrointestinal parasites in the immunocompromised. Curr. Opin. Infect. Dis. 18:427–435. 10.1097/01.qco.0000182104.40128.18 [DOI] [PubMed] [Google Scholar]
- 202.Navaneethan U, Venkatesh PG, Downs-Kelly E, Shen B. 2012. Isospora belli superinfection in a patient with eosinophilic gastroenteritis—a diagnostic challenge. J. Crohns Colitis 6:236–239. 10.1016/j.crohns.2011.08.010 [DOI] [PubMed] [Google Scholar]
- 203.Fletcher SM, Stark D, Harkness J, Ellis J. 2012. Enteric protozoa in the developed world: a public health perspective. Clin. Microbiol. Rev. 25:420–449. 10.1128/CMR.05038-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Bernard E, Delgiudice P, Carles M, Boissy C, Saint-Paul MC, Le Fichoux Y, Michiels JF, Dellamonica P. 1997. Disseminated isosporiasis in an AIDS patient. Eur. J. Clin. Microbiol. Infect. Dis. 16:699–701. 10.1007/BF01708565 [DOI] [PubMed] [Google Scholar]
- 205.Restrepo C, Macher AM, Radany EH. 1987. Disseminated extraintestinal isosporiasis in a patient with acquired immune deficiency syndrome. Am. J. Clin. Pathol. 87:536–542 [DOI] [PubMed] [Google Scholar]
- 206.Walther Z, Topazian MD. 2009. Isospora cholangiopathy: case study with histologic characterization and molecular confirmation. Hum. Pathol. 40:1342–1346. 10.1016/j.humpath.2009.01.020 [DOI] [PubMed] [Google Scholar]
- 207.Marcos LA, Gotuzzo E. 2013. Intestinal protozoan infections in the immunocompromised host. Curr. Opin. Infect. Dis. 26:295–301. 10.1097/QCO.0b013e3283630be3 [DOI] [PubMed] [Google Scholar]
- 208.Velásquez JN, Osvaldo GA, Di Risio C, Etchart C, Chertcoff AV, Perissé GE, Carnevale S. 2011. Molecular characterization of Cystoisospora belli and unizoite tissue cyst in patients with acquired immunodeficiency syndrome. Parasitology 138:279–286. 10.1017/S0031182010001253 [DOI] [PubMed] [Google Scholar]
- 209.Velásquez JN, Carnevale S, Mariano M, Kuo LH, Caballero A, Chertcoff A, Ibáñez C, Bozzini JP. 2001. Isosporosis and unizoite tissue cysts in patients with acquired immunodeficiency syndrome. Hum. Pathol. 32:500–505. 10.1053/hupa.2001.24326 [DOI] [PubMed] [Google Scholar]
- 210.Matsubayashi M, Carreno RA, Tani H, Yoshiuchi R, Kanai T, Kimata I, Uni S, Furuya M, Sasai K. 2011. Phylogenetic identification of Cystoisospora spp. from dogs, cats, and raccoon dogs in Japan. Vet. Parasitol. 176:270–274. 10.1016/j.vetpar.2010.11.008 [DOI] [PubMed] [Google Scholar]
- 211.Murphy SC, Hoogestraat DR, Sengupta DJ, Prentice J, Chakrapani A, Cookson BT. 2011. Molecular diagnosis of cystoisosporiasis using extended-range PCR screening. J. Mol. Diagn. 13:359–362. 10.1016/j.jmoldx.2011.01.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Müller A, Bialek R, Fätkenheuer G, Salzberger B, Diehl V, Franzen C. 2000. Detection of Isospora belli by polymerase chain reaction using primers based on small-subunit ribosomal RNA sequences. Eur. J. Clin. Microbiol. Infect. Dis. 19:631–634. 10.1007/s100960000322 [DOI] [PubMed] [Google Scholar]
- 213.ten Hove RJ, van Lieshout L, Brienen EA, Perez MA, Verweij JJ. 2008. Real-time polymerase chain reaction for detection of Isospora belli in stool samples. Diagn. Microbiol. Infect. Dis. 61:280–283. 10.1016/j.diagmicrobio.2008.03.003 [DOI] [PubMed] [Google Scholar]
- 214.Samarasinghe B, Johnson J, Ryan U. 2008. Phylogenetic analysis of Cystoisospora species at the rRNA ITS1 locus and development of a PCR-RFLP assay. Exp. Parasitol. 118:592–595. 10.1016/j.exppara.2007.10.015 [DOI] [PubMed] [Google Scholar]
- 215.Franzen C, Müller A, Bialek R, Diehl V, Salzberger B, Fätkenheuer G. 2000. Taxonomic position of the human intestinal protozoan parasite Isospora belli as based on ribosomal RNA sequences. Parasitol. Res. 86:669–676. 10.1007/PL00008550 [DOI] [PubMed] [Google Scholar]
- 216.He P, Li J, Gong P, Huang J, Zhang X. 2012. Cystoisospora spp. from dogs in China and phylogenetic analysis of its 18S and ITS1 gene. Vet. Parasitol. 190:254–258. 10.1016/j.vetpar.2012.05.025 [DOI] [PubMed] [Google Scholar]
- 217.Fayer R. 2004. Sarcocystis spp. in human infections. Clin. Microbiol. Rev. 17:894–902. 10.1128/CMR.17.4.894-902.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Xiang Z, Chen X, Yang L, He Y, Jiang R, Rosenthal BM, Luan P, Attwood SW, Zuo Y, Zhang YP, Yang Z. 2009. Non-invasive methods for identifying oocysts of Sarcocystis spp. from definitive hosts. Parasitol. Int. 58:293–296. 10.1016/j.parint.2009.03.004 [DOI] [PubMed] [Google Scholar]
- 219.Lalonde LF, Gajadhar AA. 2011. Detection and differentiation of coccidian oocysts by real-time PCR and melting curve analysis. J. Parasitol. 97:725–730. 10.1645/GE-2706.1 [DOI] [PubMed] [Google Scholar]
- 220.Schuster FL, Ramirez-Avila L. 2008. Current world status of Balantidium coli. Clin. Microbiol. Rev. 21:626–638. 10.1128/CMR.00021-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Nilles-Bije ML, Rivera WL. 2010. Ultrastructural and molecular characterization of Balantidium coli isolated in the Philippines. Parasitol. Res. 106:387–394. 10.1007/s00436-009-1673-9 [DOI] [PubMed] [Google Scholar]
- 222.Schuster FL, Visvesvara GS. 2004. Amebae and ciliated protozoa as causal agents of waterborne zoonotic disease. Vet. Parasitol. 126:91–120. 10.1016/j.vetpar.2004.09.019 [DOI] [PubMed] [Google Scholar]
- 223.Ferry T, Bouhour D, De Monbrison F, Laurent F, Dumouchel-Champagne H, Picot S, Piens MA, Granier P. 2004. Severe peritonitis due to Balantidium coli acquired in France. Eur. J. Clin. Microbiol. Infect. Dis. 23:393–395. 10.1007/s10096-004-1126-4 [DOI] [PubMed] [Google Scholar]
- 224.Sharma S, Harding G. 2003. Necrotizing lung infection caused by the protozoan Balantidium coli. Can. J. Infect. Dis. 14:163–166 http://www.ncbi.nlm.nih.gov/pmc/articles/PMC2094932/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Vasilakopoulou A, Dimarongona K, Samakovli A, Papadimitris K, Avlami A. 2003. Balantidium coli pneumonia in an immunocompromised patient. Scand. J. Infect. Dis. 35:144–146. 10.1080/0036554021000027023 [DOI] [PubMed] [Google Scholar]
- 226.Anargyrou K, Petrikkos GL, Suller MT, Skiada A, Siakantaris MP, Osuntoyinbo RT, Pangalis G, Vaiopoulos G. 2003. Pulmonary Balantidium coli infection in a leukemic patient. Am. J. Hematol. 73:180–183. 10.1002/ajh.10336 [DOI] [PubMed] [Google Scholar]
- 227.Norman JG, Jessop P. 1973. Balantidium coli in a cervico-vaginal smear. Med. J. Aust. 1:694–696 [DOI] [PubMed] [Google Scholar]
- 228.Martínez-Girón R, Esteban JG, Ribas A, Doganci L. 2008. Protozoa in respiratory pathology: a review. Eur. Respir. J. 32:1354–1370. 10.1183/09031936.00022008 [DOI] [PubMed] [Google Scholar]
- 229.Veira DM. 1986. The role of ciliate protozoa in nutrition of the ruminant. J. Anim. Sci. 63:1547–1560 [DOI] [PubMed] [Google Scholar]
- 230.Li M, Wang C, Wang J, Li A, Gong X, Ma H. 2009. Redescription of Balantidium polyvacuolum Li 1963 (class: Litostomatea) inhabiting the intestines of Xenocyprinae fishes in Hubei, China. Parasitol. Res. 106:177–182. 10.1007/s00436-009-1645-0 [DOI] [PubMed] [Google Scholar]
- 231.Li M, Wang J, Zhang J, Gu Z, Ling F, Ke X, Gong X. 2008. First report of two Balantidium species from the Chinese giant salamander, Andrias davidianus: Balantidium sinensis Nie 1935 and Balantidium andianusis n. sp. Parasitol. Res. 102:605–611. 10.1007/s00436-007-0795-1 [DOI] [PubMed] [Google Scholar]
- 232.Ponce-Gordo F, Fonseca-Salamanca F, Martinez-Diaz RA. 2011. Genetic heterogeneity in internal transcribed spacer genes of Balantidium coli (Litostomatea, Ciliophora). Protist 162:774–794. 10.1016/j.protis.2011.06.008 [DOI] [PubMed] [Google Scholar]
- 233.Ponce-Gordo F, Jimenez-Ruiz E, Martinez-Diaz RA. 2008. Tentative identification of the species of Balantidium from ostriches (Struthio camelus) as Balantidium coli-like by analysis of polymorphic DNA. Vet. Parasitol. 157:41–49. 10.1016/j.vetpar.2008.06.024 [DOI] [PubMed] [Google Scholar]
- 234.Grim JN, Buonanno F. 2009. A re-description of the ciliate genus and type species, Balantidium entozoon. Eur. J. Protistol. 45:174–182. 10.1016/j.ejop.2008.10.001 [DOI] [PubMed] [Google Scholar]
- 235.Mathis A, Weber R, Deplazes P. 2005. Zoonotic potential of the microsporidia. Clin. Microbiol. Rev. 18:423–445. 10.1128/CMR.18.3.423-445.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Ojuromi OT, Izquierdo F, Fenoy S, Fagbenro-Beyioku A, Oyibo W, Akanmu A, Odunukwe N, Henriques-Gil N, del Aguila C. 2012. Identification and characterization of microsporidia from fecal samples of HIV-positive patients from Lagos, Nigeria. PLoS One 7:e35239. 10.1371/journal.pone.0035239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Saigal K, Sharma A, Sehgal R, Sharma P, Malla N, Khurana S. 2013. Intestinal microsporidiosis in India: a two year study. Parasitol. Int. 62:53–56. 10.1016/j.parint.2012.09.005 [DOI] [PubMed] [Google Scholar]
- 238.Ghosh K, Weiss LM. 2009. Molecular diagnostic tests for microsporidia. Interdiscip. Perspect. Infect. Dis. 2009:926521. 10.1155/2009/926521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Sokolova OI, Demyanov AV, Bowers LC, Didier ES, Yakovlev AV, Skarlato SO, Sokolova YY. 2011. Emerging microsporidian infections in Russian HIV-infected patients. J. Clin. Microbiol. 49:2102–2108. 10.1128/JCM.02624-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Didier ES. 2005. Microsporidiosis: an emerging and opportunistic infection in humans and animals. Acta Trop. 94:61–76. 10.1016/j.actatropica.2005.01.010 [DOI] [PubMed] [Google Scholar]
- 241.Didier ES, Weiss LM. 2011. Microsporidiosis: not just in AIDS patients. Curr. Opin. Infect. Dis. 24:490–495. 10.1097/QCO.0b013e32834aa152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Galván AL, Sánchez AM, Valentín MA, Henriques-Gil N, Izquierdo F, Fenoy S, del Aguila C. 2011. First cases of microsporidiosis in transplant recipients in Spain and review of the literature. J. Clin. Microbiol. 49:1301–1306. 10.1128/JCM.01833-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Latib MA, Pascoe MD, Duffield MS, Kahn D. 2001. Microsporidiosis in the graft of a renal transplant recipient. Transpl. Int. 14:274–277. 10.1111/j.1432-2277.2001.tb00058.x [DOI] [PubMed] [Google Scholar]
- 244.Svenungsson B, Capraru T, Evengard B, Larsson R, Lebbad M. 1998. Intestinal microsporidiosis in a HIV-seronegative patient. Scand. J. Infect. Dis. 30:314–316. 10.1080/00365549850161043 [DOI] [PubMed] [Google Scholar]
- 245.Rabodonirina M, Cotte L, Radenne S, Besada E, Trepo C. 2003. Microsporidiosis and transplantation: a retrospective study of 23 cases. J. Eukaryot. Microbiol. 50(Suppl):583. [DOI] [PubMed] [Google Scholar]
- 246.Didier ES, Weiss LM. 2006. Microsporidiosis: current status. Curr. Opin. Infect. Dis. 19:485–492. 10.1097/01.qco.0000244055.46382.23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247.Lobo ML, Xiao L, Antunes F, Matos O. 2012. Microsporidia as emerging pathogens and the implication for public health: a 10-year study on HIV-positive and -negative patients. Int. J. Parasitol. 42:197–205. 10.1016/j.ijpara.2011.12.002 [DOI] [PubMed] [Google Scholar]
- 248.Stark D, van Hal S, Barratt J, Ellis J, Marriott D, Harkness J. 2009. Limited genetic diversity among genotypes of Enterocytozoon bieneusi strains isolated from HIV-infected patients from Sydney, Australia. J. Med. Microbiol. 58:355–357. 10.1099/jmm.0.006445-0 [DOI] [PubMed] [Google Scholar]
- 249.Akinbo FO, Okaka CE, Omoregie R, Dearen T, Leon ET, Xiao L. 2012. Molecular epidemiologic characterization of Enterocytozoon bieneusi in HIV-infected persons in Benin City, Nigeria. Am. J. Trop. Med. Hyg. 86:441–445. 10.4269/ajtmh.2012.11-0548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.Decraene V, Lebbad M, Botero-Kleiven S, Gustavsson AM, Löfdahl M. 2012. First reported foodborne outbreak associated with microsporidia, Sweden, October 2009. Epidemiol. Infect. 140:519–527. 10.1017/S095026881100077X [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251.Cotte L, Rabodonirina M, Chapuis F, Bailly F, Bissuel F, Raynal C, Gelas P, Persat F, Piens MA, Trepo C. 1999. Waterborne outbreak of intestinal microsporidiosis in persons with and without human immunodeficiency virus infection. J. Infect. Dis. 180:2003–2008. 10.1086/315112 [DOI] [PubMed] [Google Scholar]
- 252.Didier ES, Maddry JA, Brindley PJ, Stovall ME, Didier PJ. 2005. Therapeutic strategies for human microsporidia infections. Expert Rev. Anti Infect. Ther. 3:419–434. 10.1586/14787210.3.3.419 [DOI] [PubMed] [Google Scholar]
- 253.Stark D, Barratt JL, van Hal S, Marriott D, Harkness J, Ellis JT. 2009. Clinical significance of enteric protozoa in the immunosuppressed human population. Clin. Microbiol. Rev. 22:634–650. 10.1128/CMR.00017-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 254.van Gool T, Snijders F, Reiss P, Eeftinck Schattenkerk JK, van den Bergh Weerman MA, Bartelsman JF, Bruins JJ, Canning EU, Dankert J. 1993. Diagnosis of intestinal and disseminated microsporidial infections in patients with HIV by a new rapid fluorescence technique. J. Clin. Pathol. 46:694–699. 10.1136/jcp.46.8.694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255.Garcia LS. 2002. Laboratory identification of the microsporidia. J. Clin. Microbiol. 40:1892–1901. 10.1128/JCM.40.6.1892-1901.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256.Thellier M, Breton J. 2008. Enterocytozoon bieneusi in human and animals, focus on laboratory identification and molecular epidemiology. Parasite 15:349–358. 10.1051/parasite/2008153349 [DOI] [PubMed] [Google Scholar]
- 257.Weiss LM, Vossbrinck CR. 1998. Microsporidiosis: molecular and diagnostic aspects. Adv. Parasitol. 40:351–395. 10.1016/S0065-308X(08)60127-X [DOI] [PubMed] [Google Scholar]
- 258.Franzen C, Müller A. 1999. Molecular techniques for detection, species differentiation, and phylogenetic analysis of microsporidia. Clin. Microbiol. Rev. 12:243–285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 259.Zhu X, Wittner M, Tanowitz HB, Kotler D, Cali A, Weiss LM. 1993. Small subunit rRNA sequence of Enterocytozoon bieneusi and its potential diagnostic role with use of the polymerase chain reaction. J. Infect. Dis. 168:1570–1575. 10.1093/infdis/168.6.1570 [DOI] [PubMed] [Google Scholar]
- 260.Wang Z, Orlandi PA, Stenger DA. 2005. Simultaneous detection of four human pathogenic microsporidian species from clinical samples by oligonucleotide microarray. J. Clin. Microbiol. 43:4121–4128. 10.1128/JCM.43.8.4121-4128.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 261.Santín M, Vecino JA, Fayer R. 2010. A zoonotic genotype of Enterocytozoon bieneusi in horses. J. Parasitol. 96:157–161. 10.1645/GE-2184.1 [DOI] [PubMed] [Google Scholar]
- 262.Zhang X, Wang Z, Su Y, Liang X, Sun X, Peng S, Lu H, Jiang N, Yin J, Xiang M, Chen Q. 2011. Identification and genotyping of Enterocytozoon bieneusi in China. J. Clin. Microbiol. 49:2006–2008. 10.1128/JCM.00372-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263.Bart A, Wentink-Bonnema E, Heddema ER, Buijs J, van Gool T. 2008. Frequent occurrence of human-associated microsporidia in fecal droppings of urban pigeons in Amsterdam, the Netherlands. Appl. Environ. Microbiol. 74:7056–7058. 10.1128/AEM.01429-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 264.Liguory O, Sarfati C, Derouin F, Molina JM. 2001. Evidence of different Enterocytozoon bieneusi genotypes in patients with and without human immunodeficiency virus infection. J. Clin. Microbiol. 39:2672–2674. 10.1128/JCM.39.7.2672-2674.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 265.Breton J, Bart-Delabesse E, Biligui S, Carbone A, Seiller X, Okome-Nkoumou M, Nzamba C, Kombila M, Accoceberry I, Thellier M. 2007. New highly divergent rRNA sequence among biodiverse genotypes of Enterocytozoon bieneusi strains isolated from humans in Gabon and Cameroon. J. Clin. Microbiol. 45:2580–2589. 10.1128/JCM.02554-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 266.Espern A, Morio F, Miegeville M, Illa H, Abdoulaye M, Meyssonnier V, Adehossi E, Lejeune A, Cam PD, Besse B, Gay-Andrieu F. 2007. Molecular study of microsporidiosis due to Enterocytozoon bieneusi and Encephalitozoon intestinalis among human immunodeficiency virus-infected patients from two geographical areas: Niamey, Niger, and Hanoi, Vietnam. J. Clin. Microbiol. 45:2999–3002. 10.1128/JCM.00684-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 267.Leelayoova S, Subrungruang I, Suputtamongkol Y, Worapong J, Petmitr PC, Mungthin M. 2006. Identification of genotypes of Enterocytozoon bieneusi from stool samples from human immunodeficiency virus-infected patients in Thailand. J. Clin. Microbiol. 44:3001–3004. 10.1128/JCM.00945-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 268.Lee SY, Lee SS, Lyoo YS, Park HM. 2011. DNA detection and genotypic identification of potentially human-pathogenic microsporidia from asymptomatic pet parrots in South Korea as a risk factor for zoonotic emergence. Appl. Environ. Microbiol. 77:8442–8444. 10.1128/AEM.05343-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 269.Talabani H, Sarfati C, Pillebout E, van Gool T, Derouin F, Menotti J. 2010. Disseminated infection with a new genovar of Encephalitozoon cuniculi in a renal transplant recipient. J. Clin. Microbiol. 48:2651–2653. 10.1128/JCM.02539-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 270.Mathis A, Tanner I, Weber R, Deplazes P. 1999. Genetic and phenotypic intraspecific variation in the microsporidian Encephalitozoon hellem. Int. J. Parasitol. 29:767–770. 10.1016/S0020-7519(99)00025-9 [DOI] [PubMed] [Google Scholar]
- 271.Xiao L, Li L, Moura H, Sulaiman I, Lal AA, Gatti S, Scaglia M, Didier ES, Visvesvara GS. 2001. Genotyping Encephalitozoon hellem isolates by analysis of the polar tube protein gene. J. Clin. Microbiol. 39:2191–2196. 10.1128/JCM.39.6.2191-2196.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 272.Sak B, Kasicková D, Kvác M, Kvetonová D, Ditrich O. 2010. Microsporidia in exotic birds: intermittent spore excretion of Encephalitozoon spp. in naturally infected budgerigars (Melopsittacus undulatus). Vet. Parasitol. 168:196–200. 10.1016/j.vetpar.2009.11.012 [DOI] [PubMed] [Google Scholar]
- 273.Pandrea I, Mittleider D, Brindley PJ, Didier ES, Robertson DL. 2005. Phylogenetic relationships of methionine aminopeptidase 2 among Encephalitozoon species and genotypes of microsporidia. Mol. Biochem. Parasitol. 140:141–152. 10.1016/j.molbiopara.2004.12.006 [DOI] [PubMed] [Google Scholar]
- 274.Fedorko DP, Nelson NA, Didier ES, Bertucci D, Delgado RM, Hruszkewycz AM. 2001. Speciation of human microsporidia by polymerase chain reaction single-strand conformation polymorphism. Am. J. Trop. Med. Hyg. 65:397–401 http://www.ajtmh.org/content/65/4/397.long [DOI] [PubMed] [Google Scholar]
- 275.Polonais V, Mazet M, Wawrzyniak I, Texier C, Blot N, El Alaoui H, Delbac F. 2010. The human microsporidian Encephalitozoon hellem synthesizes two spore wall polymorphic proteins useful for epidemiological studies. Infect. Immun. 78:2221–2230. 10.1128/IAI.01225-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 276.Clark CG, van der Giezen M, Alfellani MA, Stensvold CR. 2013. Recent developments in Blastocystis research. Adv. Parasitol. 82:1–32. 10.1016/B978-0-12-407706-5.00001-0 [DOI] [PubMed] [Google Scholar]
- 277.Alfellani MA, Stensvold CR, Vidal-Lapiedra A, Onuoha ES, Fagbenro-Beyioku A, Clark CG. 2013. Variable geographic distribution of Blastocystis subtypes and its potential implications. Acta Trop. 126:11–18. 10.1016/j.actatropica.2012.12.011 [DOI] [PubMed] [Google Scholar]
- 278.Tan KS. 2008. New insights on classification, identification, and clinical relevance of Blastocystis spp. Clin. Microbiol. Rev. 21:639–665. 10.1128/CMR.00022-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 279.Tan KS, Mirza H, Teo JD, Wu B, Macary PA. 2010. Current views on the clinical relevance of Blastocystis spp. Curr. Infect. Dis. Rep. 12:28–35. 10.1007/s11908-009-0073-8 [DOI] [PubMed] [Google Scholar]
- 280.Stensvold CR, Arendrup MC, Jespersgaard C, Mølbak K, Nielsen HV. 2007. Detecting Blastocystis using parasitologic and DNA-based methods: a comparative study. Diagn. Microbiol. Infect. Dis. 59:303–307. 10.1016/j.diagmicrobio.2007.06.003 [DOI] [PubMed] [Google Scholar]
- 281.Stensvold CR, Nielsen HV, Molbak K, Smith HV. 2009. Pursuing the clinical significance of Blastocystis—diagnostic limitations. Trends Parasitol. 25:23–29. 10.1016/j.pt.2008.09.010 [DOI] [PubMed] [Google Scholar]
- 282.Roberts T, Barratt J, Harkness J, Ellis J, Stark D. 2011. Comparison of microscopy, culture, and conventional polymerase chain reaction for detection of Blastocystis sp. in clinical stool samples. Am. J. Trop. Med. Hyg. 84:308–312. 10.4269/ajtmh.2011.10-0447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 283.Mank TG, Zaat JO, Blotkamp J, Polderman AM. 1995. Comparison of fresh versus sodium acetate acetic acid formalin preserved stool specimens for diagnosis of intestinal protozoal infections. Eur. J. Clin. Microbiol. Infect. Dis. 14:1076–1081. 10.1007/BF01590942 [DOI] [PubMed] [Google Scholar]
- 284.Stensvold R, Brillowska-Dabrowska A, Nielsen HV, Arendrup MC. 2006. Detection of Blastocystis hominis in unpreserved stool specimens by using polymerase chain reaction. J. Parasitol. 92:1081–1087. 10.1645/GE-840R.1 [DOI] [PubMed] [Google Scholar]
- 285.Jones MS, Ganac RD, Hiser G, Hudson NR, Le A, Whipps CM. 2008. Detection of Blastocystis from stool samples using real-time PCR. Parasitol. Res. 103:551–557. 10.1007/s00436-008-1006-4 [DOI] [PubMed] [Google Scholar]
- 286.Poirier P, Wawrzyniak I, Albert A, El Alaoui H, Delbac F, Livrelli V. 2011. Development and evaluation of a real-time PCR assay for detection and quantification of Blastocystis parasites in human stool samples: prospective study of patients with hematological malignancies. J. Clin. Microbiol. 49:975–983. 10.1128/JCM.01392-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 287.Stensvold CR, Ahmed UN, Andersen LO, Nielsen HV. 2012. Development and evaluation of a genus-specific, probe-based, internal process controlled real-time PCR assay for sensitive and specific detection of Blastocystis. J. Clin. Microbiol. 50:1847–1851. 10.1128/JCM.00007-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 288.Denoeud F, Roussel M, Noel B, Wawrzyniak I, Da Silva C, Diogon M, Viscogliosi E, Brochier-Armanet C, Couloux A, Poulain J, Segurens B, Anthouard V, Texier C, Blot N, Poirier P, Ng GC, Tan KS, Artiguenave F, Jaillon O, Aury JM, Delbac F, Wincker P, Vivarès CP, El Alaoui H. 2011. Genome sequence of the stramenopile Blastocystis, a human anaerobic parasite. Genome Biol. 12:R29. 10.1186/gb-2011-12-3-r29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 289.Scicluna SM, Tawari B, Clark CG. 2006. DNA barcoding of Blastocystis. Protist 157:77–85. 10.1016/j.protis.2005.12.001 [DOI] [PubMed] [Google Scholar]
- 290.Yoshikawa H, Wu Z, Kimata I, Iseki M, Ali IK, Hossain MB, Zaman V, Haque R, Takahashi Y. 2004. Polymerase chain reaction-based genotype classification among human Blastocystis hominis populations isolated from different countries. Parasitol. Res. 92:22–29. 10.1007/s00436-003-0995-2 [DOI] [PubMed] [Google Scholar]
- 291.Stensvold CR. 2013. Comparison of sequencing (barcode region) and sequence-tagged-site PCR for Blastocystis subtyping. J. Clin. Microbiol. 51:190–194. 10.1128/JCM.02541-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 292.Stensvold CR, Alfellani M, Clark CG. 2012. Levels of genetic diversity vary dramatically between Blastocystis subtypes. Infect. Genet. Evol. 12:263–273. 10.1016/j.meegid.2011.11.002 [DOI] [PubMed] [Google Scholar]
- 293.Jolley KA, Maiden MC. 2010. BIGSdb: scalable analysis of bacterial genome variation at the population level. BMC Bioinformatics 11:595. 10.1186/1471-2105-11-595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 294.Stensvold CR, Christiansen DB, Olsen KE, Nielsen HV. 2011. Blastocystis sp. subtype 4 is common in Danish Blastocystis-positive patients presenting with acute diarrhea. Am. J. Trop. Med. Hyg. 84:883–885. 10.4269/ajtmh.2011.11-0005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 295.Domínguez-Márquez MV, Guna R, Muñoz C, Gómez-Muñoz MT, Borrás R. 2009. High prevalence of subtype 4 among isolates of Blastocystis hominis from symptomatic patients of a health district of Valencia (Spain). Parasitol. Res. 105:949–955. 10.1007/s00436-009-1485-y [DOI] [PubMed] [Google Scholar]
- 296.Alfellani MA, Jacob AS, Perea NO, Krecek RC, Taner-Mulla D, Verweij JJ, Levecke B, Tannich E, Clark CG, Stensvold CR. 2013. Diversity and distribution of Blastocystis sp. subtypes in non-human primates. Parasitology 140:966–971. 10.1017/S0031182013000255 [DOI] [PubMed] [Google Scholar]
- 297.Stensvold CR. 2013. Blastocystis: genetic diversity and molecular methods for diagnosis and epidemiology. Trop. Parasitol. 3:26–34. 10.4103/2229-5070.113896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 298.Scanlan PD, Stensvold CR. 2013. Blastocystis: getting to grips with our guileful guest. Trends Parasitol. 29:523–529. 10.1016/j.pt.2013.08.006 [DOI] [PubMed] [Google Scholar]
- 299.Bethony J, Brooker S, Albonico M, Geiger SM, Loukas A, Diemert D, Hotez PJ. 2006. Soil-transmitted helminth infections: ascariasis, trichuriasis, and hookworm. Lancet 367:1521–1532. 10.1016/S0140-6736(06)68653-4 [DOI] [PubMed] [Google Scholar]
- 300.de Silva NR, Brooker S, Hotez PJ, Montresor A, Engels D, Savioli L. 2003. Soil-transmitted helminth infections: updating the global picture. Trends Parasitol. 19:547–551. 10.1016/j.pt.2003.10.002 [DOI] [PubMed] [Google Scholar]
- 301.Brooker S, Hotez PJ, Bundy DA. 2010. The global atlas of helminth infection: mapping the way forward in neglected tropical disease control. PLoS Negl. Trop. Dis. 4:e779. 10.1371/journal.pntd.0000779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 302.Brooker S. 2010. Estimating the global distribution and disease burden of intestinal nematode infections: adding up the numbers—a review. Int. J. Parasitol. 40:1137–1144. 10.1016/j.ijpara.2010.04.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 303.Olsen A, van Lieshout L, Marti H, Polderman T, Polman K, Steinmann P, Stothard R, Thybo S, Verweij JJ, Magnussen P. 2009. Strongyloidiasis—the most neglected of the neglected tropical diseases? Trans. R. Soc. Trop. Med. Hyg. 103:967–972. 10.1016/j.trstmh.2009.02.013 [DOI] [PubMed] [Google Scholar]
- 304.Bisoffi Z, Buonfrate D, Montresor A, Requena-Mendez A, Munoz J, Krolewiecki AJ, Gotuzzo E, Mena MA, Chiodini PL, Anselmi M, Moreira J, Albonico M. 2013. Strongyloides stercoralis: a plea for action. PLoS Negl. Trop. Dis. 7:e2214. 10.1371/journal.pntd.0002214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 305.Marcos LA, Terashima A, Dupont HL, Gotuzzo E. 2008. Strongyloides hyperinfection syndrome: an emerging global infectious disease. Trans. R. Soc. Trop. Med. Hyg. 102:314–318. 10.1016/j.trstmh.2008.01.020 [DOI] [PubMed] [Google Scholar]
- 306.Montes M, Sawhney C, Barros N. 2010. Strongyloides stercoralis: there but not seen. Curr. Opin. Infect. Dis. 23:500–504. 10.1097/QCO.0b013e32833df718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 307.Mejia R, Nutman TB. 2012. Screening, prevention, and treatment for hyperinfection syndrome and disseminated infections caused by Strongyloides stercoralis. Curr. Opin. Infect. Dis. 25:458–463. 10.1097/QCO.0b013e3283551dbd [DOI] [PMC free article] [PubMed] [Google Scholar]
- 308.Requena-Mendez A, Chiodini P, Bisoffi Z, Buonfrate D, Gotuzzo E, Munoz J. 2013. The laboratory diagnosis and follow up of strongyloidiasis: a systematic review. PLoS Negl. Trop. Dis. 7:e2002. 10.1371/journal.pntd.0002002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 309.Liu J, Gratz J, Amour C, Kibiki G, Becker S, Janaki L, Verweij JJ, Taniuchi M, Sobuz SU, Haque R, Haverstick DM, Houpt ER. 2013. A laboratory-developed TaqMan array card for simultaneous detection of 19 enteropathogens. J. Clin. Microbiol. 51:472–480. 10.1128/JCM.02658-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 310.Cutillas C, Callejon R, de Rojas M, Tewes B, Ubeda JM, Ariza C, Guevara DC. 2009. Trichuris suis and Trichuris trichiura are different nematode species. Acta Trop. 111:299–307. 10.1016/j.actatropica.2009.05.011 [DOI] [PubMed] [Google Scholar]
- 311.Areekul P, Putaporntip C, Pattanawong U, Sitthicharoenchai P, Jongwutiwes S. 2010. Trichuris vulpis and T. trichiura infections among schoolchildren of a rural community in northwestern Thailand: the possible role of dogs in disease transmission. Asian Biomed. 4:49–60 http://abm.digitaljournals.org/index.php/abm/article/viewFile/310/211 [Google Scholar]
- 312.Liu GH, Zhou W, Nisbet AJ, Xu MJ, Zhou DH, Zhao GH, Wang SK, Song HQ, Lin RQ, Zhu XQ. 31 October 2012, posting date Characterization of Trichuris trichiura from humans and T. suis from pigs in China using internal transcribed spacers of nuclear ribosomal DNA. J. Helminthol. 10.1017/S0022149X12000740 [DOI] [PubMed] [Google Scholar]
- 313.Liu GH, Gasser RB, Su A, Nejsum P, Peng L, Lin RQ, Li MW, Xu MJ, Zhu XQ. 2012. Clear genetic distinctiveness between human- and pig-derived Trichuris based on analyses of mitochondrial datasets. PLoS Negl. Trop. Dis. 6:e1539. 10.1371/journal.pntd.0001539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 314.Nissen S, Al-Jubury A, Hansen TV, Olsen A, Christensen H, Thamsborg SM, Nejsum P. 2012. Genetic analysis of Trichuris suis and Trichuris trichiura recovered from humans and pigs in a sympatric setting in Uganda. Vet. Parasitol. 188:68–77. 10.1016/j.vetpar.2012.03.004 [DOI] [PubMed] [Google Scholar]
- 315.Ravasi DF, O'Riain MJ, Davids F, Illing N. 2012. Phylogenetic evidence that two distinct Trichuris genotypes infect both humans and non-human primates. PLoS One 7:e44187. 10.1371/journal.pone.0044187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 316.Diawara A, Drake LJ, Suswillo RR, Kihara J, Bundy DA, Scott ME, Halpenny C, Stothard JR, Prichard RK. 2009. Assays to detect beta-tubulin codon 200 polymorphism in Trichuris trichiura and Ascaris lumbricoides. PLoS Negl. Trop. Dis. 3:e397. 10.1371/journal.pntd.0000397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 317.Velasquez DE, Arvelo W, Cama VA, López B, Reyes L, Roellig DM, Kahn GD, Lindblade KA. 2011. Molecular insights for Giardia, Cryptosporidium, and soil-transmitted helminths from a facility-based surveillance system in Guatemala. Am. J. Trop. Med. Hyg. 85:1141–1143. 10.4269/ajtmh.2011.11-0437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 318.Leles D, Araujo A, Vicente AC, Iniguez AM. 2009. Molecular diagnosis of ascariasis from human feces and description of a new Ascaris sp. genotype in Brazil. Vet. Parasitol. 163:167–170. 10.1016/j.vetpar.2009.03.050 [DOI] [PubMed] [Google Scholar]
- 319.Wiria AE, Prasetyani MA, Hamid F, Wammes LJ, Lell B, Ariawan I, Uh HW, Wibowo H, Djuardi Y, Wahyuni S, Sutanto I, May L, Luty AJ, Verweij JJ, Sartono E, Yazdanbakhsh M, Supali T. 2010. Does treatment of intestinal helminth infections influence malaria? Background and methodology of a longitudinal study of clinical, parasitological and immunological parameters in Nangapanda, Flores, Indonesia (ImmunoSPIN Study). BMC Infect. Dis. 10:77. 10.1186/1471-2334-10-77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 320.Basuni M, Muhi J, Othman N, Verweij JJ, Ahmad M, Miswan N, Rahumatullah A, Aziz FA, Zainudin NS, Noordin R. 2011. A pentaplex real-time polymerase chain reaction assay for detection of four species of soil-transmitted helminths. Am. J. Trop. Med. Hyg. 84:338–343. 10.4269/ajtmh.2011.10-0499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 321.Hamid F, Wiria AE, Wammes LJ, Kaisar MM, Lell B, Ariawan I, Uh HW, Wibowo H, Djuardi Y, Wahyuni S, Schot R, Verweij JJ, van Ree R, May L, Sartono E, Yazdanbakhsh M, Supali T. 2011. A longitudinal study of allergy and intestinal helminth infections in semi urban and rural areas of Flores, Indonesia (ImmunoSPIN Study). BMC Infect. Dis. 11:83. 10.1186/1471-2334-11-83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 322.Taniuchi M, Verweij JJ, Noor Z, Sobuz SU, Lieshout L, Petri WA, Jr, Haque R, Houpt ER. 2011. High throughput multiplex PCR and probe-based detection with Luminex beads for seven intestinal parasites. Am. J. Trop. Med. Hyg. 84:332–337. 10.4269/ajtmh.2011.10-0461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 323.Nejsum P, Betson M, Bendall RP, Thamsborg SM, Stothard JR. 2012. Assessing the zoonotic potential of Ascaris suum and Trichuris suis: looking to the future from an analysis of the past. J. Helminthol. 86:148–155. 10.1017/S0022149X12000193 [DOI] [PubMed] [Google Scholar]
- 324.Leles D, Gardner SL, Reinhard K, Iniguez A, Araujo A. 2012. Are Ascaris lumbricoides and Ascaris suum a single species? Parasit. Vectors 5:42. 10.1186/1756-3305-5-42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 325.Peng W, Criscione CD. 2012. Ascariasis in people and pigs: new inferences from DNA analysis of worm populations. Infect. Genet. Evol. 12:227–235. 10.1016/j.meegid.2012.01.012 [DOI] [PubMed] [Google Scholar]
- 326.Leles D, Araujo A, Vicente AC, Iniguez AM. 2010. ITS1 intra-individual variability of Ascaris isolates from Brazil. Parasitol. Int. 59:93–96. 10.1016/j.parint.2009.10.001 [DOI] [PubMed] [Google Scholar]
- 327.Gasser RB, Cantacessi C, Campbell BE. 2009. Improved molecular diagnostic tools for human hookworms. Expert Rev. Mol. Diagn. 9:17–21. 10.1586/14737159.9.1.17 [DOI] [PubMed] [Google Scholar]
- 328.Verweij JJ, Brienen EA, Ziem J, Yelifari L, Polderman AM, Van Lieshout L. 2007. Simultaneous detection and quantification of Ancylostoma duodenale, Necator americanus, and Oesophagostomum bifurcum in fecal samples using multiplex real-time PCR. Am. J. Trop. Med. Hyg. 77:685–690 http://www.ajtmh.org/content/77/4/685.long [PubMed] [Google Scholar]
- 329.van Mens SP, Aryeetey Y, Yazdanbakhsh M, van Lieshout L, Boakye D, Verweij JJ. 2013. Comparison of real-time PCR and Kato smear microscopy for the detection of hookworm infections in three consecutive faecal samples from schoolchildren in Ghana. Trans. R. Soc. Trop. Med. Hyg. 107:269–271. 10.1093/trstmh/trs094 [DOI] [PubMed] [Google Scholar]
- 330.Ngui R, Lim YA, Chua KH. 2012. Rapid detection and identification of human hookworm infections through high resolution melting (HRM) analysis. PLoS One 7:e41996. 10.1371/journal.pone.0041996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 331.Schar F, Odermatt P, Khieu V, Panning M, Duong S, Muth S, Marti H, Kramme S. 2013. Evaluation of real-time PCR for Strongyloides stercoralis and hookworm as diagnostic tool in asymptomatic schoolchildren in Cambodia. Acta Trop. 126:89–92. 10.1016/j.actatropica.2012.12.012 [DOI] [PubMed] [Google Scholar]
- 332.Wang JX, Pan CS, Cui LW, Chen X. 2012. Application of a real-time PCR method for detecting and monitoring hookworm Necator americanus infections in southern China. Asian Pac. J. Trop. Biomed. 2:925–929. 10.1016/S2221-1691(13)60001-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 333.Wiria AE, Hamid F, Wammes LJ, Kaisar MM, May L, Prasetyani MA, Wahyuni S, Djuardi Y, Ariawan I, Wibowo H, Lell B, Sauerwein R, Brice GT, Sutanto I, van Lieshout L, de Craen AJ, van Ree R, Verweij JJ, Tsonaka R, Houwing-Duistermaat JJ, Luty AJ, Sartono E, Supali T, Yazdanbakhsh M. 2013. The effect of three-monthly albendazole treatment on malarial parasitemia and allergy: a household-based cluster-randomized, double-blind, placebo-controlled trial. PLoS One 8:e57899. 10.1371/journal.pone.0057899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 334.Jonker FA, Calis JC, Phiri K, Brienen EA, Khoffi H, Brabin BJ, Verweij JJ, van Hensbroek MB, van Lieshout L. 2012. Real-time PCR demonstrates Ancylostoma duodenale is a key factor in the etiology of severe anemia and iron deficiency in Malawian pre-school children. PLoS Negl. Trop. Dis. 6:e1555. 10.1371/journal.pntd.0001555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 335.Gasser RB, Cantacessi C, Loukas A. 2008. DNA technological progress toward advanced diagnostic tools to support human hookworm control. Biotechnol. Adv. 26:35–45. 10.1016/j.biotechadv.2007.09.003 [DOI] [PubMed] [Google Scholar]
- 336.Hu M, Chilton NB, Zhu X, Gasser RB. 2002. Single-strand conformation polymorphism-based analysis of mitochondrial cytochrome c oxidase subunit 1 reveals significant substructuring in hookworm populations. Electrophoresis 23:27–34. [DOI] [PubMed] [Google Scholar]
- 337.de Gruijter JM, Polderman AM, Dijkshoorn L, Roberts H, Ziem J, Kunwar CB, Gasser RB. 2006. AFLP fingerprinting for the analysis of genetic diversity within Necator americanus. Mol. Cell. Probes 20:317–321. 10.1016/j.mcp.2006.03.004 [DOI] [PubMed] [Google Scholar]
- 338.Schwenkenbecher JM, Albonico M, Bickle Q, Kaplan RM. 2007. Characterization of beta-tubulin genes in hookworms and investigation of resistance-associated mutations using real-time PCR. Mol. Biochem. Parasitol. 156:167–174. 10.1016/j.molbiopara.2007.07.019 [DOI] [PubMed] [Google Scholar]
- 339.Diawara A, Schwenkenbecher JM, Kaplan RM, Prichard RK. 2013. Molecular and biological diagnostic tests for monitoring benzimidazole resistance in human soil-transmitted helminths. Am. J. Trop. Med. Hyg. 88:1052–1061. 10.4269/ajtmh.12-0484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 340.Cantacessi C, Mitreva M, Jex AR, Young ND, Campbell BE, Hall RS, Doyle MA, Ralph SA, Rabelo EM, Ranganathan S, Sternberg PW, Loukas A, Gasser RB. 2010. Massively parallel sequencing and analysis of the Necator americanus transcriptome. PLoS Negl. Trop. Dis. 4:e684. 10.1371/journal.pntd.0000684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 341.Genta RM. 1989. Global prevalence of strongyloidiasis: critical review with epidemiologic insights into the prevention of disseminated disease. Rev. Infect. Dis. 11:755–767. 10.1093/clinids/11.5.755 [DOI] [PubMed] [Google Scholar]
- 342.Streit A. 2008. Reproduction in Strongyloides (Nematoda): a life between sex and parthenogenesis. Parasitology 135:285–294. 10.1017/S003118200700399X [DOI] [PubMed] [Google Scholar]
- 343.Carvalho EM, Da Fonseca Porto A. 2004. Epidemiological and clinical interaction between HTLV-1 and Strongyloides stercoralis. Parasite Immunol. 26:487–497. 10.1111/j.0141-9838.2004.00726.x [DOI] [PubMed] [Google Scholar]
- 344.Hirata T, Uchima N, Kishimoto K, Zaha O, Kinjo N, Hokama A, Sakugawa H, Kinjo F, Fujita J. 2006. Impairment of host immune response against Strongyloides stercoralis by human T cell lymphotropic virus type 1 infection. Am. J. Trop. Med. Hyg. 74:246–249 http://www.ajtmh.org/content/74/2/246.long [PubMed] [Google Scholar]
- 345.Fardet L, Genereau T, Cabane J, Kettaneh A. 2006. Severe strongyloidiasis in corticosteroid-treated patients. Clin. Microbiol. Infect. 12:945–947. 10.1111/j.1469-0691.2006.01443.x [DOI] [PubMed] [Google Scholar]
- 346.Genta RM. 1992. Dysregulation of strongyloidiasis: a new hypothesis. Clin. Microbiol. Rev. 5:345–355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 347.Keiser PB, Nutman TB. 2004. Strongyloides stercoralis in the immunocompromised population. Clin. Microbiol. Rev. 17:208–217. 10.1128/CMR.17.1.208-217.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 348.Grove DI. (ed). 1989. Strongyloidiasis: a major roundworm infection of man. Taylor & Francis, New York, NY [Google Scholar]
- 349.Liu LX, Weller PF. 1993. Strongyloidiasis and other intestinal nematode infections. Infect. Dis. Clin. North Am. 7:655–682 [PubMed] [Google Scholar]
- 350.Uparanukraw P, Phongsri S, Morakote N. 1999. Fluctuations of larval excretion in Strongyloides stercoralis infection. Am. J. Trop. Med. Hyg. 60:967–973 [DOI] [PubMed] [Google Scholar]
- 351.Siddiqui AA, Berk SL. 2001. Diagnosis of Strongyloides stercoralis infection. Clin. Infect. Dis. 33:1040–1047. 10.1086/322707 [DOI] [PubMed] [Google Scholar]
- 352.Intapan PM, Maleewong W, Wongsaroj T, Singthong S, Morakote N. 2005. Comparison of the quantitative formalin ethyl acetate concentration technique and agar plate culture for diagnosis of human strongyloidiasis. J. Clin. Microbiol. 43:1932–1933. 10.1128/JCM.43.4.1932-1933.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 353.Kramme S, Nissen N, Soblik H, Erttmann K, Tannich E, Fleischer B, Panning M, Brattig N. 2011. Novel real-time PCR for the universal detection of Strongyloides species. J. Med. Microbiol. 60:454–458. 10.1099/jmm.0.025338-0 [DOI] [PubMed] [Google Scholar]
- 354.Verweij JJ, Canales M, Polman K, Ziem J, Brienen EA, Polderman AM, van Lieshout L. 2009. Molecular diagnosis of Strongyloides stercoralis in faecal samples using real-time PCR. Trans. R. Soc. Trop. Med. Hyg. 103:342–346. 10.1016/j.trstmh.2008.12.001 [DOI] [PubMed] [Google Scholar]
- 355.Sultana Y, Jeoffreys N, Watts MR, Gilbert GL, Lee R. 2013. Real-time polymerase chain reaction for detection of Strongyloides stercoralis in stool. Am. J. Trop. Med. Hyg. 88:1048–1051. 10.4269/ajtmh.12-0437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 356.Janwan P, Intapan PM, Thanchomnang T, Lulitanond V, Anamnart W, Maleewong W. 2011. Rapid detection of Opisthorchis viverrini and Strongyloides stercoralis in human fecal samples using a duplex real-time PCR and melting curve analysis. Parasitol. Res. 109:1593–1601. 10.1007/s00436-011-2419-z [DOI] [PubMed] [Google Scholar]
- 357.Moghaddassani H, Mirhendi H, Hosseini M, Rokni M, Mowlavi G, Kia E. 2011. Molecular diagnosis of Strongyloides stercoralis infection by PCR detection of specific DNA in human stool samples. Iran. J. Parasitol. 6:23–30 http://www.ncbi.nlm.nih.gov/pmc/articles/PMC3279873/ [PMC free article] [PubMed] [Google Scholar]
- 358.Ramachandran S, Gam AA, Neva FA. 1997. Molecular differences between several species of Strongyloides and comparison of selected isolates of S. stercoralis using a polymerase chain reaction-linked restriction fragment length polymorphism approach. Am. J. Trop. Med. Hyg. 56:61–65 [DOI] [PubMed] [Google Scholar]
- 359.Labes EM, Wijayanti N, Deplazes P, Mathis A. 2011. Genetic characterization of Strongyloides spp. from captive, semi-captive and wild Bornean orangutans (Pongo pygmaeus) in Central and East Kalimantan, Borneo, Indonesia. Parasitology 138:1417–1422. 10.1017/S0031182011001284 [DOI] [PubMed] [Google Scholar]
- 360.Repetto SA, Alba Soto CD, Cazorla SI, Tayeldin ML, Cuello S, Lasala MB, Tekiel VS, Gonzalez Cappa SM. 2013. An improved DNA isolation technique for PCR detection of Strongyloides stercoralis in stool samples. Acta Trop. 126:110–114. 10.1016/j.actatropica.2013.02.003 [DOI] [PubMed] [Google Scholar]
- 361.Fürst T, Keiser J, Utzinger J. 2012. Global burden of human food-borne trematodiasis: a systematic review and meta-analysis. Lancet Infect. Dis. 12:210–221. 10.1016/S1473-3099(11)70294-8 [DOI] [PubMed] [Google Scholar]
- 362.Johansen MV, Sithithaworn P, Bergquist R, Utzinger J. 2010. Towards improved diagnosis of zoonotic trematode infections in Southeast Asia. Adv. Parasitol. 73:171–194. 10.1016/S0065-308X(10)73007-4 [DOI] [PubMed] [Google Scholar]
- 363.Fürst T, Duthaler U, Sripa B, Utzinger J, Keiser J. 2012. Trematode infections: liver and lung flukes. Infect. Dis. Clin. North Am. 26:399–419. 10.1016/j.idc.2012.03.008 [DOI] [PubMed] [Google Scholar]
- 364.Phan VT, Ersboll AK, Nguyen KV, Madsen H, Dalsgaard A. 2010. Farm-level risk factors for fish-borne zoonotic trematode infection in integrated small-scale fish farms in northern Vietnam. PLoS Negl. Trop. Dis. 4:e742. 10.1371/journal.pntd.0000742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 365.Subasinghe R. 2006. Aquaculture topics and activities. State of world aquaculture. Fisheries and Aquaculture Department, FAO, Rome, Italy: http://www.fao.org/docrep/009/a0874e/a0874e00.htm [Google Scholar]
- 366.Fürst T, Sayasone S, Odermatt P, Keiser J, Utzinger J. 2012. Manifestation, diagnosis, and management of foodborne trematodiasis. BMJ 344:e4093. 10.1136/bmj.e4093 [DOI] [PubMed] [Google Scholar]
- 367.Wang KX, Zhang RB, Cui YB, Tian Y, Cai R, Li CP. 2004. Clinical and epidemiological features of patients with clonorchiasis. World J. Gastroenterol. 10:446–448 http://www.wjgnet.com/1007-9327/full/v10/i3/446.htm [DOI] [PMC free article] [PubMed] [Google Scholar]
- 368.Suksumek N, Leelawat K, Leelawat S, Russell B, Lek-Uthai U. 2008. TaqMan real-time PCR assay for specific detection of Opisthorchis viverrini DNA in Thai patients with hepatocellular carcinoma and cholangiocarcinoma. Exp. Parasitol. 119:217–224. 10.1016/j.exppara.2008.01.018 [DOI] [PubMed] [Google Scholar]
- 369.Kim EM, Verweij JJ, Jalili A, van Lieshout L, Choi MH, Bae YM, Lim MK, Hong ST. 2009. Detection of Clonorchis sinensis in stool samples using real-time PCR. Ann. Trop. Med. Parasitol. 103:513–518. 10.1179/136485909X451834 [DOI] [PubMed] [Google Scholar]
- 370.Cai XQ, Yu HQ, Bai JS, Tang JD, Hu XC, Chen DH, Zhang RL, Chen MX, Ai L, Zhu XQ. 2012. Development of a TaqMan based real-time PCR assay for detection of Clonorchis sinensis DNA in human stool samples and fishes. Parasitol. Int. 61:183–186. 10.1016/j.parint.2011.06.010 [DOI] [PubMed] [Google Scholar]
- 371.Sanpool O, Intapan PM, Thanchomnang T, Janwan P, Lulitanond V, Doanh PN, Van Hien H, Dung DT, Maleewong W, Nawa Y. 2012. Rapid detection and differentiation of Clonorchis sinensis and Opisthorchis viverrini eggs in human fecal samples using a duplex real-time fluorescence resonance energy transfer PCR and melting curve analysis. Parasitol. Res. 111:89–96. 10.1007/s00436-011-2804-7 [DOI] [PubMed] [Google Scholar]
- 372.Sun J, Xu J, Liang P, Mao Q, Huang Y, Lv X, Deng C, Liang C, De Hoog GS, Yu X. 2011. Molecular identification of Clonorchis sinensis and discrimination with other opisthorchid liver fluke species using multiple ligation-depended probe amplification (MLPA). Parasit. Vectors 4:98. 10.1186/1756-3305-4-98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 373.Arimatsu Y, Kaewkes S, Laha T, Hong SJ, Sripa B. 2012. Rapid detection of Opisthorchis viverrini copro-DNA using loop-mediated isothermal amplification (LAMP). Parasitol. Int. 61:178–182. 10.1016/j.parint.2011.08.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 374.Le TH, Nguyen NT, Truong NH, De NV. 2012. Development of mitochondrial loop-mediated isothermal amplification for detection of the small liver fluke Opisthorchis viverrini (Opisthorchiidae; Trematoda; Platyhelminthes). J. Clin. Microbiol. 50:1178–1184. 10.1128/JCM.06277-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 375.Huang S, Zhao G, Fu B, Xu M, Wang C, Wu S, Zou F, Zhu X. 2012. Genomics and molecular genetics of Clonorchis sinensis: current status and perspectives. Parasitol. Int. 61:71–76. 10.1016/j.parint.2011.06.008 [DOI] [PubMed] [Google Scholar]
- 376.Shekhovtsov SV, Katokhin AV, Romanov KV, Besprozvannykh VV, Fedorov KP, Yurlova NI, Serbina EA, Sithithaworn P, Kolchanov NA, Mordvinov VA. 2009. A novel nuclear marker, pm-int9, for phylogenetic studies of Opisthorchis felineus, Opisthorchis viverrini, and Clonorchis sinensis (Opisthorchiidae, Trematoda). Parasitol. Res. 106:293–297. 10.1007/s00436-009-1628-1 [DOI] [PubMed] [Google Scholar]
- 377.Laoprom N, Sithithaworn P, Andrews RH, Ando K, Laha T, Klinbunga S, Webster JP, Petney TN. 2012. Population genetic structuring in Opisthorchis viverrini over various spatial scales in Thailand and Lao PDR. PLoS Negl. Trop. Dis. 6:e1906. 10.1371/journal.pntd.0001906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 378.Laoprom N, Sithithaworn P, Ando K, Sithithaworn J, Wongkham S, Laha T, Klinbunga S, Webster JP, Andrews RH. 2010. Microsatellite loci in the carcinogenic liver fluke, Opisthorchis viverrini and their application as population genetic markers. Infect. Genet. Evol. 10:146–153. 10.1016/j.meegid.2009.11.005 [DOI] [PubMed] [Google Scholar]
- 379.Ai L, Chen MX, Alasaad S, Elsheikha HM, Li J, Li HL, Lin RQ, Zou FC, Zhu XQ, Chen JX. 2011. Genetic characterization, species differentiation and detection of Fasciola spp. by molecular approaches. Parasit. Vectors 4:101. 10.1186/1756-3305-4-101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 380.Chen MX, Ai L, Zhang RL, Xia JJ, Wang K, Chen SH, Zhang YN, Xu MJ, Li X, Zhu XQ, Chen JX. 2011. Sensitive and rapid detection of Paragonimus westermani infection in humans and animals by loop-mediated isothermal amplification (LAMP). Parasitol. Res. 108:1193–1198. 10.1007/s00436-010-2162-x [DOI] [PubMed] [Google Scholar]
- 381.Pozio E, Armignacco O, Ferri F, Gomez Morales MA. 2013. Opisthorchis felineus, an emerging infection in Italy and its implication for the European Union. Acta Trop. 126:54–62. 10.1016/j.actatropica.2013.01.005 [DOI] [PubMed] [Google Scholar]
- 382.Gryseels B, Polman K, Clerinx J, Kestens L. 2006. Human schistosomiasis. Lancet 368:1106–1118. 10.1016/S0140-6736(06)69440-3 [DOI] [PubMed] [Google Scholar]
- 383.Clerinx J, Bottieau E, Wichmann D, Tannich E, Van Esbroeck M. 2011. Acute schistosomiasis in a cluster of travelers from Rwanda: diagnostic contribution of schistosome DNA detection in serum compared to parasitology and serology. J. Travel Med. 18:367–372. 10.1111/j.1708-8305.2011.00552.x [DOI] [PubMed] [Google Scholar]
- 384.Wichmann D, Poppert S, Von Thien H, Clerinx J, Dieckmann S, Jensenius M, Parola P, Richter J, Schunk M, Stich A, Zanger P, Burchard GD, Tannich E. 2013. Prospective European-wide multicentre study on a blood based real-time PCR for the diagnosis of acute schistosomiasis. BMC Infect. Dis. 13:55. 10.1186/1471-2334-13-55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 385.Hamburger J, Xu YX, Ramzy RM, Jourdane J, Ruppel A. 1998. Development and laboratory evaluation of a polymerase chain reaction for monitoring Schistosoma mansoni infestation of water. Am. J. Trop. Med. Hyg. 59:468–473 [DOI] [PubMed] [Google Scholar]
- 386.Hamburger J, He-Na, Abbasi I, Ramzy RM, Jourdane J, Ruppel A. 2001. Polymerase chain reaction assay based on a highly repeated sequence of Schistosoma haematobium: a potential tool for monitoring schistosome-infested water. Am. J. Trop. Med. Hyg. 65:907–911 http://www.ajtmh.org/content/65/6/907.long [DOI] [PubMed] [Google Scholar]
- 387.Pontes LA, Dias-Neto E, Rabello A. 2002. Detection by polymerase chain reaction of Schistosoma mansoni DNA in human serum and feces. Am. J. Trop. Med. Hyg. 66:157–162 http://www.ajtmh.org/content/66/2/157.long [DOI] [PubMed] [Google Scholar]
- 388.Pontes LA, Oliveira MC, Katz N, Dias-Neto E, Rabello A. 2003. Comparison of a polymerase chain reaction and the Kato-Katz technique for diagnosing infection with Schistosoma mansoni. Am. J. Trop. Med. Hyg. 68:652–656 http://www.ajtmh.org/content/68/6/652.long [PubMed] [Google Scholar]
- 389.Aryeetey YA, Essien-Baidoo S, Larbi IA, Ahmed K, Amoah AS, Obeng BB, van Lieshout L, Yazdanbakhsh M, Boakye DA, Verweij JJ. 2013. Molecular diagnosis of Schistosoma infections in urine samples of school children in Ghana. Am. J. Trop. Med. Hyg. 88:1028–1031. 10.4269/ajtmh.12-0571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 390.Mutapi F. 2011. Improving diagnosis of urogenital schistosome infection. Expert Rev. Anti Infect. Ther. 9:863–865. 10.1586/eri.11.101 [DOI] [PubMed] [Google Scholar]
- 391.Ibironke OA, Phillips AE, Garba A, Lamine SM, Shiff C. 2011. Diagnosis of Schistosoma haematobium by detection of specific DNA fragments from filtered urine samples. Am. J. Trop. Med. Hyg. 84:998–1001. 10.4269/ajtmh.2011.10-0691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 392.Ibironke O, Koukounari A, Asaolu S, Moustaki I, Shiff C. 2012. Validation of a new test for Schistosoma haematobium based on detection of DraI DNA fragments in urine: evaluation through latent class analysis. PLoS Negl. Trop. Dis. 6:e1464. 10.1371/journal.pntd.0001464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 393.Reinstrup L, Jorgensen A, Vennervald BJ, Kristensen TK. 2012. DNA extraction from dried Schistosoma haematobium eggs isolated on nylon filters. Trans. R. Soc. Trop. Med. Hyg. 106:270–272. 10.1016/j.trstmh.2011.12.002 [DOI] [PubMed] [Google Scholar]
- 394.ten Hove RJ, Verweij JJ, Vereecken K, Polman K, Dieye L, van Lieshout L. 2008. Multiplex real-time PCR for the detection and quantification of Schistosoma mansoni and S. haematobium infection in stool samples collected in northern Senegal. Trans. R. Soc. Trop. Med. Hyg. 102:179–185. 10.1016/j.trstmh.2007.10.011 [DOI] [PubMed] [Google Scholar]
- 395.Kjetland EF, Hove RJ, Gomo E, Midzi N, Gwanzura L, Mason P, Friis H, Verweij JJ, Gundersen SG, Ndhlovu PD, Mduluza T, Van Lieshout L. 2009. Schistosomiasis PCR in vaginal lavage as an indicator of genital Schistosoma haematobium infection in rural Zimbabwean women. Am. J. Trop. Med. Hyg. 81:1050–1055. 10.4269/ajtmh.2009.09-0081 [DOI] [PubMed] [Google Scholar]
- 396.Downs JA, Kabangila R, Verweij JJ, Jaka H, Peck RN, Kalluvya SE, Changalucha JM, Johnson WD, van Lieshout L, Fitzgerald DW. 2013. Detectable urogenital schistosome DNA and cervical abnormalities 6 months after single-dose praziquantel in women with Schistosoma haematobium infection. Trop. Med. Int. Health 18:1090–1096. 10.1111/tmi.12154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 397.Wichmann D, Panning M, Quack T, Kramme S, Burchard GD, Grevelding C, Drosten C. 2009. Diagnosing schistosomiasis by detection of cell-free parasite DNA in human plasma. PLoS Negl. Trop. Dis. 3:e422. 10.1371/journal.pntd.0000422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 398.Cnops L, Tannich E, Polman K, Clerinx J, Van Esbroeck M. 2012. Schistosoma real-time PCR as diagnostic tool for international travellers and migrants. Trop. Med. Int. Health 17:1208–1216. 10.1111/j.1365-3156.2012.03060.x [DOI] [PubMed] [Google Scholar]
- 399.Craig P, Ito A. 2007. Intestinal cestodes. Curr. Opin. Infect. Dis. 20:524–532. 10.1097/QCO.0b013e3282ef579e [DOI] [PubMed] [Google Scholar]
- 400.Ito A, Craig PS. 2003. Immunodiagnostic and molecular approaches for the detection of taeniid cestode infections. Trends Parasitol. 19:377–381. 10.1016/S1471-4922(03)00200-9 [DOI] [PubMed] [Google Scholar]
- 401.McManus DP. 2006. Molecular discrimination of taeniid cestodes. Parasitol. Int. 55(Suppl):S31–S37. 10.1016/j.parint.2005.11.004 [DOI] [PubMed] [Google Scholar]
- 402.Praet N, Verweij JJ, Mwape KE, Phiri IK, Muma JB, Zulu G, van Lieshout L, Rodriguez-Hidalgo R, Benitez-Ortiz W, Dorny P, Gabriël S. 2013. Bayesian modelling to estimate the test characteristics of coprology, coproantigen ELISA and a novel real-time PCR for the diagnosis of taeniasis. Trop. Med. Int. Health 18:608–614. 10.1111/tmi.12089 [DOI] [PubMed] [Google Scholar]
- 403.Nkouawa A, Sako Y, Li T, Chen X, Wandra T, Swastika IK, Nakao M, Yanagida T, Nakaya K, Qiu D, Ito A. 2010. Evaluation of a loop-mediated isothermal amplification method using fecal specimens for differential detection of Taenia species from humans. J. Clin. Microbiol. 48:3350–3352. 10.1128/JCM.00697-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 404.Almeida CR, Ojopi EP, Nunes CM, Machado LR, Takayanagui OM, Livramento JA, Abraham R, Gattaz WF, Vaz AJ, Dias-Neto E. 2006. Taenia solium DNA is present in the cerebrospinal fluid of neurocysticercosis patients and can be used for diagnosis. Eur. Arch. Psychiatry Clin. Neurosci. 256:307–310. 10.1007/s00406-006-0612-3 [DOI] [PubMed] [Google Scholar]
- 405.Michelet L, Fleury A, Sciutto E, Kendjo E, Fragoso G, Paris L, Bouteille B. 2011. Human neurocysticercosis: comparison of different diagnostic tests using cerebrospinal fluid. J. Clin. Microbiol. 49:195–200. 10.1128/JCM.01554-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 406.Yera H, Dupont D, Houze S, Ben M'rad M, Pilleux F, Sulahian A, Gatey C, Gay Andrieu F, Dupouy-Camet J. 2011. Confirmation and follow-up of neurocysticercosis by real-time PCR in cerebrospinal fluid samples of patients living in France. J. Clin. Microbiol. 49:4338–4340. 10.1128/JCM.05839-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 407.Wicht B, Yanagida T, Scholz T, Ito A, Jimenez JA, Brabec J. 2010. Multiplex PCR for differential identification of broad tapeworms (Cestoda: Diphyllobothrium) infecting humans. J. Clin. Microbiol. 48:3111–3116. 10.1128/JCM.00445-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 408.Guo AJ, Liu K, Gong W, Luo XN, Yan HB, Zhao SB, Hu SN, Jia WZ. 2012. Molecular identification of Diphyllobothrium latum and a brief review of diphyllobothriosis in China. Acta Parasitol. 57:293–296. 10.2478/s11686-012-0036-3 [DOI] [PubMed] [Google Scholar]
- 409.de Marval F, Gottstein B, Weber M, Wicht B. 2013. Imported diphyllobothriasis in Switzerland: molecular methods to define a clinical case of Diphyllobothrium infection as Diphyllobothrium dendriticum, August 2010. Euro Surveill. 18(3):pii=20355 http://www.eurosurveillance.org/ViewArticle.aspx?ArticleId=20355 [PubMed] [Google Scholar]
- 410.Koonmee S, Intapan PM, Yamasaki H, Sugiyama H, Muto M, Kuramochi T, Kularbkeaw J, Kanpittaya J, Maleewong W, Nawa Y. 2011. Molecular identification of a causative parasite species using formalin-fixed paraffin embedded (FFPE) tissues of a complicated human pulmonary sparganosis case without decisive clinical diagnosis. Parasitol. Int. 60:460–464. 10.1016/j.parint.2011.07.018 [DOI] [PubMed] [Google Scholar]
- 411.Wicht B, Ruggeri-Bernardi N, Yanagida T, Nakao M, Peduzzi R, Ito A. 2010. Inter- and intra-specific characterization of tapeworms of the genus Diphyllobothrium (Cestoda: Diphyllobothriidea) from Switzerland, using nuclear and mitochondrial DNA targets. Parasitol. Int. 59:35–39. 10.1016/j.parint.2009.09.002 [DOI] [PubMed] [Google Scholar]
- 412.Macnish MG, Morgan-Ryan UM, Monis PT, Behnke JM, Thompson RC. 2002. A molecular phylogeny of nuclear and mitochondrial sequences in Hymenolepis nana (Cestoda) supports the existence of a cryptic species. Parasitology 125:567–575. 10.1017/S0031182002002366 [DOI] [PubMed] [Google Scholar]
- 413.McAuliffe GN, Anderson TP, Stevens M, Adams J, Coleman R, Mahagamasekera P, Young S, Henderson T, Hofmann M, Jennings LC, Murdoch DR. 2013. Systematic application of multiplex PCR enhances the detection of bacteria, parasites, and viruses in stool samples. J. Infect. 67:122–129. 10.1016/j.jinf.2013.04.009 [DOI] [PubMed] [Google Scholar]
- 414.Amar CF, East CL, Gray J, Iturriza-Gomara M, Maclure EA, McLauchlin J. 2007. Detection by PCR of eight groups of enteric pathogens in 4,627 faecal samples: re-examination of the English case-control Infectious Intestinal Disease Study (1993-1996). Eur. J. Clin. Microbiol. Infect. Dis. 26:311–323. 10.1007/s10096-007-0290-8 [DOI] [PubMed] [Google Scholar]
- 415.Amin OM. 2002. Seasonal prevalence of intestinal parasites in the United States during 2000. Am. J. Trop. Med. Hyg. 66:799–803 http://www.ajtmh.org/content/66/6/799.long [DOI] [PubMed] [Google Scholar]
- 416.Masucci L, Graffeo R, Bani S, Bugli F, Boccia S, Nicolotti N, Fiori B, Fadda G, Spanu T. 2011. Intestinal parasites isolated in a large teaching hospital, Italy, 1 May 2006 to 31 December 2008. Euro Surveill. 16(24):pii=19891 http://www.eurosurveillance.org/ViewArticle.aspx?ArticleId=19891 [DOI] [PubMed] [Google Scholar]
- 417.Mosli M, Gregor J, Chande N, Lannigan R. 2012. Nonutility of routine testing of stool for ova and parasites in a tertiary care Canadian centre. Can. J. Microbiol. 58:653–659. 10.1139/w2012-039 [DOI] [PubMed] [Google Scholar]
- 418.Verweij JJ. Application of PCR-based methods for diagnosis of intestinal parasitic infections in the clinical laboratory. Parasitology, in press [DOI] [PubMed] [Google Scholar]
- 419.Soonawala D, van Lieshout L, den Boer MAM, Claas ECJ, Verweij JJ, Godkewitsch A, Ratering M, Visser LG. Post-travel screening of asymptomatic long-term travelers to the tropics for intestinal parasites using molecular diagnostics. Am. J. Trop. Med. Hyg., in press [DOI] [PMC free article] [PubMed] [Google Scholar]
- 420.Polage CR, Stoddard GJ, Rolfs RT, Petti CA. 2011. Physician use of parasite tests in the United States from 1997 to 2006 and in a Utah Cryptosporidium outbreak in 2007. J. Clin. Microbiol. 49:591–596. 10.1128/JCM.01806-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 421.Schuurs TA, Nguyen AJ, Stellingwerf AM, Kooistra-Smid AM, Noordhoek GT. 2010. Molecular diagnostics of gastroenteritis in clinical samples: a multicentre, quality control study in the Netherlands, abstr P1688 Abstr. 20th Eur. Congr. Clin. Microbiol. Infect. Dis. http://www.blackwellpublishing.com/eccmid20/ [Google Scholar]
- 422.Mengelle C, Mansuy JM, Prere MF, Grouteau E, Claudet I, Kamar N, Huynh A, Plat G, Benard M, Marty N, Valentin A, Berry A, Izopet J. 2013. Simultaneous detection of gastrointestinal pathogens with a multiplex Luminex-based molecular assay in stool samples from diarrhoeic patients. Clin. Microbiol. Infect. 19:E458–E465. 10.1111/1469-0691.12255 [DOI] [PubMed] [Google Scholar]
- 423.Claas EC, Burnham CA, Mazzulli T, Templeton K, Topin F. 2013. Performance of the xTAG gastrointestinal pathogen panel, a multiplex molecular assay for simultaneous detection of bacterial, viral, and parasitic causes of infectious gastroenteritis. J. Microbiol. Biotechnol. 23:1041–1045. 10.4014/jmb.1212.12042 [DOI] [PubMed] [Google Scholar]
- 424.Buss SN, Alter R, Iwen PC, Fey PD. 2013. Implications of culture-independent panel-based detection of Cyclospora cayetanensis. J. Clin. Microbiol. 51:3909. 10.1128/JCM.02238-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 425.Harrington AT, Creutzfeldt CJ, Sengupta DJ, Hoogestraat DR, Zunt JR, Cookson BT. 2009. Diagnosis of neurocysticercosis by detection of Taenia solium DNA using a global DNA screening platform. Clin. Infect. Dis. 48:86–90. 10.1086/594128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 426.Andersen LO, Vedel Nielsen H, Stensvold CR. 2013. Waiting for the human intestinal eukaryotome. ISME J. 7:1253–1255. 10.1038/ismej.2013.21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 427.Ali IK, Hossain MB, Roy S, Ayeh-Kumi P, Petri WA, Jr, Haque R, Clark CG. 2003. Entamoeba moshkovskii infections in children, Bangladesh. Emerg. Infect. Dis. 9:580–584. 10.3201/eid0905.020548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 428.Verweij JJ, Laeijendecker D, Brienen EA, van Lieshout L, Polderman AM. 2003. Detection and identification of Entamoeba species in stool samples by a reverse line hybridization assay. J. Clin. Microbiol. 41:5041–5045. 10.1128/JCM.41.11.5041-5045.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 429.Parija SC, Khairnar K. 2005. Entamoeba moshkovskii and Entamoeba dispar-associated infections in Pondicherry, India. J. Health Popul. Nutr. 23:292–295 http://www.bioline.org.br/pdf?hn05037 [PubMed] [Google Scholar]
- 430.Hamzah Z, Petmitr S, Mungthin M, Leelayoova S, Chavalitshewinkoon-Petmitr P. 2006. Differential detection of Entamoeba histolytica, Entamoeba dispar, and Entamoeba moshkovskii by a single-round PCR assay. J. Clin. Microbiol. 44:3196–3200. 10.1128/JCM.00778-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 431.Solaymani-Mohammadi S, Rezaian M, Babaei Z, Rajabpour A, Meamar AR, Pourbabai AA, Petri WA., Jr 2006. Comparison of a stool antigen detection kit and PCR for diagnosis of Entamoeba histolytica and Entamoeba dispar infections in asymptomatic cyst passers in Iran. J. Clin. Microbiol. 44:2258–2261. 10.1128/JCM.00530-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 432.Khairnar K, Parija SC, Palaniappan R. 2007. Diagnosis of intestinal amoebiasis by using nested polymerase chain reaction-restriction fragment length polymorphism assay. J. Gastroenterol. 42:631–640. 10.1007/s00535-007-2080-6 [DOI] [PubMed] [Google Scholar]
- 433.Khairnar K, Parija SC. 2007. A novel nested multiplex polymerase chain reaction (PCR) assay for differential detection of Entamoeba histolytica, E. moshkovskii and E. dispar DNA in stool samples. BMC Microbiol. 7:47. 10.1186/1471-2180-7-47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 434.Tanyuksel M, Ulukanligil M, Guclu Z, Araz E, Koru O, Petri WA., Jr 2007. Two cases of rarely recognized infection with Entamoeba moshkovskii. Am. J. Trop. Med. Hyg. 76:723–724 http://www.ajtmh.org/content/76/4/723.long [PubMed] [Google Scholar]
- 435.Fotedar R, Stark D, Beebe N, Marriott D, Ellis J, Harkness J. 2007. PCR detection of Entamoeba histolytica, Entamoeba dispar, and Entamoeba moshkovskii in stool samples from Sydney, Australia. J. Clin. Microbiol. 45:1035–1037. 10.1128/JCM.02144-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 436.Fotedar R, Stark D, Marriott D, Ellis J, Harkness J. 2008. Entamoeba moshkovskii infections in Sydney, Australia. Eur. J. Clin. Microbiol. Infect. Dis. 27:133–137. 10.1007/s10096-007-0399-9 [DOI] [PubMed] [Google Scholar]
- 437.Ayed SB, Aoun K, Maamouri N, Abdallah RB, Bouratbine A. 2008. First molecular identification of Entamoeba moshkovskii in human stool samples in Tunisia. Am. J. Trop. Med. Hyg. 79:706–707 http://www.ajtmh.org/content/79/5/706.long [PubMed] [Google Scholar]
- 438.Parija SC, Khairnar K. 2008. Mutation detection analysis of a region of 16S-like ribosomal RNA gene of Entamoeba histolytica, Entamoeba dispar and Entamoeba moshkovskii. BMC Infect. Dis. 8:131. 10.1186/1471-2334-8-131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 439.Beck DL, Dogan N, Maro V, Sam NE, Shao J, Houpt ER. 2008. High prevalence of Entamoeba moshkovskii in a Tanzanian HIV population. Acta Trop. 107:48–49. 10.1016/j.actatropica.2008.03.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 440.Parija SC, Garg A, Pushpa K, Khairnar K, Priya T. 2010. Polymerase chain reaction confirmation of diagnosis of intestinal amebiasis in Puducherry. Indian J. Gastroenterol. 29:140–142. 10.1007/s12664-010-0033-0 [DOI] [PubMed] [Google Scholar]
- 441.Nazemalhosseini Mojarad E, Nochi Z, Sahebekhtiari N, Rostami Nejad M, Dabiri H, Zali MR, Kazemi B, Haghighi A. 2010. Discrimination of Entamoeba moshkovskii in patients with gastrointestinal disorders by single-round PCR. Jpn. J. Infect. Dis. 63:136–138 http://www0.nih.go.jp/JJID/63/136.pdf [PubMed] [Google Scholar]
- 442.Levecke B, Dreesen L, Barrionuevo-Samaniego M, Ortiz WB, Praet N, Brandt J, Dorny P. 2011. Molecular differentiation of Entamoeba spp. in a rural community of Loja Province, South Ecuador. Trans. R. Soc. Trop. Med. Hyg. 105:737–739. 10.1016/j.trstmh.2011.08.010 [DOI] [PubMed] [Google Scholar]
- 443.Anuar TS, Al-Mekhlafi HM, Abdul Ghani MK, Abu Bakar E, Azreen SN, Salleh FM, Ghazali N, Bernadus M, Moktar N. 2012. Molecular epidemiology of amoebiasis in Malaysia: highlighting the different risk factors of Entamoeba histolytica and Entamoeba dispar infections among Orang Asli communities. Int. J. Parasitol. 42:1165–1175. 10.1016/j.ijpara.2012.10.003 [DOI] [PubMed] [Google Scholar]
- 444.Shahrul Anuar T, Al-Mekhlafi HM, Abdul Ghani MK, Osman E, Mohd Yasin A, Nordin A, Nor Azreen S, Md Salleh F, Ghazali N, Bernadus M, Moktar N. 2012. Prevalence and risk factors associated with Entamoeba histolytica/dispar/moshkovskii infection among three Orang Asli ethnic groups in Malaysia. PLoS One 7:e48165. 10.1371/journal.pone.0048165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 445.Anuar TS, Al-Mekhlafi HM, Abdul Ghani MK, Azreen SN, Salleh FM, Ghazali N, Bernadus M, Moktar N. 2013. Different clinical outcomes of Entamoeba histolytica in Malaysia: does genetic diversity exist? Korean J. Parasitol. 51:231–236. 10.3347/kjp.2013.51.2.231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 446.Ngui R, Angal L, Fakhrurrazi SA, Lian YL, Ling LY, Ibrahim J, Mahmud R. 2012. Differentiating Entamoeba histolytica, Entamoeba dispar and Entamoeba moshkovskii using nested polymerase chain reaction (PCR) in rural communities in Malaysia. Parasit. Vectors 5:187. 10.1186/1756-3305-5-187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 447.Wang Z, Vora GJ, Stenger DA. 2004. Detection and genotyping of Entamoeba histolytica, Entamoeba dispar, Giardia lamblia, and Cryptosporidium parvum by oligonucleotide microarray. J. Clin. Microbiol. 42:3262–3271. 10.1128/JCM.42.7.3262-3271.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 448.Haque R, Roy S, Siddique A, Mondal U, Rahman SM, Mondal D, Houpt E, Petri WA., Jr 2007. Multiplex real-time PCR assay for detection of Entamoeba histolytica, Giardia intestinalis, and Cryptosporidium spp. Am. J. Trop. Med. Hyg. 76:713–717 http://www.ajtmh.org/content/76/4/713.long [PubMed] [Google Scholar]
- 449.Wiria AE, Wammes LJ, Hamid F, Dekkers OM, Prasetyani MA, May L, Kaisar MM, Verweij JJ, Tamsma JT, Partono F, Sartono E, Supali T, Yazdanbakhsh M, Smit JW. 2013. Relationship between carotid intima media thickness and helminth infections on Flores Island, Indonesia. PLoS One 8:e54855. 10.1371/journal.pone.0054855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 450.Lee SH, Joung M, Yoon S, Choi K, Park WY, Yu JR. 2010. Multiplex PCR detection of waterborne intestinal protozoa: microsporidia, Cyclospora, and Cryptosporidium. Korean J. Parasitol. 48:297–301. 10.3347/kjp.2010.48.4.297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 451.Stark D, Al-Qassab S, Barratt JL, Stanley K, Roberts T, Marriott D, Harkness J, Ellis JT. 2011. Evaluation of multiplex tandem real-time PCR for detection of Cryptosporidium spp., Dientamoeba fragilis, Entamoeba histolytica, and Giardia intestinalis in clinical stool samples. J. Clin. Microbiol. 49:257–262. 10.1128/JCM.01796-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 452.Jex AR, Stanley KK, Lo W, Littman R, Verweij JJ, Campbell BE, Nolan MJ, Pangasa A, Stevens MA, Haydon S, Gasser RB. 2012. Detection of diarrhoeal pathogens in human faeces using an automated, robotic platform. Mol. Cell. Probes 26:11–15. 10.1016/j.mcp.2011.10.004 [DOI] [PubMed] [Google Scholar]
- 453.Tolentino-Ruiz R, Montoya-Varela D, García-Espitia M, Salas-Benito M, Gutiérrez-Escolano A, Gómez-García C, Figueroa-Arredondo P, Salas-Benito J, De Nova-Ocampo M. 2012. Development of a multiplex PCR assay to detect gastroenteric pathogens in the feces of Mexican children. Curr. Microbiol. 65:361–368. 10.1007/s00284-012-0167-7 [DOI] [PubMed] [Google Scholar]
- 454.Goldfarb DM, Dixon B, Moldovan I, Barrowman N, Mattison K, Zentner C, Baikie M, Bidawid S, Chan F, Slinger R. 2013. Nanolitre real-time PCR detection of bacterial, parasitic, and viral agents from patients with diarrhoea in Nunavut, Canada. Int. J. Circumpolar Health 72:19903. 10.3402/ijch.v72i0.19903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 455.Wessels E, Rusman LG, van Bussel MJAWM, Claas ECJ. 2014. Added value of multiplex Luminex gastrointestinal pathogen panel (xTAG GPP) testing in the diagnosis of infectious gastroenteritis. Clin. Microbiol. Infect. 20:O182–O187. 10.1111/1469-0691.12364 [DOI] [PubMed] [Google Scholar]
- 456.Arndt MB, John-Stewart G, Richardson BA, Singa B, van Lieshout L, Verweij JJ, Sangare LR, Mbogo LW, Naulikha JM, Walson JL. 2013. Impact of helminth diagnostic test performance on estimation of risk factors and outcomes in HIV-positive adults. PLoS One 8:e81915. 10.1371/journal.pone.0081915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 457.Verweij JJ, Ten Hove R, Brienen EA, van Lieshout L. 2007. Multiplex detection of Enterocytozoon bieneusi and Encephalitozoon spp. in fecal samples using real-time PCR. Diagn. Microbiol. Infect. Dis. 57:163–167. 10.1016/j.diagmicrobio.2006.08.009 [DOI] [PubMed] [Google Scholar]
- 458.Obeng BB, Aryeetey YA, de Dood CJ, Amoah AS, Larbi IA, Deelder AM, Yazdanbakhsh M, Hartgers FC, Boakye DA, Verweij JJ, van Dam GJ, van Lieshout L. 2008. Application of a circulating-cathodic-antigen (CCA) strip test and real-time PCR, in comparison with microscopy, for the detection of Schistosoma haematobium in urine samples from Ghana. Ann. Trop. Med. Parasitol. 102:625–633. 10.1179/136485908X337490 [DOI] [PubMed] [Google Scholar]
- 459.Stark D, Beebe N, Marriott D, Ellis J, Harkness J. 2005. Detection of Dientamoeba fragilis in fresh stool specimens using PCR. Int. J. Parasitol. 35:57–62. 10.1016/j.ijpara.2004.09.003 [DOI] [PubMed] [Google Scholar]
- 460.Stark D, Barratt J, Roberts T, Marriott D, Harkness J, Ellis J. 2010. Comparison of microscopy, two xenic culture techniques, conventional and real-time PCR for the detection of Dientamoeba fragilis in clinical stool samples. Eur. J. Clin. Microbiol. Infect. Dis. 29:411–416. 10.1007/s10096-010-0876-4 [DOI] [PubMed] [Google Scholar]
- 461.Banik GR, Barratt JL, Marriott D, Harkness J, Ellis JT, Stark D. 2011. A case-controlled study of Dientamoeba fragilis infections in children. Parasitology 138:819–823. 10.1017/S0031182011000448 [DOI] [PubMed] [Google Scholar]
- 462.Calderaro A, Gorrini C, Montecchini S, Peruzzi S, Piccolo G, Rossi S, Gargiulo F, Manca N, Dettori G, Chezzi C. 2010. Evaluation of a real-time polymerase chain reaction assay for the detection of Dientamoeba fragilis. Diagn. Microbiol. Infect. Dis. 67:239–245. 10.1016/j.diagmicrobio.2010.02.016 [DOI] [PubMed] [Google Scholar]
- 463.Vandenberg O, Peek R, Souayah H, Dediste A, Buset M, Scheen R, Retore P, Zissis G, van Gool T. 2006. Clinical and microbiological features of dientamoebiasis in patients suspected of suffering from a parasitic gastrointestinal illness: a comparison of Dientamoeba fragilis and Giardia lamblia infections. Int. J. Infect. Dis. 10:255–261. 10.1016/j.ijid.2005.05.011 [DOI] [PubMed] [Google Scholar]
- 464.Relman DA, Schmidt TM, Gajadhar A, Sogin M, Cross J, Yoder K, Sethabutr O, Echeverria P. 1996. Molecular phylogenetic analysis of Cyclospora, the human intestinal pathogen, suggests that it is closely related to Eimeria species. J. Infect. Dis. 173:440–445. 10.1093/infdis/173.2.440 [DOI] [PubMed] [Google Scholar]
- 465.Pieniazek NJ, Slemenda SB, da Silva AJ, Alfano EM, Arrowood MJ. 1996. PCR confirmation of infection with Cyclospora cayetanensis. Emerg. Infect. Dis. 2:357–359. 10.3201/eid0204.960415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 466.Jinneman KC, Wetherington JH, Hill WE, Adams AM, Johnson JM, Tenge BJ, Dang NL, Manger RL, Wekell MM. 1998. Template preparation for PCR and RFLP of amplification products for the detection and identification of Cyclospora sp. and Eimeria spp. oocysts directly from raspberries. J. Food Prot. 61:1497–1503 [DOI] [PubMed] [Google Scholar]
- 467.Jinneman KC, Wetherington JH, Hill WE, Omiescinski CJ, Adams AM, Johnson JM, Tenge BJ, Dang NL, Wekell MM. 1999. An oligonucleotide-ligation assay for the differentiation between Cyclospora and Eimeria spp. polymerase chain reaction amplification products. J. Food Prot. 62:682–685 [DOI] [PubMed] [Google Scholar]
- 468.Eberhard ML, da Silva AJ, Lilley BG, Pieniazek NJ. 1999. Morphologic and molecular characterization of new Cyclospora species from Ethiopian monkeys: C. cercopitheci sp.n., C. colobi sp.n., and C. papionis sp.n. Emerg. Infect. Dis. 5:651–658. 10.3201/eid0505.990506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 469.Shields JM, Olson BH. 2003. PCR-restriction fragment length polymorphism method for detection of Cyclospora cayetanensis in environmental waters without microscopic confirmation. Appl. Environ. Microbiol. 69:4662–4669. 10.1128/AEM.69.8.4662-4669.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 470.Orlandi PA, Carter L, Brinker AM, da Silva AJ, Chu DM, Lampel KA, Monday SR. 2003. Targeting single-nucleotide polymorphisms in the 18S rRNA gene to differentiate Cyclospora species from Eimeria species by multiplex PCR. Appl. Environ. Microbiol. 69:4806–4813. 10.1128/AEM.69.8.4806-4813.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 471.Hussein EM, Abdul-Manaem A, el-Attary S. 2005. Cyclospora cayetanensis oocysts in sputum of a patient with active pulmonary tuberculosis, case report in Ismailia, Egypt. J. Egypt. Soc. Parasitol. 35:787–793 [PubMed] [Google Scholar]
- 472.Helmy MM, Rashed LA, Abdel-Fattah H. 2006. Co-infection with Cryptosporidium parvum and Cyclospora cayetanensis in immunocompromised patients. J. Egypt. Soc. Parasitol. 36:613–627 [PubMed] [Google Scholar]
- 473.Hussein EM, El-Moamly A, Dawoud HA, Fahmy H, El-Shal H, Sabek NA. 2007. Real-time PCR and flow cytometry in detection of Cyclospora oocysts in fecal samples of symptomatic and asymptomatic pediatrics patients. J. Egypt. Soc. Parasitol. 37:151–170 [PubMed] [Google Scholar]
- 474.Lalonde LF, Gajadhar AA. 2008. Highly sensitive and specific PCR assay for reliable detection of Cyclospora cayetanensis oocysts. Appl. Environ. Microbiol. 74:4354–4358. 10.1128/AEM.00032-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 475.Sulaiman IM, Ortega Y, Simpson S, Kerdahi K. 24 July 2013, posting date Genetic characterization of human-pathogenic Cyclospora cayetanensis parasites from three endemic regions at the 18S ribosomal RNA locus. Infect. Genet. Evol. [Epub ahead of print.] 10.1016/j.meegid.2013.07.015 [DOI] [PubMed] [Google Scholar]
- 476.Shields JM, Joo J, Kim R, Murphy HR. 2013. Assessment of three commercial DNA extraction kits and a laboratory-developed method for detecting Cryptosporidium and Cyclospora in raspberry wash, basil wash and pesto. J. Microbiol. Methods 92:51–58. 10.1016/j.mimet.2012.11.001 [DOI] [PubMed] [Google Scholar]
- 477.Hester JD, Varma M, Bobst AM, Ware MW, Lindquist HD, Schaefer FW. 2002. Species-specific detection of three human-pathogenic microsporidial species from the genus Encephalitozoon via fluorogenic 5′ nuclease PCR assays. Mol. Cell. Probes 16:435–444. 10.1006/mcpr.2002.0442 [DOI] [PubMed] [Google Scholar]
- 478.Wolk DM, Schneider SK, Wengenack NL, Sloan LM, Rosenblatt JE. 2002. Real-time PCR method for detection of Encephalitozoon intestinalis from stool specimens. J. Clin. Microbiol. 40:3922–3928. 10.1128/JCM.40.11.3922-3928.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 479.Menotti J, Cassinat B, Sarfati C, Liguory O, Derouin F, Molina JM. 2003. Development of a real-time PCR assay for quantitative detection of Encephalitozoon intestinalis DNA. J. Clin. Microbiol. 41:1410–1413. 10.1128/JCM.41.4.1410-1413.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 480.Menotti J, Cassinat B, Porcher R, Sarfati C, Derouin F, Molina JM. 2003. Development of a real-time polymerase-chain-reaction assay for quantitative detection of Enterocytozoon bieneusi DNA in stool specimens from immunocompromised patients with intestinal microsporidiosis. J. Infect. Dis. 187:1469–1474. 10.1086/374620 [DOI] [PubMed] [Google Scholar]
- 481.Notermans DW, Peek R, de Jong MD, Wentink-Bonnema E, Boom R, van Gool T. 2005. Detection and identification of Enterocytozoon bieneusi and Encephalitozoon species in stool and urine specimens by PCR and differential hybridization. J. Clin. Microbiol. 43:610–614. 10.1128/JCM.43.2.610-614.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 482.Lejeune A, Espern A, Phung DC, Nguyen TC, Miegeville M. 2005. Presentation of the first Enterocytozoon bieneusi intestinal microsporidia case in an HIV patient, Hanoi Vietnam. Med. Mal. Infect. 35:425–426. 10.1016/j.medmal.2005.06.003 [DOI] [PubMed] [Google Scholar]
- 483.Wumba R, Longo-Mbenza B, Mandina M, Odio WT, Biligui S, Sala J, Breton J, Thellier M. 2010. Intestinal parasites infections in hospitalized AIDS patients in Kinshasa, Democratic Republic of Congo. Parasite 17:321–328. 10.1051/parasite/2010174321 [DOI] [PubMed] [Google Scholar]
- 484.Polley SD, Boadi S, Watson J, Curry A, Chiodini PL. 2011. Detection and species identification of microsporidial infections using SYBR green real-time PCR. J. Med. Microbiol. 60:459–466. 10.1099/jmm.0.026781-0 [DOI] [PubMed] [Google Scholar]
- 485.Wumba R, Longo-Mbenza B, Menotti J, Mandina M, Kintoki F, Situakibanza NH, Kakicha MK, Zanga J, Mbanzulu-Makola K, Nseka T, Mukendi JP, Kendjo E, Sala J, Thellier M. 2012. Epidemiology, clinical, immune, and molecular profiles of microsporidiosis and cryptosporidiosis among HIV/AIDS patients. Int. J. Gen. Med. 5:603–611. 10.2147/IJGM.S32344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 486.Verweij JJ, Polderman AM, Wimmenhove MC, Gasser RB. 2000. PCR assay for the specific amplification of Oesophagostomum bifurcum DNA from human faeces. Int. J. Parasitol. 30:137–142. 10.1016/S0020-7519(99)00175-7 [DOI] [PubMed] [Google Scholar]
- 487.de Gruijter JM, van Lieshout L, Gasser RB, Verweij JJ, Brienen EA, Ziem JB, Yelifari L, Polderman AM. 2005. Polymerase chain reaction-based differential diagnosis of Ancylostoma duodenale and Necator americanus infections in humans in northern Ghana. Trop. Med. Int. Health 10:574–580. 10.1111/j.1365-3156.2005.01440.x [DOI] [PubMed] [Google Scholar]
- 488.Leles D, Araujo A, Ferreira LF, Vicente AC, Iniguez AM. 2008. Molecular paleoparasitological diagnosis of Ascaris sp. from coprolites: new scenery of ascariasis in pre-Colombian South America times. Mem. Inst. Oswaldo Cruz 103:106–108. 10.1590/S0074-02762008005000004 [DOI] [PubMed] [Google Scholar]
- 489.Sato M, Sanguankiat S, Yoonuan T, Pongvongsa T, Keomoungkhoun M, Phimmayoi I, Boupa B, Moji K, Waikagul J. 2010. Copro-molecular identification of infections with hookworm eggs in rural Lao PDR. Trans. R. Soc. Trop. Med. Hyg. 104:617–622. 10.1016/j.trstmh.2010.06.006 [DOI] [PubMed] [Google Scholar]
- 490.Ngui R, Ching LS, Kai TT, Roslan MA, Lim YA. 2012. Molecular identification of human hookworm infections in economically disadvantaged communities in peninsular Malaysia. Am. J. Trop. Med. Hyg. 86:837–842. 10.4269/ajtmh.2012.11-0446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 491.Wongratanacheewin S, Pumidonming W, Sermswan RW, Pipitgool V, Maleewong W. 2002. Detection of Opisthorchis viverrini in human stool specimens by PCR. J. Clin. Microbiol. 40:3879–3880. 10.1128/JCM.40.10.3879-3880.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 492.Stensvold CR, Saijuntha W, Sithithaworn P, Wongratanacheewin S, Strandgaard H, Ornbjerg N, Johansen MV. 2006. Evaluation of PCR based coprodiagnosis of human opisthorchiasis. Acta Trop. 97:26–30. 10.1016/j.actatropica.2005.08.008 [DOI] [PubMed] [Google Scholar]
- 493.Le TH, De NV, Blair D, Sithithaworn P, McManus DP. 2006. Clonorchis sinensis and Opisthorchis viverrini: development of a mitochondrial-based multiplex PCR for their identification and discrimination. Exp. Parasitol. 112:109–114. 10.1016/j.exppara.2005.09.012 [DOI] [PubMed] [Google Scholar]
- 494.Muller B, Schmidt J, Mehlhorn H. 2007. PCR diagnosis of infections with different species of Opisthorchiidae using a rapid clean-up procedure for stool samples and specific primers. Parasitol. Res. 100:905–909. 10.1007/s00436-006-0321-x [DOI] [PubMed] [Google Scholar]
- 495.Duenngai K, Sithithaworn P, Rudrappa UK, Iddya K, Laha T, Stensvold CR, Strandgaard H, Johansen MV. 2008. Improvement of PCR for detection of Opisthorchis viverrini DNA in human stool samples. J. Clin. Microbiol. 46:366–368. 10.1128/JCM.01323-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 496.Umesha KR, Kumar S, Parvathi A, Duenngai K, Sithithaworn P, Karunasagar I, Karunasagar I. 2008. Opisthorchis viverrini: detection by polymerase chain reaction (PCR) in human stool samples. Exp. Parasitol. 120:353–356. 10.1016/j.exppara.2008.09.004 [DOI] [PubMed] [Google Scholar]
- 497.Traub RJ, Macaranas J, Mungthin M, Leelayoova S, Cribb T, Murrell KD, Thompson RCA. 2009. A new PCR-based approach indicates the range of Clonorchis sinensis now extends to central Thailand. PLoS Negl. Trop. Dis. 3:e367. 10.1371/journal.pntd.0000367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 498.Lovis L, Mak TK, Phongluxa K, Soukhathammavong P, Sayasone S, Akkhavong K, Odermatt P, Keiser J, Felger I. 2009. PCR diagnosis of Opisthorchis viverrini and Haplorchis taichui infections in a Lao community in an area of endemicity and comparison of diagnostic methods for parasitological field surveys. J. Clin. Microbiol. 47:1517–1523. 10.1128/JCM.02011-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 499.Intapan PM, Thanchomnang T, Lulitanond V, Pongsaskulchoti P, Maleewong W. 2009. Rapid molecular detection of Opisthorchis viverrini in human fecal samples by real-time polymerase chain reaction. Am. J. Trop. Med. Hyg. 81:917–920. 10.4269/ajtmh.2009.09-0275 [DOI] [PubMed] [Google Scholar]
- 500.Wongsawad C, Phalee A, Noikong W, Chuboon S, Nithikathkul C. 2012. Co-infection with Opisthorchis viverrini and Haplorchis taichui detected by human fecal examination in Chomtong District, Chiang Mai Province, Thailand. Parasitol. Int. 61:56–59. 10.1016/j.parint.2011.10.003 [DOI] [PubMed] [Google Scholar]
- 501.Li QY, Dong SJ, Zhang WY, Lin RQ, Wang CR, Qian DX, Lun ZR, Song HQ, Zhu XQ. 2009. Sequence-related amplified polymorphism, an effective molecular approach for studying genetic variation in Fasciola spp. of human and animal health significance. Electrophoresis 30:403–409. 10.1002/elps.200800411 [DOI] [PubMed] [Google Scholar]
- 502.El-Gozamy BR, Shoukry NM. 2009. Identification of Egyptian Fasciola species by PCR and restriction endonucleases digestion of the nuclear small subunit ribosomal RNA gene. J. Egypt. Soc. Parasitol. 39:429–438 [PubMed] [Google Scholar]
- 503.Ichikawa M, Itagaki T. 2010. Discrimination of the ITS1 types of Fasciola spp. based on a PCR-RFLP method. Parasitol. Res. 106:757–761. 10.1007/s00436-010-1724-2 [DOI] [PubMed] [Google Scholar]
- 504.Ai L, Dong SJ, Zhang WY, Elsheikha HM, Mahmmod YS, Lin RQ, Yuan ZG, Shi YL, Huang WY, Zhu XQ. 2010. Specific PCR-based assays for the identification of Fasciola species: their development, evaluation and potential usefulness in prevalence surveys. Ann. Trop. Med. Parasitol. 104:65–72. 10.1179/136485910X12607012373713 [DOI] [PubMed] [Google Scholar]
- 505.Ai L, Li C, Elsheikha HM, Hong SJ, Chen JX, Chen SH, Li X, Cai XQ, Chen MX, Zhu XQ. 2010. Rapid identification and differentiation of Fasciola hepatica and Fasciola gigantica by a loop-mediated isothermal amplification (LAMP) assay. Vet. Parasitol. 174:228–233. 10.1016/j.vetpar.2010.09.005 [DOI] [PubMed] [Google Scholar]
- 506.Le TH, Nguyen KT, Nguyen NT, Doan HT, Le XT, Hoang CT, De NV. 2012. Development and evaluation of a single-step duplex PCR for simultaneous detection of Fasciola hepatica and Fasciola gigantica (family Fasciolidae, class Trematoda, phylum Platyhelminthes). J. Clin. Microbiol. 50:2720–2726. 10.1128/JCM.00662-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 507.Yahiro S, Habe S, Duong V, Odermatt P, Barennes H, Strobel M, Nakamura S. 2008. Identification of the human paragonimiasis causative agent in Lao People's Democratic Republic. J. Parasitol. 94:1176–1177. 10.1645/GE-1457.1 [DOI] [PubMed] [Google Scholar]
- 508.Prasad PK, Goswami LM, Tandon V, Chatterjee A. 2011. PCR-based molecular characterization and insilico analysis of food-borne trematode parasites Paragonimus westermani, Fasciolopsis buski and Fasciola gigantica from Northeast India using ITS2 rDNA. Bioinformation 6:64–68. 10.6026/97320630006064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 509.Tantrawatpan C, Intapan PM, Thanchomnang T, Sanpool O, Janwan P, Lulitanond V, Anamnart W, Maleewong W. 2013. Application of a real-time fluorescence resonance energy transfer polymerase chain reaction assay with melting curve analysis for the detection of Paragonimus heterotremus eggs in the feces of experimentally infected cats. J. Vet. Diagn. Invest. 25:620–626. 10.1177/1040638713497944 [DOI] [PubMed] [Google Scholar]
- 510.Tantrawatpan C, Intapan PM, Janwan P, Sanpool O, Lulitanond V, Srichantaratsamee C, Anamnart W, Maleewong W. 2013. Molecular identification of Paragonimus species by DNA pyrosequencing technology. Parasitol. Int. 62:341–345. 10.1016/j.parint.2012.11.008 [DOI] [PubMed] [Google Scholar]
- 511.Gobert GN, Chai M, Duke M, McManus DP. 2005. Copro-PCR based detection of Schistosoma eggs using mitochondrial DNA markers. Mol. Cell. Probes 19:250–254. 10.1016/j.mcp.2005.01.006 [DOI] [PubMed] [Google Scholar]
- 512.Sandoval N, Siles-Lucas M, Perez-Arellano JL, Carranza C, Puente S, Lopez-Aban J, Muro A. 2006. A new PCR-based approach for the specific amplification of DNA from different Schistosoma species applicable to human urine samples. Parasitology 133:581–587. 10.1017/S0031182006000898 [DOI] [PubMed] [Google Scholar]
- 513.Gomes AL, Melo FL, Werkhauser RP, Abath FG. 2006. Development of a real time polymerase chain reaction for quantitation of Schistosoma mansoni DNA. Mem. Inst. Oswaldo Cruz 101(Suppl 1):133–136. 10.1590/S0074-02762006000900021 [DOI] [PubMed] [Google Scholar]
- 514.Lier T, Simonsen GS, Haaheim H, Hjelmevoll SO, Vennervald BJ, Johansen MV. 2006. Novel real-time PCR for detection of Schistosoma japonicum in stool. Southeast Asian J. Trop. Med. Public Health 37:257–264 http://www.tm.mahidol.ac.th/seameo/2006_37_2/03-3713.pdf [PubMed] [Google Scholar]
- 515.Lier T, Simonsen GS, Wang T, Lu D, Haukland HH, Vennervald BJ, Hegstad J, Johansen MV. 2009. Real-time polymerase chain reaction for detection of low-intensity Schistosoma japonicum infections in China. Am. J. Trop. Med. Hyg. 81:428–432 http://www.ajtmh.org/content/81/3/428.long [PubMed] [Google Scholar]
- 516.Allam AF, Kader O, Zaki A, Shehab AY, Farag HF. 2009. Assessing the marginal error in diagnosis and cure of Schistosoma mansoni in areas of low endemicity using Percoll and PCR techniques. Trop. Med. Int. Health 14:316–321. 10.1111/j.1365-3156.2009.02225.x [DOI] [PubMed] [Google Scholar]
- 517.Xu J, Rong R, Zhang HQ, Shi CJ, Zhu XQ, Xia CM. 2010. Sensitive and rapid detection of Schistosoma japonicum DNA by loop-mediated isothermal amplification (LAMP). Int. J. Parasitol. 40:327–331. 10.1016/j.ijpara.2009.08.010 [DOI] [PubMed] [Google Scholar]
- 518.Gomes LI, Marques LH, Enk MJ, Coelho PM, Rabello A. 2009. Further evaluation of an updated PCR assay for the detection of Schistosoma mansoni DNA in human stool samples. Mem. Inst. Oswaldo Cruz 104:1194–1196. 10.1590/S0074-02762009000800021 [DOI] [PubMed] [Google Scholar]
- 519.Gomes LI, Dos Santos Marques LH, Enk MJ, de Oliveira MC, Coelho PM, Rabello A. 2010. Development and evaluation of a sensitive PCR-ELISA system for detection of Schistosoma infection in feces. PLoS Negl. Trop. Dis. 4:e664. 10.1371/journal.pntd.0000664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 520.Oliveira LM, Santos HL, Goncalves MM, Barreto MG, Peralta JM. 2010. Evaluation of polymerase chain reaction as an additional tool for the diagnosis of low-intensity Schistosoma mansoni infection. Diagn. Microbiol. Infect. Dis. 68:416–421. 10.1016/j.diagmicrobio.2010.07.016 [DOI] [PubMed] [Google Scholar]
- 521.Imai K, Koibuchi T, Kumagai T, Maeda T, Osada Y, Ohta N, Koga M, Nakamura H, Miura T, Iwamoto A, Fujii T. 2011. Cerebral schistosomiasis due to Schistosoma haematobium confirmed by PCR analysis of brain specimen. J. Clin. Microbiol. 49:3703–3706. 10.1128/JCM.01073-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 522.Guo JJ, Zheng HJ, Xu J, Zhu XQ, Wang SY, Xia CM. 2012. Sensitive and specific target sequences selected from retrotransposons of Schistosoma japonicum for the diagnosis of schistosomiasis. PLoS Negl. Trop. Dis. 6:e1579. 10.1371/journal.pntd.0001579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 523.Enk MJ, Oliveira e Silva G, Rodrigues NB. 2012. Diagnostic accuracy and applicability of a PCR system for the detection of Schistosoma mansoni DNA in human urine samples from an endemic area. PLoS One 7:e38947. 10.1371/journal.pone.0038947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 524.Fung MS, Xiao N, Wang S, Carlton EJ. 2012. Field evaluation of a PCR test for Schistosoma japonicum egg detection in low-prevalence regions of China. Am. J. Trop. Med. Hyg. 87:1053–1058. 10.4269/ajtmh.2012.12-0177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 525.Carvalho GC, Marques LH, Gomes LI, Rabello A, Ribeiro LC, Scopel KK, Tibirica SH, Coimbra ES, Abramo C. 2012. Polymerase chain reaction for the evaluation of Schistosoma mansoni infection in two low endemicity areas of Minas Gerais, Brazil. Mem. Inst. Oswaldo Cruz 107:899–902. 10.1590/S0074-02762012000700010 [DOI] [PubMed] [Google Scholar]
- 526.Espirito-Santo MC, Alvarado-Mora MV, Pinto PL, Carrilho FJ, Pinho JR, Gryschek RC. 2012. Two sequential PCR amplifications for detection of Schistosoma mansoni in stool samples with low parasite load. Rev. Inst. Med. Trop. Sao Paulo 54:245–248. 10.1590/S0036-46652012000500002 [DOI] [PubMed] [Google Scholar]
- 527.Lodh N, Mwansa JC, Mutengo MM, Shiff CJ. 2013. Diagnosis of Schistosoma mansoni without the stool: detecting DNA from filtered urine comparison of three diagnostic tests to detect Schistosoma mansoni infection from filtered urine in Zambia. Am. J. Trop. Med. Hyg. 89:46–50. 10.4269/ajtmh.13-0104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 528.Gonzalez LM, Montero E, Harrison LJ, Parkhouse RM, Garate T. 2000. Differential diagnosis of Taenia saginata and Taenia solium infection by PCR. J. Clin. Microbiol. 38:737–744 http://jcm.asm.org/content/38/2/737.long [DOI] [PMC free article] [PubMed] [Google Scholar]
- 529.Gonzalez LM, Montero E, Sciutto E, Harrison LJ, Parkhouse RM, Garate T. 2002. Differential diagnosis of Taenia saginata and Taenia solium infections: from DNA probes to polymerase chain reaction. Trans. R. Soc. Trop. Med. Hyg. 96(Suppl 1):S243–S250. 10.1016/S0035-9203(02)90083-0 [DOI] [PubMed] [Google Scholar]
- 530.Gonzalez LM, Montero E, Puente S, Lopez-Velez R, Hernandez M, Sciutto E, Harrison LJ, Parkhouse RM, Garate T. 2002. PCR tools for the differential diagnosis of Taenia saginata and Taenia solium taeniasis/cysticercosis from different geographical locations. Diagn. Microbiol. Infect. Dis. 42:243–249. 10.1016/S0732-8893(01)00356-X [DOI] [PubMed] [Google Scholar]
- 531.Rodriguez-Hidalgo R, Geysen D, Benítez-Ortiz W, Geerts S, Brandt J. 2002. Comparison of conventional techniques to differentiate between Taenia solium and Taenia saginata and an improved polymerase chain reaction-restriction fragment length polymorphism assay using a mitochondrial 12S rDNA fragment. J. Parasitol. 88:1007–1011. 10.1645/0022-3395(2002)088[1007:COCTTD]2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- 532.Nunes CM, Lima LG, Manoel CS, Pereira RN, Nakano MM, Garcia JF. 2003. Taenia saginata: polymerase chain reaction for taeniasis diagnosis in human fecal samples. Exp. Parasitol. 104:67–69. 10.1016/S0014-4894(03)00111-5 [DOI] [PubMed] [Google Scholar]
- 533.Yamasaki H, Sato MO, Sako Y, Nakao M, Nakaya K, Mamuti W, Craig PS, Margono SS, Ito A. 2003. Cysticercosis/taeniasis: recent advances in serological and molecular diagnoses. Southeast Asian J. Trop. Med. Public Health 34:98–102 http://www.tm.mahidol.ac.th/seameo/2003-34-suppl-2/18-098.pdf [PubMed] [Google Scholar]
- 534.Gonzalez LM, Montero E, Morakote N, Puente S, Diaz De Tuesta JL, Serra T, Lopez-Velez R, McManus DP, Harrison LJ, Parkhouse RM, Garate T. 2004. Differential diagnosis of Taenia saginata and Taenia saginata asiatica taeniasis through PCR. Diagn. Microbiol. Infect. Dis. 49:183–188. 10.1016/j.diagmicrobio.2004.03.013 [DOI] [PubMed] [Google Scholar]
- 535.Yamasaki H, Allan JC, Sato MO, Nakao M, Sako Y, Nakaya K, Qiu D, Mamuti W, Craig PS, Ito A. 2004. DNA differential diagnosis of taeniasis and cysticercosis by multiplex PCR. J. Clin. Microbiol. 42:548–553. 10.1128/JCM.42.2.548-553.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 536.Nunes CM, Dias AK, Dias FE, Aoki SM, de Paula HB, Lima LG, Garcia JF. 2005. Taenia saginata: differential diagnosis of human taeniasis by polymerase chain reaction-restriction fragment length polymorphism assay. Exp. Parasitol. 110:412–415. 10.1016/j.exppara.2005.03.008 [DOI] [PubMed] [Google Scholar]
- 537.Mwape KE, Phiri IK, Praet N, Muma JB, Zulu G, Van den Bossche P, de Deken R, Speybroeck N, Dorny P, Gabriel S. 2012. Taenia solium infections in a rural area of eastern Zambia—a community based study. PLoS Negl. Trop. Dis. 6:e1594. 10.1371/journal.pntd.0001594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 538.Somers R, Dorny P, Nguyen VK, Dang TC, Goddeeris B, Craig PS, Vercruysse J. 2006. Taenia solium taeniasis and cysticercosis in three communities in north Vietnam. Trop. Med. Int. Health 11:65–72. 10.1111/j.1365-3156.2005.01537.x [DOI] [PubMed] [Google Scholar]
- 539.Li T, Craig PS, Ito A, Chen X, Qiu D, Qiu J, Sato MO, Wandra T, Bradshaw H, Li L, Yang Y, Wang Q. 2006. Taeniasis/cysticercosis in a Tibetan population in Sichuan Province, China. Acta Trop. 100:223–231. 10.1016/j.actatropica.2006.11.003 [DOI] [PubMed] [Google Scholar]
- 540.Myadagsuren N, Davaajav A, Wandra T, Sandar T, Ichinkhorloo P, Yamasaki H, Sako Y, Nakao M, Sato MO, Nakaya K, Ito A. 2007. Taeniasis in Mongolia, 2002-2006. Am. J. Trop. Med. Hyg. 77:342–346 http://www.ajtmh.org/content/77/2/342.long [PubMed] [Google Scholar]
- 541.Anantaphruti MT, Yamasaki H, Nakao M, Waikagul J, Watthanakulpanich D, Nuamtanong S, Maipanich W, Pubampen S, Sanguankiat S, Muennoo C, Nakaya K, Sato MO, Sako Y, Okamoto M, Ito A. 2007. Sympatric occurrence of Taenia solium, T. saginata, and T. asiatica, Thailand. Emerg. Infect. Dis. 13:1413–1416. 10.3201/eid1309.061148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 542.Mayta H, Gilman RH, Prendergast E, Castillo JP, Tinoco YO, Garcia HH, Gonzalez AE, Sterling CR, Cysticercosis Working Group in Peru 2008. Nested PCR for specific diagnosis of Taenia solium taeniasis. J. Clin. Microbiol. 46:286–289. 10.1128/JCM.01172-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 543.Hernandez M, Gonzalez LM, Fleury A, Saenz B, Parkhouse RM, Harrison LJ, Garate T, Sciutto E. 2008. Neurocysticercosis: detection of Taenia solium DNA in human cerebrospinal fluid using a semi-nested PCR based on HDP2. Ann. Trop. Med. Parasitol. 102:317–323. 10.1179/136485908X278856 [DOI] [PubMed] [Google Scholar]
- 544.Jeon HK, Chai JY, Kong Y, Waikagul J, Insisiengmay B, Rim HJ, Eom KS. 2009. Differential diagnosis of Taenia asiatica using multiplex PCR. Exp. Parasitol. 121:151–156. 10.1016/j.exppara.2008.10.014 [DOI] [PubMed] [Google Scholar]
- 545.Nkouawa A, Sako Y, Nakao M, Nakaya K, Ito A. 2009. Loop-mediated isothermal amplification method for differentiation and rapid detection of Taenia species. J. Clin. Microbiol. 47:168–174. 10.1128/JCM.01573-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 546.Gonzalez LM, Bailo B, Ferrer E, Garcia MD, Harrison LJ, Parkhouse MR, McManus DP, Garate T. 2010. Characterization of the Taenia spp HDP2 sequence and development of a novel PCR-based assay for discrimination of Taenia saginata from Taenia asiatica. Parasit. Vectors 3:51. 10.1186/1756-3305-3-51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 547.Anantaphruti MT, Okamoto M, Yoonuan T, Saguankiat S, Kusolsuk T, Sato M, Sato MO, Sako Y, Waikagul J, Ito A. 2010. Molecular and serological survey on taeniasis and cysticercosis in Kanchanaburi Province, Thailand. Parasitol. Int. 59:326–330. 10.1016/j.parint.2010.03.007 [DOI] [PubMed] [Google Scholar]
- 548.Jeon HK, Yong TS, Sohn WM, Chai JY, Hong SJ, Han ET, Jeong HG, Chhakda T, Sinuon M, Socheat D, Eom KS. 2011. Molecular identification of Taenia tapeworms by Cox1 gene in Koh Kong, Cambodia. Korean J. Parasitol. 49:195–197. 10.3347/kjp.2011.49.2.195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 549.Devleesschauwer B, Aryal A, Joshi DD, Rijal S, Sherchand JB, Praet N, Speybroeck N, Duchateau L, Vercruysse J, Dorny P. 2012. Epidemiology of Taenia solium in Nepal: is it influenced by the social characteristics of the population and the presence of Taenia asiatica? Trop. Med. Int. Health 17:1019–1022. 10.1111/j.1365-3156.2012.03017.x [DOI] [PubMed] [Google Scholar]
- 550.Jeon HK, Yong TS, Sohn WM, Chai JY, Min DY, Yun CH, Rim HJ, Pongvongsa T, Banouvong V, Insisiengmay B, Phommasack B, Eom KS. 2013. Current status of human taeniasis in Lao People's Democratic Republic. Korean J. Parasitol. 51:259–263. 10.3347/kjp.2013.51.2.259 [DOI] [PMC free article] [PubMed] [Google Scholar]