Abstract
Xylan is an abundant plant cell wall polysaccharide and is a dominant component of dietary fiber. Bacteria in the distal human gastrointestinal tract produce xylanase enzymes to initiate the degradation of this complex heteropolymer. These xylanases typically derive from glycoside hydrolase (GH) families 10 and 11; however, analysis of the genome sequence of the xylan-degrading human gut bacterium Bacteroides intestinalis DSM 17393 revealed the presence of two putative GH8 xylanases. In the current study, we demonstrate that the two genes encode enzymes that differ in activity. The xyn8A gene encodes an endoxylanase (Xyn8A), and rex8A encodes a reducing-end xylose-releasing exo-oligoxylanase (Rex8A). Xyn8A hydrolyzed both xylopentaose (X5) and xylohexaose (X6) to a mixture of xylobiose (X2) and xylotriose (X3), while Rex8A hydrolyzed X3 through X6 to a mixture of xylose (X1) and X2. Moreover, rex8A is located downstream of a GH3 gene (xyl3A) that was demonstrated to exhibit β-xylosidase activity and would be able to further hydrolyze X2 to X1. Mutational analyses of putative active site residues of both Xyn8A and Rex8A confirm their importance in catalysis by these enzymes. Recent genome sequences of gut bacteria reveal an increase in GH8 Rex enzymes, especially among the Bacteroidetes, indicating that these genes contribute to xylan utilization in the human gut.
INTRODUCTION
Xylan is an abundant plant cell wall polysaccharide that consists of a β-(1,4)-linked xylosyl backbone with various degrees of polymerization and substitution. Xylans are the major polysaccharides in cereal-derived food products, fruits, and vegetables that are consumed daily by humans. The hydrolysis and fermentation of plant cell wall polysaccharides, inclusive of xylan, are important metabolic processes that contribute approximately 10% of the caloric requirements in human hosts (1). The fermentation process yields volatile fatty acids, including butyrate, which is a compound essential for maintaining the integrity of colonic epithelial cells (2–4). Due to the heterogeneous nature of xylan, its degradation requires a combination of different enzymes, including endoxylanases, β-xylosidases, α-l-arabinofuranosidases, α-glucuronidases, ferulic acid esterases, and acetylxylan esterases (5). Humans do not possess these enzymes; therefore, degradation of xylan in the intestine is a function exclusively undertaken by commensal microorganisms (6, 7).
Among the human gut microbiota, Bacteroides spp. are the most numerically dominant xylanolytic bacteria (8, 9). Past studies have identified Bacteroides eggerthii, B. ovatus, B. fragilis, B. vulgatus, B. intestinalis, B. cellulosilyticus, and B. xylanisolvens as the major xylanolytic members of this genus (10–17). Many of the Bacteroides spp. possess xylanases, which are required to depolymerize xylans by cleaving the long polymeric chain into shorter chains of xylose units. The genome sequences of Bacteroides ovatus ATCC 8483 and Bacteroides intestinalis DSM 17393 revealed that these organisms possess the most highly expanded repertoire of glycoside hydrolase and polysaccharide lyase genes among all gut bacteria sequenced to date (18). These genes are arranged in polysaccharide utilization loci (PULs) that are specifically regulated at the transcriptional level during growth with the cognate polysaccharides (19). In B. ovatus ATCC 8483, a relatively large number of genes are regulated at the transcriptional level during growth on xylan and these genes are highly expressed in vivo (20), indicating that they are important for xylan degradation by this bacterium. Despite the large number of genes induced by xylan, biochemical evidence to define the substrate specificities of these enzymes is lacking. This information is particularly important in helping to define the metabolic potential of these abundant gut bacteria.
The majority of xylanases that have been studied derive from the glycoside hydrolase (GH) families 10 and 11, with a relative minority belonging to GH families 5, 8, 30, and 43 (21, 22). Compared to xylanases in the GH10 and GH11 families, the substrate preference and hydrolysis product profiles of xylanases in GH families 5, 8, 30, and 43 have not been extensively studied. As of January 2014, the CAZy database has 729 entries in the GH8 family, with a total of 56 enzymes listed as characterized. Among these entries, six have been shown to degrade xylan, including endoxylanases (23–26) and reducing-end xylose-releasing exo-oligoxylanases (27, 28). The genome map of B. intestinalis DSM 17393 revealed the presence of two GH family 8 genes (BACINT_04210 and BACINT_00927) (Fig. 1). BACINT_04210 is located in a polysaccharide utilization locus (PUL) consisting of 11 genes (BACINT_04220 to BACINT_ 04210). BACINT_00927 is located downstream of a predicted GH3 glycosidase (BACINT_00926). The genomic context of these genes indicates possible roles in xylan degradation.
FIG 1.
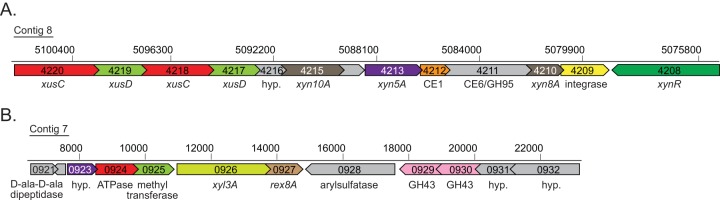
Genomic context for the two Bacteroides intestinalis GH8 genes. (A) The xyn8A gene (BACINT_04210) is located within a large xylan-specific polysaccharide utilization locus (BACINT_04220 to BACINT_04210). An integrase gene (BACINT_04209) and an xynR transcriptional regulator homolog (BACINT_04208) are also located in close proximity to the xyn8A gene. (B) The rex8A gene (BACINT_00927) is not located within a polysaccharide utilization locus but is preceded immediately by xyl3A (BACINT_00926), which encodes a GH family 3 β-xylosidase.
In this study, the protein coding sequences for BACINT_00926, BACINT_00927, and BACINT_04210 were cloned, and the proteins were expressed in Escherichia coli and purified to near homogeneity. It is hypothesized that these three genes are involved in xylan degradation, and therefore, the activities of the three proteins against xylo-oligosaccharides and natural xylans were evaluated. The important catalytic residues in the two GH8 enzymes were also evaluated by site-directed mutagenesis. Results from this study reveal that BACINT_04210 encodes an endoxylanase (Xyn8A), BACINT_00926 encodes a β-xylosidase (Xyl3A), and BACINT_00927 encodes a reducing-end xylose-releasing exo-oligoxylanase (Rex8A). Xyl3A cleaves xylobiose released by Rex8A, thus representing an alternative xylan-degrading pathway in gut bacteria involving GH8 and GH3 enzymes.
MATERIALS AND METHODS
Materials and strains.
Bacteroides intestinalis DSM 17393 (29) was obtained from the DSMZ (Braunschweig, Germany). Escherichia coli XL-10 Gold competent cells and E. coli BL-21 CodonPlus(DE3) RIL competent cells were obtained from Agilent (Santa Clara, CA). Medium viscosity wheat arabinoxylan (WAX) and xylo-oligosaccharides were obtained from Megazyme (Bray, Ireland). All other reagents were obtained from Sigma-Aldrich or Fisher Scientific.
Gene cloning, expression, and protein purification.
B. intestinalis DSM 17393 genomic DNA was extracted using the UltraClean Soil DNA isolation kit from Mo-Bio (Carlsbad, CA) according to the manufacturer's protocol. The concentrations of total DNA were quantified using the Qubit dsDNA BR assay kit (Invitrogen, Grand Island, NY). Oligonucleotide primers used for amplifying xyl3A, rex8A, and xyn8A (Table 1) were engineered to include 5′ and 3′ extensions for subsequent ligation-independent cloning (LIC). Signal peptide cleavage sites were predicted at the N terminus of each protein using SignalP v4.1 (http://www.cbs.dtu.dk/services/SignalP/) (30). Thus, to ensure that the protein accumulates within the E. coli cells, the forward primers were designed to amplify the genes beginning with the codon immediately downstream of the predicted peptidase cleavage site. The coding sequences for these three genes were amplified by PCR using the PicoMaxx high-fidelity PCR mix from Agilent, and the resulting amplicons were purified using the Wizard DNA purification kit (Promega, Madison, WI). The purified amplicons were digested using the exonuclease activity of T4 DNA polymerase, annealed with a similarly digested pET-46b vector (EMD Chemicals, Darmstadt, Germany), and introduced into E. coli XL10 Gold competent cells by chemical transformation. Individual colonies were picked and cultured overnight in lysogeny broth (LB) supplemented with ampicillin (100 μg/ml), and plasmid DNA was purified using a Plasmid Minikit from Qiagen (Valencia, CA). The cloned inserts were then sequenced to confirm the integrity of the genes (W. M. Keck Center for Comparative and Functional Genomics at the University of Illinois).
TABLE 1.
Primers used in this study
| Gene use and name | Orientation | Sequence(5′–3′)a | Desired mutation |
|---|---|---|---|
| Cloninga | |||
| xyl3A (BACINT_00926) | Forward | GACGACGACAAGATGCAACCTCCCTACAAAAACCC | |
| Reverse | GAGGAGAAGCCCGGTTATTTAACAATGACTGGTATCGCC | ||
| rex8A (BACINT_00927) | Forward | GACGACGACAAGATGGACCCGACAAAGCCCTGGGATAAAG | |
| Reverse | GAGGAGAAGCCCGGTTACTTTTCAGGGAAGATGATGCGGTAG | ||
| xyn8A (BACINT_04210) | Forward | GACGACGACAAGATGCATCCTGTTCAGGAAGACAGTAGTGGGG | |
| Reverse | GAGGAGAAGCCCGGTTACTTGATAATCCGGAAATTGCCACTGACATGC | ||
| Mutagenesisb | |||
| rex8A E90A | Forward | CATGATGTCCGCACCGCAGGTATGTCTTACGGA | Glu90Ala |
| rex8A D148A | Forward | GGCCCCGCCTCCGCCGGAGAACTTTACT | Asp148Ala |
| rex8A D286A | Forward | GATGCTTTCCGCTTCGCTTCTTGGCGTGTACCG | Asp286Ala |
| xyn8A E104A | Forward | AATCAGGATGTACGTACAGCAGGAATGTCCTATGGAATG | Glu104Ala |
| xyn8A D164A | Forward | AGGAGCCAAGTTGCGCTTCTGCTGGTGAAATTTATTTTATAACT | Asp164Ala |
| xyn8A D303A | Forward | CAAGAAGATATCAGTTTGCTGCTCTTCGCTGTGCCAT | Asp303Ala |
Underlined sequences indicate the incorporated T4 exonuclease digestion sites from the pET-46b Ek-LIC cloning kit.
Underlined sequences indicate the substituted codon. Primers were designed using the Agilent QuikChange Primer Design tool (http://www.genomics.agilent.com/primerDesignProgram.jsp).
The recombinant pET-46 EK/LIC plasmids containing the cloned genes (xyl3A, rex8A, or xyn8A) were introduced into E. coli BL-21 CodonPlus(DE3) RIL chemically competent cells (Agilent, Santa Clara, CA) by the heat shock transformation method and cultured overnight on LB agar plates supplemented with ampicillin (100 μg/ml) and chloramphenicol (50 μg/ml) at 37°C. After 12 h, a single colony was used to inoculate fresh LB (10 ml) supplemented with the same antibiotics and cultured with vigorous aeration at 37°C for 8 h. The culture was then diluted into 1 liter of fresh LB supplemented with ampicillin (100 μg/ml) and chloramphenicol (50 μg/ml) in 2.8-liter Fernbach flasks, and the cultures were incubated at 37°C with vigorous aeration by shaking at 250 rpm. When the culture reached an optical density at 600 nm (OD600) of 0.3, isopropyl β-d-thiogalactopyranoside (IPTG) was added to a final concentration of 0.1 mM, and the culture was incubated at 16°C for an additional 16 h. The cells were then harvested by centrifugation (4,000 × g, 15 min, 4°C). The cell pellets were resuspended in 30 ml of ice-cold lysis buffer (50 mM Tris-HCl, 300 mM NaCl, pH 7.5) and ruptured by two passages through an EmulsiFlex C-3 cell homogenizer from Avestin (Ottawa, Canada). The cell lysates were clarified by centrifugation at 12,000 × g for 30 min at 4°C. The recombinant proteins were then purified from the clarified lysates using Talon metal affinity resin (ClonTech, Mountain View, CA) according to the supplier's protocol with slight modifications to the binding (50 mM Tris-HCl, 300 mM NaCl, pH 7.5) and elution (50 mM Tris-HCl, 300 mM NaCl, 250 mM imidazole, pH 7.5) buffers. Aliquots of eluted fractions were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) according to Laemmli's method (31), and protein bands were visualized by staining with Coomassie brilliant blue G-250. The protein concentrations were calculated by absorbance spectroscopy at 280 nm using a NanoDrop 1000 instrument (Thermo Scientific, Waltham, MA) with extinction coefficients of 120,670 M−1 cm−1, 117,940 M−1 cm−1, and 103,625 M−1 cm−1 for Xyl3A, Rex8A, and Xyn8A, respectively.
Size exclusion chromatography.
The quaternary structures of Xyn8A and Rex8A were analyzed by size exclusion chromatography using a Superdex 200 10/300 GL size exclusion column affixed to an AKTAxpress FPLC unit. One hundred microliters of Xyn8A (1 mg/ml), Rex8A (1 mg/ml), or a gel filtration standard mixture (Bio-Rad, Hercules, CA) was loaded onto the column preequilibrated with a buffer composed of 50 mM sodium phosphate–150 mM NaCl. The pH of the buffer was adjusted to 6.0 for Rex8A and 6.5 for Xyn8A. The proteins were eluted in the same buffer at a flow rate of 0.5 ml/min. A calibration curve of molecular mass versus retention time was constructed with the gel filtration standards, and the apparent molecular masses of the two proteins were calculated by comparison of experimental retention times with calibration standards.
CD spectroscopy.
Circular dichroism (CD) spectra were obtained using a J-815 spectropolarimeter from Jasco (Easton, MD). The enzymes were exchanged into CD buffer (50 mM sodium phosphate, pH 6.0 for Rex8A or pH 6.5 for Xyn8A) using a HiPrep 26/10 desalting column affixed to an AKTAxpress FPLC unit. The proteins were then diluted in CD buffer to a final concentration of 1 μM, and the far-UV CD spectra were recorded from 260 to 190 nm with a wavelength step of 0.1 nm. The secondary-structure contents of Xyn8A and Rex8A were calculated using the DichroWeb online circular dichroism analysis server with the reference set 4 optimized for 190 to 240 nm (32).
Evaluation of hydrolysis of xylo-oligosaccharides.
The capacities of Xyn8A, Rex8A, and Xyl3A to hydrolyze xylo-oligosaccharides were tested with xylose (X1), xylobiose (X2), xylotriose (X3), xylotetraose (X4), xylopentaose (X5), and xylohexaose (X6). Substrates (5 mM) were diluted in phosphate reaction buffer (50 mM sodium phosphate, 150 mM NaCl, pH 6.0 for Rex8A or pH 6.5 for Xyn8A and Xyl3A); reactions were initiated by the addition of enzyme (0.5 μM final concentration), and the mixtures were incubated at 37°C for 16 h. Preliminary experiments revealed pH optima of 6.0 for Rex8A and 6.5 for Xyn8A and Xyl3A (see Fig. S1 in the supplemental material). Reaction mixtures were terminated by heating at 99°C for 10 min, and 1-μl aliquots of samples were spotted onto silica gel thin-layer chromatography (TLC) plates (60 Å, 250-μm thickness) (Whatman, Piscataway, NJ). The products of hydrolysis were then resolved by one ascent with a mobile phase composed of n-butanol, acetic acid, and water (10:5:1). The plates were then dried, and products were visualized by spraying with a mixture of sulfuric acid (10%, vol/vol), orcinol (0.1%, wt/vol), and methanol (50%, vol/vol) and developed by heating at 80°C for 15 min.
To evaluate the hydrolysis patterns over time, enzyme concentrations that gave near-complete hydrolysis over the 3-h reaction period for each enzyme-substrate combination were first determined. For Rex8A, the enzyme concentration used was 50 nM (X3 through X6), whereas for Xyn8A the concentration was 500 nM (X4), 20 nM (X5), or 2 nM (X6). Reaction mixtures were prepared with substrate (1 mM) and enzyme, incubated at 37°C, and were then taken at 10-, 30-, 60-, 120-, 180-, and 360-min intervals for heat inactivation. The products of hydrolysis were first diluted 50-fold and then analyzed using high-performance anion exchange chromatography (HPAEC) with a System Gold high-performance liquid chromatography (HPLC) instrument (Beckman Coulter, Fullerton, CA). The instrument was fitted with a CarboPac PA1 guard column (4 by 50 mm) and a CarboPac PA1 analytical column (4 by 250 mm) from Dionex Corporation (Sunnyvale, CA) and with a Coulochem III electrochemical detector (ESA Biosciences, Chelmsford, MA). Peak retention times and peak areas from sample chromatograms were compared to those obtained using commercially available xylo-oligosaccharides analyzed as standards.
Evaluation of hydrolysis of natural xylans.
The capacity of the enzymes to hydrolyze natural xylans (i.e., wheat arabinoxylan [WAX] and oat spelt xylan [OSX]) was assessed by dissolving WAX or OSX (2.5 mg/ml) in phosphate buffer (50 mM, 150 mM NaCl,; pH 6.0 for Rex8A or pH 6.5 for Xyn8A; final volume, 100 μl). Reactions were initiated by the addition of enzyme (final concentration, 0.5 μM), and reaction mixtures were incubated at 37°C for 16 h. The reaction mixtures were then heat inactivated at 99°C for 10 min and centrifuged for 10 min at 15,000 × g, and hydrolysis products in the supernatants were evaluated by TLC as described above. The concentrations of reducing sugars were quantified by the PAHBAH (para-hydroxybenzoic acid hydrazide) method as previously described (33).
Site-directed mutagenesis.
Mutagenesis was performed using the QuikChange Lightning Multi Site-directed mutagenesis kit (Agilent Technologies, Santa Clara, CA). Mutagenic primers were designed with the QuikChange Primer Design tool (Agilent Technologies) (Table 1). Mutations were prepared according to the QuikChange protocol with either the rex8A or the xyn8A pET-46b plasmid as the DNA template. The residual parent plasmid was then digested by incubation with DpnI overnight at 37°C, and the resulting DNA was transformed into E. coli XL10-Gold ultracompetent cells by the heat shock method. Colonies were picked and cultured, and plasmids were purified using a Plasmid Minikit (Qiagen, Valencia, CA). The purified plasmid DNA was then sequenced to ensure that the appropriate mutations were introduced and that the rest of the gene sequences remained unchanged. Expression and purification of the mutant recombinant proteins were performed as described above for the wild-type (WT) proteins.
Hydrolysis of pNP-linked sugars.
The enzyme-catalyzed hydrolysis of para-nitrophenyl (pNP)-linked monosaccharide substrates was assayed by using a thermostated Synergy II multimode microplate reader (BioTek Instruments Inc., Winooski, VT). A library of pNP substrates was screened for activity as described previously (34). The substrates (1 mM) in 100 μl phosphate buffer (50 mM sodium phosphate, 150 mM NaCl, pH 6.5) were incubated at 37°C in the presence or absence of Xyl3A (0.1 μM for pNPXyl; 0.5 μM for pNPAra; 1 μM for pNPGal and pNPGlu) for 30 min, and the amount of pNP release was determined by continuously monitoring the absorbance at 400 nm. The path length correction feature of the instrument was employed to convert the absorbance values recorded to correspond to those for a 1-cm path length. The extinction coefficient for pNP at pH 6.5 and at a wavelength of 400 nm was measured to be 3.179 mM−1 cm−1.
RESULTS
Identification of two xylan-specific GH8 genes in Bacteroides intestinalis.
Genomic analysis of Bacteroides intestinalis DSM 17393 revealed the presence of two GH8 genes, BACINT_04210 and BACINT_00927, which encode the proteins Xyn8A and Rex8A, respectively. The genomic context for these genes supports their predicted roles in xylan degradation. To illustrate, BACINT_04210 is located in a polysaccharide utilization locus (PUL) consisting of 11 genes (BACINT_04220 to BACINT_04210). Included in this PUL is a large gene cluster consisting of two tandem repeats of susC and susD orthologs (xusC and xusD) followed by a hypothetical gene (BACINT_04216) and a GH10 endoxylanase (BACINT_04215) (Fig. 1A). Downstream are a hypothetical protein (BACINT_04214), a GH5 endoxylanase (xyn5A, BACINT_04213) (17), a family 1 carbohydrate esterase (BACINT_04212), a two-domain CE6/GH95 gene (BACINT_04211), and the GH8 gene BACINT_04210. Following this cluster is a predicted integrase, an observation that suggests that this locus may be part of an integrative and conjugative element. Divergently transcribed relative to the gene cluster described above is a homolog of xynR, a hybrid two-component system regulator that regulates xylanase gene expression in Prevotella bryantii B14 (35). The second GH8 gene (BACINT_00927) is located directly downstream of a predicted GH3 glycosidase (Fig. 1B).
Both of these proteins, Xyn8A and Rex8A, possess putative signal peptides with predicted signal peptidase II cleavage sites, followed by GH8 domains. Amino acid sequence alignments of the GH8 domains demonstrated 39% amino acid sequence identity (see Fig. S1 in the supplemental material).
Cloning, expression, and purification of Xyn8A and Rex8A.
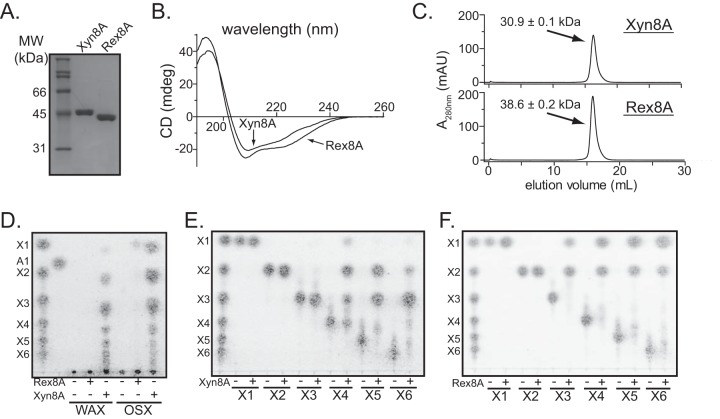
To determine whether or not the two GH8 genes encode proteins with redundant biochemical properties, they were individually cloned into an E. coli expression vector. The two proteins were then expressed as soluble recombinant hexahistidine fusion proteins for subsequent purification via cobalt immobilized metal affinity chromatography (IMAC). In a single IMAC step, the two recombinant proteins were purified to near homogeneity as assessed by SDS-PAGE (Fig. 2A).
FIG 2.
The two Bacteroides intestinalis GH8 genes encode xylan-degrading enzymes with distinct properties. (A) Purification of the two GH8 proteins, Xyn8A and Rex8A. The proteins were produced in their recombinant forms in E. coli, purified by immobilized metal affinity chromatography, resolved by 12% SDS-PAGE, and stained with Coomassie brilliant blue G250. (B) Predicted secondary structure of the two GH8 proteins. Circular dichroism (CD) spectra were obtained for Rex8A and Xyn8A in the far-UV region (190 to 260 nm). (C) Gel filtration chromatography. The sizes of purified Rex8A and Xyn8A were determined by size exclusion fast-protein liquid chromatography (FPLC). The molecular masses of Rex8A and Xyn8A were calculated from the retention time of the peak absorbance by comparison with calibration standards having known molecular masses. (D) Hydrolysis of model xylans. Rex8A and Xyn8A were incubated with wheat arabinoxylan (WAX) or oat spelt xylan (OSX) for 16 h, and the products of hydrolysis were analyzed by thin-layer chromatography (TLC). (E and F) Hydrolysis of xylo-oligosaccharides (XOS). Rex8A (E) or Xyn8A (F) was incubated with XOS of different lengths, and the products were analyzed by TLC.
Recombinant Xyn8A and Rex8A have similar secondary structures.
To compare the secondary structural compositions of the two recombinant proteins, CD spectra in the far-UV region (190 to 260 nm) were collected (Fig. 2B). The raw data were then uploaded onto the DichroWeb server, and secondary structural elements were predicted by comparison with a standardized data set. The recombinant proteins both yielded CD spectra indicating the presence of α-helix, β-sheet, and β-turn secondary structural elements. Comparison of the DichroWeb results using the Student t test revealed no significant differences in α-helices, β-sheets, β-turns, or unordered regions between the two proteins (Table 2). These data suggest that when expressed as recombinant proteins, Xyn8A and Rex8A have similar overall secondary structural properties.
TABLE 2.
Analysis of CD spectra using DichroWeba
| Protein (ORF no.) | % α-helix | % β-sheet | % β-turn | % Unordered |
|---|---|---|---|---|
| Xyn8A (BACINT_04210) | 71 ± 6 | 9 ± 3 | 9 ± 4 | 10 ± 3 |
| Rex8A (BACINT_00927) | 81 ± 2 | 6 ± 1 | 6 ± 2 | 6 ± 2 |
CD spectra were recorded in the far-UV range utilizing a J-815 CD spectropolarimeter. The buffer composition was 50 mM sodium phosphate, pH 6.0 for Rex8A or pH 6.5 for Xyn3A. The spectra were recorded from 190 to 260 nm at a scan rate of 50 nm/s and a 0.1-nm wavelength step with five accumulations. Data are from three independent experiments and are presented as means ± standard deviations of the means. The spectra were uploaded onto the DichroWeb online server and analyzed as described in Materials and Methods. ORF, open reading frame.
Xyn8A and Rex8A are both monomers in solution.
To evaluate whether the two proteins exhibited similar quaternary structures and to rule out whether the two proteins exist as aggregates, their apparent molecular weights were determined by gel filtration analysis. Both proteins eluted from the column well after the size exclusion limit, indicating that neither of the two proteins existed as large aggregates in solution. The elution volumes of the two proteins revealed apparent molecular weights slightly lower than those demonstrated by SDS-PAGE. This discrepancy suggests that the proteins may have different shapes from those of the proteins in the calibration standards. Nevertheless, these results indicate that both proteins exist as monomers in solution (Fig. 2C).
Xyn8A and Rex8A exhibit different biochemical properties.
The two proteins were incubated with either wheat arabinoxylan (WAX), oat spelts xylan (OSX), or xylo-oligosaccharides (XOS), and their capacity to degrade the substrates was assessed by analyzing the hydrolysates by TLC. In the absence of enzyme, no oligosaccharides were present in either the WAX or the OSX mixture (Fig. 2D). However, upon the addition of Xyn8A, several spots appeared, corresponding to shorter xylo-oligosaccharides being released, an activity characteristic of endoxylanase enzymes (Fig. 2D). The spots did not clearly comigrate with the standard XOS, which is likely explained by the fact that these products are xylo-oligosaccharides substituted with arabinosyl side chains. In contrast, no detectable degradation of the two polysaccharides was noted for Rex8A.
Both enzymes degraded XOS (Fig. 2E and Fig. 2F); however, Xyn8A exhibited no detectable activity toward X2 and X3. Upon incubation with xylotetraose, a mixture of X3, X2, and X1 was observed, and a mixture of X3 and X2 was seen following incubation with xylopentaose and xylohexaose. In contrast, Rex8A completely converted X3 through X6 to a mixture of X2 and X1. Most notably, the two enzymes displayed differences in the product distribution profiles for X5, with Xyn8A cleaving X5 into X2 and X3, whereas Rex8A converted X5 to X1 and X2. This result suggests that the two enzymes have different cleavage site preferences, with Xyn8A cleaving the middle glycosidic linkage within X5, and Rex8A cleaving one of the terminal glycosidic linkages in X5, producing X4 and X1 and then converting X4 to X3 and X1 and so on.
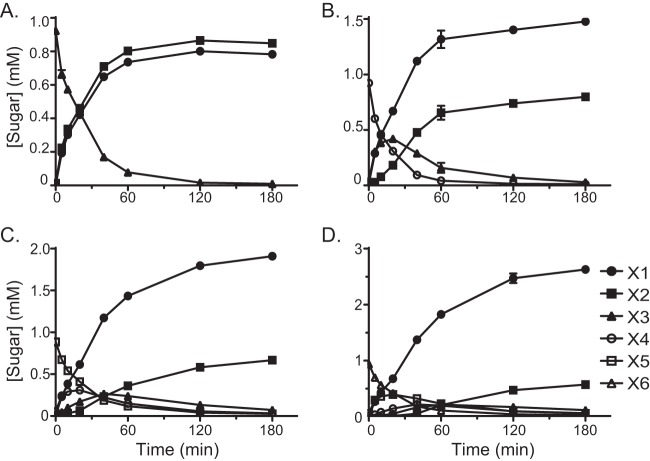
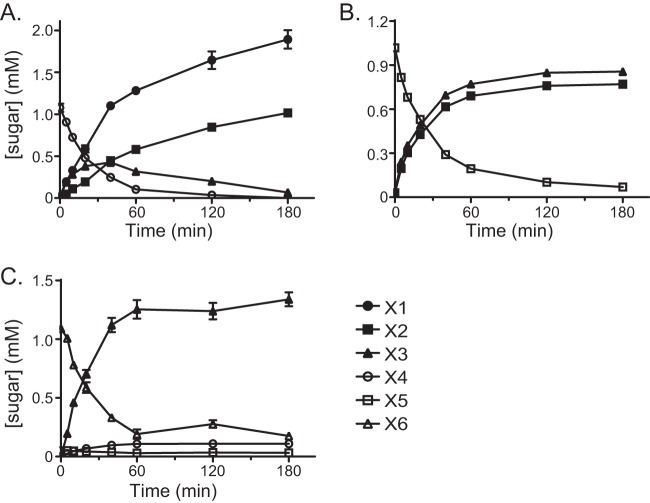
To further evaluate the hydrolytic activity of Rex8A with XOS, the enzyme was incubated with X3 through X6, and the concentrations of X1 through X6 were followed over time by HPLC. These experiments confirmed results from the TLC plates, which showed that the predominant products at the final time point of the reaction were X1 and X2 for all substrates tested (Fig. 3). Rex8A converted all of the X3 to a mixture of X1 and X2 by 180 min of incubation (Fig. 3A). Rex8A hydrolyzed all of the X4 substrate after 180 min of incubation by first accumulating X1 and X3 and further hydrolyzing the accumulated X3 to X2 and X1 (Fig. 3B). With X5 as a substrate, there was an initial accumulation of X4 and X1 and a subsequent hydrolysis of X4 to accumulate X3 (Fig. 3C). Similarly, in the hydrolysis of X6, X5 first accumulated during 20 min of incubation and was subsequently hydrolyzed to X4 and X3 and finally to X1 and X2 by the end of the 180-min incubation (Fig. 3D). At a relatively high concentration of Xyn8A (500 nM), X4 was converted first to X3 and X1, and then the X3 was eventually hydrolyzed to X2 and X1 (Fig. 4A). The enzyme exhibited much higher activity with longer oligosaccharides; therefore, the amount of enzyme added was decreased to observe the initial hydrolysis products (X5, 20 nM; X6, 2 nM). Under these conditions, X5 was converted to stoichiometric amounts of X3 and X2, whereas X6 was cleaved, accumulating mainly X3 (Fig. 4B and C).
FIG 3.
Hydrolysis of XOS by Rex8A (BACINT_00927). Rex8A was incubated with xylotriose (A), xylotetraose (B), xylopentaose (C), and xylohexaose (D), and products were analyzed at designated intervals using high-performance anion exchange chromatography (HPAEC). Abbreviations are as follows: X6, xylohexaose; X5, xylopentaose; X4, xylotetraose; X3, xylotriose; X2, xylobiose; and X1, xylose). Concentrations were determined by comparison to calibration curves constructed with known concentrations of sugars. Results are presented as means and standard deviations from three individual experiments.
FIG 4.
Hydrolysis of XOS by Xyn8A (BACINT_04210). Xyn8A was incubated with xylotetraose (A), xylopentaose (B), and xylohexaose (C), and products were analyzed at designated intervals using high-performance anion exchange chromatography (HPAEC). Abbreviations are as follows: X6, xylohexaose; X5, xylopentaose; X4, xylotetraose; X3, xylotriose; X2, xylobiose; and X1, xylose. Concentrations were determined by comparison to calibration curves constructed with known concentrations of sugars. Results are presented as means and standard deviations from three individual experiments.
Taken together, the activities seen for Xyn8A with polysaccharides and XOS indicate that this enzyme is an endoxylanase, whereas the activity for Rex8A was clearly very different from that of Xyn8A and is consistent with the activity seen for reducing-end xylose-releasing exo-oligoxylanases, which have been demonstrated for GH8 proteins (27, 28).
Site-specific mutagenesis to elucidate catalytic residues.
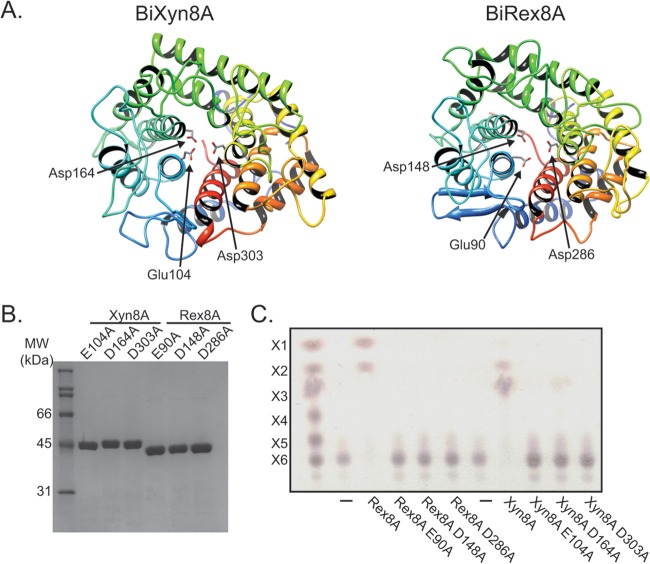
To predict amino acid residues important for catalysis, three-dimensional homology models were constructed for Rex8A and Xyn8A with the reducing-end xylose-releasing exo-oligoxylanase (Rex) from Bacillus halodurans (PDB accession number 1WU4) and the cold-adapted endoxylanase from Pseudoalteromonas haloplanktis Xyn8A (PDB accession number 1H12), respectively. These structures revealed that the three catalytic residues proposed previously for REX (27, 36) (Glu90, Asp148, Asp286) were conserved in both Rex8A and Xyn8A (Fig. 5A). Therefore, to evaluate whether these residues (Glu104, Asp164, and Asp303 for Xyn8A and Glu90, Asp148, and Asp286 for Rex8A) were also important for Xyn8A and Rex8A, these three residues were changed to alanine by site-directed mutagenesis.
FIG 5.
Mutational analysis reveals residues important for catalysis in Xyn8A (BACINT_04210) and Rex8A (BACINT_00927). The residues are Glu104, Asp164, and Asp303 for Xyn8A. The residues are Glu90, Asp148, and Asp286 for Rex8A. (A) Three-dimensional homology modeling. Homology models were built for Xyn8A and Rex8A using the ModWeb server (http://modbase.compbio.ucsf.edu/ModWeb20-html/modweb.html) with Pseudoalteromonas haloplanktis Xyn8A (PDB accession no. 1H12) and Bacillus halodurans Rex (PDB accession no. 1WU4) as the templates, respectively. (B) Purification of mutants. Three mutants for each Xyn8A and Rex8A were expressed as recombinant proteins, purified by cobalt affinity chromatography, and analyzed by SDS-PAGE. (C) Hydrolysis of xylohexaose. Wild-type and mutant proteins were incubated with xylohexaose (X6) for 16 h, and the reaction products were analyzed by thin-layer chromatography.
The three mutants for each protein were expressed in E. coli and purified by cobalt IMAC as described for the wild-type protein. All six proteins were expressed in the soluble fraction, indicating that the mutations did not cause the proteins to form insoluble aggregates. Similar to the wild-type protein, the mutants were purified after a single chromatography step (Fig. 5B). The proportions of secondary structural elements between the WT and the mutants were determined and compared by circular-dichroism (CD) spectroscopy to ensure that the change in enzyme activity was not caused by gross structural differences. Comparison of the α-helix, β-sheet, β-turn, and unordered regions using the Student t test revealed no significant differences between the wild-type and mutant forms of Rex8A and Xyn8A (see Table S1 in the supplemental material). These data demonstrate that mutation of these active-site residues did not appreciably alter the secondary structures of the proteins.
Following 16 h of incubation at 37°C with X6, only the D164A mutant of Xyn8A exhibited minor residual activity compared to the wild type, with a trace amount of X3 visualized on the TLC plate (Fig. 5C). For the three Rex8A mutants, no activity was detected (Fig. 5C). These results therefore show that these three carboxylate-containing residues are important for catalysis in both Rex8A and Xyn8A.
Identification of Xyl3A as a GH3 β-xylosidase.
Rex8A was observed to hydrolyze X3 through X6 to a mixture of xylose and xylobiose (Fig. 2F). Given that rex8A is located immediately downstream of a predicted GH3 glycosidase gene, it is possible that the xylobiose produced by Rex8A would be converted to xylose by this GH3 enzyme. Because GH3 glycosidases have a very broad substrate range, it was decided to clone this GH3 glycosidase gene and study its biochemical properties. The protein was produced recombinantly in E. coli as described for Xyn8A and Rex8A and purified to near homogeneity by cobalt IMAC (Fig. 6A). Biochemical activity assays with a library of para-nitrophenyl (pNP)-linked substrates revealed that the enzyme had highest activity with pNP-β-d-xylopyranoside as a substrate (Fig. 6B). These results suggest that the enzyme is a β-xylosidase. To further confirm this with natural substrates, the enzyme was incubated with XOS and the capacity to hydrolyze these substrates was assessed by TLC. Following incubation of the purified protein with XOS ranging in length from X2 through X6, the sole product visualized was xylose (Fig. 6C). These results indicate that BACINT_00926 encodes a GH3 β-xylosidase, and the associated enzyme was therefore named Xyl3A.
FIG 6.
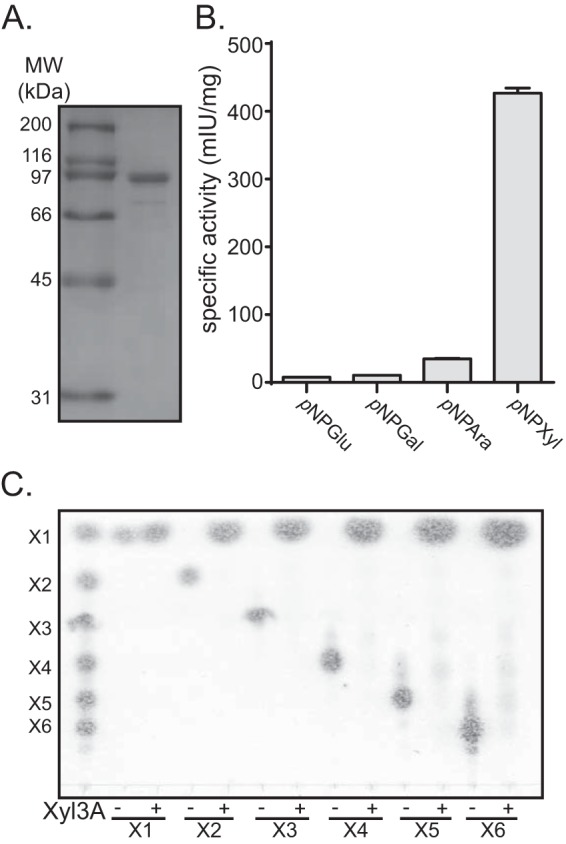
BACINT_00926 encodes a GH3 β-xylosidase, Xyl3A. (A) Expression and purification of Xyl3A. Xyl3A was expressed as a recombinant protein, purified by cobalt affinity chromatography, and analyzed by SDS-PAGE. (B) Activity with artificial substrates. Purified Xyl3A was screened with a library of pNP-linked glycans using a continuous spectrophotometric assay, and the specific activities for a subset of substrates are shown. Abbreviations are as follows: pNPAra, pNP-α-l-arabinofuranoside; pNPGal, pNP-β-d-galactopyranoside; pNPXyl, pNP-β-d-xylopyranoside; pNPGlu, pNP-β-d-glucopyranoside. (C) Hydrolysis of XOS. Xyl3A was incubated with XOS ranging in length from xylobiose (X2) to xylohexaose (X6) for 16 h, and products of hydrolysis were analyzed by thin-layer chromatography. Specific activity (mIU/mg) is displayed in units of nmol substrate consumed per minute per mg of protein.
DISCUSSION
A recent analysis of human gut microbiome reference genomes revealed that of all gut bacterial strains, Bacteroides intestinalis DSM 17393 exhibited the highest representation of individual glycoside hydrolase (GH) and polysaccharide lyase (PL) families and the second highest total number of GH and PL genes (18). The number of genes in the B. intestinalis genome targeted toward degradation of the plant cell structural polysaccharide xylan is particularly high. Coupled with previous studies of this organism (17, 37), the current data demonstrate that B. intestinalis DSM 17393 encodes at least nine endoxylanase enzymes deriving from three different GH families (i.e., GH10, GH5, and GH8).
Expansion of GH families is a characteristic of many gut bacteria, most notably those from the Bacteroidetes phylum (18, 38, 39). The current study highlights this process by revealing that B. intestinalis DSM 17393 harbors two copies of genes encoding xylan-specific GH8 enzymes with completely distinct biochemical properties. Xyn8A is a GH8 endoxylanase enzyme that targets longer xylan fragments, hydrolyzing them to shorter xylo-oligosaccharides that can subsequently be degraded by side-chain-cleaving enzymes and β-xylosidases. Rex8A, on the other hand, is likely a reducing-end xylose-releasing exo-oligoxylanase that releases xylose from the reducing end of xylo-oligosaccharides, liberating fermentable monosaccharides. The discovery of these two enzymes broadens our understanding of xylan degradation by gut bacteria and provides insight into the highly dynamic genomic assemblages that gut bacteria possess to capture energy from dietary polysaccharides.
Rex8A forms the core of a group of GH8 enzymes that derive largely from bacteria isolated from human gastrointestinal tract and rumen sources (Fig. 7A). Despite sharing high amino acid sequence similarity, the polypeptide domain architectures vary remarkably among these different organisms. Another human gut bacterium, Bacteroides eggerthii, possesses a carbohydrate esterase (CE) family 15 domain at the N terminus, while Prevotella copri, Prevotella bergensis, and Bacteroides uniformis possess N-terminal GH43/CBM6 modules (Fig. 7B). These proteins are likely to be bifunctional enzymes with two distinct active sites. CE family 15 proteins have been demonstrated to cleave the methyl ester linkage in 4-O-methyl-glucuronyl methyl esters (40, 41), an activity that is important for fermenting the glucuronic acid component of heteroxylans. GH43/CBM6 proteins are commonly found to have either β-xylosidase or arabinofuranosidase activities, both of which are associated with the degradation of xylan fragments. Therefore, it is likely that these alternative forms of GH8 Rex enzymes have evolved to possess additional enzymatic activities that are important for xylan degradation.
FIG 7.
Expansion and diversity of Rex8A homologs in the gut microbiome. (A) Phylogenetic analysis of Xyn8A (BACINT_04210) and Rex8A (BACINT_00927). The amino acid sequences for GH8 proteins annotated by the CAZy database were aligned with Xyn8A and Rex8A using ClustalW, and a neighbor-joining tree was constructed using CLC Genomics Workbench v5.0 software. Each alignment was resampled 100 times, and the bootstrap values are indicated on the internal branches. The branch length is reported as the expected number of substitutions per amino acid position. GIT, gastrointestinal tract. (B) Protein domain structure of Rex8A homologs. Domain architectures were predicted using dbCAN (48). Signal peptides and lipoprotein signal sequences were predicted using SignalP v4.1 (30) and LipoP v1.0 (43), respectively. Organism abbreviations: Bcel, Bacteroides cellulosilyticus DSM 14838; Bi, Bacteroides intestinalis; Bole, Bacteroides oleiciplenus YIT 12058; Begg, Bacteroides eggerthii DSM 20697; Buni, Bacteroides uniformis CL03T12C37; Pber, Prevotella bergensis DSM 17361; Pcop, Prevotella copri DSM 18205; Dmos, Dysgonomonas mossii DSM 22836; Prum, Prevotella ruminicola 23; Dgad, Dysgonomonas gadei ATCC BAA-286; Mpal, Mucilaginibacter paludis DSM 18603; Slin, Spirosoma linguale DSM 74; Fjoh, Flavobacterium johnsoniae UW101; Bhal, Bacillus halodurans C-125; Bsp, Bacillus sp. 10403023; Pgol, Parabacteroides goldsteinii CL02T12C30; Fsuc, Fibrobacter succinogenes subsp. succinogenes S85; Chut, Cytophaga hutchinsonii ATCC 33406; Ccel, Clostridium cellulolyticum H10; Acel, Acetivibrio cellulolyticus; Athe, Anaerolinea thermophila UNI-1; Phal, Pseudoalteromonas haloplanktis; Cthe, Clostridium thermocellum ATCC 27405.
The rex8A gene is located immediately downstream of the xyl3A gene, which encodes a GH3 β-xylosidase. This unique pairing of two genes encoding enzymes with activity against xylo-oligosaccharides is deserving of further discussion. Rex enzymes possess an active site with a minimum binding requirement for three xylose sugars that is closed off at the terminal reducing end. Catalysis then occurs between the sugars occupying the −1 and +1 subsites, liberating xylose and the remaining oligosaccharide chain (36). Importantly, the minimum requirement of binding 3 sugars for catalysis means that the shortest XOS that this enzyme can cleave is xylotriose, and therefore xylobiose is not cleaved by these enzymes. On the other hand, GH3 glycosidases have a short coin-shaped active site that contains two subsites (42); therefore, enzymes from this family efficiently cleave xylobiose (34). These two enzymes have complementary activities that may be important for the mechanism of xylan degradation employed by this organism.
The three proteins described in this study each possess an N-terminal signal peptide with a signal peptidase cleavage II site, suggesting that they are transferred across the inner membrane and anchored to a lipid moiety (43). The so-called “+2” rule implies that prolipoproteins containing an aspartic acid at the +2 position relative to the signal peptidase II cleavage site results in retention of the polypeptide on the inner membrane within the cytoplasm, whereas a serine residue directs the protein to the outer membrane (44, 45). Xyn8A, Rex8A, and Xyl3A all possess serine residues at the +2 position, which suggests that these proteins are all localized to the outer membrane.
Since GH8 Rex enzymes represent a recently discovered group of enzymes, relatively few studies have evaluated the catalytic residues for these enzymes. In the current study, we identified amino acid residues (E90, D148, and D286) that were absolutely conserved among the GH8 Rex and Xyn enzymes and made the corresponding mutations in Xyn8A and Rex8A. These mutations had very large effects on the activities of the two enzymes, despite CD spectroscopy demonstrating no significant change in secondary structure (see Table S1 in the supplemental material). These data confirm results from previous studies that show residues corresponding to E90, D148, and D286 as having roles as catalytic acid, pKa modulator, and catalytic base, respectively (46).
In summary, this study reports distinct activities for two GH8 enzymes that are present in B. intestinalis DSM 17393, a bacterium that is endemic to the human gut. Our findings reiterate the previous observation that this enzyme family contains members with considerable differences in their substrate specificity (47). Furthermore, our results suggest a xylan degradation pathway active in gut Bacteroidetes that involves endoxylanases, a reducing-end oligo-xylanase, and a β-xylosidase. These results could be of importance in understanding the pathways of xylan degradation present in other gut microorganisms harboring GH8 enzymes.
Supplementary Material
ACKNOWLEDGMENT
This project was partially supported by Agriculture and Food Research Initiative Competitive Grant no. 2012-67015-19451 from the USDA National Institute of Food and Agriculture.
Footnotes
Published ahead of print 24 January 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.03176-13.
REFERENCES
- 1.Bergman E. 1990. Energy contributions of volatile fatty acids from the gastrointestinal tract in various species. Physiol. Rev. 70:567–590 [DOI] [PubMed] [Google Scholar]
- 2.Hague A, Singh B, Paraskeva C. 1997. Butyrate acts as a survival factor for colonic epithelial cells: further fuel for the in vivo versus in vitro debate. Gastroenterology 112:1036–1040. 10.1053/gast.1997.v112.agast971036 [DOI] [PubMed] [Google Scholar]
- 3.Roediger WE. 1980. The colonic epithelium in ulcerative colitis: an energy-deficiency disease? Lancet ii:712–715 [DOI] [PubMed] [Google Scholar]
- 4.Scheppach W, Bartram HP, Richter F. 1995. Role of short-chain fatty acids in the prevention of colorectal cancer. Eur. J. Cancer 31A:1077–1080 [DOI] [PubMed] [Google Scholar]
- 5.Dodd D, Cann IK. 2009. Enzymatic deconstruction of xylan for biofuel production. Glob. Change Biol. Bioenergy 1:2–17. 10.1111/j.1757-1707.2009.01004.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Davison A, Blaxter M. 2005. Ancient origin of glycosyl hydrolase family 9 cellulase genes. Mol. Biol. Evol. 22:1273–1284. 10.1093/molbev/msi107 [DOI] [PubMed] [Google Scholar]
- 7.Morris SC. 2003. Life's solution: inevitable humans in a lonely universe. Cambridge University Press, Cambridge, United Kingdom [Google Scholar]
- 8.Hespell RB, Whitehead TR. 1990. Physiology and genetics of xylan degradation by gastrointestinal tract bacteria. J. Dairy Sci. 73:3013–3022. 10.3168/jds.S0022-0302(90)78988-6 [DOI] [PubMed] [Google Scholar]
- 9.Chassard C, Goumy V, Leclerc M, Del'homme C, Bernalier-Donadille A. 2007. Characterization of the xylan-degrading microbial community from human faeces. FEMS Microbiol. Ecol. 61:121–131. 10.1111/j.1574-6941.2007.00314.x [DOI] [PubMed] [Google Scholar]
- 10.Chassard C, Delmas E, Lawson PA, Bernalier-Donadille A. 2008. Bacteroides xylanisolvens sp. nov., a xylan-degrading bacterium isolated from human faeces. Int. J. Syst. Evol. Microbiol. 58:1008–1013. 10.1099/ijs.0.65504-0 [DOI] [PubMed] [Google Scholar]
- 11.Salyers AA, Vercellotti JR, West SE, Wilkins TD. 1977. Fermentation of mucin and plant polysaccharides by strains of Bacteroides from the human colon. Appl. Environ. Microbiol. 33:319–322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Robert C, Chassard C, Lawson PA, Bernalier-Donadille A. 2007. Bacteroides cellulosilyticus sp. nov., a cellulolytic bacterium from the human gut microbial community. Int. J. Syst. Evol. Microbiol. 57:1516–1520. 10.1099/ijs.0.64998-0 [DOI] [PubMed] [Google Scholar]
- 13.Crittenden R, Karppinen S, Ojanen S, Tenkanen M, Fagerstrom R, Matto J, Saarela M, Mattila-Sandholm T, Poutanen K. 2002. In vitro fermentation of cereal dietary fibre carbohydrates by probiotic and intestinal bacteria. J. Sci. Food Agric. 82:781–789. 10.1002/jsfa.1095 [DOI] [Google Scholar]
- 14.Weaver J, Whitehead TR, Cotta MA, Valentine PC, Salyers AA. 1992. Genetic analysis of a locus on the Bacteroides ovatus chromosome which contains xylan utilization genes. Appl. Environ. Microbiol. 58:2764–2770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Whitehead TR. 1995. Nucleotide sequences of xylan-inducible xylanase and xylosidase/arabinosidase genes from Bacteroides ovatus V975. Biochim. Biophys. Acta 1244:239–241. 10.1016/0304-4165(95)00051-C [DOI] [PubMed] [Google Scholar]
- 16.Mirande C, Mosoni P, Bera-Maillet C, Bernalier-Donadille A, Forano E. 2010. Characterization of Xyn10A, a highly active xylanase from the human gut bacterium Bacteroides xylanisolvens XB1A. Appl. Microbiol. Biotechnol. 87:2097–2105. 10.1007/s00253-010-2694-0 [DOI] [PubMed] [Google Scholar]
- 17.Dodd D, Moon YH, Swaminathan K, Mackie RI, Cann IK. 2010. Transcriptomic analyses of xylan degradation by Prevotella bryantii and insights into energy acquisition by xylanolytic Bacteroidetes. J. Biol. Chem. 285:30261–30273. 10.1074/jbc.M110.141788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kaoutari AE, Armougom F, Gordon JI, Raoult D, Henrissat B. 2013. The abundance and variety of carbohydrate-active enzymes in the human gut microbiota. Nat. Rev. Microbiol. 11:497–504. 10.1038/nrmicro3050 [DOI] [PubMed] [Google Scholar]
- 19.Martens EC, Koropatkin NM, Smith TJ, Gordon JI. 2009. Complex glycan catabolism by the human gut microbiota: the Bacteroidetes Sus-like paradigm. J. Biol. Chem. 284:24673–24677. 10.1074/jbc.R109.022848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Martens EC, Lowe EC, Chiang H, Pudlo NA, Wu M, McNulty NP, Abbott DW, Henrissat B, Gilbert HJ, Bolam DN, Gordon JI. 2011. Recognition and degradation of plant cell wall polysaccharides by two human gut symbionts. PLoS Biol. 9:e1001221. 10.1371/journal.pbio.1001221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Collins T, Gerday C, Feller G. 2005. Xylanases, xylanase families and extremophilic xylanases. FEMS Microbiol. Rev. 29:3–23. 10.1016/j.femsre.2004.06.005 [DOI] [PubMed] [Google Scholar]
- 22.Cantarel BL, Coutinho PM, Rancurel C, Bernard T, Lombard V, Henrissat B. 2009. The Carbohydrate-Active EnZymes database (CAZy): an expert resource for glycogenomics. Nucleic Acids Res. 37:D233–D238. 10.1093/nar/gkn663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yoon K-H, Yun HN, Jung KH. 1998. Molecular cloning of a Bacillus sp. KK-1 xylanase gene and characterization of the gene product. Biochem. Mol. Biol. Int. 45:337–347 [DOI] [PubMed] [Google Scholar]
- 24.Elleuche S, Piascheck H, Antranikian G. 2011. Fusion of the OsmC domain from esterase EstO confers thermolability to the cold-active xylanase Xyn8 from Pseudoalteromonas arctica. Extremophiles 15:311–317. 10.1007/s00792-011-0361-8 [DOI] [PubMed] [Google Scholar]
- 25.Brennan Y, Callen WN, Christoffersen L, Dupree P, Goubet F, Healey S, Hernandez M, Keller M, Li K, Palackal N, Sittenfeld A, Tamayo G, Wells S, Hazlewood GP, Mathur EJ, Short JM, Robertson DE, Steer BA. 2004. Unusual microbial xylanases from insect guts. Appl. Environ. Microbiol. 70:3609–3617. 10.1128/AEM.70.6.3609-3617.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lee CC, Kibblewhite-Accinelli RE, Wagschal K, Robertson GH, Wong DW. 2006. Cloning and characterization of a cold-active xylanase enzyme from an environmental DNA library. Extremophiles 10:295–300. 10.1007/s00792-005-0499-3 [DOI] [PubMed] [Google Scholar]
- 27.Honda Y, Kitaoka M. 2004. A family 8 glycoside hydrolase from Bacillus halodurans C-125 (BH2105) is a reducing end xylose-releasing exo-oligoxylanase. J. Biol. Chem. 279:55097–55103. 10.1074/jbc.M409832200 [DOI] [PubMed] [Google Scholar]
- 28.Lagaert S, Van Campenhout S, Pollet A, Bourgois TM, Delcour JA, Courtin CM, Volckaert G. 2007. Recombinant expression and characterization of a reducing-end xylose-releasing exo-oligoxylanase from Bifidobacterium adolescentis. Appl. Environ. Microbiol. 73:5374–5377. 10.1128/AEM.00722-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bakir MA, Kitahara M, Sakamoto M, Matsumoto M, Benno Y. 2006. Bacteroides intestinalis sp. nov., isolated from human faeces. Int. J. Syst. Evol. Microbiol. 56:151–154. 10.1099/ijs.0.63914-0 [DOI] [PubMed] [Google Scholar]
- 30.Petersen TN, Brunak S, von Heijne G, Nielsen H. 2011. SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat. Methods 8:785–786. 10.1038/nmeth.1701 [DOI] [PubMed] [Google Scholar]
- 31.Laemmli UK. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680–685. 10.1038/227680a0 [DOI] [PubMed] [Google Scholar]
- 32.Whitmore L, Wallace BA. 2004. DICHROWEB, an online server for protein secondary structure analyses from circular dichroism spectroscopic data. Nucleic Acids Res. 32(Suppl 2):W668–W673. 10.1093/nar/gkh371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Powell JC, Lever M. 1972. A new automated procedure for the colorimetric determination of glucose. Biochem. Med. 6:543–547. 10.1016/0006-2944(72)90008-7 [DOI] [PubMed] [Google Scholar]
- 34.Dodd D, Kiyonari S, Mackie RI, Cann IK. 2010. Functional diversity of four glycoside hydrolase family 3 enzymes from the rumen bacterium Prevotella bryantii B14. J. Bacteriol. 192:2335–2345. 10.1128/JB.01654-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Miyazaki K, Miyamoto H, Mercer DK, Hirase T, Martin JC, Kojima Y, Flint HJ. 2003. Involvement of the multidomain regulatory protein XynR in positive control of xylanase gene expression in the ruminal anaerobe Prevotella bryantii B14. J. Bacteriol. 185:2219–2226. 10.1128/JB.185.7.2219-2226.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fushinobu S, Hidaka M, Honda Y, Wakagi T, Shoun H, Kitaoka M. 2005. Structural basis for the specificity of the reducing end xylose-releasing exo-oligoxylanase from Bacillus halodurans C-125. J. Biol. Chem. 280:17180–17186. 10.1074/jbc.M413693200 [DOI] [PubMed] [Google Scholar]
- 37.Dodd D, Mackie RI, Cann IK. 2011. Xylan degradation, a metabolic property shared by rumen and human colonic Bacteroidetes. Mol. Microbiol. 79:292–304. 10.1111/j.1365-2958.2010.07473.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Flint HJ, Bayer EA, Rincon MT, Lamed R, White BA. 2008. Polysaccharide utilization by gut bacteria: potential for new insights from genomic analysis. Nat. Rev. Microbiol. 6:121–131. 10.1038/nrmicro1817 [DOI] [PubMed] [Google Scholar]
- 39.Koropatkin NM, Cameron EA, Martens EC. 2012. How glycan metabolism shapes the human gut microbiota. Nat. Rev. Microbiol. 10:323–335. 10.1038/nrmicro2746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Topakas E, Moukouli M, Dimarogona M, Vafiadi C, Christakopoulos P. 2010. Functional expression of a thermophilic glucuronyl esterase from Sporotrichum thermophile: identification of the nucleophilic serine. Appl. Microbiol. Biotechnol. 87:1765–1772. 10.1007/s00253-010-2655-7 [DOI] [PubMed] [Google Scholar]
- 41.Spanikova S, Biely P. 2006. Glucuronoyl esterase—novel carbohydrate esterase produced by Schizophyllum commune. FEBS Lett. 580:4597–4601. 10.1016/j.febslet.2006.07.033 [DOI] [PubMed] [Google Scholar]
- 42.Varghese JN, Hrmova M, Fincher GB. 1999. Three-dimensional structure of a barley beta-D-glucan exohydrolase, a family 3 glycosyl hydrolase. Structure 7:179–190. 10.1016/S0969-2126(99)80024-0 [DOI] [PubMed] [Google Scholar]
- 43.Juncker AS, Willenbrock H, Von Heijne G, Brunak S, Nielsen H, Krogh A. 2003. Prediction of lipoprotein signal peptides in Gram-negative bacteria. Protein Sci. 12:1652–1662. 10.1110/ps.0303703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Seydel A, Gounon P, Pugsley AP. 1999. Testing the ‘+2 rule' for lipoprotein sorting in the Escherichia coli cell envelope with a new genetic selection. Mol. Microbiol. 34:810–821. 10.1046/j.1365-2958.1999.01647.x [DOI] [PubMed] [Google Scholar]
- 45.Yamaguchi K, Yu F, Inouye M. 1988. A single amino acid determinant of the membrane localization of lipoproteins in E. coli. Cell 53:423–432. 10.1016/0092-8674(88)90162-6 [DOI] [PubMed] [Google Scholar]
- 46.Collins T, De Vos D, Hoyoux A, Savvides SN, Gerday C, Van Beeumen J, Feller G. 2005. Study of the active site residues of a glycoside hydrolase family 8 xylanase. J. Mol. Biol. 354:425–435. 10.1016/j.jmb.2005.09.064 [DOI] [PubMed] [Google Scholar]
- 47.Pollet A, Schoepe J, Dornez E, Strelkov SV, Delcour JA, Courtin CM. 2010. Functional analysis of glycoside hydrolase family 8 xylanases shows narrow but distinct substrate specificities and biotechnological potential. Appl. Microbiol. Biotechnol. 87:2125–2135. 10.1007/s00253-010-2659-3 [DOI] [PubMed] [Google Scholar]
- 48.Yin Y, Mao X, Yang J, Chen X, Mao F, Xu Y. 2012. dbCAN: a web resource for automated carbohydrate-active enzyme annotation. Nucleic Acids Res. 40:W445–W451. 10.1093/nar/gks479 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.