Abstract
Leptospirosis, caused by Leptospira spp., is one of the most common zoonotic diseases in the world. We tested four recombinant proteins of Leptospira interrogans, namely, rLipL21, rLoa22, rLipL32, and rLigACon4-8, to evaluate their potential for use as antigens for the diagnosis of equine leptospirosis. We employed equine sera (n = 130) that were microscopic agglutination test (MAT) negative and sera (n = 176) that were MAT positive for the 5 serovars that most commonly cause equine leptospirosis. The sensitivity and specificity of ELISA compared to MAT were 82.39% and 86.15%, respectively, for LigACon4-8, 77.84% and 92.31%, respectively, for Loa22, 77.84% and 86.15%, respectively, for LipL32, and 84.66% and 83.85%, respectively, for LipL21. When one of the two antigens was test positive, the sensitivity and specificity of ELISA were 93.75% and 78.46%, respectively, for rLigACon4-8 and LipL32, 93.18% and 76.15%, respectively, for rLigACon4-8 and LipL21, 89.77% and 80.77%, respectively, for rLigACon4-8 and Loa22, 91.48% and 78.46%, respectively, for LipL21 and Loa22, 93.75% and 76.92%, respectively, for LipL21 and LipL32, and 90.34% and 80.77%, respectively, for Loa22 and LipL32. In conclusion, we have developed an indirect ELISA utilizing rLigACon4-8, rLoa22, rLipL32, and rLipL21 as diagnostic antigens for equine leptospirosis. The use of four antigens in the ELISA was found to be sensitive and specific, the assay was easy to perform, and the results concurred with the results of the standard Leptospira MAT.
INTRODUCTION
Leptospirosis is a worldwide zoonotic disease caused by pathogenic Leptospira spp. (1, 2). Infection in humans and animals may result from direct transmission via contaminated urine or placental fluid or from indirect exposure through contaminated soil or water (3). Although sporadic cases of renal and hepatic disease have been reported, the disease has most often been associated with abortion and equine recurrent uveitis in horses (1, 2, 4). Recently, acute respiratory failure caused by Leptospira spp. was reported in foals (5). The clinical signs of equine leptospirosis are nonspecific, which hinders the clinical diagnosis of equine leptospirosis (1, 2). Attempts to establish a definitive diagnosis of equine leptospirosis by use of laboratory tests have met with equally difficulty. Due to the fastidious and slow-growing nature of Leptospira and the difficulty in observing the organism in blood, urine, or body fluids, diagnosis of leptospirosis often depends on serologic testing, which is also difficult to interpret due to the high leptospiral seroprevalence in the equine population (6). Currently, the microscopic agglutination test (MAT) is the standard reference method for the serologic diagnosis of leptospirosis (7). However, the MAT requires considerable expertise to perform and interpret, and a panel of live strains of all common serovars and locally isolated serovars needs to be maintained, which is challenging. Thus, the MAT is usually restricted to reference laboratories (8). The current interpretive criterion for the Leptospira MAT for active infection requires a 4-fold rise in titer between acute- and convalescent-phase sera (3). It is well recognized that seroconversion or increasing antibody titers in paired serum specimens provide strong evidence for true infection, but the samples need to be taken 2 to 3 weeks apart in order to see changes in titer (3), which is not practical in the clinical setting. Commercial enzyme-linked immunosorbent assay (ELISA) kits using antigens derived from a nonpathogenic Leptospira strain (e.g., Leptospira biflexa serovar Patoc) have generally been found to have lower sensitivities than that of the MAT, because the ELISA antigens do not detect all infecting serovars (9, 10). From a previous study, we found that rLigACon can be a useful antigen for indirect ELISA (11). In this study, we evaluated 3 other recombinant antigens, rLipL21, rLoa22, and rLipL32, as well as rLigACon4-8, in an attempt to improve the specificity and sensitivity of the indirect ELISA for equine leptospirosis.
MATERIALS AND METHODS
Bacterial strain.
L. interrogans serovar Pomona (NVSL 1427-35-093002) was used for this study (12). Leptospira isolates were maintained on Ellinghausen, McCullough, Johnson, and Harris (EMJH) medium at 30°C. Growth of Leptospira was monitored using dark-field microscopy.
Sera.
All equine sera were collected from 2010 to 2012 by the New York State Animal Health Diagnostic Center (AHDC), Cornell University, Ithaca, NY. These serum samples were either positive or negative by MAT for the most common serovars causing equine leptospirosis, including L. interrogans serovar Pomona, L. kirschneri serovar Grippotyphosa, L. interrogans serovar Icterohaemorrhagiae, and L. interrogans serovar Bratislava.
Cloning, expression, and purification of proteins.
pLip32L was cloned into pGEX4T2 by using the primers ATAGCGGCCGCAGGTGCTTTCGGTGGTCTG (forward) and GCCACCTTTCGGTACCTTTTTAACC (reverse). The PCR products derived from the genes encoding LipL21 (amplified with primers CCGGAATTCTGTTCCAGTACTGACACA [forward] and ATTCTCGAGTTATTGTTTGGAAACCTCTTGAGCTTTTG [reverse]) and Loa22 (amplified with primers CGCGGATCCGAAAAAAAAGAGGAATCC [forward] and ATTCTCGAGTTATTGTTGTGGTGCGGA [reverse]) were cloned into pET28 (Invitrogen), with a 6-histidine tag at the 5′ end of the inserted DNA. The oligonucleotide primer pairs were designed for each gene, with the incorporation of XhoI at the 5′ end and EcoRI at the 3′ end (restriction sites are underlined in primer sequences). The PCR products and plasmid vector were double digested with those two enzymes and then ligated. For rLigACon4-8 construction, we used forward primer A (5′-GATCCACTCCAGCAGCCTTA-3′) and forward primer B (5′-CACTCCAGCAGCCTTA-3′) (both complementary to LigA4) plus reverse primer C (5′-AGCTTAAGAATTGCGGGAGT-3′) and reverse primer D (5′-TAAGAATTGCGGGAGT-3′) (both complementary to LigA8) to generate a sticky-ended PCR product. Two pairs of primers (A-D and B-C) were used to run PCRs individually, and the PCR products were phosphorylated by using T4 polynucleotide kinase at 37°C for 2 h and then ligated into pET28 cut with BamHI and HindIII. The obtained recombinant gene was transformed into Escherichia coli DH5α as the host strain. The DNA insert of each clone was verified by DNA sequencing, and the recombinant plasmid was then transformed into E. coli BL21(DE3) (Stratagene, Santa Clara, CA) for expression. Protein expression and purification were performed as previously described (11). The concentration of purified protein was then determined using the Bradford method, and the protein was finally used for ELISA (11).
Leptospira MAT.
The MAT was used as the reference method to determine serum titers, using live L. interrogans as antigen, as previously described (11, 13).
ELISA.
Indirect ELISA was performed as previously described (11), using purified LigACon4-8, LipL32, Loa22, and LipL21 proteins.
Western blot analysis.
Western blot analysis was performed as previously described (11), using purified rLigACon4-8, rLipL32, rLoa22, and rLipL21 antigens.
Statistical analysis.
The performance of the ELISA was evaluated using the MAT as the reference method (gold standard) (11). First, we compared the ELISA results for the individual recombinant proteins to the results of the MAT. The accuracy of ELISA relative to the MAT was measured in terms of sensitivity and specificity. Sensitivity is the probability of the respective protein test results being positive given that the MAT results were positive. Specificity is the probability of the respective protein test results being negative given that the MAT results were negative. Second, we compared sensitivities and specificities by using the ELISA results for each one, two, three, or four recombinant proteins compared to those of the MAT, using the same accuracy measures.
RESULTS
Cloning, expression, and purification of recombinant proteins.
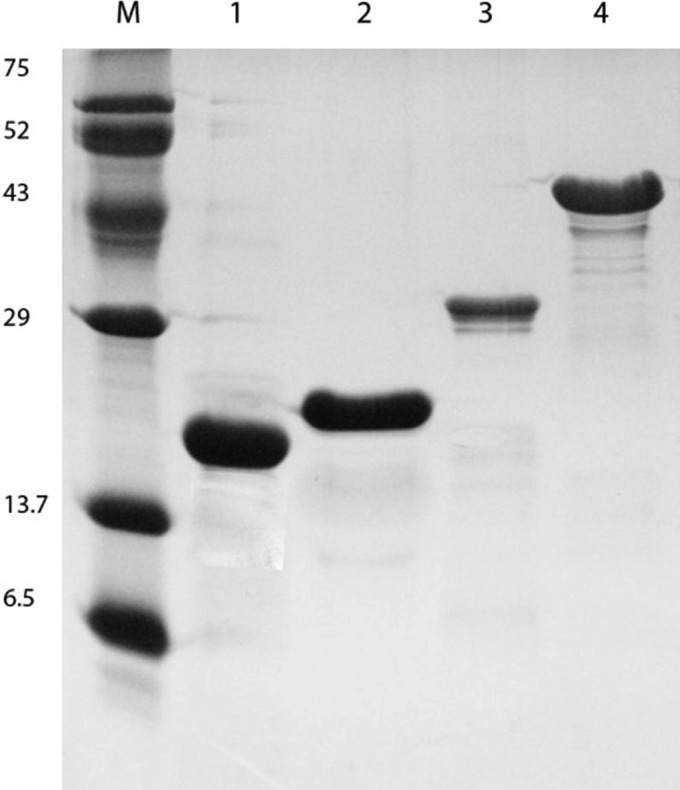
All the recombinant proteins were expressed and purified as His-tagged fusion or glutathione S-transferase (GST)-tagged proteins as previously described (11). SDS-PAGE and Coomassie blue staining of the purified recombinant proteins revealed protein bands corresponding to the expected sizes of the proteins (Fig. 1). All these proteins were expressed in soluble form, which allowed easy recovery and purification.
FIG 1.
Expression of recombinant proteins. Analysis of affinity chromatography-purified recombinant fragments was performed by Coomassie brilliant blue staining of SDS-PAGE gels. Lane M, protein marker; lane 1, LipL21; lane 2, Loa22; lane 3, LipL32; lane 4, LigACon4-8.
MAT.
Sera with titers of ≥100 against one or more serovars were considered MAT positive (11, 14). As previously reported, most seropositive cases were positive for multiple serovars (11).
Optimization of antigen concentration in ELISA.
Proteins at various concentrations (25, 50, 100, and 200 ng/well) in 100 μl coating buffer were added to each well and incubated at 4°C overnight, while the test serum concentration also varied (1:500, 1:1,000, 1:2,000, and 1:4,000 dilutions). The equine MAT-positive and -negative sera were employed as positive and negative reaction controls, respectively. A serum MAT titer of 1:800 was selected as the optimum dilution, based on its optical density at 630 nm (OD630) in the range of 0 to 1.0. For rLipL21, rLipL32, and rLoa22, a protein concentration of 100 ng/well was selected for performing the assay, while 50 ng/well was selected for the LigACon4-8protein.
Evaluation of ELISA in comparison with MAT and Western blot analysis.
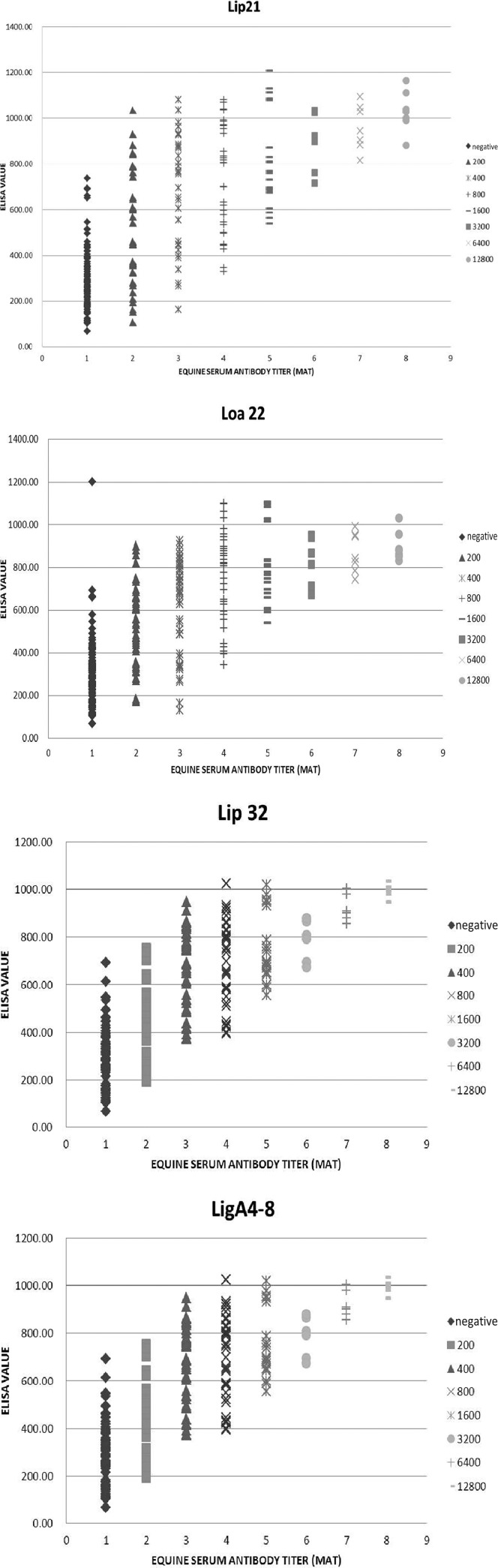
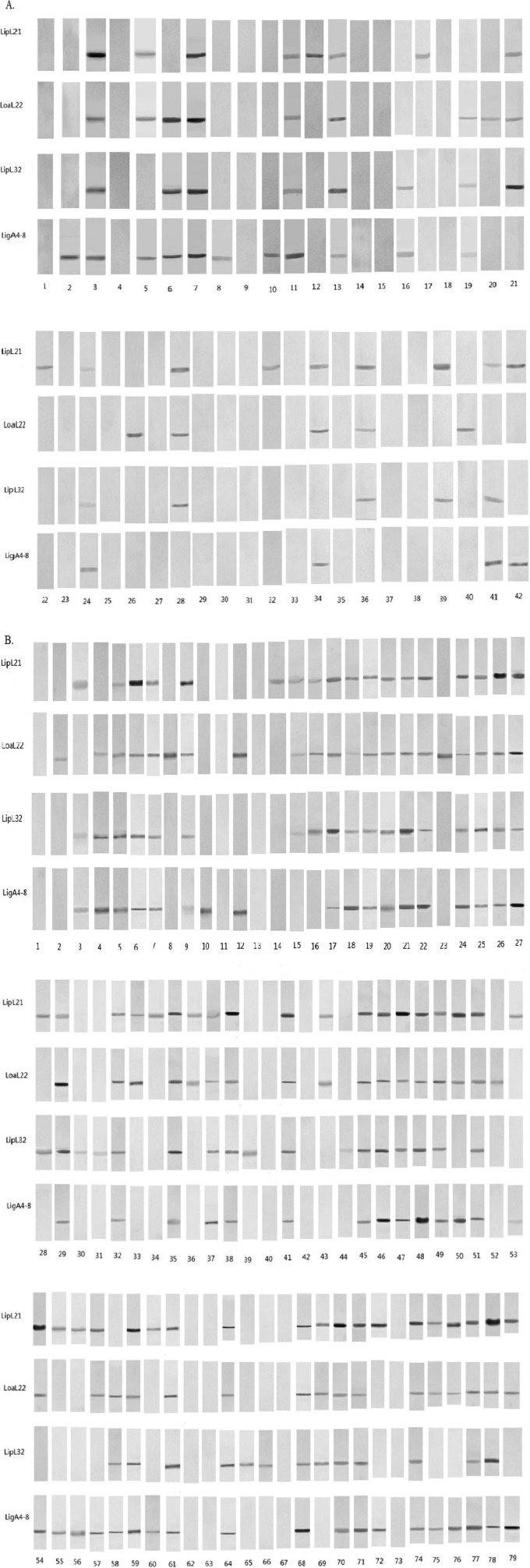
One hundred thirty negative and 176 positive serum samples were used in this experiment (306 total serum samples). All four recombinant proteins reacted with MAT-positive equine serum samples, and the results are shown in Fig. 2 and Table 1. The sensitivity and specificity of ELISA compared to the MAT were 82.39% and 86.2%, respectively, for rLigACon4-8, 77.8% and 92.3%, respectively, for rLoa22, 77.8% and 86.2%, respectively, for rLipL32, and 84.7% and 83.8%, respectively, for rLipL21 (Table 2). When two to four proteins were used and were all positive, we considered the ELISA result to be positive; the sensitivity and specificity of ELISA are shown in Table 3. The sensitivity and specificity of ELISA if one of these two to four proteins was positive and the ELISA result was considered to be positive are also shown in Table 3. Western blots of all MAT-positive and -negative samples are shown in Fig. 3. Among MAT-negative serum samples, 21, 10, 18, and 18 were ELISA positive for rLipL21, rLoa22, rLipL32, and rLigACon4-8, respectively (Table 3). Among the MAT-positive serum samples, 27, 39, 39, and 31 were ELISA negative, and 19, 25, 29, and 22 were Western blot analysis negative, for rLipL21, rLoa22, rLipL32, and rLigACon4-8, respectively (Table 4). Interestingly, five of these negative samples were positive for at least one of these four recombinant proteins (Table 5). Four of these negative serum samples had a MAT titer of 1:200, while the other had a MAT titer of 1:400.
FIG 2.
Graphs of IgG ELISA reactivities of 306 equine sera. The x axis indicates the MAT titers of the tested sera. The y axis indicates the ELISA readings (OD450).
TABLE 1.
Results of MAT, ELISA, and Western blot analyses of serum samples used in this study
| Protein | No. of serum samples |
|||||
|---|---|---|---|---|---|---|
| MAT negative | MAT and ELISA negative | MAT, ELISA, and Western blot negative | MAT positive | MAT and ELISA positive | MAT, ELISA, and Western blot positive | |
| LipL21 | 130 | 109 | 93 | 176 | 149 | 116 |
| Loa22 | 130 | 120 | 90 | 176 | 137 | 102 |
| LipL32 | 130 | 112 | 100 | 176 | 137 | 115 |
| LigACon4-8 | 130 | 112 | 97 | 176 | 145 | 116 |
TABLE 2.
Sensitivity and specificity of ELISA when a single protein was evaluated
| Protein | % Sensitivity | % Specificity |
|---|---|---|
| rLipL21 | 84.66 | 83.85 |
| rLoa22 | 77.84 | 92.31 |
| rLipL32 | 77.84 | 86.15 |
| rLigACon4-8 | 82.39 | 86.15 |
TABLE 3.
Sensitivity and specificity of ELISA when multiple proteins were evaluated
| Assay result and proteins used | % Sensitivity | % Specificity |
|---|---|---|
| Two to four proteins were evaluated, and all were positive (positive ELISA) | ||
| rLipL21 and rLoa22 | 71.02 | 97.69 |
| rLipL21 and rLipL32 | 68.75 | 93.08 |
| rLipL21 and rLigACon4-8 | 73.86 | 93.85 |
| rLoa22 and rLipL32 | 65.34 | 97.69 |
| rLoa22 and rLigACon4-8 | 70.45 | 97.69 |
| rLipL32 and rLigACon4-8 | 66.48 | 93.85 |
| rLipL21, rLoa22, and rLipL32 | 60.23 | 99.23 |
| rLipL21, rLipL32, and rLigACon4-8 | 60.80 | 96.15 |
| rLoa22, rLipL32, and rLigACon4-8 | 60.23 | 99.23 |
| rLipL21, rLoa22, LipL32, and rLigACon4-8 | 55.68 | 100.00 |
| One of two, three, or four proteins was positive, and the result was considered positive | ||
| LipL21 and Loa22 | 91.48 | 78.46 |
| LipL21 and LipL32 | 93.75 | 76.92 |
| LipL21 and LigACon4-8 | 93.18 | 76.15 |
| Loa22 and LipL32 | 90.34 | 80.77 |
| Loa22 and LigACon4-8 | 89.77 | 80.77 |
| LipL32 and LigACon4-8 | 93.75 | 78.46 |
| LipL21, Loa22, and LipL32 | 95.45 | 73.08 |
| LipL21, LipL32, and LigACon4-8 | 96.59 | 71.54 |
| LipL22, LipL32, and LigACon4-8 | 96.02 | 74.62 |
| LipL21, Loa22, LipL32, and LigACon4-8 | 97.16 | 67.69 |
FIG 3.
(A) Western blot results for sera that were MAT negative but ELISA positive. (B) Western blot results for sera that were MAT positive but ELISA negative. The numbers indicate the horse serum numbers.
TABLE 4.
Comparison of MAT-negative, ELISA- and Western blot-positive results and MAT-positive, ELISA- and Western blot-negative results
| Protein | No. of MAT-negative samples |
No. of MAT-positive samples |
||
|---|---|---|---|---|
| ELISA positive | ELISA and Western blot positive | ELISA negative | ELISA and Western blot negative | |
| LipL21 | 21 | 17 | 27 | 19 |
| Loa22 | 10 | 7 | 39 | 25 |
| LipL32 | 18 | 12 | 39 | 29 |
| LigACon4-8 | 18 | 13 | 31 | 22 |
TABLE 5.
Results of ELISA and Western blot analyses of the 79 samples that were MAT positive but ELISA negative for at least one of the four antigens
| Serum no. | ELISA result/Western blot resulta |
|||
|---|---|---|---|---|
| LipL21 | Loa22 | LipL32 | LigACon4-8 | |
| 1 | −/− | −/− | −/− | −/− |
| 2 | −/− | +/+ | −/− | +/− |
| 3 | +/+ | −/− | +/+ | +/+ |
| 4 | −/− | +/+ | +/+ | −/+ |
| 5 | −/+ | +/+ | +/+ | +/+ |
| 6 | −/+ | +/+ | +/+ | +/+ |
| 7 | −/+ | +/+ | +/+ | +/+ |
| 8 | −/− | +/+ | −/− | −/− |
| 9 | −/+ | +/+ | +/+ | +/+ |
| 10 | −/− | −/− | −/− | +/+ |
| 11 | −/− | −/− | −/− | +/− |
| 12 | −/− | +/+ | −/− | +/+ |
| 13 | −/− | −/− | −/− | −/− |
| 14 | +/+ | −/− | −/− | −/− |
| 15 | +/+ | −/+ | +/+ | −/− |
| 16 | +/+ | −/+ | +/+ | −/− |
| 17 | −/+ | +/+ | +/+ | +/+ |
| 18 | +/+ | +/+ | +/+ | −/+ |
| 19 | +/+ | −/+ | +/+ | +/+ |
| 20 | +/+ | +/+ | +/+ | −/+ |
| 21 | +/+ | +/+ | −/+ | +/+ |
| 22 | +/+ | −/+ | +/+ | +/+ |
| 23 | +/− | +/+ | −/− | −/− |
| 24 | +/+ | +/+ | +/+ | −/+ |
| 25 | +/+ | +/+ | −/+ | +/+ |
| 26 | +/+ | +/+ | +/+ | −/+ |
| 27 | +/+ | −/+ | +/+ | +/+ |
| 28 | +/+ | −/− | +/+ | −/− |
| 29 | +/+ | +/+ | +/+ | +/+ |
| 30 | −/− | −/− | +/+ | −/− |
| 31 | +/− | −/− | +/+ | −/− |
| 32 | +/+ | −/+ | +/+ | +/+ |
| 33 | +/+ | −/+ | −/− | −/− |
| 34 | +/+ | −/− | −/− | +/− |
| 35 | +/+ | +/+ | −/+ | +/+ |
| 36 | +/+ | +/+ | −/− | −/− |
| 37 | +/+ | +/+ | +/+ | −/+ |
| 38 | +/+ | +/+ | −/+ | +/+ |
| 39 | −/− | −/− | +/+ | −/− |
| 40 | −/− | −/− | −/− | −/− |
| 41 | +/+ | +/+ | +/+ | −/+ |
| 42 | −/− | −/− | −/− | −/− |
| 43 | +/+ | +/+ | −/− | −/− |
| 44 | +/− | −/− | +/+ | −/− |
| 45 | −/+ | +/+ | +/+ | +/+ |
| 46 | +/+ | +/+ | −/+ | +/+ |
| 47 | −/+ | +/+ | +/+ | +/+ |
| 48 | +/+ | +/+ | −/+ | +/+ |
| 49 | +/+ | +/+ | −/+ | +/+ |
| 50 | +/+ | +/+ | −/− | +/+ |
| 51 | +/+ | +/+ | +/+ | −/+ |
| 52 | −/− | −/+ | +/− | −/− |
| 53 | +/+ | −/− | −/− | +/+ |
| 54 | +/+ | +/+ | −/− | +/+ |
| 55 | +/+ | −/− | −/− | +/+ |
| 56 | +/+ | −/− | −/− | +/+ |
| 57 | +/+ | +/+ | −/− | +/+ |
| 58 | −/− | −/+ | +/+ | +/+ |
| 59 | −/+ | +/+ | +/+ | +/+ |
| 60 | +/+ | −/− | −/− | +/+ |
| 61 | +/+ | −/+ | +/+ | +/+ |
| 62 | −/− | −/− | −/− | +/− |
| 63 | −/− | −/− | −/− | −/− |
| 64 | +/+ | −/+ | +/+ | +/+ |
| 65 | −/− | −/− | +/+ | −/− |
| 66 | −/− | −/− | +/+ | −/− |
| 67 | +/− | −/− | −/− | +/− |
| 68 | +/+ | +/+ | +/+ | −/+ |
| 69 | +/+ | −/+ | +/+ | −/− |
| 70 | +/+ | −/+ | +/+ | +/+ |
| 71 | +/+ | −/+ | +/+ | +/+ |
| 72 | +/+ | −/− | −/− | +/+ |
| 73 | −/− | −/− | +/− | +/− |
| 74 | +/+ | +/+ | −/+ | +/+ |
| 75 | +/+ | +/+ | −/− | +/+ |
| 76 | +/+ | +/+ | −/− | +/+ |
| 77 | +/+ | +/+ | −/+ | +/+ |
| 78 | +/+ | +/+ | −/+ | +/+ |
| 79 | +/+ | +/+ | −/− | +/+ |
−, negative; +, positive.
DISCUSSION
Leptospirosis is an important zoonotic disease in the United States and throughout the world (15–17). Leptospirosis is also an important disease of horses, causing abortions and uveitis (18–20). The diagnosis of leptospirosis by MAT, bacterial culture, PCR, real-time PCR, and/or histopathological examination has been reported previously (21). Because of the serious drawbacks of these assays, numerous attempts have been made to develop an ELISA serodiagnostic test (22–30) or to develop a dual-path platform (DPP) assay (31). We previously used the rLigA protein for diagnosis of equine and canine leptospirosis (11, 32, 33). We hypothesized that the use of multiple antigens in the ELISA would improve the sensitivity and specificity of this serologic test. In this study, we used 4 different recombinant antigens, rLigACon4-8, rLipL32, rLipL21, and rLoa22, for further single-antigen ELISA evaluation of equine serum samples.
We used equine serum samples collected from the Animal Health Diagnostic Center (AHDC) at Cornell University. The AHDC indicates that five serovars occur commonly in New York State, and these are used routinely in our MAT for equine leptospirosis. From 2010 to 2012, we collected 176 MAT-positive and 130 MAT-negative equine sera for further ELISA evaluation using four antigens. The MAT targets both IgM and IgG but is skewed toward IgG (1, 34); therefore, we used the rLigACon4-8, rLipL32, rLipL21, and rLoa22 proteins as the coated antigens to establish an ELISA for improved detection of specific IgG in sera from equine patients with positive MAT titers.
A 4-fold rise in titer or seroconversion has been used as a definitive criterion for the serologic diagnosis of active leptospirosis. This requires collecting serum samples from the same animal 3 or 4 weeks later, and this delay is not practical in the clinical setting. Alternatively, a single high titer in the MAT may be taken as evidence of active infection. Therefore, the WHO Leptospirosis Burden Epidemiology Reference Group and the U.S. Centers for Disease Control and Prevention (CDC) recently defined a MAT titer of 400 in a single serum specimen as evidence supporting laboratory confirmation (35, 36). A defined positive titer is also needed for horses. However, to our knowledge, no such titer has been defined for the diagnosis of animal leptospirosis. Based on the results from this study and a previous study (11), a definition similar to that of the WHO and CDC may be applied to equine leptospirosis, i.e., a MAT titer of 400.
The use of recombinant proteins as ELISA antigens for the diagnosis of leptospirosis in humans and other mammals was reported previously (25–28, 32, 37–39). We reported the use of the Lig protein in the diagnosis of equine leptospirosis (11, 33). Hartleben et al. reported that the sensitivity and specificity of the rLipL32 ELISA for swine leptospirosis were 100% and 85.1%, respectively (26). Joseph et al. reported that the sensitivity and specificity of the rLipL21 ELISA for bovine leptospirosis were 100% and 97%, respectively (27). It has been reported that the efficiency of rLipL32 and rLoa22 in the diagnosis of human leptospirosis is 75%, whereas that of rLip21 was reported as only 68% (23). Only a few published reports detail the diagnosis of equine leptospirosis. Further studies are needed to address the diagnosis of equine leptospirosis by ELISA. Surprisingly, we found that 21, 10, 18, and 18 MAT-negative serum samples tested positive by ELISA when rLip21, rLoa22, rLip32L, and rrLigACon4-8, respectively, were used as antigens. We further evaluated these ELISA-positive serum samples by Western blot analysis and found that 17 of 21, 7 of 10, 12 of 18, and 13 of 18 of the above-mentioned samples, respectively, were also Western blot positive. This suggests that these horses were infected previously but that the MAT antibody titers to Leptospira lipopolysaccharide antigens declined to levels below the detection threshold (<1:100).
We also found that 27, 39, 39, and 31 MAT-positive serum samples were negative by ELISA when rLip21, rLoa22, rLip32L, and rrLigACon4-8, respectively, were used as antigens. However, Western blot analysis indicated that only five of these ELISA-negative samples were negative for all four recombinant antigens. All others were positive for at least one of these antigens (Table 4). It is unknown why the results were not positive for all four antigens. However, we speculate that horses infected with either different Leptospira serovars or strains have differential expression of these antigens in vivo. In conclusion, the ELISA developed in this research, utilizing rLip21, rLoa22, rLip32L, and rRLigACon4-8 as antigens, could increase the sensitivity and specificity of ELISA for detection of leptospirosis in horses. This ELISA may be able to replace or supplement the current equine MAT for the diagnosis of equine leptospirosis in the near future, after further validation with more defined equine serum samples.
ACKNOWLEDGMENTS
This work was supported in part by the Biotechnology Research and Development Corporation (BRDC) and the New York State Science and Technology Foundation and Center of Advanced Technology (CAT) to Y.-F.C. C.Y. was supported by a scholarship from the China Scholarship Council (grant 2009850564).
Footnotes
Published ahead of print 22 January 2014
REFERENCES
- 1.Palaniappan RU, Ramanujam S, Chang YF. 2007. Leptospirosis: pathogenesis, immunity, and diagnosis. Curr. Opin. Infect. Dis. 20:284–292. 10.1097/QCO.0b013e32814a5729 [DOI] [PubMed] [Google Scholar]
- 2.Faisal SM, McDonough SP, Chang YF. 2012. Leptospira: invasion, pathogenesis and persistence, p 143–172 In Embers ME. (ed), The pathogenic spirochetes: strategies for evasion of host immunity and persistence. Springer Science, New York, NY [Google Scholar]
- 3.Levett PN. 2001. Leptospirosis. Clin. Microbiol. Rev. 14:296–326. 10.1128/CMR.14.2.296-326.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Divers TJ, Byars TD, Shin SJ. 1992. Renal dysfunction associated with infection of Leptospira interrogans in a horse. J. Am. Vet. Med. Assoc. 201:1391–1392 [PubMed] [Google Scholar]
- 5.Broux B, Torfs S, Wegge B, Deprez P, van Loon G. 2012. Acute respiratory failure caused by Leptospira spp. in 5 foals. J. Vet. Intern. Med. 26:684–687. 10.1111/j.1939-1676.2012.00902.x [DOI] [PubMed] [Google Scholar]
- 6.Toyokawa T, Ohnishi M, Koizumi N. 2011. Diagnosis of acute leptospirosis. Expert Rev. Anti Infect. Ther. 9:111–121. 10.1586/eri.10.151 [DOI] [PubMed] [Google Scholar]
- 7.Ko AI, Goarant C, Picardeau M. 2009. Leptospira: the dawn of the molecular genetics era for an emerging zoonotic pathogen. Nat. Rev. Microbiol. 7:736–747. 10.1038/nrmicro2208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Woodward MJ, Swallow C, Kitching A, Dalley C, Sayers AR. 1997. Leptospira hardjo serodiagnosis: a comparison of MAT, ELISA and Immunocomb. Vet. Rec. 141:603–604 [PubMed] [Google Scholar]
- 9.Myers DM, Coltorti EA. 1978. Broadly reacting precipitating and agglutinating antigen of leptospirae. J. Clin. Microbiol. 8:580–590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fairbrother JM. 1984. Serological interrelationship of Leptospira serovar and genus-specific antigens by enzyme-linked immunosorbent assay. J. Clin. Microbiol. 20:1089–1093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yan W, Saleem MH, McDonough P, McDonough SP, Divers TJ, Chang YF. 2013. Development of an enzyme-linked immunosorbent assay using a recombinant LigA fragment comprising repeat domains 4 to 7.5 as an antigen for diagnosis of equine leptospirosis. Clin. Vaccine Immunol. 20:1143–1149. 10.1128/CVI.00245-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Palaniappan RU, McDonough SP, Divers TJ, Chen CS, Pan MJ, Matsumoto M, Chang YF. 2006. Immunoprotection of recombinant leptospiral immunoglobulin-like protein A against Leptospira interrogans serovar Pomona infection. Infect. Immun. 74:1745–1750. 10.1128/IAI.74.3.1745-1750.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cole JR, Jr, Sulzer CR, Pursell AR. 1973. Improved microtechnique for the leptospiral microscopic agglutination test. Appl. Microbiol. 25:976–980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yan W, Faisal SM, Divers T, McDonough SP, Akey B, Chang YF. 2010. Experimental Leptospira interrogans serovar Kennewicki infection of horses. J. Vet. Intern. Med. 24:912–917. 10.1111/j.1939-1676.2010.00507.x [DOI] [PubMed] [Google Scholar]
- 15.Jobbins SE, Sanderson CE, Alexander KA. 2014. Leptospira interrogans at the human-wildlife interface in northern Botswana: a newly identified public health threat. Zoonoses Public Health 61:113–123. 10.1111/zph.12052 [DOI] [PubMed] [Google Scholar]
- 16.Minor K, Mohan A. 2013. Severe leptospirosis: treatment with intravenous corticosteroids and supportive care. Am. J. Emerg. Med. 31:449.e1–449.e2. 10.1016/j.ajem.2012.06.016 [DOI] [PubMed] [Google Scholar]
- 17.Bharti AR, Nally JE, Ricaldi JN, Matthias MA, Diaz MM, Lovett MA, Levett PN, Gilman RH, Willig MR, Gotuzzo E, Vinetz JM. 2003. Leptospirosis: a zoonotic disease of global importance. Lancet Infect. Dis. 3:757–771. 10.1016/S1473-3099(03)00830-2 [DOI] [PubMed] [Google Scholar]
- 18.Verma A, Stevenson B. 2012. Leptospiral uveitis—there is more to it than meets the eye! Zoonoses Public Health 59(Suppl 2):132–141. 10.1111/j.1863-2378.2011.01445.x [DOI] [PubMed] [Google Scholar]
- 19.Timoney JF, Kalimuthusamy N, Velineni S, Donahue JM, Artiushin SC, Fettinger M. 2011. A unique genotype of Leptospira interrogans serovar Pomona type Kennewicki is associated with equine abortion. Vet. Microbiol. 150:349–353. 10.1016/j.vetmic.2011.02.049 [DOI] [PubMed] [Google Scholar]
- 20.Donahue JM, Williams NM. 2000. Emergent causes of placentitis and abortion. Vet. Clin. North Am. Equine Pract. 16:443–456 [DOI] [PubMed] [Google Scholar]
- 21.Schreier S, Doungchawee G, Chadsuthi S, Triampo D, Triampo W. 2013. Leptospirosis: current situation and trends of specific laboratory tests. Expert Rev. Clin. Immunol. 9:263–280. 10.1586/eci.12.110 [DOI] [PubMed] [Google Scholar]
- 22.Bomfim MR, Ko A, Koury MC. 2005. Evaluation of the recombinant LipL32 in enzyme-linked immunosorbent assay for the serodiagnosis of bovine leptospirosis. Vet. Microbiol. 109:89–94. 10.1016/j.vetmic.2005.05.002 [DOI] [PubMed] [Google Scholar]
- 23.Chalayon P, Chanket P, Boonchawalit T, Chattanadee S, Srimanote P, Kalambaheti T. 2011. Leptospirosis serodiagnosis by ELISA based on recombinant outer membrane protein. Trans. R. Soc. Trop. Med. Hyg. 105:289–297. 10.1016/j.trstmh.2011.01.008 [DOI] [PubMed] [Google Scholar]
- 24.Flannery B, Costa D, Carvalho FP, Guerreiro H, Matsunaga J, Da Silva ED, Ferreira AG, Riley LW, Reis MG, Haake DA, Ko AI. 2001. Evaluation of recombinant Leptospira antigen-based enzyme-linked immunosorbent assays for the serodiagnosis of leptospirosis. J. Clin. Microbiol. 39:3303–3310. 10.1128/JCM.39.9.3303-3310.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Croda J, Ramos JG, Matsunaga J, Queiroz A, Homma A, Riley LW, Haake DA, Reis MG, Ko AI. 2007. Leptospira immunoglobulin-like proteins as a serodiagnostic marker for acute leptospirosis. J. Clin. Microbiol. 45:1528–1534. 10.1128/JCM.02344-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hartleben CP, Leal FM, Monte LG, Hartwig DD, Seixas FK, Vasconcellos SA, Brihuega B, Dellagostin OA. 2013. Serological analysis by enzyme-linked immunosorbent assay using recombinant antigen LipL32 for the diagnosis of swine leptospirosis. Curr. Microbiol. 66:106–109. 10.1007/s00284-012-0237-x [DOI] [PubMed] [Google Scholar]
- 27.Joseph S, Thomas N, Thangapandian E, Singh VP, Verma R, Srivastava SK. 2012. Evaluation and comparison of native and recombinant LipL21 protein-based ELISAs for diagnosis of bovine leptospirosis. J. Vet. Sci. 13:99–101. 10.4142/jvs.2012.13.1.99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Oliveira TR, Longhi MT, de Morais ZM, Romero EC, Blanco RM, Kirchgatter K, Vasconcellos SA, Nascimento AL. 2008. Evaluation of leptospiral recombinant antigens MPL17 and MPL21 for serological diagnosis of leptospirosis by enzyme-linked immunosorbent assays. Clin. Vaccine Immunol. 15:1715–1722. 10.1128/CVI.00214-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sankar S, Harshan HM, Somarajan SR, Srivastava SK. 2010. Evaluation of a recombinant LigB protein of Leptospira interrogans serovar Canicola in an enzyme-linked immunosorbent assay for the serodiagnosis of bovine leptospirosis. Res. Vet. Sci. 88:375–378. 10.1016/j.rvsc.2009.11.004 [DOI] [PubMed] [Google Scholar]
- 30.Srimanote P, Wongdeethai N, Jieanampunkul P, Samonkiert S, Leepiyasakulchai C, Kalambaheti T, Prachayasittikul V. 2008. Recombinant ligA for leptospirosis diagnosis and ligA among the Leptospira spp. clinical isolates. J. Microbiol. Methods 72:73–81. 10.1016/j.mimet.2007.10.012 [DOI] [PubMed] [Google Scholar]
- 31.Nabity SA, Ribeiro GS, Aquino CL, Takahashi D, Damiao AO, Goncalves AH, Miranda-Filho DB, Greenwald R, Esfandiari J, Lyashchenko KP, Reis MG, Medeiros MA, Ko AI. 2012. Accuracy of a dual path platform (DPP) assay for the rapid point-of-care diagnosis of human leptospirosis. PLoS Negl. Trop. Dis. 6:e1878. 10.1371/journal.pntd.0001878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Palaniappan RU, Chang YF, Hassan F, McDonough SP, Pough M, Barr SC, Simpson KW, Mohammed HO, Shin S, McDonough P, Zuerner RL, Qu J, Roe B. 2004. Expression of leptospiral immunoglobulin-like protein by Leptospira interrogans and evaluation of its diagnostic potential in a kinetic ELISA. J. Med. Microbiol. 53:975–984. 10.1099/jmm.0.45568-0 [DOI] [PubMed] [Google Scholar]
- 33.Palaniappan RU, Chang YF, Jusuf SS, Artiushin S, Timoney JF, McDonough SP, Barr SC, Divers TJ, Simpson KW, McDonough PL, Mohammed HO. 2002. Cloning and molecular characterization of an immunogenic LigA protein of Leptospira interrogans. Infect. Immun. 70:5924–5930. 10.1128/IAI.70.11.5924-5930.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Faine S, Adher B, Bloin C, Perolat P. 1999. Leptospira and leptospirosis, 2nd ed. MedSci, Melbourne, Australia [Google Scholar]
- 35.WHO Leptospirosis Burden Epidemiology Reference Group. 13 July 2010, posting date Zoonoses and veterinary public health. WHO, Geneva, Switzerland: http://www.who.int [Google Scholar]
- 36.CDC Public Health Surveillance and Informatics Program Office. 2013. Leptospirosis (Leptospira interrogans). CDC, Atlanta, GA [Google Scholar]
- 37.Subathra M, Senthilkumar TM, Ramadass P. 2013. Recombinant OmpL1 protein as a diagnostic antigen for the detection of canine leptospirosis. Appl. Biochem. Biotechnol. 169:431–437. 10.1007/s12010-012-9973-4 [DOI] [PubMed] [Google Scholar]
- 38.Hartwig DD, Seixas FK, Cerqueira GM, McBride AJ, Dellagostin OA. 2011. Characterization of the immunogenic and antigenic potential of putative lipoproteins from Leptospira interrogans. Curr. Microbiol. 62:1337–1341. 10.1007/s00284-010-9865-1 [DOI] [PubMed] [Google Scholar]
- 39.Lin X, Chen Y, Yan J. 2008. Recombinant multiepitope protein for diagnosis of leptospirosis. Clin. Vaccine Immunol. 15:1711–1714. 10.1128/CVI.00189-08 [DOI] [PMC free article] [PubMed] [Google Scholar]