Abstract
Hydrogen peroxide production is a well-known trait of many bacterial species associated with the human body. In the presence of oxygen, the probiotic lactic acid bacterium Lactobacillus johnsonii NCC 533 excretes up to 1 mM H2O2, inducing growth stagnation and cell death. Disruption of genes commonly assumed to be involved in H2O2 production (e.g., pyruvate oxidase, NADH oxidase, and lactate oxidase) did not affect this. Here we describe the purification of a novel NADH-dependent flavin reductase encoded by two highly similar genes (LJ_0548 and LJ_0549) that are conserved in lactobacilli belonging to the Lactobacillus acidophilus group. The genes are predicted to encode two 20-kDa proteins containing flavin mononucleotide (FMN) reductase conserved domains. Reductase activity requires FMN, flavin adenine dinucleotide (FAD), or riboflavin and is specific for NADH and not NADPH. The Km for FMN is 30 ± 8 μM, in accordance with its proposed in vivo role in H2O2 production. Deletion of the encoding genes in L. johnsonii led to a 40-fold reduction of hydrogen peroxide formation. H2O2 production in this mutant could only be restored by in trans complementation of both genes. Our work identifies a novel, conserved NADH-dependent flavin reductase that is prominently involved in H2O2 production in L. johnsonii.
INTRODUCTION
Hydrogen peroxide (H2O2) production is a well-known capacity of several bacterial species that are associated with the human body. Some of these H2O2-forming species are opportunistic pathogens or pathobionts, such as Streptococcus pyogenes (1), Streptococcus mutans (2, 3), and Streptococcus pneumoniae (4, 5). Other H2O2-producing species or strains have been proposed to have probiotic properties, such as Bifidobacterium bifidum (6) and Lactobacillus johnsonii (7), or are prevalent in the commensal vaginal microbiota, such as Lactobacillus crispatus, Lactobacillus jensenii, and Lactobacillus gasseri (8).
Accumulation of H2O2 mainly occurs in species that lack the main hydrogen peroxide-scavenging enzymes, such as catalase and NADH peroxidase. Analogously, when the genes encoding these enzymes are deleted from the Escherichia coli genome, hydrogen peroxide is generated upon oxygenation and accumulates in the extracellular growth medium (9, 10). Hydrogen peroxide is mainly produced in central carbon and energy metabolism by oxidases, including pyruvate oxidase (Pox), lactate oxidase (Lox), and NADH oxidases (Nox) (11). For example, the activity of the lactate oxidase encoded by S. pyogenes is primarily responsible for H2O2 production in cells that are depleted for glucose (12, 13), whereas H2O2 production by S. pneumoniae is due to pyruvate oxidase activity (4). In several species, H2O2-producing NADH oxidases have been identified: for instance, in Thermus thermophilus (14), S. mutans (2), and Amphibacillus xylanus (15).
Species that accumulate H2O2 or other reactive oxygen species (ROS) upon exposure to molecular oxygen generally have an energy metabolism that is adapted to anaerobic environments. The proteins that catalyze the low-potential redox reactions in anaerobic energy metabolism, such as fumarate and nitrate respiration, commonly carry low-potential metal clusters and solvent-exposed flavin cofactors that readily react with oxygen and contribute to the generation of ROS. H2O2 and superoxide (O2−) belong to the strongest oxidant species and can accelerate the rate of ROS generation, the chemistry of which has been reviewed previously (16).
This study focuses on the H2O2-producing species L. johnsonii. This organism is applied as a probiotic supplement in the food industry (17). The strain was isolated from the human intestine, where it interacts with the host epithelium as well as with other microbes (18–20). The gastrointestinal (GI) tract is predominantly an anaerobic niche, but the presence of oxygen gradients in the proximity of the mucosal surfaces is well established (21, 22). Hydrogen peroxide derived from species like L. johnsonii may play a role in these environments. Several studies have speculated on the effect that H2O2 may exert on the host as well as on the microbiome. Some authors propose that it can directly damage the epithelium (23, 24) and cause cell death of other bacteria (5, 7). Others suggest that H2O2 accumulation may contribute to the maintenance of a normal and homeostatic microbiota. Especially for the vaginal microbiota, strong evidence exists that women carrying H2O2-producing lactobacilli are less prone to develop bacterial vaginosis (25, 26), which is a very common disease and an independent risk factor for the acquisition of sexually transmitted disease and preterm birth (27, 28).
Despite the data that support this hypothesis, the mechanism for the proposed homeostatic effect of H2O2-producing lactobacilli in the microbiota remains largely unknown. It has been suggested that H2O2 can contribute to the anti-inflammatory effect of commensal and probiotic bacteria through its influence on the peroxisome proliferator activated receptor γ (PPAR-γ), which plays a central role in regulation of intestinal inflammation and homeostasis (29, 30). Expression of PPAR-γ is induced in vivo and in vitro by the presence of L. crispatus and is inhibited by the addition of either catalase or glutathione, pinpointing H2O2 as the responsible factor for the observed induction (31). A recent study on development of type 1 diabetes in rats substantiated the role in immune modulation by bacterially derived hydrogen peroxide. Here, H2O2 directly affected the activity of indoleamine 2,3-dioxygenase, which is an important immune modulator (32).
Among the important microbial groups involved in H2O2 production in the vaginal and GI tract microbiota, members of the L. acidophilus group are frequently encountered. This group of lactobacilli encompasses several closely related species (33, 34), including those that are proposed to confer probiotic effects on consumers (L. johnsonii, L. gasseri, and L. acidophilus), as well as several important organisms in food fermentations (Lactobacillus delbruecki subsp. bulgaricus, Lactobacillus kefiranofaciens, and Lactobacillus helveticus). Although many studies have reported on the H2O2 production by species of the L. acidophilus group, our understanding of the enzymatic reactions and mechanisms underlying these observations remains limited to the notion that NADH and flavin are involved in the reaction (35, 36) and that it is catalyzed by a protein that is constitutively expressed (8). The enzymes that catalyze the H2O2-generating reactions remain uncharacterized, to date.
In this study, we identify a novel NADH-dependent flavin reductase as the primary source for H2O2 in anaerobically grown L. johnsonii NCC 533, a member of the L. acidophilus group, upon exposure to oxygen. The enzyme is encoded by two small consecutive genes that show high similarity and are conserved throughout the L. acidophilus group. Mutation of these genes in L. johnsonii NCC 533 led to a strain that failed to produce H2O2 upon exposure to molecular oxygen.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
L. johnsonii NCC 533 was obtained from the Nestec Culture Collection and cultured in commercial MRS medium (Merck, Whitehouse Station, NJ) (37) at 37°C under static conditions, with minimal headspace for 16 h. The deletion strains NCC 9333, NCC 9334, NCC 9337, and NCC 9359 were precultured in MRS containing 5 ng ml−1 erythromycin, while 5 ng ml−1 chloramphenicol was added for the NCC 9359 strains carrying plasmid pDP1016, pDP1017, or pDP1019.
Growth and H2O2 production.
Cells were grown overnight in closed static tubes at 37°C in LAPTg medium. (20 g liter−1 glucose, 10 g liter−1 yeast extract, 10 g liter−1 Bacto peptone, 10 g liter−1 Bacto tryptone, plus 1 g liter−1 Tween 80). This medium was used instead of the regular MRS medium for lactobacilli, because the meat extract in MRS was found to interfere with the enzymatic assay for H2O2. Cell density was determined by measurement of the optical density at 600 nm (OD600). H2O2 concentrations were determined using the phenol red assay (described below).
Cell extracts.
Bacterial cultures were grown in 1-liter bottles that were filled to the top with MRS medium, to minimize the headspace volume. Cultures were inoculated with 5-ml overnight precultures in the same medium and incubated for 24 h at 37°C with continuous stirring. Cells were harvested by centrifugation (5 min, 2,600 × g, 4°C), and the cell pellets were suspended in 50 ml of 50 mM potassium phosphate buffer (pH 7.0) with 2 mM EDTA and 25 mM NaCl. Lysozyme (Sigma-Aldrich, St. Louis, MO) was added to a final concentration of 1 mg ml−1 and incubated for 30 min at 37°C. Subsequently, cells were disrupted by 3 rounds of 1 min of sonication at 100 W (Branson Ultrasonics, Danbury, CT) and cooled on ice water. Crude cell debris was removed from the disrupted cell suspension by low-speed centrifugation (5 min, 2,600 × g, 4°C), followed by ultracentrifugation of the supernatant (60 min, 165,000 × g, 4°C), generating the cell extract (supernatant) that was used in subsequent purification steps and enzyme assays. Protein concentrations of cell extracts were determined using the MicroBCA (microbicinchoninic acid) assay kit (Thermo Fisher Scientific, Inc., Waltham, MA).
Alternatively, 50-ml overnight anaerobic cultures in MRS were centrifuged, suspended in 50 mM potassium phosphate buffer (pH 7.0), and transferred to screw-cap tubes with 100-mg zirconium beads. Cells were disrupted in 3 rounds for 20 s, and cell debris was removed by centrifugation (10 min, 21,500 × g, 4°C). Protein concentrations of cell extracts were determined using the MicroBCA assay kit (Thermo Fisher Scientific, Inc.). These extracts showed a 2-fold-lower enzyme activity overall. They were employed for analysis of the activity, Km determination, and SDS gel electrophoresis.
Protein purification: ammonium sulfate precipitation, Q column, gel filtration, and SDS gel electrophoresis.
Cell extracts were placed in a beaker at 4°C, and ammonium sulfate was added slowly under continuous stirring until reaching intermediate steps (30, 50, 70, and 90%) of saturation. Next, the cell extract was left without stirring on ice for 20 min and subsequently spun down (10 min, 12,000 × g, 4°C). The precipitate was suspended in 50 mM potassium phosphate buffer (pH 7.0) and dialyzed overnight at 4°C against 3 liters 20 mM Tris buffer (pH 8.0).
Anion-exchange chromatography was carried out with an Äkta fast protein liquid chromatography (FPLC) system (GE Healthcare, Little Chalfont, Buckinghamshire, United Kingdom) fitted with a Hi-Trap Q HP 5-ml column (GE Healthcare). As a loading buffer, 20 mM Tris (pH 8.0) was used at a rate of 4 ml min−1, and a linear gradient was applied from 0 to 1 M NaCl in 20 column volumes. The fraction showing NADH oxidase activity (see below) was concentrated to 0.2 ml using a 10K Corning Spin-XUF6 column. This concentrated fraction was subjected to further fractionation by size exclusion chromatography using a Superdex 200 HR 10/30 column (GE Healthcare) in 20 mM Tris buffer (pH 8.0) with 250 mM NaCl at 0.5 ml min−1.
For the SDS gel electrophoresis, cell extracts were boiled for 5 min with SDS sample buffer (50 mM Tris-HCl [pH 6.8], 100 mM dithiothreitol [DTT], 2% SDS, 0.1% bromophenol blue, 10% glycerol) and loaded on a 15% SDS gel with a 7% stacking gel on a Hoefer system (Thermo Scientific). The amount of sample that was applied was corrected for the variations in OD600 of the culture. The prestained marker Pageruler Plus from Fermentas (Thermo Scientific) was used. Gels were stained with PageBlue protein staining solution (Fermentas Coomassie G-250 dye).
Enzyme activity assay.
NADH-dependent flavin reductase activity in crude extract (for preparation, see above) was analyzed by determination of the NADH dissipation, as well as through the determination of the final H2O2 concentration. NADH and NADPH oxidation was measured by monitoring the absorption at 340 nm at 37°C in a 200-μl reaction mixture with 500 μM NADH or NADPH. As a flavin source, either 250 μM flavin adenine dinucleotide (FAD) or 25 μM FAD, flavin mononucleotide (FMN), or riboflavin was added. The reaction mixture was buffered by 50 mM potassium phosphate buffer at pH 7.0.
For H2O2 measurement, the same reaction mixture in a 200-μl volume was used with a lower NADH concentration (250 μM) to prevent oxygen from becoming the limiting substrate. After 10 min of incubation, the H2O2 concentration was determined with a phenol red enzymatic assay by transferring 20 μl of sample to 180 μl of a reaction mixture containing 5 μg ml−1 horseradish peroxidase (Roche, Penzberg, Germany) and 30 μM phenol red in water. After 5 min of reaction, pH was increased by the addition of 10 μl 1 M NaOH. Absorption was determined at 620 nm.
These enzymatic activity assays were employed to detect protein activity in the fractions that were obtained from the three protein purification steps. Furthermore, the assays were used to determine the enzyme activity level in cell extracts of mutant strains. In the latter instance, 25 μM FMN was used as the flavin source. Enzyme activities are expressed as specific activity per milligram of protein in the cell extract per minute and were measured in triplicate.
The specificity constant Km was determined by measuring the NADH consumption rate of the cell extract in the presence of various concentrations of FAD, FMN, and riboflavin (2.5 to 250 μM). The Km is calculated by fitting a hyperbolic curve [Vmax × Cs/(Km + Cs), where Cs is the flavin concentration] and optimization for Vmax and Km using the Solver function in Microsoft Excel (R2 > 0.96). Averages and standard deviations are calculated from technical triplicates.
Digestion, MS analysis, and protein identification.
Semipurified protein samples were digested with an in-house protocol using 1 mg of trypsin (modified to prevent autodigestion; Promega, Madison, WI) per 50 mg of protein in a 0.1 M Tris (pH 7.5) buffer following alkylation of the cysteine residues with DTT and iodoacetamide. Trypsin digestion was stopped after 16 h by the addition of 10% trifluoroacetic acid (TFA) to a final concentration of 1%. Tryptic peptides were purified using an 80-μg capacity OMIX tip (Varian, Agilent) and collected in a volume of 30 μl 50% acetonitrile (ACN)–0.1% TFA.
Mass spectrometry analysis of the peptide samples was performed with a Micromass Q-TOF1 (quadrupole time of flight) mass spectrometer (Waters, Milford, MA) coupled to a nano-liquid chromatography (nano-LC) system (LC Packings, Dionex, Sunnyvale, CA). The peptides were separated on a nano-analytical column (75-μm inside diameter [i.d.], 25-cm-length C18 PepMap; Dionex) using a gradient of 0 to 50% acetonitrile and 0.1% formic acid. The LC eluent flow of 300 nl min−1 was directly infused into the Q-TOF1 spectrometer, operating in data-dependent MS and tandem MS (MS/MS) modes. Low-energy collision-induced dissociation (CID) of selected precursor ions was used to obtain fragmentation spectra of the peptides. After processing the raw data with the Masslynx software (Micromass; Waters), the resulting peaklist (.pkl file) was used to search in the NCBInr database with MASCOT online.
The search parameters were a fixed modification of carbamidomethyl for cysteine, variable modifications of oxidized methionine, trypsin with the allowance of one missed cleavage, peptide and MS/MS tolerance of ±0.3 Da, and a peptide charge state of +1. Probability-based MASCOT scores were used to evaluate the protein identifications.
Construction of L. johnsonii deletion strains.
An overview of the mutants and plasmids used in this study can be found in Table 1. An overview of all primers can be found in Table S1 in the supplemental material. The genome sequence of L. johnsonii is available in GenBank under accession no. AE017198 (17).
TABLE 1.
List of strains and plasmids used in this study
| Strain or plasmid | Description |
|---|---|
| Strains | |
| NCC 533 | Lactobacillus johnsonii strain from the Nestec Culture Collection |
| NCC 9333 | Δpox (38) |
| NCC 9334 | ΔLJ_1826 (predicted to encode lactate oxidase; Lox) |
| NCC 9337 | ΔLJ_1254 ΔLJ_1255 (predicted to encode NADH oxidase; Nox) |
| NCC 9359 | ΔLJ_0548 ΔLJ_0549 (predicted to encode NADH flavin reductase) |
| Plasmids | |
| pDP749 | Temperature-sensitive, allele exchange plasmid for L. johnsonii NCC 533 (39) |
| pDP889 | pDP749 construct for lox (LJ_1826) deletion |
| pDP902 | pDP749 construct for nox (LJ_1254 and LJ_1255) deletion |
| pDP1010 | pDP749 construct for LJ_0548 and LJ_0549 deletion |
| pDP794 | pNZ124-based expression plasmid with LJ_0045 promoter and LJ_1125 terminator |
| pDP1016 | pDP794 with LJ_0548 expression plasmid |
| pDP1017 | pDP794 with LJ_0549 expression plasmid |
| pDP1019 | pDP794 with LJ_0548 and LJ_0549 expression plasmid |
The construction of the pox deletion strain NCC 9333 has been described previously (38). The deletion of the gene LJ_1826 encoding a predicted lactate oxidase enzyme was achieved similarly: the 5′ homology region of the LJ_1826 gene was amplified from L. johnsonii NCC 533 genomic DNA using primers A and B. The 1,077-bp amplicon was SacI-BamHI digested and cloned in SacI-BamHI-digested pDP749, yielding an intermediate plasmid. The 3′ region of the LJ_1826 gene was amplified using the primers C plus D, and the 1,170-bp amplicon was digested with PstI-KpnI and cloned in the similarly digested intermediate plasmid to yield the lox deletion plasmid pDP889. Plasmid pDP889 isolated from Lactococcus lactis was used to transform NCC 533 (39) and loop-in/loop-out gene replacement was achieved as described previously (58). The deletion was confirmed by PCR analysis, and the deletion strain was named NCC 9334.
In L. johnsonii NCC 533, NADH oxidase is predicted to be encoded by the genes LJ_1254 and LJ_1255. Deletion of LJ_1254 and LJ_1255 was achieved in the same way as for the Lox-encoding gene LJ_1826: the 1,008 bp at the 5′-end region of the LJ_1255 gene was amplified using primers E and F, and the 993 bp at the 3′-end region of the LJ_1254 gene was amplified using primers G plus H. These amplicons were cloned into pDP749 to give plasmid pDP902 and used to produce the nox deletion strain NCC 9337.
The deletion of LJ_0548 and LJ_0549 was achieved by amplification of 1,062 bp at the 5′-end region of the LJ_0548 gene with primers I plus J, and the 1,098 bp at the 3′-end region of the LJ_0549 gene was amplified using primers K plus L. These were cloned into pDP749 to give plasmid pDP1010, which was used to produce the LJ_0548 LJ_0549 deletion strain NCC 9359.
Construction of the L. johnsonii overexpression strains.
The L. johnsonii expression plasmid pDP794 was constructed as follows: the predicted bidirectional terminator situated between the LJ_1125 and LJ_1126 genes of NCC 533 was amplified using primers O and P. This 359-bp amplicon was digested with the restriction enzymes HindIII and XhoI and cloned into similarly digested pNZ124 (40) to yield pNZ124-LJ_1125 trm. The LJ_0045 d-lactate dehydrogenase promoter was amplified using NCC 533 chromosomal DNA as a template with the primers R plus S. This 215-bp amplicon was digested with the restriction enzymes BglII plus SacI and cloned into similarly digested pNZ124-LJ--_1125 trm to produce plasmid pDP794. This plasmid, with the promoter region of the LJ_0045 lactate dehydrogenase gene and a bidirectional terminator, was used for overexpression of the LJ_0548 and LJ_0549 genes.
For the construction of these overexpression plasmids, the following cloning steps were performed: For pDP1016, the gene LJ_0548 was amplified using primers T plus V. The 597-bp amplicon was digested with SphI and HindIII and cloned into SphI- plus HindIII-digested pDP794 to give plasmid pDP1016. For pDP1017, the LJ_0549 gene was amplified using primers U plus W. The 583-bp amplicon was digested with SphI and HindIII and cloned into SphI- plus HindIII-digested pDP794 to give plasmid pDP1017. For pDP1019, the genes LJ_0548 and LJ_0549 were amplified using primers T plus W, and the 1,132-bp amplicon was digested with SphI and HindIII and cloned into SphI- plus HindIII-digested pDP794 to give plasmid pDP1019. These cloning procedures yielded plasmids on which expression of LJ_0548 and/or LJ_0549 is controlled by the strong ldh promoter. Genetic maps of plasmids pDP1016, pDP1017, and pDP1019 were created using Clone Manager (see Fig. S1 in the supplemental material).
Growth in batch culture.
Aerotolerance of NCC 9359 was compared to that of wild-type L. johnsonii in continuously stirred vessels with 400 ml MRS medium. Batches were sparged with specific gas mixtures containing 5% CO2 and either no oxygen (0% oxygen [anaerobic]) or normal oxygen levels (20% oxygen [aerobic]). Cultures were grown at 37°C with continuous mixing (ca. 200 rpm), and pH was maintained at 6.5 by automated 4 M NaOH titration. Cell densities were determined by measuring the optical density at 600 nm (OD600). The maximum specific growth rate was determined by fitting an exponential trend line through the data points with a minimal R2 of 0.99.
Organic acid measurement by HPLC.
Extracellular metabolite concentrations were determined as described previously (41) using high-pressure liquid chromatography (HPLC) (LKB and Pharmacia, Oregon City, OR) with a chromatograph fitted with a Rezex organic acid analysis column (Phenomenex, Torrance, CA) at 45°C and an RI 1530 refractive index detector (Jasco, Easton, MD). The mobile phase consisted of a 7.2 mM H2SO4 solution. Chromatograms were analyzed using AZUR chromatography software (St. Martin D'Heres, France).
Statistical analysis.
Statistical significance was determined using a Student's two-tailed t test for unequal or equal variance. An F test was employed to verify whether variances could be considered equal (P > 0.05) or unequal (P < 0.05).
RESULTS
H2O2 accumulation results in premature growth stagnation during aerobic growth of L. johnsonii NCC 533.
To assess the growth behavior of L. johnsonii NCC 533 under anaerobic and aerobic conditions, LAPTg medium was inoculated with an overnight culture and incubated at 37°C either in a static tube with minimal headspace (anaerobic) or under continuous shaking with 10 volumes of headspace (aerobic). Growth rates of aerobic and anaerobic cultures were similar up to an OD600 of 1.0 (Fig. 1A). However, aerobic cultures accumulated up to 1 mM H2O2 during growth (Fig. 1B), leading to growth stagnation at an approximate density of OD600 of 1.5. This growth stagnation could be completely abolished by the addition of 0.5 mg ml−1 catalase to the medium, which prevented the accumulation of H2O2. These findings show that oxidative stress resulting from endogenous H2O2 production is the main cause for the observed growth arrest of L. johnsonii NCC 533 under aerobic conditions.
FIG 1.
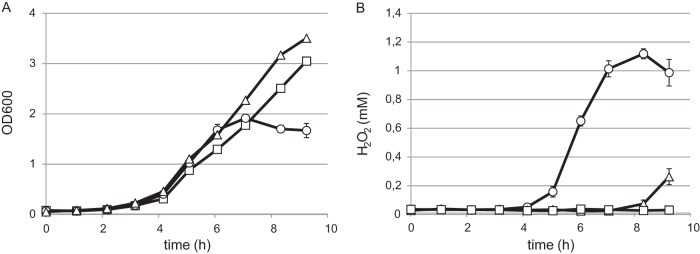
Growth and hydrogen peroxide concentration of L. johnsonii NCC 533 in LAPTg medium under anaerobic conditions (square symbols), aerobic conditions alone (circular symbols), or aerobic conditions with 0.5 mg ml−1 catalase added to the medium (triangular symbols). Culture densities were determined by optical density measurement at 600 nm (A), and H2O2 concentrations were determined by the phenol red enzymatic assay (B). The data represent duplicate experiments ± standard errors of the means.
H2O2 production is not dependent on predicted pyruvate oxidase-, lactate oxidase-, or NADH oxidase-encoding genes.
The main contributor to H2O2 production in lactic acid bacteria (LAB) has been proposed to be the oxygen-dependent lactate utilization pathway, which oxidizes lactate via pyruvate and acetyl-phosphate to acetate, generating CO2, ATP, NADH, and H2O2 (1, 42). The redox balance in this pathway is proposed to be restored by dissipation of the NADH via an NADH-oxidase-dependent reaction that generates either H2O2 or water. The oxygen-dependent lactate utilization pathway thereby encompasses three potential H2O2-producing reactions: (i) pyruvate oxidation (catalyzed by the Pox enzyme), (ii) lactate oxidation (catalyzed by the Lox enzyme), and (iii) NADH oxidation (catalyzed by the Nox enzyme).
To assess the contribution to the observed H2O2 production of the genes predicted to encode these enzymes in Lactobacillus johnsonii, mutant derivatives of the wild-type strain were constructed that lack the lactate oxidase-encoding gene LJ_1826 (NCC 9334), the pyruvate oxidase-encoding gene LJ_1853 (NCC 9333), or the NADH oxidase-encoding genes LJ_1254 and LJ_1255 (NCC 9337). The H2O2 production capacity of the mutants was compared to that of the wild-type strain. To this end, all strains were grown anaerobically (static cultures) in LAPTg medium to an OD600 of ∼0.7 and subsequently transferred to aerobic conditions (shake flask incubation). H2O2 production was measured after 1 and 2 h of incubation. Deletion of the predicted pox, lox, or nox genes did not significantly affect the level of hydrogen peroxide production after 1 h in these strains in comparison with the level produced by the parental strain NCC 533 (Fig. 2) (all P values of >0.05). Exposure to oxygen for 2 h resulted in small differences between hydrogen peroxide levels between the strains: the Δpox mutant produced less H2O2 (0.48 mM versus 0.53 mM in the wild type; P < 0.05), and the Δnox produced more H2O2 (0.56 mM; P < 0.05). It appears justified to conclude that the oxidative lactate utilization pathway is not responsible for the greater part of the H2O2 production, suggesting that an alternative metabolic conversion may account for the H2O2 production. This suggestion is in agreement with the previous observation that in the presence of oxygen, no substantial production and excretion of acetate occur (38).
FIG 2.
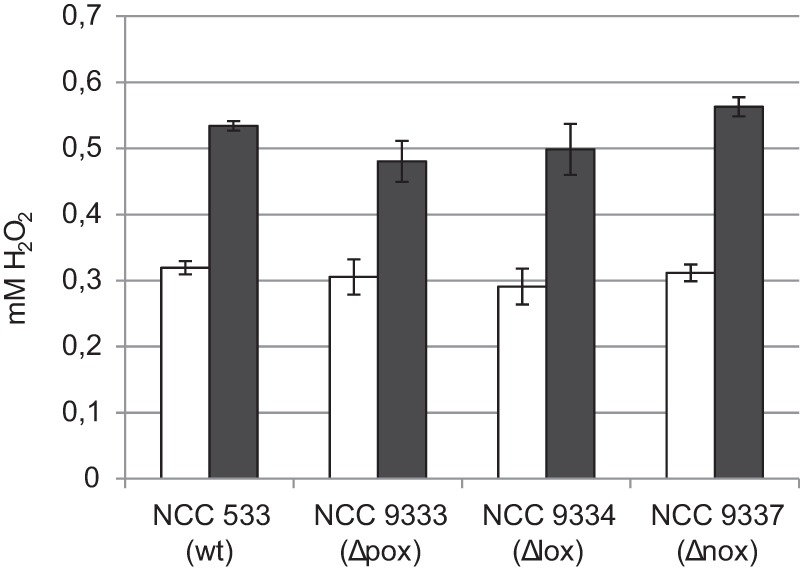
H2O2 production of L. johnsonii NCC 533 (wild type [wt]) and the derivatives NCC 9333 (Δpox), NCC 9334 (Δlox), and NCC 9337 (Δnox). Anaerobic logarithmic-phase cultures were transferred to a shake flask and incubated at 37°C. After 1 h (open bars) and 2 h (closed bars), cells were removed by centrifugation, and H2O2 concentrations were determined in the culture medium, using the phenol red-peroxidase enzymatic assay. Data represent the average of three independent experiments.
Previously, it has been suggested that L. delbrueckii, a close relative of L. johnsonii, produces H2O2 via an NADH-dependent reaction that is enhanced by the addition of a flavin source (8, 35, 36). To identify the protein and gene involved in such proposed enzymatic reaction in L. johnsonii, we initiated its purification, using enzyme activity assays to track the enzyme during purification.
Cell extracts of L. johnsonii NCC 533 contain NADH-dependent flavin reductase activity.
Cell extracts of L. johnsonii NCC 533 contain NADH consumption activity when a flavin compound is added as a supplement to the assay's reaction mixture (Fig. 3A). The activity requires the addition of either FAD, FMN, or riboflavin and does not show any activity with NADPH instead of NADH. Addition of a 10-fold-lower FMN concentration resulted in a significantly lower enzymatic rate (0.21 and 0.12 μmol/mg protein/min; P < 0.05). Following this observation, we further explored enzyme kinetics with different flavin sources at various concentrations. Michaelis-Menten-like kinetics were observed when the flavin concentration is varied (see Materials and Methods for the method of Km calculation), indicating that flavin is a direct substrate for the enzyme. The Kms of free flavin do not significantly differ for the various flavins used: 30 ± 8 μM for FMN, 64 ± 21 μM for FAD, and 41 ± 10 μM for riboflavin (P > 0.05).
FIG 3.
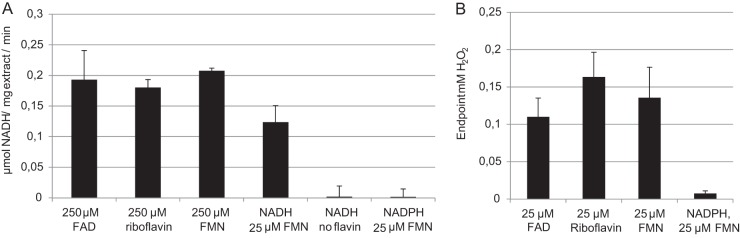
Typical NADH-dependent flavin reductase activity in L. johnsonii cell extract. NADH consumption rates were measured by absorption at 340 nm (A). Endpoint H2O2 concentrations were determined using the phenol red assay (B). Either 500 μM NADH was used, replaced by 500 μM NADPH where indicated (A), or 250 μM NADH was replaced by 250 μM NADPH where indicated (B). A 250 μM or 25 μM concentration of FAD, FMN, or riboflavin was added as a flavin source. The protein concentration in the cell-free extracts was determined by the MicroBCA assay. Data represent the average of technical triplicates ± standard deviation and are representative of cell extracts derived in comparable experiments.
The reaction is likely to involve a 2-electron transfer reaction since a considerable amount of H2O2 is formed as an end product, regardless of the flavin form that is added. When the FMN level in the assay is lowered to 25 μM, the hydrogen peroxide concentration exceeds the flavin concentration more than 5-fold (135 ± 33 μM H2O2), indicating that the free flavin is recycled during the reaction (Fig. 3B). Altogether, these observations allow the classification of the protein(s) responsible for the measured activity as an NADH-dependent flavin reductase.
Purification and identification of the NADH-dependent flavin reductase activity.
In order to identify the protein(s) responsible for the NADH-dependent flavin reductase activity, cell extract of wild-type L. johnsonii strain NCC 533 was subjected to the following purification steps: (i) ammonium sulfate precipitation, (ii) Q column chromatography, and (iii) gel filtration (see Materials and Methods for details). After the ammonium sulfate precipitation, the 50% to 70% fractions clearly display the most NADH consumption. After the subsequent fractionation of this fraction using anion-exchange chromatography, the highest activity clearly eluted after 83 and 86 ml (see Fig. S2 in the supplemental material). These fractions were combined and were subsequently further separated by size exclusion chromatography (Superdex2000). Only a single fraction eluted during this chromatography step that displayed clear H2O2 production in the enzymatic assay (Fig. 4). The sizes of the enzymes in this fraction were, based on elution time, estimated to be ∼18 kDa. The enzyme or enzymes in this fraction that showed NADH flavin reductase activity were only partially purified, since multiple bands were apparent when it was loaded on SDS gel (results not shown). The active fraction obtained was digested with trypsin and analyzed using LC-MS/MS, using the fraction preceding this active fraction (and containing no activity) as a comparative negative control. In the active fraction, 25 L. johnsonii proteins could be assigned with a probability-based score of P < 0.05. The protein that was predicted with the highest probability was the hypothetical protein LJ_0548, to which 8 peptides could be assigned with a total coverage of 63%, indicating a high abundance in the fraction. Three peptides could be assigned to the hypothetical protein LJ_0549 (33% sequence coverage), which is encoded by LJ_0549, the gene neighboring LJ_0548. Noteworthy, the predicted protein sequences of LJ_0548 (accession no. Q74HL7) and LJ_0549 (accession no. Q74HL8) both contain a conserved FMN reductase domain, supporting the role of these gene products in the NADH-dependent flavin reductase activity. Furthermore, no peptides belonging to these two proteins were detected in the fraction that did not show any NADH flavin reductase activity but eluted close to the active fraction (negative control). Taken together, these observations pointed toward the involvement of the LJ_0548 and LJ_0549 genes in the NADH flavin reductase activity of cell extracts of L. johnsonii.
FIG 4.
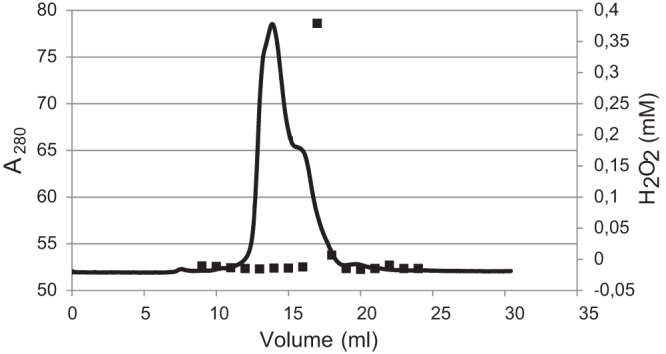
Size exclusion chromatogram (Superdex200) of the final purification step. The protein concentration is determined by absorption at 280 nm (black line). The eluting proteins were collected in fractions of 1 ml and tested for NADH flavin reductase activity by addition of 500 μM NADH and 250 μM FAD. The H2O2 concentration was determined after 10 min (symbols). The column was calibrated using a protein with a known size.
A mutant derivative of L. johnsonii NCC 533 was constructed that lacks the genes LJ_0548 and LJ_0549 (NCC 9359). The cell extracts of this mutant strain completely lacked the flavin-dependent NADH reductase activity that was detected in the extracts obtained from the wild-type strain, nor could any H2O2 be detected in the reaction mixture supplemented with the mutant strain extract. These results indicate that LJ_0548 and/or LJ_0549 encode the NADH-dependent flavin reductase activity. The LJ_0548 LJ_0549 deletion strain (NCC 9359) was complemented by providing one or both of the deleted genes in trans on a plasmid under expression control of the strong, constitutive d-lactate dehydrogenase gene promoter (ldhDp) (LJ_0045). Cell extracts derived from the NCC 9359 strain harboring either the LJ_0548 (pDP1016) or LJ_0549 (pDP1017) expression plasmid did not show any additional bands on an SDS protein gel. These cell extracts also did not show any significant NADH-dependent flavin reductase activity. Conversely, the extract derived from the strain harboring the plasmid expressing both the LJ_0548 and LJ_0549 genes (pDP1019) displayed an additional band of 20 kDa on an SDS gel (see Fig. S3 in the supplemental material) and an NADH consumption rate that was more than 7-fold higher than the rate measured in extracts derived from the wild-type strain (Fig. 5A). The level of H2O2 production driven by the extract derived from the strain overexpressing LJ_0548 and LJ_0549 was comparable to the level produced by the extract from the wild type, which reflects the maximal level of H2O2 production that can be obtained in this assay as a consequence of the limited amount of NADH provided in the reaction mixture (Fig. 5B). These results show that the LJ_0548-LJ_0549 operon encodes the observed NADH-dependent flavin reductase activity.
FIG 5.
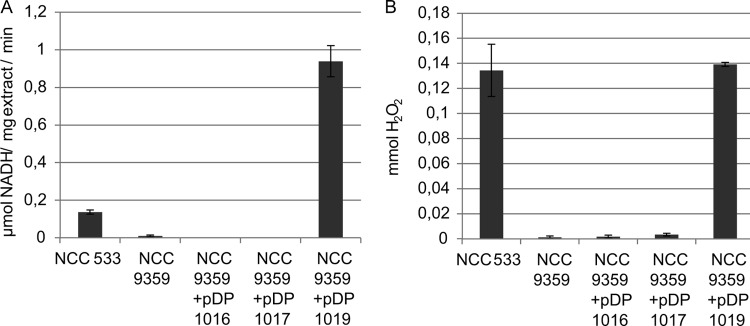
Typical NADH-dependent flavin reductase activity in cell extracts of wild-type L. johnsonii NCC 533 and its LJ_0548 LJ_0549 (NCC 9359) mutant derivative, with and without complementation by plasmid-borne expression of one (pDP1016 and pDP1017) or both (pDP1019) of the deleted genes. Standard assay conditions were employed, containing 500 μM NADH and 25 μM FMN (NADH consumption rates) and 250 μM NADH and 25 μM FMN (to determine H2O2 concentration). NADH consumption rates were measured by absorption at 340 nm (A). H2O2 concentrations were determined after 10 min of reaction using the phenol red assay (panel B). Protein concentrations in the cell extracts were determined by MicroBCA assay. Data represent the average of three technical replicates ± standard deviation and are representative for activity measured in multiple (>3) cell extracts.
H2O2 production of the LJ_0548 LJ_0549 deletion strain.
Having identified LJ_0548 and LJ_0549 as coding for the enzymes responsible for NADH-dependent flavin reductase activity in the cell extracts of L. johnsonii NCC 533, we studied the in vivo contribution of this activity to the aerobic physiology of this bacterium by comparing the wild type to its nfr deletion derivative NCC 9359 (ΔLJ_0548 ΔLJ_0549). The maximum specific growth rate of the mutant in the shake flask was similar to that of the wild type, and its metabolism remained homolactic (result not shown). However, when exposed to stronger aeration (750 ml/min, 75% N2, 20% O2, 5% CO2), the nfr deletion derivative displayed a reduced growth rate compared to wild-type strain, whereas exposure to the anaerobic gas mixture equivalent of this regimen (750 ml/min 95% N2–5% CO2) did not result in a difference between the nfr mutant and its wild-type counterpart (see Fig. S4 in the supplemental material).
To test hydrogen peroxide production, the wild-type strain (NCC 533) and its LJ_0548 LJ_0549 deletion derivatives were grown anaerobically to the mid-logarithmic phase (OD600 of ∼0.8) at 37°C and were then transferred to aerobic (shake flask) conditions. H2O2 in the spent medium of each of the cultures was assessed after 1 h of oxygen exposure (Fig. 6). Notably, the LJ_0548-LJ_0549 deletion (strain NCC 9359) resulted in complete loss of the capacity to produce hydrogen peroxide under these conditions, while substantial amounts of this reactive oxygen molecule were detected in the NCC 533 culture exposed to the same conditions. Moreover, the NCC 9359 mutant strain that was in trans complemented with plasmid-borne expression of either LJ_0548 (pDP1016) or LJ_0549 (pDP1017) did not produce detectable H2O2 levels, whereas the strain complemented with plasmid-borne expression of both LJ_0548 and LJ_0549 (pDP1019) displayed a restored H2O2 production capacity comparable to that observed in the parental strain NCC 533. These data confirm that the L. johnsonii NCC 533 NADH-dependent flavin reductase is encoded by the LJ_0548-LJ_0549 cluster and that this activity is the major H2O2-producing system expressed under the conditions employed here.
FIG 6.
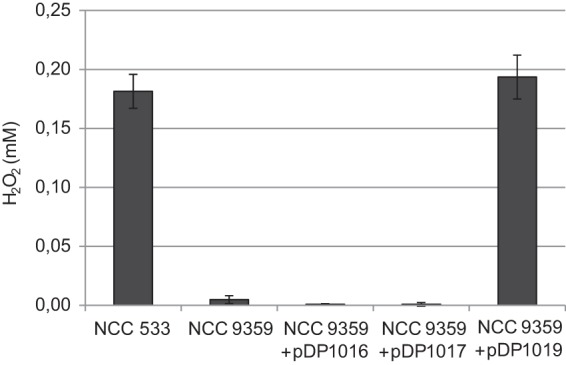
H2O2 production in L. johnsonii NCC 533 and NCC 9359, with or without complementation of LJ_0548 and LJ_0549 under aerobic conditions. Anaerobic logarithmic-phase cultures of wild-type Lactobacillus johnsonii and its deletion derivative were transferred to a shake flask and incubated at 37°C. After 1 h, cells were removed by centrifugation, and H2O2 concentrations were determined in the culture medium, using the phenol red assay. Data represent the average of three independent experiments ± standard deviation.
In order to test if LJ_0548 and LJ_0549 play a role in anaerobic fumarate respiration of L. johnsonii, the external metabolite profiles of wild-type NCC 533 and NCC 9359 (the LJ_0548 LJ_0549 deletion derivative) were compared. After 7 h of anaerobic growth in MRS medium supplemented with 10 mM fumarate, cells were removed by centrifugation, and external metabolites were analyzed using HPLC. No change in the concentration of fumarate was observed, and no succinate formation was detected. Both aerobic and anaerobic metabolism of the mutant strains remained entirely homolactic (results not shown).
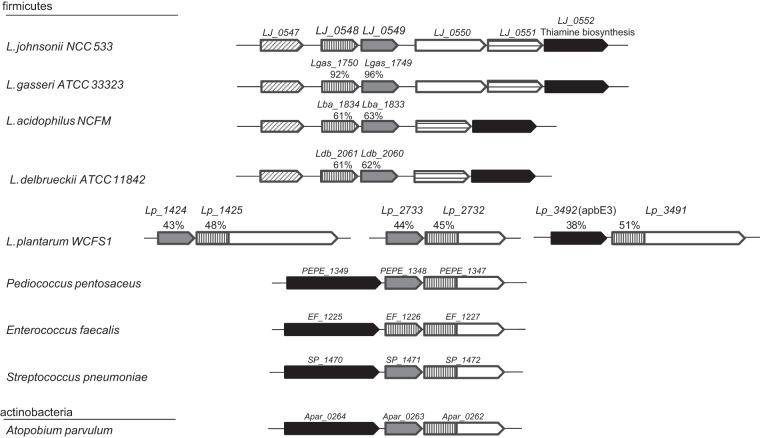
Sequence analysis of the LJ_0548 and LJ_0549 genes and their conservation among bacteria.
Using the wealth of genomic sequence availability, the prevalence and context of the LJ_0548 and LJ_0549 single genes as well as the combination of the two consecutive genes were analyzed using diverse genome comparison tools.
The LJ_0548 and LJ_0549 genes are predicted to encode proteins of 178 and 184 residues, respectively, that share substantial similarity (40% identity and 59% similarity at the amino acid sequence level). The protein domain signature recognition module Interproscan (43) revealed that both genes have a highly conserved FMN reductase domain (PFAM domain 03358), covering the N-terminal 145 residues. In the PANTHER classification system program (44), LJ_0548 was classified as a chromate reductase, a group of enzymes that have been annotated as such due to their potential use in chromate bioremediation (45). The ortholog in Pseudomonas putida has been shown to catalyze the transfer of electrons from NADH to the quinone pool (46). The crystal structure of the E. coli gene annotated as chromate reductase clearly demonstrates the amino acids that constitute the flavin binding site (47). A ClustalW multiple sequence alignment shows that 4 out of the 11 residues of this binding site are conserved in both LJ_0548 and LJ_0549: i.e., Ser18, Asn20, Glu82, and Ser117 (see Fig. S5 in the supplemental material).
The LJ_0548-LJ_0549 locus in L. johnsonii NCC 533 appears to be conserved in all other members of the L. acidophilus group. Examination of the genetic context in more distant species using the STRING module (48) revealed that the closest homologues of LJ_0548 and LJ_0549 are encountered as consecutive genes in several different species (Fig. 7), including L. plantarum, and several species belonging to the Streptococcus, Enterococcus, and Pediococcus genera. In these examples, the first of these two genes is similar in sequence and size to either LJ_0548 or LJ_0549 and is followed by a second, larger gene, of which the N-terminal residues (∼200) are homologues to LJ_0548. This type of arrangement is also present in a more distant species from the Actinobacteraceae class, Atopobium parvulum, which is a species often found in the human oral cavity (49). In all aforementioned species, the homologues of the products of LJ_0548 and LJ_0549 are annotated as fumarate reductases, NADH dehydrogenases, or flavin reductases, but to the best of our knowledge there are no experimental data to support these annotations.
FIG 7.
Genetic context conservation of LJ_0548 and LJ_0549 found using the STRING module (48).
DISCUSSION
Our work has identified a novel NADH-dependent FMN reductase in L. johnsonii NCC 533 that is encoded by two adjacent genes (LJ_0548 and LJ_0549) and acts as the major H2O2-producing system in this bacterium. L. johnsonii is a lactic acid bacterium that belongs to the phylogenetically closely related L. acidophilus group (34, 50), which includes several strains of Lactobacillus species that are marketed as probiotic supplements, like L. acidophilus, L. johnsonii, L. jensenii, L. crispatus, and L. gasseri, but also encompasses the well-known yoghurt bacterium L. delbrueckii. It has been established that these species endure oxidative stress as a consequence of endogenously produced H2O2 (36, 51, 52). The enzymes identified here have characteristics that are in agreement with those observed previously in strains of the L. acidophilus group, including that the enzyme reaction consumes NADH, uses flavin as a cofactor, is constitutively expressed, and is not produced in response to molecular oxygen (8, 35, 36). In addition, the maximal reaction rate measured in cell extracts of L. johnsonii is in the same order of magnitude as the rate previously reported for L. delbrueckii (36).
Apparent Michaelis-Menten kinetics were observed when the concentration of flavin was varied in the enzymatic assay. However, the hydrogen peroxide produced in the assay reaches a higher level than the total initial concentration of flavin added to the assay, indicating that free oxidized flavin serves as a substrate in this reaction, but the reduced flavin is subsequently oxidized and reused. This is in agreement with the behavior of reduced flavins that spontaneously react with oxygen, yielding hydrogen peroxide.
Despite the recognition of this enzyme activity in various L. acidophilus group species, the molecular characteristics and genetic determinant(s) of this activity have not been described to date. Therefore, our study is the first to identify and characterize this novel enzyme family and its encoding genes. The enzyme is shown to be responsible for the major H2O2 production in an industrially relevant member of the L. acidophilus group, L. johnsonii. This capacity has previously been proposed to influence gut homeostasis and anti-inflammatory activity of this group of organisms (31), and the identification of the responsible genes and the construction of the corresponding deletion strain may accelerate the establishment of this presumed function in vivo, which is assumed to play a role in the bacterium's protective effect against vaginal disease (25).
The Vmax and Km values of the different flavins that were tested in the enzymatic assay did not differ strongly. The Km values for flavins were found in the range of 30 to 50 μM. Intracellular flavin concentrations in E. coli and Shewanella oneidensis were reported in the order of 0.5 μmol/g of protein (53), which in combination with the “rule-of-thumb” estimate of ∼200 g/liter as the concentration of intracellular protein in prokaryotes (54) implies that the intracellular flavin concentration would be ∼100 μM. The partitioning of flavin bound to protein and available as an electron acceptor for the proteins we describe here is unknown. One study on intracellular free FAD concentration in Amphibacillus xylanus finds 13 μM using HPLC (55). Such a flavin concentration in L. johnsonii would be sufficient for its proposed in vivo role. Given the clear confirmation of the in vivo role of Nfr by the physiology of the nfr deletion mutants, we consider it a valid conclusion that analogous to the previously found concentration in A. xylanus, the free flavin levels in L. johnsonii suffice for significant hydrogen peroxide production.
Surprisingly, the genes that are considered to be responsible for H2O2 production in other LAB were shown to not contribute significantly to this phenotype in L. johnsonii, since the genes predicted to encode lactate, pyruvate, or NADH oxidases could be deleted without consequences for the H2O2 production in this species. The homologs of the products of LJ_0548 and LJ_0549 have been annotated as fumarate reductase, NADH dehydrogenase, or NAD(P)H-dependent FMN reductase. The LJ_0548- and LJ_0549-encoded proteins are predicted to be small flavoproteins that are highly similar (40%). The size of the denatured protein components in the cell extract of the LJ_0548- and LJ_0549-overexpressing strain on the SDS gel is 20 kDa, corresponding to the size of the gene product of either LJ_0548 or LJ_0549, as inferred from their gene sequence. Its elution from the gel filtration column suggests that the active protein has a size of ∼18 kDa, which would mean that only one of either LJ_0548 or LJ_0549 would be required for activity. However, for complementation of the LJ_0548 LJ_0549 deletion strain, both genes appear to be required, whereas in trans complementation with a plasmid harboring either one of the genes failed to result in detectable protein expression (SDS-PAGE) or functional complementation. Although we can conclude that both genes are required to produce the functional enzyme, its exact composition remains unclear.
The observation that the deletion derivative NCC 9359 produces small amounts of H2O2 upon prolonged exposure to oxygen indicates that besides the NADH flavin reductase identified here, other H2O2-producing enzymes may exist in this species. Nevertheless, the enzyme identified here appears to be the major contributor to the H2O2 production capacity in this species. Possibly, the additional H2O2-producing reactions involve oxidases, like the aforementioned pyruvate, lactate, and NADH oxidase, which may contribute to H2O2 production upon extended oxygen exposure. However, it is unlikely that the conditions of oxygenation used in this study, in terms of both its duration and/or oxygen tension, will be encountered in the GI tract, which is thought to be the natural habitat of L. johnsonii. We propose therefore that the constitutive flavin reductase is the primary source of H2O2 in an environment where microbes predominantly encounter anaerobic (intestinal lumen) conditions and only sporadically encounter lower and more variable concentrations of oxygen when they are present in closer proximity to the intestine mucosa. In contrast, prolonged exposure to aerobic conditions and/or higher oxygen tensions can occur during industrial processing, which may elicit the activation of alternative H2O2 production reactions, as our preliminary observations imply.
L. johnsonii has been proposed to have lost numerous genes and pathways during its adaptation to the nutrient-rich environment of the intestinal tract (17). Nevertheless, the newly discovered NADH-dependent flavin reductase appears to be constitutively expressed, suggesting that it plays an important role in the lifestyle of L. johnsonii in its natural environment. Since lactate fermentation from glucose is entirely redox neutral, it is unclear in what metabolic step the NADH is generated that is consumed in the reaction catalyzed by the LJ_0548-LJ_549 enzyme. We hypothesize that the additional electrons are generated in the metabolism of one of the many vitamins, peptides, and amino acids that are consumed by L. johnsonii in addition to glucose.
Although the results presented here do not rule out that this newly identified flavin reductase serves a metabolic purpose in which H2O2 is a side product, we suggest that the production of H2O2 in itself has a biological function. For example, it may contribute to the antimicrobial capacities of L. johnsonii that may be of great importance for the organism to maintain its niche/position within the densely populated microbiota (7). Moreover, H2O2 may serve as a chemical signal in host-microbe interactions, as it has been proposed to influence PPAR-γ, one of the major regulators of inflammation in the intestinal epithelium (31). Alternatively, the reduced aerotolerance of the nfr deletion derivative compared to its wild-type counterpart suggests that the reaction catalyzed by this enzyme may enable L. johnsonii to prevent or reduce oxidative stress. If the flavins that are reduced by these proteins form the most readily oxidized parts in the cytoplasm and can effectively capture oxygen, the activity of these flavins may prevent other, more damaging effects of oxygen, such as the direct oxidation of iron-sulfur clusters (56, 57) or the formation of semiquinones (46). Also in L. johnsonii, the controlled production of H2O2 may be preferred over the uncontrollable other effects oxygen might exert. The role that this NADH-dependent flavin reductase plays in oxidative stress, hydrogen peroxide scavenging, and aerotolerance of L. johnsonii is the subject of further studies.
Supplementary Material
ACKNOWLEDGMENTS
The work we present has been funded by the Nestlé Research Centre, Nestec, Ltd. (Switzerland), and is the result of a collaboration between NIZO food research (The Netherlands) and the Nestlé Research Centre. David Pridmore was employed by Nestlé Research Centre and affiliated with the Kluyver Centre for Genomics of Industrial Fermentation (Delft, The Netherlands) for the duration of his contribution to this article. Christof Gysler is currently employed by the Nestlé Research Centre. The members of the Nestlé Research Centre have contributed to this article by supplying the wild-type and mutant strains and through regular discussion. Rosanne Hertzberger and Michiel Kleerebezem are affiliated with NIZO food research.
We hereby explicitly state that there are no patents, products in development, or marketed products to declare.
We acknowledge Anne-Cécile Pittet for technical assistance with construction of the mutants and Filipe Branco dos Santos for valuable input on the purification of the NADH flavin reductase activity.
Footnotes
Published ahead of print 31 January 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.04272-13.
REFERENCES
- 1.Seki M, Iida K, Saito M, Nakayama H, Yoshida S. 2004. Hydrogen peroxide production in Streptococcus pyogenes: involvement of lactate oxidase and coupling with aerobic utilization of lactate. J. Bacteriol. 186:2046–2051. 10.1128/JB.186.7.2046-2051.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Poole LB, Higuchi M, Shimada M, Calzi ML, Kamio Y. 2000. Streptococcus mutans H2O2-forming NADH oxidase is an alkyl hydroperoxide reductase protein. Free Radic. Biol. Med. 28:108–120. 10.1016/S0891-5849(99)00218-X [DOI] [PubMed] [Google Scholar]
- 3.Thomas EL, Pera KA. 1983. Oxygen metabolism of Streptococcus mutans: uptake of oxygen and release of superoxide and hydrogen peroxide. J. Bacteriol. 154:1236–1244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pericone CD, Park S, Imlay JA, Weiser JN. 2003. Factors contributing to hydrogen peroxide resistance in Streptococcus pneumoniae include pyruvate oxidase (SpxB) and avoidance of the toxic effects of the Fenton reaction. J. Bacteriol. 185:6815–6825. 10.1128/JB.185.23.6815-6825.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pericone CD, Overweg K, Hermans PW, Weiser JN. 2000. Inhibitory and bactericidal effects of hydrogen peroxide production by Streptococcus pneumoniae on other inhabitants of the upper respiratory tract. Infect. Immun. 68:3990–3997. 10.1128/IAI.68.7.3990-3997.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kawasaki S, Satoh T, Todoroki M, Niimura Y. 2009. b-type dihydroorotate dehydrogenase is purified as a H2O2-forming NADH oxidase from Bifidobacterium bifidum. Appl. Environ. Microbiol. 75:629–636. 10.1128/AEM.02111-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pridmore RD, Pittet AC, Praplan F, Cavadini C. 2008. Hydrogen peroxide production by Lactobacillus johnsonii NCC 533 and its role in anti-Salmonella activity. FEMS Microbiol. Lett. 283:210–215. 10.1111/j.1574-6968.2008.01176.x [DOI] [PubMed] [Google Scholar]
- 8.Martin R, Suarez JE. 2010. Biosynthesis and degradation of H2O2 by vaginal lactobacilli. Appl. Environ. Microbiol. 76:400–405. 10.1128/AEM.01631-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Korshunov S, Imlay JA. 2010. Two sources of endogenous hydrogen peroxide in Escherichia coli. Mol. Microbiol. 75:1389–1401. 10.1111/j.1365-2958.2010.07059.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Seaver LC, Imlay JA. 2004. Are respiratory enzymes the primary sources of intracellular hydrogen peroxide? J. Biol. Chem. 279:48742–48750. 10.1074/jbc.M408754200 [DOI] [PubMed] [Google Scholar]
- 11.Condon S. 1987. Responses of lactic acid bacteria to oxygen. FEMS Microbiol. Rev. 46:269–280. 10.1111/j.1574-6968.1987.tb02465.x [DOI] [Google Scholar]
- 12.Seki M, Iida K, Saito M, Nakayama H, Yoshida S. 2004. Hydrogen peroxide production in Streptococcus pyogenes: involvement of lactate oxidase and coupling with aerobic utilization of lactate. J. Bacteriol. 186:2046–2051. 10.1128/JB.186.7.2046-2051.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kietzman CC, Caparon MG. 2010. CcpA and LacD1 affect temporal regulation of Streptococcus pyogenes virulence genes. Infect. Immun. 78:241–252. 10.1128/IAI.00746-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hecht HJ, Erdmann H, Park HJ, Sprinzl M, Schmid RD. 1995. Crystal structure of NADH oxidase from Thermus thermophilus. Nat. Struct. Biol. 2:1109–1114. 10.1038/nsb1295-1109 [DOI] [PubMed] [Google Scholar]
- 15.Niimura Y, Nishiyama Y, Saito D, Tsuji H, Hidaka M, Miyaji T, Watanabe T, Massey V. 2000. A hydrogen peroxide-forming NADH oxidase that functions as an alkyl hydroperoxide reductase in Amphibacillus xylanus. J. Bacteriol. 182:5046–5051. 10.1128/JB.182.18.5046-5051.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Imlay JA. 2003. Pathways of oxidative damage. Annu. Rev. Microbiol. 57:395–418. 10.1146/annurev.micro.57.030502.090938 [DOI] [PubMed] [Google Scholar]
- 17.Pridmore RD, Berger B, Desiere F, Vilanova D, Barretto C, Pittet AC, Zwahlen MC, Rouvet M, Altermann E, Barrangou R, Mollet B, Mercenier A, Klaenhammer T, Arigoni F, Schell MA. 2004. The genome sequence of the probiotic intestinal bacterium Lactobacillus johnsonii NCC 533. Proc. Natl. Acad. Sci. U. S. A. 101:2512–2517. 10.1073/pnas.0307327101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Neeser JR, Granato D, Rouvet M, Servin A, Teneberg S, Karlsson KA. 2000. Lactobacillus johnsonii La1 shares carbohydrate-binding specificities with several enteropathogenic bacteria. Glycobiology 10:1193–1199. 10.1093/glycob/10.11.1193 [DOI] [PubMed] [Google Scholar]
- 19.Bernet MF, Brassart D, Neeser JR, Servin AL. 1994. Lactobacillus acidophilus LA 1 binds to cultured human intestinal cell lines and inhibits cell attachment and cell invasion by enterovirulent bacteria. Gut 35:483–489. 10.1136/gut.35.4.483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Granato D, Perotti F, Masserey I, Rouvet M, Golliard M, Servin A, Brassart D. 1999. Cell surface-associated lipoteichoic acid acts as an adhesion factor for attachment of Lactobacillus johnsonii La1 to human enterocyte-like Caco-2 cells. Appl. Environ. Microbiol. 65:1071–1077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Marteyn B, West NP, Browning DF, Cole JA, Shaw JG, Palm F, Mounier J, Prévost M, Sansonetti P, Tang CM. 2010. Modulation of Shigella virulence in response to available oxygen in vivo. Nature 465:355–358. 10.1038/nature08970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.He G, Shankar RA, Chzhan M, Samouilov A, Kuppusamy P, Zweier JL. 1999. Noninvasive measurement of anatomic structure and intraluminal oxygenation in the gastrointestinal tract of living mice with spatial and spectral EPR imaging. Proc. Natl. Acad. Sci. U. S. A. 96:4586–4591. 10.1073/pnas.96.8.4586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huycke MM, Abrams V, Moore DR. 2002. Enterococcus faecalis produces extracellular superoxide and hydrogen peroxide that damages colonic epithelial cell DNA. Carcinogenesis 23:529–536. 10.1093/carcin/23.3.529 [DOI] [PubMed] [Google Scholar]
- 24.Ding SZ, O'Hara AM, Denning TL, Dirden-Kramer B, Mifflin RC, Reyes VE, Ryan KA, Elliott SN, Izumi T, Boldogh I, Mitra S, Ernst PB, Crowe SE. 2004. Helicobacter pylori and H2O2 increase AP endonuclease-1/redox factor-1 expression in human gastric epithelial cells. Gastroenterology 127:845–858. 10.1053/j.gastro.2004.06.017 [DOI] [PubMed] [Google Scholar]
- 25.Eschenbach DA, Davick PR, Williams BL, Klebanoff SJ, Young-Smith K, Critchlow CM, Holmes KK. 1989. Prevalence of hydrogen peroxide-producing Lactobacillus species in normal women and women with bacterial vaginosis. J. Clin. Microbiol. 27:251–256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hawes SE, Hillier SL, Benedetti J, Stevens CE, Koutsky LA, Wolner-Hanssen P, Holmes KK. 1996. Hydrogen peroxide-producing lactobacilli and acquisition of vaginal infections. J. Infect. Dis. 174:1058–1063. 10.1093/infdis/174.5.1058 [DOI] [PubMed] [Google Scholar]
- 27.Donati L, Di Vico A, Nucci M, Quagliozzi L, Spagnuolo T, Labianca A, Bracaglia M, Ianniello F, Caruso A, Paradisi G. 2010. Vaginal microbial flora and outcome of pregnancy. Arch. Gynecol. Obstet. 281:589–600. 10.1007/s00404-009-1318-3 [DOI] [PubMed] [Google Scholar]
- 28.Schwebke JR. 2005. Abnormal vaginal flora as a biological risk factor for acquisition of HIV infection and sexually transmitted diseases. J. Infect. Dis. 192:1315–1317. 10.1086/462430 [DOI] [PubMed] [Google Scholar]
- 29.Desreumaux P, Dubuquoy L, Nutten S, Peuchmaur M, Englaro W, Schoonjans K, Derijard B, Desvergne B, Wahli W, Chambon P, Leibowitz MD, Colombel JF, Auwerx J. 2001. Attenuation of colon inflammation through activators of the retinoid X receptor (Rxr)/peroxisome proliferator-activated receptor gamma (Pparγ) heterodimer. A basis for new therapeutic strategies. J. Exp. Med. 193:827–838. 10.1084/jem.193.7.827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Su CG, Wen X, Bailey ST, Jiang W, Rangwala SM, Keilbaugh SA, Flanigan A, Murthy S, Lazar MA, Wu GD. 1999. A novel therapy for colitis utilizing PPAR-γ ligands to inhibit the epithelial inflammatory response. J. Clin. Invest. 104:383–389. 10.1172/JCI7145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Voltan S, Martines D, Elli M, Brun P, Longo S, Porzionato A, Macchi V, D'Inca R, Scarpa M, Palu G, Sturniolo GC, Morelli L, Castagliuolo I. 2008. Lactobacillus crispatus M247-derived H2O2 acts as a signal transducing molecule activating peroxisome proliferator activated receptor-gamma in the intestinal mucosa. Gastroenterology 135:1216–1227. 10.1053/j.gastro.2008.07.007 [DOI] [PubMed] [Google Scholar]
- 32.Valladares R, Bojilova L, Potts AH, Cameron E, Gardner C, Lorca G, Gonzalez CF. 2013. Lactobacillus johnsonii inhibits indoleamine 2,3-dioxygenase and alters tryptophan metabolite levels in BioBreeding rats. FASEB J. 27:1711–1720. 10.1096/fj.12-223339 [DOI] [PubMed] [Google Scholar]
- 33.Felis GE, Dellaglio F. 2007. Taxonomy of lactobacilli and bifidobacteria. Curr. Issues Intest. Microbiol. 8:44–61 [PubMed] [Google Scholar]
- 34.Berger B, Pridmore RD, Barretto C, Delmas-Julien F, Schreiber K, Arigoni F, Brüssow H. 2007. Similarity and differences in the Lactobacillus acidophilus group identified by polyphasic analysis and comparative genomics. J. Bacteriol. 189:1311–1321. 10.1128/JB.01393-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yi X, Kot E, Bezkorovainy A. 1998. Properties of NADH oxidase from Lactobacillus delbrueckii ssp bulgaricus. J. Sci. Food Agric. 78:527–534 [Google Scholar]
- 36.Marty-Teysset C, de la Torre F, Garel J. 2000. Increased production of hydrogen peroxide by Lactobacillus delbrueckii subsp. bulgaricus upon aeration: involvement of an NADH oxidase in oxidative stress. Appl. Environ. Microbiol. 66:262–267. 10.1128/AEM.66.1.262-267.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.de Man JC, Rogosa M, Sharpe MEA. 1960. A medium for the cultivation of lactobacilli. J. Appl. Bacteriol. 23:130–135. 10.1111/j.1365-2672.1960.tb00188.x [DOI] [Google Scholar]
- 38.Hertzberger RY, Pridmore RD, Gysler C, Kleerebezem M, Teixeira de Mattos MJ. 2013. Oxygen relieves the CO2 and acetate dependency of Lactobacillus johnsonii NCC 533. PLoS One 8:e57235. 10.1371/journal.pone.0057235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.van der Kaaij H, Desiere F, Mollet B, Germond JE. 2004. l-Alanine auxotrophy of Lactobacillus johnsonii as demonstrated by physiological, genomic, and gene complementation approaches. Appl. Environ. Microbiol. 70:1869–1873. 10.1128/AEM.70.3.1869-1873.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Platteeuw C, Simons G, de Vos WM. 1994. Use of the Escherichia coli beta-glucuronidase (gusA) gene as a reporter gene for analyzing promoters in lactic acid bacteria. Appl. Environ. Microbiol. 60:587–593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bekker M, de Vries S, Ter Beek A, Hellingwerf KJ, de Mattos MJ. 2009. Respiration of Escherichia coli can be fully uncoupled via the nonelectrogenic terminal cytochrome bd-II oxidase. J. Bacteriol. 191:5510–5517. 10.1128/JB.00562-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Goffin P, Muscariello L, Lorquet F, Stukkens A, Prozzi D, Sacco M, Kleerebezem M, Hols P. 2006. Involvement of pyruvate oxidase activity and acetate production in the survival of Lactobacillus plantarum during the stationary phase of aerobic growth. Appl. Environ. Microbiol. 72:7933–7940. 10.1128/AEM.00659-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zdobnov EM, Apweiler R. 2001. InterProScan—an integration platform for the signature-recognition methods in InterPro. Bioinformatics 17:847–848. 10.1093/bioinformatics/17.9.847 [DOI] [PubMed] [Google Scholar]
- 44.Mi H, Muruganujan A, Thomas PD. 2013. PANTHER in 2013: modeling the evolution of gene function, and other gene attributes, in the context of phylogenetic trees. Nucleic Acids Res. 41:D377–D386. 10.1093/nar/gks1118; 10.1093/nar/gks1118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ackerley DF, Gonzalez CF, Park CH, Blake R, Keyhan M, Matin A. 2004. Chromate-reducing properties of soluble flavoproteins from Pseudomonas putida and Escherichia coli. Appl. Environ. Microbiol. 70:873–882. 10.1128/AEM.70.2.873-882.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gonzalez CF, Ackerley DF, Lynch SV, Matin A. 2005. ChrR, a soluble quinone reductase of Pseudomonas putida that defends against H2O2. J. Biol. Chem. 280:22590–22595. 10.1074/jbc.M501654200 [DOI] [PubMed] [Google Scholar]
- 47.Eswaramoorthy S, Poulain S, Hienerwadel R, Bremond N, Sylvester MD, Zhang YB, Berthomieu C, Van Der Lelie D, Matin A. 2012. Crystal structure of ChrR—a quinone reductase with the capacity to reduce chromate. PLoS One 7:e36017. 10.1371/journal.pone.0036017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jensen LJ, Kuhn M, Stark M, Chaffron S, Creevey C, Muller J, Doerks T, Julien P, Roth A, Simonovic M, Bork P, von Mering C. 2009. STRING 8—a global view on proteins and their functional interactions in 630 organisms. Nucleic Acids Res. 37:D412–D416. 10.1093/nar/gkn760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Copeland A, Sikorski J, Lapidus A, Nolan M, Del Rio TG, Lucas S, Chen F, Tice H, Pitluck S, Cheng JF, Pukall R, Chertkov O, Brettin T, Han C, Detter JC, Kuske C, Bruce D, Goodwin L, Ivanova N, Mavromatis K, Mikhailova N, Chen A, Palaniappan K, Chain P, Rohde M, Goker M, Bristow J, Eisen JA, Markowitz V, Hugenholtz P, Kyrpides NC, Klenk HP, Detter JC. 2009. Complete genome sequence of Atopobium parvulum type strain (IPP 1246). Stand. Genomic Sci. 1:166–173. 10.4056/sigs.29547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pot B, Hertel C, Ludwig W, Descheemaeker P, Kersters K, Schleifer KH. 1993. Identification and classification of Lactobacillus acidophilus, L. gasseri and L. johnsonii strains by SDS-PAGE and rRNA-targeted oligonucleotide probe hybridization. J. Gen. Microbiol. 139:513–517. 10.1099/00221287-139-3-513 [DOI] [PubMed] [Google Scholar]
- 51.Talwalkar A, Kailasapathy L. 2004. The role of oxygen in the viability of probiotic bacteria with reference to Lactobacillus acidophilus and Bifidobacterium spp. Curr. Issues Intest. Microbiol. 5:1–8 [PubMed] [Google Scholar]
- 52.Villegas E, Gilliland SE. 2006. Hydrogen peroxide production by Lactobacillus delbrueckii subsp. lactis I at 5°C. J. Food Sci. 63:1070–1074. 10.1111/j.1365-2621.1998.tb15857.x [DOI] [Google Scholar]
- 53.von Canstein H, Ogawa J, Shimizu S, Lloyd JR. 2008. Secretion of flavins by Shewanella species and their role in extracellular electron transfer. Appl. Environ. Microbiol. 74:615–623. 10.1128/AEM.01387-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ellis RJ. 2001. Macromolecular crowding: an important but neglected aspect of the intracellular environment. Curr. Opin. Struct. Biol. 11:114–119. 10.1016/S0959-440X(00)00172-X [DOI] [PubMed] [Google Scholar]
- 55.Ohnishi K, Niimura Y, Yokoyama K, Hidaka M, Masaki H, Uchimura T, Suzuki H, Uozumi T, Kozaki M, Komagata K. 1994. Purification and analysis of a flavoprotein functional as NADH oxidase from Amphibacillus xylanus overexpressed in Escherichia coli. J. Biol. Chem. 269:31418–31423 [PubMed] [Google Scholar]
- 56.Pan N, Imlay JA. 2001. How does oxygen inhibit central metabolism in the obligate anaerobe Bacteroides thetaiotaomicron? Mol. Microbiol. 39:1562–1571. 10.1046/j.1365-2958.2001.02343.x [DOI] [PubMed] [Google Scholar]
- 57.Martins BM, Dobbek H, Cinkaya I, Buckel W, Messerschmidt A. 2004. Crystal structure of 4-hydroxybutyryl-CoA dehydratase: radical catalysis involving a [4Fe-4S] cluster and flavin. Proc. Natl. Acad. Sci. U. S. A. 101:15645–15649. 10.1073/pnas.0403952101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Denou E, Pridmore RD, Berger B, Panoff JM, Arigoni F, Brüssow H. 2008. Identification of genes associated with the long-gut-persistence phenotype of the probiotic Lactobacillus johnsonii strain NCC533 using a combination of genomics and transcriptome analysis. J. Bacteriol. 190:3161–3168. 10.1128/JB.01637-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.